Abstract
Regeneration is vital for many organisms, enabling them to repair injuries and adapt to environmental changes. The mechanisms underlying regeneration are complex and involve coordinated events at the cellular and molecular levels. Moreover, while some species exhibit remarkable regenerative capabilities, others, like mammals, have limited regenerative potential. Central to this process is the regulation of gene expression, and among the numerous genes involved, MYC emerges as a regulator of relevant processes during regeneration with roles conserved in several species, including Drosophila. This mini-review aims to provide valuable insights into the regeneration process in flies, focusing on significant organs where the role of MYC has been identified: from the imaginal discs, where MYC regulates cell growth, structure, and proliferation, to the gut, where it maintains the balance between renewal and differentiation of stem cells, and the central nervous system, where it influences the activities of neural stem cells and the interaction between glia and neuronal cells. By emphasizing the molecular mechanisms regulated by MYC, its significance in controlling regeneration mechanisms, and its conserved role in flies, we aim to offer valuable insights into the utility of Drosophila as a model for studying regeneration. Moreover, unraveling MYC’s function in Drosophila during regeneration may help translate findings into the mechanisms underlying human tissue repair.
Keywords: MYC, regeneration, imaginal discs, epithelial cells, gut, neurons and glia, Drosophila
1 Regeneration
The ability to regenerate and restore lost body parts after injury reflects key physiological pathways governed by developmental processes; regeneration capacity is widespread in animals and, in some species, has been lost during evolution, contributing to the variations in regenerative capacities across species (Losner et al., 2021). While remarkable abilities are observed in cnidarians, crustaceans, salamanders, and certain vertebrates, humans have limited regenerative potential (Wells and Watt, 2018), underscoring the need to understand molecular mechanisms of tissue and organ development for regenerative medicine.
Animal regeneration is categorized into five types: 1) structural regeneration, seen in the distal regrowth of appendages in vertebrates and arthropods; 2) organ regeneration, where damaged organs restore their mass; 3) tissue regeneration, responding to damaged epithelial or epidermis; 4) whole-body regeneration, involving the regrowth of an organism’s central axis; and 5) cellular regeneration, such as the regrowth of severed nerve axons (Bely and Nyberg, 2010). Regeneration, depending on tissue and damage types, involves distinct steps, including wound healing, the formation of a proliferative blastema, cellular differentiation, and tissue patterning. The blastema, comprised of progenitor cells responsible for the regeneration process, is formed temporarily at the injury site and undergoes morphogenesis through cell migration and proliferation to regenerate the missing organ (King and Newmark, 2012; Slack, 2017). Additionally, immune cells at the injury site play a crucial role in debris clearance and secretion of signaling molecules, initiating specific cellular proliferation and differentiation processes necessary for thriving tissue regeneration (Julier et al., 2017). Despite the progress made in understanding tissue regeneration, identifying novel signaling pathways that govern reprogramming mechanisms remains a significant challenge. Consequently, simple animal models are indispensable for gaining a deeper understanding of these intricate processes.
Although Drosophila does not possess the extensive regenerative abilities of some other species, its advanced genetic technology, previously used to uncover the complex genetic networks governing development, framework, which connects body parts and identity genes (such as the Hox genes), as well as pattern formation components (like Hedgehog, Decapentaplegic (Dpp), and Wingless (Wg) analogous to vertebrate Wnt), can now be utilized to investigate the molecular basis of regeneration (Fox et al., 2020). Here, we review the role of MYC in regeneration models such as wing imaginal discs, gut, and neuronal-glia cells, where processes like cell growth, division, and apoptosis may depend critically on MYC’s function.
2 Drosophila MYC
The MYC/MAX/MAD network in Drosophila stands out for its lack of redundancy, as the Drosophila genome contains a single gene for each component (Gallant, 2006). Despite being only 26% identical to its human counterpart, the Drosophila MYC protein shares highly conserved functional domains such as Box I and II, the degron sequences, and the basic-helix-loop-helix leucine zipper (bHLH/LZ) domain, to mediate MYC: MAX heterodimers that bind the E-box sequences on target genes (Orian et al., 2003; Hulf et al., 2005). The discovery that MYC mutants, also called diminutive, are composed of smaller cells (Johnston et al., 1999) paved the way for genetic experiments that revealed MYC’s role in controlling growth and ribosomal biogenesis. The similarity in phenotypes between MYC mutants and those of the insulin (InR/IRS/chico) (Bohni et al., 1999) and Target of Rapamycin (TOR/S6K) (Montagne et al., 1999) pathways has contributed to unveiling how growth pathways influence MYC activity in flies (Bellosta and Gallant, 2010; Parisi et al., 2011). These studies revealed the control of MYC protein stability by growth factors signaling through the phosphorylation of conserved domains (degrons) by Ras-ERK/MAPK and GSK3ß kinases, confirming this pathway of MYC protein degradation in flies (Galletti et al., 2009; Schwinkendorf and Gallant, 2009). Furthermore, MYC levels increase during starvation in the fat body, a metabolic tissue that parallels the function of vertebrate adipose tissue and the liver (Teleman et al., 2008; Parisi et al., 2013). Indeed, we showed that MYC increases metabolic processes like glycolysis and glutaminolysis during nutrient starvation (Parisi et al., 2013; de la Cova et al., 2014) and promotes the catabolic process autophagy in the fat cells, leading to survival (Nagy et al., 2013; Paiardi et al., 2017).
MYC’s control over ribosome biogenesis is highlighted by its coordination of RNA polymerases I, II, and III activities. MYC facilitates the recruitment of RNA polymerase I to rDNA, ensuring proper rRNA synthesis with the transcription of ribosomal proteins (Destefanis et al., 2020). MYC’s role in regulating ribosomal biogenesis led to the discovery of its role in cell competition; a physiological process initially observed in flies heterozygous for the Minute ribosomal proteins (Morata and Ripoll, 1975). In this process, cells with higher MYC levels outcompete unfit neighboring cells (with lower MYC), leading to their apoptosis (de la Cova et al., 2004; Moreno and Basler, 2004). This property of MYC was later demonstrated in the development of vertebrates (Claveria et al., 2013; Ellis et al., 2019), and it may underscore a role for MYC in mechanisms of tissue repair and regeneration across diverse organisms (Gogna et al., 2015; Yusupova and Fuchs, 2023).
3 Organ-specific regeneration: the wing imaginal discs, gut, and neural cells, three models to study regeneration
3.1 Wing imaginal discs
Imaginal discs in Drosophila larvae are sac-like structures of epithelial tissue (Figure 1A) and they are the precursors of adult organs. Due to their accessibility and the availability of a wide range of genetic tools, imaginal discs have become, in the last decade, an invaluable tissue for studying regeneration. They also provide an excellent platform for analyzing evolutionarily conserved pathways identified in the regeneration (Hariharan and Serras, 2017; Fox et al., 2020). Early studies on regeneration demonstrated that when imaginal wing discs were cut into small pieces and transplanted into either adult female abdomen, which served as natural culture chambers, or young larvae, they regenerated to their correct size and shape (Bergantinos et al., 2010b; Worley and Hariharan, 2022). This indicated the ability of the discs to resume proliferation and regenerate the missing part. These pioneering experiments demonstrated the regenerative potential of imaginal discs and unveiled their plasticity. In addition, fragments of discs cultured through prolonged transplantation cell-fate changes such as leg-to-wing, leading to the regeneration of alternative organs, in a phenomenon called transdetermination. This phenomenon demonstrates the capacity of Drosophila imaginal cells to be reprogrammed to various lineages (McClure and Schubiger, 2007). The refinement of surgical ablation of imaginal discs facilitated the exploration of regeneration during larval and pupal development. This technique revealed the critical role of cell division and the timing of ablation during development in shaping the regeneration timing (Diaz-Garcia and Baonza, 2013). More advanced technology was developed using genetic tools to induce apoptosis in specific domains of the disc and monitor tissue recovery, utilizing the binary UAS/Gal4 system (Brand and Perrimon, 1993). This widely used technique was adapted to study regeneration by temporally inducing the expression of apoptotic genes in the wing disc, regulated by the temperature-sensitive allele Gal80ts, an inhibitor of Gal4 (Figure 1B) (Smith-Bolton et al., 2009; Bergantinos et al., 2010a). Furthermore, the UAS/Gal4 system was combined with an engineered LexA-LexAop system, enabling precise temporal induction of cell death (Santabarbara-Ruiz et al., 2015). These methods allowed the identification of crucial genes involved in blastema formation, including Wg, a key regulator of regeneration in many species, and MYC (Smith-Bolton et al., 2009; Worley et al., 2012). Indeed, MYC was found to be upregulated in the proliferating cells surrounding the blastema, and its reduction partially impeded regeneration in the wing pouch (Smith-Bolton et al., 2009). Subsequent research demonstrated that MYC reduction, combined with reaper ablation, significantly hindered regeneration in the wing disc. Conversely, under the same conditions, MYC overexpression improved both the size and morphology of the adult wings, confirming its crucial role in the regeneration process (Harris et al., 2020). Additionally, MYC has been identified to regulate Yorkie (Yki), the unique Drosophila ortholog of YAP/TAZ, a component of the Hippo tumor suppressor pathway, in a feed-back mechanism that restrains the growth of the imaginal discs (Neto-Silva et al., 2010; Ziosi et al., 2010). In mammals, the Hippo-YAP/TAZ pathway regulates regeneration by controlling cell proliferation, apoptosis, and stem cell maintenance to ensure proper tissue growth and repair (Moya and Halder, 2019). Thus, MYC’s regulation of Yorkie (Yki) could be crucial for balancing cell proliferation and tissue growth in response to damage. This coordination is vital for developmental processes and organ growth, where MYC and the Hippo pathway are key players (de la Cova et al., 2004; Pan, 2007). Cells at the regeneration site stimulate proliferation through non-autonomous mechanisms such as apoptosis-induced proliferation (AiP), compensating for the apoptotic zones by triggering cell proliferation (Fogarty and Bergmann, 2017). The mechanisms controlling AiP are still under investigation; however, one hypothesis is that the release of ROS by the dying cells activates the ROS-sensitive kinase 1 (Ask1), expressed during regeneration, and its signal attenuated by Akt1/PKB/InR in living cells surrounding the blastema modulates moderate JNK/p38 signaling, which is crucial for controlling apoptosis in the regenerative response (Santabarbara-Ruiz et al., 2019; Esteban-Collado et al., 2021). Recent single-cell transcriptomics analysis of blastema from wing imaginal discs identified Ets21C, a transcription factor that controls patterning and organ development. This factor is induced by cell damage and is essential for the expression of genes crucial for regeneration (Worley et al., 2022). Interestingly, our RNA sequencing data reveals that both Ets21C and MYC are upregulated in wing disc cells undergoing apoptosis induced by proteotoxic stress (not published), suggesting that their expression may share components in the stress response pathways still to be investigated.
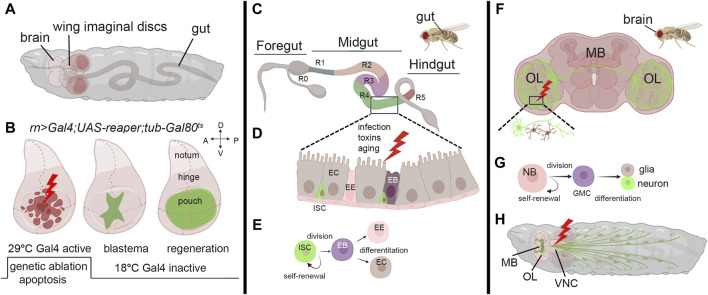
FIGURE 1.
Models to study regeneration. (A) Schematic view of third instar larvae indicating the brain, wing imaginal discs, and the gut. (B) Third instar wing imaginal discs in which apoptosis is induced in the pouch using a specific Gal4-promoter. (Left) The induction of the apoptotic gene occurs through a controlled temperature switch. At 18°C, Gal80 binds to Gal4, repressing its activity and preventing its expression. However, when the temperature is switched to 29°C, Gal80 expression is suppressed, releasing Gal4 from inhibition and initiating the expression of the apoptotic gene and cell death (Hariharan and Serras, 2017). (Middle) After a few hours, animals are switched to the permissive temperature of 18°C to block apoptosis, allowing regeneration to occur with the formation of the blastema (green) that expands until a fully recovered pouch is obtained (Right). (C) Representation of the adult gut with the zone that characterizes its function (R0-5) (Buchon et al., 2013). (D) Model of the midgut epithelium where regeneration occurs upon injury. Cells are color-coded as in panel (E), where the stem-cell niche is represented: ISC: Intestinal Stem Cell, EB: Enteroblast, EE: Enteroendocrine cell, EC: Enterocyte. (F) Schematic representation of the adult brain indicating the most common structures MB: mushroom body, OL: Optical Lobe. The inset represents a site of injury with neuron and glial cells represented in green and brown. (G) Representation of neuroblasts division. Neuroblasts (NB) divide asymmetrically, generating a ganglion mother cell (GMC), which then divides to produce a postmitotic neuron or glial cell (Homem and Knoblich, 2012). (H) Schematic representation of a third instar larva indicating the mushroom body (MB), the Optical lobe (OL) and the neurons (green). In red is a common site for injury in the Ventral Neural Cord (VNC). The figure was created using BioRender Premium, license (XV26VCD8GB), and further refined using Adobe Photoshop for its final appearance.
Finally, we would like to briefly address the critical role of the steroid hormone ecdysone during regeneration and its relation with MYC. Ecdysone controls cellular and specific pathways that regulate physiological organ growth and developmental timing (Andersen et al., 2013). Ecdysone is produced by the prothoracic gland (PG) at specific development times to regulate larval molting and metamorphosis (Edgar, 2006; Tennessen and Thummel, 2011). In the regeneration process, ecdysone levels determine the timing after which larvae terminate their window of regenerative potential by controlling the state of epithelial cell progenitors through regulating the transcription factors chinmo and broad (Narbonne-Reveau and Maurange, 2019; Karanja et al., 2022). Moreover, the release of ecdysone by the PG is indirectly controlled by the dying cells in the regenerating discs that secrete Dilp8, a peptide belonging to the insulin/relaxin-like growth factor family, which binds to the LGR3 receptor in the brain. This inhibits the release of ecdysone from the PG (Colombani et al., 2015; Vallejo et al., 2015) and slows down the development, allowing the damaged cells of the discs to complete their regeneration process (Blanco-Obregon et al., 2022; Karanja et al., 2022). Moreover, the physiological reduction of ecdysone at specific development points corresponds to an increase in MYC in the fat body (FB) (Delanoue et al., 2010). MYC in the FB favors the storage of nutrients (fat and sugars) and activates survival pathways such as autophagy to survive starvation (Parisi et al., 2013; Paiardi et al., 2017). It is known that regeneration in wing discs is affected by pathways regulated by nutrients (Esteban-Collado et al., 2021), and animals allowed to regenerate in starvation do not complete this process (Figure 2). The observation that animals in starvation have a reduced ability to regenerate suggests that non-autonomous signals from the FB are necessary to complete this process. Although MYC is upregulated in the FB of starved animals (Teleman et al., 2008; Parisi et al., 2013), the impaired regeneration observed under starvation conditions indicates that the upregulation of endogenous MYC activity in the FB is insufficient to sustain regeneration. Alternatively, starvation may prevent the storage of nutrients in the FB or hinder the production/secretion of factors necessary for regeneration.
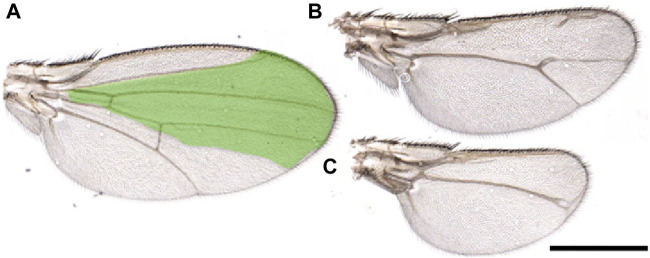
FIGURE 2.
Starvation affects wing regeneration. (A-C) Wings from animals that underwent regeneration while subjected to amino acid starvation. Reaper was temporarily induced in the spalt domain (green), three days after egg laying in larvae of the genotype: SpaltPE-Gal4/tub-Gal80 ts ; UAS-rpr. Animals were kept in a starvation medium (PBS/20% sucrose) until eclosion. (A) Wings from flies not expressing reaper. (B, C) or in which reaper was induced. These images highlight the morphological defects observed in the wings due to the incomplete regeneration process. Scale Bar 1 mm.
3.2 Gut
Research on Drosophila gut regeneration offers valuable insights into repair mechanisms relevant to regenerative medicine, given the similarities in tissue composition, anatomy, and physiological functions with the human intestine. To investigate regeneration, various methods are employed to induce stress and cell damage, such as chemical exposure (e.g., Dextran Sulfate Disodium (DSS), bacterial infection, heat stress, oxidative stress (e.g., H2O2), and mechanical damage (Apidianakis and Rahme, 2011; Zhang and Edgar, 2022) Drosophila gut comprises an anterior, middle, and posterior hindgut (Figure 1C); however, regeneration primarily occurs in the midgut, where the Intestinal Stem Cells (ISCs) generate a niche initiated by Notch (Ohlstein and Spradling, 2007). These cells divide asymmetrically and give rise to a new ISC and an Enteroblast (EB) that will differentiate into Enterocytes (ECs) or Enteroendocrine cells (EE) in the absence of cell division (Figures 1D,E) (Mathur et al., 2010; Amcheslavsky et al., 2014). Wg is necessary to maintain ISCs self-renewal and is the balance between Notch and Wg signaling that controls the equilibrium between the proliferation and differentiation of ISCs (Zhang and Edgar, 2022). MYC plays a crucial role in mediating gut fitness both in ISCs and in ECs. MYC activity is essential for their differentiation and proliferation and acts downstream of stress-dependent and growth factor pathways such as JAK-STAT, Wg, Hippo, and EGFR (Ren et al., 2013). MYC is also crucial in maintaining gut health in response to different diet conditions. A nutrient-rich diet suppresses MYC in ECs, increasing cell death and gut permeability and shortening lifespan. Conversely, dietary restriction boosts MYC, enhancing EC fitness, gut integrity, and lifespan (Akagi et al., 2018). This may occur through MYC-inducing cell competition, which is crucial for maintaining the fitness of adult enterocytes (ECs), especially during dietary changes. Interestingly, this is similar to what was previously described in intestinal ISCs for Minute genes, many of which are MYC targets, where both ISC and differentiated Minute/+ cells were eliminated through cell competition to promote the proliferation and self-renewal of wild-type stem cells (Kolahgar et al., 2015). Recent evidence also reveals the role of MYC as a regulator of the amino acid transporter arcus (acs) in ECs (Socha et al., 2023). This signal is coordinated with the activation of the insulin pathway that favors aminoacidic absorption and ECs recovery after bacterial-mediated toxin damage, suggesting another active role for MYC in the gut to favor the regeneration of these cells.
3.3 Neuronal cells
Drosophila’s neural stem cells (NSCs), or neuroblasts, are pivotal for brain development. They exhibit remarkable plasticity, transitioning between quiescent and active states in response to environmental cues or injury. This dynamic regulation underscores their importance in maintaining brain homeostasis and promoting tissue repair. Neuroblasts (NB) play a crucial role in larval development, undergoing asymmetric division to generate neuroblasts and smaller ganglion mother cells (GMC). These GMCs divide further to produce post-mitotic neurons or glial cells (Figure 1G) (Homem and Knoblich, 2012; Otsuki and Brand, 2020). The neurons establish identities via proneural and selector genes, resulting in four classes (I-IV) of dendritic arborization (da) sensory neurons. Class IV-ddaC neurons, known for their intricate dendritic arbors sensitive to mechanical stimuli, serve as models for dendrite repair and the study of neurodevelopmental disorders (Grueber et al., 2007; Liu et al., 2023).
Methods for investigating neuronal regeneration during development include gently crushing the larval segmental nerve to maintain larval viability or employing laser ablation (Figure 1H). This approach involves labeling specific axon patterns using GFP expressed by neuronal-specific promoters, facilitating the visualization of cells during regeneration events throughout larval development (Pfeiffer et al., 2008). In adult flies, few models exist for studying neuronal regeneration. Experimental stab lesions to either the optic lobes (OL) or the central brain result in local neurogenesis days after injury (Figure 1F). This response was attributed to dormant neural progenitor cells (qNPs) activation (Moreno et al., 2015; Crocker et al., 2021). Glial cells respond to nervous system damage by increasing their number and changing morphology after neuronal cell death. This process is conserved across regions of the peripheral nervous system and involves Dpp and Hh signaling, with the JNK pathway contributing to glial migration (Velarde et al., 2021). Glial cells exhibit an immune response like microglia, expressing the phagocytic receptor draper (drpr), crucial for axon regeneration and debris clearance. While macrophages aid central nervous system (CNS) regeneration in vertebrates, their role in Drosophila neural injury remains unclear (Losada-Perez et al., 2021).
Recent discoveries highlight the crucial role of NSCs in maintaining and regenerating adult brain tissues (Li and Hidalgo, 2020). In contrast to adult mammals, Drosophila NSCs can be activated by different diets or exercises initiated by larval hatching. However, the mechanisms by which NSCs transition between quiescence and activation remain elusive (Ding et al., 2020). Brain injuries in adult flies are thought to trigger the recruitment of quiescent neural progenitors (qNPs) near the injury site, facilitated by damage-responsive neuroglial clusters (DNGCs). These clusters stimulate the proliferation of distant qNPs, thereby expanding the zone of stem cell activation through the reactivation of dormant qNPs (Moreno et al., 2015; Crocker et al., 2021). Since previous research has shown that a ubiquitous pulse of MYC promotes qNP division (Fernandez-Hernandez et al., 2013), it is possible that MYC could induce growth factors in qNPs through injury-induced secretion, allowing these cells to survive and proliferate. MYC has also emerged as a non-autonomous regulator of metabolism in retinal ganglion glial cells, where using a model of reprogrammed glial cells that activate PI3K and EGFR pathways (RGCPE), MYC activity was shown relevant for the regeneration of neurons by mediating pro-regeneration metabolic pathways in glia (Li et al., 2020), including the glia-neuron lactate shuttle essential for neuronal survival (Volkenhoff et al., 2015). This highlights its important role in inducing nonautonomous signals that control axon regeneration.
4 Discussion
Studying regeneration in Drosophila has unveiled complex cellular and molecular mechanisms guiding tissue repair and organ regeneration across species. Although tissues display differing regenerative abilities, common pathways and principles govern regeneration. The pivotal role of MYC emphasizes its importance in regulating fundamental conserved processes, connecting metabolism and growth, influencing cell competition, and highlighting regeneration’s complexity. Insights from Drosophila research hold potential for future advancements in regenerative medicine. Further exploring molecular mechanisms across organisms is fundamental to developing novel therapeutic strategies to enhance human tissue repair and organ regeneration.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Open access funding was provided by the grant “5%1000” from the University of Trento to PB. Spanish Ministerio de Ciencia, Innovación y Universidades PID2021-123300NB-100 to FS.
Author contributions
FS: Writing–review and editing, Conceptualization. PB: Writing–original draft, Writing–review and editing, Conceptualization.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Akagi K., Wilson K. A., Katewa S. D., Ortega M., Simons J., Hilsabeck T. A., et al. (2018). Dietary restriction improves intestinal cellular fitness to enhance gut barrier function and lifespan in D. melanogaster . PLoS Genet. 14, e1007777. 10.1371/journal.pgen.1007777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A., Song W., Li Q., Nie Y., Bragatto I., Ferrandon D., et al. (2014). Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Rep. 9, 32–39. 10.1016/j.celrep.2014.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen D. S., Colombani J., Leopold P. (2013). Coordination of organ growth: principles and outstanding questions from the world of insects. Trends Cell Biol. 23, 336–344. 10.1016/j.tcb.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Apidianakis Y., Rahme L. G. (2011). Drosophila melanogaster as a model for human intestinal infection and pathology. Dis. Model Mech. 4, 21–30. 10.1242/dmm.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P., Gallant P. (2010). Myc function in Drosophila. Genes Cancer 1, 542–546. 10.1177/1947601910377490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely A. E., Nyberg K. G. (2010). Evolution of animal regeneration: re-emergence of a field. Trends Ecol. Evol. 25, 161–170. 10.1016/j.tree.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Bergantinos C., Corominas M., Serras F. (2010a). Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 137, 1169–1179. 10.1242/dev.045559 [DOI] [PubMed] [Google Scholar]
- Bergantinos C., Vilana X., Corominas M., Serras F. (2010b). Imaginal discs: renaissance of a model for regenerative biology. Bioessays 32, 207–217. 10.1002/bies.200900105 [DOI] [PubMed] [Google Scholar]
- Blanco-Obregon D., El Marzkioui K., Brutscher F., Kapoor V., Valzania L., Andersen D. S., et al. (2022). A Dilp8-dependent time window ensures tissue size adjustment in Drosophila. Nat. Commun. 13, 5629. 10.1038/s41467-022-33387-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., Andruss B. F., et al. (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97, 865–875. 10.1016/s0092-8674(00)80799-0 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. 10.1242/dev.118.2.401 [DOI] [PubMed] [Google Scholar]
- Buchon N., Osman D., David F. P., Fang H. Y., Boquete J. P., Deplancke B., et al. (2013). Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738. 10.1016/j.celrep.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Claveria C., Giovinazzo G., Sierra R., Torres M. (2013). Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500, 39–44. 10.1038/nature12389 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Boulan L., Boone E., Romero N., Virolle V., et al. (2015). Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr. Biol. 25, 2723–2729. 10.1016/j.cub.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Crocker K. L., Marischuk K., Rimkus S. A., Zhou H., Yin J. C. P., Boekhoff-Falk G. (2021). Neurogenesis in the adult Drosophila brain. Genetics 219, iyab092. 10.1093/genetics/iyab092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cova C., Abril M., Bellosta P., Gallant P., Johnston L. A. (2004). Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116. 10.1016/s0092-8674(04)00214-4 [DOI] [PubMed] [Google Scholar]
- De La Cova C., Senoo-Matsuda N., Ziosi M., Wu D. C., Bellosta P., Quinzii C. M., et al. (2014). Supercompetitor status of Drosophila myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 19, 470–483. 10.1016/j.cmet.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R., Slaidina M., Leopold P. (2010). The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev. Cell 18, 1012–1021. 10.1016/j.devcel.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Destefanis F., Manara V., Bellosta P. (2020). Myc as a regulator of ribosome biogenesis and cell competition: a link to cancer. Int. J. Mol. Sci. 21, 4037. 10.3390/ijms21114037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Garcia S., Baonza A. (2013). Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ . Proc. Natl. Acad. Sci. U.S.A. 110, 13032–13037. 10.1073/pnas.1220543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W. Y., Huang J., Wang H. (2020). Waking up quiescent neural stem cells: molecular mechanisms and implications in neurodevelopmental disorders. PLoS Genet. 16, e1008653. 10.1371/journal.pgen.1008653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A. (2006). How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907–916. 10.1038/nrg1989 [DOI] [PubMed] [Google Scholar]
- Ellis S. J., Gomez N. C., Levorse J., Mertz A. F., Ge Y., Fuchs E. (2019). Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature 569, 497–502. 10.1038/s41586-019-1199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban-Collado J., Corominas M., Serras F. (2021). Nutrition and PI3K/Akt signaling are required for p38-dependent regeneration. Development 148, dev197087. 10.1242/dev.197087 [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernandez I., Rhiner C., Moreno E. (2013). Adult neurogenesis in Drosophila. Cell Rep. 3, 1857–1865. 10.1016/j.celrep.2013.05.034 [DOI] [PubMed] [Google Scholar]
- Fogarty C. E., Bergmann A. (2017). Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ. 24, 1390–1400. 10.1038/cdd.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. T., Cohen E., Smith-Bolton R. (2020). Model systems for regeneration: Drosophila. Development 147, dev173781. 10.1242/dev.173781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P. (2006). Myc/Max/Mad in invertebrates: the evolution of the Max network. Curr. Top. Microbiol. Immunol. 302, 235–253. 10.1007/3-540-32952-8_9 [DOI] [PubMed] [Google Scholar]
- Galletti M., Riccardo S., Parisi F., Lora C., Saqcena M. K., Rivas L., et al. (2009). Identification of domains responsible for ubiquitin-dependent degradation of dMyc by glycogen synthase kinase 3beta and casein kinase 1 kinases. Mol. Cell Biol. 29, 3424–3434. 10.1128/MCB.01535-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogna R., Shee K., Moreno E. (2015). Cell competition during growth and regeneration. Annu. Rev. Genet. 49, 697–718. 10.1146/annurev-genet-112414-055214 [DOI] [PubMed] [Google Scholar]
- Grueber W. B., Ye B., Yang C. H., Younger S., Borden K., Jan L. Y., et al. (2007). Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134, 55–64. 10.1242/dev.02666 [DOI] [PubMed] [Google Scholar]
- Hariharan I. K., Serras F. (2017). Imaginal disc regeneration takes flight. Curr. Opin. Cell Biol. 48, 10–16. 10.1016/j.ceb.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. E., Stinchfield M. J., Nystrom S. L., Mckay D. J., Hariharan I. K. (2020). Damage-responsive, maturity-silenced enhancers regulate multiple genes that direct regeneration in Drosophila. Elife 9, e58305. 10.7554/eLife.58305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C., Knoblich J. A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297–4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., et al. (2005). Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell Biol. 25, 3401–3410. 10.1128/MCB.25.9.3401-3410.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P. (1999). Drosophila myc regulates cellular growth during development. Cell 98, 779–790. 10.1016/s0092-8674(00)81512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier Z., Park A. J., Briquez P. S., Martino M. M. (2017). Promoting tissue regeneration by modulating the immune system. Acta Biomater. 53, 13–28. 10.1016/j.actbio.2017.01.056 [DOI] [PubMed] [Google Scholar]
- Karanja F., Sahu S., Weintraub S., Bhandari R., Jaszczak R., Sitt J., et al. (2022). Ecdysone exerts biphasic control of regenerative signaling, coordinating the completion of regeneration with developmental progression. Proc. Natl. Acad. Sci. U.S.A. 119, e2115017119. 10.1073/pnas.2115017119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. S., Newmark P. A. (2012). The cell biology of regeneration. J. Cell Biol. 196, 553–562. 10.1083/jcb.201105099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolahgar G., Suijkerbuijk S. J., Kucinski I., Poirier E. Z., Mansour S., Simons B. D., et al. (2015). Cell competition modifies adult stem cell and tissue population dynamics in a JAK-STAT-dependent manner. Dev. Cell 34, 297–309. 10.1016/j.devcel.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Sami A., Noristani H. N., Slattery K., Qiu J., Groves T., et al. (2020). Glial metabolic rewiring promotes axon regeneration and functional recovery in the central nervous system. Cell Metab. 32, 767–785. 10.1016/j.cmet.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Hidalgo A. (2020). Adult neurogenesis in the Drosophila brain: the evidence and the void. Int. J. Mol. Sci. 21, 6653. 10.3390/ijms21186653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao Y., Zou W. (2023). Molecular mechanisms of neurite regeneration and repair: insights from C. elegans and Drosophila. Cell Regen. 12, 12. 10.1186/s13619-022-00155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada-Perez M., Garcia-Guillen N., Casas-Tinto S. (2021). A novel injury paradigm in the central nervous system of adult Drosophila: molecular, cellular and functional aspects. Dis. Model Mech. 14, dmm044669. 10.1242/dmm.044669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losner J., Courtemanche K., Whited J. L. (2021). A cross-species analysis of systemic mediators of repair and complex tissue regeneration. NPJ Regen. Med. 6, 21. 10.1038/s41536-021-00130-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D., Bost A., Driver I., Ohlstein B. (2010). A transient niche regulates the specification of Drosophila intestinal stem cells. Science 327, 210–213. 10.1126/science.1181958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclure K. D., Schubiger G. (2007). Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int. J. Biochem. Cell Biol. 39, 1105–1118. 10.1016/j.biocel.2007.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J., Stewart M. J., Stocker H., Hafen E., Kozma S. C., Thomas G. (1999). Drosophila S6 kinase: a regulator of cell size. Science 285, 2126–2129. 10.1126/science.285.5436.2126 [DOI] [PubMed] [Google Scholar]
- Morata G., Ripoll P. (1975). Minutes: mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 42, 211–221. 10.1016/0012-1606(75)90330-9 [DOI] [PubMed] [Google Scholar]
- Moreno E., Basler K. (2004). dMyc transforms cells into super-competitors. Cell 117, 117–129. 10.1016/s0092-8674(04)00262-4 [DOI] [PubMed] [Google Scholar]
- Moreno E., Fernandez-Marrero Y., Meyer P., Rhiner C. (2015). Brain regeneration in Drosophila involves comparison of neuronal fitness. Curr. Biol. 25, 955–963. 10.1016/j.cub.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya I. M., Halder G. (2019). Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20, 211–226. 10.1038/s41580-018-0086-y [DOI] [PubMed] [Google Scholar]
- Nagy P., Varga A., Pircs K., Hegedus K., Juhasz G. (2013). Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster . PLoS Genet. 9, e1003664. 10.1371/journal.pgen.1003664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne-Reveau K., Maurange C. (2019). Developmental regulation of regenerative potential in Drosophila by ecdysone through a bistable loop of ZBTB transcription factors. PLoS Biol. 17, e3000149. 10.1371/journal.pbio.3000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R. M., De Beco S., Johnston L. A. (2010). Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell. 19, 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992. 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Orian A., Van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., et al. (2003). Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17, 1101–1114. 10.1101/gad.1066903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki L., Brand A. H. (2020). Quiescent neural stem cells for brain repair and regeneration: lessons from model systems. Trends Neurosci. 43, 213–226. 10.1016/j.tins.2020.02.002 [DOI] [PubMed] [Google Scholar]
- Paiardi C., Mirzoyan Z., Zola S., Parisi F., Vingiani A., Pasini M. E., et al. (2017). The stearoyl-CoA desaturase-1 (Desat1) in Drosophila cooperated with myc to induce autophagy and growth, a potential new link to tumor survival. Genes (Basel) 8, 131. 10.3390/genes8050131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. (2007). Hippo signaling in organ size control. Genes Dev. 21, 886–897. 10.1101/gad.1536007 [DOI] [PubMed] [Google Scholar]
- Parisi F., Riccardo S., Daniel M., Saqcena M., Kundu N., Pession A., et al. (2011). Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo . BMC Biol. 9, 65. 10.1186/1741-7007-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F., Riccardo S., Zola S., Lora C., Grifoni D., Brown L. M., et al. (2013). dMyc expression in the fat body affects DILP2 release and increases the expression of the fat desaturase Desat1 resulting in organismal growth. Dev. Biol. 379, 64–75. 10.1016/j.ydbio.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., Murphy C., et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 105, 9715–9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Shi Q., Chen Y., Jiang A., Ip Y. T., Jiang H., et al. (2013). Drosophila Myc integrates multiple signaling pathways to regulate intestinal stem cell proliferation during midgut regeneration. Cell Res. 23, 1133–1146. 10.1038/cr.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santabarbara-Ruiz P., Esteban-Collado J., Perez L., Viola G., Abril J. F., Milan M., et al. (2019). Ask1 and Akt act synergistically to promote ROS-dependent regeneration in Drosophila. PLoS Genet. 15, e1007926. 10.1371/journal.pgen.1007926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santabarbara-Ruiz P., Lopez-Santillan M., Martinez-Rodriguez I., Binagui-Casas A., Perez L., Milan M., et al. (2015). ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 11, e1005595. 10.1371/journal.pgen.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinkendorf D., Gallant P. (2009). The conserved Myc box 2 and Myc box 3 regions are important, but not essential, for Myc function in vivo . Gene 436, 90–100. 10.1016/j.gene.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Slack J. M. (2017). Animal regeneration: ancestral character or evolutionary novelty? EMBO Rep. 18, 1497–1508. 10.15252/embr.201643795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bolton R. K., Worley M. I., Kanda H., Hariharan I. K. (2009). Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 16, 797–809. 10.1016/j.devcel.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socha C., Pais I. S., Lee K. Z., Liu J., Liegeois S., Lestradet M., et al. (2023). Fast drosophila enterocyte regrowth after infection involves a reverse metabolic flux driven by an amino acid transporter. iScience 26, 107490. 10.1016/j.isci.2023.107490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M. (2008). Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7, 21–32. 10.1016/j.cmet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., Thummel C. S. (2011). Coordinating growth and maturation - insights from Drosophila. Curr. Biol. 21, R750–R757. 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo D. M., Juarez-Carreno S., Bolivar J., Morante J., Dominguez M. (2015). A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science 350, aac6767. 10.1126/science.aac6767 [DOI] [PubMed] [Google Scholar]
- Velarde S. B., Quevedo A., Estella C., Baonza A. (2021). Dpp and Hedgehog promote the glial response to neuronal apoptosis in the developing Drosophila visual system. PLoS Biol. 19, e3001367. 10.1371/journal.pbio.3001367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenhoff A., Weiler A., Letzel M., Stehling M., Klambt C., Schirmeier S. (2015). Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 22, 437–447. 10.1016/j.cmet.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Wells J. M., Watt F. M. (2018). Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. 10.1038/s41586-018-0073-7 [DOI] [PubMed] [Google Scholar]
- Worley M. I., Everetts N. J., Yasutomi R., Chang R. J., Saretha S., Yosef N., et al. (2022). Ets21C sustains a pro-regenerative transcriptional program in blastema cells of Drosophila imaginal discs. Curr. Biol. 32, 3350–3364.e6. 10.1016/j.cub.2022.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley M. I., Hariharan I. K. (2022). Imaginal disc regeneration: something old, something new. Cold Spring Harb. Perspect. Biol. 14, a040733. 10.1101/cshperspect.a040733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley M. I., Setiawan L., Hariharan I. K. (2012). Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46, 289–310. 10.1146/annurev-genet-110711-155637 [DOI] [PubMed] [Google Scholar]
- Yusupova M., Fuchs Y. (2023). To not love thy neighbor: mechanisms of cell competition in stem cells and beyond. Cell Death Differ. 30, 979–991. 10.1038/s41418-023-01114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Edgar B. A. (2022). Insect gut regeneration. Cold Spring Harb. Perspect. Biol. 14, a040915. 10.1101/cshperspect.a040915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M., Baena-Lopez L. A., Grifoni D, Froldi F., Pession A., Garoia F., et al. (2010). dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]