Abstract
Although foamy viruses (FVs) are endemic among nonhuman primates, FV infection among humans is rare. Recently, simian foamy virus (SFV) infection was reported in 4 of 231 individuals occupationally exposed to primates (1.8%). Secondary transmission to spouses has not been seen, suggesting that while FV is readily zoonotic, humans may represent dead-end hosts. Among different simian species, SFV demonstrates significant sequence diversity within the U3 region of the long terminal repeat (LTR) and 3′ accessory open reading frames (ORFs). To examine if persistent human SFV infection and apparent lack of secondary transmission are associated with genetic adaptations in FV regulatory regions, we conducted sequence analysis of the LTR, internal promoter, ORF-1, and ORF-2 on a tissue culture isolate and peripheral blood mononuclear cell samples from a human infected with SFV of African green monkey origin (SFV-3). Compared to the prototype SFV-3 sequence, the LTR, internal promoter, and FV transactivator (ORF-1) showed sequence conservation, suggesting that FV zoonosis is not dependent on host-specific adaptation to these transcriptionally important regions. However, ORF-2 contains a number of deleterious mutations predicted to result in premature termination of protein synthesis. ORF-2 codes in part for the 60-kDa Bet fusion protein, proposed to be involved in the establishment of persistent cellular SFV infections. These results suggest that persistent human infection by SFV and reduced transmissibility may be influenced by the absence of a functional ORF-2.
Foamy viruses (FVs) (Spumavirinae) represent a unique genus of retrovirus, endemic in a number of mammalian species, including nonhuman primates, cats, rodents, and cows (20, 25, 36). Extremely cytopathic in vitro, FVs have yet to be associated with any disease pathology in their natural hosts. FVs appear to transmit readily via infectious saliva, resulting in infection rates as high as 70 to 90% in some species of nonhuman primates (28). Despite common evolution and cohabitation with lower primates, FV infection is not endemic in humans. Although an FV referred to as human foamy virus (HFV) was reportedly isolated from cultures of a nasopharnygeal carcinoma taken from a Kenyan patient (1, 12), the lack of evidence of antibodies to HFV in widespread and diverse human populations and the high degree of sequence relatedness between HFV and the strain of simian foamy virus (SFV) found in chimpanzees has led to suggestions that HFV may not be of human origin (3, 20, 21, 41, 42). It has recently been reported that humans occupationally exposed to nonhuman primates demonstrate a substantial prevalence (1.8%) of SFV infection, suggesting that SFV is readily zoonotic (19). Consistent with the lack of widespread FV infection in human populations, there is no evidence of secondary transmission to exposed spouses, suggesting that humans may represent dead-end hosts for SFV infection.
Known human retroviruses likely arose from the zoonotic transmission and subsequent adaptation of retroviruses found in Old World primates (10, 14, 17). The considerable period of time between the identification of each of these human retroviruses and the initial zoonotic events responsible for their establishment in humans, precludes the investigation of the earliest adaptive changes which occur during persistent infection of the new human host. The identification of SFV-infected individuals provides a unique opportunity to examine changes associated with retroviruses after zoonotic transmission from the natural host into humans. Evidence of SFV infection in all four cases reported among occupationally exposed animal handlers included seropositivity and proviral DNA detection by PCR (19). An SFV-3-like virus was subsequently isolated from one individual who was severely bitten by an African green monkey prior to 1975 and who demonstrated seropositivity since 1995, the earliest time point for which material was available (19). The derivation of this isolate, designated SFVHU-1, confirms long-term persistent infection of this individual with a replication-competent SFV. Here, we present evidence suggesting that while the long terminal repeat (LTR) and the orf-1 accessory gene of SFV-3 demonstrate significant sequence stability upon cross-species infection, the orf-2/bet gene does not appear to be required for the establishment of a persistent infection in this individual.
The FV genome codes for Gag, Pol, and Env structural proteins and for two or three accessory genes located between env and the 3′ LTR (13, 32, 33). Unique among retroviruses is the presence of an internal promoter located within the 3′ end of the env gene which functions to independently drive the expression of the accessory open reading frames (ORFs) (7, 22, 23). The 5′ proximal FV accessory gene (orf-1) codes for the FV Tas (or Bel-1) transactivator protein which functions to facilitate FV transcription through cis-acting response elements found in both the LTR U3 region as well as the internal promoter (13, 29, 31–33). The function of the second accessory ORF is currently unknown, although a 44-kDa protein believed to be encoded by orf-2 has been identified in cells infected with HFV (15). Alternate mRNA splicing results in a third HFV accessory protein generated by the fusion of the N-terminal 88 amino acids of orf-1 with the complete orf-2. The resulting fusion protein of approximately 60 kDa, referred to as Bet, is expressed at high levels within infected cells (4, 13, 39). Deletion analysis has revealed that only orf-1 is mandatory for replication competence (34). The deletion of orf-2/bet, although permissive for viral replication, reduces virus replication in vitro (4, 46, 47). While it has been speculated that orf-2/bet may be required for viral replication in vivo, animal studies involving deletion mutant viruses have not been reported to date. Bet protein secreted from cells persistently infected with HFV can subsequently be taken up by uninfected cells, while cells which stably express Bet are resistant to FV-induced lysis, suggesting that Bet may behave as a virokine by impairing viral infection (16, 35). Bet expression in the absence of structural gene expression has been associated with persistent HFV infection, while HFV mutant viruses, which lack an intact Bet gene, are apparently unable to establish chronic infections in vitro (16, 35, 37). Together these results imply a role for Bet in the establishment and control of viral persistence.
SFV endemic to different species of simians demonstrate the greatest level of genome sequence diversity within the U3 region of the LTR and 3′ ORFs (13). To examine if persistent human SFV infection and the apparent lack of secondary transmission are associated with genetic adaptations in FV regulatory regions, we conducted sequence analysis of the LTR, internal promoter, orf-1, and orf-2 of SFVHU-1. The complete nucleotide sequence for SFV-3 has previously been published, enabling comparisons to be made with this zoonotic form of SFV-3 (30). The SFVHU-1 template DNA used for sequence analysis was derived from infected Cf2Th canine thymocyte cultures. Infected cells were lysed in 10 mM Tris-HCl (pH 8.3), 0.05% Triton X-100, and 100 μg of proteinase K/ml by incubation at 56°C for 1 h. Twenty microliters of infected Cf2Th lysate was used in a 100-μl PCR volume containing 2 mM MgCl2, 200 mM dinucleoside triphosphates, 1 U of Taq polymerase, and 100 ng each of oligonucleotide primer. PCR was performed in a Perkin-Elmer thermocycler 9600. Reaction conditions include an initial denaturization step at 94°C for 2 min, followed by 35 cycles with incubations at 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 5 min.
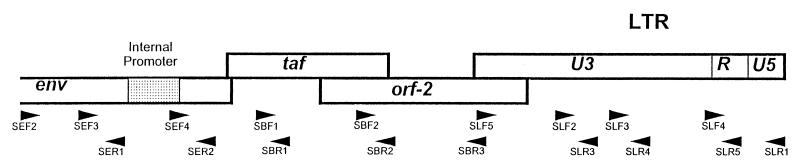
Primers were synthesized based on SFV-3 sequence to amplify the LTR, envelope, orf-1, and orf-2 regions of SFVHU-1 (Fig. 1). A series of overlapping amplicons were generated and sequenced with the following primer pairs: SLF2/SLR4, SLF3/SLR5, SLF2/SLR1, SLF2/SLR5, SLF3/SLR1, SLF5/SLR3, SEF2/SER1, SEF4/SBR1, SEF3/SER2, SEF4/SBR1, SBF1/SBR2, SBF2/SBR3, SBF2/SLR3, and SBF2/SLR4. The PCR fragments were either sequenced directly or cloned into the pGEM T easy vector from Promega (Madison, Wis.). At least two independent clones and/or amplicons were sequenced in each region. Sequencing was performed using either ABI Prism Dye Terminator or the Big Dye Terminator Cycle Sequencing Ready Reaction Kit as specified by the manufacturer (PE Applied Biosystems, Foster City, Calif.). An additional sequencing primer, SB5F3, 5′ GGATCTATTGGTCATTGTGC 3′ (nucleotides [nt] 11179 to 11198), was used for sequencing orf-2. The GenBank accession numbers of other FVs used for comparison and analysis include: SFV-3 (M74895), SFV-1 (M33561), HFV (Y07725), and SFVcpz (U04327). Sequence analysis was performed with the Wisconsin Package, version 9.1, (Genetics Computer Group, Madison, Wis.).
FIG. 1.
Sequencing of the accessory genes and the regulatory regions of SFVHU-1. The locations of the primers used for PCR and sequencing are shown above. The primers and their locations based on SFV-3 base numbers are as follows: SLF5, 5′ CCCAGGAAAAGGATTATTGG 3′ (SFV-3 nt 31 to 50/11434 to 11453); SLF2, 5′ AACGACTGAGTGACATGAAG 3′ (nt 11940 to 11960); SLF3, 5′ GCACAGTAAATTAAGCTAGCAG 3′ (nt 12232 to 12253); SLF4, 5′ ACTGCTCGCTGCGTCGAGAG 3′ (nt 12746 to 12766); SLR3, 5′ CTAGTGGCTCCTATTGAGAG 3′ (nt 11974 to 11993); SLR4, 5′ ATTAAAGGGATTCGAACTAC 3′ (nt 12280 to 12299); SLR5, 5′ TTACCAAGCCTGGAGAGACTCG 3′ (nt 12770 to 12791); SLR1, 5′ TCCTTAAAGAATTCCACCTC 3′ (nt 13066 to 13086); SEF2, 5′ AATGATGAAAGGTTACAACAAGG 3′ (nt 8865 to 8887); SEF3, 5′ TAGGTCATCTTGTTGAGTCAGCTGG 3′ (nt 9242 to 9265); SEF4, 5′ AAAGATCAGATTGAAAGAGC 3′ (nt 9735 to 9754); SER1, 5′ TCACAAATCACATAATCTTG 3′ (nt 9369 to 9380); SER2, 5′ GTTACCTATGCCTTGAAGAGC 3′ (nt 9858 to 9878); SBR1, 5′ TATATAGTCCACAAAGAATAAG 3′ (nt 10292 to 10313); SBF1, 5′ AGAAATTGGGTTCCTGATCC 3′ (nt 10241 to 10260); SBF2, 5′ ATGTCTGGAGGACCCTTCTGG3′ (nt 10820 to 10841); SBR2, 5′ CCTAATTTTCACTAGGCCCAG 3′ (nt 10878 to 10898); SBR3, 5′ CTTCCATGCTGAGGTCCATAAGC 3′ (nt 11368 to 11390).
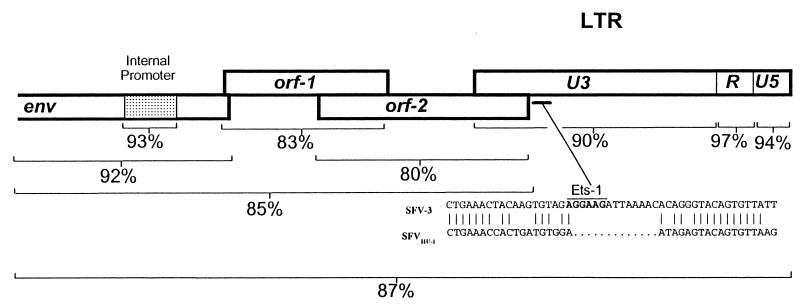
SFV-3 has two accessory genes located between env and the 3′ LTR, similar to what has been reported for SFV-1 and differing from the three accessory ORFs described for HFV (30, 44). The first accessory ORF of SFV-3 encodes a 298-amino-acid protein which has been shown to function as the viral transactivator (26, 29, 31). Amino acid identity between SFV-3 orf-1 and the equivalent tas gene of SFV-1 and HFV is 52 and 38%, respectively. Sequence homologies between the transactivators of SFVHU-1 and SFV-3 are 83% identical at the nucleic acid level and 81% identical at the amino acid level (Fig. 2). The ratio of synonymous to nonsynonymous mutations between orf-1 of SFVHU-1 and that of SFV-3 is 0.2, suggesting that this region has most likely been placed under negative selective pressures to remain unchanged after zoonosis. Together, these data suggest that zoonosis and persistent human infection do not require or result in significant changes within the viral transactivator.
FIG. 2.
Nucleic acid identity between SFVHU-1 and SFV-3. The sequence similarities between SFVHU-1 and SFV-3 vary over different regions. The cis-acting regulatory regions in the LTR and internal promoter show the highest identities, while orf-2 is the least conserved.
The SFV-3 orf-1 transactivator mediates viral transcription through response elements located within the internal promoter and U3 region of the LTR (31). The LTRs of FV tend to be large, ranging in length from 1,600 to 1,700 bp (13). SFVs endemic to different species of simians demonstrate a high degree of sequence conservation within the R and U5 regions of the LTR, while sequence divergence tends to be predominate within the U3 region (13). Moreover, the U3 region of HFV is particularly prone to deletion mutations during both in vitro as well as in vivo replication, suggesting that deletions within this region of the LTR may be associated with viral persistence during chronic HFV infection (8, 40). SFVHU-1 maintains a high degree of identity to SFV-3 in both the LTR and internal promoters (Fig. 2). At 92% identity, the LTRs of SFVHU-1 and SFV-3 are more similar than what has been reported for two independent African green monkey SFV isolates (6), implying that this cross-species infection into a human does not require significant changes within the LTR.
While the U3 region of the SFV-3 LTR is reportedly devoid of consensus sequence motifs for known cellular transcription factors (31), the LTR of HFV contains several Jun/Fos binding AP-1 sites as well as two Ets-1 sites located at bases 325 to 431 and bases 364 to 369. These Ets-1 sites bind a class of lymphocyte-specific transcription factors believed to confer a lymphotropic character on the promoter of HFV (40). Deletion of these sites in HFV results in diminished tissue culture kinetics in the H9 human T-cell line (40). Reanalysis of the published SFV-3 LTR sequence identifies the conservation of these Ets-1 sites at roughly the same location in the U3 of SFV-3, as well as an additional third Ets-1 site at bases 963 to 968. Point mutations in SFVHU-1 have abolished the first Ets-1 site, and a 13-bp deletion corresponding to bases 397 to 409 of SFV-3 has resulted in the loss of the second Ets-1 site in the U3 of SFVHU-1 (Fig. 2). The third putative Ets-1 site persists.
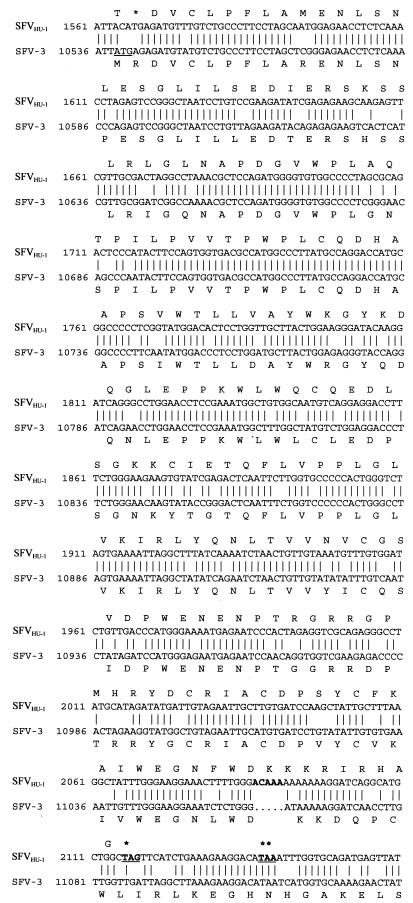
SFV-3 orf-2 is predicted to code for a protein of 388 amino acids, with 54% amino acid identity to orf-2 of SFV-1 and 38% identity to the analogous gene product in HFV. Overall, this region of SFVHU-1 shows close nucleic acid identity to that of SFV-3 (80%), supporting the proposed SFV-3 origin of the SFVHU-1 isolate (Fig. 2). However, orf-2 of SFVHU-1 has a number of deleterious mutations predicted to result in a loss of the ability of orf-2 to code for a functional protein. An ATG to ACA mutation in the predicted start codon is expected to result in a loss of the normal start of translation for SFVHU-1 orf-2. However, even in the absence of these mutations, a 5-base insertion at position 11063 and the resulting frameshift mutation, coupled with downstream mutations, results in the generation of a premature stop codon at position 183 (Fig. 3).
FIG. 3.
orf-2 of SFVHU-1 contains a number of deleterious mutations predicted to result in the loss of a functional orf-2 and Bet protein. The orf-2 coding regions of SFVHU-1 and SFV-3 are aligned with their amino acid translations above and below, respectively. The predicted ATG start site of SFV-3 is underlined. The 5-base insertion in SFVHU-1 is in bold type. In bold type and underlined are the stop codons read as a result of the frameshift caused by either the 5-base or 4-base insertion identified in SFVHU-1 (∗) and the infected individual’s PBMCs (∗∗), respectively. The primers used for PCR and sequencing of orf-2 from the PBMCs of the infected individual are as follows: 51F1, 5′ GTTATTCATGAACCTATGCC 3′ (SFV-3 nt 10427 to 10446); 51F2, 5′ GGATTATGGCTAAAAATGGG 3′ (nt 10469 to 10488); 51R2, 5′ GCACCAGGAATTGAGTCCCG 3′ (nt 10853 to 10872); 51R1, 5′ AATTTTCACTAGACCCAGTG 3′ (nt 10876 to 10895); 52F1, 5′ CCTGGAACCTCCGAAATGGC 3′ (nt 10793 to 10812); 52F2, 5′ GTGGCAATGTCAGGAGGACC 3′ (nt 10814 to 10833); 52R2, 5′ TATGGATAAGGTCTAGATTC 3′ (nt 11157 to 11176); 52R1, 5′ TCAACTCTCATTTTAACTTG 3′ (nt 11217 to 11236).
Based on the SFVHU-1 sequence, two sets of nested primers were designed to amplify selected regions of orf-2 present in the peripheral blood mononuclear cells (PBMCs) from the infected individual. Nested PCR was performed with 5 μl of the primary reaction mixture used in a 100-μl reaction mixture as described above. Primer pairs 51F1/51R1 and 51F2/51R2 amplified a region encompassing the start of orf-2. Primer pairs 52F1/52R1 and 52F2/52R2 amplified the section of orf-2 containing the deletion described above. The nested primers were used to sequence the PCR fragments generated. Primer sequences are given in the legend of Fig. 3.
All the mutations identified in the orf-2 region of SFVHU-1 were confirmed in PBMCs from the infected individual, with the exception that the 5-bp insertion in SFVHU-1 is represented by a 4-bp insertion at the same site, resulting in the generation of a translational stop codon at position 191. These data were generated by eight independent sequencing reactions performed on two separate PCRs using PBMCs from different time points. The identification of similar mutations within SFVHU-1 isolated from this individual suggests that they are associated with a replication-competent virus rather than a defective form of FV. At this time it is unclear if the 1-bp difference in insertion size is indicative of the chance isolation of a minor quasispecies in the infected individual or if the additional single base insertion was acquired during isolation or tissue culture passaging of the virus. Since PBMCs were the only tissue available for analysis, it is not possible to preclude the existence of a viral species with a functional orf-2 in other tissues.
Proteins analogous to Bet have been described for SFV-3 (24). Mutation in orf-2 of SFVHU-1 is predicted to result in the truncation of the entire orf-2 portion of the SFVHU-1 Bet protein because of the generation of a stop codon. Definitive analysis of the translational pattern of the accessory genes of SFVHU-1 awaits the development of specific immunologic reagents. With the material available, it is not possible to determine whether these changes are required for, or result from, human infection by SFV. However, they do indicate that human infection with FV can occur in the absence of a complete complement of accessory genes. These regions are being examined in additional SFV-infected individuals to further elucidate the role of SFV accessory genes in zoonosis and persistent infection.
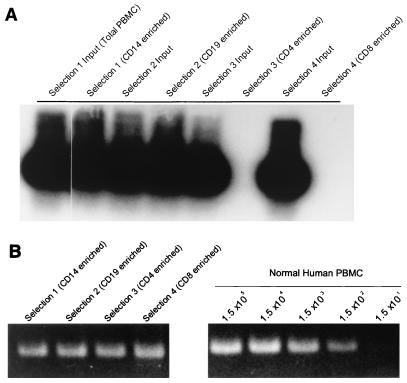
To characterize the cellular reservoirs for persistent FV infection within the PBMCs of this individual, monocyte, B-cell, CD4 T-cell, and CD8 T-cell subsets were positively selected by using immunomagnetic beads directly conjugated with antibodies to CD19, CD14, CD4, and CD8 (Dynal). Prior to selection, PBMCs were stained with fluorescent-tagged antibodies and analyzed by FACScan Cell Quest software (Becton-Dickinson) to determine the percentage of cells in the total population. Approximate cell numbers were calculated based on the percentage of cells and the total cell count. Directly conjugated immunomagnetic beads were used in 10-fold excess of the approximate cell count to positively select each PBMC subset. Void volume from the positive selection was retained for the positive selection of the next subset. PBMCs bound to immunomagnetic beads were washed once with phosphate-buffered saline containing 1% fetal bovine serum and lysed for PCR at a concentration of 6 × 106 cells per ml. This process was repeated until each specific subset was selected, beginning with CD19 selection, followed by CD14 selection, CD4 selection, and finally CD8 selection. After each positive selection, approximately 2 × 105 cells from the void volume were stained with fluorescent-tagged antibodies directed against the population previously selected to confirm depletion of the population. Samples were stored at −70°C for PCR analysis of SFV proviral pol sequences as described elsewhere (2, 19). PCR analysis revealed the presence of SFV proviral pol gene sequences in both monocytes and B cells from the infected individual, with the absence of SFV in both CD4+ and CD8+ T lymphocytes (Fig. 4A). Analysis of human beta-globulin gene sequences ensured that for each cellular subset, amplifiable DNA was present and intact (Fig. 4B). These results differ from what has been reported in other studies, where positive selection on different aliquots of PBMCs identified CD8+ lymphocytes as the major cellular reservoir for FV in African green monkeys, chimpanzees, and two human infections (43). While FV was found exclusively in the CD8 cells of infected humans, differential infection of B cells, monocytes, and polymorphonuclear leukocytes was reported for infected monkeys, suggesting that other cell types may be permissive to SFV infection in vivo. SFVHU-1 differs from SFV-3 in that it cannot grow in the SupT.1 human T-cell line (data not shown). The small changes in the LTR may be responsible for the lack of SFVHU-1 infection of T cells in vitro and in vivo. Transfection experiments using LTR/reporter gene constructs may be necessary to confirm this.
FIG. 4.
Detection of FV DNA in immunomagnetic bead-enriched peripheral blood leukocyte populations. (A) Nested PCR amplification of a 153-bp FV pol sequence demonstrates that the major cellular reservoirs for persistent FV infection within this individual are CD19+ (B lymphocytes) and CD14+ (monocyte). No FV DNA was detectable in CD4+ and CD8+ T lymphocytes. (B) PCR amplification of a human beta-globulin sequence in the enriched cell populations from the SFV-infected individual and titrated normal human PBMCs implies that all PCRs in panel A contain equivalent numbers of cells (approximately 1,500 cells).
The demonstration of persistent human infection by SFV in the absence of an intact orf-2/bet has implications for the proposed use of FVs as viral vectors for gene therapy and novel recombinant vaccine applications (5, 9, 18, 27, 45). The dispensability of orf-2/bet for HFV replication in vitro has led to the development of several replication-competent FV vector systems in which the orf-2/bet region is replaced with foreign DNA (38). To date, the in vivo growth characteristics of these vectors have not been reported. Our data suggest that replication-competent FV vectors lacking an intact orf-2/bet may be capable of establishing long-term infections in humans. Recently, a number of retrovirus vectors based on HIV-1 have been reported in the literature, although vector stability and safety have remained a concern (11). In addition to the lack of disease in and secondary transmission from people infected with SFV, the apparent long-term stability of regulatory elements during persistent human infection would also be advantageous for vector applications based on SFV.
Nucleotide sequence accession number.
The GenBank accession number for the SFVHU-1 sequences is AF18200.
Acknowledgments
This work was supported in part by the Emerging Infectious Diseases Fellowship Program administered by the Centers for Disease Control and Prevention and the Association of Public Health Laboratories.
REFERENCES
- 1.Achong B G, Mansell P W A, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. [PubMed] [Google Scholar]
- 2.Allan J S, Broussard S R, Michaels M G, Starzl T E, Leighton K L, Whitehead E M, Comuzzie A G, Lanford R E, Leland M M, Switzer W M, Heneine W. Amplification of simian retroviral sequences from human recipients of baboon liver transplants. AIDS Res Hum Retroviruses. 1998;14:821–824. doi: 10.1089/aid.1998.14.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali M, Taylor G P, Pitman R J, Parker D, Rethwilm A, Cheingsong-Popov R, Weber J N, Bieniasz P D, Bradley J, McClure M O. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res Hum Retroviruses. 1996;12:1473–1483. doi: 10.1089/aid.1996.12.1473. [DOI] [PubMed] [Google Scholar]
- 4.Baunach G, Maurer B, Hahn H, Kranz M, Rethwilm A. Functional analysis of human foamy virus accessory reading frames. J Virol. 1993;67:5411–5418. doi: 10.1128/jvi.67.9.5411-5418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Irlwein O, Aguzzi A, Rethwilm A, McClure M O. Gene transfer using replication-defective human foamy virus vectors. Virology. 1997;235:65–72. doi: 10.1006/viro.1997.8658. [DOI] [PubMed] [Google Scholar]
- 6.Broussard S R, Comuzzie A G, Leighton K L, Leland M M, Whitehead E M, Allan J S. Characterization of new simian foamy viruses from African nonhuman primates. Virology. 1997;237:349–359. doi: 10.1006/viro.1997.8797. [DOI] [PubMed] [Google Scholar]
- 7.Campbell M, Renshaw-Gegg L, Renne R, Luciw P A. Characterization of the internal promoter of simian foamy viruses. J Virol. 1994;68:4811–4820. doi: 10.1128/jvi.68.8.4811-4820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Celis-Kosmas J, Coronel A, Grigorian I, Emanoil-Ravier R, Tobaly-Tapiero J. Non-random deletions in human foamy virus long terminal repeat during viral infection. Arch Virol. 1997;142:1237–1246. doi: 10.1007/s007050050155. [DOI] [PubMed] [Google Scholar]
- 9.Dittmer U, Brooks D M, Hasenkrug K J. Characterization of a live-attenuated retroviral vaccine demonstrates protection via immune mechanisms. J Virol. 1998;72:6554–6558. doi: 10.1128/jvi.72.8.6554-6558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doolittle R F. Immunodeficiency viruses: the simian-human connection. Nature. 1989;339:338–339. doi: 10.1038/339338a0. [DOI] [PubMed] [Google Scholar]
- 11.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein M A, Achong B G, Ball G. Further observations on a human syncytial virus from a nasopharyngeal carcinoma. J Natl Cancer Inst. 1974;53:681–688. doi: 10.1093/jnci/53.3.681. [DOI] [PubMed] [Google Scholar]
- 13.Flugel R M. The molecular biology of the human spumavirus. In: Cullen B R, editor. Human retroviruses. Oxford, United Kingdom: IRL Press; 1993. pp. 193–214. [Google Scholar]
- 14.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 15.Giron M-L, Rozain F, Debons-Guillemin M C, Canivet M, Peries J, Emanoil-Ravier R. Human foamy virus polypeptides: identification of env and bel gene products. J Virol. 1993;67:3596–3600. doi: 10.1128/jvi.67.6.3596-3600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giron M-L, de The H, Saib A. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J Virol. 1998;72:4906–4910. doi: 10.1128/jvi.72.6.4906-4910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goubau P, Vandamme A M, Desmyter J. Questions on the evolution of primate T-lymphotropic viruses raised by molecular and epidemiological studies of divergent strains. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S242–S247. doi: 10.1097/00042560-199600001-00036. [DOI] [PubMed] [Google Scholar]
- 18.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri S, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T M. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 20.Herchenroder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 21.Herchenroder O, Turek R, Neumann-Haefelin D, Rethwilm A, Schneider J. Infectious proviral clones of chimpanzee foamy virus (SFVcpz) generated by long PCR reveal close functional relatedness to human foamy virus. Virology. 1995;214:685–689. doi: 10.1006/viro.1995.0086. [DOI] [PubMed] [Google Scholar]
- 22.Kang Y, Blair W S, Cullen B R. Identification and functional characterization of a high-affinity Bel-1 DNA binding site located in the human foamy virus internal promoter. J Virol. 1998;72:504–511. doi: 10.1128/jvi.72.1.504-511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y, Cullen B R. Derivation and functional characterization of a consensus DNA binding sequence for the Tas transcriptional activator of simian foamy virus type 1. J Virol. 1998;72:5502–5509. doi: 10.1128/jvi.72.7.5502-5509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lochelt M, Flugel R M. The molecular biology of human and primate spumaretroviruses. In: Levy J A, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 25.Loh P C. Spumaviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1992. pp. 361–397. [Google Scholar]
- 26.Mergia A, Shaw K E, Pratt-Lowe E, Barry P A, Luciw P A. Identification of the simian foamy virus transcriptional transactivator gene (taf) J Virol. 1991;65:2903–2909. doi: 10.1128/jvi.65.6.2903-2909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nestler U, Heinkelein M, Lucke M, Meixensberger J, Scheurlen W, Kretschmer A, Rethwilm A. Foamy virus vectors for suicide gene therapy. Gene Ther. 1997;4:1270–1277. doi: 10.1038/sj.gt.3300561. [DOI] [PubMed] [Google Scholar]
- 28.Neumann-Haefelin D, Fleps U, Renne R, Schweizer M. Foamy viruses. Intervirology. 1993;35:196–207. doi: 10.1159/000150310. [DOI] [PubMed] [Google Scholar]
- 29.Renne R, Fleps U, Luciw P A, Neumann-Haefelin D. Transactivation of the two promoters of SFV-3 by different mechanisms. Virology. 1996;221:362–367. doi: 10.1006/viro.1996.0387. [DOI] [PubMed] [Google Scholar]
- 30.Renne R, Friedl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3) Virology. 1992;186:597–608. doi: 10.1016/0042-6822(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 31.Renne R, Mergia A, Renshaw-Gegg L W, Neumann-Haefelin D, Luciw P A. Regulatory elements in the long terminal repeat (LTR) of simian foamy virus type 3 (SFV-3) Virology. 1993;192:365–369. doi: 10.1006/viro.1993.1045. [DOI] [PubMed] [Google Scholar]
- 32.Rethwilm A. Regulation of foamy virus gene expression. Curr Top Microbiol Immunol. 1995;193:1–24. doi: 10.1007/978-3-642-78929-8_1. [DOI] [PubMed] [Google Scholar]
- 33.Rethwilm A. Unexpected replication pathways of foamy viruses. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S248–S253. doi: 10.1097/00042560-199600001-00037. [DOI] [PubMed] [Google Scholar]
- 34.Saib A, Peries J, de The H. A defective human foamy provirus generated by pregenome splicing. EMBO J. 1993;12:4439–4444. doi: 10.1002/j.1460-2075.1993.tb06129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saib A, Koken M H M, van der Spek P, Peries J, de The H. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J Virol. 1995;69:5261–5268. doi: 10.1128/jvi.69.9.5261-5268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saib A, de The H. Molecular biology of the human foamy virus. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S254–S260. doi: 10.1097/00042560-199600001-00038. [DOI] [PubMed] [Google Scholar]
- 37.Saib A, Neves M, Giron M-L, Guillemin M-C, Valla J, Peries J, Canivet M. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology. 1997;228:263–268. doi: 10.1006/viro.1996.8383. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt M, Niewiesk S, Heeney J, Aguzzi A, Rethwilm A. Mouse model to study the replication of primate foamy viruses. J Gen Virol. 1997;78:1929–1933. doi: 10.1099/0022-1317-78-8-1929. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt M, Herchenroder O, Heeney J, Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer K-O, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res Hum Retroviruses. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 42.Tobaly-Tapiero J, Santillana-Hayat M, Giron M-L, Guillemin M C, Rozain F, Peries J, Emanoil-Ravier R. Molecular differences between two immunologically related spumaretroviruses: the human prototype HSRV and the chimpanzee isolate SFV6. AIDS Res Hum Retroviruses. 1990;6:951–957. doi: 10.1089/aid.1990.6.951. [DOI] [PubMed] [Google Scholar]
- 43.Von Laer D, Neumann-Haefelin D, Heeney J L, Schweizer M. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology. 1996;221:240–244. doi: 10.1006/viro.1996.0371. [DOI] [PubMed] [Google Scholar]
- 44.Weissenberger J, Flugel R M. Identification and characterization of the Bel 3 protein of human foamy virus. AIDS Res Hum Retroviruses. 1994;10:595–600. doi: 10.1089/aid.1994.10.595. [DOI] [PubMed] [Google Scholar]
- 45.Wu M, Chari S, Yanchis T, Mergia A. cis-acting sequences required for simian foamy virus type 1 vectors. J Virol. 1998;72:3451–3454. doi: 10.1128/jvi.72.4.3451-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wybier-Franqui J, Tobaly-Tapiero J, Coronel A, Giron M-L, Chopin-Robert C, Peries J, Emanoil-Ravier R. Human foamy virus DNA forms and expression in persistently infected Dami megakaryocytic cells. AIDS Res Hum Retroviruses. 1995;11:829–836. doi: 10.1089/aid.1995.11.829. [DOI] [PubMed] [Google Scholar]
- 47.Yu S E, Linial M J. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6621. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]