Abstract
Radiology narrative reports often describe characteristics of a patient’s disease, including its location, size, and shape. Motivated by the recent success of multimodal learning, we hypothesized that this descriptive text could guide medical image analysis algorithms. We proposed a novel vision-language model, ConTEXTual Net, for the task of pneumothorax segmentation on chest radiographs. ConTEXTual Net extracts language features from physician-generated free-form radiology reports using a pre-trained language model. We then introduced cross-attention between the language features and the intermediate embeddings of an encoder-decoder convolutional neural network to enable language guidance for image analysis. ConTEXTual Net was trained on the CANDID-PTX dataset consisting of 3196 positive cases of pneumothorax with segmentation annotations from 6 different physicians as well as clinical radiology reports. Using cross-validation, ConTEXTual Net achieved a Dice score of 0.716±0.016, which was similar to the degree of inter-reader variability (0.712±0.044) computed on a subset of the data. It outperformed vision-only models (Swin UNETR: 0.670±0.015, ResNet50 U-Net: 0.677±0.015, GLoRIA: 0.686±0.014, and nnUNet 0.694±0.016) and a competing vision-language model (LAVT: 0.706±0.009). Ablation studies confirmed that it was the text information that led to the performance gains. Additionally, we show that certain augmentation methods degraded ConTEXTual Net’s segmentation performance by breaking the image-text concordance. We also evaluated the effects of using different language models and activation functions in the cross-attention module, highlighting the efficacy of our chosen architectural design.
Keywords: Multimodal, Fusion, Segmentation, Pneumothorax
Introduction
Radiology is a multimodal field. Picture archiving and communication systems (PACS) contain medical images and accompanying reports generated by radiologists. These reports serve as the official record of the reading physicians’ interpretations for radiological exams, playing an essential role in communicating findings to patients and their healthcare teams. Additionally, such reports offer radiologists invaluable context regarding prior imaging results when interpreting follow-up exams. For example, in reading a current set of images, radiologists often review the patient’s prior images and reports to ascertain the location and extent of the disease. This allows for monitoring disease evolution over time and assessing the effectiveness of treatments. The comparative review process is instrumental in identifying new developments or subtle changes in a patient’s condition that could otherwise go unnoticed without such historical reference for comparison. Although reviewing past exams can be time-consuming, its value in many diagnostic applications is undeniable.
A clinical scenario where repeated images are acquired is the management of patients diagnosed with pneumothorax. Pneumothorax is a condition in which air accumulates in the space between the lung and chest wall. Due to its life-threatening nature, rapid detection and intervention are crucial to prevent severe morbidity or mortality. If the pneumothorax is large or increasing in size, a chest tube must be inserted [1]. Therefore, monitoring its changes is essential. Pneumothorax can be challenging to detect and may not be evident on follow-up images. As a result, physicians often consult prior images and reports as part of their standard workflow. This workflow could be significantly enhanced through the use of automatic pneumothorax segmentation tools.
Prior works have explored the application of deep learning techniques to assist several medical tasks such as disease classification [2, 3], image retrieval [4, 5], and disease detection [6]. Motivated by the recent success of multimodal vision-language models [7, 8], we aim to leverage information in clinical reports to improve medical image analysis. Models like ConVIRT [9] and GLoRIA [10] have used image-report pairs and contrastive learning objectives to learn vision representations, which have shown promising results in downstream classification tasks. However, these models only utilized text for pre-training, without integrating it into the model to guide image analysis. Other approaches employed image and text encoders to identify instances of pneumonia and placed bounding boxes around them [11], but did not provide pixel-level segmentation. LAVT [12] produced pixel-wise predictions from image-text pairs, specifically for referring image segmentation of household items rather than for use in medical imaging. LVIT [13] is a vision-language model that took chest x-ray (CXR) images and text as inputs and generated segmentation masks for patients diagnosed with coronavirus disease 2019 (COVID-19). Nevertheless, instead of utilizing real free-form radiology reports, the work synthesized the text inputs from the ground-truth labels, which casts ambiguity on whether physician-generated text can enhance image analysis. Additionally, these existing works do not address language model choice, which data augmentations are suitable for multimodal vision-language segmentation, and a series of other methodological questions that we attempt to address in this study.
In this work, we aimed to apply multimodal learning to integrate physician reports into the task of pneumothorax segmentation on chest radiographs and explore the augmentations, model architecture, and training methodologies for use in the medical domain. To this end, we developed an algorithm that maps concepts from language space into medical image space so that descriptive text can be used to guide image segmentation. Instead of bounding box detection, we pursued fine-grained segmentation, as it provides delineation of boundaries, and allows for morphological analysis and quantitative measurements (e.g., volume) of the disease. Compared to non-medical segmentation tasks, pneumothorax segmentation poses distinct challenges, including limited image availability, and the expertise and overall cost required for annotation. Our approach of directly incorporating physician-generated text into the image analysis has the benefit of improving segmentation accuracy by allowing the model to leverage the physician’s expertise. Moreover, this language integration paves the way for real-time, physician-guided disease assessment.
The paper organizes itself as follows: Section “Methods” delves into the methodology of our proposed vision-language model, ConTEXTual Net. Section “Experimental Setting” details the experimental setup. We present the segmentation results of our model and ablation analysis in “Results” section. Lastly, we discuss our findings and conclusions.
Methods
Model Architecture
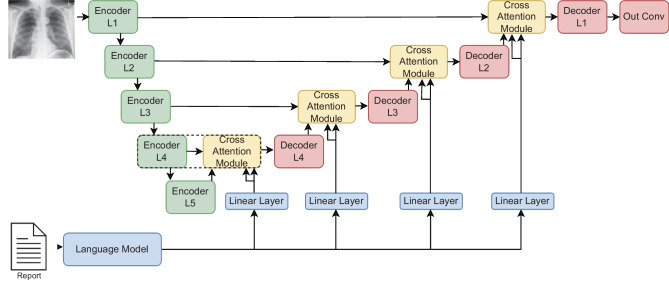
Figure 1 shows the architecture of ConTEXTual Net, and the open-source project1 provides the implementation details. A U-Net encoding scheme, shown in green, extracts vision features from the image, while a pre-trained language model, shown in blue, extracts language features. This approach leverages the ability of the U-Net to contour disease and the ability of transformer-based language models to extract semantically rich vectors that can be used to help localize the disease. Within the U-Net, each vision encoder layer is a stack of two sub-layers, where each sub-layer is a convolution followed by batch normalization and ReLU activation. Each encoder layer feeds its output as a skip connection to a cross-attention module, depicted in yellow in Fig. 1, and described in the “Language Cross-Attention” section. Meanwhile, the encoder output is also downsampled via max pooling and fed to the next encoder layer. The output of the cross-attention module is subsequently fed to a decoder layer, which is also a stack of two convolution sub-layers with batch normalization and ReLU (similar to the encoder layer) and then upsampled as inputs to the next cross-attention module. The last layer of the decoder is a convolution, which reduces the channels to a single output channel and is used for pixel-level prediction.
Fig. 1.
ConTEXTual Net combines a U-Net and a transformer architecture. It uses the encoder layers of the U-Net to extract visual representations and uses a pre-trained language model (bottom-left) to extract language representations. The architecture is modular such that any language model that creates word-level vector representations can be used. It then performs cross-attention between the modalities and finally uses the decoder layers (top-right) of the U-Net to predict the segmentation masks. The cross-attention module (dotted box) is further detailed in Fig 2
To integrate language into the model, a pre-trained language model is used to encode the text report into contextualized text embeddings. These embeddings are further projected by a linear layer and fed as inputs to the cross-attention modules. The model is trained using supervised learning with a cross-entropy loss on the prediction and ground-truth segmentation labels. Unless explicitly stated otherwise, we freeze the pre-trained language model during training, thereby reducing the number of parameters that must be learned and reducing GPU memory requirements.
Language Cross-Attention
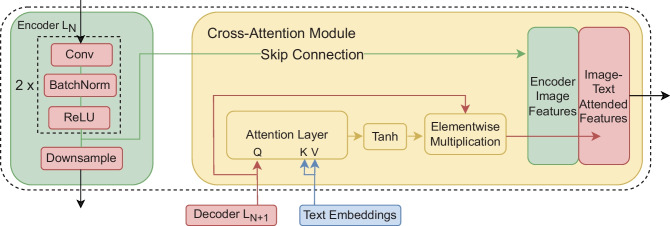
ConTEXTual Net uses a cross-attention module to integrate the embeddings from the language model into the vision-based segmentation model. We inject the text embeddings into the decoding side of a U-Net. Conceptually, the text embeddings contain semantic information about the presence and location of the disease and can be used to guide the U-Net segmentation model. The cross-attention between the text embeddings and the decoded feature maps produces a pixel-wise attention map. This pixel-wise attention map then gets fed into a Tanh activation function to normalize values between and 1. The normalized pixel-wise attention map is then multiplied pixel-wise with the query feature map. The pixel-wise attention map, , is obtained by
| 1 |
where is the query vectors flattened from the upsampled feature map of size in c channels, are the projected text embeddings of a report of l words, and , , and are the learnable weights that project the query, key and value vectors to the same dimensional space. Here, we select the projected text dimension to match the number of channels c at each decoder layer of the U-Net. After matrix multiplication this yields a matrix that is which is then unflattened into with dimensions Subsequently, the cross-attention module output is calculated as
| 2 |
where is the element-wise multiplication of the input feature map and the normalized pixel attention map , and is the attention-weighted feature map. Empirically, we found that the Tanh activation performs better than other activation functions.
Fig. 2.
The cross-attention module takes in the upsampled feature maps, which are used as the query, and the projected word-level embeddings from the language model, which are used as both the key and value. It calculates pixel-level attention maps which are used to weight the decode feature maps
Augmentations
Data augmentation is the process of altering training data to synthetically increase dataset size so that the model better generalizes to new situations. For multimodal models, these data augmentations must preserve the concordance between the text description and image. For example, if horizontal image flips are used, the descriptions of “left” and “right” no longer correspond to the image. This can be addressed by picking augmentations invariant to the other modality or augmenting both modalities to retain concordance. In this work, we focused on finding augmentations invariant to the other modality.
We considered the following set of image augmentations: horizontal flip 50% of the time, 30% of the time choosing one from RandomContrast, RandomGamma, and RandomBrightness, and again 30% of the time choosing from ElasticTransform, GridDistortion, and OpticalDistortion and lastly ShiftScaleRotate, all of which have been used previously for pneumothorax segmentation [14, 15]. Out of those, horizontal flipping was the only augmentation that we found to break the image-text concordance and was thus left out from all experiments unless otherwise stated. All augmentations were implemented using the Albumentations library [16].
Text augmentations were experimented with to improve the model’s generalizability to different writing styles. Specifically, two augmentations were used: sentence shuffling and synonym replacement. In sentence shuffling, the text is split into sentences and randomly rearranged. For radiology reports, sentences are generally self-contained with few inter-sentence dependencies, and we expect sentence shuffling to have little to no effect on the meaning. For synonym replacement, we used RadLex [17], a radiology ontology that contains definitions and synonyms for radiology-specific terms. During train time, each word in the report with a RadLex-listed synonym was replaced with that synonym 15% of the time.
Experimental Setting
CANDID-PTX Dataset
We developed and evaluated ConTEXTual Net using the CANDID-PTX dataset. The CANDID-PTX dataset consists of 19,237 chest radiographs with reports and segmentations of pneumothoraces, acute rib fractures, and intercostal chest tubes. We focused on pneumothorax in this study. There are 3196 positive cases of pneumothorax with segmentation annotations labeled by six different physicians. A second annotator checks each individual physician annotation for validity; this checked annotation is treated as ground truth for the purposes of our study. It is important to note that the physician who dictated the original report was different from the physician responsible for labeling.
The segmentation performance is evaluated using Dice scores given by
| 3 |
which compares two observers’ predictions (i.e., model vs annotation or one physician annotator vs another). The inter-rater Dice similarity scores between the six physicians ranged from 0.64 to 0.85 on a test set of 73 randomly selected images meant to evaluate inter-annotator variability. The primary annotator labeled 92.7% of the images in the dataset and had a mean Dice similarity score of 0.712 when compared to the other five physicians. The dataset was originally collected with approval from an ethics committee with waiver of informed consent and was made available under a data use agreement [18].
Model Comparison
To show the efficacy of our architectural design, we compared ConTEXTual Net against methods from previous studies. Specifically, we trained a U-Net model [19] that shares the same architecture with the vision-only component of ConTEXTual Net. We also included a baseline model that was built on the standard U-Net, with the encoder being replaced by Resnet50 [20]. Additionally, we compared against a U-Net with the encoder weights initialized from the GLoRIA model, which was pre-trained using multimodal contrastive learning with approximately 200k image-report pairs of chest x-rays [10]. More modern transformer segmentation methods also include Swin UNETR [21], which uses a Swin transformer as an encoder in a U-Net fashion. We also compare against a top-performing convolutional model nnUnet [22]. Lastly, we finetuned LAVT [12], which is a state-of-the-art vision-language model, for the task of text-guided pneumothorax segmentation. LAVT uses a Swin transformer to encode the image information and a BERT model to encode the language and fuses these via a pixel-word attention module.
Language Models
ConTEXTual Net uses language models to extract and encode important statements from the reports. In this study, we compared T5-Large’s encoder [23], RoBERTa-Large [24], RadBERT [25] and BERT [26], as the language encoders. T5-Large is a larger model with 770M parameters, RoBERTa-Large contains 354M parameters, and RadBERT and Bert are considerably smaller at 125M and 110M parameters, respectively. All models are transformer-based, but their training datasets and tasks differ. T5 is trained on the c4 dataset, which contains roughly 300GB of text and is intended to be capable of diverse tasks, including text classification, question answering, machine translation, and abstractive summarization. RoBERTa-Large and Bert are trained via masked language modeling, but RoBERta-Large was trained on a 161GB text corpus, whereas BERT’s training corpus was 16GB. RadBERT is initialized from RoBERTa-Base but is trained with roughly 4M additional radiology reports from the U.S. Department of Veterans Affairs.
The reports were fed into the language models, and the hidden state vectors were used as our report representations. The report representation has dimensions of (token length embedding dimension) for T5-Large and RoBERTa-Large and for RadBERT and BERT. This report representation then goes through a projection head to lower the embedding dimensionality of the hidden state vector to match the number of channels in the encoder feature maps. This is a necessary step for the language cross-attention to work at multiple levels in the U-Net decoder. Language models were imported via HuggingFace library [27].
Ablation Analysis
We performed ablations studies to determine the additive value of augmentations as well as ConTEXTual Net’s components. Ablation studies were performed in three settings: without any augmentations, with image augmentations only, and with both image and text augmentations. We additionally tested ConTEXTual Net with the full language encoder and cross-attention modules, but the input text was replaced with an empty string to help quantify the effects of the physician text.
Along with these experiments, we probed using different language models, activation functions, integration points, and unfreezing schedules. RoBERTa-Large, RadBERT, and Bert were swapped with the T5 encoder to examine how sensitive ConTEXTual Net is to the language model used. To determine the best activation function in the cross-attention module, we tested using no activation function, ReLU, Sigmoid, and the hyperbolic tangent function. We examined the impact of only using a single attention module at the four different levels of the network. This is meant to probe whether textual information should be inserted early in the image analysis or at later stages. We also explored the consequences of unfreezing the final two layers of the language model at various stages during training. For the unfreezing experiments, RadBERT was used as it is a smaller model, meaning we could unfreeze more of its layers without running into memory problems. All language model, activation function, and attention module integration experiments were performed with vision augmentations only.
Model Hyperparameters
All ConTEXTual Net models were trained with the AdamW optimizer, a learning rate of 5e-5, and 100 epochs with a binary cross-entropy loss. The native image dimensions of were used. The model which did best on the validation set was used on the cross-validation test set. We report the average and standard deviation of 5-fold Monte Carlo cross-validation with 80% in training, 10% in validation, and 10% in testing for each fold. All models were trained on NVIDIA A100 GPUs.
Results
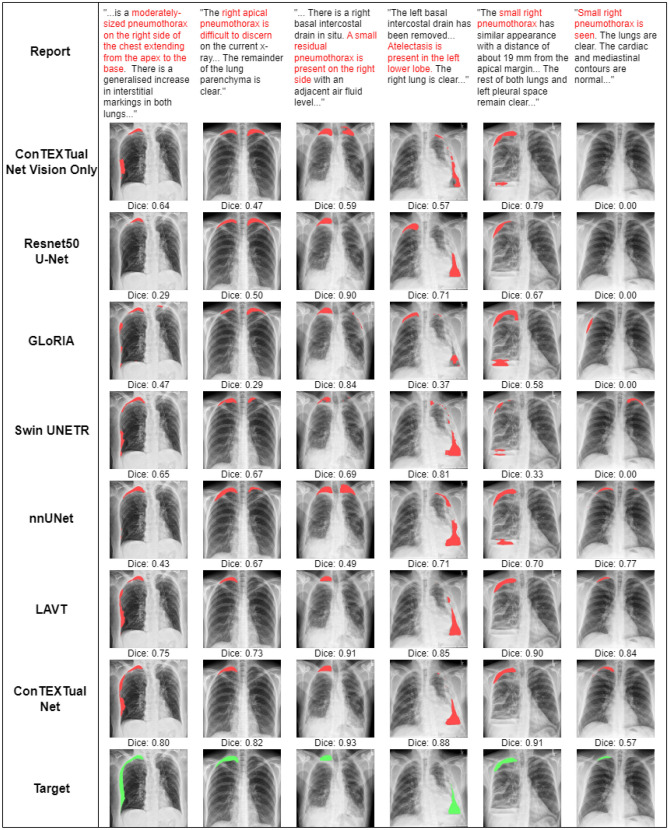
Table 1 shows a comparison of Dice scores for all models and the primary physician annotator as compared to the other physician annotators. Overall, the best-performing configuration of ConTEXTual Net (Dice 0.716 ± 0.016) outperformed the baseline U-Net (0.680 ± 0.014), the transformer-based vision model Swin UNETR (0.670 ± 0.015), the best vision-only model nnUnet (0.694 ± 0.016) and LAVT (0.706 ± 0.009) and performed similarly to the primary labeling physician (0.712 ± 0.044). Example images with results are shown in Fig. 3.
Table 1.
Model comparisons
| Model type | AVG Dice | SD |
|---|---|---|
| ConTEXTual vision-only U-Net | 0.680 | 0.014 |
| Resnet50 U-Net [20] | 0.677 | 0.015 |
| GLoRIA [10] | 0.686 | 0.014 |
| Swin UNETR [21] | 0.670 | 0.015 |
| nnUNet [22] | 0.694 | 0.016 |
| LAVT [12] | 0.706 | 0.009 |
| ConTEXTual Net | 0.716 | 0.016 |
| Primary Physician Annotator [18] | 0.712 | 0.044 |
The bolding is commonly used to denote the best-performing model and is bolded for quick reference
Fig. 3.
Predictions from all seven evaluated models, as well as the physician labeled target, and a portion of the text is shown. Note how ConTEXTual Net is able to use descriptions like “extending from the apex to the base” (first column) and avoid false positives (columns 2–5). The last column demonstrates how multimodal models are able to use the text to detect subtle disease described in the report; interestingly, nnUNet also performs well in this case, while all the other vision models fail to detect the apical pneumothorax
We report results from ablation analyses in Table 2. Due to the joint dependencies of ConTEXTual Net’s components and the data augmentations used during training, we report results separately based on the types of data augmentations.
Table 2.
Ablation study of ConTEXTual Net
| Model type | AVG Dice | SD |
|---|---|---|
| No augmentations | ||
| Baseline U-Net | 0.649 | 0.014 |
| ConTEXTual Net | 0.668 | 0.010 |
| Vision augmentations | ||
| Baseline U-Net | 0.680 | 0.014 |
| ConTEXTual Net | 0.716 | 0.016 |
| ConTEXTual Net with flipping | 0.675 | 0.016 |
| ConTEXTual Net w/o reports | 0.671 | 0.019 |
| Text augmentations | ||
| No text augmentations | 0.716 | 0.016 |
| Synonym Replacement | 0.705 | 0.008 |
| Sentence Shuffle | 0.713 | 0.023 |
| Synonym + Sentence Shuffle | 0.714 | 0.014 |
| Language models | ||
| ConTEXTual Net (T5) | 0.716 | 0.016 |
| ConTEXTual Net (RoBERTa-Large) | 0.713 | 0.010 |
| ConTEXTual Net (RadBERT) | 0.716 | 0.022 |
| ConTEXTual Net (BERT) | 0.713 | 0.020 |
| Activation functions | ||
| ConTEXTual Net (Tanh) | 0.716 | 0.016 |
| ConTEXTual Net (ReLU) | 0.698 | 0.027 |
| ConTEXTual Net (Sigmoid) | 0.710 | 0.010 |
| ConTEXTual Net (No Activation) | 0.704 | 0.011 |
| Cross-attention integration | ||
| Attention Module L4 | 0.712 | 0.019 |
| Attention Module L3 | 0.709 | 0.013 |
| Attention Module L2 | 0.685 | 0.021 |
| Attention Module L1 | 0.679 | 0.011 |
| Unfreezing language model | ||
| Unfreeze at start | 0.704 | 0.011 |
| Unfreeze at 25th epoch | 0.712 | 0.014 |
| Unfreeze at 50th epoch | 0.716 | 0.020 |
| Unfreeze at 75th epoch | 0.716 | 0.010 |
| Frozen | 0.716 | 0.022 |
Bold values denote the highest-performing configuration of ConTEXTual Net
Vision Augmentations
We evaluated the added value of ConTEXTual Net’s cross-attention module with different vision-based augmentation methods. Without vision augmentations, ConTEXTual Net (0.668 ± 0.010) only slightly outperformed the baseline U-Net (0.649 ± 0.014). When the vision augmentations were applied, ConTEXTual Net’s (0.716 ± 0.016) relative performance compared to the baseline U-Net (0.680 ± 0.014) increases to a 3.6 Dice point improvement. The performance of ConTEXTual Net significantly decreases (0.671 ± 0.019) when the reports are replaced with padding tokens (i.e., an empty string was used as input), which indicates that the reports are indeed helping guide the segmentation. When horizontal image flipping is applied, ConTEXTual Net’s (0.675 ± 0.016) performance decreases by 4.1 Dice points and is comparable to using the empty string as input. This suggests the model learned to ignore the text when augmentations break the image-text correspondence. All other vision augmentations improved the model performance.
Text Augmentations
Although text augmentation represents a promising approach to vision-language data augmentations, text augmentations did not lead to gains in segmentation performance, as shown in Table 2. Using RadLex-based synonym replacement (0.705 ± 0.008) resulted in a decrease in performance of 1.1 dice points. Sentence shuffle (0.713 ± 0.023) led to increased variance across runs. Using both augmentations with the T5 encoder (0.714 ± 0.014) had minimal effect on the model’s performance.
Language Models
We evaluated using RoBERTa-Large (0.713 ± 0.010), RadBERT (0.716 ± 0.022), and BERT (0.713 ± 0.020) instead of T5 as the language encoder while using vision augmentations. This had a negligible impact on performance, suggesting that ConTEXTual Net is robust to the language model used.
Activation Functions
The use of different activation functions was investigated using T5 as our language encoder with vision augmentations and without text augmentations. It was found that the hyperbolic tangent activation function (0.716 ± 0.016) performed the best, outperforming the sigmoid function (0.710 ± 0.010). Using ReLU was found to decrease performance (0.698 ± 0.027) when compared to not using any activation function (0.704 ± 0.018).
Integration of Cross-attention Modules at Different Levels
We found that the lower the attention was integrated, the better the model performed. The L4 (i.e., decoder layer 4) cross-attention module (0.712 ± 0.019) performed slightly better than the L3 cross-attention module (0.709 ± 0.013). Moving from the L3 to L2 cross-attention module (0.685 ± 0.021) precipitated the most significant drop in performance of 2.4 dice points. Finally, the L1 cross-attention module performed slightly worse than the L2 cross-attention module.
Unfreezing of the Language Model
Unfreezing the language model too early in the training process led to decreased performance. The most significant impact is observed when the language model is unfrozen at the beginning of training (0.704 ± 0.011), which results in a decrease of 1.2 dice points. By the 25th epoch, unfreezing the language model (0.712 ± 0.014) had a much smaller effect, decreasing the performance by 0.4 dice points. Unfreezing the language model had no effect after the 50th epoch, with all models having an average dice score of 0.716.
Discussion
In this work, we proposed a method to extract information from clinical text reports and integrate it into a medical image segmentation algorithm. Our multimodal approach led to a 3.6 Dice point improvement over a traditional vision-only U-Net and achieved accuracy that matched physician performance. Additionally, we demonstrated the importance of maintaining image-text concordance when performing data augmentation and showed that early fusion leads to more accurate predictions for multimodal segmentation.
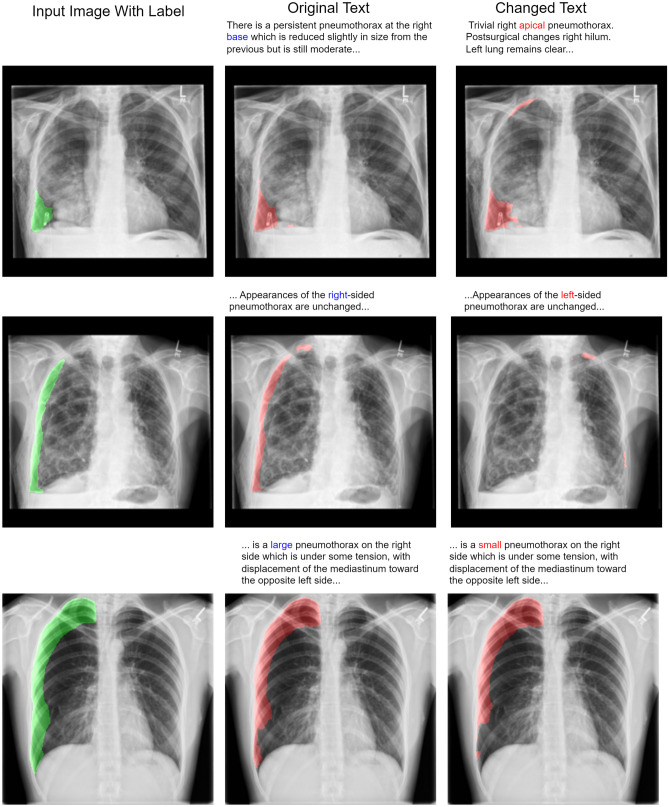
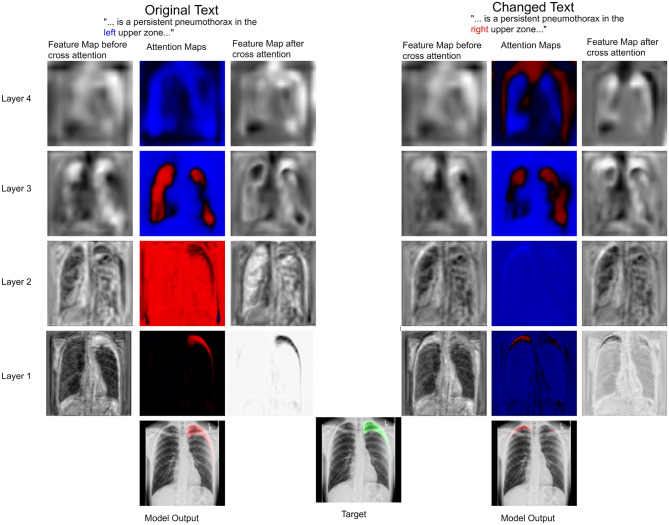
We showed the feasibility of using language from radiology reports to guide the output of segmentation algorithms. To illustrate how text can guide pixel-level predictions, we can change the input text and observe how the segmentation model reacts. For example, in Fig. 4, we changed the word “right” to “left”, “large pneumothorax” to “small pneumothorax”, and the location descriptor from “base” to “apical”, and observed corresponding changes in the output segmentation map. We also showed that flipping an image during data augmentation can negatively impact model performance. In Fig. 5, we show that changing the text from “left” pneumothorax to “right” pneumothorax alters the attention maps from the cross-attention module, resulting in changes in the corresponding feature maps and model output.
Fig. 4.
The same input image with different text is fed into the multimodal model. In the top row, an incorrect report describing an apical pneumothorax is used as input with an image, demonstrating that location descriptors like “apical” and “base” carry relevant information for segmentation. In the middle row, we show an example of an image and text with the term “right” changed to “left”. This illustrates the model’s sensitivity at the word level. In the bottom row, we changed the term “large” to “small”, which resulted in a reduction of segmented pixels by 10%. Note that “left” and “right” correspond to the patient’s “left” and “right”
Fig. 5.
The same image is fed to the model with only a single word changed in the text report: “left” was switched to “right”. Feature maps of a selected channel at different layers of the U-Net are shown when two different text inputs are used. Shown are the input feature maps to the language cross-attention module, the attention output by the cross-attention module after the Tanh activation, and the feature maps after the pixel-wise multiplication. In this case, it can be seen how the language changes the attention maps and guides the feature maps to reflect this change in the language. In the original text, the attention maps suppress the pixels in the right lung and portions of the left lower zone. In contrast, the attention maps in the case of the altered text suppress the pixels in the left lung but fail to suppress this signal completely, and the result is a prediction of both a right and a small left pneumothorax
ConTEXTual Net compared favorably to other vision-language models. The GLoRIA model, which only used text reports for pre-training, performed similarly to the baseline U-Net. This is not unexpected, as the purported advantage of GLoRIA is its performance in low-label settings, with limited advantages when datasets are sufficiently large [10]. Our model slightly outperformed LAVT. LAVT was developed for segmenting the object(s) described in the text caption. Text captions were, on average, 1.6 words to 8.4 words long in the LAVT study, depending on the dataset. In contrast, our study had a single segmentation target, which was pneumothorax, and the physician-generated text that described the disease was, on average, much longer at 63.7 words. Another key difference in methods is our use of a CNN as the vision encoder, whereas LAVT used a Swin vision transformer. This design choice is driven by the lower number of images in medical databases compared to the natural image databases LAVT is trained on and the observation that CNNs are typically more sample efficient [28]. Experimentally, this can be seen in the poor performance of Swin UNETR when compared to the other convolutional-based models.
Our cross-attention integration experiments reveal a crucial insight into the integration of language with the vision encoder. Notably, we observed that integrating language at the lower levels of the U-Net improved model performance. This suggests that prioritizing early integration may be a strategic design choice. Cross-attention at a single layer may be a pragmatic approach to optimize resource utilization.
Additionally, we show that prematurely unfreezing the language model can negatively affect the model’s performance. We hypothesize that the relatively large losses incurred early on in training cause large changes to the language model weights, hurting the language model’s ability to extract useful features from the text. By waiting until the model produces more accurate predictions with lower losses and then unfreezing the language model, the degradation in performance can be avoided. Lastly, our experiments show limited to no gains from unfreezing the language model’s parameters, so by keeping them frozen, the computational graph created by Pytorch is minimized, saving memory and speeding up training.
ConTEXTual Net was insensitive to the language model used to encode the radiology reports, but it was sensitive to the data augmentation methods and the activation function used within the cross-attention module. We found that Tanh, by both bounding the output of the cross-attention and preserving the negative values, performed better than other activation functions in the cross-attention module. LAVT likewise found that Tanh was the best activation function [12].
Multimodal medical image segmentation algorithms have several potential applications in radiology workflows. A primary motivation for this work was to address the challenge of reviewing follow-up imaging exams. For example, many patients who present with pneumothorax get an initial chest X-ray and then receive another chest X-ray within 3–6 h to monitor the pneumothorax [29]. This means radiologists must visually compare the size and extent of the disease on previous chest X-rays. Multimodal models such as ConTEXTual Net could help with tasks involving longitudinal assessment. Vision-language segmentation models may also enable segmentation based on physician dictation, which could enable voice-guided disease quantification. While a practical limitation of ConTEXTual Net is that it requires language as input, which precludes its use on exams that a physician has not yet reviewed, it does provide a mechanism by which physicians can work together with AI to produce better outputs. These types of models can help to address patient and clinician concerns about the use of autonomous AI in medicine.
In this study, we only analyzed positive cases of pneumothorax. Prior studies on pneumothorax segmentation included both positive and negative cases. For example, in the SIIM-ACR Pneumothorax Segmentation challenge, models that erroneously placed regions of interest in cases that were negative for pneumothorax were assigned a Dice score of 0. Models that correctly refused to place regions of interest in negative cases were assigned a Dice score of 1 [15]. Since the SIIM-ACR dataset had mostly negative cases, simply predicting negative on all cases resulted in a Dice score of around 0.79. We did not include negative cases because the input text to our multimodal model explicitly states whether the case is negative or positive. A language model could easily classify the cases as positive or negative based on the report [18]. Therefore, it is not appropriate to compare our multimodal model, which has access to the report text, to other vision models that do not have a way to use the text. Consequently, it is not easy to directly compare the results of our multimodal study to other vision-only studies on pneumothorax segmentation. Instead, we compared ConTEXTual Net’s performance to the degree of inter-physician variability on the same dataset. We found that ConTEXTual Net’s segmentation accuracy was comparable to the variability between physician contours.
There are several limitations of this proof-of-concept study. First, the study was trained and evaluated using a single dataset. This limits our ability to know how generalizable the ConTEXTual Net architecture is to other related tasks. Despite the availability of large medical imaging datasets that have images and radiology reports (e.g., MIMIC-CXR [30]) or that have images and segmentation labels (e.g., SIIM Kaggle [31]), there is a scarcity of large datasets containing all three elements. An additional limitation is that most of the samples in the CANDID-PTX dataset were labeled by a single physician. While the original CANDID-PTX study did analyze inter-observer variability, our model’s pneumothorax segmentations likely reflect the tendencies of the primary annotator.
In conclusion, we demonstrated the feasibility of using vision-language models to enhance medical image segmentation and showed that descriptive language can guide medical image analysis algorithms.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Zach Huemann, Xin Tie, Junjie Hu, Tyler Bradshaw. The first draft of the manuscript was written by Zach Huemann and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Tyler Bradshaw received funding through a master research agreement from GE Healthcare. Research reported in this publication was supported by the National Institute Of Biomedical Imaging And Bioengineering of the National Institutes of Health under Award Number R01EB033782. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declarations
Ethics Approval
Institutional Review Board approval was obtained. Informed consent was waived by IRB due to minimal risk to subjects.
Competing Interests
Tyler Bradshaw, Zachary Huemann, and Junjie Hu have a patent pending for Automatic Image Quantification From Physician-Generated Reports. Zachary Huemann received Nvidia RTX6000 GPU as an Academic Hardware Grant in support of this project.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Paul Zarogoulidis, Ioannis Kioumis, Georgia Pitsiou, Konstantinos Porpodis, Sofia Lampaki, Antonis Papaiwannou, Nikolaos Katsikogiannis, Bojan Zaric, Perin Branislav, Nevena Secen, et al. Pneumothorax: from definition to diagnosis and treatment. Journal of thoracic disease, 6(Suppl 4):S372, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pranav Rajpurkar, Jeremy Irvin, Kaylie Zhu, Brandon Yang, Hershel Mehta, Tony Duan, Daisy Ding, Aarti Bagul, Curtis Langlotz, Katie Shpanskaya, Matthew P. Lungren, and Andrew Y. Ng. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning, 2017.
- 3.Saban Öztürk and Tolga Çukur. Focal modulation based end-to-end multi-label classification for chest x-ray image classification. In 31st Signal Processing and Communications Applications Conference, SIU 2023, Istanbul, Turkey, July 5-8, 2023, pages 1–4. IEEE, 2023.
- 4.Şaban Öztürk, Emin Çelik, and Tolga Çukur. Content-based medical image retrieval with opponent class adaptive margin loss. Information Sciences, 637:118938, 2023. 10.1016/j.ins.2023.118938 [DOI] [Google Scholar]
- 5.Şaban Öztürk, Adi Alhudhaif, and Kemal Polat. Attention-based end-to-end cnn framework for content-based x-ray image retrieval. Turkish Journal of Electrical Engineering and Computer Sciences, 2021:2680-2693, 10 2021. [Google Scholar]
- 6.Ekin Tiu, Ellie Talius, Pujan Patel, Curtis P. Langlotz, Andrew Y. Ng, and Pranav Rajpurkar. Expert-level detection of pathologies from unannotated chest X-ray images via self-supervised learning. Nature Biomedical Engineering, 6(12):1399–1406, September 2022. [DOI] [PMC free article] [PubMed]
- 7.Aditya Ramesh, Prafulla Dhariwal, Alex Nichol, Casey Chu, and Mark Chen. Hierarchical text-conditional image generation with clip latents. arXiv preprint arXiv:2204.06125, 2022.
- 8.Chitwan Saharia, William Chan, Saurabh Saxena, Lala Li, Jay Whang, Emily Denton, Seyed Kamyar Seyed Ghasemipour, Burcu Karagol Ayan, S Sara Mahdavi, Rapha Gontijo Lopes, et al. Photorealistic text-to-image diffusion models with deep language understanding. arXiv preprint arXiv:2205.11487, 2022.
- 9.Yuhao Zhang, Hang Jiang, Yasuhide Miura, Christopher D Manning, and Curtis P Langlotz. Contrastive learning of medical visual representations from paired images and text. arXiv preprint arXiv:2010.00747, 2020.
- 10.Shih-Cheng Huang, Liyue Shen, Matthew P Lungren, and Serena Yeung. Gloria: A multimodal global-local representation learning framework for label-efficient medical image recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 3942–3951, 2021.
- 11.Riddhish Bhalodia, Ali Hatamizadeh, Leo Tam, Ziyue Xu, Xiaosong Wang, Evrim Turkbey, and Daguang Xu. Improving pneumonia localization via cross-attention on medical images and reports. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 571–581. Springer, 2021.
- 12.Zhao Yang, Jiaqi Wang, Yansong Tang, Kai Chen, Hengshuang Zhao, and Philip H. S. Torr. LAVT: Language-Aware Vision Transformer for Referring Image Segmentation, April 2022. arXiv:2112.02244 [cs].
- 13.Zihan Li, Yunxiang Li, Qingde Li, You Zhang, Puyang Wang, Dazhou Guo, Le Lu, Dakai Jin, and Qingqi Hong. Lvit: language meets vision transformer in medical image segmentation. arXiv preprint arXiv:2206.14718, 2022. [DOI] [PubMed]
- 14.Aimoldin Anuar. SIIM-ACR Pneumothorax Segmentation. https://github.com/sneddy/pneumothorax-segmentation, 2019.
- 15.Ayat Abedalla, Malak Abdullah, Mahmoud Al-Ayyoub, and Elhadj Benkhelifa. Chest x-ray pneumothorax segmentation using u-net with efficientnet and resnet architectures. PeerJ Computer Science, 7:e607, 2021. 10.7717/peerj-cs.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander Buslaev, Vladimir I Iglovikov, Eugene Khvedchenya, Alex Parinov, Mikhail Druzhinin, and Alexandr A Kalinin. Albumentations: fast and flexible image augmentations. Information, 11(2):125, 2020.
- 17.Curtis P Langlotz. Radlex: a new method for indexing online educational materials, 2006. [DOI] [PubMed]
- 18.Sijing Feng, Damian Azzollini, Ji Soo Kim, Cheng-Kai Jin, Simon P Gordon, Jason Yeoh, Eve Kim, Mina Han, Andrew Lee, Aakash Patel, et al. Curation of the candid-ptx dataset with free-text reports. Radiology: Artificial Intelligence, 3(6), 2021. [DOI] [PMC free article] [PubMed]
- 19.Olaf Ronneberger, Philipp Fischer, and Thomas Brox. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer, 2015.
- 20.Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. Deep Residual Learning for Image Recognition, December 2015. arXiv:1512.03385 [cs].
- 21.Ali Hatamizadeh, Vishwesh Nath, Yucheng Tang, Dong Yang, Holger Roth, and Daguang Xu. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images, January 2022. arXiv:2201.01266 [cs, eess].
- 22.Fabian Isensee, Paul F. Jaeger, Simon A. A. Kohl, Jens Petersen, and Klaus H. Maier-Hein. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods, 18(2):203–211, February 2021. [DOI] [PubMed]
- 23.Colin Raffel, Noam Shazeer, Adam Roberts, Katherine Lee, Sharan Narang, Michael Matena, Yanqi Zhou, Wei Li, Peter J Liu, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res., 21(140):1–67, 2020.
- 24.Yinhan Liu, Myle Ott, Naman Goyal, Jingfei Du, Mandar Joshi, Danqi Chen, Omer Levy, Mike Lewis, Luke Zettlemoyer, and Veselin Stoyanov. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692, 2019.
- 25.An Yan, Julian McAuley, Xing Lu, Jiang Du, Eric Y. Chang, Amilcare Gentili, and Chun-Nan Hsu. RadBERT: Adapting Transformer-based Language Models to Radiology. Radiology: Artificial Intelligence, 4(4):e210258, July 2022. [DOI] [PMC free article] [PubMed]
- 26.Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. BERT: pre-training of deep bidirectional transformers for language understanding. CoRR, abs/1810.04805, 2018.
- 27.Thomas Wolf, Lysandre Debut, Victor Sanh, Julien Chaumond, Clement Delangue, Anthony Moi, Pierric Cistac, Tim Rault, Rémi Louf, Morgan Funtowicz, et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations, pages 38–45, 2020.
- 28.Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, and Neil Houlsby. AN IMAGE IS WORTH 16X16 WORDS: TRANSFORMERS FOR IMAGE RECOGNITION AT SCALE. 2021.
- 29.Zhigang Li, Haidong Huang, Qiang Li, Konstantinos Zarogoulidis, Ioanna Kougioumtzi, Georgios Dryllis, Ioannis Kioumis, Georgia Pitsiou, Nikolaos Machairiotis, Nikolaos Katsikogiannis, et al. Pneumothorax: observation. Journal of Thoracic Disease, 6(Suppl 4):S421, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alistair EW Johnson, Tom J Pollard, Seth J Berkowitz, Nathaniel R Greenbaum, Matthew P Lungren, Chih-ying Deng, Roger G Mark, and Steven Horng. Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Scientific data, 6(1):1–8, 2019. [DOI] [PMC free article] [PubMed]
- 31.Alexey Tolkachev, Ilyas Sirazitdinov, Maksym Kholiavchenko, Tamerlan Mustafaev, and Bulat Ibragimov. Deep learning for diagnosis and segmentation of pneumothorax: the results on the kaggle competition and validation against radiologists. IEEE Journal of Biomedical and Health Informatics, 25(5):1660–1672, 2020. 10.1109/JBHI.2020.3023476 [DOI] [PubMed] [Google Scholar]