Abstract
Synthetic data generation has emerged as a promising solution to overcome the challenges which are posed by data scarcity and privacy concerns, as well as, to address the need for training artificial intelligence (AI) algorithms on unbiased data with sufficient sample size and statistical power. Our review explores the application and efficacy of synthetic data methods in healthcare considering the diversity of medical data. To this end, we systematically searched the PubMed and Scopus databases with a great focus on tabular, imaging, radiomics, time-series, and omics data. Studies involving multi-modal synthetic data generation were also explored. The type of method used for the synthetic data generation process was identified in each study and was categorized into statistical, probabilistic, machine learning, and deep learning. Emphasis was given to the programming languages used for the implementation of each method. Our evaluation revealed that the majority of the studies utilize synthetic data generators to: (i) reduce the cost and time required for clinical trials for rare diseases and conditions, (ii) enhance the predictive power of AI models in personalized medicine, (iii) ensure the delivery of fair treatment recommendations across diverse patient populations, and (iv) enable researchers to access high-quality, representative multimodal datasets without exposing sensitive patient information, among others. We underline the wide use of deep learning based synthetic data generators in 72.6 % of the included studies, with 75.3 % of the generators being implemented in Python. A thorough documentation of open-source repositories is finally provided to accelerate research in the field.
Keywords: Synthetic data generation, Data privacy, Healthcare, Artificial intelligence, Tabular data, Imaging data, Radiomics data, Time-series data, Omics data, Multimodal data
Graphical Abstract
1. Introduction
The exponential growth in digital health technologies, such as electronic health records (EHRs), wearable health devices, genomic sequencing, medical imaging, mobile health application, and telemedicine, leads to a vast amount of daily generated data which can significantly enhance healthcare outcomes through advanced analytics and artificial intelligence (AI) [1], [2]. However, the sensitive nature of patient data limits their accessibility and poses significant obstacles in research and development [3], [4], [5]. Synthetic data are artificially generated data that can mimic real-world data without compromising the identity of the individuals. Thus, synthetic data offer a unique way to leverage the wealth of health information while preserving patient privacy with respect to regulations like the Health Insurance Portability and Accountability Act (HIPAA) in the U.S. or the General Data Protection Regulation (GDPR) in Europe. The value of synthetic data in healthcare is of great importance. Synthetic data can be used to improve the performance of AI models, to accelerate drug discovery through simulated clinical trials, to improve data accessibility by completing existing data and increasing data volume and to protect privacy by reproducing the original data avoiding any personally identifiable information (PPI) [6], [7], [8], [9], [10]. Thus, synthetic data do not only secure patient anonymity but also allow researchers to overcome barriers in data availability which empowers them to conduct a wide range of experiments and simulations without the risk of exposing the patients’ identity. Furthermore, synthetic data can facilitate the development of more diverse and accessible data to improve the generalizability of the AI models across diverse populations. This is particularly crucial in the case where data can be skewed or underrepresented in the context of harmful bias (e.g. age, race, gender), where synthetic data generation can be utilized as a mitigation methodology.
Data privacy is a critical concern in the healthcare domain considering the sensitive nature of personal health information [3], [4], [5], [11]. Any kind of data misuse or data breach can have severe implications for patients which in turn obscures their trust in AI systems. Synthetic data can ensure that personal identifiers are completely absent, thereby safeguarding patient confidentiality, while allowing researchers to harness meaningful knowledge. This is particularly important when developing AI models, where access to large scale data is crucial to ensure their increased accuracy and reliability. Considering that the more data the researchers’ access, the higher the risk of exposing sensitive information, the use of synthetic data mitigates any risk of exposing the real patient data. Through this way, the access to high-quality data is democratized, a fact that accelerates innovations in AI and data science. In addition, the use of synthetic data can lead to more robust and generalized AI models that perform well across various demographics and conditions, thereby improving their equity and effectiveness. On the other hand, synthetic data must maintain a balance between realism and privacy. This balance is critical especially in the healthcare sector, where the predictive accuracy of the AI models has significant effects on patient outcomes. Harmful biases which are often introduced in real data such as gender identity and sexual orientation, cultural and religious beliefs, language and communication barriers, geographic location, occupational hazards, and health insurance status, can be mitigated by creating balanced data that reflect the diversity of the affected populations [12].
Synthetic data can serve as a substitute for real data when training AI models. But how can we generate synthetic data? Synthetic data can be generated by capturing the statistical properties of the real data to create new data points with similar properties. According to the literature, a variety of methods has been proposed for the generation of high-quality synthetic tabular, imaging, radiomics, time-series, and omics data, which are categorized into: (i) statistical-based methods, like the multivariate normal distribution (MVND) and bootstrapping to generate virtual populations for hypertension drug programs [13], (ii) probabilistic-based methods, like the Stochastic Block Models (SBM) [14] to integrate multi‑omics data with consistent (common) and differential cluster patterns and the time-evolving graphs with metastability [15] to validate methods for capturing temporary changes in the time-evolving graphs for human microbiome analysis, (iii) machine-learning based methods like the tree ensembles [16], [17], [18] for data augmentation to improve the performance of disease progression and risk stratification models for cardiovascular and autoimmune diseases, the Gaussian Mixture Models (GMM) [19], [20], [21] to generate large-scale virtual populations, at reduced complexity, for in silico clinical trials, and the Hidden Markov Models (HMMs) [22] to generate realistic synthetic behavior-based sensor data for activity recognition in smart homes, and (iv) deep-learning based methods, which dominate the literature, like the virtual autoencoders (VAEs) to generate synthetic PPG signals [23] and myriad variations of the generative adversarial networks (GANs) like the Adaptive Deconfounding Synthetic GAN (ADS-GAN) to generate high-fidelity privacy-conscious synthetic patient data for causal effect estimation with multiple treatments [24], the Conditional GAN (CGAN) to generate realistic synthetic tabular data for benchmarking [25], the Wasserstein GAN with Gradient Penalty (WGAN-GP) to generate synthetic radiomics data from RT and CT images [26], the Copula GAN (CopulaGAN) for the generation of digital twins [27], the Multi-label Time Series GAN (MTGAN) to generate EHRs and simultaneously improve the quality of uncommon disease generation [28], the Transformer-Based Time Series GAN (TTS-GAN) to generate human heartbeat signals, timesteps, accelerator values, and sinusoidal waves [29], the Cycle-Consistent GAN (CycleGAN) [30], [31], [32], [33], and the Dual-Discriminator Conditional GAN (DDcGAN) for Multi-resolution PET and MR image fusion [34], among many others. However, two fundamental key aspects should be taken into consideration prior to the training of any synthetic data generator: (i) data anonymization, and (ii) data fidelity. Data anonymization refers to the process of removing personally identifiable information from the data, so that the patients remain anonymous whereas data fidelity refers to the degree to which synthetic data “mimic” the real data using a variety of metrics like the goodness of fit, correlation, and the Kullback-Leibler divergence, among many others [4], [6], [16]. High fidelity is vital to ensure that synthetic data can reliably replace real data without compromising data integrity.
Multimodality in healthcare refers to the use of multiple forms of data inputs (modalities) to aid in decision-making and patient care. These modalities can include tabular data (e.g., demographics, laboratory examinations, therapies, conditions), imaging data (e.g., CT, MRI, PET; and image based quantitative features which are referred to as radiomics), time-series data (e.g., ECG, EEG, PPG), and omics data (e.g., genomics, proteomics, lipidomics, metabolomics), among others, each providing different perspectives on patient health. The integration of these diverse data types presents unique challenges in data analysis, but it also offers a more holistic view of patient health leading to better outcomes. Synthetic data have a crucial role in this interplay since they can provide large and diverse data. However, privacy is an important factor which is not guaranteed by data fidelity. To this end, best practices should be adopted for data protection, clearer standards for assessing identifiability, and proportionate regulatory approaches to facilitate innovation while ensuring privacy. Thus, the availability of high-quality synthetic data can enable researchers to develop multimodal AI models. Furthermore, synthetic data can enable the simulation of complex patient scenarios that might not be frequently encountered in real datasets, thereby enhancing the robustness of healthcare systems against rare but critical conditions. Moreover, by utilizing synthetic data, researchers can bypass many logistical and ethical hurdles that occur during the aggregation and analysis of multimodal data, thus accelerating the pace of research. Ultimately, the use of synthetic data can significantly advance personalized medicine, improving treatment efficacy and patient outcomes while upholding stringent data privacy standards.
The current review aims to provide a thorough analysis of synthetic data generation methodologies, open-source repositories with codes and synthetic data to drive innovation and address common challenges more effectively across various healthcare domains, as well as, to improve the impact of synthetic data in targeted medical research and practice. The primary objectives of this review are the following: (i) to provide a better understanding of the methods that are used to generate synthetic tabular, imaging, omics, time-series data in healthcare, (ii) to provide open source repositories to implement these methods, (iii) to explore applications and benefits of using synthetic data in healthcare, (iv) to evaluate the impact of synthetic data on patient privacy and regulatory compliance, (v) to highlight the challenges and limitations of synthetic data, and (vi) to suggest future directions for research and development in this area.
2. Methods
2.1. Review process
We conducted a systematic review of the existing literature based on the PubMed and Scopus databases to ensure a robust thematic analysis of the different use cases on synthetic data generation technologies in healthcare based on high quality peer reviewed journals and international conferences. Our analysis focuses on five main types of data: tabular data, imaging data, radiomics data (image-based quantitative features), time-series data, and omics data. A special case on multimodal synthetic data generation cases was also investigated. A custom Python script was developed to automate the retrieval process. The script iterates over each year from 2015 to 2024 to apply the respective search query for each data type and retrieves the count of publications per year. The Scopus API (https://api.elsevier.com/content/search/scopus) and the PubMed API (https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi) were utilized to send HTTP requests with specific queries and additional parameters to obtain the total number of results. The function iterates over each year within the defined range (from 2015 to 2024), updating the query to include the publication date for that year, and retrieves the count of publications. These counts are stored in a dictionary for each data type. Once the metadata have been collected, the total counts per year and the final counts for each data type are calculated and saved into CSV files for further analysis.
Six individual database queries were designed and executed with a focus on the retrieval of studies which are related to the use/generation of: (i) synthetic tabular data (or virtual data or virtual population) by excluding papers related to imaging, text, videos, and time series data, to better capture advances in the healthcare domain focusing on clinical and lifestyle data (e.g. demographics, conditions, therapies, patient history), (ii) synthetic imaging data with a focus on the GANs or similar deep-learning architectures for the generation of synthetic medical images while excluding text, tabular data, videos, time series to tailor the query for medical imaging applications, (iii) synthetic radiomics data by exploring studies within the radiomics field involving the extraction of large amounts of features from medical images using data-characterization algorithms, (iv) synthetic time-series data by identifying papers with a focus on the generation and use of longitudinal, temporal data, and various biosignals like EEG, ECG, PPG, MEG, wearables, and vital sensors, (v) synthetic omics data by excluding text, tabular data, demographics, videos, imaging, and time series data with a focus on diverse biological fields like genomics, proteomics, and metabolomics, among others, and (vi) synthetic multimodal data by specifically targeting papers that combines multiple data modalities such as omics and imaging, time series and clinical data, imaging and clinical data, time series and imaging.
2.2. PRISMA flowchart
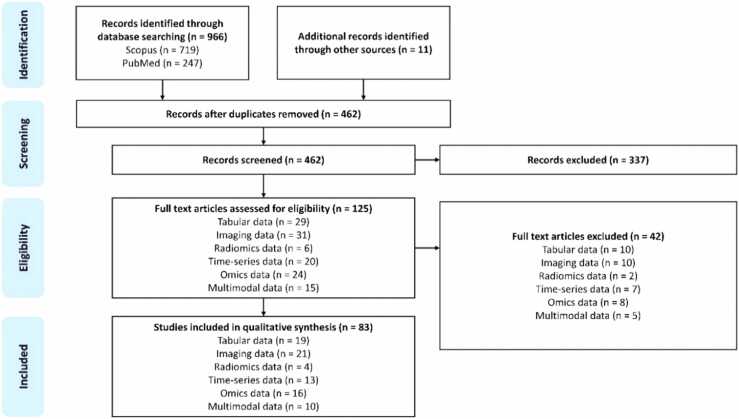
The PRISMA flowchart of the study is presented in Fig. 1 to summarize the multi-phase process of identifying, screening, assessing, and including studies in the review. The identification stage involves the collection of records through extensive database searches (966 records; 719 from Scopus and 247 from PubMed, and 11 additional records from other sources). After compiling these records, the screening phase follows to remove duplicate records due to overlapping indexing in the two databases yielding 462 records. The unique records then underwent an initial screening based on their titles and abstracts to quickly filter out clearly irrelevant studies by 4 independent researchers. The eligibility phase involves a detailed examination process, where full-text articles of the screened records were assessed against predefined criteria to ensure that only the studies that truly fit the review’s scope and quality requirements are included.
Fig. 1.
PRISMA flowchart for the systematic review including the database searches, the number of abstracts screened, and the full texts retrieved.
To this end, the number of full-text articles assessed was 124; tabular data = 29, imaging data = 30, radiomics data = 6, time-series data = 20, omics data = 24, multimodal data = 15). From those, 42 were excluded by filtering out articles which were: (i) not related to the fields of engineering, mathematics, and computer science, (ii) written in a non-English language, (iii) pre-prints. The final phase lists the 82 studies that passed the eligibility criteria. Those studies are presented as part of the qualitative synthesis of this review.
2.3. The synthetic data generation workflow
Fig. 2 depicts the core stages of the synthetic data generation workflow. It consists of four stages, including: (i) data acquisition, (ii) data preparation, (iii) data modeling, and (iv) data quality evaluation. The first stage involves the retrieval and management of real data. This includes ensuring proper permissions, data governance, and privacy compliance to handle sensitive information responsibly. The second stage involves the curation and transformation of the real data to make them suitable for modeling. This stage is crucial to make the data suitable for subsequent modeling. It involves handling missing values, normalizing data, and possibly augmenting the dataset to enhance its quality and representativeness. The third stage involves the development of models (statistical, probabilistic, machine learning, deep learning) to generate synthetic data that mimic the properties of the real data. The objective is to create synthetic datasets that retain the essential characteristics and patterns of the original data without revealing any sensitive information. The final stage focuses on the assessment of the generated synthetic data quality to ensure that they meet the required standards of fidelity, privacy, and utility.
Fig. 2.
The four stages of the synthetic data generation workflow.
3. Results
3.1. Summary of trends in the field
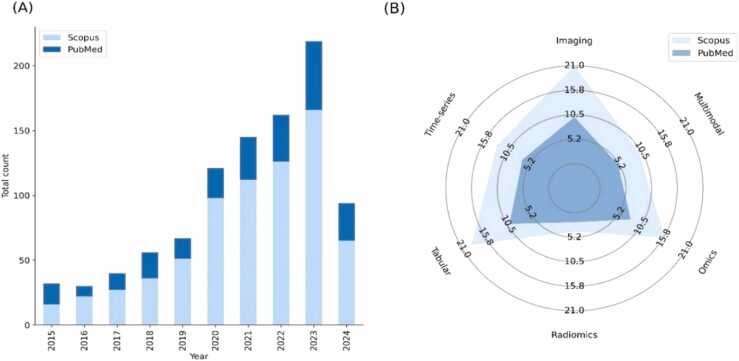
Fig. 3 summarizes the trends of the existing studies on synthetic data generation technologies in healthcare, from 2015 to mid. 2024 which highlights the growing interest within the field. More specifically, Fig. 3(A) illustrates the total number (counts) of publications per year, from 2015 to 2024, as indexed by PubMed and Scopus. It shows a significant increase in the number of publications over the years, where a significant rise is observed in 2023. Fig. 3(B) presents the distribution of the publications across various data types which are involved in synthetic data generation, including tabular, imaging, radiomics, time-series, omics, and multimodal cases. Each axis shows the count of publications from Scopus and PubMed, suggesting that synthetic imaging and tabular data generation are the most researched areas, whereas the time-series, omics, radiomics and multimodal data generation studies are fewer.
Fig. 3.
An overview of: (A) the total number of synthetic data generation studies in healthcare per year by PubMed and Scopus, and (B) the final number of studies across different data types (five main data types and multimodal data cases) by PubMed and Scopus.
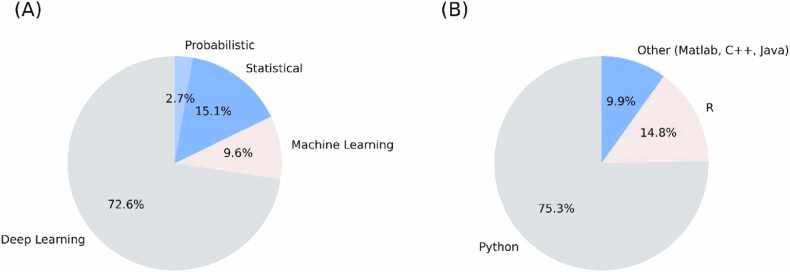
On the other hand, the types of methods and the programming languages which are used for synthetic data generation are depicted in Fig. 4 for the studies presented in Fig. 3(B), including publication trends, data type usage, methodological approaches, and programming languages. Deep learning appears to be the predominant method for synthetic data generation at 72.6 % of the studies, followed by statistical methods at 15.1 %, machine learning at 9.6 %, and probabilistic methods at 2.7 %. According to Fig. 4(B) Python is the most widely used language for 75.3 % of the studies, followed by R for 14.8 %, and other languages like C+ +, Java, and Matlab for 9.9 %.
Fig. 4.
Overview of methods and programming languages used for synthetic data generation in healthcare: (A) Types of methods used in the studies, (B) Programming languages used for the implementation.
3.2. Synthetic data generation methods and implementations per data type
3.2.1. Tabular data
The current methods for synthetic tabular data generation (including deep demographics, laboratory examinations, medical conditions, therapies, lifestyle data) can be grouped into statistical- and probabilistic-based, machine learning (ML)-based, and deep learning (DL)-based. The statistical- and probabilistic-based methods utilize statistical or probabilistic models to generate synthetic data based on the statistical distributions and relationships of the variables in the real data. Examples of such methods (Table 1) include bootstrapping, the multivariate normal distribution (MVND) and the log MVND [13], [18], [35], the Bayesian models [36], [37], [38], the vine copula models [39], the probabilistic Bayesian networks [18], [38], [40], and the Bayesian (hierarchical) generalized linear models (hGLM) [37]. According to Table 1, these methods have been used for: (i) the simulation of covariates in clinical trials, (ii) the generation of high-fidelity, large scale patient data, (iii) disease progression modeling, (iv) data augmentation to enhance the performance of disease classification and risk stratification models, and (v) the simulation of augmented clinical trials. The ML-based methods can overcome the statistical assumptions for specific distributions in the real data by capturing complex patterns. Examples of such methods (Table 1) include the supervised and unsupervised tree ensembles, the radial basis function (RBF)-based artificial neural networks (ANNs) [18], [36], the state-transition machines [41], [42], the sequential decision tree-based synthesizers [27], [43], [44], [45], the Gaussian Mixture Models (GMM), the Gaussian Mixture Models with Bayesian inference (BGMM) and the BGMM with optimal components estimation (BGMM-OCE) [19], [20]. These methods have been widely used (Table 1) for: (i) data augmentation for disease progression, (ii) transforming clinical patient data and modeling of disease progression, which are applied in various contexts including digital twin generation and replicability evaluation, (iii) large scale virtual population generation for in silico clinical trials. The DL-based methods leverage multi-layer artificial neural network (ANN) architectures to better capture nonlinearities and complex data interactions. Examples of such methods (Table 1) include different variations of the GANs), such as, the Adaptive Deconfounding Synthetic GAN (ADS-GAN), the Conditional GAN (CGAN), the Wasserstein GAN (WGAN) [24], [25], the Copula GAN (CopulaGAN), the Conditional Tabular GAN (CTGAN), the Medical GAN (MedGAN), and the RadialGAN (radial basis functions within a GAN), as well as, the Tabular Variational Autoencoder (TVAE), the Variational Autoencoders (VAEs), and the Tabular Denoising Diffusion Probabilistic Model (TabDDPM) [27], [43], [44], [46], [47]. These methods (Table 1) have been used for: (i) privacy-conscious synthetic data generation for clinical decision support, (ii) generating synthetic populations and digital twins, and (iii) improving the predictive performance on minority groups.
Table 1.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic tabular data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [13] | Simulation of covariates for clinical trials | Bootstrapping, MVND | R | https://cran.r-project.org/web/packages/mice/index.html | Summary statistics, RPE, RB, CC |
| [24] | Privacy-conscious synthetic data generation for causal effect estimation in treatment analysis | ADS-GAN | Python | https://github.com/tensorflow/tensorflow | WD, CC, SC, KRC, ε-identifiability |
| [36] | Data augmentation for disease classification and risk stratification | Bayesian models, tree ensembles, RBF-based ANNs | R, Python | https://cran.r-project.org/web/packages/semiArtificial/index.html | GOF, KLD, CC |
| [39] | To generate realistic virtual patient data in pharmacometrics | Vine copula models | R | https://github.com/vanhasseltlab/copula_vps | RECC, mean, standard deviation, median RE, PRC, DPs |
| [20] | Virtual population generation for in-silico clinical trials in HCM | BGMM | Python | https://github.com/scikit-learn/scikit-learn | CC, GOF, KLD |
| [40] | Synthetic dataset generation using Bayesian methods for clinical applications | Probabilistic Bayesian networks | OpenMarkov software | - | - |
| [19] | To generate high-quality, large-scale synthetic data at reduced computational complexity | BGMMO-CE | Python | https://github.com/vpz4/BGMM-OCE | cV, GOF, KLD, CC |
| [27] | Digital twin generation for personalized clinical trials | TabularSimulationBase, GaussianCopula, CopulaGAN, TVAE, CTGAN, MedGAN | Python | https://github.com/RyanWangZf/PyTrial | CC, WD |
| [43] | A comparative analysis of five distinct approaches for creating virtual data populations from individuals suffering from chronic coronary disorders |
Tabular Preset, Gaussian Copula, GANs, CTGAN, VAEs | Python | https://github.com/sdv-dev/SDV | KS test, CS test, CC, CSD |
| [37] | A Bayesian hierarchical method for combining in silico and in vivo data onto an augmented clinical trial with binary endpoints. | Bayesian (hierarchical) generalised linear models (hGLM) | R | https://cran.r-project.org/web/packages/rstan/index.html | KS test, CS test, CC |
| [41] | To develop a pipeline for transforming clinical patient data to conform with a model designed using OBO Foundry ontologies using synthetic data |
State-transition machines | Java | https://github.com/synthetichealth/synthea | GOF, CC, KS test, CS test |
| [18] | To predict disease progression for patients diagnosed with HCM during a 10-year period using synthetic data | MVND, log-MVND, RBF-based ANNs, tree ensembles, Bayesian networks | R, Python | https://cran.r-project.org/web/packages/deal/index.html, https://cran.r-project.org/web/packages/semiArtificial/index.html, scipy | CC, KS test, CS test |
| [25] | To develop realistic synthetic datasets suitable for validating digital health applications with a focus on clinical decision support systems. | CGAN, WGAN | Python | - | CC, JSI, GOF |
| [46] | To examine the usability of synthetic data in decision support systems, with a focus on data quality and security | CTGAN | Python | https://github.com/sdv-dev/SDV | - |
| [44] | To overcome the lack of high-fidelity datasets and ensure patient’s privacy | CTGAN, Gaussian Copula | Python | sklearn, imblearn, sdv | PCD, MMD, KLD |
| [42] | To develop a model of novel coronavirus (COVID-19) disease progression and treatment | State-transition machines | Java | https://github.com/synthetichealth/synthea | CC, KS test, CS test |
| [47] | To improve predictive performance on minority groups | RadialGAN, TabDDPM, CTGAN, TVAE | Python | https://github.com/vanderschaarlab/synthcity | JSD, WD, KLD, KS test, MMD, K-anonymity, K-map, L-diversity |
| [38] | To generate high-fidelity synthetic patient data based on UK primary care patient data | Bayesian networks | R | https://github.com/zhenchenwang/latent_model | - |
| [45] | To evaluate the replicability of analyses using synthetic data | Sequential decision tree-based synthesizer, GANs | Python, R | https://osf.io/vsku2/ | BEC |
The metrics which are used to measure synthetic tabular data fidelity and quality, include descriptive statistics (mean, median, standard deviation, variance-covariance, range, and proportions for categorical data), and more straightforward metrics, such as, the relative predictor error (RPE), the relative bias (RB), the Wasserstein distance (WD), the Pearson’s correlation coefficient (CC), the Spearman correlation (SC), the Kendall’s rank coefficient (KRC), the goodness of fit (GOF), the KL divergence (KLD), the relative error (RE), the polynomial regression coefficients (PRC), and the density plots (DPs). These metrics assess how well the synthetic data preserve the statistical properties, feature relationships and context of the real data. Additional statistical measures like the KS test (Kolmogorov-Smirnov test), the CS test (Chi-Squared test), the cosine similarity distance (CSD), the Jaccard similarity index (JSI), the pairwise correlation difference (PCD), the maximum mean discrepancy (MMD), the coefficient of variation (cV), the Jensen-Shannon distance (JSD), and the bias-eliminated coverage (BEC) are employed to ensure the synthetic data fidelity. Privacy metrics focus on ensuring the synthetic data does not compromise individual privacy, using measures, such as, the ε-identifiability, the K-anonymity, the K-map, and the L-diversity to evaluate re-identification risks. The majority of the metrics for the evaluation of the tabular data fidelity and quality, as well as, for the other types of data which are described next, are presented in [7].
3.2.2. Imaging data
The current advances in synthetic medical imaging data generation mainly rely on the deployment of GANs and several proposed variations of the GANs, as well as, on DL-oriented, specialized algorithms. GANs play a critical role in image synthesis. Examples (Table 2) include the Enhanced Balancing GAN, which is utilized for generating minority class images in imbalanced datasets [48], and other forms of GANs such as the Attention-based GAN [49], the CycleGAN [30], [31], [32], [33], and the Dual-Discriminator Conditional GAN (DDcGAN) [34] applied in tasks ranging from medical image enhancement to cross-modality image synthesis. Other specialized GAN variants, such as, the Progressively Growing GANs [50] and the Style Distribution GAN (SD-GAN) [51] focus on generating clinically realistic X-rays and transferring style distributions in images, respectively. Other DL-oriented approaches include a variety of neural network architectures beyond traditional GANs, such as, the Conditional Variational Autoencoder [52] and the Contrastive Diffusion Model [53] which are notable for their performance in generating realistic, high-resolution images and fine-detail PET reconstruction. Furthermore, Vision Transformers [54], have shown their potential in fast MRI reconstruction by harnessing the capabilities of transformer models, which have been successful in natural language processing but lately used on image classification and segmentation tasks. Furthermore, the Ensemble of Convolutional Neural Networks [55], which is a combination of DL approaches, enhances the detection of out-of-distribution objects in imaging data, which is crucial for reliable medical diagnosis. Similarly, Normalizing Flows [56] have been employed to mitigate the effects of CT acquisition and reconstruction anomalies, providing more accurate and consistent imaging outputs. Finally, a pythonic library containing multiple pre-trained GAN-based models (CT-GAN, WGAN, SinGAN, PGGAN, FastGAN, pix2pix) has been also reported, named medigan [57], to allow researchers to access, generate, and benefit from synthetic medical imaging data, including mammographies, brain MRI, endoscopy, chest X-ray, cardiac MRI, and breast DCE-MRI, among others.
Table 2.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic imaging data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [48] | Enhanced Balancing GAN for Minority Class Image Generation | Enhanced Balancing GAN | Python | https://github.com/GH920/improved-bagan-gp | FID, IS |
| [55] | Efficient Data Augmentation Network for Out-of-Distribution Image Detection | Ensemble of Convolutional Neural Networks | Python | https://github.com/majic0626/Data-Augmentation-Network | - |
| [49] | Blind Degradation Modelling for High-Resolution Medical Images (BliMSR) | Attention-based GAN | Python | https://github.com/Samiran-Dey/BliMSR | - |
| [52] | Conditional Variational Autoencoder with Balanced Pre-training for GANs | Conditional Variational Autoencoder, GAN | Python | https://github.com/alibraytee/CAPGAN | FID, SSIM |
| [53] | Contrastive Diffusion Model with Auxiliary Guidance for Coarse-to-Fine PET Reconstruction | Contrastive Diffusion Model | Python | https://github.com/Show-han/PET-Reconstruction | PSNR, SSIM, NMSE |
| [30] | Correction of Out-of-Focus Microscopic Images by Deep Learning | CycleGAN | Python | https://github.com/jiangdat/COMI | PSNR, SSIM, CC |
| [56] | CTFlow: Mitigating Effects of CT Acquisition and Reconstruction with Normalizing Flows | Normalizing Flows | Python | https://github.com/hsu-lab/ctflow | PSNR, SSIM, LPIPS |
| [34] | Dual-Discriminator Conditional GAN for Multi-Resolution Image Fusion (DDcGAN) | Dual-Discriminator Conditional GAN | Python | https://github.com/jiayi-ma/DDcGAN | entropy, mean gradient, spatial frequency, PSNR, SSIM, CC, VIF |
| [31] | Endoscopic Ultrasound Image Synthesis Using a Cycle-Consistent Adversarial Network | Cycle-Consistent Adversarial Network | - | https://ebonmati.github.io/ | FID |
| [32] | DC-cycleGAN: Bidirectional CT-to-MR synthesis from unpaired data | CycleGAN | Python | https://github.com/JiayuanWang-JW/DC-cycleGAN | PSNR, SSIM, MAE |
| [54] | Fast MRI Reconstruction: How Powerful Transformers Are? | Vision Transformer | Python | https://github.com/ayanglab/SwinGANMR | PSNR, SSIM, FID |
| [58] | Flow-Based Visual Quality Enhancer for Super-Resolution Magnetic Resonance Spectroscopic Imaging | Flow-Based Network | Python | https://github.com/dsy199610/Flow-Enhancer-SR-MRSI | PSNR, SSIM, LPIPS |
| [59] | HQG-Net: Unpaired Medical Image Enhancement with High-Quality Guidance | Combination of Enlighten & Still GANs | Python | https://github.com/ChunmingHe/HQG-Net | PSNR, average gradient, ENIQE, BRISQUE |
| [60] | Image Augmentation Using a Task-Guided Generative Adversarial Network for Age Estimation on Brain MRI | Task-Guided GAN | Python | https://github.com/ruizhe-l/tgb-gan | MSE, MAE |
| [61] | On Data Augmentation for GAN Training | Data Augmentation for GANs | Python | https://github.com/sutd-visual-computing-group/dag-gans | FID, IS, KLD |
| [50] | Evaluating the Clinical Realism of Synthetic Chest X‑Rays Generated Using Progressively Growing GANs | Progressively Growing GANs | Python | https://github.com/BradSegal/CXR_PGGAN | FID, Human eYe Perceptual Evaluation |
| [51] | SD-GAN: A Style Distribution Transfer Generative Adversarial Network | Style Distribution GAN (SD-GAN) | Python | https://github.com/tasleem-hello/SD-GAN/tree/SD-GAN | PSNR, SSIM |
| [62] | Self-Supervised Visual Representation Learning for Histopathological Images | CS-CO: hybrid self-supervised visual representation learning method tailored for H&E-stained histopathological images | Python | https://github.com/easonyang1996/CS-CO | - |
| [63] | Slice Profile Estimation From 2D MRI Acquisition Using Generative Adversarial Networks | GAN | Python, Docker | https://github.com/shuohan/espreso | MAE, PSNR, SSIM |
| [33] | StainGAN: Stain Style Transfer for Digital Histological Images | CycleGAN | Python | https://github.com/xtarx/StainGAN | PSNR, SSIM, FSIM, CC |
| [57] | medigan: A complete pythonic library with multiple pre-trained GANs for the generation of synthetic medical imaging data (mamographies, brain MRI, endoscopy, chest X-ray, cardiac MRI, breast DCE-MRI) | CDGAN, CycleGAN, WGAN-GP, C-DCGAN, PGGAN, FastGAN, SinGAN, pix2pix | Python | https://github.com/RichardObi/medigan | FID |
The metrics which are widely deployed to assess the synthetic imaging data fidelity and quality include the Frechet Inception Distance (FID) and the Inception Score (IS), which assess the similarity of synthetic data to real data by comparing feature distributions and evaluating the performance of image classifiers. The Structural similarity index measure (SSIM), the peak signal to noise ratio (PSNR), the normalized mean squared error (NMSE), and the mean average error (MAE) are also used to quantify the visual and statistical similarity between synthetic and real images. Furthermore, the Learned Perceptual Image Patch Similarity (LPIPS), the entropy, the mean gradient, the spatial frequency, the correlation coefficient, and the Visual Information Fidelity (VIF) are further used to assess the perceptual and statistical properties of the synthetic imaging data. Additional metrics, such as, the Natural Image Quality Evaluator (ENIQE), the Blind Reference Image Spatial Quality Evaluator (BRISQUE), the mean squared error (MSE), and the feature similarity index for image quality assessment (FSIM) can provide more detailed evaluations of the synthetic image quality.
3.2.2.1. Radiomics data (Image-based quantitative features)
Radiomics data consist of quantitative features which are extracted by medical images. They formulate a critical subfield of medical imaging data. According to Table 3, most of the studies which focus on synthetic radiomics data generation are mainly DL-based using methods, such as, the WGAN-GP [26], the CTGAN [52], the TVAE and the Copula GAN to offer enhanced flexibility and capacity to capture complex data distributions. On the other hand, the tabular Preset and the Gaussian Copula are the two statistical methods that have been used for synthetic radiomics data generation, relying on the statistical properties of the real-world training data [64]. These methods harness the power of adversarial networks to learn the underlying data distribution and generate synthetic data that closely resemble real-world radiomic features. Several attempts have been also reported towards the generation of synthetic radiomic images like the RadSynth [64] which is a deep CNN-based model that produces synthetic GLCM (Grey Level Co-occurrence Matrix) entropy images.
Table 3.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic imaging data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [26] | To apply the WGAN-GP algorithm to generate radiomics data. | WGAN-GP | Python | https://github.com/EmilienDupont/wgan-gp | t-SNE |
| [66] | Developed a CNN model to efficiently generate radiomics data. | RadSynth | - | - | CC, BA plot |
| [65] | To combine MRI-Based Radiomics with DL-based data augmentation for differentiating IDH-mutant grade 4 astrocytomas from IDH-wild-type glioblastomas. | CTGAN | R, Python | https://github.com/sdv-dev/CTGAN, https://github.com/kasaai/ctgan?tab=readme-ov-file | - |
| [64] | To evaluate the potential of synthetic radiomic data generation in addressing data scarcity in radiomics/ radiogenomics models. | Tabular Preset, Gaussian Copula, TVAE, CTGAN, Copula GAN | Python | https://github.com/sdv-dev/SDV | CS test, basic statistical correlation test |
The metrics which are used to measure the fidelity and quality of the synthetic radiomics data, include the Distributed Stochastic Neighbor Embedding (t-SNE), which is a dimensionality reduction technique used to visualize high-dimensional data and assess clustering and distribution similarities between synthetic and real data. The correlation coefficient (CC) is also used to measure the linear relationship between real and synthetic data. The Bland-Altman (BA) plot is used to compare two measurement techniques by plotting the differences between synthetic and real data against their averages, helping to identify any systematic differences. In addition, the Chi-Square (CS) test is often deployed to compare the distributions of categorical variables in synthetic and real data, assessing how well the synthetic data matches the distribution of real data. Basic statistical correlation tests further evaluate the preservation of statistical properties in synthetic data.
3.2.3. Time series data
The methods for synthetic time series data generation (including electrocardiogram (ECG), photoplethysmographic (PPG), sensor-based measurements, longitudinal observations, and other biosignals) can be split into statistical- and probabilistic-based, ML-based, and DL-based. The statistical-based methods rely on several statistical principles and probabilistic models. One noticeable approach is the Guided Evolutionary Synthesizer (GES), which integrates genetic algorithms, concept maps, and randomness operators [67]. Another significant statistical-based method is the statistical feature space selection, which involves identifying critical features and using them for representative sampling [23]. The Synthetic Acute Syndromes Creator (SASC) utilizes summary statistics and internal correlations, maintaining cross-patient consistency [68]. Additionally, SASC utilizes random generation under constraints focusing on single-parameter distributions and their relative correlations [68]. The above-mentioned approaches (Table 4) demonstrate the adaptability and robustness of statistical-based methods in generating synthetic time series data. According to Table 4, these methods have been widely used for: (i) adversarial learning on biosignal data, (ii) generating synthetic data considering metadata as part of the generation process, (iii) augmenting sensor-based data, (iv) synthesizing time series EHR data and tackling the imbalance of uncommon diseases, (v) multivariate time series generation, (vi) employing existing generative models to produce medical time series, (vii) generating realistic synthetic time series data sequences of arbitrary length and (viii) generating ECG data.
Table 4.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic time-series data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [70] | Develop a platform for providing synthetic data considering metadata as part of the time series generation process. | WGAN-GP, DGAN | - | - | PRD plots, DLA, Autocorrelation, MAE, CC |
| [74] | Generate synthetic ECG data utilizing diffusion-based techniques. | SSSD-ECG model based on the DiffWave architecture, WaveGAN* , Pulse2Pulse | Python | https://github.com/AI4HealthUOL/SSSD-ECG | Utilizing a reference model for assessing the realism of the synthetic data |
| [75] | Novel generative model for medical time series generation. | Causal Recurrent Variational AutoEncoder (CRVAE) | Python | https://github.com/hongmingli1995/CR-VAE | MMD, MSE |
| [67] | Framework for bias analysis in healthcare time series data | Guided Evolutionary Synthesizer (GES) | - | - | Bias score for bias mitigation |
| [71] | A Generative Adversarial Network (GAN) architecture for sensor-based health data augmentation | TS-GAN | - | - | Discriminator loss, MMD, t-SNE and PCA |
| [23] | Generation of synthetic PPG data using an in-silico cardiac model | Variational Autoencoder (VAE) | - | - | Mainly based on classification/prediction performance |
| [68] | An efficient approach for generating longitudinal observational patients cohorts | Classical statistical distribution, Summary statistics, Internal correlations | R | https://github.com/Fraunhofer-ITMP/SASC | Correlation plots between the correlations of real and synthetic data |
| [28] | Generate time series EHR data and imbalance uncommon diseases. | Multi-label Time series GAN (MTGAN) | Python | https://github.com/LuChang-CS/MTGAN | GT, JSD, ND |
| [72] | A novel framework for multivariate time series generation | COmmon Source CoordInated GAN (COSCI-GAN) | Python | https://github.com/aliseyfi75/COSCI-GAN | AED, WD, MAE, Frobenius norm, SC, KRC |
| [73] | Employing existing generative models to produce medical time series | HealthGAN, Wasserstein GAN, TimeGAN | Python | https://bitbucket.org/mvdschaar/mlforhealthlabpub/src/master/alg/timegan/ | AHEC, Welsch t-test |
| [29] | A transformer-based GAN generating realistic synthetic time series data sequences of arbitrary length | TTS-GAN | Python | https://github.com/imics-lab/tts-gan | t-SNE, PCA, ACS, JSD |
| [69] | A broad analysis on adversarial learning on biosignal data | GAN, Adversarial AutoEncoder, Modality Transfer GAN | Python | https://github.com/theekshanadis/biosignalGANs | Mainly based on classification/prediction performance |
| [22] | Synthetic time series data generation that is composed of nested sequences | Combination of HMM and regression algorithms, Time series distance measures | Python | https://github.com/jb3dahmen/SynSys-Updated | AED, DTW |
The ML-based methods for synthetic time series data generation vary from conventional supervised learning algorithms to advanced AI modeling. An example of such a method (Table 4) is the two-level Hidden Markov Models (HMMs) with regression learners [22], where the first-level HMM generates realistic sequences of activities, while the second one creates sensor events reflective of those activities. Regression learners apply statistical regression to capture time gaps and the duration of each activity, ensuring accurate representation of time series data. The latter is typically more flexible and adaptive compared to statistical-based methods. ML-based methods are widely used for generating synthetic time series data composed of nested sequences. On the other hand, DL-based methods lie in the core of synthetic generation of healthcare related time-series data. They often rely on GANs [69], which consist of a trained generator on the real dataset that produces the synthetic data and a discriminator that evaluates its reliability. One notable approach is the Wasserstein Generative Adversarial Network with Gradient Penalty (WGAN-GP) [70], which enhances traditional GANs by stabilizing training and improving convergence. DoppelGANger (DGAN) [70] introduces a unique approach by generating metadata with a Multi-Layer Perceptron (MLP). Time Series Generative Adversarial Network (TS-GAN) [71] focuses on Long Short-Term Memory (LSTM) networks to maintain temporal dependencies. Other GAN-based methods include the Multi-label Time series Generative Adversarial Network (MTGAN) [28], designed to generate synthetic data with multiple labels, and the COmmon Source CoordInated Generative Adversarial Network (COSCI-GAN) [72], which manages inter-channel correlations to preserve relationships between time series. HealthGAN [73], built on the Wasserstein GAN architecture, targets healthcare applications, while the Transformer-Based Time Series Generative Adversarial Network (TTS-GAN) [29] employs the transformer model's self-attention mechanism. The Modality Transfer Generative Adversarial Network [69] uses GANs to generate synthetic time series data by transferring modalities. In addition to GAN-based approaches, other DL algorithms contribute to synthetic time series data generation such as the diffusion-based conditional models, combined with structured state space models (SSSMs) [74], the causal recurrent variational autoencoder (CR-VAE) [75], the Variational Autoencoders (VAEs) [23] and the Adversarial Autoencoders (AAEs) [69]. The above-mentioned DL-based methods showcase the adaptability and potential of DL in synthetic time series data generation.
The utilized metrics to assess the synthetic time-series generated data quality and fidelity include a variety of statistical, visual, and performance-based measures. Metrics and visualization techniques, including the Distribution (PRD) plots, the Data Labelling Analysis (DLA), the Autocorrelation, the Mean Absolute Error (MAE), and the correlation coefficient are used to evaluate how well the synthetic data preserves the distribution and relationships present in the real data. Utilizing a reference model assesses the realism of synthetic data by comparing model performance on synthetic versus real data. The Maximum Mean Discrepancy (MMD) and the Mean Squared Error (MSE) are used to measure the difference in distributions and errors between synthetic and real data. The bias score evaluates the effectiveness of synthetic data in mitigating biases. The discriminator loss in generative models, the visual inspection using t-SNE and PCA, and the correlation plots provide insights into the synthetic data's visual and structural quality. Additional metrics, such as, the Generated Disease Types (GT), the Jensen-Shannon Divergence (JSD), the Normalized Distance (ND), the Average Euclidean Distance (AED), the Wasserstein Distance (WD), the Frobenius norm, the Spearman’s ρ, and the Kendall’s τ are used to further quantify the similarity in statistical properties. Moreover, metrics like the Average Hourly Energy Consumption (AHEC), the Welsch t-test, the Average Cosine Similarity (ACS), and the Dynamic Time Warping (DTW) are also deployed in specific domain applications. Furthermore, classification and prediction performance-based metrics are crucial for evaluating the practical utility of synthetic data in predictive modeling.
3.2.4. Omics data
According to the literature, the majority of the existing synthetic omics generation approaches rely heavily on established statistical principles and models to simulate multi-omics data (e.g. transcriptomics, metabolomics, proteomics, gene expression). Examples of such methods (Table 5) include the randomly selected and randomly permuted enriched pathways [76], causal feature clusters [77], the random covariance method (RCM) and the Cascade method [78], probabilistic modeling [79], random generation from uniform distributions [80], MVND [81], power law degree distribution [76], random perturbations [82], the simulated linear test (s-test) [83], the stochastic Block Models (SBM) [14] and the time-evolving graphs with metastability based on stochastic differential equations [15]. According to Table 5, these methods have been used to: (i) produce semi-synthetic metabolomics data preserving underlying distributions, the statistical assumptions based on the number of pathways, clusters, (ii) validate stratified causal discovery approaches in synthetic omics data, (iii) simulate gene expression data, accounting for additive biases, (iv) to model real data distributions in metabolomics and other omics data, (v) generate network topologies for tumor and normal cells in co-expression networks, (vi) mimic realistic complexities in multi-omics heterogeneous data analysis, (vii) improve proteomics data analysis through synthetic data generation, (viii) overcome challenges in multi-omics data integration, (ix) study human microbiome dynamics, (x) generate synthetic transcriptomics data reflecting specific trends, and (xi) model complex multi-omics data related to cancer. DL-based methods have been also deployed (Table 5), but to a smaller extent, including the WGAN-GP [84], the omicsGAN [85], the virtual Autoencoders (VAEs), and the Deep Boltzmann Machines (DBMs) [86] to: (i) address class imbalance problems in high-dimensional microarray and lipidomics data, (ii) enhance disease phenotype predictions, and (iii) enhance the interpretability of complex omics data patterns and interactions.
Table 5.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic omics data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [87] | To evaluate the performance of single-sample pathway analysis (ssPA) methods on semi-synthetic COVID-19 metabolomics data | Randomly selected and randomly permuted enriched pathways to produce semi-synthetic metabolomics data preserving the underlying distributions (both joint and marginal) | Python, R | https://github.com/cwieder/py-ssPA | Classification performance/ prediction metrics, OC |
| [88] | To demonstrate the benefit of grouping molecules into pathways using semi-synthetic COPD and COVID-19 metabolomics, proteomics and transcriptomics data | Python, R | https://github.com/cwieder/PathIntegrate | Classification performance/ prediction metrics, Sensitivity to Low Signal-to-Noise Signals, Significance of Pathway Feature VIP or MB-VIP Value |
|
| [84] | To address the class imbalance problem in high-dimensional microarray and lipidomics data using synthetic data | WGAN-GP | Python | https://github.com/sjcusworth/GAN_Scripts | Welch’s t-test, standard deviation, mean difference in scores, distance metric on generator loss |
| [77] | To validate a stratified causal discovery approach using synthetic omics data | Statistical assumptions based on the number of pathways, clusters, causal feature clusters | Matlab | https://github.com/MehrdadMansouri/Aristotle | Classification performance/ prediction metrics |
| [14] | To overcome the challenges posed by the integration of multi-omics data (miRNA, DNA methylation, gene expression) in five different types of cancer using synthetic data for validation | Stochastic Block Model (SBM) | Matlab | https://github.com/hamas200/MVCPM | Classification performance/ prediction metrics |
| [15] | To study the dynamic processes of the human microbiome using synthetic data for validation | Time-evolving graphs with metastability using a model based on stochastic differential equations | C+ +, Python | https://github.com/k-melnyk/graphKKE | Classification performance/ prediction metrics, CC, visual inspection of temporal patterns |
| [78] | To simulate real-world gene expression data, including the effects of additive biases | Random covariance method (RCM), Cascade method | Matlab | https://github.com/evcphd/C-SHIFT | Classification performance/ prediction metrics, CC |
| [85] | To enhance the prediction of disease phenotypes by generating synthetic data that better reflect the underlying biological mechanisms | omicsGAN (uses two Wasserstein GANs with a gradient penalty (wGAN-GP)) | Python | https://github.com/CompbioLabUCF/omicsGAN | Classification performance/ prediction metrics, Student’s t-test, Kaplan-Meier Survival Plots and Log-Rank Test P-values, heat maps and bar graphs, comparing the empirical correlations and normalization performance |
| [79] | To identify the biological relevance of different variables in metabolomics, transcriptomics and proteomics data using synthetic data for validation | Probabilistic modeling of the real data distributions given mass-to-charge ratios, peak intensities and noise levels | R | https://bitbucket.org/cesaremov/targetdecoy_mining/src/master/ | Classification performance/ prediction metrics |
| [89] | To identify and analyze gene expression profiles with distinct spatial patterns based on synthetic spatial transcriptomics data | Image based (uses a black and white image to create a structured grid of gene expression values) and Turing based (uses mathematical models to simulate Turing patterns) | Python | https://github.com/almaan/sepal | Diffusion time, entropy, pattern families |
| [80] | To recover significant circadian and non-circadian trends from transcriptomic data using synthetic data for validation | Random generation from uniform distributions with given parameters (e.g., slope, phase sight, growth rate, equilibrium shift) and value ranges | R | https://github.com/delosh653/MOSAIC | Distance between correlation matrices, heat maps to visualize the relative error between the correlation matrices of real and synthetic data after normalization |
| [86] | To analyze patterns and interactions of complex omics data (single-cell RNA-Seq data) using synthetic data to enhance the interpretability of biological processes | Variational Autoencoders (VAEs), Deep Boltzmann Machines (DBMs), log-linear models | Python | https://github.com/ssehztirom/Exploring-generative-deep-learning-for-omics-data-by-using-log-linear-models | Discrimination ability between different cell types by varying the number of selected genes for annotation, DBI, Robustness Against Dichotomization, NMF) |
| [81] | To enable multi-insight data visualization using synthetic and simulated multi-omics data (mRNA expression, DNA methylation) related to ovarian and breast cancer |
Multivariate normal distribution to model methylation, gene and protein expression data | R | https://cran.r-project.org/web/packages/InterSIM/index.html | Classification performance/ prediction metrics, Student’s t-test |
| [76] | To generate gene/protein co-expression networks specifically for tumor cells | A power law degree distribution is used to randomly generate tumor and normal network topologies | R | https://github.com/petraf01/TSNet | Classification performance/ prediction metrics, standard deviation, Welch’s t-test |
| [83] | To improve the analysis of proteomics data, particularly in experiments where technical variation plays a significant role due to small sample sizes | A simulated linear test (s-test) using adaptive Gauss-Hermite quadrature to generate synthetic data | R, Matlab | https://tvpham.github.io/stest/ | s-test, RMSE, Log-Likelihood Ratio Test, cV, Gauss-Hermite Quadrature |
| [82] | To evaluate a multi-omics heterogeneous data (methylation, gene expression and miRNA expression) analysis method using synthetic data for validation | Statistical method using structured and random perturbations to mimic realistic complexities | Python | https://github.com/yangzi4/iNMF | MDS, Frobenius norm, Classification performance/ prediction metrics |
The metrics which are used to measure the fidelity and quality of synthetic omics data include a wide range of performance-based, statistical, and visual techniques to ensure the synthetic data closely mirrors the real data. Performance metrics such as recall, precision, AUC (Area Under the Curve), adjusted rand index (ARI), overlap coefficient (OC), and variable importance in projection (VIP) are used to evaluate classification performance, clustering similarity, and feature significance. The sensitivity to low signal-to-noise signals and the significance of pathway features are assessed to ensure robustness. Statistical tests like the Welch’s t-test, the standard deviation, the mean difference in scores, the Student’s t-test, the Kaplan-Meier survival plots, and the Log-Rank Test P-values provide comparative analysis between synthetic and real data distributions. Correlation coefficients are crucial for preserving linear relationships between variables. Visual inspection techniques, including heat maps and bar graphs, are often employed to compare empirical correlations and normalization performance. Advanced metrics, such as, the distance between correlation matrices, the Frobenius norm, the diffusion time, the entropy, and the pattern families are used to assess temporal and structural fidelity. Additional metrics like the Davies-Bouldin index (DBI), the robustness against dichotomization, and comparisons with non-negative matrix factorization (NMF) are deployed to measure clustering quality and robustness. The module detection score (MDS) is used to evaluate the detection of similar patterns in the synthetic data.
3.2.5. Multimodal data
Table 6 presents significant efforts that have been made in the literature towards synthetic multimodal data generation. Most of these efforts focused on the development of AI-based methods including the Multimodal Neural Ordinary Differential Equations (MultiNODEs) [90], the CycleGAN [91], LSTM-based encoder-decoder models [92], the RAGAN combined with Modified U-Net and Multi-Branch Convolutional Neural Network [93], the CTAB-GAN+ alongside normalizing flows (NFlow) [94], the Temporally Correlated Multimodal Generative Adversarial Networks (TC-MultiGAN) with Document Sequence Generators [95], CMSG-Net in comparison with Pix2pix [96], the TGAN [97] which combines cGAN and CycleGAN, an End-to-end MultImodal X-ray genERative model (EMIXER) [98], and the PromptEHR [99] compared against a suite of LSTM and GPT-2 based models. The applications of these methods are diverse and focused on enhancing the utility and privacy of healthcare data. Those include the generation of: (i) synthetic patient-level data that integrate static and longitudinal elements, (ii) multimodal 4D datasets for medical image registration, the generation of synthetic text and tabular data for electronic health records, (iii) missing MRI modalities to complete clinical datasets, mimicking real clinical trial data, (iv) real-time multimodal electronic health records, (v) MRI synchronous images from single modalities, (vi) pseudo-medical images between various imaging modalities, (vii) synthetic X-ray images and corresponding textual reports, and (viii) synthetic EHRs. These advancements underscore the pivotal role of synthetic data in improving data availability while ensuring privacy in healthcare settings.
Table 6.
A summary of the scope, algorithms, programming languages, open-source codes or libraries, and metrics to measure synthetic data quality which are used by the studies that focus on the generation of synthetic multimodal data.
| Study | Scope | Statistical approaches / algorithm (s) used | Programming language/ Software | Open-source codes or libraries used | Metrics used to measure synthetic data fidelity/privacy |
|---|---|---|---|---|---|
| [90] | To generate synthetic patient-level data using a novel approach which integrates both static and longitudinal data | Multimodal Neural Ordinary Differential Equations (MultiNODEs) | Python | https://github.com/SCAI-BIO/MultiNODEs | JSD, CSA, MTC, Classification performance/ prediction metrics |
| [91] | To overcome the limitation of sparse annotated data in medical image registration by synthesizing multimodal 4D datasets (CT, CBCT, and MR images) | CycleGAN | - | - | MAE, SSIM, FSIM, EPR, EGR, NPS, CC, NM, HistCC, DSC |
| [92] | To generate synthetic free-text and tabular data in electronic health records (EHRs) using deep learning algorithms to enhance data sharing and privacy | Encoder-decoder models based on LSTM RNNs | Python | https://github.com/scotthlee/nrc | Classification performance/ prediction metrics, COR |
| [93] | To generate missing MRI modalities (T1, T1ce, FLAIR) from existing T2 modality images to address the issue of incomplete multimodal datasets in clinical settings | RAGAN, Modified U-Net, Multi-Branch Convolutional Neural Network | Python | tensorflow and keras libraries | PSNR, SSIM, FSIM, EPR, EGR, NPS, NCC, DSC |
| [94] | To generate synthetic clinical, laboratory, genetic data mimicking real AML patient data from clinical trials | CTAB-GAN+ and normalizing flows (NFlow) | Python | https://github.com/waldemar93/synthetic_data_pipeline | Summary statistics, log-transformed correlation score, Kaplan-Meier-Divergence, PLC |
| [95] | Synthetic data generation of real-time multimodal electronic health and physical records (MHR, wearable biometric and behavioral data, and self-assessment surveys in the standard FHIR format) | Temporally Correlated Multimodal GAN (TC-MultiGAN), Document Sequence Generator (DSG) | Python | https://github.com/GATEKEEPER-OU/synthetic-data | WD, KS test, JSD, PCD |
| [96] | MRI synchronous construction from a single T1-weight (T1) image for MRIgRT synthetic CT (sCT) image generation | CMSG-Net compared against Pix2pix, CUT, TransUNet, ResViT, SE2SD-Net | Python | pytorch | MAE, NRMSE, PSNR, SSIM |
| [97] | To synthesize pseudo-medical images between multimodal datasets (CBCT -> CT, CBCT -> MRI, MRI -> CT) | TGAN (cGAN and CycleGAN) | Python | tensorflow | PSNR, SSIM, MAE, NMI, Dose Distribution and Gamma Analysis |
| [98] | To generate synthetic X-ray images and corresponding text reports | End-to-end MultImodal X-ray genERative model (EMIXER) |
Python | pytorch | Classification performance/ prediction metrics, BLEU 1-4, CIDEr Score, FID |
| [99] | To generate synthetic EHRs (including numerical and categorical data as well as text) | PromptEHR (based on language models) compared against LSTM+MedGA, SynTEG, LSTM+MLP and GPT-2 | Python | https://github.com/RyanWangZf/PromptEHR | Perplexity, Recall@ 10 and Recall@ 20, t-test, Wilcoxon test, Fisher’s exact test |
The metrics used to measure synthetic multimodal data fidelity and quality, in this context, include a diverse array of performance, statistical, and visual measures. The Jensen-Shannon divergence (JSD) and the correlation structure analysis (CSA) are used to evaluate the distributional similarities and correlations between synthetic and real data, while performance metrics like the AUC and the median trajectory comparison (MTC) provide insights into the overall predictive performance and temporal alignment. The Mean Absolute Error (MAE), the structural Similarity Index Measure (SSIM), the Feature Similarity Index Measure (FSIM), the Edge Preservation Ratio (EPR), the Edge Generation Ratio (EGR), the Noise Power Spectrum (NPS), the Noise Magnitude (NM), the Histogram Correlation Coefficient (HistCC), and the Dice Similarity Coefficient (DSC) are used to assess various aspects of image quality and feature preservation. Classification/prediction performance metrics, such as, the recall, the F1 score, the accuracy, the crude odds ratios (COR), and the removal of Personally Identifiable Information (PII) are crucial to ensure both accuracy and privacy. The PSNR, the SSIM, the FSIM, and other noise-related metrics evaluate the visual and structural fidelity of synthetic data. Statistical measures, including mean, median, standard deviation, log-transformed correlation scores, and Kaplan-Meier-Divergence, alongside the Privacy Leakage Coefficient (PLC), provide an indication of data integrity and privacy. The Wasserstein distance, the KS test, and the distance pairwise correlation further measure the statistical similarity between datasets. Additional metrics like normalized mutual information (NMI), the dose distribution, the gamma analysis, the BLEU scores, the CIDEr score, the Fréchet Inception Distance (FID), the perplexity, and the recall@ 10 and recall@ 20 are also used to assess both the fidelity and utility of synthetic data along with statistical tests, such as, the t-test, the Wilcoxon test, and the Fisher’s exact test.
4. Discussion
A thorough overview of the above-mentioned synthetic data generators utilized in the assessed studies are presented in Table 7. The table presents also the advantages and weaknesses of each methodological approach. The advantages and weaknesses of each synthetic data generation approach are defined on the basis of diverse criteria, such as, implementation simplicity, computational efficiency, flexibility in handling non-linear data, robustness in modeling complex dependencies, effectiveness in addressing class imbalance, and suitability for the healthcare domain. The evaluations draw on insights from recent literature reviews and empirical studies, highlighting both the potential and limitations of various synthetic data generation methods [9], [100], [101], [102]. In the case of probabilistic-based models, bootstrapping and MVND offer a straightforward implementation but might not capture complex data dependencies adequately. Vine Copula Models stand out for their ability to model intricate dependencies between variables, although they are complex to set up and interpret. SSM are well-suited for modeling sequential data and transitions, particularly in applications with clear state definitions, but are limited to such specific scenarios. Bayesian Networks are characterized by their powerful probabilistic modeling and inference capabilities which incorporate causal relationships, though they may struggle with big data and require complex structuring. In the field of omics, the SBM effectively models complex relationships and community structures yet demands precise parameter tuning and can be computationally demanding. Similarly, the RCM and the Cascade Method aim to simulate realistic gene expression data, including various biases, but might oversimplify and not capture all underlying biological complexities.
Table 7.
A thorough report of the advantages and weaknesses of the synthetic data generation algorithms deployed in the studies from Table 1, Table 2, Table 3, Table 4, Table 5, Table 6.
| No | Algorithm [Indicative study] | Type of method | Supported type (s) of data | Advantages | Weaknesses | Programming language |
|---|---|---|---|---|---|---|
| 1 | Bootstrapping, MVND[13] | Statistical | Tabular | Simple to implement, robust statistical foundations. | May not capture complex dependencies in data. | R, Python |
| 2 | ADS-GAN[24] | Deep learning | Tabular | Good for generating privacy-preserving synthetic data. | Requires careful tuning to prevent mode collapse. | Python |
| 3 | Bayesian models[40] | Machine learning | Tabular | Flexible, good for non-linear data, incorporates uncertainty. | Computationally intensive, requires substantial data. | R, Python |
| 4 | Tree ensembles[16] | Machine learning | Tabular | Combine multiple decision trees to improve the robustness of the generated data. | Training can be computationally expensive, especially with large datasets and a high number of trees, leading to longer processing times and higher resource usage. | R, Python |
| 5 | RBF-based ANNs[103] | Deep learning | Tabular | Suitable for generating high-quality synthetic data that accurately reflects the underlying patterns in the original dataset. | Scalability issues as the number of data points increases, leading to higher computational costs and potential difficulties in managing large datasets. | R, Python |
| 6 | Vine Copula Models[39] | Statistical | Tabular | Excellent at modeling complex dependencies between variables. | Complex to set up and interpret. | R |
| 7 | BGMM[19] | Machine learning | Tabular | Efficient at clustering and density estimation. | Sensitive to the initialization and number of components. | Python |
| 8 | BGMMO-CE[19] | Machine learning | Tabular | Optimized for computational efficiency. | May lose some nuances of data complexity. | Python |
| 9 | TabularSimulationBase[27] | Deep learning | Tabular | Versatile and capable of generating diverse synthetic datasets. | Can be challenging to tune multiple models effectively. | Python |
| 10 | GaussianCopula[43] | Statistical | Tabular | Effectively captures complex dependencies between multiple variables, allowing for a more accurate representation of multivariate relationships. | Assumption of normality. | Python |
| 11 | CopulaGAN[27] | Deep learning | Tabular | Leverages the flexibility of copula models to capture complex dependencies between variables and the generative power of GANs to produce realistic synthetic data. | Training can be computationally intensive, requiring significant computational resources and time, especially for high-dimensional datasets. | Python |
| 12 | TVAE[47] | Deep learning | Tabular | Specifically designed to model tabular data, capturing complex relationships. | Training can be complex and computationally intensive, requiring careful tuning of hyperparameters and sufficient computational resources to achieve optimal performance. | Python |
| 13 | MedGAN[57] | Deep learning | Tabular | It can generate realistic synthetic healthcare data, including high-dimensional EHRs. | Need for substantial computational resources and expertise in fine-tuning GAN models. | Python |
| 14 | Tabular Preset[64] | Deep learning | Tabular, Radiomics | Handles high-dimensional data well. | High complexity and computational demand. | Python |
| 15 | Bayesian (hierarchical) Generalized Linear Models (hGLM)[37] | Machine learning | Tabular | Excellent for data with hierarchical structures. | Requires extensive computational resources. | R |
| 16 | State-transition Machines[41] | Statistical | Tabular | Good for modeling sequential data and transitions. | Limited to applications with clear state transitions. | Java |
| 17 | CGAN[25] | Deep learning | Tabular | Advanced GAN models capable of generating highly realistic data. | Training stability can be an issue. | Python |
| 18 | CTGAN[46] | Deep learning | Tabular | Specialized for tabular data, helps mitigate class imbalance. | Requires careful hyperparameter tuning. | Python |
| 19 | RadialGAN[47] | Deep learning | Tabular | Cutting-edge methods for detailed synthetic data generation. | Complex architectures that require significant training. | Python |
| 20 | TabDDPM[47] | Deep learning | Tabular | Models the data generation process through a series of diffusion steps, capturing complex data distributions and dependencies accurately. | Iterative nature of denoising diffusion models, requires significant computational resources and time to train, especially on large datasets. | Python |
| 21 | Bayesian Networks[38] | Machine learning | Tabular | Powerful for probabilistic modeling and inference. | Graph structure may be hard to specify with limited data. | R, Software |
| 22 | Sequential Decision Tree-Synthesizer[45] | Deep learning | Tabular | Flexible and scalable to different data types. | Complexity increases with data dimensionality. | R, Python |
| 23 | Enhanced Balancing GAN[48] | Deep learning | Imaging | Specifically designed to address class imbalance in image data. | Potentially limited to specific image-related tasks. | Python |
| 24 | Ensemble of Convolutional Neural Networks[55] | Deep learning | Imaging | Effective for robust image analysis and out-of-distribution data. | Requires significant computational power and data for training. | Python |
| 25 | Attention-GAN[49] | Deep learning | Imaging | Capable of capturing intricate details in image synthesis. | May be prone to overfitting on small datasets. | Python |
| 26 | Conditional Variational Autoencoder[52] | Deep learning | Imaging | Combines the strengths of CVAE and GAN for improved generation. | Complex to implement and tune. | Python |
| 27 | Contrastive Diffusion Model[53] | Deep learning | Imaging | Excels at generating high-resolution, fine-grained images. | Computationally demanding and requires tuning. | Python |
| 28 | Normalizing Flows[56] | Deep learning | Imaging | Offers exact likelihood computation and invertibility. | Requires careful design to ensure effective flow architectures. | Python |
| 29 | Dual-Discriminator Conditional GAN[34] | Deep learning | Imaging | Enhances detail and realism in multi-resolution image fusion. | May introduce high complexity and training difficulty. | Python |
| 30 | Vision Transformer[54] | Deep learning | Imaging | Harnesses the power of transformers for image processing. | Requires large datasets and extensive training time. | Python |
| 31 | Flow-Based Network[58] | Deep learning | Imaging | Useful for enhancing visual quality in super-resolution tasks. | Relatively new with potentially unexplored limitations. | Python |
| 32 | Task-Guided GAN[60] | Deep learning | Imaging | Tailors the generation process to specific tasks, enhancing utility. | Task-specific tuning can limit general application. | Python |
| 33 | Progressively Growing GANs[50] | Deep learning | Imaging | Allows for gradual building of image resolution, enhancing detail. | High resource consumption and complex training dynamics. | Python |
| 34 | Style Distribution GAN (SD-GAN)[51] | Deep learning | Imaging | Focuses on transferring and blending diverse style features. | Managing style variations effectively can be challenging. | Python |
| 35 | CS-CO (Self-Supervised Learning)[62] | Deep learning | Imaging | Self-supervised learning method for histopathological images. | Limited by the quality and variation of unlabeled data. | Python |
| 36 | SinGAN[57] | Deep learning | Imaging | Capable of generating high-quality images from a single training image. | May not perform well with complex scenes containing multiple objects. | Python |
| 37 | FastGAN[57] | Deep learning | Imaging | Faster and more efficient training compared to traditional GANs. | Limited research and applications compared to more established GAN models. | Python |
| 38 | PGGAN[57] | Deep learning | Imaging | Can generate very high-resolution images (e.g., 1024 ×1024). | Training can be computationally intensive and time-consuming. | Python |
| 39 | pix2pix[57] | Deep learning | Imaging | Effective for image-to-image translation tasks. | Can suffer from mode collapse, where the generator produces limited diversity in outputs. | Python |
| 40 | WGAN-GP[57] | Deep learning | Radiomics | Effective at generating realistic samples and stable training. | May require extensive computational resources. | Python |
| 41 | RadSynth[66] | Deep learning | Radiomics | Specifically designed for radiomic image synthesis. | Limited information available; potential specificity to tasks. | Software |
| 42 | SSSD-ECG[74] | Deep learning | Time-series | Specifically tailored for synthetic ECG data generation. | Specifically tailored for synthetic ECG data generation. | Python |
| 43 | DiffWave[74] | Deep learning | Time-series | Particularly effective in generating high-fidelity synthetic data by leveraging the power of diffusion models to produce realistic and high-quality outputs. | Requires significant computational power and expertise in deep learning and diffusion model techniques to achieve optimal results. | Python |
| 44 | WaveGAN[74] | Deep learning | Time-series | Effective for applications that require realistic and coherent audio data generation, such as speech and music synthesis. | Can be unstable and requires careful tuning of hyperparameters. | Python |
| 45 | Pulse2Pulse[74] | Deep learning | Time-series | Tailored for generating realistic physiological pulse signals, such as ECG or PPG data. | Requires extensive hyperparameter tuning and significant computational resources to accurately capture the nuances of physiological signals. | Python |
| 46 | Causal Recurrent Variational AutoEncoder (CRVAE)[75] | Deep learning | Time-series | Excels at generating time-series data with underlying causality. | Potentially complex to implement and requires substantial data. | Python |
| 47 | Guided Evolutionary Synthesizer (GES)[67] | Deep learning | Time-series | Adaptable to different bias scenarios in time-series data. | May require expert knowledge to configure and operate. | Python |
| 48 | TS-GAN[71] | Deep learning | Time-series | Tailored for sensor- health data augmentation. | Specific to sensor data, may not generalize across domains. | Python |
| 49 | Variational Autoencoder (VAE)[23] | Deep learning | Time-series | Good for modeling distribution of data for simulation. | Sometimes struggles with the quality of generated samples. | Python |
| 50 | Multi-label Time series GAN (MTGAN)[28] | Deep learning | Time-series | Effective for handling time series data with multiple labels. | Requires careful tuning and extensive dataset preparation. | Python |
| 51 | COmmon Source CoordInated GAN (COSCI-GAN)[72] | Deep learning | Time-series | Innovative for generating multivariate time series. | New approach with potential untested scenarios. | Python |
| 52 | HealthGAN, Wasserstein GAN, TimeGAN[73] | Deep learning | Time-series | Advanced suite of models for comprehensive time series generation. | High computational demand and complexity. | Python |
| 53 | Transformer-Based GAN (TTS-GAN)[29] | Deep learning | Time-series | Utilizes transformer architectures for high fidelity synthesis. | Requires extensive computational resources and data. | Python |
| 54 | HMM and Regression Algorithms[22] | Machine learning | Time-series | Effective for capturing sequences and transitions in time series data. | Complex integration of multiple modeling techniques. | Python |
| 55 | Randomly Selected and Randomly Permuted Enriched Pathways[87],[88] | Statistical | Omics | Efficiently preserves the statistical distributions for semi-synthetic metabolomics data analysis. | Limited to the statistical properties available in the data; may not introduce novel biological insights. | R, Python |
| 56 | Stochastic Block Model (SBM)[14] | Probabilistic | Omics | Effectively models complex relationships and community structures within multi-omics data. | Requires careful parameter tuning and can be computationally intensive. | Matlab |
| 57 | Time-evolving Graphs with Metastability[15] | Probabilistic | Omics | Captures dynamic processes effectively, useful for studying temporal changes in microbiomes. | Complex to implement and requires understanding of differential equations and graph theory. | C+ +, Python |
| 58 | Random Covariance Method (RCM)[78] | Statistical | Omics | Simulates real-world gene expression data including various biases, enhancing realism. | Potentially oversimplified, might not capture all underlying biological complexities. | Matlab |
| 59 | Cascade Method[78] | Statistical | Omics | Effective in handling hierarchical or sequential processes by breaking down complex problems into simpler, smaller stages, which can improve the accuracy and manageability of modeling efforts. | Errors can accumulate and propagate through the stages of the cascade, potentially leading to reduced overall accuracy and reliability in the final synthetic data generation if not carefully managed. | Matlab |
| 60 | omicsGAN[85] | Deep learning | Omics | Utilizes advanced GAN technology to generate high-fidelity omics data, improving phenotype prediction. | GANs can be challenging to train and require large amounts of data to avoid mode collapse. | Python |
| 61 | Image-based and Turing-based Methods[89] | Deep learning | Omics | Innovative use of visual and mathematical models to simulate spatial gene expression patterns. | May require specific expertise in both image processing and mathematical modeling. | Python |
| 62 | Random Generation from Uniform Distributions[80] | Statistical | Omics | Simple and effective for generating data with specified statistical properties. | Lacks complexity, might not be suitable for capturing non-linear relationships or interactions. | R |
| 63 | Deep Boltzmann Machines (DBMs)[86] | Deep learning | Omics | Capable of capturing complex and high-dimensional data distributions. | Computationally intensive and challenging due to the need for layer-wise pre-training and fine-tuning. | Python |
| 64 | Power Law Degree Distribution[76] | Statistical | Omics | Useful for generating network topologies that mimic natural biological networks. | Assumes network connectivity that follows a power law, which might not be appropriate for all types of biological data. | R |
| 65 | Simulated Linear Test (s-test)[83] | Statistical | Omics | Adapts well to small sample sizes and can handle technical variations in proteomics data. | Specific to scenarios with small sample sizes and may not generalize to larger or different datasets. | R, Matlab |
| 66 | Structured and Random Perturbations[82] | Statistical | Omics | Allows for the generation of complex multi-omics data, enhancing the realism and applicability of synthetic datasets. | Requires careful calibration to ensure the perturbations reflect realistic biological variability. | Python |
| 67 | Multimodal Neural Ordinary Differential Equations (MultiNODEs)[90] | Deep learning | Multimodal | Integrates static and longitudinal data effectively for patient-level data synthesis. | Requires careful configuration and understanding of both differential equations and neural networks. | Python |
| 68 | CycleGAN[91] | Deep learning | Multimodal | Excellent for image-to-image translation tasks without needing paired data, useful in medical imaging. | Can struggle with maintaining consistency in synthesized images where there is a large variation between input modalities. | Python |
| 69 | Encoder-decoder models based on LSTM RNNs[92] | Deep learning | Multimodal | Effective for generating coherent and contextually relevant text and tabular data. | May face challenges with very long sequences or extremely diverse datasets. | Python |
| 70 | RAGAN, Modified U-Net, Multi-Branch Convolutional Neural Network[93] | Deep learning | Multimodal | Combines multiple advanced techniques to fill missing MRI modalities, enhancing dataset completeness. | Complex to train and requires substantial computational resources. | Python |
| 71 | CTAB-GAN+ and Normalizing Flows (NFlow)[94] | Deep learning | Multimodal | Allows for detailed control over the statistical properties of synthetic data, suitable for clinical and laboratory data simulation. | Configuration and tuning can be complex, and understanding statistical underpinnings is essential. | Python |
| 72 | Temporally Correlated Multimodal Generative Adversarial Network (TC-MultiGAN)[95] | Deep learning | Multimodal | Tailored for generating time-correlated multimodal datasets, particularly in dynamic and real-time environments. | Can be challenging to synchronize multiple data streams effectively. | Python |
| 73 | Document Sequence Generator (DSG)[95] | Deep learning | Multimodal | Particularly useful for tasks involving document and text data generation by capturing complex temporal dependencies within sequences. | Requires substantial computational power and careful tuning of model parameters to achieve high-quality and coherent text generation. | Python |
| 74 | CMSG-Net[96] | Deep learning | Multimodal | A robust set of tools for MRIgRT synthetic CT image generation, utilizing both established and cutting-edge techniques. | Each model brings its own set of parameters and complexities, potentially complicating integration and optimization. | Python |
| 75 | TGAN[97] | Deep learning | Multimodal | Enables effective synthesis of medical images between different modalities, addressing the scarcity of annotated medical images. | Requires careful adjustment to ensure high fidelity and avoid artifacts common in synthesized images. | Python |
| 76 | End-to-end MultImodal X-ray generative model (EMIXER)[98] | Deep learning | Multimodal | Specifically designed to generate synthetic X-ray images along with corresponding textual reports, enhancing data utility for training AI models. | Integrating text and image generation smoothly can be technically challenging and requires extensive data for training. | Python |
| 77 | PromptEHR (based on language models)[99] | Deep learning | Multimodal | Utilizes advanced language models to generate synthetic EHRs, enabling a high degree of realism and complexity. | Balancing the generation of coherent and realistic EHRs while ensuring privacy can be difficult. | Python |
Bayesian Models and Tree Ensembles are noted for their flexibility and effectiveness in handling non-linear data patterns. They excel at incorporating uncertainty into predictions, which is crucial for decision-making processes, where risk assessment is significant. However, their performance is restricted by the scale of the data, and they are computationally demanding, a fact that limits their use in real-time or resource-constrained environments. On the other hand, the Radial Basis Function (RBF)-based ANNs are designed to handle complex, non-linear interactions within data. They offer a powerful mechanism for pattern recognition and classification tasks but require significant computational resources, particularly in tuning and training phases. The BGMM algorithm is efficient for clustering and for density estimation. It offers a probabilistic framework that helps to determine the number of components (clusters) in a dataset. The primary challenges with BGMM involve: (i) the sensitivity to initial parameters, and (ii) the selection of the number of Gaussian components, which can significantly affect the model's performance.
The Probabilistic BNs, which are built on probabilistic reasoning, are excellent for causal inference and they are particularly useful in fields like epidemiology and genetics where understanding causal relationships is crucial. The downside lies in their complexity in structure and computational demands, especially in the case of big data which may slow down the inference process. In omics studies, the SBM effectively models complex relationships and community structures within biological data. It requires precise parameter tuning and substantial computational power, which may limit its practical application in resource-constrained settings. In addition, the RCM and the Cascade method aim to simulate realistic gene expression data, considering various biases to enhance the realism of synthetic datasets. They might simplify complex biological interactions, but they might miss some underlying dynamics.
Table 7 also presents various DL-based generators, each designed to handle specific challenges. These methods leverage the capabilities of DL to learn complex patterns, and to generate synthetic data in an effective way. To this end, the ADS-GAN, the CTGAN, and other GAN-based variations are particularly effective for generating synthetic tabular data that preserve privacy. However, although they are known for their ability to handle high-dimensional data, they require hyperparameter tuning to avoid issues like mode collapse. The Enhanced Balancing GAN and the Attention-GAN are designed for imaging data to tackle crucial problems, such as, class imbalance and contrast enhancement during image synthesis. Although they are powerful for capturing intricate details, they may be prone to overfitting, especially in small datasets. The Contrastive Diffusion Model and the Flow-Based Network model excel in generating high-resolution and fine-grained images. They offer precise likelihood computation and are effective in tasks like super-resolution, though they require significant computational resources and careful tuning to perform effectively.
VAEs and Transformer-Based Models are widely used for modeling time-series and imaging data. Transformer-based models, like the Vision Transformers, utilize attention mechanisms to handle big data but they require extensive training time and resources. Multimodal approaches, such as, the CycleGAN, and the Encoder-decoder models are ideal for image-to-image translation tasks without the need of paired data. Furthermore, encoder-decoder models based on LSTM RNNs can effectively generate coherent text and tabular data. Although these methods manage to synthesize data robustly across different modalities, they often struggle to maintain consistency or handle very long sequences. In medical imaging, models like TGAN and CMSG-Net are widely used to synthesize medical images that can address the scarcity of annotated images and thus enhance data utility for training AI models. In omics, methods like omicsGAN and Probabilistic Modeling utilize advanced GAN technology and probabilistic approaches to simulate complex biological data patterns, improving phenotype prediction but requiring large datasets to avoid overfitting.
There is no doubt that synthetic data generation has been the point of interest in a broad spectrum of studies under the healthcare domain. However, they often require significant computational resources and configuration to optimize their performance and utility. The DL-based generators demonstrate a broad capability to generate, enhance, and analyze data in healthcare. They are marked by their ability to handle complex and high-dimensional data, but often at the cost of high computational demand and the need for extensive data and model tuning. The ML-based generators are robust and capable of modeling complex, non-linear relationships and are computational efficient. Their effectiveness often comes at the cost of increased computational requirements and complexity in tuning and operation which necessitates their optimized implementation to maximize their potential by effectively reducing resource constraints. Probabilistic models are characterized by their ability to incorporate uncertainty into the modeling process. However, they often require careful design and parameter tuning and can be computationally intensive with limited scalability when handling complex data.
In addition, synthetic data can significantly contribute to the principles of trustworthy AI (TwAI) by enhancing privacy, fairness, and robustness. The generation of synthetic data that can “mimic” the real data without containing any personal or sensitive information, safeguards individuals' privacy and mitigates the risks of data breaches. Synthetic data can also be used to correct biases which are present in real-world data (e.g. by populating unprivileged groups to reduce demographic disparities), thereby promoting fairness and reducing discrimination in AI models. Moreover, synthetic data can enable the development of robust AI models by offering diverse and high-quality data for augmentation, as well as reducing vulnerabilities to adversarial attacks.
5. Conclusion and future directions
The current review reveals a noteworthy and exponentially increasing number of studies which focus on the development and deployment of synthetic data generation technologies in healthcare across various data modalities, including tabular, imaging, radiomics, time-series, and omics. These studies make use of synthetic data to not only address privacy concerns but also to enhance the availability and diversity of the real data which is crucial for training AI-driven diagnostic and predictive models to improve patient outcomes and to support healthcare research. In addition, the current work presents the advantages and weaknesses of a variety of statistical, probabilistic, ML and DL based synthetic data generators. Great emphasis was given to reporting open-source tools to promote collaborative efforts within the research community to accelerate advancements in the field.
The ability to synthesize tabular, imaging, radiomics, time-series, and omics data is essential for the development of robust AI models that can deliver more accurate and personalized healthcare solutions. Towards this direction, there has been a reported increase in the use of statistical and probabilistic methods, machine learning methods, and deep learning methods for generating synthetic data with improved fidelity and utility. For tabular data, statistical methods like MVND and bootstrapping, and probabilistic methods like Bayesian Models are widely used for generating synthetic distributions that preserve the underlying statistical properties of the real data. These methods are valuable for simulations in clinical trials and disease progression modeling. Machine learning methods, such as, GMM and tree ensembles can effectively capture complex patterns within the data, aiding in the generation of large-scale virtual populations for in silico clinical trials. DL-based methods like GANs and VAEs have been utilized to enhance privacy-conscious data generation, supporting applications such as clinical decision support and predictive modeling. For imaging data, CycleGAN and Enhanced Balancing GAN are instrumental in generating synthetic medical images, including functionalities to address minority classes in datasets or to perform style transfer between different imaging modalities. Conditional Variational Autoencoder (CVAE) and Attention-based GANs are deployed for specific tasks like image augmentation and high-resolution image synthesis, showcasing their adaptability in handling varied imaging data challenges. In radiomics data, WGAN-GP and CTGAN have been employed to generate synthetic radiomic data, which are crucial for training models to differentiate between various medical conditions using radiomic features. Copula GAN and Diffusion-based Models are cutting-edge methods enhancing the capacity to generate realistic and statistically coherent radiomic images and features.
For time-series data, Wasserstein GAN with Gradient Penalty (WGAN-GP) and Multi-label Time Series GAN (MTGAN) offer robust solutions for generating realistic time-series data, crucial for medical applications where temporal dynamics are essential. Transformer-Based GANs and Causal Recurrent Variational Autoencoders (CR-VAEs) underscore the evolution towards using complex architectures to maintain temporal dependencies and enhance the fidelity of synthetic time-series datasets. In omics data, Randomly Selected Pathways and Causal Feature Clusters are statistical-based methods which are used for generating synthetic omics data, which are vital for addressing issues like class imbalance and enhancing disease phenotype predictions. OmicsGAN and DBMs are advanced deep learning methods focusing on the generation of complex omics datasets, facilitating better interpretations of intricate biological processes. As for multimodal data generation, MultiNODEs and TC-MultiGAN illustrate the integration of various data types through advanced neural networks, tackling challenges in multimodal data synthesis like generating comprehensive electronic health records or synthetic MRI images. CycleGAN and End-to-End Multimodal X-ray Generative Model (EMIXER) demonstrate the versatility of GANs in creating synthetic datasets that span multiple medical imaging modalities and integrating imaging with textual data.
The variety of methods that has been discussed highlights a significant advancement in the field of synthetic data generation, tailored to diverse needs across different types of medical data. Each algorithm or tool brings specific strengths to the table, addressing the challenges posed by the vast and varied data landscape in healthcare. Despite the advancements, there are ongoing challenges which are related to the quality, representativeness, and ethical use of synthetic data. Challenges, such as, data fidelity, potential biases introduced in the generated data, and the need for big, diverse data for AI model training remain critical areas for improvement. Future research in the field is needed to continue to explore these technologies, particularly focusing on improving the accuracy, reliability, and ethical aspects of synthetic data generation. This will not only enhance the robustness of the AI models but also ensure their applicability in real-world medical settings, ultimately leading to better patient outcomes and more efficient healthcare systems. Furthermore, future research should focus on improving the fidelity of synthetic data to ensure that they can mimic real-world data. This includes the development of more sophisticated models that can capture complex dependencies and interactions within the real data. Addressing biases in synthetic data generation is another critical factor to ensure fairness and equity in the AI models. Emphasis should be given to identifying and mitigating potential biases, particularly in data with underrepresented populations. As healthcare data continues to grow, scalable and efficient synthetic data generation methods are needed with reduced computational complexity while maintaining high-quality outcomes. Emphasis should also be given on the improvement and refinement of ethical guidelines and regulatory frameworks for the use of synthetic data in healthcare to ensure transparency in data generation and strict adherence to privacy standards.
CRediT authorship contribution statement
Vasileios C. Pezoulas: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Conceptualization. Dimitrios Zaridis: Writing – original draft, Methodology, Investigation. Eugenia Mylona: Writing – original draft, Methodology, Investigation. Christos Androutsos: Writing – original draft, Methodology, Investigation. Dimitrios Fotiadis: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Conceptualization. Kosmas Apostolidis: Writing – original draft, Methodology, Investigation. Nikolaos S. Tachos: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
The research work has received funding from the European Commission under GA 101135932 (FAITH Project).
References
- 1.Shilo S., Rossman H., Segal E. Axes of a revolution: challenges and promises of big data in healthcare. Nat Med. 2020;vol. 26(1):29–38. doi: 10.1038/s41591-019-0727-5. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal R., Prabakaran S. Big data in digital healthcare: lessons learnt and recommendations for general practice. Heredity. 2020;vol. 124(4):525–534. doi: 10.1038/s41437-020-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appenzeller A., Leitner M., Philipp P., Krempel E., Beyerer J. Privacy and utility of private synthetic data for medical data analyses. Appl Sci. 2022;vol. 12(23):12320. doi: 10.3390/app122312320. [DOI] [Google Scholar]
- 4.S.M. Bellovin, P.K. Dutta, N. Reitinger, Privacy and Synthetic Datasets, vol. 22.
- 5.Yale A., Dash S., Dutta R., Guyon I., Pavao A., Bennett K.P. Generation and evaluation of privacy preserving synthetic health data. Neurocomputing. 2020;vol. 416:244–255. doi: 10.1016/j.neucom.2019.12.136. [DOI] [Google Scholar]
- 6.Gonzales A., Guruswamy G., Smith S.R. Synthetic data in health care: a narrative review. PLOS Digit Health. 2023;vol. 2(1) doi: 10.1371/journal.pdig.0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murtaza H., Ahmed M., Khan N.F., Murtaza G., Zafar S., Bano A. Synthetic data generation: state of the art in health care domain. Comput Sci Rev. 2023;vol. 48 doi: 10.1016/j.cosrev.2023.100546. [DOI] [Google Scholar]
- 8.J. Jordon et al., “Synthetic Data -- what, why and how?” arXiv, May 06, 2022. Accessed: May 28, 2024. [Online]. Available: 〈http://arxiv.org/abs/2205.03257〉.
- 9.Figueira A., Vaz B. Survey on synthetic data generation, evaluation methods and GANs. Mathematics. 2022;vol. 10(15):2733. doi: 10.3390/math10152733. [DOI] [Google Scholar]
- 10.O. Mendelevitch, “Review of Methods and Experimental Results”.
- 11.Cheng V., Suriyakumar V.M., Dullerud N., Joshi S., Ghassemi M. Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, Virtual Event. ACM; Canada: 2021. Can You Fake It Until You Make It?: Impacts of Differentially Private Synthetic Data on Downstream Classification Fairness; pp. 149–160. [DOI] [Google Scholar]
- 12.Ferrara E. Fairness and bias in artificial intelligence: a brief survey of sources, impacts, and mitigation strategies. Sci. 2023;vol. 6(1):3. doi: 10.3390/sci6010003. [DOI] [Google Scholar]
- 13.Smania G., Jonsson E.N. Conditional distribution modeling as an alternative method for covariates simulation: Comparison with joint multivariate normal and bootstrap techniques. CPT Pharmacomet Syst Pharmacol. 2021;vol. 10(4):330–339. doi: 10.1002/psp4.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AL-kuhali H.A., et al. Multiview clustering of multi-omics data integration by using a penalty model. BMC Bioinforma. 2022;vol. 23(1):288. doi: 10.1186/s12859-022-04826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnyk K., Klus S., Montavon G., Conrad T.O.F. GraphKKE: graph Kernel Koopman embedding for human microbiome analysis. Appl Netw Sci. 2020;vol. 5(1):96. doi: 10.1007/s41109-020-00339-2. [DOI] [Google Scholar]
- 16.Pezoulas V.C., Grigoriadis G.I., Tachos N.S., Barlocco F., Olivotto I., Fotiadis D.I. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) IEEE; 2020. Generation of virtual patient data for in-silico cardiomyopathies drug development using tree ensembles: a comparative study; pp. 5343–5346. [DOI] [PubMed] [Google Scholar]
- 17.Robnik-Šikonja M. Dataset comparison workflows. Int J Data Sci. 2018;vol. 3(2):126–145. [Google Scholar]
- 18.Pičulin M., et al. Disease progression of hypertrophic cardiomyopathy: modeling using machine learning. JMIR Med Inform. 2022;vol. 10(2) doi: 10.2196/30483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pezoulas V.C., Tachos N.S., Gkois G., Olivotto I., Barlocco F., Fotiadis D.I. Bayesian inference-based gaussian mixture models with optimal components estimation towards large-scale synthetic data generation for in silico clinical trials. IEEE Open J Eng Med Biol. 2022 doi: 10.1109/OJEMB.2022.3181796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pezoulas V.C., Grigoriadis G.I., Tachos N.S., Barlocco F., Olivotto I., Fotiadis D.I. 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) IEEE; 2021. Variational Gaussian Mixture Models with robust Dirichlet concentration priors for virtual population generation in hypertrophic cardiomyopathy: a comparison study; pp. 1674–1677. [DOI] [PubMed] [Google Scholar]
- 21.Amudala S., Ali S., Najar F., Bouguila N. 2019 IEEE Symposium Series on Computational Intelligence (SSCI), Xiamen. IEEE; China: 2019. Variational Inference of Finite Generalized Gaussian Mixture Models; pp. 2433–2439. [DOI] [Google Scholar]
- 22.Dahmen J., Cook D. SynSys: a synthetic data generation system for healthcare applications. Sensors. 2019;vol. 19(5):1181. doi: 10.3390/s19051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazumder O., Banerjee R., Roy D., Bhattacharya S., Ghose A., Sinha A. Synthetic PPG signal generation to improve coronary artery disease classification: study with physical model of cardiovascular system. IEEE J Biomed Health Inform. 2022;vol. 26(5):2136–2146. doi: 10.1109/JBHI.2022.3147383. [DOI] [PubMed] [Google Scholar]
- 24.Shi J., Wang D., Tesei G., Norgeot B. Generating high-fidelity privacy-conscious synthetic patient data for causal effect estimation with multiple treatments. Front Artif Intell. 2022;vol. 5 doi: 10.3389/frai.2022.918813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arvanitis T.N., White S., Harrison S., Chaplin R., Despotou G. A method for machine learning generation of realistic synthetic datasets for validating healthcare applications. Health Inform J. 2022;vol. 28(2) doi: 10.1177/14604582221077000. 146045822210770. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., et al. GAN-based one dimensional medical data augmentation. Soft Comput. 2023;vol. 27(15):10481–10491. doi: 10.1007/s00500-023-08345-z. [DOI] [Google Scholar]
- 27.Das T., Wang Z., Sun J. in Proceedings of the 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining. ACM; Long Beach CA USA: 2023. TWIN: Personalized Clinical Trial Digital Twin Generation; pp. 402–413. [DOI] [Google Scholar]
- 28.Lu C., Reddy C.K., Wang P., Nie D., Ning Y. Multi-label clinical time-series generation via conditional GAN. IEEE Trans Knowl Data Eng. 2024;vol. 36(4):1728–1740. doi: 10.1109/TKDE.2023.3310909. [DOI] [Google Scholar]
- 29.X. Li, V. Metsis, H. Wang, A.H.H. Ngu, TTS-GAN: A Transformer-based Time-Series Generative Adversarial Network. arXiv, Jun. 26, 2022. Accessed: May 23, 2024. [Online]. Available: 〈http://arxiv.org/abs/2202.02691〉.
- 30.Zhang C., et al. Correction of out-of-focus microscopic images by deep learning. Comput Struct Biotechnol J. 2022;vol. 20:1957–1966. doi: 10.1016/j.csbj.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimwood A., et al. In: Noble J.A., Aylward S., Grimwood A., Min Z., Lee S.-L., Hu Y., editors. vol. 12967. Cham: Springer International Publishing; 2021. Endoscopic Ultrasound Image Synthesis Using a Cycle-Consistent Adversarial Network, in Simplifying Medical Ultrasound; pp. 169–178. (Lecture Notes in Computer Science). vol. 12967. vol. 12967. [DOI] [Google Scholar]
- 32.Wang J., Wu Q.M.J., Pourpanah F. DC-cycleGAN: bidirectional CT-to-MR synthesis from unpaired data. Comput Med Imaging Graph. 2023;vol. 108 doi: 10.1016/j.compmedimag.2023.102249. [DOI] [PubMed] [Google Scholar]
- 33.Shaban M.T., Baur C., Navab N., Albarqouni S. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) IEEE; Venice, Italy: 2019. Staingan: Stain Style Transfer for Digital Histological Images; pp. 953–956. [DOI] [Google Scholar]
- 34.Ma J., Xu H., Jiang J., Mei X., Zhang X.-P. DDcGAN: a dual-discriminator conditional generative adversarial network for multi-resolution image fusion. IEEE Trans Image Process. 2020;vol. 29:4980–4995. doi: 10.1109/TIP.2020.2977573. [DOI] [PubMed] [Google Scholar]
- 35.Pezoulas V., Tachos N., Fotiadis D. Generation of virtual patients for in silico cardiomyopathies drug development. 2019 IEEE 19th Int Conf Bioinforma Bioeng (BIBE) 2019:671–674. doi: 10.1109/BIBE.2019.00126. [DOI] [PubMed] [Google Scholar]
- 36.Pezoulas V.C., et al. A computational pipeline for data augmentation towards the improvement of disease classification and risk stratification models: a case study in two clinical domains. Comput Biol Med. 2021;vol. 134 doi: 10.1016/j.compbiomed.2021.104520. [DOI] [PubMed] [Google Scholar]
- 37.Kiagias D., Russo G., Sgroi G., Pappalardo F., Juárez M.A. Bayesian augmented clinical trials in TB therapeutic vaccination. Front Med Technol. 2021;vol. 3 doi: 10.3389/fmedt.2021.719380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tucker A., Wang Z., Rotalinti Y., Myles P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. Npj Digit Med. 2020;vol. 3(1):147. doi: 10.1038/s41746-020-00353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwep L.B., Guo T., Nagler T., Knibbe C.A.J., Meulman J.J., Van Hasselt J.G.C. Virtual patient simulation using copula modeling. Clin Pharmacol Ther. 2024;vol. 115(4):795–804. doi: 10.1002/cpt.3099. [DOI] [PubMed] [Google Scholar]
- 40.Kharya S., Soni S., Swarnkar T. Generation of synthetic datasets using weighted bayesian association rules in clinical world. Int J Inf Technol. 2022;vol. 14(6):3245–3251. doi: 10.1007/s41870-022-01081-x. [DOI] [Google Scholar]
- 41.H. Freedman, M.A. Miller, H. Williams, C. J. S. Jr, “Scaling and Querying a Semantically Rich, Electronic Healthcare Graph”.
- 42.Walonoski J., et al. Synthea™ Novel coronavirus (COVID-19) model and synthetic data set. Intell -Based Med. 2020;vol. 1–2 doi: 10.1016/j.ibmed.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koloi A., et al. 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) IEEE; Sydney, Australia: 2023. A comparison study on creating simulated patient data for individuals suffering from chronic coronary disorders; pp. 1–4. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Almeida A.J., et al. Synthetic patient data generation and evaluation in disease prediction using small and imbalanced datasets. IEEE J Biomed Health Inform. 2023;vol. 27(6):2670–2680. doi: 10.1109/JBHI.2022.3196697. [DOI] [PubMed] [Google Scholar]
- 45.El Emam K., Mosquera L., Fang X., El-Hussuna A. An evaluation of the replicability of analyses using synthetic health data. Sci Rep. Mar. 2024;vol. 14(1):6978. doi: 10.1038/s41598-024-57207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohaj O., Paralič J., Kushnir D., Vanko J.I. 2024 IEEE 22nd World Symposium on Applied Machine Intelligence and Informatics (SAMI) IEEE; Stará Lesná, Slovakia: 2024. Usability of a synthetically generated dataset for decision support; pp. 000435–000440. [DOI] [Google Scholar]
- 47.Z. Qian and R. Davis, Synthcity: a benchmark framework for diverse use cases of tabular synthetic data.
- 48.Huang G., Jafari A.H. Enhanced balancing GAN: minority-class image generation. Neural Comput Appl. 2023;vol. 35(7):5145–5154. doi: 10.1007/s00521-021-06163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dey S., Basuchowdhuri P., Mitra D., Augustine R., Saha S.K., Chakraborti T. In: Waiter G., Lambrou T., Leontidis G., Oren N., Morris T., Gordon S., editors. vol. 14122. Springer Nature Switzerland; Cham: 2024. BliMSR: Blind Degradation Modelling for Generating High-Resolution Medical Images; pp. 64–78. (in Lecture Notes in Computer Science). Medical Image Understanding and Analysis, vol. 14122. Medical Image Understanding and Analysis, vol. 14122. [DOI] [Google Scholar]
- 50.Segal B., Rubin D.M., Rubin G., Pantanowitz A. Evaluating the clinical realism of synthetic chest X-rays generated using progressively growing GANs. SN Comput Sci. 2021;vol. 2(4):321. doi: 10.1007/s42979-021-00720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kausar T., Lu Y., Kausar A., Ali M., Yousaf A. SD-GAN: a style distribution transfer generative adversarial network for covid-19 detection through X-ray images. IEEE Access. 2023;vol. 11:24545–24560. doi: 10.1109/ACCESS.2023.3253282. [DOI] [Google Scholar]
- 52.Yao Y., et al. 2022 IEEE 9th International Conference on Data Science and Advanced Analytics (DSAA) IEEE; Shenzhen, China: 2022. Conditional Variational Autoencoder with Balanced Pre-training for Generative Adversarial Networks; pp. 1–10. [DOI] [Google Scholar]
- 53.Han Z., et al. In: Greenspan H., Madabhushi A., Mousavi P., Salcudean S., Duncan J., Syeda-Mahmood T., Taylor R., editors. vol. 14229. Springer Nature Switzerland; Cham: 2023. Contrastive Diffusion Model with Auxiliary Guidance for Coarse-to-Fine PET Reconstruction, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2023; pp. 239–249. (Lecture Notes in Computer Science). vol. 14229. vol. 14229. [DOI] [Google Scholar]
- 54.Huang J., Wu Y., Wu H., Yang G. 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) IEEE; Glasgow, Scotland, United Kingdom: 2022. Fast MRI Reconstruction: How Powerful Transformers Are? pp. 2066–2070. [DOI] [PubMed] [Google Scholar]
- 55.Lin C.-H., Lin C.-S., Chou P.-Y., Hsu C.-C. An efficient data augmentation network for out-of-distribution image detection. IEEE Access. 2021;vol. 9:35313–35323. doi: 10.1109/ACCESS.2021.3062187. [DOI] [Google Scholar]
- 56.Wei L., Yadav A., Hsu W. In: Greenspan H., Madabhushi A., Mousavi P., Salcudean S., Duncan J., Syeda-Mahmood T., Taylor R., editors. vol. 14226. Springer Nature Switzerland; Cham: 2023. CTFlow: mitigating effects of computed tomography acquisition and reconstruction with normalizing flows; pp. 413–422. (Lecture Notes in Computer Science). Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, vol. 14226. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, vol. 14226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osuala R., et al. medigan: a Python library of pretrained generative models for medical image synthesis. J Med Imaging. 2023;vol. 10 doi: 10.1117/1.JMI.10.6.061403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong S., et al. In: Mukhopadhyay A., Oksuz I., Engelhardt S., Zhu D., Yuan Y., editors. vol. 13609. Springer Nature Switzerland; Cham: 2022. Flow-Based Visual Quality Enhancer for Super-Resolution Magnetic Resonance Spectroscopic Imaging, in Deep Generative Models; pp. 3–13. (Lecture Notes in Computer Science). vol. 13609. vol. 13609. [DOI] [Google Scholar]
- 59.He C., et al. HQG-Net: unpaired medical image enhancement with high-quality guidance. IEEE Trans Neural Netw Learn Syst. 2024:1–15. doi: 10.1109/TNNLS.2023.3315307. [DOI] [PubMed] [Google Scholar]
- 60.Li R., Bastiani M., Auer D., Wagner C., Chen X. In: Papież B.W., Yaqub M., Jiao J., Namburete A.I.L., Noble J.A., editors. vol. 12722. Springer International Publishing; Cham: 2021. Image Augmentation Using a Task Guided Generative Adversarial Network for Age Estimation on Brain MRI; pp. 350–360. (Lecture Notes in Computer Science). Medical Image Understanding and Analysis, vol. 12722. Medical Image Understanding and Analysis, vol. 12722. [DOI] [Google Scholar]
- 61.Tran N.-T., Tran V.-H., Nguyen N.-B., Nguyen T.-K., Cheung N.-M. On data augmentation for GAN training. IEEE Trans Image Process. 2021;vol. 30:1882–1897. doi: 10.1109/TIP.2021.3049346. [DOI] [PubMed] [Google Scholar]
- 62.Yang P., Hong Z., Yin X., Zhu C., Jiang R. In: De Bruijne M., Cattin P.C., Cotin S., Padoy N., Speidel S., Zheng Y., Essert C., editors. vol. 12902. Springer International Publishing; Cham: 2021. Self-supervised Visual Representation Learning for Histopathological Images; pp. 47–57. (in Lecture Notes in Computer Science). Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, vol. 12902. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, vol. 12902. [DOI] [Google Scholar]
- 63.Han S., Carass A., Schar M., Calabresi P.A., Prince J.L. 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) IEEE; Nice, France: 2021. Slice Profile Estimation From 2D MRI Acquisition Using Generative Adversarial Networks; pp. 145–149. [DOI] [Google Scholar]
- 64.Ahmadian M., et al. Overcoming data scarcity in radiomics/radiogenomics using synthetic radiomic features. Comput Biol Med. 2024;vol. 174 doi: 10.1016/j.compbiomed.2024.108389. [DOI] [PubMed] [Google Scholar]
- 65.Hosseini S., et al. MRI-based radiomics combined with deep learning for distinguishing IDH-mutant WHO grade 4 astrocytomas from IDH-wild-type glioblastomas. Cancers. 2023;vol. 15(3):951. doi: 10.3390/cancers15030951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parekh V.S., Jacobs M.A. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) IEEE; Venice, Italy: 2019. Radiomic Synthesis Using Deep Convolutional Neural Networks; pp. 1114–1117. [DOI] [Google Scholar]
- 67.Dakshit S., Dakshit S., Khargonkar N., Prabhakaran B. Bias analysis in healthcare time series (BAHT) decision support systems from meta data. J Healthc Inform Res. 2023;vol. 7(2):225–253. doi: 10.1007/s41666-023-00133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khorchani T., Gadiya Y., Witt G., Lanzillotta D., Claussen C., Zaliani A. SASC: a simple approach to synthetic cohorts for generating longitudinal observational patient cohorts from COVID-19 clinical data. Patterns. 2022;vol. 3(4) doi: 10.1016/j.patter.2022.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dissanayake T., Fernando T., Denman S., Sridharan S., Fookes C. Generalized generative deep learning models for biosignal synthesis and modality transfer. IEEE J Biomed Health Inform. 2023;vol. 27(2):968–979. doi: 10.1109/JBHI.2022.3223777. [DOI] [PubMed] [Google Scholar]
- 70.Isasa I., et al. Effect of incorporating metadata to the generation of synthetic time series in a healthcare context. 2023 IEEE 36th Int Symp Comput-Based Med Syst (CBMS) 2023:910–916. doi: 10.1109/CBMS58004.2023.00341. [DOI] [Google Scholar]
- 71.Yang Z., Li Y., Zhou G. TS-GAN: time-series GAN for sensor-based health data augmentation. ACM Trans Comput Healthc. 2023;vol. 4(2):1–21. doi: 10.1145/3583593. [DOI] [Google Scholar]
- 72.A. Seyfi, J.-F. Rajotte,R.T. Ng, Generating multivariate time series with COmmon Source CoordInated GAN (COSCI-GAN).
- 73.Dash S., Yale A., Guyon I., Bennett K.P. In: Michalowski M., Moskovitch R., editors. vol. 12299. Springer International Publishing; Cham: 2020. Medical Time-Series Data Generation Using Generative Adversarial Networks; pp. 382–391. (in Lecture Notes in Computer Science). Artificial Intelligence in Medicine, vol. 12299. Artificial Intelligence in Medicine, vol. 12299. [DOI] [Google Scholar]
- 74.Alcaraz J.M.L., Strodthoff N. Diffusion-based conditional ECG generation with structured state space models. Comput Biol Med. 2023;vol. 163 doi: 10.1016/j.compbiomed.2023.107115. [DOI] [PubMed] [Google Scholar]
- 75.Li H., Yu S., Principe J. Causal recurrent variational autoencoder for medical time series generation. Proc AAAI Conf Artif Intell. 2023;vol. 37(7):8562–8570. doi: 10.1609/aaai.v37i7.26031. [DOI] [Google Scholar]
- 76.Petralia F., Wang L., Peng J., Yan A., Zhu J., Wang P. A new method for constructing tumor specific gene co-expression networks based on samples with tumor purity heterogeneity. Bioinformatics. 2018;vol. 34(13):i528–i536. doi: 10.1093/bioinformatics/bty280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mansouri M., Khakabimamaghani S., Chindelevitch L., Ester M. Aristotle: stratified causal discovery for omics data. BMC Bioinforma. 2022;vol. 23(1):42. doi: 10.1186/s12859-021-04521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chunikhina E., Logan P., Kovchegov Y., Yambartsev A., Mondal D., Morgun A. The C-SHIFT algorithm for normalizing covariances. IEEE/ACM Trans Comput Biol Bioinform. 2023;vol. 20(1):720–730. doi: 10.1109/TCBB.2022.3151840. [DOI] [PubMed] [Google Scholar]
- 79.Ovando-Vázquez C., Cázarez-García D., Winkler R. Target–Decoy MineR for determining the biological relevance of variables in noisy datasets. Bioinformatics. 2021;vol. 37(20):3595–3603. doi: 10.1093/bioinformatics/btab369. [DOI] [PubMed] [Google Scholar]
- 80.De Los Santos H., Bennett K.P., Hurley J.M. MOSAIC: a joint modeling methodology for combined circadian and non-circadian analysis of multi-omics data. Bioinformatics. 2021;vol. 37(6):767–774. doi: 10.1093/bioinformatics/btaa877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fanaee-T H., Thoresen M. Multi-insight visualization of multi-omics data via ensemble dimension reduction and tensor factorization. Bioinformatics. 2019;vol. 35(10):1625–1633. doi: 10.1093/bioinformatics/bty847. [DOI] [PubMed] [Google Scholar]
- 82.Yang Z., Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2016;vol. 32(1):1–8. doi: 10.1093/bioinformatics/btv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pham T., Jimenez C. Simulated linear test applied to quantitative proteomics. Bioinformatics. 2016;vol. 32(17):i702–i709. doi: 10.1093/bioinformatics/btw440. [DOI] [PubMed] [Google Scholar]
- 84.Cusworth S., Gkoutos G.V., Acharjee A. A novel generative adversarial networks modelling for the class imbalance problem in high dimensional omics data. BMC Med Inform Decis Mak. 2024;vol. 24(1):90. doi: 10.1186/s12911-024-02487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahmed K.T., Sun J., Cheng S., Yong J., Zhang W. Multi-omics data integration by generative adversarial network. Bioinformatics. 2021;vol. 38(1):179–186. doi: 10.1093/bioinformatics/btab608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hess M., Hackenberg M., Binder H. Exploring generative deep learning for omics data using log-linear models. Bioinformatics. 2020;vol. 36(20):5045–5053. doi: 10.1093/bioinformatics/btaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wieder C., Lai R.P.J., Ebbels T.M.D. Single sample pathway analysis in metabolomics: performance evaluation and application. BMC Bioinforma. 2022;vol. 23(1):481. doi: 10.1186/s12859-022-05005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wieder C., et al. PathIntegrate: Multivariate modelling approaches for pathway-based multi-omics data integration. PLOS Comput Biol. 2024;vol. 20(3) doi: 10.1371/journal.pcbi.1011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andersson A., Lundeberg J. sepal: identifying transcript profiles with spatial patterns by diffusion-based modeling. Bioinformatics. 2021;vol. 37(17):2644–2650. doi: 10.1093/bioinformatics/btab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wendland P., Birkenbihl C., Gomez-Freixa M., Sood M., Kschischo M., Fröhlich H. Generation of realistic synthetic data using multimodal neural ordinary differential equations. Npj Digit Med. 2022;vol. 5(1):122. doi: 10.1038/s41746-022-00666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bauer D.F., et al. Generation of annotated multimodal ground truth datasets for abdominal medical image registration. Int J Comput Assist Radiol Surg. 2021;vol. 16(8):1277–1285. doi: 10.1007/s11548-021-02372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee S.H. Natural language generation for electronic health records. Npj Digit Med. 2018;vol. 1(1):63. doi: 10.1038/s41746-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang Y., Zhang S., Chi J. Multi-modal brain tumor data completion based on reconstruction consistency loss. J Digit Imaging. 2023;vol. 36(4):1794–1807. doi: 10.1007/s10278-022-00697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eckardt J.-N., et al. Mimicking clinical trials with synthetic acute myeloid leukemia patients using generative artificial intelligence. Npj Digit Med. 2024;vol. 7(1):76. doi: 10.1038/s41746-024-01076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haleem M.S., Ekuban A., Antonini A., Pagliara S., Pecchia L., Allocca C. Deep-learning-driven techniques for real-time multimodal health and physical data synthesis. Electronics. 2023;vol. 12(9):1989. doi: 10.3390/electronics12091989. [DOI] [Google Scholar]
- 96.Zhou X., et al. Multimodality MRI synchronous construction based deep learning framework for MRI-guided radiotherapy synthetic CT generation. Comput Biol Med. 2023;vol. 162 doi: 10.1016/j.compbiomed.2023.107054. [DOI] [PubMed] [Google Scholar]
- 97.Sun H., et al. Research on new treatment mode of radiotherapy based on pseudo-medical images. Comput Methods Prog Biomed. 2022;vol. 221 doi: 10.1016/j.cmpb.2022.106932. [DOI] [PubMed] [Google Scholar]
- 98.S. Biswal, P. Zhuang, A. Pyrros, N. Siddiqui, S. Koyejo, J. Sun, EMIXER: End-to-end Multimodal X-ray Generation via Self-supervision. arXiv, Jan. 15, 2021. Accessed: May 23, 2024. [Online]. Available: 〈http://arxiv.org/abs/2007.05597〉.
- 99.Z. Wang and J. Sun, “PromptEHR: Conditional Electronic Healthcare Records Generation with Prompt Learning.” arXiv, Oct. 11, 2022. Accessed: May 23, 2024. [Online]. Available: 〈http://arxiv.org/abs/2211.01761〉.
- 100.Paulin G., Ivasic‐Kos M. Review and analysis of synthetic dataset generation methods and techniques for application in computer vision. Artif Intell Rev. 2023;vol. 56(9):9221–9265. doi: 10.1007/s10462-022-10358-3. [DOI] [Google Scholar]
- 101.Y. Lu et al., Machine Learning for Synthetic Data Generation: A Review. arXiv, Jun. 30, 2024. Accessed: Jul. 03, 2024. [Online]. Available: 〈http://arxiv.org/abs/2302.04062〉.
- 102.X. Guo and Y. Chen, Generative AI for Synthetic Data Generation: Methods, Challenges and the Future.” arXiv, Mar. 06, 2024. Accessed: Jul. 03, 2024. [Online]. Available: 〈http://arxiv.org/abs/2403.04190〉.
- 103.Robnik-Sikonja M. Data generators for learning systems based on RBF networks. IEEE Trans Neural Netw Learn Syst. 2016;vol. 27(5):926–938. doi: 10.1109/TNNLS.2015.2429711. [DOI] [PubMed] [Google Scholar]