Abstract
Many neurotransmitter receptors activate G proteins through exchange of GDP for GTP. The intermediate nucleotide-free state has eluded characterization, due largely to its inherent instability. Here we characterize a G protein variant associated with a rare neurological disorder in humans. GαoK46E has a charge reversal that clashes with the phosphate groups of GDP and GTP. As anticipated, the purified protein binds poorly to guanine nucleotides yet retains wild-type affinity for G protein βγ subunits. In cells with physiological concentrations of nucleotide, GαoK46E forms a stable complex with receptors and Gβγ, impeding effector activation. Further, we demonstrate that the mutant can be easily purified in complex with dopamine-bound D2 receptors, and use cryo-electron microscopy to determine the structure, including both domains of Gαo, without nucleotide or stabilizing nanobodies. These findings reveal the molecular basis for the first committed step of G protein activation, establish a mechanistic basis for a neurological disorder, provide a simplified strategy to determine receptor-G protein structures, and a method to detect high affinity agonist binding in cells.
Subject terms: Neuroscience, G protein-coupled receptors, Mutation, Cryoelectron microscopy
Many neurotransmitters act on receptors coupled to GTP-binding G proteins. Here authors report the structure and activity of a mutant that locks the nucleotide-free and receptor-bound state of the G protein, leading to a rare neurological disorder.
Introduction
Many clinically important drugs, environmental signals, hormones, and neurotransmitters elicit their effects through G protein-coupled receptors (GPCRs). Receptors of this class represent one of the largest gene families, and by far the largest class of drug targets, and includes members with little or no shared sequence similarity. In contrast, G proteins are highly conserved in structure and function, and in mammals consist of just four subfamilies (Gs, Gi/o, Gq/11, and G12/13) encoded by 16 individual genes.
The ability of such widely divergent receptors to activate a comparatively small number of G proteins indicates a shared mechanism of activation1,2. Broadly speaking, agonist binding to receptors leads to conformational changes in the G protein α subunit, release of GDP, binding to GTP, and dissociation of Gα-GTP from the Gβγ dimer3,4. Either of the dissociated subunits can modulate the activity of downstream effector proteins, including adenylyl cyclase and phospholipase Cβ5,6, among others. In addition, Gβγ dimers regulate potassium channels, calcium channels, phosphatidylinositol 3-kinases and MAPKs7. Agonist-induced signaling is terminated after Gα hydrolyzes GTP and the subunits reassemble; in most cases, this inactivation step is accelerated by regulators of G protein signaling (RGS) proteins3. Thus, receptors serve as signal discriminators and nucleotide exchange factors, RGS proteins serve as timing devices and G proteins serve as determinants of output specificity.
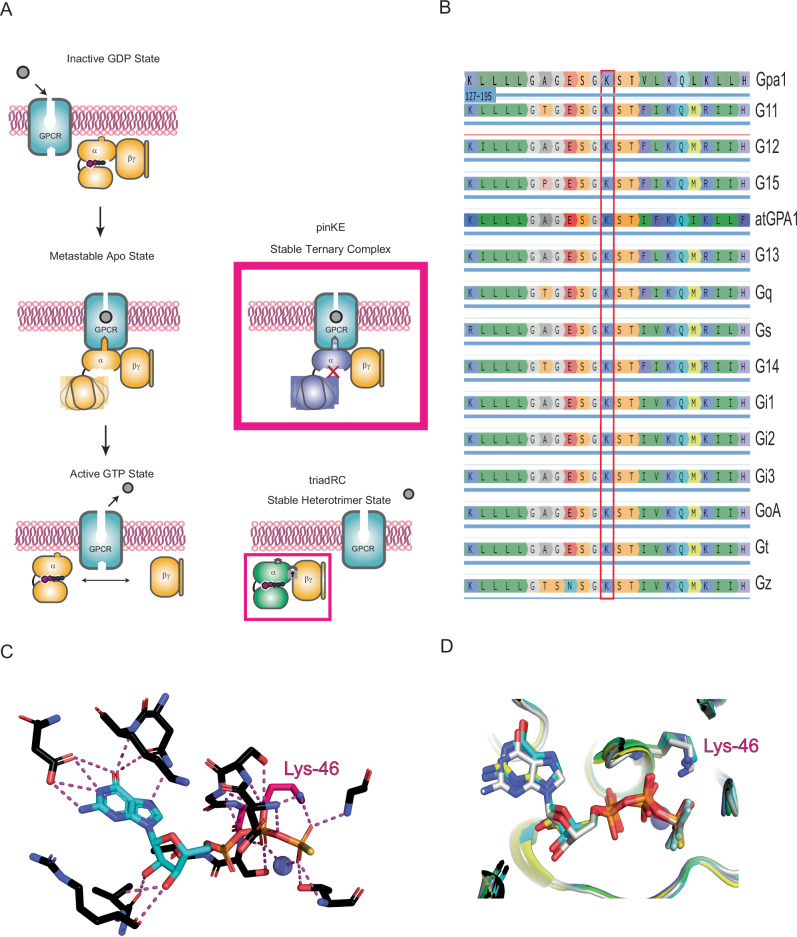
Activated GPCRs can stabilize the G protein in the nucleotide-free (apo) state, thereby facilitating GTP-GDP exchange8–10. The G protein nucleotide exchange step is likely accomplished by opening the cleft between the two domains of a given Gα protein in conjunction with docking of the α5 helix to the receptor11,12. Direct evidence of such inter-domain movement was first provided through double electron-electron resonance experiments on rhodopsin-activated Gαi13 and Gαs14, as well as through hydrogen-deuterium exchange studies15,16, and X-ray crystallographic studies of the β2-adrenergic receptor bound to the Gs protein heterotrimer17. In every study, Gβγ remained associated with Gα in the apo state. However, upon binding to GTP, the G protein subunits dissociate and are free to activate downstream effectors. Thus, the process of G protein activation is comprised of three discrete binding and unbinding transformations: (i) GPCR-dependent unbinding of GDP and stabilization of the nucleotide-free form of Gα, (ii) GTP-binding-dependent conformational changes in Gα, and (iii) unbinding of Gα-GTP from the Gβγ subunits (Fig. 1A).
Fig. 1. The phosphate-interacting and neutralizing lysine (pinK) controls one of three binding and unbinding transformations in Gα.
A Agonist binding leads to stabilization of the nucleotide-free form of the G protein heterotrimer, while GTP binding leads to conformational changes in Gα and unbinding of Gα-GTP from the Gβγ subunits. Mutations disrupt GTP binding (pinKE) and GTP-dependent subunit dissociation (triadRC). B The conserved phosphate interacting and neutralizing lysine (pinK) is present in all human Gα proteins as well as Gα proteins from the yeast S. cerevisiae (Gpa1) and plant A. thaliana (AtGPA1). C pinK (Lys-46 in Gαo) forms polar interactions with GTP. D Overlay of multiple Gα structures from PDB ID: 1GIA (Gi1, green), 1AZT (Gs, cyan), 1TND (Gt, blue), 3C7K (Go, gray), and 1ZCA (G12, yellow), showing that pinK bridges the β and γ phosphates of GTP.
Previous studies have characterized mutations that disrupt each of the transformations.
Lambert and colleagues described two types of Gα mutants18,19, both of which bind stably with agonist-occupied receptor but fail to release GDP20. One strategy involved inserting four amino acids and extending the α5 helix, mimicking the translation that normally occurs during receptor engagement. Under these circumstances receptor-G protein subtype selectivity was diminished20. Mutations at the catalytic glutamine (required for GTP hydrolysis21,22) impose an ensemble of activated states that lead to sustained effector activation23. Mutations at a conserved arginine, part of the Gly-Arg-Glu (G-R-E) triad, prevent the release of Gβγ even in the GTP-bound state (Fig. 1A)24.
Following a recent comprehensive screen of mutations associated with a neurological disorder in humans25, we identified a substitution (GαoK46E) that seemed likely to prevent guanine nucleotide binding, and thereby impose the first transformation state of the activation process (Fig. 1A)26,27. Of the 55 mutations tested, GαoK46E most strongly suppressed signaling in cells. This phosphate-interacting and neutralizing Lys (“pinK”) is conserved in all G proteins (Fig. 1B), as well as in most protein kinases28. By swapping a positively charged side chain with one that is negatively charged, and at a position buried within the nucleotide-binding pocket, the Lys-to-Glu (“pinKE”) substitution is likely to repel the phosphate groups of GDP and GTP (Fig. 1C, D). At the same time, the presence of the glutamate may help to secure the protein in the apo state, since the negatively charged residue may take the place of the negatively charged phosphates.
Here, we demonstrate that the pinKE mutation is incompatible with proper nucleotide binding but preserves the ability of the protein to bind receptors and Gβγ, as determined through detailed protein structure and function analysis. Consequently, this mutant inhibits signaling, in a dominant-negative fashion, as determined through cell-based activity measures. Our investigations reveal a likely mechanism of action for a rare neurological disorder and provide a tool for the isolation and characterization of G proteins in complex with agonist-occupied receptors.
Results
Dominant negative inhibition of Gβγ signaling
In a screen of 55 Gαo mutations, each associated with a neurological disorder in humans25, we identified a subset of 29 mutations that suppress Gβγ dissociation in cells. Because these mutants inhibit signaling in the presence of wild-type Gα, they may be regarded as dominant negative mutants29,30. Of the 55 variants tested, the best-performing mutant had a charge substitution at a conserved lysine that bridges the β and γ phosphates of GTP. Replacement of the lysine with a negatively charged glutamate (GαoK46E) is likely to be incompatible with GDP or GTP binding, due to charge-charge repulsion. Therefore, we postulated that GαoK46E persists in the nucleotide-free state and, consequently, remains associated with Gβγ and receptor. To test this hypothesis, we used bioluminescence resonance energy transfer (BRET) to monitor Gα-Rluc8 dissociation from Gβγ-GFP2 following GPCR activation (Fig. 2A)25,31. Lys46 is conserved in all human Gα members (position G.H1.01 using the CGN nomenclature1; Fig. 1B). To determine if its function is conserved, we introduced pinKE mutations into 10 human Gα-Rluc8 subtypes and the two major splice variants of Gαo. As controls we used wild-type Gα-Rluc and a mutation at a conserved arginine (triadRC, position G.H2.04)27,32,33, which is necessary for GTP-dependent subunit dissociation24. We co-transfected HEK293T cells with these G protein variants and with canonical receptors known to couple to each subtype; the μ-opioid receptor (MOR, couples to Gαi1, Gαi2, Gαi3, Gαz, GαoA, and GαoB), the dopamine D2 receptor (D2R, couples to GαoA and GαoB), the neurotensin 1 receptor (NT1R, couples to Gαq, Gα11, Gα13 and Gα15), and the β2-adrenergic receptor (β2AR, couples to Gαs) (Supplementary Fig. 1)31. For all ten subtypes tested, the triadRC substitutions led to diminished subunit dissociation (Fig. 2A and Supplementary Fig. 1A), consistent with the known defect in GTP-dependent conformational changes24. The pinKE substitutions likewise diminished responses (Fig. 2A and Supplementary Fig. 1A, B). Notably, a subset of pinKE mutants displayed significantly reduced luminescence, basal BRET ratios, or both, indicating poor expression or association, respectively (Supplementary Fig. 1C and 1D). While GαoA appeared to express more poorly than the other subtypes tested (Supplementary Fig. 1C), it bound particularly well to Gβγ under basal conditions (Supplementary Fig. 1D). Together, these data indicate that Lys46 is needed for proper Gβγ dissociation.
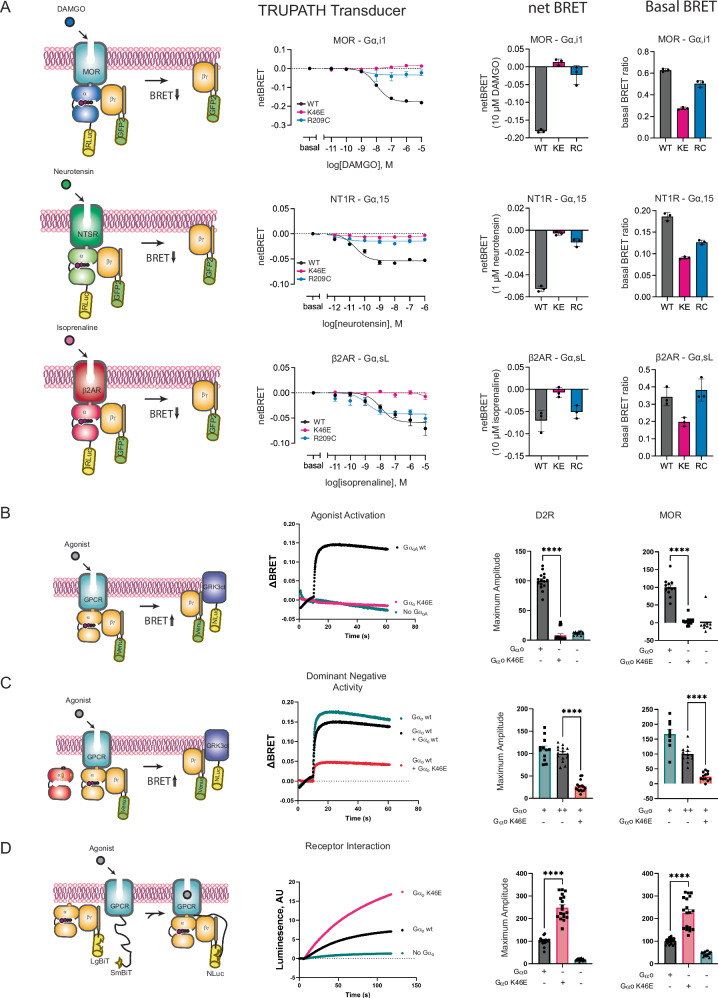
Fig. 2. The pinKE mutation impedes G protein subunit dissociation.
A Agonist-induced dissociation of Gα and Gβγ subunits (left). HEK293T cells transfected with the indicated receptor, WT or mutant Gα-RLuc8 donor, Gβ and Gγ-GFP acceptor proteins (middle). Representative concentration-response measurements using the μ-opioid (MOR; Gi1), neurotensin (NT1R; Gq, G13), or β2-adrenergic receptor (β2AR; Gs) receptors, presented as fold decrease in dynamic range measured by comparing the energy transfer from donor to acceptor and reported as ΔBRET (GFP/Rluc8 per well minus Basal BRET, or GFP/Rluc8, at lowest dose of agonist) in comparison to basal activity, presented as raw BRET values prior to stimulus (right). Data are means ± SEM from 3 independent experiments, 2 measurements each. B Agonist-induced dissociation of Gβγ subunits (left). HEK293FT cells transfected with MOR or D2R, either empty vector (pcDNA3.1) or vector with wild-type or mutant Gαo, masGRK3ct-Nluc-HA donor and Venus-Gβγ acceptor proteins. Representative time-course measurements for D2R after dopamine addition, presented as ΔBRET (ratio of emission by Venus at 535 nm and Nluc at 475 nm; recorded prior to agonist stimulation and subtracted from the experimental BRET values, middle). Effect of mutations, quantified as maximum amplitude relative to wild type (right). Data are means ± SEM from 4 (vector) or 5 (Gαo or GαoK46E) independent experiments, 3 measurements each. C Dominant-negative inhibition of subunit release (left). HEK293 cells transfected as in B but with the addition of an equal amount of wild-type Gαo (middle), done by transfecting equivalent amounts of mutant and wild-type DNA. Effect of mutations quantified as maximum amplitude (right). BRET data are means ± SEM from 4 (vector) or 5 (Gαo or GαoK46E) independent experiments, 3 measurements each. D Agonist-induced association of GPCRs and G proteins (left). HEK293FT cells transfected with MOR or D2R fused to myc-SmBiT, Gαo, Gβ fused to LgBiT, and Gγ. Representative time-course measurements after dopamine addition leading to reconstitution of functional Nluc, presented as arbitrary luminesence units (ΔAU, middle). Effect of mutations, quantified as maximum amplitude (right). Data are means ± SEM from 6 independent experiments, 3 measurements each. Statistical analysis performed with 2-tailed unpaired t test; ****p < 0.0001. Source data are provided as a Source Data file.
The BRET method used above provides a direct and quantitative readout of distance between donor and acceptor, in this case bound to Gα and Gβγ, and therefore is well suited for comparison of the pinKE substitutions in multiple Gα subtypes. Using this method, we demonstrated that pinKE impedes the release of Gβγ 25, in the manner of the triad arginine mutant24. Whereas the triadRC mutant assembles with Gβγ even in the GTP-bound state, however, we postulated that pinKE would be predominantly in the nucleotide-free state and would thus fail to dissociate from receptors as well as from Gβγ.
While the diminished BRET signal can reflect diminished proximity, it is also dependent on the dipole angle of the fluorophores within each Gα subtype. In addition, it is possible that some G proteins do not fully dissociate following receptor activation. As an additional test of our hypothesis, we used an alternative BRET method, one that measures the ability of the mutant to inhibit receptor-mediated activation and in competition with wild-type G protein. Once again, we compared the response to dopamine as well as to μ-opioid ligands, but in this case we monitored the ability of free Gβγ to bind the C-terminal domain of the G protein-coupled receptor kinase (GRK3ct)34–36. As shown in Fig. 2B, wild type - but not the pinKE mutant – form of GαoA (hereafter Gαo) released Gβγ in response to agonists, as indicated by the relative increase in fluorescence over time. We then tested the ability of pinKE to suppress signaling by wild-type Gαo, coexpressed in the same cell. As shown in Fig. 2C, an extra copy of Gαo dampened the response while pinKE largely abrogated the response (see also Supplementary Fig. 1E). Thus, and as anticipated by our model, GαoK46E suppressed the release of Gβγ, and did so both in the absence and presence of wild-type Gαo.
The results shown above indicate that GαoK46E forms a non-productive complex with Gβγ. Our model was that it also binds stably to agonist-bound receptors, and by extension with Gβγ. To test this feature of the model we used another protein proximity assay, one that relies on complementation between SmBit-tagged receptor and LgBit-tagged Gβ. Assembly of receptor and Gβγ, and therefore of SmBit with LgBit, reconstitutes luciferase activity. In this case, binding of receptor to GαoK46E was higher by at least two-fold, in comparison with wild-type Gαo (Fig. 2D). This is in contrast to GαoR209C, which binds to receptors in the manner of the wild-type G protein24,27,32,33,37. We conclude that both mutants sequester Gβγ, but pinKE alone sequesters receptors.
Inhibition of effector function
Our cell-based analysis revealed that GαoK46E inhibits receptor-mediated G protein activation and does so in a genetically dominant manner. Given that GαoK46E was identified in individuals with a severe neurological condition38, and given that Gαo is the most abundant G protein in the brain and central nervous system39,40, we next sought to establish its function towards a physiologically relevant effector system. We turned to the inwardly rectifying potassium (GIRK) channel, which like Gαo is abundant in the brain. Upon binding to free Gβγ, GIRK channel opening leads to potassium flux. The resulting current provides a readout of G protein heterotrimer dissociation (Fig. 3A).
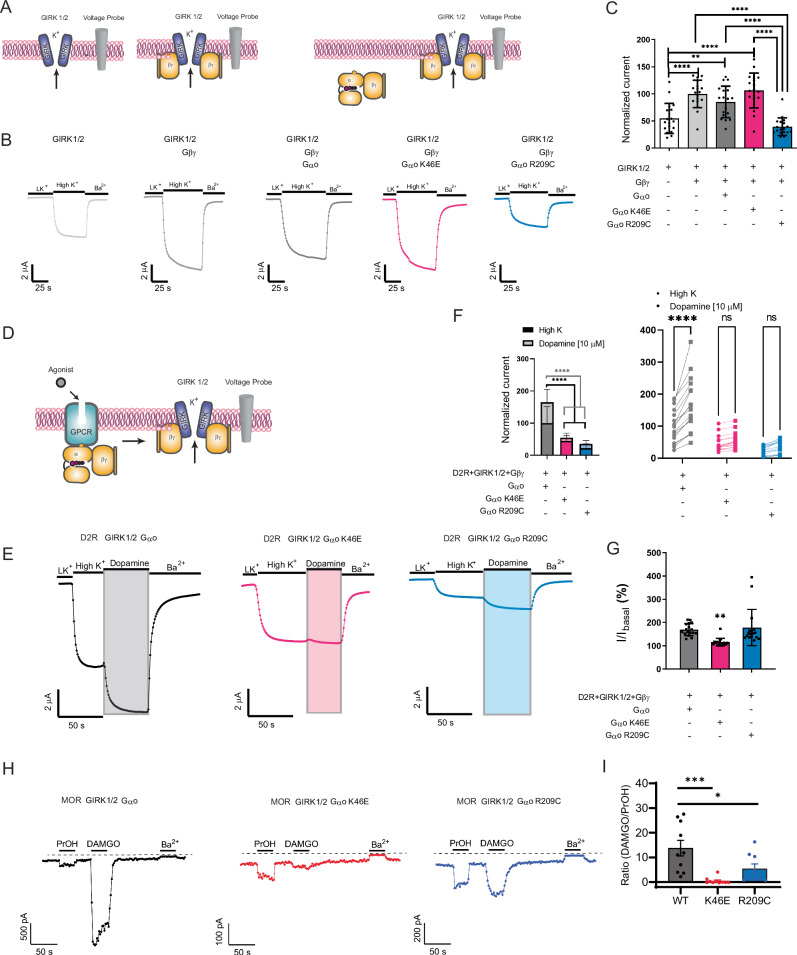
Fig. 3. The pinKE mutation impedes Gβγ-mediated effector activation.
A GIRK1/GIRK2 channel current without (left) and with Gβγ (middle) or Gαβγ (right). B Representative two-electrode voltage clamp (TEVC) recordings in Xenopus laevis oocytes expressing GIRK1/2, Gβ1γ2, and either Gαo (black), GαoK46E (red) or GαoR209C (blue) following treatment with high potassium solution (High K+) and barium (Ba2+), which directly activate and inactivate the channel, respectively. C Maximum voltage differences. Normalized current, normalized to the GIRK 1/2 + Gβγ condition (100%). Data are means ± SD from 3 independent experiments with 6 measurements each; ****p < 0.0001; **p = 0.0083. D GIRK1/GIRK2 channel current with Gαβγ and receptor. E Representative TEVC recordings in Xenopus laevis oocytes expressing dopamine D2 receptor, GIRK1/2, Gβ1γ2, and either Gαo (black), GαoK46E (red), or GαoR209C (blue) following treatment with high potassium solution (High K+) and barium (Ba2+), as well as dopamine, which activates the D2 receptor. F Normalized potassium- and dopamine-induced currents. Data are means ± SD from 3 independent experiments, with 6, 6, and 5 measurements each; ****p < 0.0001. G Normalized currents induced by dopamine divided by currents induced by high potassium (I/Ibasal). Data from (F) are means ± SD; **p = 0.0039. H Representative current recordings from HEK293T cells expressing MOR and the indicated Gαo subunit, showing activation with 1-propanol (PrOH), activation with DAMGO or inhibition with BaCl2. Each solution was delivered for 30 s. I Bar plot shows the ratio of DAMGO-activated to PrOH-activated current for each Gαo subunit. Data are means ± SD from 9 (R209C) or 10 (WT, K46E) independent experiments; ***p = 0.0003; *p = 0.0221. Statistical analysis in C and G performed with one-way ANOVA Tukey’s test. Statistical analysis in F performed by two-way ANOVA with Tukey’s multiple comparison (left) and Sidak pairwise comparison (right) tests. Statistical analysis in I performed with one-way ANOVA Dunett’s test. Source data are provided as a Source Data file.
To corroborate previous findings, we introduced to Xenopus laevis oocytes the GIRK1 and GIRK2 channels, followed by Gβγ and the Gαo protein variants (Fig. 3A). To establish the functionality of the GIRK channel, we measured the current in response to potassium, which provides a measure of basal (unstimulated) channel opening, and in response to barium, which inhibits channel current. Introduction of Gβγ alone activated the channel, and that activity was reduced by addition of GαoR209C, presumably due to sequestration of Gβγ. In contrast, the signal was unaffected by the GαoK46E mutant and wild-type proteins (Fig. 3B, C). When we introduced the dopamine D2 receptor and treated the cells with a saturating concentration of agonist (Fig. 3D), both mutants reduced the effect of potassium, and pinKE largely abrogated the effect of dopamine (Fig. 3E–G). These results are consistent with our initial screen25, and support the model that the triadRC mutant sequesters Gβγ on its own while pinKE does so only in complex with the activated receptor.
To verify and expand our analysis, we used a human cell expression system (HEK293T) and measured activation of GIRK1/2 channels in cells co-expressed with either Gαo, GαoK46E or GαoR209C. We compared μ-opioid receptor activation with the direct activation of GIRK channels by alcohol (100 mM 1-propanol, PrOH). Alcohol-dependent activation bypasses G protein activation of GIRK channels41,42, thus enabling a comparison between G protein-activated and directly-activated channels. The barium-inhibited current was not significantly different when comparing the wild type and mutant G proteins (Supplementary Fig. 1F), suggesting similar levels of GIRK expression. In contrast, we observed a significant reduction in the receptor-activated current, as compared to PrOH-activated current, with GαoR209C and an even stronger reduction with GαoK46E (Fig. 3H, I). Indeed, a large proportion (7/10) of recorded cells had PrOH-activated currents but negligible ( < 1 pA) responses to DAMGO, suggesting a dominant negative effect. These data support the conclusion that both GαoK46E and GαoR209C mutants sequester Gβγ, but GαoK46E does so only in the presence of activated receptor. Accordingly, differences in Fig. 3C, F could be due to stabilization by receptor of the G protein heterotrimer. Stated differently, triadRC acts as a Gβγ-specific dominant negative mutant and pinKE is a receptor-specific dominant negative mutant.
pinKE binds to Gβγ but not guanine nucleotides
Our cell-based measurements indicate that the pinKE variant can sequester receptors, possibly because it is locked in the nucleotide-free state. To test this directly, we purified GαoK46E, as well as Gαo and GαoR209C, and used three different measures of GTP binding and hydrolysis. First, we determined the melting temperature of purified protein using fast determination of quantitative cysteine reactivity (fQCR). In this method, buried cysteine residues that become exposed when the protein unfolds are covalently labeled with a fluorogenic compound. Previously we used fQCR to show that G proteins are more stable when bound to GTPγS than GDP24,43, and we anticipated that the thermostability would be further reduced for a nucleotide-free mutant44. Accordingly, we purified wild type and mutant variants of Gαo and monitored labeling as a function of temperature. As shown in Fig. 4A, whereas the wild-type protein was more stable when bound to GTPγS than GDP, GαoR209C showed no stabilization in the presence of GTPγS. The GαoK46E variant had a substantially lower Tm, and likewise showed no differences when comparing GTPγS and GDP. Only at 1000-fold higher concentrations of GDP did we observe a Tm close to that of the wild-type protein (Fig. 4B).
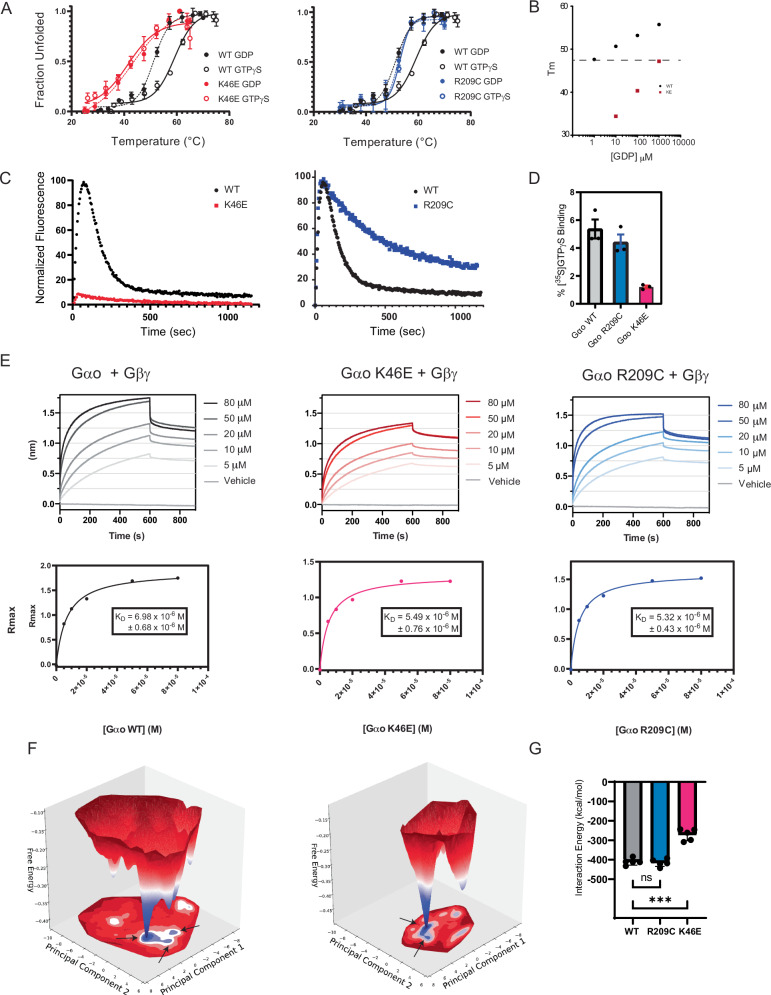
Fig. 4. The pinKE mutation preserves high affinity binding to Gβγ but not guanine nucleotides.
A Thermostability of purified Gαo (black), GαoK46E (red), and GαoR209C (blue) equilibrated in GTPγS (solid lines) or GDP (dashed lines). Tm values were quantified by fitting a two-state model of thermal unfolding. B Comparison Gαo and GαoK46E binding (Tm) to GDP. C Combination of purified Gαo, GαoK46E, and GαoR209C with BODIPY-GTP to monitor binding (increase in fluorescence) and hydrolysis (decrease in fluorescence). Normalized fluorescence, defined as percentage of maximum signal after subtracting the starting signal, for each experimental run. D Percent Gαo bound to GTPγ[35S] determined using the ratio of the measured activity per sample (cpm/pmol Gαo) to the average total specific activity of 35S added to each sample (cpm/pmol). E Purified biotinylated Gβ and Gγ immobilized on streptavidin were combined with the indicated concentration of purified Gαo (left), GαoK46E (middle), and GαoR209C (right). Binding is reported as a shift in the interference pattern (nanometers, nm). Rmax, defined as the absolute signal in nm after subtracting the starting (buffer) control. F Free energy surface of Gαo-apo system (left) and GαoK46E -apo system (right). Shown are representative structures from the local minima indicated by arrows on the free energy surface. G Interaction energy of GTP with proteins in the Gαo-GTP, GαoR209C-GTP, and GαoK46E -GTP systems. Data in A and D are means ± SEM, from 3 independent experiments, 3 measurements each. Data in B are means of 2 independent experiments. Data in C are representative of 3 or more independent experiments. Data in E are representative of 2 independent experiments, 2 measurements each. Data in G are means ± SEM, from 5 independent experiments; ***p = 0.0003. Source data are provided as a Source Data file.
Second, we tracked the ability of all three proteins to bind and hydrolyze GTP. To that end, we monitored the increase in fluorescence that accompanies the binding of BODIPY-GTP to protein, followed by the loss of fluorescence as BODIPY-GTP is reduced to BODIPY-GDP45,46. By this measure, we showed that the GTP analog bound to wild-type protein and GαoR209C, but not to GαoK46E (Fig. 4C). GTP was slowly hydrolyzed by GαoR209C, which is usually indicative of a hyperactive G protein. However, we have shown previously that substitutions of the triad-arginine suppress Gβγ dissociation, even in the GTP-bound state24. Thus, the slow rate of hydrolysis would not affect the levels of free Gα and Gβγ in the cell. Finally, to confirm that lack of binding was not an artifact of the BODIPY moiety, we measured the association of radiolabeled GTPγ35S to the mutant and wild-type forms of Gαo. Once again, nucleotide bound well to Gαo and GαoR209C but not to GαoK46E (Fig. 4D).
The lack of cooperativity in our thermostability profiles suggested that the pinKE mutant in the monomeric state is in a highly dynamic, and possibly molten, globular form. To determine whether GαoK46E was properly folded, we tested its ability to bind Gβγ directly, using bio-layer interferometry (BLI). We combined biotin-Gβγ, immobilized onto Streptavidin-coated biosensor tips, with a solution containing GDP and purified Gαo, GαoK46E, or GαoR209C. After 10 min the tips were transferred to an identical solution but lacking Gα. By this measure, the wild type and mutant proteins exhibited similar association and dissociation kinetics, indicating that the binding affinities were the same for each (Fig. 4E). Taken together, our results indicate that both GαoR209C and GαoK46E stabilize the G protein heterotrimer but do so by different mechanisms. Whereas GαoK46E binds poorly to guanine nucleotides, GαoR209C binds to nucleotides but fails to release Gβγ.
To gain mechanistic insights into the residues that contribute to nucleotide binding affinity, we performed molecular dynamics simulations (MDS) on the monomeric form of wild-type Gαo and the two mutants, GαoK46E and GαoR209C, with and without GTP bound. Using the aggregated trajectories from the MDS, we performed principal component analysis (PCA) and projected the free energy in the first two top-weighted principal components (PC1 and PC2). Figure 4F shows the free energy surface for monomeric Gαo and GαoK46E, along with the structures representing the minima in the free energy surfaces. The GαoK46E mutant had fewer free energy minima, indicating less flexibility than that of the wild-type Gαo. The structures representing the minima revealed that the Ras-like domain is more flexible in the mutant protein than it is in the wild type. To understand the residues in Gαo that contribute to GTP binding, we calculated the interaction energy (sum of electrostatic and van der Waals interaction energies) of GTP with the whole of Gαo, GαoK46E, or GαoR209C. As seen in Fig. 4G, we observed significantly reduced interaction energy between GTP and the GαoK46E mutant, as compared to wild-type Gαo or GαoR209C. We then examined the contact frequency (percentage of MD snapshots that make contact with GTP) of various residues in the nucleotide-binding site (Supplementary Fig. 2). This analysis revealed that residues Glu-43, Ser-44, Gly-45, Lys-46, Lys-181, and Thr-182 weaken their contact frequency by more than 30% in the GαoK46E mutant as compared to wild-type Gαo. The total interaction energy of the α5 helix with the Ras domain is weaker in GαoK46E than in wild-type Gαo; similarly, the structure of the previously described GDP-bound Gαo−4A mutant shows that the α5 helix is detached from the Ras domain18. These data suggest that the K46E and −4A mutants operate by similar mechanisms. In summary, our MDS analysis reveals how the Lys-46-Glu substitution affects the structure and dynamics of Gαo, and provides a rationale for the observed reduction in GTP-binding affinity.
pinKE preserves a high-affinity agonist-bound form of the receptor
Agonist binding to receptor stabilizes the nucleotide-free form of the G protein and, conversely, the nucleotide-free G protein confers increased affinity of receptors for agonists (but not for antagonists)47. Because pinKE binds poorly to nucleotide, we anticipated that it would lock the receptor in the activated, agonist-bound conformation. We initially tested this using a saturation binding assay to determine affinity for radioactive agonist, comparing Gαo and GαoK46E in the presence and absence of the GTP analog GppCp. To ensure proper coupling and a 1:1 stoichiometry, we used a fusion of the μ-opioid receptor and Gαo (MOR-Gαo). As anticipated by our model, MOR-GαoK46E bound agonist with high affinity, even in the presence of GppCp; in contrast, MOR-Gαo displayed decreased affinity for agonist with addition of GppCp (Fig. 5A–C, black vs red traces). Furthermore, we purified the full-length dopamine D2 receptor gene (without any mutations, D2R) co-expressed in sf9 insect cells with GαoK46E, Gβ1, and Gγ2, in the presence and absence of apyrase, an enzyme that degrades ATP/GTP to AMP/GMP, using the full agonist dopamine to form the complex. Size exclusion and SDS gel analysis revealed that complex formation is achieved and stable during cell lysis throughout the multi-day purification process, even in the absence of apyrase (Supplementary Fig. 3). Taken together, these results indicate that GαoK46E imposes the high affinity agonist-binding conformation of the receptor, and does so even in the presence of guanine nucleotide.
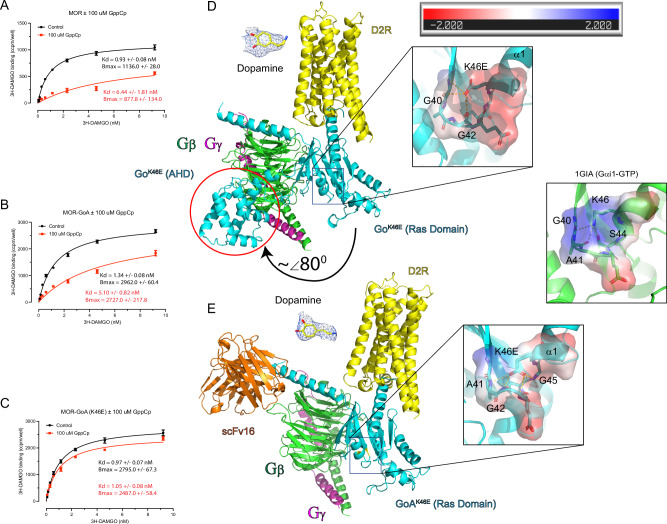
Fig. 5. The pinKE mutation stabilizes the high affinity agonist-bound form of the receptor.
A Saturation binding assays using [3H] DAMGO in the absence (black) and presence (red curve) of nonhydrolyzable GTP (GppCp) for μ-opioid receptor (MOR), 15 μg/well. B As in A, but for MOR fusion with Gαο, 15 μg/well. C As in A, but for MOR fusion with GαοK46E,12 μg/well. D Cryo-EM structure of the dopamine D2 receptor (D2R), GαοK46E and Gβγ complex. Insets show representative EM density for dopamine and the K46E region of Gαο (note occluded pocket and charge reversal). Distance lines are <4 Å. Black directional arrow, movement of the all-helical domain (AHD, enclosed by a red circle) away from Ras-like domain. Inset, electrostatic potential calculated using the APBS plugin for PyMol. Displayed is the colored surface with a range of ±2 KbT/ec. E Cryo-EM structure of D2R, GαοK46E, Gβγ, and scFv16 complex. Insets show representative EM density for dopamine and the Lys-46 region in Gαi1 (PDB ID: 1GIA, note positive charge of region). Distance lines are <4 Å. Inset, electrostatic potential calculated using the APBS plugin for PyMol. Displayed is the colored surface with a range of ±2 KbT/ec. Data in A–C are presented as means ± SEM from independent experiments, 3 measurements each. Statistical differences of Kd values derived from concentration-response assays were determined using the Extra sum-of-squares F test function in GraphPad Prism 9.0 comparing ±100 μM GppCp: (A) p < 0.0001; F(1,185) = 35.5 (B), p < 0.0001; F(1,188) = 62.2; (C) p = 0.5185; F(1,186) = 0.4185. In panels D and E, superposition of loop region residues G40-S47 Cα reveal an RMSD of 1.094 Å and 0.598 Å when comparing Gαi1-GTP (PDB ID: 1GIA) with D2R-GoK46E and D2R-GoK46E + scFv16, respectively. Source data are provided as a Source Data file.
Our results indicate that a single amino acid substitution confers stable association of Gαo with agonist-bound receptor and the Gβγ subunits. Accordingly, the pinKE mutant could be used for structure determination of receptors bound to G proteins. To test this, we solved the structure of the mutant complex using cryo-EM. For these studies we used the full-length dopamine D2 receptor (D2R) coexpressed with GαoK46E, Gβ1, and Gγ2, in the presence and absence of the Gi/o protein-stabilizing single chain antibody scFv1648. scFv16 binds to the N-terminus of the α subunit and is used in structural studies to prevent dissociation of the G protein ternary complex48. The full agonist dopamine was used to form the ligand-bound receptor-G protein complexes. We determined the structures of the dopamine-bound D2R-GαoK46E complex, with and without scFv16, with overall resolutions of 3.3 Å and 3.2 Å, respectively (Fig. 5D, E; Supplementary Figs. 4 and 5; and Supplementary Table 1).
The two structures (D2R-GoK46E +/− scFv16) were readily superimposable and with only discreet differences, having a root mean square difference (RMSD) of 0.69 Å for all aligned Cα’s. The EM density was well resolved for the Ras-like domain along with the C-terminal α helix of each G protein complex. However, the EM density for the all-helical domain (AHD) in the scFv16 structure was much weaker in intensity and less resolved when compared to the structure in the absence of scFv16, at the same EM density contour level, and was not modeled into the structure. It is important to note that the scFv16 in our complex did not appear to clash with the AHD and did not prevent complex formation (scFv16 was co-expressed with receptor and G protein subunits). In the D2R-GoK46E (- scFv16) structure, Gα was in the open conformation with the AHD interacting with the outermost β strand of the WD3 repeat of the Gβ1 subunit. The position of the AHD in our structure is most similar to that seen in the NTR1 neurotensin receptor and Gi1 protein complex (PDB ID: 7L0Q)49, in which the αA helix of the AHD is almost perpendicular with the Gβ1 subunit (~80° angle relative to the Gα αN helix). In contrast, in the β2-adrenergic receptor-Gs protein complex crystal structure (PDB ID: 3SN6)17, the AHD is positioned further from the Ras-like domain, possibly due to the presence of a stabilizing nanobody at the domain interface or due to crystal packing or both; the AHD αA helix is parallel with the Gs N-terminus (~0° angle relative to the Gα αN helix) and has no apparent interaction with the WD3 repeat of the Gβ1 subunit (Supplementary Fig. 6).
In the high-resolution crystal structures of the heterotrimeric Gi1 protein complex (PDB ID: 1GP2) bound to GDP50 and the Gαi1 subunit bound to GTP (PDB ID: 1GIA), the conserved Lys-46 is engaged in a charged interaction with the β phosphate group of the bound GDP/GTP. Additionally, the Gαi1 lysine is engaged in charged interactions with the Ser-44 hydroxyl and the backbone carbonyl of residues Gly-40 and Ala-41 of the loop region between sheet β1 and helix α1, essentially creating a positively charged pocket in which the GDP phosphate groups reside (Fig. 1). Conversely, in our D2R-Go K46E structure, there is no bound nucleotide while the Glu-46 is engaged in weak charged interactions with backbone carbonyl and backbone amide of Gly-40 through Gly-45 loop residues. Superposition with the same region in the Gαi1 structure, along with electrostatic analysis, revealed that the K46E substitution causes a slight rearrangement of the aforementioned loop (RMSD of 1.094 Å and 0.598 Å when comparing Gαi1-GTP (PDB ID: 1GIA) with D2R-GoK46E and D2R-GoK46E + scFv16, respectively) and is shifted toward the front of the pocket along with a charge reversal to a negatively charged pocket (Fig. 5D, E). Thus, the pinKE mutant occludes and electrostatically repels GDP, providing a structural explanation for the properties of the mutant with respect to nucleotide affinity and structural stability.
To date, there exist three active state structures of D2R in complex with a G protein (PDB ID: 6VMS, 7JVR, and 8IRS). However, these structures are in complex with Gi1, not Go, and bound with the agonists rotigotine or bromocriptine, not dopamine. Superposition of the receptors from our D2R-GoK46E structure (without scFv16) and from D2R-Gi1 (PDB ID: 8IRS) reveals an RMSD of 1.50 Å of all aligned α carbons (Fig. 6A). Most of the divergence between the two structures is at the extracellular ends of the transmembrane (TM) domains and in the extracellular loops. In comparison to D2R-Gi1, TM5 and TM6 in the D2R-Go K46E structure are more ordered, have more α helical content and are extended by 3 and 5 residues, respectively. In contrast, the orthosteric binding site residues superpose well in the two structures. The bound ligands occupy a similar area and engage overlapping residues, even though rotigotine is larger than dopamine and thus occupies more buried surface area (bsa) (272 Å2 versus 131 Å2, Fig. 6B and Supplementary Table 2). Furthermore, a comparison of our dopamine-bound D2R complex with the dopamine-bound D1R structure (PDB ID: 7F1O) revealed that the binding-site residues are conserved in these receptor subtypes (Supplementary Table 2).
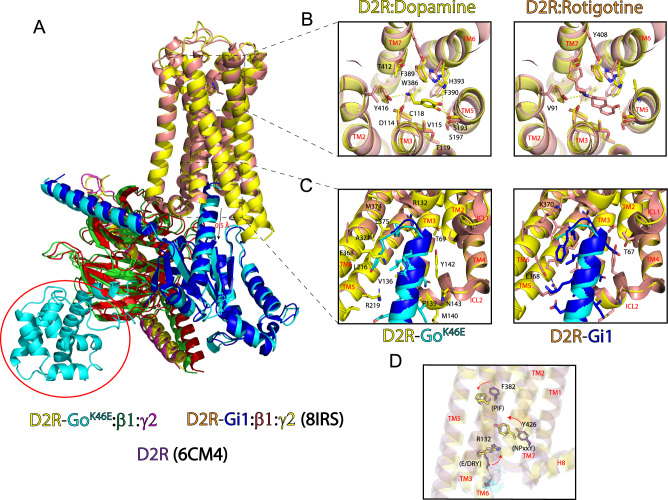
Fig. 6. Structural comparison of D2R-GoK46E with D2R-Gi1 reveals conserved features.
A Superposition of D2R-Go K46E bound with dopamine and D2R-Gi1 bound with rotigotine (PDB ID: 8IRS) reveals an RMSD of 1.50 Å between the two receptors. GαοK46E is shifted downward approximately 0.5 Å relative to Gαο. B Top-down view of the extracellular region and orthosteric binding sites of D2R-Go K46E and D2R-Gi1. Stick figures are residues that interact with either dopamine or rotigotine. Yellow distance lines are <4 Å. Stick figures of residues and bound ligand correspond to the receptor complex. Left panel, stick figures are shared residues; right panel, residues specific for rotigotine. TMs are labeled in red. C Comparison of intracellular region of D2R-GoK46E and D2R-Gi1. Stick figures are interacting residues from D2R and the respective G protein C-terminal tails. Stick figures of residues correspond to the receptor complex. Left panel, stick figures of shared residues; right panel, additional residues specific for Gi1. TMs and intracellular loops (ICLs) are labeled in red. D Structural comparison of GPCR activation motifs in the D2R inactivate state (PDB ID: 6CM4) and those of D2R-GoK46E and D2R-Gi1. Stick figures, key residues in the PIF, E/DRY, and NPxxY activation motifs. Arrows, direction of movement in going from the inactive to the active state. Coloring is consistent with the corresponding D2R.
Comparison of the intracellular interface of D2R-GoK46E and D2R-Gi1 revealed an approximate 0.5 Å off-center shift of the two G protein heterotrimer complexes, a difference that reverberates through the complex (Fig. 6A). For the D2R-GoK46E complex, Gαo contributes 12 sidechain residues and 915 Å2 of bsa to the interface; the C-terminal α-helix alone contributes 9 out of 12 residues and 773 Å2 of the total bsa (Supplementary Table 3). For the D2R-Gi1 complex, Gαi1 contributes 16 sidechain residues and 940 Å2 of bsa to the interface; the C-terminal α-helix contributes 12 out of 16 residues and 808 Å2 of the total bsa (Supplementary Table 4). Conversely, the D2R receptor contributes 16 and 19 sidechain residues to the Go and Gi1 protein interfaces, respectively, 14 of which are shared between the two structures (Fig. 6C).
Activation motif changes in GPCRs signify a transition from the inactive to active state, and vice versa. Rearrangement of the phenylalanine in the PIF connector region, rotation of the tyrosine in the tyrosine toggle switch (NPxxY), and disruption of an ionic-lock in the E/DRY motif have all been reported to be important for receptor activation, but can be receptor- and ligand-specific51. The PIF connector motif is likely to have an important role in connecting the agonist binding pocket to downstream conformational rearrangements required for receptor activation17. The NPxxY and E/DRY motifs have been proposed as stabilizing elements of an active conformation52,53. These motifs are superimposed in the activated D2R-GoK46E and D2R-Gi1 structures, and far less so when compared to the inactive D2R receptor structure (PDB ID: 6CM4)54 (Fig. 6D). Thus, structurally speaking, the single substitution in GoAK46E has no apparent negative impact on ligand binding or activation of D2R. We conclude that the pinKE mutation locks the receptor in the agonist-bound, transitory pre-activated state.
Discussion
G proteins are among the most studied of all enzymes. Detailed biochemical and structural analyses have revealed the molecular transitions leading to subunit dissociation and the conversion of GTP to GDP. In contrast, little is known about the molecular properties of the apo form of the protein, which is needed to exchange GDP for GTP. In cells, it exists only transiently, during activation by agonist-occupied receptors. In vitro, it is unstable and has proven difficult to isolate and characterize biochemically55. Here we describe a Gα variant that favors the nucleotide-free conformation of the protein yet binds with normal affinity to Gβγ and remains in a non-productive complex with receptor. Thus, pinKE sustains the first committed step of G protein activation.
Looking forward, we expect that pinKE will help to accelerate structure determination of G protein ternary complexes. Prior efforts have relied on the simultaneous assembly of agonist, receptor, and G protein heterotrimer, with removal of guanine nucleotides using the enzyme apyrase. This approach was used to solve the first crystal structure of a receptor-G protein complex, a breakthrough that provided details about conformational changes in Gα during nucleotide release17, and that provided important mechanistic details of a critical, but ephemeral, step of the G protein activation cycle. Indeed, these rearrangements are by far the most dramatic of the various conformational changes in the G protein, including those leading to subunit dissociation or engagement with effectors and RGS proteins. Subsequent studies used variants of Gα containing 4-8 mutations (none of which target the pinK or triadR)56,57, shown previously to destabilize nucleotide binding58,59. The structure of the all-helical domain was not well resolved in those efforts. In comparison, we have determined the structure of the receptor and G protein heterotrimer, including much of the all-helical domain, using a single well-characterized substitution in Gα.
The pinKE allele may also prove useful for high-throughput drug screening programs. Just as agonist binding stabilizes the apo form of the G protein, the apo form of the G protein confers increased affinity of receptors for agonists (but not for antagonists). By directly comparing binding of ligands in cells that express pinKE with those that express wild-type G protein, it should be possible to rapidly differentiate agonists from antagonists, since only agonists will bind preferentially to receptors in cells expressing the mutant allele. Any drug screening programs may be carried out experimentally as well as computationally, in conjunction with molecular docking programs and with virtual chemical libraries60.
In this work, we establish a potential mechanistic basis for human disease. The Gαo K46E and Gαo R209C mutations have been implicated in a pathogenic condition characterized by seizures, movement disorders, intellectual disability, and developmental delay37,40,61–64. One individual with the K46E mutation exhibited focal, tonic, spasm, and tonic spasm seizures beginning 6 h after birth38. That individual died in her sleep at 23 months of age. Individuals with the R209C mutation are far less likely to have seizures but do exhibit involuntary movement, difficulty speaking, as well as intellectual and developmental delay65,66. In our analysis, both mutations lead to sustained assembly of Gαo with Gβγ, and in both cases result in diminished activation of effector channels. However, our analysis reveals that these two mutants have distinct molecular properties; whereas the pinKE mutant binds poorly to nucleotide and, consequently, forms a non-productive complex with receptors and Gβγ, the triadRC mutant binds strongly to the Gβγ subunits, regardless of bound nucleotide24. Given that both residues are conserved in all G proteins, the mechanisms detailed here are likely to be universal for all subtypes and species. More broadly, our mechanistic analyses of Gαo variants illustrate how small changes at the molecular level (e.g. a single amino acid substitution) can lead to broader consequences at the cellular and organismal level. Such insights may eventually reveal potential treatments for the associated neurological disorders in humans. Just as a detailed understanding of the molecular basis for ligand binding has led to important advances in receptor pharmacology, a better understanding of nucleotide exchange mechanisms could reveal strategies to compensate for the genetic defects that cause disease. Potential treatments might include ligands that bind to and regulate GPCRs23, G proteins67–71, and G protein-regulated potassium channels72. However, and most importantly, the mechanistic differences between the pinKE and triadRC mutants indicate that different treatment strategies will likely be needed to treat what has long been considered a single disease entity.
Methods
Mutagenesis and plasmid construction
Primestar Max (Takara Bio; R045A) mutagenesis was performed using primer pairs, each containing 5 bp 5’ and 20-30 bp 3’ to the relevant codon, and are provided in Supplementary Table 5.
Complete sequences for parent plasmids generated in this study are provided in Supplementary Information: PET-SUMO-Gαo for purification, pMAX-Gαo for GIRK assays in X. oocyes, pcDNA3.1-Gαo for GIRK assays in HEK293 cells, MOR-Gαo fusion for radioligand binding, and pFastbac-Gαo for cryo-electron microscopy.
Bioluminescence resonance energy transfer
Basal association and receptor-mediated dissociation of Gα and Gβγ were determined by BRET31. HEK293T cells (ATCC; CRL-11268) were maintained at 37 oC in Dulbecco’s Modified Eagle Medium (DMEM) (Corning; 10-017-CV) supplemented with 10% FBS (GIBCO; A3382001), 100 I.U./mL penicillin, and 100 mg/mL streptomycin. Cells were then co-transfected with a 1:1:1:1 ratio of plasmids containing the human receptor, Gα (multiple isoforms, wild-type and mutant constructs) fused to Renilla luciferase 8 (Rluc8), Gγ (multiple isoforms) fused to GFP2 and Gβ (multiple isoforms) using TransIT-2020 (Mirus) transfection reagent following manufacturer guidelines. The next day, cells were plated into poly-L-lysine coated 96-well white clear bottom plates (Greiner Bio-One; 655098) at a density of ~60,000 cells/well in DMEM supplemented with 1% dialyzed FBS, 100 I.U./mL penicillin, and 100 mg/mL streptomycin. The following day, cells were washed with assay buffer (Hank’s Balanced Salt Solution containing 20 mM (4-(2-hydroxyethyl)−1-piperazineethanesulfonic acid) (HEPES), 0.1% bovine serum albumin, and 0.01% ascorbic acid, pH 7.4,), then incubated with 60 μL assay buffer containing 7.5 μM coelentrazine 400a (Nanolight; 340). After 10 min in the dark, 30 μL (3X concentration) of agonist in drug buffer was added to each well. After another 10 min in the dark the plates were serially read five times using a PheraSTAR FSX plate reader with 395 nm (Rluc8-coelenterazine 400a) and 510 nm (GFP2) filters for 1 sec per well. Measurements from the final read were used for all analyses. The BRET ratio was calculated as the ratio between acceptor (GFP) and donor (Rluc8) emission values. ΔBRET values were determined by subtracting the BRET ratio from cells treated with vehicle from the entire dataset. ΔBRET data for each dose-response curve were fit using the log(inhibitor) vs. response (three parameter) function in GraphPad Prism (GraphPad Software Inc., San Diego, CA) with the top fit parameter constrained to 0. Expression of Gα was verified by western blotting.
Basal and receptor-mediated association of Gβ1γ2 and masGRK3ct was determined by BRET36. HEK293FT cells (Thermo Fisher Scientific; R70007) were grown at 37 °C in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific;11965-092) + 10% fetal bovine serum (Millipore Sigma; 12303 C), minimum Eagle’s medium non-essential amino acids (Thermo Fisher Scientific;11140-050), 1 mM sodium pyruvate34,36. Approximately 50,000 cells per well were distributed in 96-well flat-bottom white microplates (Greiner Bio-One; 655073) and transfected with plasmids encoding the human Flag-tagged dopamine D2 receptor (DRD2)73 or human μ-opioid receptor (OPRM1)74, as well as masGRK3ct-Nluc (comprised of the C-terminal 141 amino acids of GRK3, an N-terminal peptide directing myristoylation, the HA epitope tag and Nano luciferase)36, human Gγ2 fused to residues 1-155 Venus (Venus 1-155-Gγ2)34, human Gβ1 fused to Venus 156-239 (Venus 156-239-Gβ1)34, and human Gαο or GαοK46E (constructs described above) in a 1:1:1:1:2 ratio, carried out using Lipofectamine LTX Reagent with PLUS (Thermo Fisher Scientific; 15338100). After 16-24 hr the cells were washed once with BRET buffer (Dulbecco’s Phosphate-Buffered Saline (PBS) + 0.5 mM MgCl2 and 0.1% glucose) and resuspended in 50 mL BRET buffer. Fifty μL of the Nluc substrate furimazine (Promega; N1120) in BRET buffer was added per well, as directed by the manufacturer, followed by 50 μL (3X concentration) of 100 μM dopamine (Millipore Sigma; H8502) or 10 μM DAMGO (Adipogen; AG-CP3-0005V-M001). The plates were read using a PHERAstar FSX microplate reader (BMG Labtech) equipped with two emission photomultiplier tubes, allowing detection of two emissions simultaneously with a highest possible resolution of 20 ms per data point. All measurements were performed at 37oC. The BRET signal was determined by calculating the ratio of the light emitted by the Venus-Gβ1γ2 (535 nm ± 30 nm) over the light emitted by the masGRK3ct-Nluc-HA (475 nm ± 30 nm). The average baseline value (basal BRET ratio, with no Gα transfected) recorded prior to agonist stimulation was subtracted from the experimental BRET signal values.
Basal association and receptor-mediated dissociation of receptors and Gβ1γ2 was determined using the NanoBiT assay37. Briefly, 4,000,000 HEK293FT cells in 60 mm dishes transfected with plasmids encoding either the human dopamine D2 receptor (D2R)37 or human μ-opioid receptor (MOR)74, each fused to the myc epitope tag and SmBiT, as well as human Gβ1 fused to LgBiT37 and human Gγ2 (cDNA Resource Center; GNG0200000). Transfected cells were harvested, washed in PBS, 5 mM EDTA, collected by centrifugation at 500 xg for 5 min resuspended in BRET buffer and distributed in 96-well plates as explained above. Furimazine (Promega; N1120, 750x diluted Nano-Glo substrate) in BRET buffer was added to transfected cells in 96-well plates and after 1 min to stabilize the brightness of luminescence before addition of dopamine or DAMGO. Luminescence was measured using a PHERAstar FSX plate reader at 37 oC.
No commonly misidentified cell lines were used in the study. HEK293 cell lines were from commercial sources and not independently authenticated. All cell lines tested negative for mycoplasma contamination.
GIRK channel activity measurements
Two-electrode voltage clamp (TEVC) methods. Complementary DNA constructs used for electrophysiology experiments were human GIRK1 (pGEMSH vector), mouse GIRK2 (pXOOM vector), mouse dopamine D2 receptor (pXOOM vector), bovine Gβ1 (pGEMSH vector), bovine Gγ2 (pGEMSH vector)75, and human Gαo, GαoK46E or GαoR209C (pMAX vector, described above). Xenopus laevis oocytes were isolated and microinjected with in vitro synthesized circular RNAs (0.125 to 2.5 ng per oocyte)76. To allow for sufficient protein expression, injected oocytes were incubated at 18 °C for 2 days. The following day, a GeneClamp 500 amplifier (Axon Instruments) was used to measure whole-cell oocyte currents. The microelectrodes had resistances of 0.5 to 1 MΩ using a 3 M KCl solution in 1.4% agarose. For TEVC measurements, oocytes were held at 0 mV (EK), and a ramp protocol with a command potential from −80 to +80 mV was used to monitor the currents. The current amplitude was measured at the end of a sweep of 0.4 s. All currents were analyzed when steady state was reached. To establish a baseline for TEVC recordings, oocytes were perfused with a low K+ (LK) solution ND96 (2 mM KCl, 96 mM NaCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES–Na pH 7.4). A high potassium (High K + ) solution ND96K (96 mM KCl, 10 mM HEPES–K pH 7.4, 1 mM MgCl2, and 1.8 mM CaCl2) was used to measure basal current. The oocyte chamber was perfused with 3 mM BaCl2 (Ba2+) in ND96K to block the current. The barium-sensitive current was used exclusively for statistical analysis. Ten μM dopamine in ND96K was used to determine the effect of Gαo wild type and mutants on agonist-induced activity of GIRK channels.
Basal and receptor-mediated activation of GIRK1/GIRK2 channels was determined by whole-cell patch-clamp recordings42,77, HEK293T (ATCC; CRL-1573) cells were maintained in Dulbecco’s Modified Eagle Medium (Life Technologies; 11965-118) supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% (v/v) heat-inactivated fetal bovine serum (HI-FBS; Thermo Fisher Scientific, 10438026). Lipofectamine 3000 (Thermo Fisher Scientific; L3000015) was used in accordance with the manufacturer’s protocol to transiently transfect cDNAs for rodent GIRK1-GIRK2 tandem dimer (200 ng)77, eYFP (50 ng)77, human MOR (400 ng)78, and one of the following Gαo subunits, human Gαo, GαoK64E or GαoR209C (400 ng) (constructs described above). All genes were cloned into pcDNA3 vector. 48-72 h post-transfection, cells were plated onto Matrigel-coated (0.4% v/v) 12 mm glass coverslips (Electron Microscopy Sciences; 72290-03) in 24 well plate (VWR; 10062-896) for whole-cell patch-clamp recordings. Whole-cell recordings were made with the investigator blinded to the Gα type, and then unblinded after completing the recordings.
Whole-cell patch-clamp recordings were acquired using an Axopatch 200B amplifier and pClamp software (Molecular Devices)42,77. Whole-cell series resistance compensation was applied and used to measure the cell size (pF). Patch-clamp experiments were conducted in 20 K external solution, containing 10 mM HEPES pH 7.4, 140 mM NaCl, 20 mM KCl, 2 mM MgCl2, 0.5 mM CaCl2 (~318 mOsm). Borosilicate glass electrodes (Warner Instruments) had resistances of 3-4 MΩ and were filled with internal solution containing 10 mM HEPES pH 7.4, 20 mM NaCl, 130 mM KCl, 5.46 mM MgCl2, 5 mM EGTA, 2.56 mM K2ATP, and 0.3 mM LiGTP ( ~ 300 mOsm). 10 μM DAMGO, 100 mM PrOH, and 1 mM BaCl2 were prepared in the external solution on the day of the experiment.
Currents were measured (10 kHz sampling and 1 kHz filter) from eYFP-positive cells using a voltage protocol of a holding potential of −40 mV, followed by a 60 ms step to −100 mV and a voltage ramp to 50 mV over 200 ms. Solutions were applied locally for 30 s with a rapid perfusion pipet in the following order: 20 K, 20 K + PrOH, 20 K, 20 K + DAMGO, 20 K, 20 K + BaCl2, and 20 K. The maximal inward current was measured at −100 mV. Except for DAMGO, the average current was calculated during the 30 s drug application. For DAMGO, the maximal inward current was calculated before the onset of MOR desensitization. To adjust for differences in GIRK channel expression, MOR activation was compared to G protein-independent alcohol activation41, and calculated as a ratio of DAMGO-induced to PrOH-induced currents (IDAMGO – IBasal)/(IPrOH – IBasal). Barium-sensitive inhibition of GIRK current density was calculated as (IBasal – IBa2+)/pF. Cells not expressing a propanol-activated current were excluded from the analysis. Statistical tests performed using one-way ANOVA followed by Dunnett’s post hoc test (GraphPad Prism 9.0).
Expression and purification of Gαo
Bacterial expression was used for the production of recombinant full-length human Gαo79,23. Rosetta (Novagen; 70954) or RIPL (Agilent; 230280) cells were transformed with PET-SUMO-Gαo (human, wild type and mutant) plasmid (derived from PET-SUMO-Gαi179) and grown at 37 °C in 2X Luria Broth to saturation. Once at OD600 = 0.6–0.8, protein expression was induced by adding 600 μM isopropyl-β-d-1-thiogalactoside. The culture temperature was lowered to 16 °C and shaken overnight. The subsequent purification was carried out at 4 °C for all steps. The saturated cell culture was collected by centrifugation at 5400×g for 30 min. Cells were lysed by sonication and clarified by centrifugation at 26,900×g for 45 min. Lysates (30 mL each) were incubated with 5 mL (10 mL 50/50 slurry) of HisPur™ Ni-NTA Resin (Thermo Fisher Scientific; 88221) or ProBond Ni-chelating resin (Invitrogen; R80101) and 10 mM imidazole for 1 h. The resin was washed twice with phosphate buffer pH 7.0 (25 mM H2KO4P, 25 mM HK2O4P, 500 mM (tris(2-carboxyethyl)phosphine) (TCEP), 50 μM GDP, 50 μM MgCl2) then twice with phosphate buffer + 10 mM imidazole. Protein was eluted with phosphate buffer + 250 mM imidazole. The SUMO tag was cleaved by addition of 0.5 mg ULP1 protease and the eluate was dialyzed overnight in 4 L of phosphate buffer. The next day, cleaved protein was collected by reverse-nickel chromatography.
Expression and purification of Gβγ
Recombinant baculovirus was used for the production of recombinant rat biotinylated Gβ180 and bovine Gγ281, for BLI analysis81. Briefly, High 5 cells (Invitrogen; B85502) were grown in SF900 II serum-free medium (Thermo-Fisher; 10902104) in suspension culture at 27 oC with continuous shaking. 2 × 106 cells/mL were infected with high titer Gβ1 and Gγ2 baculoviruses. After 48 hr the cells were collected by centrifugation at 2600×g, resuspended in 50 mL of lysis buffer (50 mM HEPES pH 8.0, 3 mM MgCl2, 50 mM NaCl, 10 mM 2-mercaptoethanol (2-ME), 1 mM EDTA, 50 μM 1 mL/L of protease inhibitor cocktail (Sigma-Aldrich; P-2714), and freeze-thawed using liquid nitrogen four times and maintained after at 4 °C. Samples were then diluted to 100 mL and centrifuged at 50,000 xg for 30 min. After discarding the supernatant, the pellet was resuspended in 5 mL membrane wash buffer (50 mM HEPES pH 8.0, 3 mM MgCl2, 50 mM NaCl, 50 μM 1 mL/L of protease inhibitor cocktail, 10 mM 2-ME, 50 μM GDP) and transferred to a dounce homogenizer. Upon resuspension, the volume was brought to 60 mL with additional membrane wash buffer. Using a cholate solution (Sigma-Aldrich; 229101)82, cholate was added to a final concentration of 1% and allowed to extract for 1 h with stirring. Following cholate extraction, the membranes were centrifuged at 125,000 xg for 45 min. The resulting supernatant was diluted to 5 times the initial volume by slowly adding Buffer A (20 mM HEPES pH 8.0, 1 mM MgCl2, 100 mM NaCl, 10 mM 2-ME, 50 μM 1 mL/L of protease inhibitor cocktail, 50 μM GDP, 0.5% Lubrol (C12E10) (Sigma-Aldrich; P9769) to the stirring solution.
The diluted solution was loaded onto a Ni-NTA agarose column (Qiagen; 1018244) equilibrated with Buffer A. The column was then washed with 100 mL Buffer B (20 mM HEPES pH 8.0, 1 mM MgCl2, 300 mM NaCl, 10 mM 2-ME, 50 μM 1 mL/L of protease inhibitor cocktail, 10 mM imidazole, 10 μM GDP, 0.5% lubrol). Following this, the column was warmed tmo room temperature (RT) for 15 min and washed with an additional 8 mL of RT Buffer B. The column was then washed with 32 mL of Buffer C (20 mM HEPES pH 8.0, 100 mM NaCl, 50 mM MgCl2, 10 mM NaF, 30 μM AlCl3, 50 μM 1 mL/L of protease inhibitor cocktail, 20 mM imidazole, 10 μM GDP, 0.5% Lubrol) at room temperature. The column was returned to 4 °C, where it was then washed with 10 mL Buffer D (20 mM HEPES pH 8.0, 50 mM NaCl, 0.5% Lubrol, 50 μM 1 mL/L of protease inhibitor cocktail, 20 mM imidazole). Following this, the column was washed with three column volumes of Buffer D (50 mM imidazole). Finally, 2 mL fractions were eluted from the Ni-NTA column with Buffer D (250 mM imidazole). The purified yield was typically 1 mg of Gβ1γ2/L of cell culture83.
Thermostability measurements
The fast determination of Quantitative Cysteine Reactivity (fQCR) assay was used to determine Tm values for purified Gαo proteins43. Briefly, 10 μL of 30 μM protein, 10 μL of 1 mM GDP or GTPγS, 10 μL of 500 mM 4-fluoro-7-sulfamoylbenzofurazan (ABD-F) cysteine-reactive dye (Abcam; 91366-65-3), and 170 μL phosphate buffer pH 7.0 was added to 12-strip PCR tubes. Samples were subjected to a 40-degree temperature gradient in a Biometra TProfessional Thermocycler for 3 min and fluorescence values were collected on a PHERAstar plate reader (BMG Labtech) at 400 nm excitation, 500 nm emission.
Nucleotide loading and hydrolysis
To measure GTP binding and hydrolysis45,84, BODIPYFL-GTP (ThermoFisher Scientific, Invitrogen; G12411) (50 nM) equilibrated in hydrolysis buffer (50 mM HEPES pH 7, 10 mM MgCl2, 25 mM NaCl) was combined with purified Gαo protein (3 μM) in a 1 mL cuvette for 60 sec. Fluorescence was measured using a Perkin-Elmer Luminescence Spectrometer (502 nm excitation, 511 nm emission) and analyzed using the FLWinLab software package (Perkin-Elmer).
To measure GTPγ35S binding39, Gαo was diluted to 1 μM in a 50 mM HEPES pH 8.0, 1 mM EDTA, 0.1% C12 E10 (lubrol), and 1 mM dithiothreitol (DTT) in polypropylene reaction tubes on ice. A blank sample was prepared similarly without Gαo. An equal volume of binding mix (50 mM HEPES pH 8.0, 1 mM EDTA, 1 mM DTT, 40 mM MgCl2, 200 mM NaCl, 2 μM GTPγ35S, and 300,000 – 800,000 cpm GTPγ35S) was added to each sample, and placed in a 30 °C circulating water bath for 60 min. After incubation, samples were applied to 0.45 µm nitrocellulose filters under vacuum (Cytiva Protran BA85 Nitrocellulose Blotting Membrane; 10402578). Each sample tube was washed three times with 2 mL wash buffer (20 mM Tris-Cl pH 8.0, 100 mM NaCl, 25 mM MgCl2), applying each wash to the filter. The filter was washed three additional times with 2 mL fresh wash buffer. Filter membranes were air dried and samples measured for 35S activity on a scintillation counter.
Biolayer interferometry
Binding of Gα to Gβγ was determined by Biolayer Interferometry (BLI) using Octet Red96 (Fortebio; 18-5019)85. Briefly, purified biotinylated Gβγ (3 mg/mL) was incubated for 15 min with streptavidin biosensors in PBST-NGM (25 mM KPO4 pH 7.0, 50 mM NaCl, 0.1% Tween, 50 μM GDP + 5 mM MgCl2). Purified Gαο (untagged) was diluted into PBST-NGM and then mixed with Gβγ-loaded sensors for 10 min (association) and then protein-free buffer (dissociation) at 25 oC. Nonspecific binding was measured using biosensors that were exposed to buffer alone. Baseline subtraction and Gβγ loading normalization were done in Excel. Kinetic Analysis was done using GraphPad Prism.
Computational methods
Starting structural models and molecular dynamics simulations
The structural model of monomeric GTP-bound Gαo protein with Mg2+ ion was built using the monomeric GTP bound mouse Gαo crystal structure (PDB ID: 3C7K) as template and using the homology modeling method in the Prime module of Maestro software from Schrodinger [https://www.schrodinger.com/products/maestro]. The ligand-free apo-state of the monomeric Gαo protein was obtained by removing GTP from the modeled structure; the Mg2+ ion is retained in all the simulations since it plays an important role in G protein activity. Point mutations to generate the structures of GαoK46E and GαoR209C were performed using Maestro Biologics suite. The side chain packing was done for all the residues with 5 Å of the mutated residue position including the mutated residues using Maestro Prime suite. All structures were subjected to energy minimization using conjugate gradient method with a convergence cutoff of 0.1 kcal/mol/Å. Input files for molecular dynamics (MD) simulations were generated using CHARMM-GUI86. Each monomeric Gαo protein was solvated in explicit TIP3P water molecules in a cubic box (9.5 nm × 9.5 nm × 9.5 nm) with 0.15 M of KCl for maintaining the physiological condition. We used software GROMACS87 (Version 2021.3) with all-atom CHARMM36 force field88 to perform MD simulations. MD simulations were performed at 310 K coupled to a temperature bath with a relaxation time of 0.1 ps89. Pressure of the systems was calculated with molecular virial and was held constant by a weak coupling to a pressure bath with a relaxation time of 0.5 ps. Equilibrium bond length and geometry of water molecules were constrained using the SHAKE algorithm90. The short-range electrostatic and van der Waals interactions were estimated every 2 fs using a charged group pair list with cutoff of 8 Å between centers of geometry of charged groups. Long-range van der Waals interactions were calculated using a cutoff of 14 Å and long-range electrostatic interactions were treated with the particle mesh Ewald method91. Temperature was kept constant at 310 K by applying the Nose-Hoover thermostat92. Desired pressure for all systems were achieved by using Parrinello-Rahman barostat with a pressure relaxation time of 2 ps93. Before production runs, all systems were subjected to a 5000-step steepest descent energy minimization to remove bad contacts94. After minimization, the systems were heated up to 310 K under constant temperature-volume ensemble (NVT). The simulations were saved every 200 ps for analysis. The protein, Mg2+ ion, and nucleotide were subjected to positional constraints under a harmonic force constant of 1000 kJ/(mol*nm2) during the NVT step while solvent molecules were free to move. The systems then were further equilibrated using constant pressure ensemble (NPT), in which the force constant applied to the protein, Mg2+ ion, and nucleotide were gradually reduced from 5 kJ/(mol*nm2) to zero in six steps of 5 ns each. An additional 50 ns of unconstrained simulations were performed, making it a total of 80 ns NPT equilibration prior to production runs. We performed five production runs of 1000 ns each using five different initial velocities for every system. Therefore, we had 5 μs long MD trajectory for wild type and K46E monomeric Gαo protein of GTP-bound state or apo state, and 5 μs long MD trajectory for R209C monomeric Gαo protein of GTP-bound state only. The convergence of the MD simulations was ascertained by plotting the RMSD in the coordinates of the Cα atoms as a function of time showing lowering of fluctuations (Supplementary Fig. 2C).
Calculation of residue contact map of protein with nucleotide
For GαoWT and GαoK46E systems that contain nucleotide, the last 200 ns from five independent molecular dynamics simulation runs were merged into one concatenated trajectory. The concatenated trajectory was sampled every 2 ns. The sampled trajectory was fed to GetContacts script available on GitHub [https://github.com/getcontacts/getcontacts]. For specific parameters, the interaction type flag “itype” was set to “all”, and the two atom group flags for contacts were “protein” and “resname GTP”. Contact results were further processed in Python environment: briefly, contacts with van der Waals force were excluded, and contact frequency of every contact was calculated. Contact frequencies between GαoWT system and GαoK46E system were compared, and contacts with more than 30% frequency decreases from the GαoWT system to GαoK46E system were selected and depicted.
Calculation of interaction energy of nucleotide with the protein
For each system (GαoWT, GαoK46E, and GαoR209C), we aggregated the last 200 ns of each simulation run to calculate the interaction energy of GTP with the Gαo protein. Total non-bond interaction energy (van der Waals + Columbic) between Gαo protein and GTP (shown in Fig. 4G) was calculated using gmx energy module of GROMACS. The interaction energy was calculated for every snapshot and averaged for the last 200 ns in all five simulation runs for every system.
Principal Component Analysis, free energy surface generation, and representative structures
For apo systems of WT and K46E, five full-length independent molecular dynamics simulation runs were merged into one concatenated trajectory. Principal component analysis was performed on each concatenated trajectory using gmx covar module of GROMACS with covariance matrix of Cα atoms of all residues. The first two principal components (PC1 and PC2) of every system were extracted using gmx anaeig module of GROMACS and imported into Python as a data-frame using Python Pandas package. The kernel density value of the data-frame was calculated using Python scikit-learn package. The multiplicative inverse of the kernel density values was used as the Z-axis values to plot the free energy surface. Both the 3-D and 2-D density maps were plotted using Python Matplotlib package. Representative structures of local and global minima were extracted by first clustering the points using K-mean clustering method and extracting cluster centers using K-medoids method from Python scikit-learn package.
Radioligand binding assays
Saturation binding assays were performed using membranes from Expi293™cells (ThermoFisher; A14527) transiently expressing wild-type and mutant MOR-Gαo fusion. Binding assays were set up in 96-well plates in standard binding buffer (50 mM HEPES, pH 7.4, 50 mM NaCl, 20 mM MgCl2, 0.5 mM EDTA, ±100 μM GppCp). Saturation binding assays with 0.5–10 nM [3H]-DAMGO (Perkin-Elmer, specific activity 51.7 Ci/mmol; NET902250UC) in standard binding buffer were performed to determine equilibrium dissociation constant (Kd) and Bmax, whereas 10 μM final concentration of morphine was used to define nonspecific binding. All reactions were incubated for 1.5 h at room temperature in the dark and terminated by rapid vacuum filtration onto chilled 0.3% PEI-soaked GF/C UniFilter plates (Perkin-Elmer; 6055690) followed by three quick washes with cold washing buffer (50 mM Tris HCl, pH 7.4). The UniFilter plates were dried in an oven before adding 20 µl/well Microscint™-O cocktail (Perkin-Elmer; 6013611). Radioactivity counts were determined using a Wallac Trilux MicroBeta counter in the form of corrected count per min (ccpm) with counting efficiency of 40% (Perkin-Elmer). Results were analyzed using GraphPad Prism 9.0 using “One site -- Total and nonspecific binding”. Statistical differences (p < 0.05) of pKd values derived from concentration-response assays were determined by using the Extra sum-of-squares F test (p < 0.05) function in GraphPad Prism 9.0 comparing control versus 100 μM GppCp.
Construction and expression of the receptor-G protein complex
The Bac-to-Bac baculovirus expression system (Invitrogen; 10359016) was used to generate the recombinant baculovirus for protein expression. Baculoviruses corresponding to N-terminal Maltose Binding Protein (MBP) - human D2 receptor (MBP-D2R), human GoAK46E, Gβ1γ2 were co-expressed by infecting Sf9 cells at a density of 2×106 cells per mL at MOI ratio of 3:1.5:1.5 ±scFv16 at an MOI of 1, respectively. Cells were harvested by centrifugation at 3500×g for 15 min 48 h post-infection and stored at −80 °C for future use.
Receptor-G protein complex purification
The cell pellet of the D2R-GoAK46E complex was thawed on ice and incubated with 20 mM HEPES pH 7.5, 100 mM NaCl, 20 mM MgCl2, 20 mM KCl, 5 mM CaCl2, proteinase inhibitor, 40 units apyrase, 50 μM dopamine, and 100 μM TCEP at room temperature. After 1.5 h, the cell suspension was homogenized, membrane was collected by centrifugation at 100,000×g for 35 min and solubilized using 20 mM HEPES pH 7.5, 100 mM NaCl, 10% (w/v) glycerol, 0.5% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace; NG310), 0.05% (w/v) cholesteryl hemisuccinate (CHS), 20 mM MgCl2, 20 mM KCl, 5 mM CaCl2, proteinase inhibitor, 40 units apyrase, 50 μM dopamine, and 100 μM TCEP for 4 h at 4 °C. The solubilized proteins were isolated by ultra-centrifugation at 100,000×g for 45 min and then incubated overnight at 4 °C with TALON IMAC resin (Takara; 635653) and 20 mM imidazole. The resin was collected the next day and washed with 25 column volumes 20 mM HEPES pH 7.5, 100 mM NaCl, 20 mM imidazole, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, 50 μM dopamine, and 100 μM TCEP. The protein was then eluted using the same buffer supplemented with 250 mM imidazole. Eluted protein was subjected to TEV protease (in-house) overnight at 4 °C and a ratio of 1:10 (1 mg receptor: 0.1 mg TEV). The protein was then concentrated and subjected to size-exclusion chromatography on a Superdex 200 Increase 10/300 column (GE Healthcare; 289909944) that was pre-equilibrated with 20 mM HEPES pH 7.5, 100 mM NaCl, 20 mM MgCl2, 50 μM dopamine, 0.00075% (w/v) LMNG, 0.00025 (w/v) glyco-diosgenin (GDN, Anatrace; GDN101), 0.00075% (w/v) CHS, and 100 μM TCEP. Peak fractions were pooled and concentrated to 5 mg/mL for making of cryoEM grids. For complex formation in the presence of ATP/GTP the above protocol was followed without apyrase. Complexes were also subjected to purification after the TALON column using an Anti-flag column (Genscript; L00432) and eluted with 500 μg/mL Flag peptide (Genscript; RP10586) prior to TEV cleavage and S200 column.
CryoEM data collection, 3D reconstitution, model building and refinement
For cryo-EM data collection and processing of the D2R-GoAK46E complex95, samples (3.2 µl) were applied to glow-discharged Quantifoil R1.2/1.3 Au300 holey carbon grids (Ted Pella; 658-300-AU) individually and were flash-frozen in a liquid ethane/propane (40/60) mixture using a Vitrobot Mark IV (FEI) set at 4 °C and 100% humidity with a blot time range from 3.0 to 4.5 sec. Images were collected using a 200 keV Talos Artica with a Gatan K3 direct electron detector at a physical pixel size of 0.88 Å. Micrograph recorded movies were automatically collected using SerialEM using a multishot array96. Data were collected at an exposure dose rate of ~15 electrons/pixel/sec as recorded from counting mode. Images were recorded for ~1.7–2.7 s in 60 subframes to give a total exposure dose of ~45 electrons per Å2. Following manual inspection and curation of the micrographs, particles were selected using the Blob particle picker and initial 2D classification yielded templates for subsequent template picking. After one round of two-dimensional classification and selection in cryoSPARC, a subset of the selected particles was used as a training set for Topaz and the particles were re-picked from the micrographs using Topaz97 and subjected to two-dimensional classification and three-dimensional classification. The select classified picked particle coordinates were next merged yielding a subset of unique particles that survived 2D classification (i.e. duplicates were removed with a radius of 25 pixels). All subsequent three-dimensional classification and refinement steps were performed within cryoSPARC98. Multiple rounds of multi-terence refinement resolved the final stack of particles that produced a map with a resolution reported in Supplementary Table 1 (by FSC using the 0.143 Å cut-off criterion)99 after Global CTF refinement and post-processing including soft masking, B-factor sharpening in cryoSPARC to generate the post-processed sharpened map. Alternative post-sharpening was performed on the two half-maps using deepEMhancer100. For more details see Supplementary Table 1.
Maps from deepEMhancer were used for map building, refinement, and subsequent structural interpretation. The D2R cryo-EM structure (PDB ID: 8IRS) was used as the initial model and docked into the cryo-EM map using Chimera101 followed by initial rigid body and simulated annealing then iterative manual adjustment in COOT102 and Phenix.real_space_refine in Phenix103. A composite map was generated by merging two local refines maps, one for the AHD domain and one for the entire complex, via the Chimera. This composite map displayed main and side chain densities for most part, which enabled placing of the all helical domain. The final refined model for the full D2R-GαoK46E complex also matches the consensus map. Model statistics were validated using Molprobity. Structural figures were prepared using Chimera or Pymol [https://pymol.org/2/]. Coordinates of D2R complexes and EM maps have been deposited in the Protein Data Bank and the Electron Microscopy Data Bank under the accession codes 8U02 (EMD-41776 and 8TZQEMD-41766) for the DR2-GαoK46E and DR2-GαοAK46E-scFv16, respectively. Interface analysis was accomplished using Contact and Areaimol of the CCP4 Suite104 along with the PISA Server105 [http://www.ebi.ac.uk/pdbe/prot_int/pistart.html].
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Funded by NIH grants F32NS124758 (W.G.L.), F31AA027949 (I.P.), R01GM117923 and R01LM013876 (N.V.), R35GM127303 (A.V.S.), R01AA018734 and U10AA008401 (P.A.S.), R01HL059949 (D.E.L.), RO1DA036596 (K.A.M.), R01MH112205 (B.L.R.), and R35GM118105 (H.G.D). The authors wish to thank Emily Brinson and Harris Teeter for motivating the GIRK channel work.
Author contributions
K.M.K., B.E.K., N.J.K., W.G.L., M.C., S.M., I.P., E.G.O., T.J.L., W.W., N.M., X-P.H., J.F.F., N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: conceptualization. K.M.K., B.E.K., N.J.K., W.G.L., M.C., S.M., I.P., E.G.O., T.J.L., W.W., N.M., X-P.H., J.F.F., N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: formal analysis. W.G.L., I.P., N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: funding acquisition. K.M.K., B.E.K., N.J.K., W.G.L., M.C., S.M., I.P., E.G.O., T.J.L., W.W., N.M., X-P.H., and J.F.F.: investigation. N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: project administration. N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: Supervision. K.M.K., B.E.K., N.J.K., W.G.L., M.C., S.M., I.P., E.G.O., T.J.L., W.W., N.M., X-P.H., J.F.F., N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: validation. K.M.K., B.E.K., N.J.K., W.G.L., M.C., S.M., I.P., E.G.O., T.J.L., W.W., N.M., X-P.H., J.F.F., N.V., A.V.S., P.A.S., D.E.L., K.A.M., B.L.R., and H.G.D.: writing – original draft. K.M.K. and H.G.D.: writing – review & editing
Peer review
Peer review information
Nature Communications thanks Moritz Bunemann, Qiuyan Chen, Francesco Raimondi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Complete sequences for plasmids generated in this study are provided in Supplementary Information: PET-SUMO-Gαo for purification, pMAX-Gαo for GIRK assays in X. oocyes, pcDNA3.1-Gαo for GIRK assays in HEK293 cells, MOR-Gαo fusion for radioligand binding, and pFastbac-Gαo for cryo-electron microscopy. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes 8U02 and EMD-41776 for the DR2-GαoK46E, and 8TZQ and EMD-41766 for DR2-GαοAK46E-scFv16. Source data supporting Fig. 2 (BRET), Fig. 3 (potassium current), Fig. 4A and B (thermostability), Fig. 4C (BODIPY fluorescence), Fig. 4D (radiolabeled GTP binding), Fig. 4E (BLI), Fig. 4F (Principal Component analysis), and Fig. 5A–C (radioligand binding) are provided with this paper. Source data are provided with this paper.
Competing interests
B.L.R. is an SAB member of Escient, Septerna and Lassogen and UNC has licensed technology from his lab to many biotechnology and pharmaceutical companies. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kevin M. Knight, Brian E. Krumm.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50964-z.
References
- 1.Flock, T. et al. Universal allosteric mechanism for Galpha activation by GPCRs. Nature524, 173–179 (2015). 10.1038/nature14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flock, T. et al. Selectivity determinants of GPCR-G-protein binding. Nature545, 317–322 (2017). 10.1038/nature22070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprang, S. R. Invited review: activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers105, 449–462 (2016). 10.1002/bip.22836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilger, D., Masureel, M. & Kobilka, B. K. Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol.25, 4–12 (2018). 10.1038/s41594-017-0011-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muralidharan, K., Van Camp, M. M. & Lyon, A. M. Structure and regulation of phospholipase Cβ and ε at the membrane. Chem. Phys. Lipids235, 105050 (2021). 10.1016/j.chemphyslip.2021.105050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halls, M. L. & Cooper, D. M. F. Adenylyl cyclase signalling complexes - pharmacological challenges and opportunities. Pharm. Ther.172, 171–180 (2017). 10.1016/j.pharmthera.2017.01.001 [DOI] [PubMed] [Google Scholar]
- 7.Smrcka, A. V. & Fisher, I. G-protein βγ subunits as multi-functional scaffolds and transducers in G-protein-coupled receptor signaling. Cell Mol. Life Sci.76, 4447–4459 (2019). 10.1007/s00018-019-03275-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire, M. E., Van Arsdale, P. M. & Gilman, A. G. An agonist-specific effect of guanine nucleotides on binding to the beta adrenergic receptor. Mol. Pharmacol.12, 335–339 (1976). [PubMed] [Google Scholar]
- 9.De Lean, A., Stadel, J. M. & Lefkowitz, R. J. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J. Biol. Chem.255, 7108–7117 (1980). 10.1016/S0021-9258(20)79672-9 [DOI] [PubMed] [Google Scholar]
- 10.Kwok-Keung Fung, B. & Stryer, L. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. Proc. Natl Acad. Sci. USA77, 2500–2504 (1980). 10.1073/pnas.77.5.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oldham, W. M., Van Eps, N., Preininger, A. M., Hubbell, W. L. & Hamm, H. E. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat. Struct. Mol. Biol.13, 772–777 (2006). 10.1038/nsmb1129 [DOI] [PubMed] [Google Scholar]
- 12.Liu, X. et al. Structural insights into the process of GPCR-G protein complex formation. Cell177, 1243–1251.e1212 (2019). 10.1016/j.cell.2019.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Eps, N. et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc. Natl Acad. Sci. USA108, 9420–9424 (2011). 10.1073/pnas.1105810108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dror, R. O. et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science348, 1361–1365 (2015). 10.1126/science.aaa5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, K. Y. et al. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature477, 611–615 (2011). 10.1038/nature10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westfield, G. H. et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc. Natl Acad. Sci. USA108, 16086–16091 (2011). 10.1073/pnas.1113645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature477, 549–555 (2011). 10.1038/nature10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaya, A. I. et al. A conserved hydrophobic core in Gαi1 regulates g protein activation and release from activated receptor. J. Biol. Chem.291, 19674–19686 (2016). 10.1074/jbc.M116.745513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nehmé, R. et al. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS ONE12, e0175642 (2017). 10.1371/journal.pone.0175642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang, W., Lu, S., Xu, X., Wu, G. & Lambert, N. A. The role of G protein conformation in receptor-G protein selectivity. Nat. Chem. Biol.19, 687–694 (2023). 10.1038/s41589-022-01231-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landis, C. A. et al. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature340, 692–696 (1989). 10.1038/340692a0 [DOI] [PubMed] [Google Scholar]
- 22.Graziano, M. P. & Gilman, A. G. Synthesis in Escherichia coli of GTPase-deficient mutants of Gs alpha. J. Biol. Chem.264, 15475–15482 (1989). 10.1016/S0021-9258(19)84854-8 [DOI] [PubMed] [Google Scholar]
- 23.Hewitt, N. et al. Catalytic site mutations confer multiple states of G protein activation. Sci. Signal16, eabq7842 (2023). 10.1126/scisignal.abq7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight, K. M. et al. A universal allosteric mechanism for G protein activation. Mol. Cell81, 1384–1396.e1386 (2021). 10.1016/j.molcel.2021.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight, K. M. et al. Molecular annotation of G protein variants in a neurological disorder. Cell Rep.42, 113462 (2023). 10.1016/j.celrep.2023.113462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slepak, V. Z. et al. Random mutagenesis of G protein alpha subunit G(o)alpha. Mutations altering nucleotide binding. J. Biol. Chem.268, 21889–21894 (1993). 10.1016/S0021-9258(20)80624-3 [DOI] [PubMed] [Google Scholar]
- 27.Apanovitch, D. M., Iiri, T., Karasawa, T., Bourne, H. R. & Dohlman, H. G. Second site suppressor mutations of a GTPase-deficient G-protein a- subunit. Selective inhibition of bg-mediated signaling. J. Biol. Chem.273, 28597–28602 (1998). 10.1074/jbc.273.44.28597 [DOI] [PubMed] [Google Scholar]
- 28.Vetter, I. R. & Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science294, 1299–1304 (2001). 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- 29.Herskowitz, I. Functional inactivation of genes by dominant negative mutations. Nature329, 219–222 (1987). 10.1038/329219a0 [DOI] [PubMed] [Google Scholar]
- 30.Barren, B. & Artemyev, N. O. Mechanisms of dominant negative G-protein alpha subunits. J. Neurosci. Res.85, 3505–3514 (2007). 10.1002/jnr.21414 [DOI] [PubMed] [Google Scholar]
- 31.Olsen, R. H. J. et al. TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat. Chem. Biol. 10.1038/s41589-020-0535-8 (2020). [DOI] [PMC free article] [PubMed]
- 32.Farfel, Z. et al. Pseudohypoparathyroidism, a novel mutation in the betagamma-contact region of Gsalpha impairs receptor stimulation. J. Biol. Chem.271, 19653–19655 (1996). 10.1074/jbc.271.33.19653 [DOI] [PubMed] [Google Scholar]
- 33.Iiri, T., Farfel, Z. & Bourne, H. R. Conditional activation defect of a human Gsalpha mutant. Proc. Natl Acad. Sci. USA94, 5656–5661 (1997). 10.1073/pnas.94.11.5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollins, B., Kuravi, S., Digby, G. J. & Lambert, N. A. The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal21, 1015–1021 (2009). 10.1016/j.cellsig.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert, N. A. et al. Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc. Natl Acad. Sci. USA107, 7066–7071 (2010). 10.1073/pnas.0912934107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuho, I. et al. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal.8, ra123 (2015). 10.1126/scisignal.aab4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muntean, B. S. et al. Gαo is a major determinant of cAMP signaling in the pathophysiology of movement disorders. Cell Rep.34, 108718 (2021). 10.1016/j.celrep.2021.108718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, X. et al. Phenotypes of GNAO1 variants in a Chinese cohort Front. Neurol.12, 662162 (2021). 10.3389/fneur.2021.662162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternweis, P. C. & Robishaw, J. D. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J. Biol. Chem.259, 13806–13813 (1984). 10.1016/S0021-9258(18)89817-9 [DOI] [PubMed] [Google Scholar]
- 40.Huff, R. M., Axton, J. M. & Neer, E. J. Physical and immunological characterization of a guanine nucleotide- binding protein purified from bovine cerebral cortex. J. Biol. Chem.260, 10864–10871 (1985). 10.1016/S0021-9258(19)85162-1 [DOI] [PubMed] [Google Scholar]
- 41.Aryal, P., Dvir, H., Choe, S. & Slesinger, P. A. A discrete alcohol pocket involved in GIRK channel activation. Nat. Neurosci.12, 988–995 (2009). 10.1038/nn.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodhinathan, K. & Slesinger, P. A. Molecular mechanism underlying ethanol activation of G-protein-gated inwardly rectifying potassium channels. Proc. Natl Acad. Sci. USA110, 18309–18314 (2013). 10.1073/pnas.1311406110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isom, D. G. et al. Protons as second messenger regulators of G protein signaling. Mol. Cell51, 531–538 (2013). 10.1016/j.molcel.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goricanec, D. et al. Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding. Proc. Natl Acad. Sci. USA113, E3629–E3638 (2016). 10.1073/pnas.1604125113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McEwen, D. P., Gee, K. R., Kang, H. C. & Neubig, R. R. Fluorescent BODIPY-GTP analogs: real-time measurement of nucleotide binding to G proteins. Anal. Biochem.291, 109–117 (2001). 10.1006/abio.2001.5011 [DOI] [PubMed] [Google Scholar]
- 46.Willard, F. S., Kimple, A. J., Johnston, C. A. & Siderovski, D. P. A direct fluorescence-based assay for RGS domain GTPase accelerating activity. Anal. Biochem.340, 341–351 (2005). 10.1016/j.ab.2005.02.015 [DOI] [PubMed] [Google Scholar]
- 47.DeVree, B. T. et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature535, 182–186 (2016). 10.1038/nature18324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maeda, S. et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun.10.1038/s41467-018-06002-w (2018). [DOI] [PMC free article] [PubMed]
- 49.Zhang, M. et al. Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs. Nat. Struct. Mol. Biol.28, 258–267 (2021). 10.1038/s41594-020-00554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wall, M. A. et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell83, 1047–1058 (1995). 10.1016/0092-8674(95)90220-1 [DOI] [PubMed] [Google Scholar]
- 51.Hilger, D. The role of structural dynamics in GPCR-mediated signaling. FEBS J.288, 2461–2489 (2021). 10.1111/febs.15841 [DOI] [PubMed] [Google Scholar]
- 52.Huang, W. et al. Structural insights into µ-opioid receptor activation. Nature524, 315–321 (2015). 10.1038/nature14886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filipek, S. Molecular switches in GPCRs. Curr. Opin. Struct. Biol.55, 114–120 (2019). 10.1016/j.sbi.2019.03.017 [DOI] [PubMed] [Google Scholar]
- 54.Wang, S. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature555, 269–273 (2018). 10.1038/nature25758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferguson, K. M., Higashijima, T., Smigel, M. D. & Gilman, A. G. The influence of bound GDP on the kinetics of guanine nucleotide binding to G proteins. J. Biol. Chem.261, 7393–7399 (1986). 10.1016/S0021-9258(17)38404-1 [DOI] [PubMed] [Google Scholar]
- 56.Draper-Joyce, C. J. et al. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature558, 559–563 (2018). 10.1038/s41586-018-0236-6 [DOI] [PubMed] [Google Scholar]
- 57.Liang, Y. L. et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature555, 121–125 (2018). 10.1038/nature25773 [DOI] [PubMed] [Google Scholar]
- 58.Iiri, T., Bell, S. M., Baranski, T. J., Fujita, T. & Bourne, H. R. A Gsalpha mutant designed to inhibit receptor signaling through Gs. Proc. Natl Acad. Sci. USA96, 499–504 (1999). 10.1073/pnas.96.2.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berlot, C. H. A highly effective dominant negative alpha s construct containing mutations that affect distinct functions inhibits multiple Gs-coupled receptor signaling pathways. J. Biol. Chem.277, 21080–21085 (2002). 10.1074/jbc.M201330200 [DOI] [PubMed] [Google Scholar]
- 60.Wacker, D., Stevens, R. C. & Roth, B. L. How ligands illuminate GPCR molecular pharmacology. Cell170, 414–427 (2017). 10.1016/j.cell.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galosi, S. et al. Motor, epileptic, and developmental phenotypes in genetic disorders affecting G protein coupled receptors-cAMP signaling. Front. Neurol.13, 886751 (2022). 10.3389/fneur.2022.886751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura, K. et al. De Novo mutations in GNAO1, encoding a Gαo subunit of heterotrimeric G proteins, cause epileptic encephalopathy. Am. J. Hum. Genet.93, 496–505 (2013). 10.1016/j.ajhg.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larasati, Y. A. et al. Restoration of the GTPase activity and cellular interactions of Gα. Sci. Adv.8, eabn9350 (2022). 10.1126/sciadv.abn9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng, H. et al. Movement disorder in GNAO1 encephalopathy associated with gain-of-function mutations. Neurology89, 762–770 (2017). 10.1212/WNL.0000000000004262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saitsu, H. et al. Phenotypic spectrum of GNAO1 variants: epileptic encephalopathy to involuntary movements with severe developmental delay. Eur. J. Hum. Genet.24, 129–134 (2016). 10.1038/ejhg.2015.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Domínguez-Carral, J. et al. Severity of GNAO1-related disorder correlates with changes in g-protein function. Ann. Neurol.94, 987–1004 (2023). 10.1002/ana.26758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishimura, A. et al. Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc. Natl Acad. Sci. USA107, 13666–13671 (2010). 10.1073/pnas.1003553107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takasaki, J. et al. A novel Galphaq/11-selective inhibitor. J. Biol. Chem.279, 47438–47445 (2004). 10.1074/jbc.M408846200 [DOI] [PubMed] [Google Scholar]
- 69.Schrage, R. et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun.6, 10156 (2015). 10.1038/ncomms10156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong, X. F. et al. Total synthesis and structure-activity relationship studies of a series of selective G protein inhibitors. Nat. Chem.8, 1035–1041 (2016). 10.1038/nchem.2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dai, S. A. et al. State-selective modulation of heterotrimeric Gαs signaling with macrocyclic peptides. Cell185, 3950–3965.e3925 (2022). 10.1016/j.cell.2022.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, Y. et al. The small molecule GAT1508 activates brain-specific GIRK1/2 channel heteromers and facilitates conditioned fear extinction in rodents. J. Biol. Chem.295, 3614–3634 (2020). 10.1074/jbc.RA119.011527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Celver, J., Sharma, M. & Kovoor, A. D(2)-Dopamine receptors target regulator of G protein signaling 9-2 to detergent-resistant membrane fractions. J. Neurochem.120, 56–69 (2012). 10.1111/j.1471-4159.2011.07559.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hauser, A. S. et al. Pharmacogenomics of GPCR drug targets. Cell172, 41–54.e19 (2018). 10.1016/j.cell.2017.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He, C., Zhang, H., Mirshahi, T. & Logothetis, D. E. Identification of a potassium channel site that interacts with G protein betagamma subunits to mediate agonist-induced signaling. J. Biol. Chem.274, 12517–12524 (1999). 10.1074/jbc.274.18.12517 [DOI] [PubMed] [Google Scholar]
- 76.Chan, K. W., Sui, J. L., Vivaudou, M. & Logothetis, D. E. Control of channel activity through a unique amino acid residue of a G protein-gated inwardly rectifying K+ channel subunit. Proc. Natl Acad. Sci. USA93, 14193–14198 (1996). 10.1073/pnas.93.24.14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horvath, G. A. et al. Gain-of-function KCNJ6 mutation in a severe hyperkinetic movement disorder phenotype. Neuroscience384, 152–164 (2018). 10.1016/j.neuroscience.2018.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mutneja, M., Berton, F., Suen, K. F., Lüscher, C. & Slesinger, P. A. Endogenous RGS proteins enhance acute desensitization of GABA(B) receptor-activated GIRK currents in HEK-293T cells. Pflug. Arch.450, 61–73 (2005). 10.1007/s00424-004-1367-1 [DOI] [PubMed] [Google Scholar]
- 79.Maly, J. & Crowhurst, K. A. Expression, purification and preliminary NMR characterization of isotopically labeled wild-type human heterotrimeric G protein alphai1. Protein Expr. Purif.84, 255–264 (2012). 10.1016/j.pep.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 80.Ghosh, M., Peterson, Y. K., Lanier, S. M. & Smrcka, A. V. Receptor- and nucleotide exchange-independent mechanisms for promoting G protein subunit dissociation. J. Biol. Chem.278, 34747–34750 (2003). 10.1074/jbc.C300271200 [DOI] [PubMed] [Google Scholar]
- 81.Kozasa, T. & Gilman, A. G. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. Characterization of alpha 12 and inhibition of adenylyl cyclase by alpha z. J. Biol. Chem.270, 1734–1741 (1995). 10.1074/jbc.270.4.1734 [DOI] [PubMed] [Google Scholar]
- 82.Tanabe, S., Kreutz, B., Suzuki, N. & Kozasa, T. Regulation of RGS-RhoGEFs by Galpha12 and Galpha13 proteins. Methods Enzymol.390, 285–294 (2004). 10.1016/S0076-6879(04)90018-3 [DOI] [PubMed] [Google Scholar]
- 83.Kozasa, T. & Gilman, A. G. Protein kinase C phosphorylates G12 alpha and inhibits its interaction with G beta gamma. J. Biol. Chem.271, 12562–12567 (1996). 10.1074/jbc.271.21.12562 [DOI] [PubMed] [Google Scholar]
- 84.Jones, J. C., Jones, A. M., Temple, B. R. & Dohlman, H. G. Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Gα proteins. Proc. Natl Acad. Sci. USA109, 7275–7279 (2012). [DOI] [PMC free article] [PubMed]
- 85.Seneviratne, A. M., Burroughs, M., Giralt, E. & Smrcka, A. V. Direct-reversible binding of small molecules to G protein betagamma subunits. Biochim. Biophys. Acta1814, 1210–1218 (2011). 10.1016/j.bbapap.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem.29, 1859–1865 (2008). 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 87.Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput.4, 435–447 (2008). 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- 88.Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods14, 71–73 (2017). 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. V., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys.81, 3684–3690 (1984). 10.1063/1.448118 [DOI] [Google Scholar]
- 90.Andersen, H. C. Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys.52, 24–34 (1983). 10.1016/0021-9991(83)90014-1 [DOI] [Google Scholar]
- 91.Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys.98, 10089–10092 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- 92.Evans, D. J. & Holian, B. L. The Nose–Hoover thermostat. J. Chem. Phys.83, 4069–4074 (1985). 10.1063/1.449071 [DOI] [Google Scholar]
- 93.Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys.52, 7182–7190 (1981). 10.1063/1.328693 [DOI] [Google Scholar]
- 94.Petrova, S. S. & Solov’ev, A. D. The origin of the method of steepest descent. Historia Math.24, 361–375 (1997). 10.1006/hmat.1996.2146 [DOI] [Google Scholar]
- 95.Peck, J. V., Fay, J. F. & Strauss, J. D. High-speed high-resolution data collection on a 200 keV cryo-TEM. IUCrJ9, 243–252 (2022). 10.1107/S2052252522000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol.152, 36–51 (2005). 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 97.Bepler, T., Kelley, K., Noble, A. J. & Berger, B. Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun.11, 5208 (2020). 10.1038/s41467-020-18952-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). 10.1038/s41592-020-00990-8 [DOI] [PubMed] [Google Scholar]
- 99.Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol.333, 721–745 (2003). 10.1016/j.jmb.2003.07.013 [DOI] [PubMed] [Google Scholar]
- 100.Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol.4, 874 (2021). 10.1038/s42003-021-02399-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–1612 (2004). 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 102.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr.60, 2126–2132 (2004). 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 103.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr.66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr.59, 1131–1137 (2003). 10.1107/S0907444903008126 [DOI] [PubMed] [Google Scholar]
- 105.Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol.372, 774–797 (2007). 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete sequences for plasmids generated in this study are provided in Supplementary Information: PET-SUMO-Gαo for purification, pMAX-Gαo for GIRK assays in X. oocyes, pcDNA3.1-Gαo for GIRK assays in HEK293 cells, MOR-Gαo fusion for radioligand binding, and pFastbac-Gαo for cryo-electron microscopy. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes 8U02 and EMD-41776 for the DR2-GαoK46E, and 8TZQ and EMD-41766 for DR2-GαοAK46E-scFv16. Source data supporting Fig. 2 (BRET), Fig. 3 (potassium current), Fig. 4A and B (thermostability), Fig. 4C (BODIPY fluorescence), Fig. 4D (radiolabeled GTP binding), Fig. 4E (BLI), Fig. 4F (Principal Component analysis), and Fig. 5A–C (radioligand binding) are provided with this paper. Source data are provided with this paper.