Abstract
Sinojackia Hu represents the first woody genus described by Chinese botanists, with all species classified as endangered ornamental plants endemic to China. Their characteristic spindle-shaped fruits confer high ornamental value to the plants, making them favored in gardens and parks. Nevertheless, the fruits likely pose a germination obstacle, contributing to the endangered status of this lineage. Here we report the chromosome-scale genome of S. xylocarpa, and explore the mechanisms underlying its endangered status, as well as its population dynamics throughout evolution. Population genomic analysis has indicated that S. xylocarpa experienced a bottleneck effect following the recent glacial period, leading to a continuous population reduction. Examination of the pericarp composition across six stages of fruit development revealed a consistent increase in the accumulation of lignin and fiber content, responsible for the sturdiness of mature fruits’ pericarps. At molecular level, enhanced gene expression in the biosynthesis of lignin, cellulose and hemicellulose was detected in pericarps. Therefore, we conclude that the highly lignified and fibrotic pericarps of S. xylocarpa, which inhibit its seed germination, should be its threatening mechanism, thus proposing corresponding strategies for improved conservation and restoration. This study serves as a seminal contribution to conservation biology, offering valuable insights for the study of other endangered ornamental plants.
Introduction
Sinojackia Hu (Ericales: Styracaceae) represents the first woody genus discovered and described by Chinese plant taxonomists [1]. It is a rare Chinese endemic genus comprising merely seven species: S. henryi (Dummer) Merr., S. huangmeiensis J. W. Ge & X. H. Yao, S. microcarpa Tao Chen & G. Y. Li, S. oblongicarpa Tao Chen & T. R. Cao, S. rehderiana Hu, S. sarcocarpa L. Q. Luo and S. xylocarpa Hu [2]. All seven species are listed as a Grade II endangered protected plant in China, mainly due to their small population size and rare, scattered distributions (http://www.iplant.cn/rep/protlist).
S. xylocarpa Hu is the representative and first-discovered species in the genus, known as the ‘jacktree’ in honor of the botanist John George Jack [1]. It is a flowering deciduous shrub or dwarf tree, reaching heights of about 4.6~6.1 metres and a diameter of nearly 10 centimetres at breast height. S. xylocarpa has a highly aesthetic and ornamental value, primarily due to its egg-shaped woody fruits (drupes) that resemble a balanced weight set (‘Chengtuo’ in Chinese). In autumn, the abundant egg-shaped fruits hang in cascading clusters, adding to its distinctive ornamental value, which can enhance the beauty of courtyards or parks. Additionally, during the spring flowering period, the tree is covered with abundant pure white blossoms (Fig. 1a). However, the extant population size of S. xylocarpa is very limited in nature, and its distribution is also extremely fragmented in the subtropical zone of Eastern China (e.g., Jiangsu and Zhejiang Provinces). Its small population size and narrow distribution are mainly attributed to its low germination rate, probably caused by the recalcitrant seeds [3]. Therefore, S. xylocarpa is classified as vulnerable (VU) in the International Union for Conservation of Nature and Natural Resources (IUCN) Red List of Threatened Species (https://www.iucnredlist.org/), attempting to raise public attention and promote protection. Botanical gardens, the primary protectors of endangered plants, have taken prioritizing action, introducing wild S. xylocarpa individuals into botanical gardens in Eastern China, such as Nanjing Botanical Garden Mem. Sun Yat-Sen and Shanghai Chenshan Botanical Garden, for decades.
Figure 1.
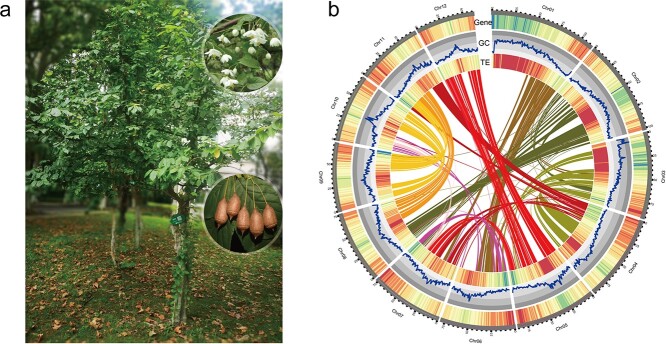
Morphology and high-quality genome assembly of the Sinojackia xylocarpa. a Flowering and fruiting branches of S. xylocarpa. b Genome features across 12 chromosomes of S. xylocarpa. From the outermost to innermost circles are chromosome ideograms, gene density (from blue to red), GC content, TE (transposable elements) density (from blue to red), and collinear genomic blocks.
Such strategies can certainly benefit the conservation of S. xylocarpa to a certain extent, potentially saving the species from extinction. In other words, by understanding the mechanism underlying the poor germination rate of S. xylocarpa seeds, we may be able to propose a highly targeted and efficient conservation plan to facilitate its sustainable recovery. Anatomically, S. xylocarpa seeds have extremely rigid external structure, consisting of indehiscent exocarp, corky mesocarp, lignified endocarp, and hard seed coat. The tough outer structure not only protects the seeds of S. xylocarpa but also renders them difficult to germinate within 2 to 3 years (http://www.efloras.org/). Unfortunately, the molecular mechanism underlying the mechanical barriers of the fruit pericarp to germination remains largely unknown, mainly due to the absence of a high-quality nuclear genome assembly for S. xylocarpa.
The reference genome is a fundamental resource for assessing, protecting, and restoring the biodiversity of endangered plants [4], including Ostrya rehderiana [5], Davidia involucrata [6], and Acer yangbiense [7, 8]. Genomic comparative analysis of the endangered ironwood O. rehderiana and its non-endangered relative Ostrya chinensis revealed the genomic effects of population collapse in this species [5]. Genomic analysis of the endangered dove-tree D. involucrata demonstrated that its endangered status might be primarily influenced by genomic factors, genetic diversity and population structure [6]. Whole-genome resequencing of 105 individuals from ten extant A. yangbiense populations revealed that their small population size might be related to low genetic diversity, repeated bottleneck events, and deleterious mutation load [8]. Genetic diversity of endangered plant species is crucial for conserving their genetic resources [9]. At present, our knowledge of the phylogeny and genetic diversity of Sinojackia and Styracaceae is limited to chloroplast genomes [10–14] and microsatellite markers [15–17].
Many plants in the order Ericales possess high economic and ornamental values, for instance, Rhododendron simsii (azalea) [18], Actinidia eriantha (kiwifruit) [19], Diospyros oleifera (persimmon) [20] and Camellia sinensis (tea) [21]. As a result, the genomes of these valuable plants have already been sequenced; however, no genomes of Styracaceae species are available. Here, we reported a chromosome-level assembly of the S. xylocarpa genome, which is the first sequenced nuclear genome in Styracaceae. Based on the high-quality reference genome, we conducted comprehensive analyses combining bioinformatics and experiments. Our study provides evidence from anatomy, gene expression, and population genomics to explore the mechanisms for the germination barriers of S. xylocarpa seeds and its narrow distribution. This work establishes a genomic foundation for investigating the molecular biology and evolution of S. xylocarpa, as well as for its biodiversity maintenance and restoration.
Results
Sequencing, assembly, and annotation of the S. xylocarpa genome
The genomic survey for the S. xylocarpa was conducted using Illumina data, and the estimated results by k-mer analysis (k-mer = 19) suggests a genome size of approximately 1.01 Gb, with a heterozygosity of around 0.91% (Fig. S1, see online supplementary material). To obtain a high-quality genome of S. xylocarpa, we employed a sequencing strategy combining high-fidelity (HiFi) long reads with a depth of nearly 17× (16.97 Gb) and Hi-C data of approximately 91× depth (98.6 Gb). The HiFi long reads were initially de novo assembled into 2138 contigs with a total length of 1072 Mb (N50 = 1.5 Mb). Subsequently, these contigs were ordered and oriented into 1168 scaffolds (N50 = 78.8 Mb) using Hi-C data. In total, 982 contigs (986 Mb) were anchored to 12 pseudochromosomes, representing for 91.98% of the entire S. xylocarpa genome (Fig. S2, see online supplementary material). Therefore, the resulting S. xylocarpa assembly is consistent with the estimated genome and should have achieved the chromosome-level in terms of continuity (Fig. 1b and Table 1).
Table 1.
Statistics of the S. xylocarpa genome assembly and annotation
| Statistic | Size |
|---|---|
| Genome size (Gb) | 1.072 |
| Number of scaffolds | 1168 |
| Scaffold length (bp) | 1072,438,512 |
| Scaffold N50 size (bp) | 78 843 817 |
| Scaffold N90 size (bp) | 61 418 785 |
| Number of contigs | 2138 |
| Contigs length (bp) | 1072,341,512 |
| Contigs N50 size (bp) | 1 500 000 |
| Contigs N90 size (bp) | 269 899 |
| Number of genes | 40 924 |
| Tandem repeats rate (%) | 6.98 |
| Transposable elements rate (%) | 53.75 |
| Complete BUSCO score (%) | 94.86 |
| GC (%) | 39.25 |
Gene annotation for the S. xylocarpa genome predicted a total of 40 924 protein-coding gene models using three gene prediction approaches: transcriptome-based, homology-based, and ab initio prediction. Among the predicted genes, 26 707 (65.26%) supported by transcriptome data, and 39 703 genes (97.02%) could be functionally annotated (Table S1, see online supplementary material). Our annotation identifies 1531 (94.86%) of the embryophyta benchmarking universal single-copy orthologs (BUSCOs) in the S. xylocarpa genome, with 1368 (84.76%) in single-copy and 163 (10.10%) in duplicate, indicating a high quality of gene prediction of S. xylocarpa genome.
Repeat sequences were also annotated, comprising 53.75% (576 Mb) of the assembly. Retroelements are the most abundant components (45.13%) among the repetitive sequences, with Gypsy and Copia comprising 21.39% and 8.46% of the genome (Table S2, see online supplementary material), respectively. Additionally, we identified 2111 transfer RNAs, 5315 ribosomal RNAs, 168 microRNAs, 152 small nuclear RNAs, and 129 small nucleolar RNAs in the S. xylocarpa genome.
Phylogenetic position of Ericales and relationships of lineages within Ericales
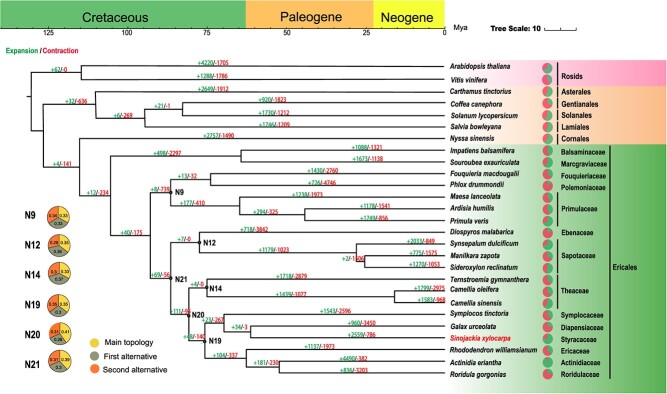
A total of 1291 low-copy orthologous genes were extracted from 27 angiosperm genomes and used to construct the sequence dataset for phylogenetic analyses. The concatenated amino acid dataset supported Ericales as the sister group of Cornales, another basal asterid lineage, with strong bootstrap support (100%, Fig. 2). The result based on the coalescent method was congruent (Fig. S3, see online supplementary material), suggesting a robust sister relationship for the two basal asterid orders.
Figure 2.
Genome evolution analysis of Sinojackia xylocarpa. Expansion and contraction of gene families and phylogenetic relationships and divergence times between S. xylocarpa and other plant species. The light green numbers represent the numbers of expanded gene families, and the red numbers represent the numbers of contracted gene families.
S. xylocarpa was resolved as the sister to Galax urceolata, a species in the Diapensiaceae family, and the two corresponding taxa diverged approximately 61.18 million years ago (Mya). The two species were sisters to Symplocos tinctoria (Symplocaceae) (Fig. 2). The monophyly of these three families has been robust and consistent across phylogenetic inferences based on organellar and nuclear data [22, 23]. This monophyletic group was further recovered as the sister to another lineage comprising Roridulaceae, Actinidiaceae, and Ericaceae. However, the relationships between some other lineages in Ericales remain ambiguous due to an ancient rapid radiation [23], with multiple lineages collapsed into a polytomy [24], including Theaceae, Sapotaceae, Ebenaceae, and Primulaceae. Our concatenated super-matrix provided hypotheses for these unresolved lineages, with Theaceae being sister to Pentaphylacaceae, Sapotaceae being sister to Ebenaceae, and Primulaceae being sister to the Polemoniaceae-Fouquieriaceae lineage. However, these inferences were all weakly supported and sometimes conflicted with the results from the coalescent analyses (Fig. S3, see online supplementary material). Incongruence and/or weak support in phylogenetic reconstruction may be due to incomplete lineage sorting (ILS), a consequence of random distribution of ancestral allelic polymorphism in derived lineages through rapid radiation [25]. Therefore, we examined the single gene trees regarding the relationships of these unresolved lineages. As expected, none of the relationships were predominant among all single gene trees, with nearly equal support for alternative relationships (Fig. 2; Fig. S3 , S4, and Table S3, see online supplementary material), suggesting substantial likely ILS during the evolution of Ericales.
Whole-genome duplication (WGD) events and karyotype evolution of S. xylocarpa
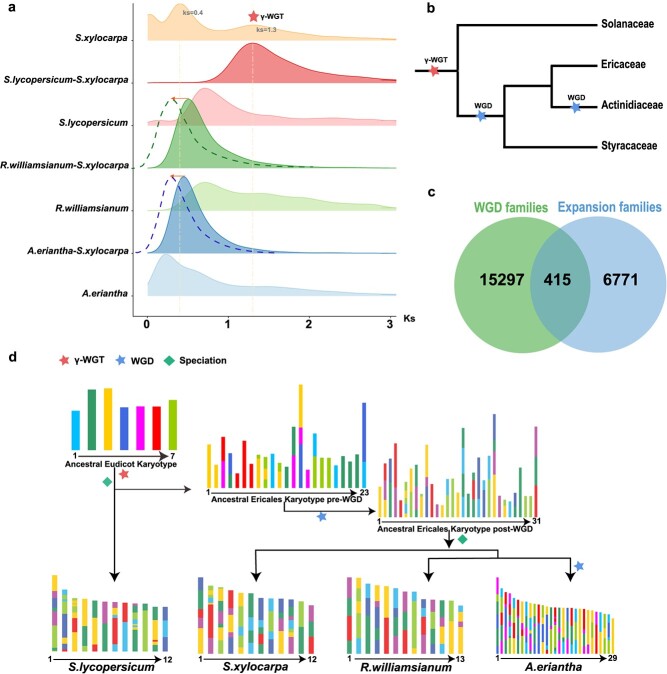
Previous genomic studies identified several independent whole-genome duplications (WGDs) in different Ericales taxa [18, 26, 27]. By comparing collinear genomic blocks (Fig. S5, see online supplementary material) and calculating the synonymous substitution rate (KS) of all paralogs in collinear regions (anchored paralogs) in the S. xylocarpa genome, we identified two Ks signature peaks approximately 1.3 and 0.4 (Fig. 3a), respectively, suggesting two rounds of WGD events.
Figure 3.
Whole-genome duplication in the Sinojackia xylocarpa genome. a Distribution of overall synonymous substitution levels (Ks) for paralogs found in syntenic blocks of Actinidia eriantha, Rhododendron williamsianum, S. xylocarpa, and Solanum lycopersicum and for orthologs between R. williamsianum and S. xylocarpa and between A. eriantha and S. lycopersicum. The yellow dotted line indicates two WGD events in S. xylocarpa. The Ks distribution of S. xylocarpa showed two peaks, one at approximately 0.3 (WGD 2) and another at approximately 1.3 (WGD 1). The arrows in different colors indicate overestimations (to the right) of the divergence events and point to the Ks values after corrections of different substitution rates based on that in S. xylocarpa. The dotted curves also show the orthologous distributions after substitution rate corrections. b Genome duplication in Ericales and outgroup. A red star indicates the core-eudicot whole-genome triplication (γ-WGT) event that occurs in conjunction with other species. Two blue stars indicate a WGD event that is commonly experienced in Ericales and an Actinidiaceae-specific WGD, respectively. c The Venn diagram shows the relationships among WGD families and expanded families in the S. xylocarpa genome. d Chromosome karyotype evolution from AEK to S. xylocarpa. A red star indicates a WGT event. A blue star indicates a WGD event. Green rhombus indicates speciation event. Different color blocks represent different ancestral chromosomes.
To accurately date the two WGDs occurred in S. xylocarpa, we compared the orthologous Ks distributions of S. xylocarpa with that of tomato (Solanum lycopersicum), azaleas (Rhododendron williamsianum) and kiwifruit (A. eriantha) (Fig. 3a). The Ks peak value of 1.3 for S. xylocarpa should correspond to the whole-genome triplication (WGT, γ) shared by all eudicots [27]. Therefore, it is logical that the peak of γ-WGT is slightly larger than the peak representing the divergence between S. xylocarpa and tomato. The more recent polyploidy event identified in S. xylocarpa should be a WGD, as indicated by the 2:1 ratio of collinear blocks (Fig. S5 and S6, see online supplementary material). However, the differential Ks among S. xylocarpa, azaleas, and kiwifruit resulted in an overestimate for the divergences between S. xylocarpa and the other two Ericlaes taxa. We therefore conducted a relative rate test to adjust the synonymous substitution, thus recognizing the more recent WGD in S. xylocarpa to be likely shared by Styracaceae, Diapensiaceae, and Symplocaceae (Fig. 3b).
When examining the retained paralogous genes (15712) after the recent WGD in S. xylocarpa, we found that they are partially overlapping with the expanded gene families (2559 families, 7186 genes, Fig. 3c) since the divergence with G. urceolata. GO analysis of these retained paralogs indicated that they are mainly enriched in ‘response to chemical and organic substance’, ‘developmental, catabolic and biological process’, and ‘organic substance catabolic and organonitrogen compound metabolic process’ with respect to ‘biological progress’, ‘ligase, transporter, acyltransferase, and kinase activity’, and ‘ATP binding’ with respect to ‘molecular function’ (Fig. S7, see online supplementary material).
WGDs are important evolutionary events that profoundly influence the genome evolution of organisms. Therefore, we reconstructed the process of karyotype variation of S. xylocarpa from the common ancestor of eudicots (Fig. 3d). The ancestral eudicot karyotype (AEK) was inferred to possess seven ancestral chromosomes before the γ-WGT [28]. Afterward, the triplicated AEK (21 chromosomes) underwent at least 10 chromosome fusions, 18 breaks, and six losses to form the karyotype in the common ancestor of S. xylocarpa, azaleas, and kiwifruit before their shared WGD (23 chromosomes). The recent WGD doubled the chromosome number, and 15 fusions, 0 breaks, and 0 losses reduced the chromosome number to 31 in the ancestor. Finally, extensive karyotype variations occurred (19 fusions, 0 breaks and 0 losses), leading to the current karyotype in S. xylocarpa.
Population genetic analyses of S. Xylocarpa
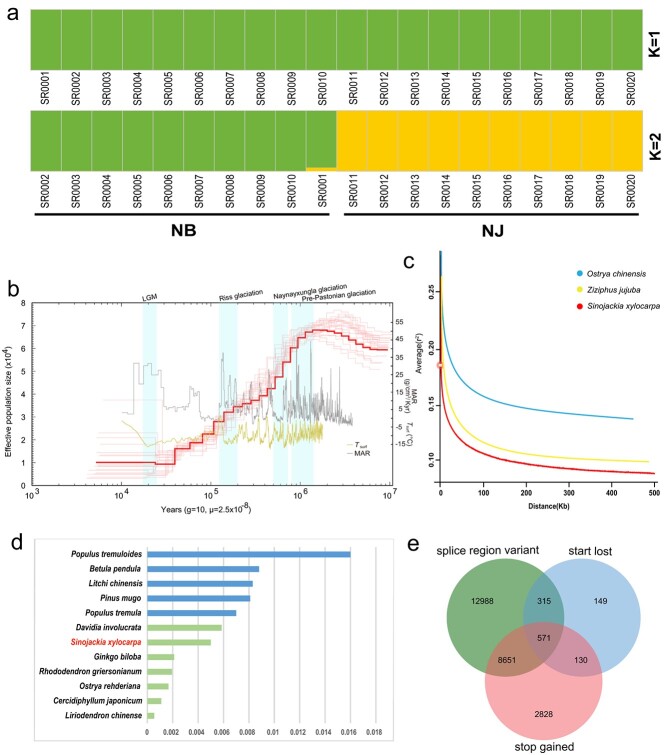
Natural populations of S. xylocarpa were found only in Nanjing, Jiangsu Province (118.39°N, 32.05°E) and Ningbo, Zhejiang Province (121.40°N, 30.08°E). We collected ten individuals from each of the two populations (Fig. S8, see online supplementary material) and conducted whole-genome resequencing on all 20 samples and obtained raw data with an average sequencing depth of 18.6×. Using a stringent GATK pipeline and strict filtering, we obtained a total of 43 817 152 single-nucleotide polymorphisms (SNPs), with approximately 65.79% of SNPs located in the intergenic regions and the remaining approximately 34.21% found in genic regions (Table S4, see online supplementary material). Phylogenetic analysis and principal component analysis (PCA) both revealed the clear divergence between the Ningbo population (Zhejiang Province) and the Nanjing population (Jiangsu Province) (Figs. S9 and S10, see online supplementary material). Based on the results of ADMIXTURE analysis, K = 1 was the best-fitted model (Table S5, see online supplementary material), indicating the small effective population size (Ne) usually possessed by the endangered species. However, the two populations (Nanjing and Ningbo) of S. xylocarpa were distinctly separated when K = 2 (Fig. 4a), consistent with the results from the PCA and the phylogenetic tree. We then estimated the historical changes of Ne; all sequenced S. xylocarpa individuals from the two populations showed a continuous decreasing trend over the recent one million years (Fig. 4b; Fig. S10 and Table S6, see online supplementary material), revealing the experienced bottleneck effect that was likely involved in the recent glacial periods. The gene flow analysis indicated the existence of bidirectional genetic communications between the two populations in NJ and NB, as well as a more predominant flow from NB to NJ (Fig. S12 and Table S7, see online supplementary material). This indicated that before the glacial period S. xylocarpa might have a continuous distribution area with frequent genetic exchange between different populations. During the glacial periods, its gene flow might be gradually restricted by habitat fragmentation and anthropogenic activities [29].
Figure 4.
Genetic diversity and demographic history of Sinojackia xylocarpa. a Admixture analysis with the number of clusters (K) ranging from 1 to 2. b Demographic history of S. xylocarpa. The last glacial maximum (LGM), Riss glaciation, the Naynayxungla glaciation, and Pre-Pastonian glaciation are highlighted in light-blue vertical bars. c Linkage disequilibrium (LD) decay of the S. xylocarpa populations and the two non-endangered species, including Ostrya chinensis and Ziziphus jujuba. d The observed genetic diversity (π) of S. xylocarpa and the remaining 17 species. e The Venn diagram of genes having three detrimental mutations, including splice region variant, loss of start codon, or gain of stop codons.
S. xylocarpa had a relatively low genome-wide nucleotide diversity (π) value (5 × 10−3), similar to other endangered plants, such as O. rehderiana (1.66 × 10−3) [5], D. involucrata (5.85 × 10−3) [6], and A. yangbiense (3.13 × 10−3) [8] (Fig. 4d), suggesting common genomic characteristics among endangered plants. Theoretically, the low genomic diversity appears to be an inevitable consequence of being endangered species, due to their small population size. This was also illustrated by the slow linkage disequilibrium (LD) decay within S. xylocarpa, in which half of the maximum r2 was not attained until ~500 kb. S. xylocarpa showed faster LD decay than two non-endangered species, including O. chinensis [5] and Ziziphus jujube [30] (Fig. 4c). We further estimated the inbreeding level via runs of homozygosity (ROH) (Fig. S13, see online supplementary material). The short ROHs occupied the majority (>99%) of the proportion, indicating the historical population bottleneck within S. xylocarpa.
Finally, we identified a total of 25 653 loss-of-function variations (Fig. 4e; Table S8, see online supplementary material) in S. xylocarpa. Among them, the variations belonging to start lost, stop gained, and splice region variant were 1165, 12 205, and 22 542, respectively. We found that 26 genes in the lignin synthesis pathway produced deleterious variations, with the most abundant being the splice region variant (Table S9, see online supplementary material). For identification of environment-associated genetic variants, we used different climate and soil factors to explore the environmental adaptation of S. xylocarpa. The results revealed that six environmental factors were most correlated with its environmental adaptation, with three temperature-related factors, namely annual mean temperature, isothermality, and mean temperature of warmest quarter, and three others related to soil conditions, namely soil clay content, cation exchange capacity, and soil organic carbon content have a significant impact on the growth of S. xylocarpa (Fig. S14 and Table S10, see online supplementary material).
Highly lignified pericarps leading to the germination obstacle for S. xylocarpa
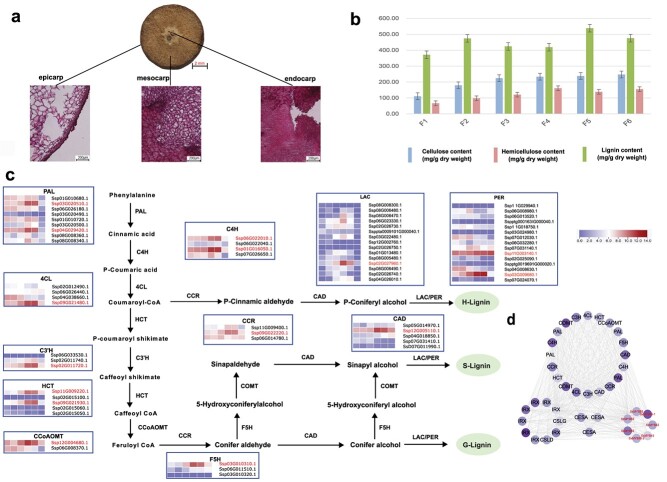
The stony pericarps can impose mechanical constraints on seed germination. A previous study proposed that the hard endocarp is likely the reason that inhibits the germination of S. xylocarpa seeds [3], which leads to the current small population size and extremely narrow distribution. To verify this hypothesis, we conducted an anatomical and staining experiment towards the longitudinal section of S. xylocarpa fruits. We observed that the seeds in the center were wrapped by hard and thick fruit pericarps composed of firm woody tissues and fibres, which is obviously the result of massive lignin and cellulose deposition (Fig. 5a). We subsequently measured the content of lignin, cellulose, and hemicellulose in pericarps at different developmental stages of fruits and found that the S. xylocarpa pericarps have continued to accumulate lignin, cellulose, and hemicellulose during the developmental process (Fig. 5b). This accumulation is likely the reason for the solid pericarps. Regarding the molecular mechanism, since the gene copy number of the LBP in S. xylocarpa does not significantly outnumber other species (Table S19, see online supplementary material), the underlying mechanism is likely related to gene expressions. When examining the lignin biosynthesis pathway (LBP), it is noticeable that some genes in the LBP showed a similar increasing expression pattern (gene IDs in red, Fig. 5c;Fig. S15, see online supplementary material), which is consistent with the trend of lignin content accumulation during fruit development. These genes include two PALs (phenylalanine ammonia lyases), two C4Hs (cinnamate 4-hydroxylases), two HCTs (shikimate hydroxycinnamoyl transferases), one C3H (p-coumarate 3-hydroxylase), one 4CL (4-coumaric acid: coenzyme A ligase), one COMT (catechol-O-Methyltransferase), one F5H (ferulate-5-hydroxylase), one CCoAOMT (caffeoyl-CoA 3-O-methyltransferase), one CCR (cinnamoyl-coenzyme A reductase), and one CAD (carbamoyl-phosphate synthetase), covering every catalysing step. There are other copies of LBP genes that did not show an increasing pattern or were expressed at low levels during fruit development. For example, one SxCAD4/5 copy (Ssp12G005110.1) displayed an increased and high expression, but the other copy (Ssp05G014970.1) was lowly expressed (Fig. 5c). We infer that these copies may be responsible for lignin biosynthesis in other organs/tissues, but not in pericarps, with different organ/tissue expressing specificity, or they may simply represent functional redundancy. Additionally, all LBP genes have undergone strong purifying selection during evolution (Table S11, see online supplementary material).
Figure 5.
The lignin biosynthesis pathways in Sinojackia xylocarpa fruit pericarp. (a) The cross-sections of S. xylocarpa pericarp. Lignin distributions of epicarp, mesocarp, and endocarp (from left to right) in S. xylocarpa pericarp were visualized via Safranin-O staining. (b) Mean values of cellulose, hemicellulose, and lignin contents of S. xylocarpa fruits during six different developmental stages. (c) Genes in the biosynthesis pathway of lignin and their expressions in pericarps. The genes marked in red are named as genes in the red module of the WGCNA analysis. (d) Gene regulatory networks associated with cellulose, hemicellulose, and lignin synthesis in the red module by WGCNA analysis, including the R2R3-MYB gene family involved in the regulation of lignin synthesis.
Naturally, in our constructed co-expression networks, the LBP genes showing a coincident increasing pattern were classified into the same module (red) by WGCNA (Fig. 5d; Table S12, see online supplementary material), suggesting that they are all potentially associated with lignin biosynthesis in S. xylocarpa pericarps. The high expression of some LBP genes may be associated with their tandemly arranged physical arrays on chromosomes, e.g., one 4CL, one HCT, and one CCR on chromosome 9, one CCoAOMT and one CAD on chromosome 12 (Fig. S16, see online supplementary material), and such gene clusters are probably beneficial for transcriptional efficiency of genes in the same pathway. Interestingly, in the same module, we can also find CesA (cellulose synthase) and IRX (Irregular Xylem 7) genes responsible for cellulose and hemicellulose biosynthesis, which also displayed an increasing pattern during pericarp development and can explain the cellulose and hemicellulose accumulation in the process. Of all genes in the module, Ssp11G009220.1 (HCT) and Ssp01G009950.1 (IRX) were estimated as hub genes with extensive connections to other genes (Table S12, see online supplementary material), suggesting that lignin and hemicellulose biosynthesis are important physiological activities during pericarp development. These results indicate the accuracy and concordance between molecular evidence and phenotypes in this study. The genes in the red co-expression module were mainly enriched in ‘RNA modification’, ‘nucleic acid phosphodiester bond hydrolysis’, ‘xylan biosynthetic process’, and ‘xylan metabolic process’ with respect to ‘biological progress’, and ‘hydrolase activity, acting on ester bonds’, ‘nuclease activity’, ‘endonuclease activity’, and ‘RNA binding’ with respect to ‘molecular function’ (Fig. S17, see online supplementary material). In addition, the XP-CLR analysis suggested that two genes associated with lignin and hemicellulose biosynthesis are under selective sweeps, i.e., Ssp03G010310.1 (F5H) and Ssp01G009950.1 (IRX7) (Fig. S18 and Table S13, see online supplementary material), suggesting the preservation of their functional roles in the pathway during evolution.
Previous studies identified a number of transcription factors (TFs) regulating lignin biosynthesis in plants [31–34], and they all belong to the R2R3-MYB family. Therefore, we took a further look into the co-expression network, and tried to identify key regulators that may govern the LBP genes during pericarp development. Firstly, phylogenetic analysis incorporating S. xylocarpa and Arabidopsis thaliana MYB genes were performed to determine all R2R3-MYBs in S. xylocarpa (Fig. S19, see online supplementary material), then 33 TFs closely connected to the LBP genes in the co-expression module were identified (Fig. S19, see online supplementary material), and finally they were cross-compared with the R2R3-MYBs to screen out the most promising regulators, which in our analysis, included seven genes, namely SxMYB20 (Ssp07G007490.1), SxMYB42 (Ssp10G023140.1), SxMYB62 (Ssp08G018180.1), SxMYB83 (Ssp10G002340.1), SxMYB85 (Ssp09G003180.1), SxMYB86–1 (Ssp09G018570.1), and SxMYB86–2 (Ssp11G007090.1) (Fig. 5d). These TFs may be the reason that triggers the high expression of LBP genes.
Discussion
To enhance plant conservation efforts for the threatened ornamental Sinojackia species, we assembled a high-quality genome of jacktree and conducted genomic resequencing of individuals in rare natural distribution habitats in Eastern China. To explore the phylogenetic positions of the jacktree and Ericales, we reconstructed a phylogenetic tree using the high-quality genome of the jacktree and another 26 angiosperms. However, our result is discordant with that of the Angiosperm Phylogeny Group IV (APG IV) [24], which recovers Cornales and Ericales as the successive sister groups to lamiids-campanulids. In fact, this discordance reflects the cyto-nuclear conflict, as plastid genome-based phylogenetic inferences mostly supported the same topology as APG IV [35–37], while recently growing nuclear evidences all favored the sister relationship between Cornales and Ericales [18, 38–40]. These data will serve as the foundation to the reconstruction of the evolutionary history and exploring the endangered mechanisms of species.
The egg-shaped fruits exhibit the distinctive morphological characteristics of S. xylocarpa, which are primarily appreciated for their ornamental value. The pericarp serves to protect the seeds from environmental factors and predators, which is a common preservation strategy of plants. For example, pericarp thickness is a defensive traits of Camellia japonica against its seed predator (Curculio camelliae) [41]. Similarly, it is very possible that the thickened S. xylocarpa pericarps might have played a protective role for seeds during the glacial age. However, it might become a factor limiting seed germination after the glacial period, as the seed germination is thus limited by the mechanical constraints of its pericarps [3]. Deleterious variations were detected in the LBP genes, yet these genes have maintained their functionality and exhibit high expression levels. While deleterious mutations typically compromise gene function, these genes exhibit evidence of strong purifying selection, indicating the influence of contradicting selective forces that preserve their functional integrity. The thickened pericarps due to the highly expressed LBP genes may be adaptive during the recent glacial age, which began about two Mya and lasted till now, with multiple subglacial and interglacial periods. Thus, the evolution pressure requires the retained functions of the LBP genes, in case of the coming subglacial period.
Nonetheless, the hard and lignified fruit pericarps (Fig. 5a) likely contribute significantly to the small population and limited natural distribution of this species. Firstly, the heavy, indurate, and inedible fruits are challenging to disperse over long distances by winds or animals, and animals are unlikely to find them palatable. Secondly, during seed germination, the highly lignified fruit pericarps are difficult to break, requiring a significant biomechanical force, likely resulting in a low seed germination rate [3]. We investigated the genetic mechanism underlying the development of its hard and lignified fruit pericarps. Our biomass analytical assay reveals that S. xylocarpa fruits exhibit increasing levels of lignin, as well as cellulose and semi-cellulose during fruit development and ripening. These findings support our hypothesis that the woody and fibrous content of S. xylocarpa pericarps is negatively correlated with their germination ratio [42], and that fruits valued for their ornamental characteristics create a virtual barrier to successive seed germination in this species.
Lignin biosynthesis is a crucial biological process in woody plants, with pathway enzyme genes present in all seed plants [43], and the resulting products often accumulating in various organs (e.g., seed coats, petioles, stems, and roots) [44, 45]. However, it appears that only S. xylocarpa fruit pericarps specifically accumulate high levels of lignin, likely related to the high expression of some, but not all LBP genes during fruit development (Fig. 5c). Because each LBP gene has multiple copies, largely due to tandem duplications (Fig. S19, see online supplementary material), it is possible that different copies have undergone functional divergence, leading to organ/tissue-specific catalytic activities. These highly expressed copies are likely the results of subfunctionalization and neofunctionalization, and are probably responsible for lignin biosynthesis specifically in fruit pericarps.
In addition to the functionally diverged enzyme genes themselves, their specific expression in fruit pericarps may also be influenced by regulatory elements. Previous studies have shown that TFs such as MYB, NAC (NAM/ATAF/CUC), and ARF (auxin response factor) play a regulatory role in lignin biosynthesis [46, 47]. For instance, Ding et al. found that MYB9, MYB60, and MYB91 might participate in the regulation of PAL, C4H, 4CL, CCoAOMT, and COMT in the seed coat of Brassica napus L. [48]. Through co-expression network analysis, we identified seven R2R3-MYB genes that are closely connected to the highly expressed LBP genes in S. xylocarpa fruit pericarps, suggesting they may function as potential regulators for these genes. Another explanation for the high expression of LBP genes could be attributed to the physical arrangement of upstream and downstream genes in the same signalling pathway, as they often form gene clusters, which is proposed to be functionally beneficial to transcriptional efficiency. Coincidentally, we observed the two clustered loci, collectively comprising five LBP genes (one 4CL, one HCT and one CCR on chromosome 9, one CCoAOMT and one CAD on chromosome 12, Fig. S16, see online supplementary material), and more importantly, these LBP genes are highly expressed in fruit pericarps (Fig. 5c), which strongly supports our hypothesis.
While the germination barriers of S. xylocarpa seeds can be explained based on the evidence from our experiments and multi-omics analyses, the ultimate question of how to efficiently conserve this endangered plant remains unanswered. Population genomic analysis not only reconstructed the evolutionary history but also provided insights into this ultimate question. Following the divergence of Nanjing and Ningbo populations, Nanjing continued to experience population size reduction, while Ningbo has maintained (or slightly recovered) its population size (Fig. 4b), resulting in Ningbo having a larger population size and a richer genetic diversity than Nanjing today. This discrepancy between the two populations may be attributed to different habitat conditions. Nanjing is an inland city, while Ningbo is located on the coast of the west Pacific and closer to the subtropics (Fig. S7, see online supplementary material), so the warm and moist air in Ningbo might soften S. xylocarpa fruit pericarps, thereby facilitating the germination process. Additionally, the two sharp declines in population size of S. xylocarpa correlate with the decline in atmospheric surface air temperature (Tsurf) and the escalation of the Chinese loess mass accumulation rate (MAR) (Fig. 4b), suggesting that its population evolutionary history was likely influenced by climatic changes and human activities. Based on these analytical results, we propose a strategy of providing additional artificial watering to the soil for in situ conservation. For ex situ conservation, priority can be given to botanical gardens in the coast areas of South and East China. Furthermore, the two populations (Nanjing and Ningbo) were genetically separated. Artificial cross-breeding is recommended as a means to increase its genetic diversity.
The interaction between plants and microbes can be considered as another conservation strategy. Certain microbes have the ability to degrade lignin in seed coat in an environmentally friendly and efficient way [49]. For instance, specific mycorrhizal fungi are required for the seed germination of threatened Paphiopedilum orchids [50]. S. xylocarpa seeds remain buried in soils for more than two years before germination. If there are specific microbes capable of degrading lignified S. xylocarpa fruit pericarps, we could isolate these microbes and apply them to the soils where S. xylocarpa fruits are buried for further propagation. This strategy aims to improve the germination rate of S. xylocarpa and can be adopted together with other strategies.
In summary, this work provides essential genomic data for the endangered ornamental plant jacktree, which serves as a valuable source for studying the evolutionary history and the endangerment mechanisms of this plant. Based on this data, a proper conservation and restoration plan has been proposed [4]. The molecular basis underlying high lignification in the S. xylocarpa pericarps suggests a direction for screening and developing new cultivars with low lignin content fruit pericarps, which may be better candidates for re-introduction into the wild fields to further enlarge the colony of S. xylocarpa. Owing to the similarity in biological features (e.g., lignified pericarps) among Sinojackia species, this study provides a starting point for exploring the causes of endangerment in other Sinojackia species. Additionally, this study provides a reference strategy in conservation biology, particularly for studies exploring the mechanisms of endangered ornamental woody plants.
Materials and methods
Genome survey and assembly
An individual of S. xylocarpa was cultivated on campus at Nanjing Forestry University (118.81°N, 32.08°E), Nanjing, Jiangsu Province, China. The plant material was selected for genome survey and genome assembly, and its fresh leaves were utilized for genomic DNA isolation. For the survey of the S. xylocarpa genome, the extracted DNA was served as establishing two paired-end (PE) libraries, each with a 300 bp insert. Sequencing of each PE library was conducted to generate ~30 Gb reads via Illumina NovaSeq 6000. For de novo genome assembly of S. xylocarpa, the extracted DNA was subjected to sequencing on a PacBio Sequel II system to yield HiFi long-read data, averaging 17.8 kb in length. For chromosome anchoring, Illumina NovaSeq 6000 was adopted to sequence a prepared Hi-C library [51], resulting in 150 bp PE reads.
The genome survey was conducted using GenomeScope (v2.0) [52] based on 60 Gb Illumina reads from two PE libraries. To construct a genome assembly, contigs were derived from de novo assembly of PacBio HiFi long-reads with the assistance of hifiasm (v0.12) [53]. The resulting contigs were then anchored onto 12 pseudochromosomes by using both HiC-Pro (v2.10.0) [54] and LACHESIS [55] with Hi-C datasets. Alignment of Hi-C reads against these resultant contigs was performed using BWA (v0.7.10-r789) [56].
Genome assembly assessment was processed in term of DNA read mapped rate and annotation completeness. Mapping of Illumina reads to the S. xylocarpa genome assembly was carried out using BWA. CEGMA (v2.5) [57] and BUSCO (v4.0.6) [58] were employed to appraise the gene completeness within the S. xylocarpa assembly, with results summarized in Tables S14 and S15, respectively (see online supplementary material).
Genome annotation
The S. xylocarpa genome was explored for protein-encoding genes searched with a synergistic application of transcriptome-based, sequence similarity-based and de-novo predicting approaches. In the transcriptomic evidence-based prediction method, GeneMarkS-T (v5.1) was utilized to infer the gene models from the transcripts of reference-guided assembly. This process involved alignment of RNA-Seq reads against the S. xylocarpa genome by HISAT2 (v2.0.4) [59], followed by genome-based assembly with using StringTie (v1.2.3) [60]. For the homology-based technique, these reference gene models were drawn from four species, namely Actinidia chinensis, A. thaliana, R. simsii, and Nyssa sinensis, using GeMoMa (v1.7) [61]. For ab initio prediction methods, the de-novo gene models were archived by a combination of Augustus (v2.4) [62] and SNAP from Korf Lab’s GitHub. Finally, the above three sets of gene models were merged using the EVidenceModeler (v1.1.1) [63], followed by refinement with PASA (v2.0.2) [64].
Protein-encoding genes were aligned and annotated with Diamond (v0.9.24) [65], with searches conducted on NCBI NR (Non-redundant) database, as well as SwissProt/TrEMBL and EggNOG (v5.0) [66]. Additionally, HMMER (v3.1) was adopted to delineate protein domains by searching against Pfam (v33.1) [67].
Non-coding RNAs (e.g., miRNA, rRNA, tRNA, snoRNA, and snRNA) were determined in the S. xylocarpa genome, and the utilized methods and databases were summarized in Table S16 (see online supplementary material). Transposon elements (TEs) were identified in the S. xylocarpa genome using a combination of RepeatModeler and RepeatMasker under RepBase database (v19.06). Annotation of terminal repeat retrotransposons (LTR-RTs) in the S. xylocarpa genome was accomplished using both LTRharvest [68] and LTR_finder [69]. TRF (v4.09.1) from Benson Lab’s GitHub and MISA (v2.1) were utilized to delineate tandem repeats in the S. xylocarpa assembly.
Phylogenetic analysis and estimation of divergence time
OrthoFinder (v2.5.4) [70] was applied to delineate orthologous gene clusters between S. xylocarpa and 26 other angiosperms. The low-copy orthologous genes were screened based on three criteria: (i) presence in ≥75% of genomes, (ii) ≤5 copies per orthologous gene group per species, and (iii) selection of the longest copy for phylogenetic analysis. A total of 1291 low-copy orthologous genes were predicted between them, and 13 145 homologous groups were identified in S. xylocarpa. Multiple sequence alignment (MSA) was accomplished using low-copy orthologous proteins via MUSCLE (v3.8.1551) [71], followed by constructing a well-supported maximum likelihood (ML) tree by IQ-TREE (v1.6.12) under the best-fitting model (JTT + F + R6) [72]. The MCMCTree program belonging to the PAML (v4.9) [73] package was utilized to date the divergence times of S. xylocarpa and the remaining 26 angiosperms. Three dated ages were chosen from TimeTree3 as standard normal priors, aligning with the speciation intervals for A. thaliana and Vitis vinifera (109–123 Mya), S. xylocarpa and Souroubea exauriculata (89–118 Mya), and Ardisia humilis with Primula veris (42–79 Mya). CAFE 5 [74] was employed to deduce the expansion and contraction patterns occurring in orthologous gene families across S. xylocarpa and the 26 other angiosperms.
Whole-genome duplication (WGD) events investigatory and ancestral karyotypes reconstruction
WGD software [75] was utilized to delineate the distribution of paralog ages, as indicated by synonymous substitutions per synonymous site (Ks) values. The MCL package was adopted to reconstruct gene family memberships using all potential paralogs which were deduced via all-vs-all protein BLAST [76], imposing an e-value of 10−10. MAFFT [77] was exploited for multi-alignment of each family. FastTree [78] was chosen to delineate a phylogenetic tree per gene families with n*(n-1)/2 ≤ ‘max airwise’. CODEML implemented in the PAML (v4.9) package [73] was employed to calculate ML-based Ks values for each gene pair. Mixture modelling was done for all inferred WGDs with the BGMM (Bayesian Gaussian Mixture Models) method. The WGDI (v0.6.2) [79] was employed to achieve collinear segment pairs. All syntenic blocks were determined with WGDI under ‘P-value = 0.05’ and the improved collinearity mode. The Ks pipeline in WGDI was adopted to infer the Ks value for each anchoring gene pair within a syntenic block, and the block mode was utilized to draw the Ks dotplot of all anchor pairs. The KsPeaks pipeline in WGDI was applied to delineate the Ks median value for each syntenic block. Finally, the Ks distributions were summarized and visualized using the R package ggplot2 (v3.5.0).
The pattern of chromosomal evolution within the order Ericales was reconstructed by utilizing well-defined polyploidization events and established phylogenetic relationships. Initially, we utilized WGDI [79] based on adjacent conserved collinear blocks to facilitate intra- or inter-genome comparisons, resulting in collinear dotplots annotated with Ks values. By including suitable outgroups and applying maximum parsimony rooted in well-defined phylogenetic relationships, we reconstructed ancestral karyotypes at every node within the phylogenetic tree of the Ericales. Lastly, by juxtaposing the acquired ancestral chromosomes with present-day species and elucidating the chromosomal evolution pattern of the Ericales.
Scanning electron microscopy and determination of lignin content
The drupes of S. xylocarpa at six developmental stages were harvested from trees located at the Xinzhuang campus of Nanjing Forestry University (118.81° N, 32.08° E). The drupes were collected every 1 month from April to September 2022, and the harvested samples were quickly frozen in liquid nitrogen and subsequently transferred to a −80°C refrigerator for storage. The content of lignin at six developmental stages was measured using a JC2203-M kit (JC DTECT Biotechnologies Co., Ltd, Nanjing, China) with triplicate samples per stage.
The fruits in the ripening stage were harvested for their morphological observation. The drupe samples were sliced into 20 μm thick sections consisting of exocarp, mesocarp, and endocarp, using a TU-213 sliding microtome (YAMATO, Saitama, Japan). A 1% aqueous Safranine O solution was utilized to stain the 20 μm-thick slices. The photographs of these qualified samples were acquisited on a BX51 microscope (Olympus, Tokyo, Japan), and processed with STL-IMCS software.
Transcriptome sequencing of pericarps and evolution analysis
The drupes were collected at six developmental stages with three biological replicates from S. xylocarpa trees cultivated at Nanjing Forestry University’s Xinzhuang campus (118.81°N, 32.08°E). Illumina RNA-Seq libraries were prepared from the RNAs that were isolated with RNAprep Pure Plant Kit (Tiangen, Beijing, China) and their sequencing was accomplished on Illumina NovaSeq 6000.
Raw reads were trimmed to remove adaptors, and low-quality reads (the reads with N ratio greater than 10% or whose base with Phread quality score [Q] ≤10 accounts for more than 50% of the whole reads) were discarded. These trimmed reads were mapped against the S. xylocarpa genome using Bowtie2 [80]. The calculation and normalization of gene expression levels were conducted via the FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) method [81]. RSEM software [82] was then used to perform the calculation of FPKM values. Gene expression heatmaps were visualized using TBtools [83]. BLASTP [76] was used to identify lignin synthase genes (including PAL, C4H, C3H, F5H, 4CL, CCR, CAD, COMT, and CCoAOMT) from the protein sequences of S. xylocarpa. The lignin synthase protein sequences of A. thaliana, used as the query, were obtained from Uniprot [84, 85] (Table S17, see online supplementary material). The protein sequences of lignin synthases from A. thaliana were aligned against all the protein sequence of S. xylocarpa, with an e-value <10−5, to obtain the potential lignin synthase protein sequences of S. xylocarpa. The conserved domains and conserved motifs in these candidate lignin synthase of S. xylocarpa were identified and inspected using TBtools and CD-search. To identify MYB candidates in S. xylocarpa, we downloaded the Hidden Markov Model (HMM) profile of MYB (PF00249) and used it as the query (P < 0.001) to search the S. xylocarpa protein sequences. A BLASTP search using A. thaliana MYB sequences from TAIR (https://www.arabidopsis.org/) as queries was accomplished with an e-value <10−5. The MYB family was finally determined by the protein sequences of the conserved domain. ClustalW was chosen for MSA [86]. IQ-TREE2 was hired to build ML trees under the ‘-alrt 1000 -B 1000’ parameter [87].
Genome resequencing and SNP calling
To conduct the population genomic analysis within the S. xylocarpa population, leaves of 20 wild samples were collected from Southeast China for genome resequencing. The natural distribution information for these samples is provided in Table S18 (see online supplementary material). Genome resequencing was conducted on the DNBSEQ-T7 sequencer to generate 150 bp PE reads with an average depth of ~18.6×.
All raw reads were trimmed with the software fastp (v0.12.4) [88] to eliminate low quality bases and adaptors, and the clean reads were then mapped to the S. xylocarpa genome using bwa-mem (v0.7.17) [89]. The SAM format files were processed using SAMtools (v1.15.1) [90] for sorting and merging. Picard (v2.25.0) was used for assigning read group information, including library, lane, and sample identity. GATK (v4.2.0.0) [91, 92] was adopted to predict SNPs. Subsequently, all these SNPs were annotated by ANNOVAR [93] with the ‘—neargene 2000’ option to define the length of upstream and downstream regions’ surrounding genes. The subsequent analyses were conducted using a set of 43 817 152 high-quality SNPs that had been identified. We utilized Plink (v1.90b6.21) for performing PCA (principal component analysis) and evaluating genomic diversity (π) [94]. We inferred the construction of the neighbour-joining (NJ) phylogenetic tree using Phylip (v3.697) [95] based on SNPs from genomic resequencing of 20 individuals. Population ancestry information was then calculated by Admixture (v1.3.0) [96], setting the K values from 1–6 for different numbers of clusters. PopLDdecay was employed to delineate the pattern of LD decay [97]. Long runs of homozygosity (ROHs) were sought using Plink (v1.90b6.21) within 50-SNP windows, where no heterozygous markers were allowed. ROHs were detected in regions longer than 10 kb with a minimum of 50 SNPs. ROHs were classified into different categories based on their lengths [98].
Demographic inference and identification of deleterious mutations
The PSMC model was employed with the settings [-N25 -t15 -r5 -p ‘4 + 25*2 + 4 + 6’] to depict ancient demographic history [99]. The mutation rate (u) was estimated using the formula u = Ks/2 T. The estimated divergence time (T) was about 62 Mya between S. xylocarpa and G. urceolata. The generation time was assumed to be 10 years based on the average time taken from seed to seed, as observed, and thus u = [0.31/ [(2 × 62e6)] × 10] = 2.5 e−8 (the substitutions per site per generation). To assess the accumulation of extreme deleterious mutations in the scale tree, the ‘-lof’ parameter of SNPEFF was used to annotate SNPs that lead to loss of function (LOF). XP-CLR (v1.0) was used to estimate the XP-CLR score for detecting signals of selective sweeps in the S. xylocarpa genome between two populations, Nanjing and Ningbo [100].
The joint site frequency spectrum (SFS) of S. xylocarpa from NB and NJ was utilized for estimating the parameters of evolutionary scenario. Various models of historical events were applied to the joint SFS of NB and NJ (Fig. S12, see online supplementary material). For convergence assurance, each model underwent 50 runs with varying starting points and the model with the highest likelihood was selected. The fastsimcoal2 program was adopted to analyse the gene flow between two subpopulations under the parameters [-m -0 -C 10 -n 100 000 -L 40 -s0 -M -q] [101]. Additionally, environmental adaptation analysis between two genetic groups (NB and NJ) was conducted using latent factor mixed models (LFMM) [102] and redundancy analysis (RDA) [103]. Nineteen climatic factors were downloaded from WorldClim (https://worldclim.org/) and 27 soil-related environmental variables were acquired from the National Earth System Science Data Center (https://www.geodata.cn).
Supplementary Material
Acknowledgements
We acknowledge Shi-Jing Sun at College of Materials Science and Engineering, Nanjing Forestry University for assistance in lignin determination. We also thank Xiao-Gang Xu at College of Life Sciences, Nanjing Forestry University, and Dao-Liang Yan at School of Forestry and Biotechnology Zhejiang A&F University, for help in collecting plant samples. We sincerely thank Wen-Tai Dai from the Institute of Soil Science, Chinese Academy of Sciences, for his assistance in soil data collection. We also appreciate the advice on data analysis provided by Jie-Yu Wang at China National Genebank (CNGB), Shenzhen, Cheng-ao Yang at College of Horticulture, Nanjing Agricultural University, and Yu-Peng Sang at College of Life Sciences, Sichuan University. We also acknowledge the data support from ‘National Earth System Science Data Center (https://www.geodata.cn)’. This work was financially supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author contributions
Y.-F.D. and J.-Y.X. conceived and designed the project. X.-F.W., S.Z., Y.-X.L., D.-W.Z., Z.-F.W., and J.G. conducted bioinformatic analyses. Y.-X.L., D.-W.Z., and S.-L.L. performed anatomical experiments and lignin determination. S.-L.L., Y.-F.S., Y.Y., X.-G.Y., and M.Z. collected the plant materials. S.Z., X.-F.W., Y.-X.L., Z.-F.W., and J.-Y.X. drafted the manuscript. All authors contributed to and approved the final manuscript.
Data availability
Assembled genome, DNA resequencing, and RNA sequencing data for S. xylocarpa are accessible from NGDC (National Genomics Data Center, https://ngdc.cncb.ac.cn/) under BioProject no. PRJCA016116, consisting of GWH (genome warehouse) no. GWHCBFM00000000, and GSA (genome sequence archive) no. CRA010572 and CRA016108, respectively. All relevant data can be found within the manuscript and its supporting materials.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Contributor Information
Sheng Zhu, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Xue-Fen Wei, College of Horticulture, Bioinformatics Center, Academy for Advanced Interdisciplinary Studies, Nanjing Agricultural University, Nanjing 210095, China.
Yu-Xin Lu, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Dao-Wu Zhang, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Ze-Fu Wang, College of Ecology and Environment, Nanjing Forestry University, Nanjing 210037, China.
Jing Ge, College of Horticulture, Bioinformatics Center, Academy for Advanced Interdisciplinary Studies, Nanjing Agricultural University, Nanjing 210095, China.
Sheng-Lian Li, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Yan-Feng Song, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Yong Yang, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Xian-Gui Yi, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Min Zhang, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
Jia-Yu Xue, College of Horticulture, Bioinformatics Center, Academy for Advanced Interdisciplinary Studies, Nanjing Agricultural University, Nanjing 210095, China.
Yi-Fan Duan, Co-innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China.
References
- 1. Hu HH. Sinojackia, a new genus of Styracaceae from southeastern China. J Arnold Arbor. 1928;9:130–1 [Google Scholar]
- 2. Xiao-Hong Y, Qi-Gang Y, Ji-Wen G. et al. A new species of Sinojackia (Styracaceae) from Hubei, Central China. Novon. 2007;17:138–40 [Google Scholar]
- 3. Wu Y, Bao WQ, Hu H. et al. Mechanical constraints in the endosperm and endocarp are major causes of dormancy in Sinojackia xylocarpa Hu (Styracaceae) seeds. J Plant Growth Regul. 2023;42:644–57 [Google Scholar]
- 4. Theissinger K, Fernandes C, Formenti G. et al. How genomics can help biodiversity conservation. Trends Genet. 2023;39:545–59 [DOI] [PubMed] [Google Scholar]
- 5. Yang Y, Ma T, Wang Z. et al. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat Commun. 2018;9:5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y, Ma T, Zhang L. et al. Genomic analyses of a ‘living fossil’: the endangered dove-tree. Mol Ecol Resour. 2020;20:756–69 [DOI] [PubMed] [Google Scholar]
- 7. Yang J, Wariss HM, Tao L. et al. De novo genome assembly of the endangered Aceryangbiense, a plant species with extremely small populations endemic to Yunnan Province, China. Gigascience. 2019;8:giz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma Y, Liu D, Wariss HM. et al. Demographic history and identification of threats revealed by population genomic analysis provide insights into conservation for an endangered maple. Mol Ecol. 2022;31:767–79 [DOI] [PubMed] [Google Scholar]
- 9. Formenti G, Theissinger K, Fernandes C. et al. The era of reference genomes in conservation genomics. Trends Ecol Evol. 2022;37:197–202 [DOI] [PubMed] [Google Scholar]
- 10. Yao X, Ye Q, Fritsch PW. et al. Phylogeny of Sinojackia (Styracaceae) based on DNA sequence and microsatellite data: implications for taxonomy and conservation. Ann Bot. 2008;101:651–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai XL, Landis JB, Wang HX. et al. Plastome structure and phylogenetic relationships of Styracaceae (Ericales). BMC Ecol Evol. 2021;21:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang LL, Zhang Y, Yang YC. et al. The complete chloroplast genome of (Ericales: Styracaceae), an endangered plant species endemic to China. Conserv Genet Resour. 2018;10:51–4 [Google Scholar]
- 13. Fan J, Fu QC, Liang Z. Complete chloroplast genome sequence and phylogenetic analysis of an endemic plant in Southwest China. Mitochondrial DNA Part B-Resources. 2019;4:1350–1 [Google Scholar]
- 14. Dong HJ, Wang HY, Li YL. et al. The complete chloroplast genome sequence of (Styracaceae). Mitochondrial DNA Part B-Resources. 2020;5:715–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao X, Zhang J, Ye Q. et al. Fine-scale spatial genetic structure and gene flow in a small, fragmented population of Sinojackia rehderiana (Styracaceae), an endangered tree species endemic to China. Plant Biol (Stuttg). 2011;13:401–10 [DOI] [PubMed] [Google Scholar]
- 16. Zhang JJ, Ye QG, Yao XH. et al. Microsatellite diversity and mating system of Sinojackia xylocarpa (Styracaceae), a species extinct in the wild. Biochem Syst Ecol. 2010;38:154–9 [Google Scholar]
- 17. Zhong TL, Zhao GW, Lou YF. et al. Genetic diversity analysis of Sinojackia microcarpa, a rare tree species endemic in China, based on simple sequence repeat markers. J For Res. 2019;30:847–54 [Google Scholar]
- 18. Yang FS, Nie S, Liu H. et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat Commun. 2020;11:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang W, Sun X, Yue J. et al. Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping. Gigascience. 2019;8:giz027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu QG, Xu Y, Yang Y. et al. The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic Res. 2019;6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang F, Zhang B, Wen D. et al. Chromosome-scale genome assembly of Camellia sinensis combined with multi-omics provides insights into its responses to infestation with green leafhoppers. Front Plant Sci. 2022;13:1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan M, Fritsch PW, Moore MJ. et al. Plastid phylogenomics resolves infrafamilial relationships of the Styracaceae and sheds light on the backbone relationships of the Ericales. Mol Phylogenet Evol. 2018;121:198–211 [DOI] [PubMed] [Google Scholar]
- 23. Larson DA, Walker JF, Vargas OM. et al. A consensus phylogenomic approach highlights paleopolyploid and rapid radiation in the history of Ericales. Am J Bot. 2020;107:773–89 [DOI] [PubMed] [Google Scholar]
- 24. The Angiosperm Phylogeny Group, Chase MW, Christenhusz MJM. et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20 [Google Scholar]
- 25. Kapli P, Yang ZH, Telford MJ. Phylogenetic tree building in the genomic age. Nat Rev Genet. 2020;21:428–44 [DOI] [PubMed] [Google Scholar]
- 26. Xia EH, Zhang HB, Sheng J. et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10:866–77 [DOI] [PubMed] [Google Scholar]
- 27. Wang X, Gao Y, Wu X. et al. High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol J. 2021;19:2544–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murat F, Armero A, Pont C. et al. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat Genet. 2017;49:490–6 [DOI] [PubMed] [Google Scholar]
- 29. Storfer A. Gene flow and endangered species translocations: a topic revisited. Biol Conserv. 1999;87:173–80 [Google Scholar]
- 30. Guo M, Zhang Z, Cheng Y. et al. Comparative population genomics dissects the genetic basis of seven domestication traits in jujube. Hortic Res. 2020;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. An C, Sheng L, Du X. et al. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic Res. 2019;6:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiao B, Zhao X, Lu W. et al. The R2R3 MYB transcription factor MYB189 negatively regulates secondary cell wall biosynthesis in Populus. Tree Physiol. 2019;39:1187–200 [DOI] [PubMed] [Google Scholar]
- 33. Sun Y, Ren S, Ye S. et al. Identification and functional characterization of PtoMYB055 involved in the regulation of the lignin biosynthesis pathway in Populus tomentosa. Int J Mol Sci. 2020;21:4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Q, Wang L, Wang Z. et al. The regulation of cell wall lignification and lignin biosynthesis during pigmentation of winter jujube. Hortic Res. 2021;8:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore MJ, Soltis PS, Bell CD. et al. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107:4623–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li HT, Yi TS, Gao LM. et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants. 2019;5:461–70 [DOI] [PubMed] [Google Scholar]
- 37. Li HT, Luo Y, Gan L. et al. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 2021;19:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeng L, Zhang N, Zhang Q. et al. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017;214:1338–54 [DOI] [PubMed] [Google Scholar]
- 39. One Thousand Plant Transcriptomes Initiative . One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang L, Su D, Chang X. et al. Phylogenomic insights into deep phylogeny of angiosperms based on broad nuclear gene sampling. Plant Commun. 2020;1:100027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toju H, Sota T. Phylogeography and the geographic cline in the armament of a seed-predatory weevil: effects of historical events vs. natural selection from the host plant. Mol Ecol. 2006;15:4161–73 [DOI] [PubMed] [Google Scholar]
- 42. Fang L, Xu X, Li J. et al. Transcriptome analysis provides insights into the non-methylated lignin synthesis in Paphiopedilum armeniacum seed. BMC Genomics. 2020;21:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue JY, Li Z, Hu SY. et al. The Saururus chinensis genome provides insights into the evolution of pollination strategies and herbaceousness in magnoliids. Plant J. 2023;113:1021–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Q, Luo L, Zheng L. Lignins: biosynthesis and biological functions in plants. Int J Mol Sci. 2018;19:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emonet A, Hay A. Development and diversity of lignin patterns. Plant Physiol. 2022;190:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007;12:64–70 [DOI] [PubMed] [Google Scholar]
- 47. Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet. 2010;44:337–63 [DOI] [PubMed] [Google Scholar]
- 48. Ding Y, Yu S, Wang J. et al. Comparative transcriptomic analysis of seed coats with high and low lignin contents reveals lignin and flavonoid biosynthesis in Brassica napus. BMC Plant Biol. 2021;21:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Atiwesh G, Parrish CC, Banoub J. et al. Lignin degradation by microorganisms: A review. Biotechnol Prog. 2022;38:e3226 [DOI] [PubMed] [Google Scholar]
- 50. Zeng S, Huang W, Wu K. et al. In vitro propagation of Paphiopedilum orchids. Crit Rev Biotechnol. 2016;36:521–34 [DOI] [PubMed] [Google Scholar]
- 51. Rao SS, Huntley MH, Durand NC. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng H, Concepcion GT, Feng X. et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Servant N, Varoquaux N, Lajoie BR. et al. HiC-pro: an optimized and flexible pipeline for hi-C data processing. Genome Biol. 2015;16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burton JN, Adey A, Patwardhan RP. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31:1119–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7 [DOI] [PubMed] [Google Scholar]
- 58. Simao FA, Waterhouse RM, Ioannidis P. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2 [DOI] [PubMed] [Google Scholar]
- 59. Kim D, Paggi JM, Park C. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pertea M, Kim D, Pertea GM. et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Keilwagen J, Hartung F, Grau J. GeMoMa: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol. 2019;1962:161–77 [DOI] [PubMed] [Google Scholar]
- 62. Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics. 2019;65:e57 [DOI] [PubMed] [Google Scholar]
- 63. Haas BJ, Salzberg SL, Zhu W. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Haas BJ, Delcher AL, Mount SM. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huerta-Cepas J, Szklarczyk D, Heller D. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mistry J, Chuguransky S, Williams L. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ou S, Jiang N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob DNA. 2019;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nguyen LT, Schmidt HA, Haeseler A. et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91 [DOI] [PubMed] [Google Scholar]
- 74. Mendes FK, Vanderpool D, Fulton B. et al. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 2021;36:5516–8 [DOI] [PubMed] [Google Scholar]
- 75. Zwaenepoel A, Van de Peer Y. Wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Camacho C, Coulouris G, Avagyan V. et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rozewicki J, Li S, Amada KM. et al. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 2019;47:W5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun P, Jiao B, Yang Y. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol Plant. 2022;15:1841–51 [DOI] [PubMed] [Google Scholar]
- 80. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Florea L, Song L, Salzberg SL. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2013;2:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen C, Chen H, Zhang Y. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202 [DOI] [PubMed] [Google Scholar]
- 84. Xiao R, Zhang C, Guo X. et al. MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int J Mol Sci. 2021;22:3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yao T, Feng K, Xie M. et al. Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front Plant Sci. 2021;12:704697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Oliver T, Schmidt B, Nathan D. et al. Using reconfigurable hardware to accelerate multiple sequence alignment with ClustalW. Bioinformatics. 2005;21:3431–2 [DOI] [PubMed] [Google Scholar]
- 87. Minh BQ, Schmidt HA, Chernomor O. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen S, Zhou Y, Chen Y. et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. 2013; arXiv:1303.3997, preprint: not peer reviewed [Google Scholar]
- 90. Danecek P, Bonfield JK, Liddle J. et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McKenna A, Hanna M, Banks E. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. DePristo MA, Banks E, Poplin R. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Purcell S, Neale B, Todd-Brown K. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences. Seattle: University of Washington; 2005: [Google Scholar]
- 96. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang C, Dong SS, Xu JY. et al. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–8 [DOI] [PubMed] [Google Scholar]
- 98. Yi HQ, Wang JY, Wang J. et al. Genomic insights into inter- and intraspecific mating system shifts in Primulina. Mol Ecol. 2022;31:5699–713 [DOI] [PubMed] [Google Scholar]
- 99. Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Excoffier L, Dupanloup I, Huerta-Sanchez E. et al. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Frichot E, François O. LEA: An R package for landscape and ecological association studies. Methods Ecol Evol. 2015;6:925–9 [Google Scholar]
- 103. van den Wollenberg AL. Redundancy analysis an alternative for canonical correlation analysis. Psychometrika. 1977;42:207–19 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.