Abstract
Deep learning can exceed dermatologists’ diagnostic accuracy in experimental image environments. However, inaccurate segmentation of images with multiple skin lesions can be seen with current methods. Thus, information present in multiple-lesion images, available to specialists, is not retrievable by machine learning. While skin lesion images generally capture a single lesion, there may be cases in which a patient’s skin variation may be identified as skin lesions, leading to multiple false positive segmentations in a single image. Conversely, image segmentation methods may find only one region and may not capture multiple lesions in an image. To remedy these problems, we propose a novel and effective data augmentation technique for skin lesion segmentation in dermoscopic images with multiple lesions. The lesion-aware mixup augmentation (LAMA) method generates a synthetic multi-lesion image by mixing two or more lesion images from the training set. We used the publicly available International Skin Imaging Collaboration (ISIC) 2017 Challenge skin lesion segmentation dataset to train the deep neural network with the proposed LAMA method. As none of the previous skin lesion datasets (including ISIC 2017) has considered multiple lesions per image, we created a new multi-lesion (MuLe) segmentation dataset utilizing publicly available ISIC 2020 skin lesion images with multiple lesions per image. MuLe was used as a test set to evaluate the effectiveness of the proposed method. Our test results show that the proposed method improved the Jaccard score 8.3% from 0.687 to 0.744 and the Dice score 5% from 0.7923 to 0.8321 over a baseline model on MuLe test images. On the single-lesion ISIC 2017 test images, LAMA improved the baseline model’s segmentation performance by 0.08%, raising the Jaccard score from 0.7947 to 0.8013 and the Dice score 0.6% from 0.8714 to 0.8766. The experimental results showed that LAMA improved the segmentation accuracy on both single-lesion and multi-lesion dermoscopic images. The proposed LAMA technique warrants further study.
Keywords: Melanoma, Dermoscopy, Deep learning, Image segmentation, Data augmentation
Introduction
An estimated 97,610 new cases of invasive melanoma and 89,070 in situ melanoma are expected to be diagnosed in 2023 in the USA [1]. Dermoscopy is a powerful imaging technique that assists dermatologists in early skin cancer detection, resulting in improved diagnostic accuracy compared to visual inspection by a domain expert [2–4].
Recent advancements in deep learning techniques [5–12] have proven immensely beneficial in medical image analysis and have been successfully applied to various medical imaging problems [13–16]. In skin cancer, deep learning techniques in combination with dermoscopy have demonstrated higher diagnostic accuracy than experienced dermatologists [13, 17–20]. Pathan et al. published a recent review describing handcrafted and deep learning (DL) techniques for computer-aided skin cancer diagnosis [21]. Recent studies show that the fusion of handcrafted and deep learning techniques boosts the diagnostic accuracy of skin cancer [20, 22–26]. The handcrafted features usually require the lesion borders to facilitate their computation [23, 24]. The precise calculation of handcrafted lesion features depends on accurate detection of the lesion border. Therefore, lesion segmentation is a crucial step in computer-aided skin cancer diagnosis.
Skin lesion segmentation methods have been developed using both conventional image processing approaches [27–29] and deep learning techniques [30–35, 57–61]. While traditional methods showed promising results on small sets, they tend to perform poorly on challenging conditions such as low contrast between lesion and background, lesions with different colors, and images containing artifacts like hair, ruler marks, gel bubbles, and ink marks. In contrast, deep learning techniques, specifically convolutional neural networks (CNN), seem to perform better in detecting lesion borders in the presence of these challenges.
Data augmentation is a widely used technique in deep learning to artificially increase the size of the training dataset and improve the generalization of the model. Augmentation involves randomly applying image transformations such as rotation, translation, scaling, flipping, and color jittering. In addition to these basic transformations, various data augmentation strategies have been developed for image classification, such as cutout [36], mixup [37, 38], and cutmix [39]. Cutout randomly masks out rectangular regions of the input image, which encourages the model to rely on other parts of the image and improves its robustness to occlusion. Mixup generates new training samples by linearly interpolating between pairs of images and their corresponding labels, which improves the model’s ability to generalize to new input distributions. Cutmix combines the cutout and mixup methods by replacing a rectangular region in one image with a corresponding region from another, which can improve the model’s ability to learn fine-grained features while preserving the overall structure of the input. Although these methods have proven successful for image classification, they cannot be adopted for skin lesion segmentation as they do not consider the location or geometry of the objects (lesions).
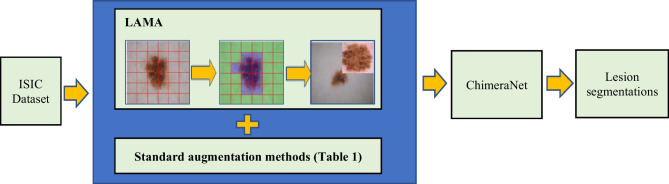
This study introduces a novel lesion-aware mixup augmentation (LAMA) method for skin lesion segmentation in dermoscopic images. The LAMA method can generate a new synthetic multi-lesion image by mixing two or more single-lesion images from the training set. Unlike Cutmix, LAMA randomly selects two or more lesions from a set of lesions and pastes the new lesions in the non-lesion area of an image. To find the non-lesion area in an image, it applies a multi-level patch generation and qualification process, which is faster, as it needs to execute only once at the beginning of training. Also, the lesion size in dermoscopic images varies greatly, with some lesions covering the whole image while others might cover only a small part of the image. To avoid excessive resizing of lesions during mixing, LAMA categorizes the lesions into groups based on their sizes, with each group corresponding to a specific level of non-lesion patches. LAMA samples the patch level or patch size while mixing based on the histogram distribution of images having non-lesion patches at each patch level in the training set. We evaluated the proposed method on a benchmark ISIC 2017 [40] skin lesion segmentation dataset. Also, as none of the publicly available datasets or the datasets used in previous studies considered multiple lesions per image, we created a new multi-lesion (MuLe) segmentation dataset using the publicly available ISIC 2020 [41] melanoma classification dataset to evaluate the effectiveness of the proposed method on real examples having multiple lesions in each image. The experimental results showed that the LAMA method combined with ChimeraNet [35, 42], a U-Net [11] model with EfficientNet [12] encoder, improved the segmentation performance on both the multi-lesion MuLe dataset and the single-lesion ISIC 2017 dataset (Fig. 1). Source code and data are available at https://github.com/nhorangl/LAMA.
Fig. 1.
Overall workflow for the LAMA technique in this article
Lesion-Aware Mixup Augmentation (LAMA)
This section describes our proposed lesion-aware mixup augmentation (LAMA) method for skin lesion segmentation. It generates a new synthetic multi-lesion image by mixing two or more skin lesion images to train the deep neural network. LAMA randomly selects one or more lesions from a training set of single-lesion images and pastes them on the non-lesion area of an image. To find a non-lesion region in the image, LAMA uses a multi-level patch generation and qualification process. As skin lesion size varies greatly, the lesion patches are categorized into groups based on their sizes, with each group corresponding to a specific level of non-lesion patches. This grouping avoids an excessive resizing of lesion patches when mixed with non-lesion patches. The following subsections present the steps for the LAMA method.
Finding Non-lesion Patches
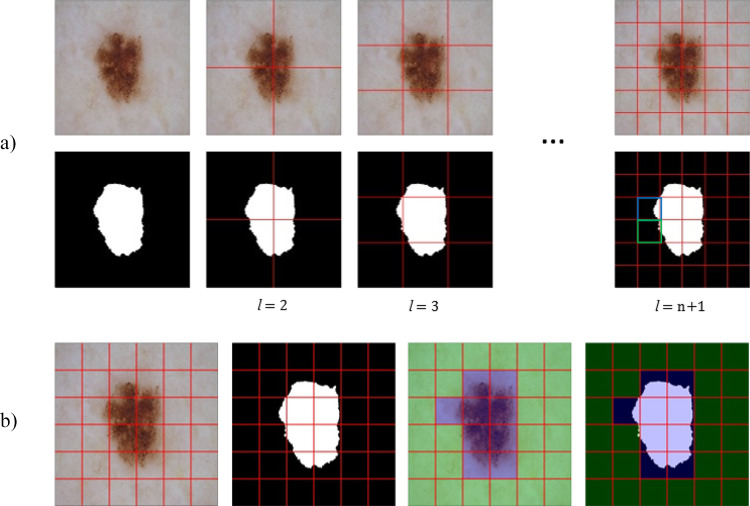
The candidate patches with various sizes are generated from a non-lesion region of a lesion image, as shown in Fig. 2. A patch is randomly selected from a pool of candidate patches to paste a new lesion. For an image , where C = number of colors (3) and a lesion mask , we define different patch levels 1 and generate non-overlapping patches of sizes for each patch level where patch height and patch width .
Fig. 2.
Non-lesion patch generation process. a Creating patches of size at different patch levels from 1. b Finding non-lesion patches at patch level . Non-lesion patches that satisfy the criteria and are overlayed with green color. The patches overlayed with blue color do not satisfy the criteria and cannot receive a new lesion. In the rightmost mask in a, the patch with blue overlay has ), and the patch with green overlay has
To minimize occlusion of an existing lesion by the new lesion, a patch image and patch mask will be only added to a pool of candidate non-lesion patches if it satisfies two criteria:
-
(i)The patch lesion area is less than 10% of total patch area,
-
(ii)The patch lesion area is less than 5% of total lesion area,
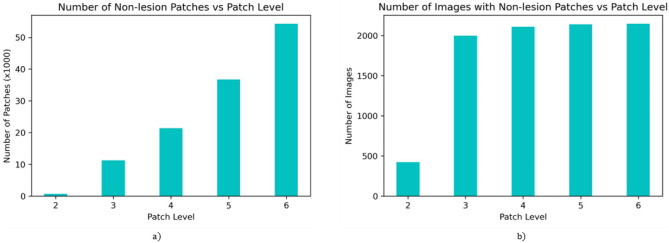
As the patch count is proportional to a square of patch level and a lesion usually covers a large part of the image, most candidate non-lesion patches will belong to higher patch levels, i.e., smaller patch sizes, as shown in Fig. 3a. The distribution of images having non-lesion patches vs. patch level in Fig. 3b shows that only a few images have larger non-lesion patches. Here, we have not selected as it will produce non-lesion patches too small for accurate representation of the lesion.
Fig. 3.
Distribution of non-lesion patches on ISIC 2017 lesion segmentation training set. a Number of non-lesion patches and b number of images having non-lesion patches for different patch levels ( = 1–5)
Lesion Groups
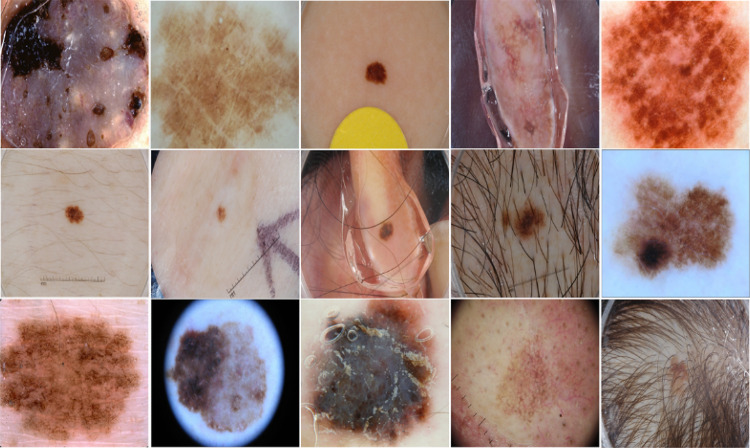
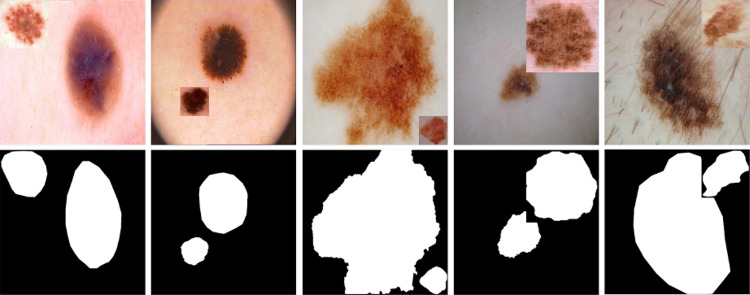
Skin lesions have different shapes and sizes. Some lesions cover most of the image, while others might occupy a tiny part of the image, as shown in Fig. 4. To avoid excessive resizing of lesions during mixing, all lesions from a training set are grouped into groups based on their sizes, with each group corresponding to a specific patch level. A lesion from group only mixes with patch level . We compute lesion size in terms of the area of a rectangular bounding box enclosing the lesion. As the non-lesion patches in each patch level are highly imbalanced, as shown in Fig. 3a, we divide the lesions into groups based on the histogram of images having non-lesion patches for patch levels, which is more balanced, as shown in Fig. 3b. The histogram of lesions per group, , is shown in Fig. 5. The weights for each group are computed based on , which are further used as a weighted probability during the random selection of a lesion group.
Fig. 4.
Skin lesion images in ISIC 2017 lesion segmentation dataset with varying lesion shapes, sizes, and colors
Fig. 5.
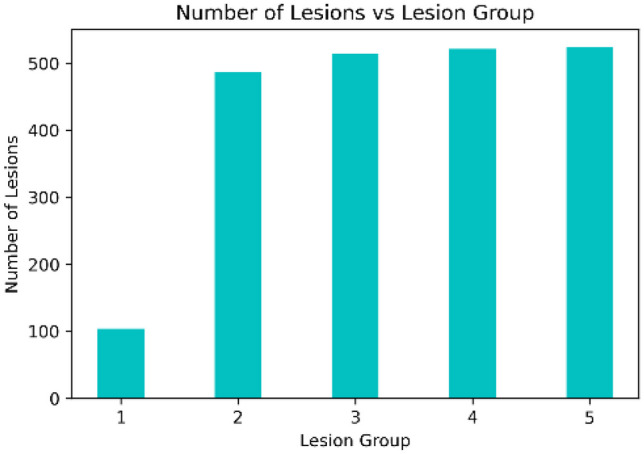
Number of lesions per group in groups ()
Mixing a New Lesion
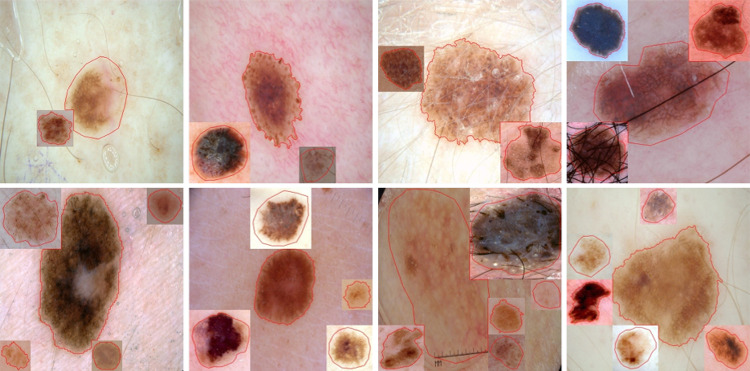
In order to paste a new lesion in a non-lesion patch, we randomly select a lesion group with a weighted probability based on the lesion group distribution in Fig. 5. Then, a new lesion is randomly selected from the lesion group . The selected lesion is cropped using a rectangular bounding box with extended height or width (extended by 5%) to ensure some background around the lesion. The cropped lesion is further augmented using random image transformations such as rotate by 90°, transpose, vertical or horizontal flip, and Gaussian noise. Next, we randomly select a non-lesion patch from the patch level to paste the augmented lesion crop. The augmented lesion crop needs to be resized to match the size of a non-lesion patch. Finally, the lesion is pasted on the selected non-lesion patch location, and a new training image is created with mixed lesions, as shown in Fig. 6
Fig. 6.
Augmented lesion images and masks after mixing a single extra lesion using our proposed lesion-aware mixup augmentation (LAMA) method. Some part of the existing lesion can be occluded as in last two columns
Adding Multiple Lesions
When adding more lesions in a mixed image, we use a heatmap mask to track a non-lesion region so that multiple lesions are not pasted in the same non-lesion patch location. In this mask, a pixel value of 0 represents a non-lesion pixel, and a pixel value of 1 represents a lesion pixel. With each new lesion addition, we update the heatmap by setting a pixel of to 1 if that pixel belongs to a lesion. Then, we update a pool of candidate non-lesion patches for all patch levels based on the heatmap . The non-lesion patch will be removed from the pool if it does not satisfy an additional criterion:
-
(iii)The patch heatmap area is less than 5% of the total patch area,
While adding multiple lesions, we vary the number of new lesions by randomly selecting a number between 1 and the maximum number of new lesions (= 5) rather than using a fixed number, as shown in Fig. 7.
Fig. 7.
Augmented images after mixing multiple lesions from the proposed method. The lesion boundary (red line) shows that new lesions do not significantly occlude other lesions. The number of additional lesions varies between 1 and 5
The complete details of the proposed algorithm are also given in Algorithms 1, 2, and 3.
Algorithm 1.

LAMA
Algorithm 2.

Policy for creating a pool of candidate non-lesion patches
Algorithm 3.

Policy for lesion selection in LAMA
Experiments
In this section, we evaluate the performance of a proposed method on the ISIC skin lesion datasets. For our experiments, we used a ChimeraNet architecture from the previous studies [35, 42] that achieved a state-of-art performance on skin lesion segmentation. The architecture uses a pretrained EfficientNet [12] as an encoder and a squeeze-and-excitation residual (SERes) structure as a decoder. In addition, the decoder network repeatedly uses a transposed convolution to upsample the feature maps from the encoder to generate a final segmentation mask the same size as the input image. In this study, we used ChimeraNet (basic), a simplified version of the model, by removing the SERes structures while only keeping the transposed convolutions in the decoder network.
As data augmentation has been an essential step for deep network training and uses more than one image transformation, we combined our augmentation method with other image transforms shown in Table 1. Other techniques, such as batch normalization [43], dropout, and early stopping, are also used to train the network efficiently. The training hyperparameters are shown in Table 2.
Table 1.
Other image transformations for an additional data augmentation
| Method | Parameter values |
|---|---|
| Rotate | (− 90°, 90°) |
| Horizontal and vertical shift | (− 0.15, 0.15) |
| Scaling | (− 0.15, 0.15) |
| Brightness | (− 0.15, 0.15) |
| Contrast | (− 0.15, 0.15) |
| Hue (HSV) | (− 10, 10) |
| Saturation (HSV) | (− 20, 20) |
| Value (HSV) | (− 10, 10) |
| Motion blur | 5 |
| Median blur | 5 |
| Gaussian blur | 5 |
| Gaussian noise | var_limit = (5, 25) |
| Optical distortion | distort_limit = 1 |
| Grid distortion | num_steps = 6, distort_limit = 1 |
| CLAHE | clip_limit = 4 |
| Transpose | |
| Horizontal and vertical flip | |
| RGB shift | |
| Channel shuffle |
Table 2.
Training hyperparameters
All experiments were implemented using Keras with a Tensorflow backend in Python 3 and trained using a 32 GB Nvidia V100 graphics card. We use the Albumentations [44] library for the additional image transforms for data augmentation.
Multiple-Lesion Segmentation
The current publicly available skin lesion segmentation datasets have only one lesion per image. We created a new multi-lesion (MuLe) dataset by randomly selecting 203 images from ISIC skin lesion classification images that might have multiple lesions. Our domain expert annotator, a pathologist in training, manually created the ground truth segmentation masks for these images, and the ground truths were verified by a dermatologist (W.V.S.). As this dataset is small, we only used it as a hold-out test set to evaluate the performance of our proposed method. To train the network, we used the publicly available ISIC 2017 [40] skin lesion segmentation dataset, with one lesion per image. It provides 2750 dermoscopic skin lesion images with ground truth lesion segmentation masks — 2000 training, 150 validation, and 600 test images. As the ISIC 2017 dataset provides a single train-validation split, we combined the official training and validation sets of ISIC2017 to create a single training set of 2150 images to run fivefold cross-validation experiments. All the images were resized to 540 × 540 using a bilinear interpolation method. During training, we only applied our proposed method on the training set to generate the synthetic multi-lesion images using the single-lesion images.
Figure 8 shows the validation loss plot for the DL models trained with or without our proposed LAMA method. The loss plot shows that the model with LAMA has a lower loss than the baseline model. The baseline model without LAMA trained for 21 epochs and exited early when the validation loss stopped improving, while the model with LAMA trained longer with improved validation loss.
Fig. 8.
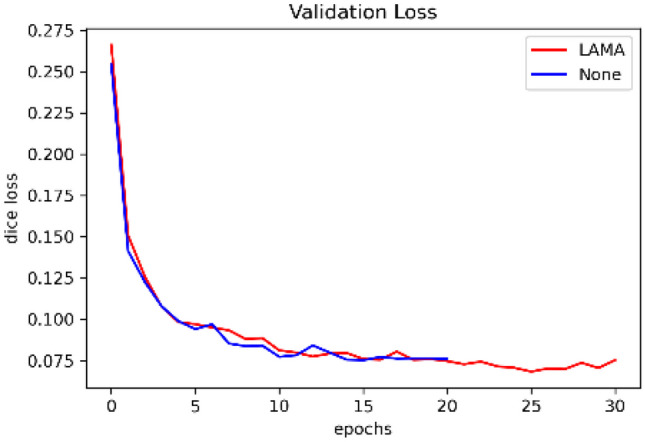
Validation loss plot for ISIC 2017 skin lesion segmentation dataset. The lesion segmentation model trained with the proposed LAMA method has a potential for lower loss than the plain model as the epoch increases
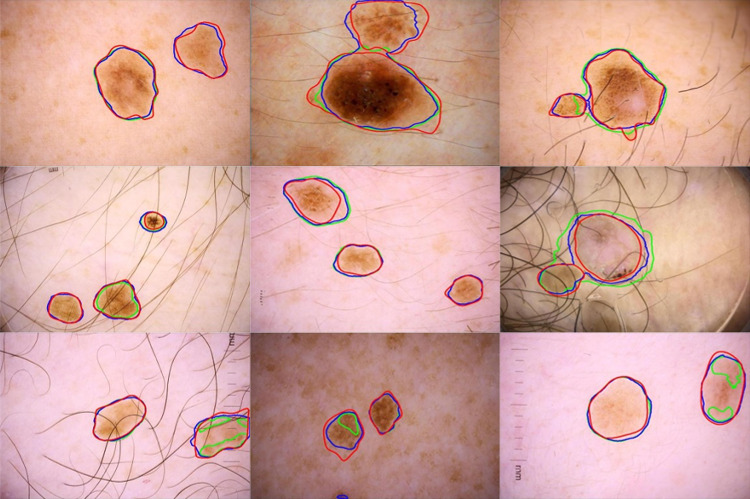
Table 3 shows the performance of the proposed method on 206 MuLe multi-lesion test images trained on the ISIC 2017 lesion segmentation dataset. The ChimeraNet model with LAMA improved overall segmentation performance on real multi-lesion images, although the model was trained using only the synthetic multi-lesion images. The most significant improvements were for recall of 0.8149 to 0.8846, Jaccard score of 0.687 to 0.7444, and Dice score of 0.7923 to 0.8321. Precision fell from 0.8551 to 0.8435. Figure 9 shows the segmentation results of the ChimeraNet model combined with or without the proposed LAMA method. The proposed method successfully finds the multiple lesions even though the model was trained using the synthetic multi-lesion images.
Table 3.
Performance of LAMA on multi-lesion (MuLe) test images
| Method |
Jaccard |
Dice |
Accuracy |
Precision |
Recall |
|---|---|---|---|---|---|
| ChimaraNet (baseline) | 0.687 | 0.7923 | 0.9666 | 0.8551 | 0.8149 |
| ChimeraNet + LAMA | 0.7440 | 0.8321 | 0.9733 | 0.8435 | 0.8846 |
Fig. 9.
Segmentation results of the proposed method on multi-lesions (MuLe) test set. Overlay of ground truth boundary (red), predicted boundary with LAMA (blue), and predicted boundary without LAMA (green), i.e., the baseline model, on skin lesion images. The baseline single-lesion model [35] errors include failure to detect a portion of a lesion or the entire lesion in some cases
ISIC 2017 Skin Lesion Segmentation
In this section, we evaluate the lesion segmentation performance of the proposed method on 600 ISIC 2017 test images. All the images in this official test set have a single lesion per image. We use the same trained model in the “ Experiments” section which was trained using the synthetic multi-lesion training images. Table 4 shows that the ChimeraNet model with the proposed LAMA method improved all measurements — Jaccard score from 0.7947 to 0.8013, Dice score from 0.8714 to 0.8766, and accuracy from 0.944 to 0.948. The proposed method shows robustness by achieving a Jaccard score of 0.8013, comparable to state-of-the-art methods, even though the model was trained using synthetic multi-lesion images.
Table 4.
Performance comparison with other lesion segmentation methods on 600 ISIC 2017 test images
| Methods | Year | Jaccard | Dice | Accuracy |
|---|---|---|---|---|
| Al-Masni et al. [30] | 2018 | 0.771 | 0.871 | 0.940 |
| Tschandl et al. [31] | 2019 | 0.768 | 0.851 | |
| Yuan and Lo [32] | 2019 | 0.765 | 0.849 | 0.934 |
| Navarro et al. [47] | 2019 | 0.769 | 0.854 | 0.955 |
| Xie et al. [33] | 2020 | 0.783 | 0.862 | 0.938 |
| Ozturk and Ozkaya [34] | 2020 | 0.783 | 0.886 | 0.953 |
| Shan et al. [48] | 2020 | 0.763 | 0.846 | 0.937 |
| Kaymak et al. [49] | 2020 | 0.725 | 0.841 | 0.939 |
| Nguyen et al. [50] | 2020 | 0.781 | 0.861 | |
| Zafar et al. [51] | 2020 | 0.772 | 0.858 | |
| Goyal et al. [52] | 2020 | 0.793 | 0.871 | |
| Tong et al. [53] | 2021 | 0.742 | 0.926 | |
| Chen et al. [54] | 2022 | 0.8036 | 0.8704 | 0.947 |
| Ashraf et al. [55] | 2022 | 0.8005 | ||
| Lama et al. [35] | 2023 | 0.807 | 0.880 | 0.948 |
| ChimeraNet (baseline) | 0.7947 | 0.8714 | 0.944 | |
| ChimeraNet + LAMA | 0.8013 | 0.8766 | 0.948 |
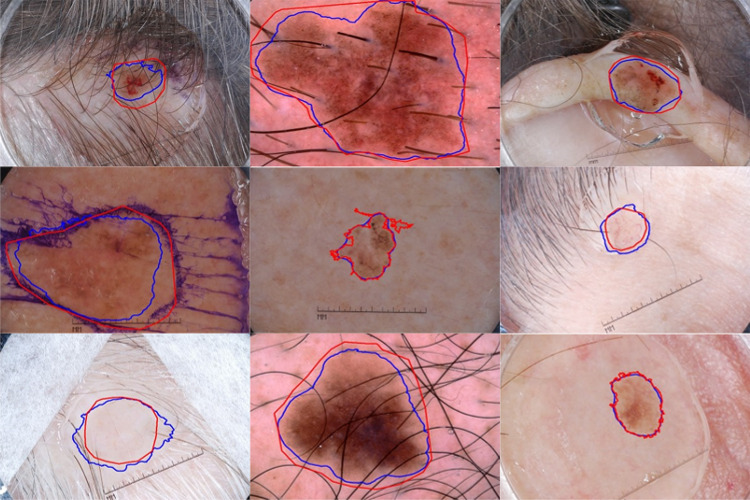
. In Fig. 10, we show the segmentation results of the proposed method on ISIC 2017 test images. The segmentation results show that the proposed method accurately segments the skin lesion even in challenging conditions like images with artifacts or low contrast.
Fig. 10.
Segmentation results of the proposed method on ISIC 2017 test set. Overlays of ground truth lesion boundary (red) and predicted lesion boundary (blue) on skin lesion images. LAMA finds lesion borders accurately in challenging images like low skin-and-lesion contrast and the presence of artifacts like hair, ruler marks, and ink marks
Discussion
This study demonstrated that our proposed lesion-aware mixup augmentation (LAMA) method helps train a robust skin lesion segmentation model. Our LAMA method successfully generated synthetic multi-lesion images by mixing two or more lesion images to train the convolutional neural network. As the currently available public ISIC skin lesion segmentation datasets have only one lesion per image, we created a new multi-lesion (MuLe) test dataset to evaluate the effectiveness of the proposed method on real examples that have one or more lesions in each image. The ChimeraNet model trained with synthetic multi-lesion images successfully detected the lesion boundaries of multiple lesions in real images. LAMA improved the Jaccard score from 0.687 to 0.744, the Dice score from 0.7923 to 0.8321, and recall from 0.8149 to 0.8846 on 203 MuLe test images. Also, the LAMA method improved the Jaccard score of 0.7947 to 0.8013 and the Dice score of 0.8714 to 8766 when evaluated on the ISIC 2017 skin lesion segmentation test set. Although the model was trained using synthetic images to detect multiple lesions per image, the model achieved results comparable to state-of-the-art methods on single-lesion test images. LAMA segmentation improvements are most noticeable on challenging images, as shown in Fig. 7.
Data augmentation methods like Mixup [37] and Cutmix [39] have improved the generalization capability of a deep neural network by providing a regularization effect during training. Mixup is specifically designed for the image classification task and cannot be adopted for image segmentation. The proposed LAMA method is very similar to Cutmix, as both randomly replace the patches from one image with patches from another. However, CutMix does not consider the location and geometry of the objects (for example, lesions in our case) while selecting the patches of the images. Also, it mixes only two images, so it cannot generate a synthetic image with more than two lesions for our problem. The proposed LAMA solves these shortcomings of Cutmix by creating the lesion patches using the bounding box covering the whole lesion and then pasting one or more lesions in the non-lesion area of the image. LAMA finds the non-lesion region of the image using a multi-level patch generation and selection scheme. This process is optimized, running only once at the beginning of training and using the same set for all epochs. The other challenge for mixing skin lesions is due to a large variability in lesion sizes. To avoid extreme resizing of lesions during mixing, all lesion patches are grouped into groups based on their sizes, and each group corresponds to one of the patch levels (or patch sizes). This ensures that the lesion and non-lesion patches have a similar size. In addition, the proposed LAMA method augments the lesion patches before pasting them on the non-lesion area of an image such that it helps to generate more variation of synthetic images.
Although the significant color variability in skin or background in skin lesion images can cause a noticeable distinction between the original image background and newly added patches when these images are mixed, as shown in Fig. 7, the experimental results have revealed that deep neural networks can successfully detect the actual boundaries of lesions and are not affected by such sharp demarcations.
We fully expected that synthetic images overlaid without corrective techniques, with color and texture features poorly matched with the original image, should not benefit CNN training. Synthetic images lack aspects of the actual images and could lead training astray because they contain elements that bias the model. That these synthetic images improved segmentation results, even on single-lesion images, was unexpected. However, the model appears to have ignored these discontinuities and was still able to discern critical lesion features used for segmentation, despite the problematic blending and borders that look artificial to the human observer. We hypothesize that the sliding window used by the CNN used a small attention field that could ignore the artificial discontinuities and accomplish the goal of these experiments: to solve the multiple lesion problem. The goal was not to blend in lesions realistically, but to find multiple lesions in a single image. The proposed method can train the segmentation of multiple lesions, potentially supplying additional information to machine learning systems. The ugly duckling sign [56], for example, shows that a given lesion differs from the patient’s other lesions. Most applicable to clinical images, this sign may also appear in dermoscopy images. Machine learning needs such information. The LAMA technique can synthesize multi-lesion samples from single-lesion images to train for the segmentation of multiple lesions, potentially providing new information for machine learning.
The limitation of this method is the use of non-overlapping patches during the non-lesion region-finding process. The non-overlapping method generates only a limited number of patch locations. For example, at level , it only generates four patches of size 270 × 270. More patches can be generated using an overlapping scanning window of 270 × 270 at level with a stride lower than 270 (say stride = 32, i.e., the distance between the centers of two adjacent patches is 32 pixels).
The proposed technique adds only parameters related to patch area and patch levels (the “Finding Non-lesion Patches” section), three parameters beyond the two-dozen deep learning training parameters routinely employed (the “ Experiments” section). Therefore, the LAMA model’s complexity is minimally increased beyond the standard single-lesion model for deep-learning image segmentation [35]. Generalizability is expected with the model tested on 600 publicly available test image disjoint from the training set by achieving state-of-the-art results.
Conclusion
In this study, we proposed a novel lesion-aware mixup augmentation (LAMA) method to train a robust deep neural network for skin lesion segmentation in dermoscopic images. None of the previous studies on skin lesion segmentation considered more than one lesion in dermoscopic skin lesion images. Recent skin lesion imaging techniques fail to provide guidance in approaching multiple-lesion images [57–59]. Therefore, we created a new multi-lesion (MuLe) segmentation dataset using publicly available ISIC skin lesion images to evaluate our proposed method. The proposed LAMA method effectively produced synthetic multi-lesion images by utilizing a training set of single-lesion images and their corresponding ground truth masks. The experimental results show that the ChimeraNet lesion segmentation model trained with LAMA not only successfully detects multiple lesions on real-life examples but also enhances the segmentation performance of single-lesion images. Further study of the LAMA technique is warranted.
Author Contribution
We confirm that all authors contributed to the study conception and design. The experiments were conducted by Norsang Lama. Data collection, analysis, and validation were performed by all authors. The first draft of the manuscript was written by Norsang Lama, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
National Institutes of Health, R43 CA153927- 01, William Van Stoecker, R44 CA101639-02A2, William Van Stoecker.
Data Availability
The test set, publicly available, is referenced on the description of the image data sets in the Introduction.
Declarations
Ethics Approval
NA.
Consent to Participate
NA.
Consent for Publication
NA.
Conflict of Interest
The authors declare no competing interests.
Disclaimer
Its contents are solely the authors’ responsibility and do not necessarily represent the official views of the NIH or NSF.
Footnotes
Article Highlights
• Current skin lesion segmentation algorithms lack accuracy for images with multiple lesions.
• LAMA is an algorithm for generating synthetic skin lesion patches for training image segmentation.
• LAMA yields multiple-lesion training images with minimal lesion overlap.
• Results show improved segmentation accuracy for both single- and multiple-lesion images.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.R. L. Siegel, K. D. Miller, N. S. Wagle, and A. Jemal, Cancer statistics, 2023, CA Cancer J Clin, vol. 73, no. 1, pp. 17–48, 2023, 10.3322/caac.21763. 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 2.H. Pehamberger, M. Binder, A. Steiner, and K. Wolff, In vivo epiluminescence microscopy: Improvement of early diagnosis of melanoma, Journal of Investigative Dermatology, vol. 100, no. 3 SUPPL., pp. S356–S362, 1993, 10.1038/jid.1993.63. 10.1038/jid.1993.63 [DOI] [PubMed] [Google Scholar]
- 3.H. P. Soyer, G. Argenziano, R. Talamini, and S. Chimenti, Is Dermoscopy Useful for the Diagnosis of Melanoma?, Arch Dermatol, vol. 137, no. 10, pp. 1361–1363, Oct. 2001, 10.1001/archderm.137.10.1361. 10.1001/archderm.137.10.1361 [DOI] [PubMed] [Google Scholar]
- 4.R. P. Braun, H. S. Rabinovitz, M. Oliviero, A. W. Kopf, and J. H. Saurat, Pattern analysis: a two-step procedure for the dermoscopic diagnosis of melanoma, Clin Dermatol, vol. 20, no. 3, pp. 236–239, May 2002, 10.1016/S0738-081X(02)00216-X. 10.1016/S0738-081X(02)00216-X [DOI] [PubMed] [Google Scholar]
- 5.A. Krizhevsky, I. Sutskever, and G. Hinton, ImageNet Classification with Deep Convolutional Neural Networks, in Advances in Neural Information and Processing Systems (NIPS), 2012, pp. 1097–1105.
- 6.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, and Z. Wojna, Rethinking the inception architecture for computer vision, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 2818–2826.
- 7.K. Simonyan and A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556, 2014.
- 8.I. Goodfellow et al., Generative adversarial networks, Commun ACM, vol. 63, no. 11, pp. 139–144, 2020. 10.1145/3422622 [DOI] [Google Scholar]
- 9.K. He, X. Zhang, S. Ren, and J. Sun, Deep residual learning for image recognition, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
- 10.A. Dosovitskiy et al., An image is worth 16x16 words: Transformers for image recognition at scale, arXiv preprint arXiv:2010.11929, 2020.
- 11.O. Ronneberger, P. Fischer, and T. Brox, U-Net: Convolutional Networks for Biomedical Image Segmentation. [Online]. Available: http://lmb.informatik.uni-freiburg.de/
- 12.M. Tan and Q. Le, Efficientnet: Rethinking model scaling for convolutional neural networks, in International conference on machine learning, 2019, pp. 6105–6114.
- 13.A. Esteva et al., Dermatologist-level classification of skin cancer with deep neural networks, Nature, vol. 542, no. 7639, pp. 115–118, 2017, 10.1038/nature21056. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.V. Gulshan et al., Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs, JAMA, vol. 316, no. 22, pp. 2402–2410, 2016. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 15.S. Sornapudi et al., Deep learning nuclei detection in digitized histology images by superpixels, J Pathol Inform, vol. 9, no. 1, p. 5, 2018. 10.4103/jpi.jpi_74_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.G. Litjens et al., A survey on deep learning in medical image analysis, Med Image Anal, vol. 42, pp. 60–88, 2017, 10.1016/j.media.2017.07.005. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 17.L. K. Ferris et al., Computer-aided classification of melanocytic lesions using dermoscopic images, J Am Acad Dermatol, vol. 73, no. 5, pp. 769–776, Nov. 2015, 10.1016/J.JAAD.2015.07.028. 10.1016/J.JAAD.2015.07.028 [DOI] [PubMed] [Google Scholar]
- 18.M. A. Marchetti et al., Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images, J Am Acad Dermatol, vol. 78, no. 2, pp. 270-277.e1, Feb. 2018, 10.1016/j.jaad.2017.08.016. 10.1016/j.jaad.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.H. A. Haenssle et al., Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists, Annals of Oncology, vol. 29, no. 8, pp. 1836–1842, 2018, 10.1093/annonc/mdy166. 10.1093/annonc/mdy166 [DOI] [PubMed] [Google Scholar]
- 20.N. C. F. Codella et al., Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images, IBM J. Res. Dev., vol. 61, no. 4–5, pp. 5:1–5:15, Jul. 2017. 10.1147/JRD.2017.2708299.
- 21.S. Pathan, K. G. Prabhu, and P. C. Siddalingaswamy, Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review, Biomed Signal Process Control, vol. 39, pp. 237–262, Jan. 2018, 10.1016/J.BSPC.2017.07.010. 10.1016/J.BSPC.2017.07.010 [DOI] [Google Scholar]
- 22.T. Majtner, S. Yildirim-Yayilgan, and J. Y. Hardeberg, Combining deep learning and hand-crafted features for skin lesion classification, 2016 6th International Conference on Image Processing Theory, Tools and Applications, IPTA 2016, 2017. 10.1109/IPTA.2016.7821017.
- 23.N. Codella, J. Cai, M. Abedini, R. Garnavi, A. Halpern, and J. R. Smith, Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images BT - Machine Learning in Medical Imaging, L. Zhou, L. Wang, Q. Wang, and Y. Shi, Eds., Cham: Springer International Publishing, 2015, pp. 118–126.
- 24.I. González-Díaz, DermaKNet: Incorporating the Knowledge of Dermatologists to Convolutional Neural Networks for Skin Lesion Diagnosis, IEEE J Biomed Health Inform, vol. 23, no. 2, pp. 547–559, 2019, 10.1109/JBHI.2018.2806962. 10.1109/JBHI.2018.2806962 [DOI] [PubMed] [Google Scholar]
- 25.J. R. Hagerty et al., Deep Learning and Handcrafted Method Fusion: Higher Diagnostic Accuracy for Melanoma Dermoscopy Images, IEEE J Biomed Health Inform, vol. 23, no. 4, pp. 1385–1391, 2019, 10.1109/JBHI.2019.2891049. 10.1109/JBHI.2019.2891049 [DOI] [PubMed] [Google Scholar]
- 26.A. K. Nambisan et al., Improving Automatic Melanoma Diagnosis Using Deep Learning-Based Segmentation of Irregular Networks, Cancers (Basel), vol. 15, no. 4, 2023. 10.3390/cancers15041259. [DOI] [PMC free article] [PubMed]
- 27.G. Celebi, Emre M.; Wen, Quan; Iyatomi, Hitoshi; Shimizu, Kouhei; Zhou, Huiyu; Schaefer, A State-of-the-Art on Lesion Border Detection in Dermoscopy Images, in Dermoscopy Image Analysis, J. S. Celebi, M. Emre; Mendonca, Teresa; Marques, Ed., Boca Raton: CRC Press, 2015, pp. 97–129. [Online]. Available: 10.1201/b19107
- 28.N. K. Mishra et al., Automatic lesion border selection in dermoscopy images using morphology and color features, Skin Research and Technology, vol. 25, no. 4, pp. 544–552, 2019. 10.1111/srt.12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. E. Celebi, H. Iyatomi, G. Schaefer, and W. v Stoecker, Lesion border detection in dermoscopy images, Computerized Medical Imaging and Graphics, vol. 33, no. 2, pp. 148–153, 2009. 10.1016/j.compmedimag.2008.11.002. [DOI] [PMC free article] [PubMed]
- 30.M. A. Al-masni, M. A. Al-antari, M. T. Choi, S. M. Han, and T. S. Kim, Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks, Comput Methods Programs Biomed, vol. 162, pp. 221–231, 2018, 10.1016/j.cmpb.2018.05.027. 10.1016/j.cmpb.2018.05.027 [DOI] [PubMed] [Google Scholar]
- 31.P. Tschandl, C. Sinz, and H. Kittler, Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation, Comput Biol Med, vol. 104, pp. 111–116, 2019, 10.1016/j.compbiomed.2018.11.010. 10.1016/j.compbiomed.2018.11.010 [DOI] [PubMed] [Google Scholar]
- 32.Y. Yuan and Y. C. Lo, Improving Dermoscopic Image Segmentation With Enhanced Convolutional-Deconvolutional Networks, IEEE J Biomed Health Inform, vol. 23, no. 2, pp. 519–526, 2019, 10.1109/JBHI.2017.2787487. 10.1109/JBHI.2017.2787487 [DOI] [PubMed] [Google Scholar]
- 33.F. Xie, J. Yang, J. Liu, Z. Jiang, Y. Zheng, and Y. Wang, Skin lesion segmentation using high-resolution convolutional neural network, Comput Methods Programs Biomed, vol. 186, p. 105241, 2020, 10.1016/j.cmpb.2019.105241. 10.1016/j.cmpb.2019.105241 [DOI] [PubMed] [Google Scholar]
- 34.Ş. Öztürk and U. Özkaya, Skin Lesion Segmentation with Improved Convolutional Neural Network, J Digit Imaging, vol. 33, no. 4, pp. 958–970, 2020, 10.1007/s10278-020-00343-z. 10.1007/s10278-020-00343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.N. Lama, J. Hagerty, A. Nambisan, R. J. Stanley, and W. V. Stoecker, Skin Lesion Segmentation in Dermoscopic Images with Noisy Data, J Digit Imaging, 2023, 10.1007/s10278-023-00819-8. 10.1007/s10278-023-00819-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.T. DeVries and G. W. Taylor, Improved regularization of convolutional neural networks with cutout, arXiv preprint arXiv:1708.04552, 2017.
- 37.H. Zhang, M. Cisse, Y. N. Dauphin, and D. Lopez-Paz, mixup: Beyond empirical risk minimization, arXiv preprint arXiv:1710.09412, 2017.
- 38.Y. Tokozume, Y. Ushiku, and T. Harada, Between-class learning for image classification, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 5486–5494.
- 39.S. Yun, D. Han, S. J. Oh, S. Chun, J. Choe, and Y. Yoo, Cutmix: Regularization strategy to train strong classifiers with localizable features, in Proceedings of the IEEE/CVF international conference on computer vision, 2019, pp. 6023–6032.
- 40.N. C. F. Codella et al., Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC), Proceedings - International Symposium on Biomedical Imaging, vol. 2018-April, pp. 168–172, 2018. 10.1109/ISBI.2018.8363547.
- 41.V. Rotemberg et al., A patient-centric dataset of images and metadata for identifying melanomas using clinical context, Sci Data, vol. 8, no. 1, p. 34, 2021. 10.1038/s41597-021-00815-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.N. Lama et al., ChimeraNet: U-Net for Hair Detection in Dermoscopic Skin Lesion Images, J Digit Imaging, no. 0123456789, 2022. 10.1007/s10278-022-00740-6. [DOI] [PMC free article] [PubMed]
- 43.S. Ioffe and C. Szegedy, Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift.
- 44.A. Buslaev, V. I. Iglovikov, E. Khvedchenya, A. Parinov, M. Druzhinin, and A. A. Kalinin, Albumentations: fast and flexible image augmentations, Information, vol. 11, no. 2, p. 125, 2020. 10.3390/info11020125 [DOI] [Google Scholar]
- 45.D. P. Kingma and J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980, 2014.
- 46.C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin, and M. Jorge Cardoso, Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations, in Deep learning in medical image analysis and multimodal learning for clinical decision support, Springer, 2017, pp. 240–248. [DOI] [PMC free article] [PubMed]
- 47.F. Navarro, M. Escudero-Viñolo, and J. Bescós, Accurate Segmentation and Registration of Skin Lesion Images to Evaluate Lesion Change, IEEE J Biomed Health Inform, vol. 23, no. 2, pp. 501–508, 2019, doi: 10.1109/ and Sustainable Development, GTSD 2020, pp. 366–371, 2020. 10.1109/GTSD50082.2020.9303084. [DOI] [PubMed]
- 48.P. Shan, Y. Wang, C. Fu, W. Song, and J. Chen, Automatic skin lesion segmentation based on FC-DPN, Comput Biol Med, vol. 123, no. April, p. 103762, 2020. 10.1016/j.compbiomed.2020.103762. [DOI] [PubMed]
- 49.R. Kaymak, C. Kaymak, and A. Ucar, Skin lesion segmentation using fully convolutional networks: A comparative experimental study, Expert Syst Appl, vol. 161, p. 113742, 2020, 10.1016/j.eswa.2020.113742. 10.1016/j.eswa.2020.113742 [DOI] [Google Scholar]
- 50.D. K. Nguyen, T. T. Tran, C. P. Nguyen, and V. T. Pham, Skin Lesion Segmentation based on Integrating EfficientNet and Residual block into U-Net Neural Network, Proceedings of 2020 5th International Conference on Green Technology Access, vol. 10, no. September, pp. 94007–94018, 2022. 10.1109/ACCESS.2022.3204280.
- 51.K. Zafar et al., Skin lesion segmentation from dermoscopic images using convolutional neural network, Sensors (Switzerland), vol. 20, no. 6, pp. 1–14, 2020, 10.3390/s20061601. 10.3390/s20061601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. Goyal, A. Oakley, P. Bansal, D. Dancey, and M. H. Yap, Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods, IEEE Access, vol. 8, pp. 4171–4181, 2020, 10.1109/ACCESS.2019.2960504. 10.1109/ACCESS.2019.2960504 [DOI] [Google Scholar]
- 53.X. Tong, J. Wei, B. Sun, S. Su, Z. Zuo, and P. Wu, Ascu-net: Attention gate, spatial and channel attention u-net for skin lesion segmentation, Diagnostics, vol. 11, no. 3, 2021. 10.3390/diagnostics11030501. [DOI] [PMC free article] [PubMed]
- 54.P. Chen, S. Huang, and Q. Yue, Skin Lesion Segmentation Using Recurrent Attentional Convolutional Networks, IEEE JBHI.2018.2825251.
- 55.H. Ashraf, A. Waris, M. F. Ghafoor, S. O. Gilani, and I. K. Niazi, Melanoma segmentation using deep learning with test-time augmentations and conditional random fields, Sci Rep, vol. 12, no. 1, pp. 1–16, 2022, 10.1038/s41598-022-07885-y. 10.1038/s41598-022-07885-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A. Scope, et al. The ugly duckling sign: agreement between observers. Archives of Dermatology, vol. 144, no. 1, pp. 58–64. 10.1001/archdermatol.2007.15 [DOI] [PubMed]
- 57.M. Zafar, M. I. Sharif, M. I. Sharif, S. Kadry, S. A. C. Bukhari, and H. T. Rauf, Skin Lesion Analysis and Cancer Detection Based on Machine/Deep Learning Techniques: A Comprehensive Survey. Life (Basel), 2023 Jan 4;13(1):146. 10.3390/life13010146. 10.3390/life13010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. M. Hosny, D. Elshoura, E. R. Mohamed, E. Vrochidou and G. A. Papakostas, Deep Learning and Optimization-Based Methods for Skin Lesions Segmentation: A Review, in IEEE Access, vol. 11, pp. 85467-85488, 2023. 10.1109/ACCESS.2023.3303961. 10.1109/ACCESS.2023.3303961 [DOI] [Google Scholar]
- 59.G. Subhashini & A. Chandrasekar (2023) Hybrid deep learning technique for optimal segmentation and classification of multi-class skin cancer, The Imaging Science Journal, 10.1080/13682199.2023.2241794 10.1080/13682199.2023.2241794 [DOI] [Google Scholar]
- 60.BAGHERI, FATEMEH; TAROKH, MOHAMMAD JAFAR; and ZIARATBAN, MAJID (2022) Skin lesion segmentation by using object detection networks, DeepLab3+, and active contours, Turkish Journal of Electrical Engineering and Computer Sciences: Vol. 30: No. 7, Article 2. 10.55730/1300-0632.3951
- 61.Öztürk, Ş., Özkaya, U. Skin Lesion Segmentation with Improved Convolutional Neural Network. J Digit Imaging33, 958–970 (2020). 10.1007/s10278-020-00343-z 10.1007/s10278-020-00343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The test set, publicly available, is referenced on the description of the image data sets in the Introduction.