Abstract
Background
Genomic fusions are potent oncogenic drivers across cancer types and many are targetable. We demonstrate the clinical performance of DNA-based comprehensive genomic profiling (CGP) for detecting targetable fusions.
Materials and Methods
We analyzed targetable fusion genes in >450 000 tissue specimens profiled using DNA CGP (FoundationOne CDx, FoundationOne). Using a de-identified nationwide (US-based) non–small cell lung cancer (NSCLC) clinico-genomic database, we assessed outcomes in patients with nonsquamous NSCLC (NonSqNSCLC) who received matched therapy based on a fusion identified using DNA CGP. Lastly, we modeled the added value of RNA CGP for fusion detection in NonSqNSCLC.
Results
We observed a broad diversity of fusion partners detected with DNA CGP in conjunction with targetable fusion genes (ALK, BRAF, FGFR2, FGFR3, NTRK1/2/3, RET, and ROS1). In NonSqNSCLC with oncogenic ALK, NTRK, RET, and ROS1 fusions detected by DNA CGP, patients treated with a matched tyrosine kinase inhibitor had better real-world progression-free survival than those receiving alternative treatment regimens and benefit was observed regardless of the results of orthogonal fusion testing. An estimated 1.3% of patients with NonSqNSCLC were predicted to have an oncogenic driver fusion identified by RNA, but not DNA CGP, according to a model that accounts for multiple real-world factors.
Conclusion
A well-designed DNA CGP assay is capable of robust fusion detection and these fusion calls are reliable for informing clinical decision-making. While DNA CGP detects most driver fusions, the clinical impact of fusion detection is substantial for individual patients and exhaustive efforts, inclusive of additional RNA-based testing, should be considered when an oncogenic driver is not clearly identified.
Keywords: DNA sequencing, RNA sequencing, gene fusion, next-generation sequencing, lung cancer
Genomic fusions are potent oncogenic drivers across cancer types, and many are targetable. This study demonstrated the clinical performance of a well-designed DNA-based comprehensive genomic profiling assay for detecting targetable fusions.
Implications for Practice.
When specific design elements and analytics are incorporated, DNA-based next-generation sequencing (NGS) assays are capable of detecting clinically important fusions, including novel structures and fusion partners. In certain scenarios, RNA-based NGS may extend or complement DNA-based NGS for fusion detection, and may be of substantial benefit when (1) a DNA NGS assay is not optimally designed or (2) in tumors where an oncogenic driver alteration has not been detected at the DNA level. Given the increased technical challenges inherent to RNA as an analyte, it is important that DNA assays are designed for maximal fusion detection.
Introduction
Two key developments in the clinical management of patients with solid tumors include the mainstream availability of comprehensive genomic profiling (CGP) and the identification of oncogenic drivers amenable to small molecule inhibition using targeted therapy.1-4 In no indication has this transformation been more impactful than in non–small cell lung cancer (NSCLC), where the identification of numerous oncogenic drivers and a rapidly growing landscape of matched targeted therapies has extended overall survival for a high percentage of patients.5,6 Even still, it is estimated that approximately 30% of NSCLC adenocarcinoma lack a well-defined oncogenic driver alteration.7-10
Historically, CGP has been performed on DNA derived from tumor specimens. However, as the importance of oncogenic fusions as cancer drivers has been realized,11,12 particularly in NSCLC,13 attention has turned to RNA as an important analyte for the detection of driver fusions that might be missed using DNA-based assays.14,15 Given such reports, both the National Comprehensive Cancer Network (NCCN) and the American Society of Clinical Oncology (ASCO) recommend pursuing RNA-based fusion profiling for driver-negative NSCLC as determined by initial DNA-based next-generation sequencing (NGS).16,17 Potential advantages of RNA-based fusion detection include the ability to determine whether a fusion product is expressed at the RNA level and avoidance of often complex methodology required to sequence introns and over large chromosomal regions. However, major limitations of RNA profiling are the lower stability, quality, and quantity of the RNA analyte compared to DNA which can result in high sequencing failure rates,18-20 especially for formalin-fixed archival material.21 Thus, there are advantages and disadvantages to either testing methodology.
The current realities of CGP testing necessitate that many patients rely on DNA-based CGP for fusion detection, and it is thus critical that DNA-based assays are optimally designed to identify these important driver events which can significantly influence treatment decisions and patient outcomes. Moreover, it is of value to identify the clinical scenarios in which patients are most likely to benefit from additional RNA-based testing. In this study, we demonstrate that robust detection of fusions is possible using carefully designed DNA-based sequencing methodologies and that fusions detected using high-quality DNA CGP assays are associated with clinical benefit from matched targeted agents. We also attempt to quantify the potential added value of RNA CGP for fusion detection following DNA CGP for patients with nonsquamous NSCLC (NonSqNSCLC).
Materials and Methods
Comprehensive Genomic Profiling
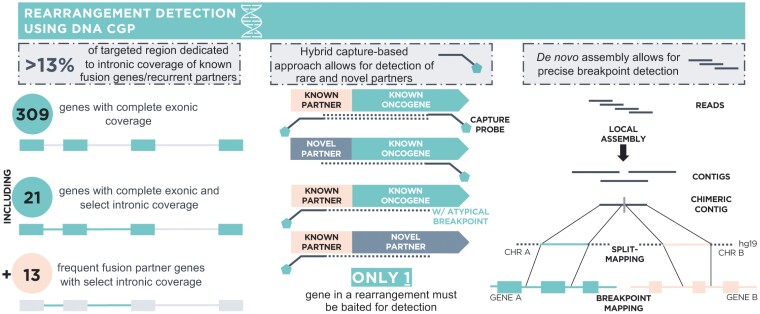
Comprehensive genomic profiling using FoundationOne CDx (F1CDx) or FoundationOne (F1) was performed on hybridization-captured, adaptor ligation-based libraries using DNA extracted from formalin-fixed paraffin-embedded (FFPE) tumor in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited, New York State-approved laboratory (Foundation Medicine, Inc., Cambridge, MA). Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817). Samples were interrogated for alterations in 324 (F1CDx) or 406 (F1) cancer-associated genes.22 Alteration types detected by the assay include base substitutions, short insertions and deletions, copy number amplifications and homozygous deletions, and large genomic rearrangements, as well as microsatellite instability and tumor mutational burden (TMB). TMB was determined on up to 1.24 Mb of sequenced DNA.23 A number of deliberate assay design features (Figure 1; Supplementary Table S1 with detailed description in Results) support rearrangement detection.
Figure 1.
F1CDx assay design enables robust DNA-based rearrangement detection. Design features of F1CDx that support rearrangement detection include (1) select intronic coverage (Supplementary Table S1) for N = 34 genes including both common fusion genes (N = 21) and frequent fusion partners (N = 13), (2) the use of a hybrid capture-based sequencing approach which only requires that one gene involved in a fusion be baited allowing for detection of known as well as rare and novel fusion partners, and (3) the use of a de novo assembly approach in the bioinformatics pipeline which facilitates precise breakpoint detection compared to reference-based methods.
Study Cohorts
Genomic-Only Cohort: Pan-Solid Tumor Fusion Partner Diversity Analysis Utilizing Foundation Medicine Genomic Database
In the Foundation Medicine (FMI) genomic database, we interrogated pan-solid tumor tissue biopsy specimens submitted for CGP (Foundation Medicine, Inc.) using F1CDx or F1 during routine clinical care between August 2014 and December 2022. For patients with multiple tissue CGP results, a single specimen was chosen on the basis of quality metrics. Patients with predicted gene fusions (in-strand and in-frame rearrangements involving distinct target and partner genes) were queried and compared to fusions reported in the AACR Project Genomics Evidence Neoplasia Information Exchange (GENIE)24,25 database (v13.1). AACR Project GENIE is an international pan-cancer registry collecting clinical-grade cancer genomic data together with clinical outcomes data for >140 000 patients (v13.1 release). Only assays/samples with structural variant profiling in GENIE were included and data from GENIE institutions that utilize FMI testing was excluded. Profiling of hematological malignancies and profiling with heme-specific panels were also excluded. GENIE data used in this study included structural variant profiling for 97 592 patients with solid tumors using 18 different assays/assay versions submitted by 7 academic institutions. This included 96 552 patients (98.9%) with DNA-only profiling, 929 (0.9%) with DNA/RNA profiling, and 111 (0.1%) with RNA-only profiling. Note that NTRK genes were profiled in most, but not all, patients (97 106/97 592, 99.5%), whereas all other analyzed genes were covered on all included assays.
Clinico-Genomic Cohort: NSCLC Genomic Driver Landscape and Outcomes Analysis on Targeted Therapy Utilizing Flatiron Health-Foundation Medicine Clinico-Genomic Database
To assess the adequacy of DNA-only fusion detection to support clinical decision-making, we studied a cohort of patients with confirmed diagnosis of NSCLC included in the nationwide US-based Flatiron Health (FH)-FMI de-identified NSCLC clinico-genomic database (CGDB). All patients underwent genomic testing using FMI CGP assays. De-identified clinical data originated from approximately 280 US cancer clinics (approximately 800 sites of care). Retrospective longitudinal clinical data were derived from electronic health records, comprising patient-level structured and unstructured data, curated via technology-enabled abstraction of clinical notes and radiology/pathology reports, which were linked to genomic data derived from FMI testing by de-identified, deterministic matching.26 The data were de-identified and subject to obligations to prevent re-identification and protect patient confidentiality. Clinical data included demographics, self-reported smoking status, clinical and laboratory features, timing of treatment exposure, and mortality. Institutional Review Board approval of the study protocol was obtained prior to the study conduct and included a waiver of informed consent.
Patient records were included in this study if they received an F1CDx or F1 CGP report between August 2014 and December 2022 and the histological and genomic characteristics of their disease (as assessed by chart abstraction and the tissue CGP report, respectively) were consistent with NonSqNSCLC. Clinical characteristics for the NonSqNSCLC cohort (N = 10 761) are presented in Supplementary Table S2. A subset of patients received additional non-FMI biomarker testing (“orthogonal testing”) as part of routine care using a variety of testing methodologies (eg, fluorescence in situ hybridization [FISH], immunohistochemistry [IHC], polymerase chain reaction [PCR], and NGS). Where orthogonal testing results were available, these have been included in the dataset for analysis limited to KRAS, EGFR, BRAF, MET, ALK, RET, ROS1, and NTRK1/2/3.
Analysis
Real-World Progression-Free Survival
Real-world progression-free survival (rwPFS) was calculated from the start of treatment to the first progression date >14 days after treatment start or to death. Real-world progression events were abstracted from patient charts using a clinician-anchored approach supported by radiology report data, as previously described.27 Mortality information in the dataset has been externally validated in comparison to the National Death Index.28 Patients were censored at their last clinic note date if no progression or death was observed. Median rwPFS values were estimated in months with 95% CI.
In these analyses, oncologist-defined, rule-based lines of therapy were classified into those including a fusion-specific FDA-approved TKI for NSCLC (ie, alectinib, brigatinib, ceritinib, crizotinib, entrectinib, larotrectinib, lorlatinib, selpercatinib, and pralsetinib) or not. These rules allow early switches from non-TKI-containing into TKI-containing lines of therapy without being considered the start of a new line (eg, after receipt of biomarker test results not available at the start of therapy). Analysis was limited to the first line of therapy in the advanced setting such that a patient who received a non-TKI in their first line and received a TKI in a later line would be considered a non-TKI patient for this analysis.
Statistical Analysis and Interpretation
Differences in time-to-event outcomes were assessed with the log-rank test and univariable and multivariable Cox proportional hazard models. Multivariable Cox models included adjustment for clinical characteristics that could impact prognosis including line number, socioeconomic status, practice type, gender, race, stage at diagnosis, self-reported smoking history, age, ECOG performance status, opioid prescription immediately prior to therapy (a proxy for burden of disease), and presence of metastases pre-therapy (in bone, central nervous system, liver, adrenal, or other). Missing values for adjustment covariables were not imputed, causing some patients to be excluded from multivariable analyses. R version 4.2.2 software was used for all statistical analyses.
RNA CGP Fusion Detection Modeling
We calculated the number of patients with driver fusion-positive NonSqNSCLC expected to be uniquely identified using RNA CGP following DNA CGP (fusionsRNA.only) as a function of several real-world factors (equation 1):
| (1) |
where prevfusions is the estimated prevalence of driver fusions in the unselected patient population with NonSqNSCLC, θDNA is the probability of detecting an oncogenic driver on DNA CGP, is the probability of DNA assay technical success, and is the probability of RNA assay technical success conditional on DNA assay technical success. To account for possible selection bias toward a higher underlying fusion prevalence in the DNA CGP driver-negative patient population, we included a prevalence multiplier (selection.biasRNA) in sensitivity analyses. In addition to our base case calculation, we conducted both one-way deterministic and probabilistic sensitivity analyses to assess the influence of our base case assumptions and to characterize uncertainty in the estimate of RNA-only identified fusions. Estimates and justifications for the base case parameter assumptions and the deterministic and probabilistic sensitivity analyses are summarized in Supplementary Table S3. We assumed perfect test characteristics for RNA fusion detection as a conservative assumption. Our base estimates and sensitivity values for the prevalence of ALK,11,29-34RET,11,33-36ROS1,11,30,33,34,37 and NTRK11,34,38,39 fusions in patients with NSCLC were based on the published literature. While BRAF, EGFR, FGFR, NRG1, and other fusions do occur in NSCLC, these are rare events such that the combined prevalence of NCCN fusion drivers (ALK + NTRK + RET + ROS1) was expected to produce a reasonable estimate of the overall prevalence of oncogenic driver fusions in NSCLC. The probability of detecting a driver alteration in NonSqNSCLC using DNA CGP was determined using FMI DNA CGP results for patients in CGDB based on a select list of NSCLC receptor tyrosine kinase (RTK)/mitogen-activated protein kinase (MAPK) driver alterations (Supplementary Table S4) as described in Results. The probability of DNA/RNA sequencing success was based on FoundationOne Heme historical data with the upper bound based on real-world experience with DNA NGS reflexed to RNA NGS testing in a study by Benayed et al.14 The base case calculation assumes no selection bias for fusions in the DNA CGP-negative patient population (ie, a prevalence multiplier of 1×). The upper bound (1.2× the base case fusion prevalence) was derived from the percentage of fusion-positive patients identified using DNA CGP and RNA CGP in the study by Benayed et al.14 The minimum and maximum values for the parameters in equation 1 (Supplementary Table S3) were used to generate deterministic one-way sensitivity values where a single parameter was modified from its base case value to either the minimum or maximum value. The parameter distributions specified in Supplementary Table S3 were used to simultaneously draw parameter values for all 5 parameters across 10 000 simulations to characterize a joint uncertainty estimate.
Results
FMI DNA CGP Detects Fusions With a Rich Diversity of Fusion Partners
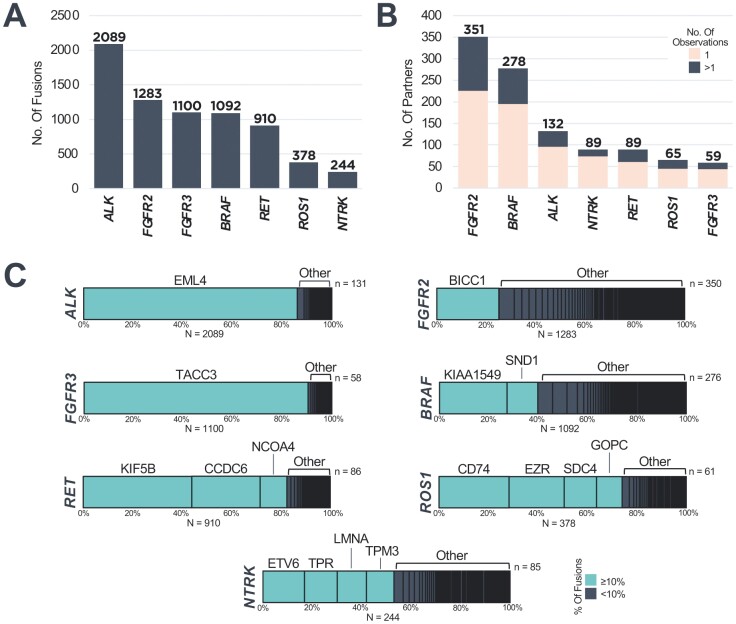
We analyzed genomic profiling data from 459 751 patients with diverse cancer types to characterize the landscape of targetable oncogenic fusions detected in tissue biopsies using a DNA CGP assay purposefully designed for rearrangement detection (Figure 1). We identified predicted fusions in ALK (n = 2089), BRAF (n = 1092), FGFR2 (n = 1283), FGFR3 (n = 1100), NTRK1/2/3 (n = 244), RET (n = 910), and ROS1 (n = 378) across all solid tumors (Fig. 2A). We observed a rich diversity of fusion partners, both previously reported and novel, across target genes (Fig. 2B, 2C; Supplementary Fig. S1; Supplementary Table S5). We used the AACR Project GENIE database (v13.1)24,25—inclusive of 97 592 patients with structural variant profiling using primarily DNA-based assays—as a point of reference for our partner diversity analysis. The most recurrent partners for each target gene were common among the data sets: EML4 with ALK; KIAA1549 with BRAF; BICC1 and TACC2 with FGFR2; TACC3 with FGFR3; ETV6, TPR, LMNA, and TPM3 with NTRK1/2/3; KIF5B, CCDC6, and NCOA4 with RET; and CD74, EZR, SDC4, and GOPC with ROS1 (Supplementary Fig. S1; Supplementary Table S5). Due to the relative size of the FMI DNA CGP cohort, a plethora of unique partner genes (observed as partner genes in the FMI but not the GENIE cohort) was detected (ALK n = 104, BRAF n = 232, FGFR2 n = 295, FGFR3 n = 44, NTRK n = 76, RET n = 71, ROS1 n = 50; Supplementary Fig. S1; Supplementary Table S5). With FMI DNA CGP, 64%-83% of fusion partners detected for each gene were observed in only a single sample, highlighting the capability of DNA CGP for the detection of fusions involving rare and novel partners (Supplementary Fig. S2A). In addition, detection of a subset of NRG1 fusions (n = 67, 0.02% of the cohort, N = 316 152) is enabled through baiting of common partner genes CD74 and SDC4 on F1CDx (Supplementary Fig. S2B).
Figure 2.
FMI DNA CGP detects fusions with a rich diversity of fusion partners. (A) Number of fusions detected involving ALK, BRAF, FGFR2, FGFR3, NTRK1/2/3, RET, and ROS1 across solid tumors. (B) Number of partner genes observed with ALK, BRAF, FGFR2, FGFR3, NTRK1/2/3, RET, and ROS1 across solid tumors. (C) Distribution of partner genes detected in association with each indicated fusion gene across solid tumors. Recurrent partners accounting for ≥10% of detected fusions are indicated in teal.
Targetable Fusions in Patients With NonsqNSCLC Detected Using FMI DNA CGP Gain More Benefit From Matched Therapy Than Patients With Fusions Detected Using Other Testing Methods
We compared the clinical benefit from targeted therapy using matched tyrosine kinase inhibitors (TKI) for oncogenic fusions detected using tissue-based FMI DNA CGP and/or an orthogonal assay (eg, FISH, IHC, PCR, and non-FMI NGS). In the total cohort of patients analyzed who received orthogonal fusion testing and were assessed for progression after starting first-line therapy for advanced disease (N = 3959; Table 1; Supplementary Fig. S3), the most common orthogonal testing method used was FISH followed by non-FMI liquid biopsy DNA NGS, with a lesser degree of IHC, PCR, and RNA-based NGS testing performed (Fig. 3A). Similar trends were observed when limiting to the cohorts in which FMI DNA CGP and orthogonal diagnostic assay results were discordant (Supplementary Fig. S4).
Table 1.
Clinical characteristics of patients with NonSqNSCLC stratified by fusion testing results.
| Clinical characteristics | Orthogonal testing performed (N = 3959) | FMI DNA CGP−/Orthog− (N = 3564) | FMI DNA CGP−/Orthog+ (N = 54) | FMI DNA CGP+/Orthog− (N = 148) | FMI DNA CGP+/Orthog+ (N = 193) |
|---|---|---|---|---|---|
| Age at 1L start (years), Median (IQR) | 65.9 (10.6) | 66.7 (10.2) | 64.0 (9.99) | 61.2 (11.5) | 56.1 (12.6) |
| Sex, n (%) | |||||
| Female | 2237 (56.5%) | 2017 (56.6%) | 29 (53.7%) | 88 (59.5%) | 103 (53.4%) |
| Male | 1722 (43.5%) | 1547 (43.4%) | 25 (46.3%) | 60 (40.5%) | 90 (46.6%) |
| Self-reported race, n (%) | |||||
| Asian | 163 (4.1%) | 143 (4.0%) | 1 (1.9%) | 3 (2.0%) | 16 (8.3%) |
| Black or African American | 257 (6.5%) | 233 (6.5%) | 2 (3.7%) | 10 (6.8%) | 12 (6.2%) |
| Hispanic or Latino | 5 (0.1%) | 5 (0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Other race | 583 (14.7%) | 525 (14.7%) | 4 (7.4%) | 24 (16.2%) | 30 (15.5%) |
| White | 2687 (67.9%) | 2414 (67.7%) | 43 (79.6%) | 100 (67.6%) | 130 (67.4%) |
| Unknown/not documented | 264 (6.7%) | 244 (6.9%) | 4 (7.4%) | 11 (7.4%) | 5 (2.6%) |
| AJCC stage at diagnosis, n (%) | |||||
| I | 355 (9.0%) | 339 (9.5%) | 2 (3.7%) | 7 (4.7%) | 7 (3.6%) |
| II | 238 (6.0%) | 225 (6.3%) | 4 (7.4%) | 3 (2.0%) | 6 (3.1%) |
| III | 694 (17.5%) | 635 (17.8%) | 11 (20.4%) | 24 (16.2%) | 24 (12.4%) |
| IV | 2598 (65.6%) | 2296 (64.4%) | 36 (66.7%) | 112 (75.7%) | 154 (79.8%) |
| Other/unknown/not documented | 74 (1.9%) | 69 (1.9%) | 1 (1.9%) | 2 (1.4%) | 2 (1.0%) |
| Smoking history, n (%) | |||||
| History of smoking | 2958 (74.7%) | 2789 (78.3%) | 40 (74.1%) | 59 (39.9%) | 70 (36.3%) |
| No history of smoking | 998 (25.2%) | 772 (21.7%) | 14 (25.9%) | 89 (60.1%) | 123 (63.7%) |
| Unknown/not documented | 3 (0.1%) | 3 (0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Practice type, n (%) | |||||
| Academic | 534 (13.5%) | 451 (12.7%) | 9 (16.7%) | 30 (20.3%) | 44 (22.8%) |
| Academic/community | 372 (9.4%) | 326 (9.2%) | 10 (18.5%) | 13 (8.8%) | 23 (11.9%) |
| Community | 3053 (77.1%) | 2787 (78.2%) | 35 (64.8%) | 105 (70.9%) | 126 (65.3%) |
| Socioeconomic status (SES), n (%) | |||||
| 1—Lowest SES | 472 (11.9%) | 433 (12.1%) | 10 (18.5%) | 13 (8.8%) | 16 (8.3%) |
| 2 | 647 (16.3%) | 598 (16.8%) | 7 (13.0%) | 17 (11.5%) | 25 (13.0%) |
| 3 | 818 (20.7%) | 724 (20.3%) | 6 (11.1%) | 42 (28.4%) | 46 (23.8%) |
| 4 | 886 (22.4%) | 805 (22.6%) | 16 (29.6%) | 24 (16.2%) | 41 (21.2%) |
| 5—Highest SES | 850 (21.5%) | 750 (21.0%) | 12 (22.2%) | 36 (24.3%) | 52 (26.9%) |
| Unknown | 286 (7.2%) | 254 (7.1%) | 3 (5.6%) | 16 (10.8%) | 13 (6.7%) |
| ECOG PS at 1L start, n (%) | |||||
| 0 | 1265 (32.0%) | 1121 (31.5%) | 19 (35.2%) | 53 (35.8%) | 72 (37.3%) |
| 1 | 1654 (41.8%) | 1506 (42.3%) | 27 (50.0%) | 57 (38.5%) | 64 (33.2%) |
| 2 | 369 (9.3%) | 345 (9.7%) | 3 (5.6%) | 10 (6.8%) | 11 (5.7%) |
| 3 | 84 (2.1%) | 78 (2.2%) | 0 (0.0%) | 2 (1.4%) | 4 (2.0%) |
| Unknown | 587 (14.8%) | 514 (14.4%) | 5 (9.3%) | 26 (17.6%) | 42 (21.8%) |
| 1L therapy class, n (%) | |||||
| No TKI Received | 3761 (95.0%) | 3551 (99.6%) | 40 (74.1%) | 101 (68.2%) | 69 (35.8%) |
| Received TKI | 198 (5.0%) | 13 (0.4%) | 14 (25.9%) | 47 (31.8%) | 124 (64.2%) |
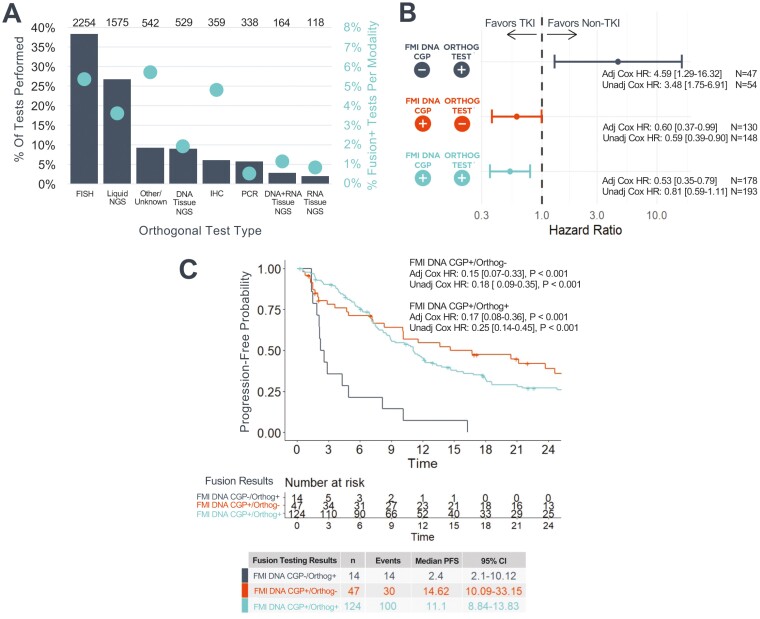
Figure 3.
Targetable fusions detected using FMI DNA CGP are associated with improved clinical benefit from matched therapy compared to fusions detected using other testing methods. (A) Distribution of orthogonal testing modalities (N = 5879 tests; left axis, gray bars) and results (right axis, teal dots) from 3959 patients with NonSqNSCLC who were assessed for progression and underwent both FMI DNA CGP and additional fusion testing. Each testing modality is only counted once per patient. However, a single patient could be counted toward multiple modalities if a patient underwent multiple types of orthogonal testing such that the sum of all bars may exceed 100%. Ns above bars indicate the number of patients who underwent each type of testing. The fusion-positive percentage was calculated based on the total number of tests performed in each category (N = 7299 tests across all modalities), even if multiples of the same testing modality were performed for a single patient. (B) Forest plot depicting the adjusted rwPFS HR of receiving a matched TKI in the 1L for NonsqNSCLC subcohorts defined by concordance/discordance between FMI DNA CGP and orthogonal fusion testing results. (C) Among patients who received 1L-matched TKI, patients who were negative for ALK, NTRK, RET, and ROS1 fusions on FMI DNA CGP had shorter rwPFS than patients who were positive by FMI DNA CGP. Unadjusted Kaplan-Meier plot is shown. All analyses are indexed to the start of 1L therapy. In addition to univariable Cox model HRs, adjusted HRs are presented for a multivariable Cox model that includes established prognostic variables (Supplementary Fig. S6). A small number of cases were excluded from multivariable analyses due to missingness of covariables. Abbreviations: FMI DNA CGP, Foundation Medicine Tissue DNA comprehensive genomic profiling; HR, hazard ratio; TKI, tyrosine kinase inhibitor; rwPFS, real-world progression-free survival; 1L, first advanced line of Tx.
Among 193 patients for whom oncogenic fusions in ALK, NTRK1/2/3, RET, or ROS1 were detected on both FMI and orthogonal assays and for whom rwPFS data were available, those who received a matched TKI in the advanced first line of therapy had nominally (nonsignificantly) better rwPFS than those who received an alternative treatment regimen (univariable HR 0.81, 95% CI 0.59-1.11, P = .19; Supplementary Fig. S5A). After adjustment for known prognostic factors in a multivariable Cox model, this difference was statistically significant (multivariable HR 0.53, 95% CI 0.35-0.79, P = .002; Fig. 3B; Supplementary Fig. S6A).
Among the 148 patients whose ALK, NTRK, RET, and ROS1 fusions were detected by FMI DNA CGP but were not observed by orthogonal testing, patients treated with a matched TKI had better rwPFS than those receiving an alternative treatment regimen (univariable HR 0.59, 95% CI 0.39-0.90, P = .012; Multivariable HR 0.60, 95% CI 0.37-0.99, P = .05; Fig. 3B; Supplementary Fig. S5B; Supplementary Fig. S6B). However, among the 54 patients for whom FMI DNA CGP did not detect an actionable fusion in ALK, NTRK, RET, or ROS1 when an orthogonal assay did detect a fusion, patients who received a matched TKI had worse rwPFS than those who did not (univariable HR 3.48, 95% CI 1.75-6.91, P < .001; multivariable HR 4.59, 95% CI 1.29-16.32, P = .019; Fig. 3B; Supplementary Fig. S5C; Supplementary Fig. S6C).
Across all patients who received a matched TKI and had at least one test which detected an ALK, NTRK, RET, or ROS1 fusion (FMI DNA CGP and/or orthogonal, n = 185), those with fusions detected by FMI DNA CGP had significantly longer rwPFS than those with fusions not detected by FMI DNA CGP, regardless of the result of orthogonal testing (relative to FMI DNA CGP−/orthogonal+: FMI DNA CGP+/orthogonal+ univariable HR 0.25, 95% CI 0.14-0.45, P < .001; FMI DNA CGP+/orthogonal− univariable HR 0.18, 95% CI 0.09-0.35, P < .001; Fig. 3C). These results were more pronounced when applying a multivariable Cox model adjusting for various known prognostic factors (Supplementary Fig. S6D).
Expected Detection of Clinically Actionable Fusions Using Complementary RNA CGP in NonSqNSCLC
To quantify the added value of RNA CGP following DNA CGP for detecting clinically important driver fusions in NonSqNSCLC, we used a mathematical model which takes into account several real-world factors. We started with the assumption that patients with an oncogenic driver detected on DNA CGP are unlikely to have an additional driver identified on RNA CGP since drivers in NSCLC tend to be mutually exclusive. To estimate the probability of identifying an oncogenic driver on DNA CGP, we analyzed genomic profiling data from 10 761 patients with NonSqNSCLC who received tissue-based FMI DNA CGP. We stratified the cohort by self-reported smoking status given established genomic differences between these 2 NSCLC subpopulations10,40,41 to inform our sensitivity analyses (Table 2; Supplementary Fig. S3). We explored the prevalence of fusion and nonfusion oncogenic driver alterations in RTK genes and the MAPK pathway. Notably, this included all patients with actionable driver alterations as defined in the NCCN guidelines for NSCLC16 as well as additional known biological drivers (Supplementary Table S4). As defined, an oncogenic driver alteration was not detected in 12% of never-smokers and 33% of ever-smokers with NonSqNSCLC (Supplementary Fig. S7). These prevalence estimates were used to inform one parameter of the model (θDNA), the probability of detecting an oncogenic driver on DNA CGP.
Table 2.
Clinico-genomic characteristics of patients with NonSqNSCLC stratified by smoking history.
| Clinico-genomic characteristics | Smoking status known (N = 8747) | Former or current smoker (N = 7081) | Never-smoker (N = 1666) | P |
|---|---|---|---|---|
| Age at diagnosis (years), median (IQR) | 67.0 (60.0;74.0) | 68.0 (60.0;74.0) | 66.0 (57.0;74.0) | <.001 |
| Age at diagnosis (categorical), n (%) | ||||
| <45 | 192 (2.2%) | 97 (1.4%) | 95 (5.7%) | <.001 |
| ≥45 | 8555 (97.8%) | 6984 (98.6%) | 1571 (94.3%) | |
| Sex, n (%) | ||||
| Female | 4801 (54.9%) | 3682 (52.0%) | 1119 (67.2%) | <.001 |
| Male | 3946 (45.1%) | 3399 (48.0%) | 547 (32.8%) | |
| Self-reported race, n (%) | ||||
| Asian | 301 (3.4%) | 126 (1.8%) | 175 (10.5%) | <.001 |
| Black or African American | 595 (6.8%) | 480 (6.8%) | 115 (6.9%) | |
| Hispanic or Latino | 8 (0.1%) | 6 (0.1%) | 2 (0.1%) | |
| Other race | 1207 (13.8%) | 978 (13.8%) | 229 (13.7%) | |
| White | 5807 (66.4%) | 4819 (68.1%) | 988 (59.3%) | |
| Unknown/not documented | 829 (9.5%) | 672 (9.5%) | 157 (9.4%) | |
| AJCC stage at diagnosis, n (%) | ||||
| I | 1055 (12.1%) | 893 (12.6%) | 162 (9.7%) | <.001 |
| II | 700 (8.0%) | 602 (8.5%) | 98 (5.9%) | |
| III | 1639 (18.7%) | 1394 (19.7%) | 245 (14.7%) | |
| IV | 5093 (58.2%) | 3984 (56.3%) | 1109 (66.6%) | |
| Other/unknown/not documented | 260 (3.0%) | 208 (2.9%) | 52 (3.1%) | |
| Practice type, n (%) | ||||
| Academic | 1051 (12.0%) | 784 (11.1%) | 267 (16.0%) | <.001 |
| Academic/community | 498 (5.7%) | 383 (5.4%) | 115 (6.9%) | |
| Community | 7198 (82.3%) | 5914 (83.5%) | 1284 (77.1%) | |
| Socioeconomic status, n (%) | ||||
| 1—Lowest SES | 1199 (13.7%) | 1049 (14.8%) | 150 (9.0%) | <.001 |
| 2 | 1519 (17.4%) | 1298 (18.3%) | 221 (13.3%) | |
| 3 | 1755 (20.1%) | 1446 (20.4%) | 309 (18.5%) | |
| 4 | 1811 (20.7%) | 1430 (20.2%) | 381 (22.9%) | |
| 5—Highest SES | 1694 (19.4%) | 1238 (17.5%) | 456 (27.4%) | |
| Unknown | 769 (8.8%) | 620 (8.8%) | 149 (8.9%) | |
| ECOG PS at diagnosis, n (%) | ||||
| 0 | 1768 (20.2%) | 1416 (20.0%) | 352 (21.1%) | <.001 |
| 1 | 1839 (21.0%) | 1545 (21.8%) | 294 (17.6%) | |
| 2 | 479 (5.5%) | 423 (6.0%) | 56 (3.4%) | |
| 3+ | 121 (1.4%) | 106 (1.5%) | 15 (0.9%) | |
| Unknown | 4540 (51.9%) | 3591 (50.7%) | 949 (57.0%) | |
| Tumor mutational burden (Mut/Mb), n (%) | ||||
| TMB < 10 | 5690 (65.2%) | 4121 (58.3%) | 1569 (94.5%) | <.001 |
| TMB (10-20) | 2029 (23.2%) | 1964 (27.8%) | 65 (3.9%) | |
| TMB ≥ 20 | 1011 (11.6%) | 985 (13.9%) | 26 (1.6%) | |
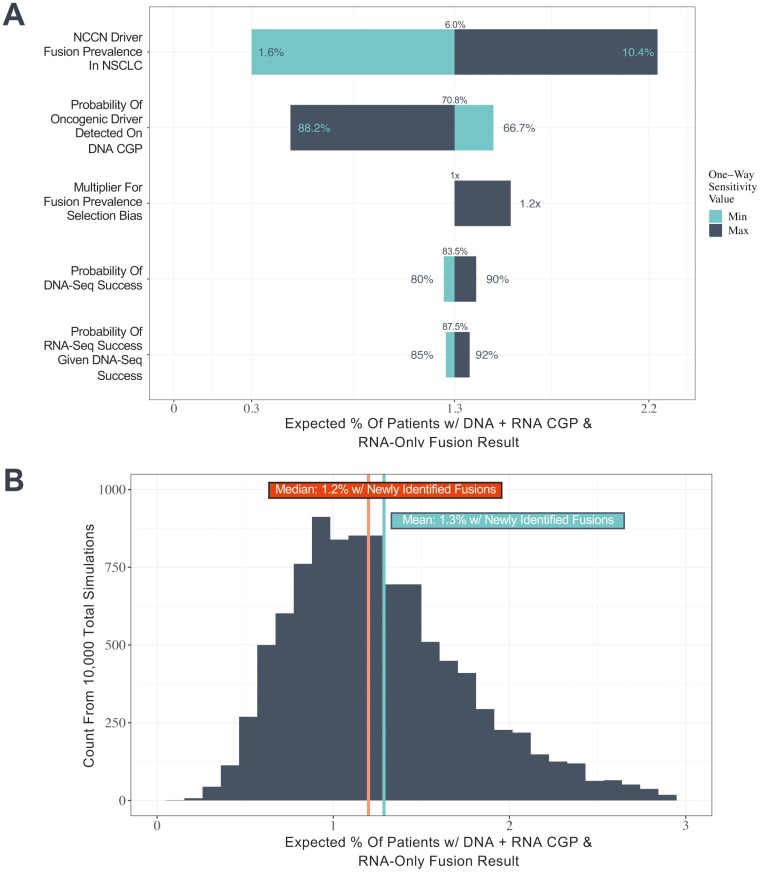
Our modeling estimates that approximately 1.3% of patients with NSCLC are expected to have an actionable driver fusion uniquely identifiable on RNA CGP (Fig. 4A). This estimate assumes (1) an underlying driver fusion prevalence of 6.0% in NSCLC based on the reported prevalence of ALK, RET, ROS1, and NTRK fusions in NSCLC and (2) a 70.8% probability of detecting an oncogenic driver using DNA CGP in the unselected patient population with NSCLC. (The full set of assumptions, parameter sources, and calculation approach are available in Methods, Supplementary Table S3, and Supplementary Fig. S8.) To determine the most influential parameters underlying the base case estimate of 1.3%, we conducted one-way sensitivity analyses (Fig. 4A). The most influential parameters were (1) underlying NSCLC fusion prevalence and (2) the probability of a driver alteration being detected on DNA CGP. However, even with an increase in the population-level fusion prevalence estimate from the base case of 6% to the maximum of 10%, we estimate a newly identified fusion will be detected on RNA CGP in only 2.2% of patients. Across all minimum and maximum potential parameter values for the one-way sensitivity analysis, this estimate ranged from 0.34% to 2.22% (Supplementary Table S6). We also conducted a probabilistic sensitivity analysis which accounts for uncertainty in all input parameters simultaneously (Fig. 4B; see distribution details in Supplementary Table S3 and individual parameter draws in Supplementary Fig. S9). Across 10 000 simulations, 95% of estimates were at or below 2.2% (minimum 0.1%, maximum 5.9%).
Figure 4.
1.3% of patients with NonSqNSCLC are expected to have an actionable fusion detected by RNA CGP that is not detected by DNA CGP. We conducted (A) one-way sensitivity analyses on key model parameters including the prevalence of oncogenic driver fusions in ALK, NTRK, RET, and ROS1 in NSCLC, the probability an oncogenic driver is detected in NSCLC using DNA CGP, the probability of successful DNA and RNA sequencing, and the potential selection bias for fusion-positive patients among those that do not have oncogenic driver alterations detected on DNA CGP and (B) a probabilistic sensitivity analysis incorporating parameter uncertainty jointly with mean and median estimates across 10 000 simulations indicated.
Discussion
FMI DNA CGP assays employ a multifaceted approach to enable robust DNA-based rearrangement detection (Figure 1). In addition to baiting breakpoint hotspot introns in important fusion genes, the platform also baits breakpoint hotspot introns in previously reported fusion partner genes. Intentional baiting of specific partners allows for the detection of fusions involving clinically important oncogenes that are difficult to bait directly,14,42 eg, detection of a subset of NRG1 fusions is achieved through baiting of CD74 and SDC4 and detection of a subset of NTRK3 fusions is achieved through baiting of ETV6. The platform includes select intronic targeting for 34 genes (Supplementary Table S1). In total, over 13% of the targeted region is dedicated to intronic coverage for the express purpose of rearrangement detection. Another feature of the assay that supports rearrangement detection is the use of a hybrid capture-based sequencing approach which requires that only one gene involved in a fusion be baited for detection, whereas its partner can be any gene in the genome even if not explicitly targeted by the assay. This allows for the detection of fusions involving recurrent or rare known partners as well as novel partners. Together with select partner gene baiting, this approach also supports the detection of known oncogenes with rare or novel breakpoints outside of baited hotspot intronic regions and oncogenes with hotspot intronic regions that cannot be efficiently baited (eg, NRG1 and NTRK3). The bioinformatics pipeline used for fusion calling also contributes to the capabilities of the assay for rearrangement detection. Initially, clusters of discordant read pairs are used to identify potential genomic rearrangement candidates. These candidates are further refined using a de novo local genomic assembly approach.22,43 This method facilitates precise breakpoint detection compared to reference-based methods because rearrangement breakpoints often feature novel sequences introduced by DNA repair mechanisms that cannot be accurately mapped. More accurate breakpoint detection allows for better predictions regarding the functional effect of rearrangement events.
Together, these design features enable robust detection of rearrangements and fusions with a DNA-based assay. We observed a broad diversity of both common and novel fusion partners associated with clinically actionable fusion genes including ALK, BRAF, FGFR2/3, NTRK1/2/3, RET, and ROS1 using FMI DNA CGP. Moreover, we established that ALK, NTRK, RET, and ROS1 fusions detected using FMI DNA CGP were highly correlated with favorable outcomes on matched TKI, whereas fusions solely detected using orthogonal methods were not, illustrating that FMI DNA CGP is a robust modality for fusion detection compared to other methods, consistent with prior reports.44,45
While most highly recurrent fusion partners were common to both the FMI and AACR GENIE datasets, one glaring discrepancy was the high frequency of FGFR2-NPM1 fusions reported in GENIE. While FGFR2-NPM1 rearrangements were detected using FMI DNA CGP, they were considered variants of uncertain significance. The FGFR2-NPM1 rearrangements detected on FMI DNA CGP had breakpoints in FGFR2 intron 9 without retention of the kinase domain and showed a cancer type distribution distinct from known pathogenic FGFR2 fusions, ie, found in NSCLC, colorectal (CRC), and other cancers but only infrequently in cholangiocarcinoma or other pancreaticobiliary cancers (data not shown). Furthermore, there is no published functional characterization of FGFR2-NPM1 fusions to our knowledge. While these fusion events have been described in a prior study, the breakpoints and domain organization of these fusions were not detailed and they likewise were found in cancer types not typically associated with FGFR2 fusions (CRC N = 2, large cell lung carcinoma N = 1).46 Notably, all FGFR2-NPM1 fusions reported in GENIE were from a single institution, likely reflecting differences in reporting rules between institutions/assays. Information about the intronic breakpoints for these fusions was not provided. The bioinformatic pipeline used for rearrangement calling/curation and manual review/interpretation are also key factors affecting assay reliability as not all structural variants are potentially pathogenic. Thus, the dependability of DNA-based rearrangement detection is reliant on both the technical capabilities of the assay as well as the reporting rules.
Both DNA and RNA CGP have advantages and shortcomings for fusion detection. While well-designed DNA CGP assays can detect a wide diversity of rearrangements and fusions, RNA CGP is better poised to detect certain rearrangement events. Whereas DNA CGP is capable of robust detection of canonical fusions with recurrent breakpoints since established breakpoint hotspots can be intentionally baited, RNA CGP can detect atypical fusions with breakpoints outside of hotspot regions. RNA CGP can also detect emergent fusions in genes not included on established DNA panels (eg, NRG1) due to technical limitations associated with baiting of certain fusion genes, eg, challenges related to long intronic regions which are prohibitive of efficient tiling as well as repetitive intronic sequence elements.14,42 Notably, detection of fusions involving these genes using a DNA CGP assay is still possible through baiting of fusion partners, eg, a subset of NRG1 fusions are detected using FMI DNA CGP due to baiting of partner genes CD74 and SDC4. RNA CGP is also ideal for identifying diagnostic fusions not commonly covered on DNA panels (eg, in sarcomas47 and salivary gland tumors48-50) as the multitude and cancer type specificity of such fusions makes it infeasible to bait intronic regions of all involved genes due to panel size restrictions. In addition, RNA profiling can clarify the functional significance of detected rearrangements since expression at the mRNA level is confirmed, whereas expression of fusions detected through DNA profiling is merely predicted, complicating interpretation of noncanonical rearrangements.51 However, the technical difficulty of working with RNA is a significant limitation to RNA CGP.18-21 In addition to general issues of stability and quality, a low level of mRNA expression in cells can limit the detection of alterations resulting in false negatives.20 Notably, DNA CGP can also be performed on circulating tumor DNA from liquid biopsies when tissue material is not readily available,52-54 whereas RNA profiling using liquid biopsies is not widely offered.55
It is estimated that approximately 30% of NSCLC adenocarcinoma lack a well-defined oncogenic driver alteration.7-10 Using tissue-based FMI DNA CGP, we observed that approximately 12%-33% of NonSqNSCLC tumors lack a clear oncogenic driver with driver alterations enriched in never-smokers (88%) compared to ever-smokers (67%), consistent with prior studies.10 However, it is important to clarify that the absence of a known oncogenic driver on DNA CGP does not necessarily equate to a missed driver, eg, a known oncogenic driver fusion that was not detectable on DNA CGP but may be detectable on RNA CGP. We expect such cases to represent a much smaller percentage of the overall NSCLC population with the remainder of driver-negative cases explained by as yet undiscovered biology.9 Accounting for several real-world factors associated with DNA/RNA CGP and the reported landscape of driver fusions in NSCLC, we estimate that approximately 1.3% of patients with NonSqNSCLC would be expected to have a driver fusion identified on RNA CGP but not on DNA CGP. This estimate is consistent with prior studies in which (1) reflex to RNA CGP after driver-negative DNA CGP identified an additional 1.1% of patients with NSCLC with targetable fusions14 and (2) concurrent DNA- and RNA-seq identified 0.3% of additional patients with actionable fusions across 20 cancer types.56 While this yield may appear relatively low in all-comers, RNA CGP may be of greater value in select subpopulations, eg, non-smokers with NSCLC10 and driver-negative NSCLC with low TMB.14 In our study, the estimated increase in fusion detection afforded by RNA CGP represents approximately 4.4% of cases that would otherwise have been classified as driver-negative NonSqNSCLC.
Limitations of this study include the use of CGP results from tissue biopsies submitted during routine clinical care which introduces certain biases, compounded by comparison to an external database consisting of multiple patient cohorts each with its own inherent biases. While AACR Project GENIE data represent multiple assays from multiple institutions which complicates comparison to results from a single assay, the GENIE database is one of the largest repositories of publicly available clinically annotated genomic data in the world, including data from leading academic medical centers with established high-quality in-house NGS platforms, as well as other commercial assays, and thus represents a reasonable point of reference for our analysis. Additionally, our mathematical modeling approach was based on a number of assumptions and our parameter estimates were based on data/studies with their own respective sets of limitations. A more accurate estimate would require formal analysis of samples analyzed with both DNA and RNA CGP which is beyond the scope of the current study but of high interest for future investigations.
Conclusion
This study shows the robust capability of DNA CGP to detect clinically important fusions. There should be high confidence in the reliability of fusion calls from a well-designed DNA CGP assay to inform treatment decisions. However, variation in assay performance is affected by differences in design strategies, validation standards, and reporting rules among manufacturers and academic laboratories and not all DNA CGP assays are equally reliable. While we predicted an additional 1%-2% of patients with actionable driver fusions could potentially be detected with the addition of RNA CGP to a DNA CGP assay in NonSqNSCLC, the actual population prevalence in different tumor types and clinical scenarios must be studied in large cohorts in which DNA and RNA are profiled concurrently and would necessarily be dependent on the capabilities of the included assays. While the expected yield of additional fusion detection using RNA CGP may be low in the unselected cancer population, the potential clinical benefit of detecting an actionable fusion driver for an individual patient is significant and every effort should be made to identify these drivers. Therefore, reflex to RNA CGP should be considered when DNA CGP does not reveal a clear oncogenic driver.
Supplementary Material
Acknowledgments
This analysis was supported by Foundation Medicine, Inc., Cambridge, MA, USA.
Contributor Information
Philip C Mack, Center for Thoracic Oncology, Tisch Cancer Institute, Icahn School of Medicine, Mount Sinai, New York, NY, USA.
Rachel B Keller-Evans, Foundation Medicine, Inc., Cambridge, MA, USA.
Gerald Li, Foundation Medicine, Inc., Cambridge, MA, USA.
Katherine T Lofgren, Foundation Medicine, Inc., Cambridge, MA, USA.
Alexa B Schrock, Foundation Medicine, Inc., Cambridge, MA, USA.
Sally E Trabucco, Foundation Medicine, Inc., Cambridge, MA, USA.
Justin M Allen, Foundation Medicine, Inc., Cambridge, MA, USA.
Khaled Tolba, Foundation Medicine, Inc., Cambridge, MA, USA.
Geoffrey R Oxnard, Foundation Medicine, Inc., Cambridge, MA, USA.
Richard S P Huang, Foundation Medicine, Inc., Cambridge, MA, USA.
Funding
This study was funded by Foundation Medicine, Inc.
Conflict of Interest
P.C.M. is on the advisory board for Guardant Health, has received honoraria from Amgen, and consults for Vivace Therapeutics. R.B.K.-E., G.L., K.T.L., A.B.S., S.E.T., J.M.A., K.T., and R.S.P.H. are employees of Foundation Medicine, a wholly owned subsidiary of Roche, and have an equity interest in Roche. G.R.O. is an employee of Lilly and has an equity interest in Roche.
Author Contributions
Conception/design: R.B.K.-E., G.L., K.T., G.R.O., R.S.P.H. Provision of study material or patients: R.S.P.H. Collection and/or assembly of data: R.B.K., G.L., K.T.L. Data analysis and interpretation: P.C.M., R.B.K., G.L., K.T.L., A.B.S., J.M.A., K.T., G.R.O., R.S.H.P. Manuscript writing and final approval of manuscript: All authors.
Data Availability
The authors declare that all relevant aggregate data supporting the findings of this study are available within the article and its supplementary information files. The data that support the findings of this study originated from Flatiron Health, Inc. and Foundation Medicine, Inc. Requests for data sharing by license or by permission for the specific purpose of replicating results in this manuscript can be submitted to PublicationsDataaccess@flatiron.com and cgdb-fmi@flatiron.com. In accordance with the Health Insurance Portability and Accountability Act, we do not have IRB approval or patient consent to share individualized patient genomic data, which contains potentially identifying or sensitive patient information and cannot be reported in a public data repository. Foundation Medicine is committed to collaborative data analysis and has well established and widely used mechanisms by which qualified researchers can query our core genomic database of >700 000 de-identified sequenced cancers. Academic researchers can submit a proposal to the Foundation Medicine Data Collaborations Committee and, if approved, the researcher/institution will be required to complete a Data Usage Agreement. More information and mechanisms for data access can be obtained by contacting the corresponding author or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com. This study made use of publicly available packages with R version 4.2.2. Code used to perform statistical analyses and generate figures is available upon request.
References
- 1. Bailey MH, Tokheim C, Porta-Pardo E, et al. ; MC3 Working Group. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173(2):371-385.e18. 10.1016/j.cell.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berger MF, Mardis ER.. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15(6):353-365. 10.1038/s41571-018-0002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakravarty D, Solit DB.. Clinical cancer genomic profiling. Nat Rev Genet. 2021;22(8):483-501. 10.1038/s41576-021-00338-8 [DOI] [PubMed] [Google Scholar]
- 4. Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017(1):1-16. 10.1200/po.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M, Herbst RS, Boshoff C.. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. 2021;27(8):1345-1356. 10.1038/s41591-021-01450-2 [DOI] [PubMed] [Google Scholar]
- 6. Howlader N, Forjaz G, Mooradian MJ, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383(7):640-649. 10.1056/NEJMoa1916623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Devarakonda S, Morgensztern D, Govindan R.. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16(7):e342-e351. 10.1016/S1470-2045(15)00077-7 [DOI] [PubMed] [Google Scholar]
- 8. Chen R, Manochakian R, James L, et al. Emerging therapeutic agents for advanced non-small cell lung cancer. J Hematol Oncol. 2020;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrot-Zhang J, Yao X, Devarakonda S, et al. ; TCGA Research Network. Whole-genome characterization of lung adenocarcinomas lacking alterations in the RTK/RAS/RAF pathway. Cell Rep. 2021;34(8):108784. 10.1016/j.celrep.2021.108784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mack PC, Klein MI, Ayers KL, et al. Targeted next-generation sequencing reveals exceptionally high rates of molecular driver mutations in never-smokers with lung adenocarcinoma. Oncologist. 2022;27(6):476-486. 10.1093/oncolo/oyac035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape & therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845-4854. 10.1038/onc.2014.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao Q, Liang WW, Foltz SM, et al. ; Fusion Analysis Working Group. Driver fusions & their implications in the development & treatment of human cancers. Cell Rep. 2018;23(1):227-238.e3. 10.1016/j.celrep.2018.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suda K, Mitsudomi T.. Emerging oncogenic fusions other than ALK, ROS1, RET, and NTRK in NSCLC and the role of fusions as resistance mechanisms to targeted therapy. Transl Lung Cancer Res. 2020;9(6):2618-2628. 10.21037/tlcr-20-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25(15):4712-4722. 10.1158/1078-0432.CCR-19-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen D, Hondelink LM, Solleveld-Westerink N, et al. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J Thorac Oncol. 2020;15(6):1000-1014. 10.1016/j.jtho.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 16. Ettinger DS, Wood DE, Aisner DL, et al. National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology - Non-Small Cell Lung Cancer Version 3.2023. 2023. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf [DOI] [PubMed]
- 17. Chakravarty D, Johnson A, Sklar J, et al. Somatic genomic testing in patients with metastatic or advanced cancer: ASCO provisional clinical opinion. J Clin Oncol. 2022;40(11):1231-1258. 10.1200/JCO.21.02767 [DOI] [PubMed] [Google Scholar]
- 18. Heyer EE, Blackburn J.. Sequencing strategies for fusion gene detection. Bioessays. 2020;42(7):e2000016. 10.1002/bies.202000016 [DOI] [PubMed] [Google Scholar]
- 19. Bruno R, Fontanini G.. Next generation sequencing for gene fusion analysis in lung cancer: a literature review. Diagnostics (Basel). 2020;10(8):521. 10.3390/diagnostics10080521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bekaii-Saab TS, Bridgewater J, Normanno N.. Practical considerations in screening for genetic alterations in cholangiocarcinoma. Ann Oncol. 2021;32(9):1111-1126. 10.1016/j.annonc.2021.04.012 [DOI] [PubMed] [Google Scholar]
- 21. Rapoport BL, Troncone G, Schmitt F, Nayler S.. Fast facts: comprehensive genomic profiling. 2020. 10.1159/isbn.978-3-318-06819-1 [DOI]
- 22. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. AACR Project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pugh TJ, Bell JL, Bruce JP, et al. ; AACR Project GENIE Consortium, Genomics and Analysis Working Group. AACR project GENIE: 100,000 cases and beyond. Cancer Discov. 2022;12(9):2044-2057. 10.1158/2159-8290.CD-21-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non–small cell lung cancer using a clinicogenomic database. JAMA. 2019;321(14):1391-1399. 10.1001/jama.2019.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffith SD, Tucker M, Bowser B, et al. Generating real-world tumor burden endpoints from electronic health record data: comparison of RECIST, radiology-anchored, and clinician-anchored approaches for abstracting real-world progression in non-small cell lung cancer. Adv Ther. 2019;36(8):2122-2136. 10.1007/s12325-019-00970-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Q, Gossai A, Monroe S, Nussbaum NC, Parrinello CM.. Validation analysis of a composite real‐world mortality endpoint for patients with cancer in the United States. Heal Serv Res. 2021;56(6):1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15(9):3143-3149. 10.1158/1078-0432.CCR-08-3248 [DOI] [PubMed] [Google Scholar]
- 30. Bergethon K, Shaw AT, Ou SI, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863-870. https://doi.org/10.1200/JCO.2011.35.6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. To KF, Tong JHM, Yeung KSF, et al. Detection of ALK rearrangement by immunohistochemistry in lung adenocarcinoma and the identification of a novel EML4-ALK variant. J Thorac Oncol. 2013;8(7):883-891. 10.1097/JTO.0b013e3182904e22 [DOI] [PubMed] [Google Scholar]
- 32. Selinger CI, Rogers TM, Russell PA, et al. Testing for ALK rearrangement in lung adenocarcinoma: a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol. 2013;26(12):1545-1553. 10.1038/modpathol.2013.87 [DOI] [PubMed] [Google Scholar]
- 33. Pan Y, Zhang Y, Li Y, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84(2):121-126. 10.1016/j.lungcan.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 34. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C.. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375-377. 10.1038/nm.2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang R, Hu H, Pan Y, et al. RET Fusions define a unique molecular and clinicopathologic subtype of non–small-cell lung cancer. J Clin Oncol. 2012;30(35):4352-4359. 10.1200/JCO.2012.44.1477 [DOI] [PubMed] [Google Scholar]
- 37. Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non–small cell lung cancer. Clin Cancer Res. 2012;18(17):4570-4579. 10.1158/1078-0432.CCR-12-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okamura R, Boichard A, Kato S, et al. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol. 2018;2018(2):1-20. 10.1200/po.18.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosen EY, Goldman DA, Hechtman JF, et al. TRK Fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin Cancer Res. 2020;26(7):1624-1632. 10.1158/1078-0432.CCR-19-3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim HR, Lim SM, Kim HJ, et al. The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol. 2013;24(9):2364-2370. 10.1093/annonc/mdt220 [DOI] [PubMed] [Google Scholar]
- 41. Kim MH, Shim HS, Kang DR, et al. Clinical and prognostic implications of ALK and ROS1 rearrangements in never-smokers with surgically resected lung adenocarcinoma. Lung Cancer. 2014;83(3):389-395. 10.1016/j.lungcan.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 42. Treangen TJ, Salzberg SL.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2012;13(1):36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Compeau PEC, Pevzner PA, Tesler G.. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29(11):987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ali SM, Hensing T, Schrock AB, et al. Comprehensive genomic profiling identifies a subset of crizotinib‐responsive ALK‐rearranged non‐small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016;21(6):762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schrock AB, Madison R, Rosenzweig M, et al. Patients with NSCLCs harboring internal inversions or deletion rearrangements of the ALK gene have durable responses to ALK kinase inhibitors. Lung Cancer (Auckl). 2020;11:33-39. 10.2147/LCTT.S239675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Helsten T, Elkin S, Arthur E, et al. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22(1):259-267. 10.1158/1078-0432.CCR-14-3212 [DOI] [PubMed] [Google Scholar]
- 47. Gounder MM, Agaram NP, Trabucco SE, et al. Clinical genomic profiling in the management of patients with soft tissue and bone sarcoma. Nat Commun. 2022;13:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brayer KJ, Frerich CA, Kang H, Ness SA.. Recurrent fusions in MYB and MYBL1 define a common, transcription factor–driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 2016;6(2):176-187. 10.1158/2159-8290.CD-15-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martins C, Fonseca I, Roque L, et al. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18(8):1048-1055. 10.1038/modpathol.3800386 [DOI] [PubMed] [Google Scholar]
- 50. Jee KJ, Persson M, Heikinheimo K, et al. Genomic profiles and CRTC1–MAML2 fusion distinguish different subtypes of mucoepidermoid carcinoma. Mod Pathol. 2013;26(2):213-222. 10.1038/modpathol.2012.154 [DOI] [PubMed] [Google Scholar]
- 51. Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM.. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7(129):129. 10.1186/s13073-015-0252-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15(9):e0237802. 10.1371/journal.pone.0237802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee JK, Hazar-Rethinam M, Decker B, et al. The Pan-Tumor Landscape Of Targetable Kinase Fusions In Circulating Tumor DNA. Clin Cancer Res. 2021;28(4):728-737. 10.1158/1078-0432.ccr-21-2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Husain H, Pavlick DC, Fendler BJ, et al. Tumor fraction correlates with detection of actionable variants across > 23,000 circulating tumor DNA samples. JCO Precis Oncol. 2022;6(6):e2200261. 10.1200/PO.22.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bardelli A, Pantel K.. Liquid biopsies, what we do not know (Yet). Cancer Cell. 2017;31(2):172-179. 10.1016/j.ccell.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 56. Michuda J, Park BH, Cummings AL, et al. Use of clinical RNA-sequencing in the detection of actionable fusions compared to DNA-sequencing alone. J Clin Oncol. 2022;40(16_suppl):3077-3077. 10.1200/jco.2022.40.16_suppl.307735442716 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant aggregate data supporting the findings of this study are available within the article and its supplementary information files. The data that support the findings of this study originated from Flatiron Health, Inc. and Foundation Medicine, Inc. Requests for data sharing by license or by permission for the specific purpose of replicating results in this manuscript can be submitted to PublicationsDataaccess@flatiron.com and cgdb-fmi@flatiron.com. In accordance with the Health Insurance Portability and Accountability Act, we do not have IRB approval or patient consent to share individualized patient genomic data, which contains potentially identifying or sensitive patient information and cannot be reported in a public data repository. Foundation Medicine is committed to collaborative data analysis and has well established and widely used mechanisms by which qualified researchers can query our core genomic database of >700 000 de-identified sequenced cancers. Academic researchers can submit a proposal to the Foundation Medicine Data Collaborations Committee and, if approved, the researcher/institution will be required to complete a Data Usage Agreement. More information and mechanisms for data access can be obtained by contacting the corresponding author or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com. This study made use of publicly available packages with R version 4.2.2. Code used to perform statistical analyses and generate figures is available upon request.