Summary
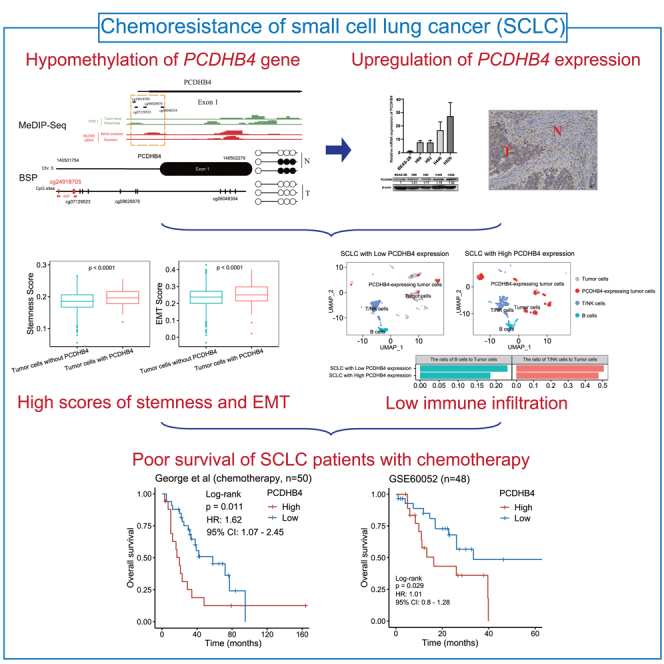
Platinum-based chemo-resistance is the major issue for the treatment of small cell lung cancer (SCLC). The integrative analysis of multi-omics data is a reliable approach for discovering novel biomarkers associated with chemo-resistance. Here, multi-omics integrative analysis and Cox regression found that higher expression of PCDHB4 was associated with poorer survival of SCLC patients who received chemotherapy. PCDHB4 gene was hypomethylated and upregulated in SCLC, which was validated in the levels of promoter methylation, mRNA, and protein expression. Mechanistically, using bulk RNA-seq data, functional enrichment analysis indicated that higher PCDHB4 expression was associated with lower immune infiltration. The analysis of single-cell RNA-seq (scRNA-seq) found that SCLC cells with PCDHB4 expression exhibited the characteristics of stemness and EMT. In addition, the high expression and hypomethylation of PCDHB4 were also significantly associated with poor survival in lung squamous cell carcinoma. In summary, PCDHB4 is a potential prognostic biomarker of platinum-based chemotherapy in SCLC.
Subject areas: cancer systems biology, cancer, transcriptomics
Graphical abstract
Highlights
-
•
High expression of PCDHB4 is associated with poor survival of SCLC
-
•
PCDHB4 gene is hypomethylated and upregulated in SCLC
-
•
PCDHB4 promotes SCLC proliferation and chemo-resistance
-
•
High PCDHB4 expression is associated with low immune infiltration, stemness, and EMT
Cancer systems biology; Cancer; Transcriptomics.
Introduction
Small cell lung cancer (SCLC) is a very aggressive neuroendocrine tumor. Platinum-based chemotherapy (cisplatin or carboplatin) is the standard treatment for SCLC. The response of SCLC patients to chemotherapy is variable.1,2 The prediction of therapeutic response and prognosis is still difficult, so it is necessary to develop new prognostic biomarkers in SCLC.
Recently, comprehensive bioinformatics analyses have been used to classify patients and identify biomarkers to predict therapeutic response and prognosis in SCLC.3 By analyzing RNA-seq data of SCLC tumors, a study classified SCLC into 4 subtypes: SCLC-A, SCLC-N, SCLC-P, and SCLC-I.4 When applying the 4 subtypes into the IMpower133 clinical trial, the SCLC-P subtype was found to have the worst prognosis in the carboplatin/etoposide treatment group.5 Via single-cell RNA-seq (scRNA-seq) of circulating tumor cell-derived xenografts (CDX), Stewart et al. revealed that intratumoral heterogeneity was increased after the onset of chemo-resistance in SCLC.6 A scRNA-seq study found that an SCLC subpopulation with high PLCG2 expression was associated with stem-like and pro-metastatic features, as well as worse overall survival (OS).7 RNA-seq of SCLC patient-derived xenografts (PDX) identified SLFN11, whose expression was decreased in chemo-resistant SCLC.8
The identification of abnormal DNA methylation in human cancer is expected to discover new diagnostic markers that are more stable than RNA samples.9 Increasing evidence has demonstrated that DNA methylation plays an important role in SCLC development and drug resistance. A microarray study of CpG methylation identified SCLC-specific DNA methylation sites in neural cell fate-specifying transcription factor genes (NEUROD1, HAND1, ZNF423, and REST). The methylation of these genes could result in a differentiation defect of neuroendocrine cells, thus promoting the transition toward SCLC.10 An integrated study of DNA methylation array and drug sensitivity in SCLC cell lines identified the increased promoter methylation of SLFN11 that correlated with resistance to DNA damaging agents.11
Aberrant DNA methylation is associated with abnormal gene expression. However, the integrated analysis of DNA methylation and gene expression in SCLC has not been studied thus far. In this study, by performing a comprehensive data analysis of gene expression, DNA methylation, drug sensitivity, and patient survival, we identified the PCDHB4 gene, and its expression and CpG methylation were validated as novel prognostic biomarkers in SCLC, as well as the other subtype of lung cancer, lung squamous cell carcinoma (LUSC).
Results
Identification of cisplatin resistance-associated genes that may be mediated by DNA methylation in small cell lung cancer cells
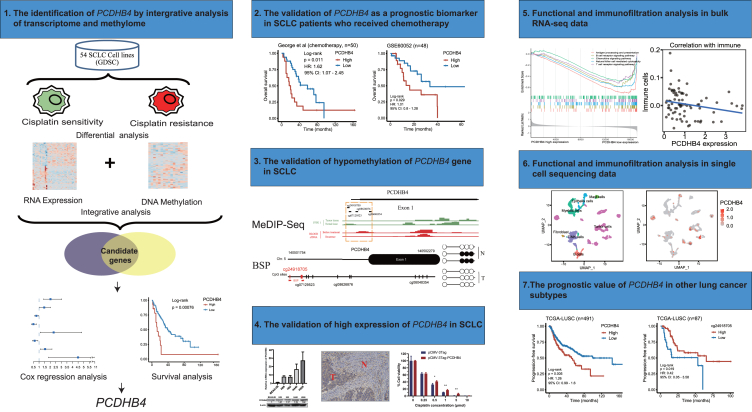
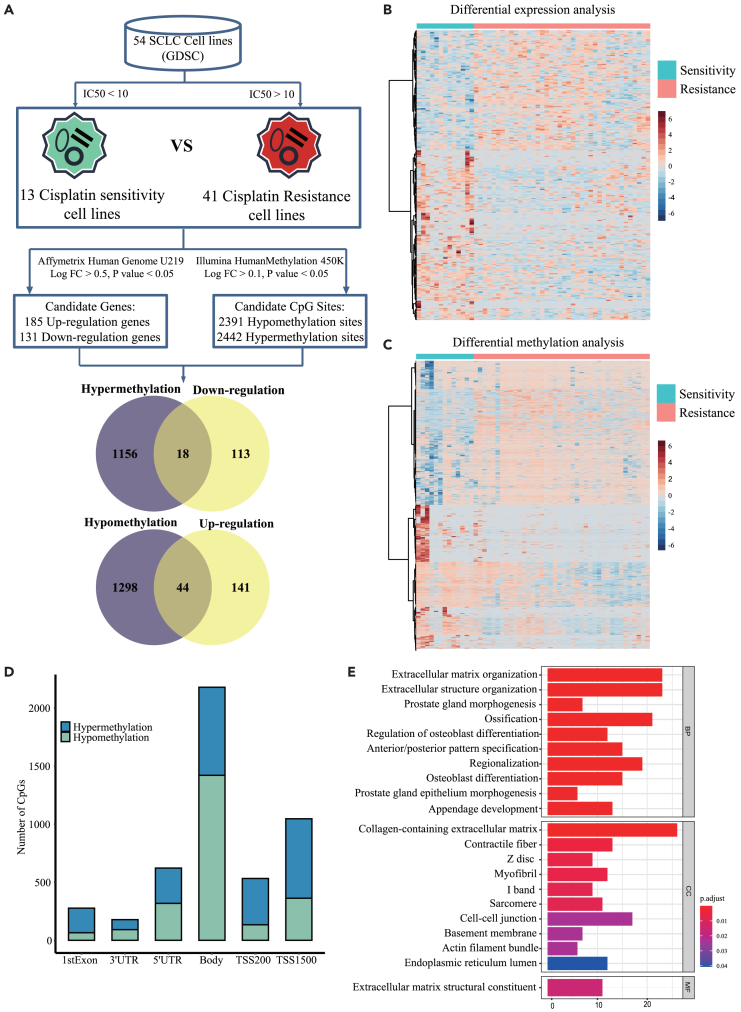
The workflow of the study is shown in Figure 1. To identify cisplatin resistance-associated genes that may be regulated by DNA methylation, we first collected gene expression data and cisplatin IC50 values in 54 SCLC cell lines from GDSC. SCLC cell lines were divided into 13 cisplatin-sensitive cell lines (IC50 < 10) and 41 cisplatin-resistant cell lines (IC50 > 10) (Figure 2A; Table S1). We mined differentially expressed genes between cisplatin-sensitive and cisplatin-resistant SCLC cells. According to the cut-off of p < 0.05 and |logFC|≥0.5, 316 differentially expressed genes were identified, including 185 up-regulated genes and 131 down-regulated genes (Figures 2A and 2B). We then collected DNA methylation data in SCLC cell lines from GDSC and analyzed the differentially methylated sites between cisplatin-sensitive and cisplatin-resistant SCLC cells. According to the cut-off of p < 0.05 and |logFC|≥0.1, a total of 4833 differentially methylated CpG sites were detected, of which 2442 sites were hypermethylated and 2391 were hypomethylated (Figures 2A and 2C). The 4833 differential CpG sites were located in different regions of genes, including the first exon (5.7%), 3′ UTR (3.7%), 5′ UTR (12.9%), gene body (45.1%), TSS1500 (21.6%) and TSS200 (11%) (Figure 2D). Generally, the hypermethylation and hypomethylation of the promoter in a gene correspond to its low and high expression, respectively. We intersected the down-regulated/hypermethylated genes as well as the up-regulated/hypomethylated genes. The Venn diagram identified 62 genes, including 18 down-regulated/hypermethylated genes and 44 up-regulated/hypomethylated genes (Figure 2A; Table S2). GO analysis revealed that the top 10 terms of the 316 differentially expressed genes were enriched in extracellular organization and cell-cell junction (Figure 2E). In summary, we identified 62 cisplatin resistance-associated genes whose expression may be regulated by DNA methylation in SCLC cells.
Figure 1.
The workflow of the study
Figure 2.
Integrative analysis of differential DNA methylation and gene expression between cisplatin-sensitive and cisplatin-resistant SCLC cell lines
(A) The workflow of bioinformatics analysis. The integrative analysis identifies 18 down-regulated/hypermethylated genes and 44 up-regulated/hypomethylated genes.
(B) Heatmap of the differentially expressed genes between cisplatin-sensitive and cisplatin-resistant SCLC cell lines.
(C) Heatmap of the differentially methylated CpG sites between cisplatin-sensitive and cisplatin-resistant SCLC cell lines.
(D) The distribution of the differentially methylated CpG sites across gene regions.
(E) GO analysis of the differentially expressed genes (top 10 terms).
Identification of the PCDHB4 gene, whose high expression is associated with poor prognosis in small cell lung cancer patients
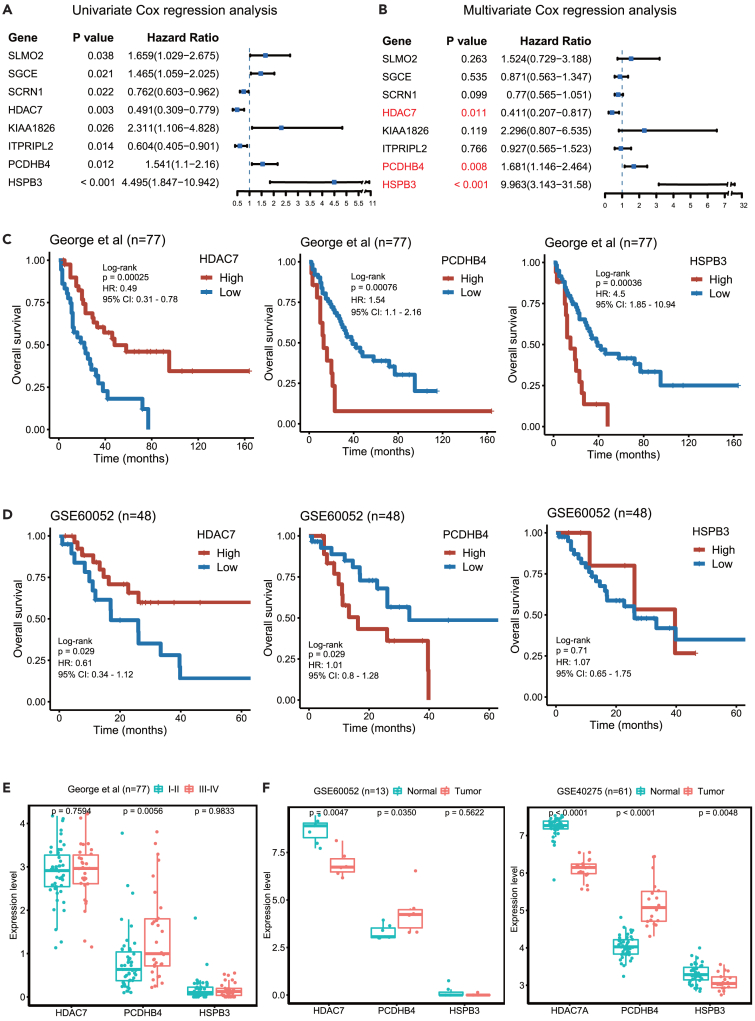
Since chemo-resistance is related to poor prognosis, we further applied Cox regression analysis to identify the genes associated with the survival of SCLC patients. We used a previously published dataset that included the RNA-seq and survival data of 77 SCLC patients.12 Using the 62 cisplatin resistance-associated genes, univariate Cox regression analysis identified 8 genes (SLMO2, SGCE, SCRN1, HDAC7, KIAA1826, ITPRIPL2, PCDHB4, and HSPB3), and their expression was significantly associated with the OS in SCLC patients (Figure 3A). Furthermore, multivariate Cox regression analysis indicated that the expression of the HDAC7, PCDHB4, or HSPB3 gene was an independent prognostic factor in SCLC patients (Figure 3B). Kaplan-Meier analysis showed that high expression of the PCDHB4 or HSPB3 gene was significantly associated with poor OS in SCLC patients, while low expression of the HDAC7 gene was significantly associated with poor OS in SCLC patients (Figure 3C).
Figure 3.
The PCDHB4 gene is associated with SCLC prognosis
(A) Univariate Cox regression identifies 8 genes associated with the OS of SCLC patients.
(B) Multivariate Cox regression identifies 3 independent prognostic genes (HDAC7, PCDHB4, and HSPB3).
(C) Kaplan-Meier curves in 77 SCLC patients based on the expression of HDAC7, PCDHB4, or HSPB3 using the George dataset.
(D) Kaplan-Meier curves in 48 SCLC patients based on the expression of HDAC7, PCDHB4, or HSPB3 using an independent validation dataset GEO: GSE60052.
(E) Boxplot showing the expression of HDAC7, PCDHB4, and HSPB3 in different stages of SCLC. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(F) Boxplot showing the expression of HDAC7, PCDHB4, and HSPB3 in SCLC tumors and adjacent normal tissues. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
Furthermore, we validated the three genes (HDAC7, PCDHB4, and HSPB3) using an independent SCLC dataset (GEO: GSE60052). Kaplan-Meier analysis demonstrated that SCLC patients with higher PCDHB4 expression or lower HDAC7 expression exhibited worse OS. There was no significant difference in the OS between the SCLC patients with high and low HSPB3 expression (Figure 3D).
In the George dataset, the expression of PCDHB4 was significantly higher in late-stage SCLC tumors than in early-stage SCLC tumors, while the expression of HDAC7 and HSPB3 was not significantly different between late-stage and early-stage SCLC tumors (Figure 3E). We then examined the expression of HDAC7, PCDHB4, and HSPB3 in SCLC tumors and normal tissues. In both the GEO: GSE60052 and GEO: GSE40275 datasets, the expression of PCDHB4 was significantly higher, and the expression of HDAC7 was significantly lower in SCLC tumors compared with adjacent normal tissues (Figure 3F). The expression of HSPB3 is significantly lower in SCLC tumors in the GEO: GSE40275 dataset, but not in the GEO: GSE60052 dataset (Figure 3F). Therefore, in all tested datasets, the PCDHB4 expression is higher in SCLC tumors, and its higher expression in tumors exhibits a worse prognosis in SCLC patients.
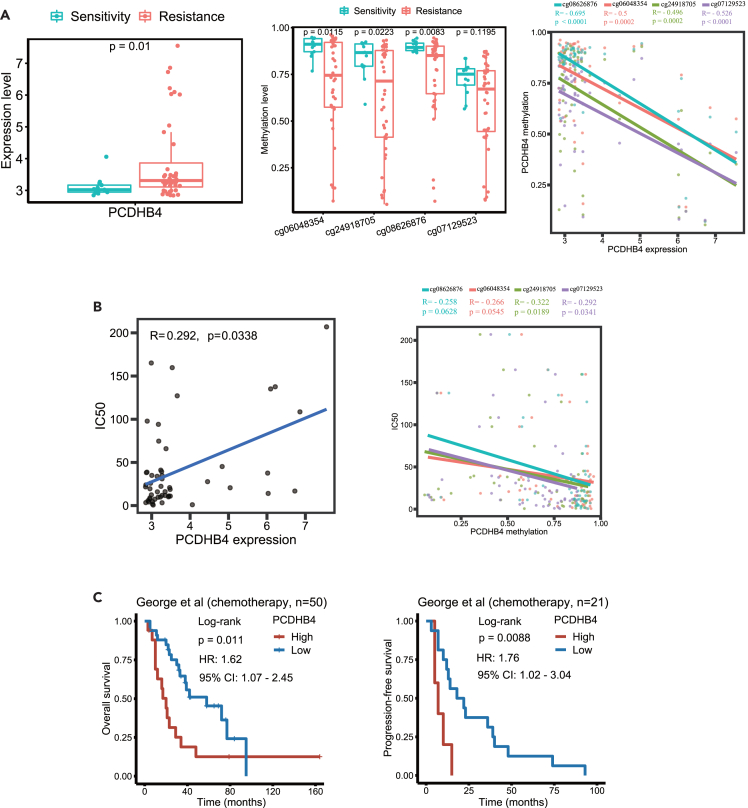
Hypomethylation of CpG sites in the PCDHB4 gene correlates with its high expression and cisplatin resistance in small cell lung cancer
We further assessed whether PCDHB4 gene expression correlated with DNA methylation in its promoter region. In the GDSC database, the expression of PCDHB4 was significantly higher in cisplatin-resistant SCLC cell lines (Figure 4A). Among the 4 CpG sites (cg06048354, cg24918705, cg08626876, and cg07129523) in the PCDHB4 promoter region, the methylation levels of the 3 CpG sites (cg06048354, cg24918705 and cg08626876) were significantly lower in cisplatin-resistant SCLC cells (Figure 4A). In 54 SCLC cell lines, the methylation levels of the 4 CpG sites were negatively correlated with the expression of PCDHB4 (Figure 4A).
Figure 4.
The correlation of PCDHB4 methylation with its expression and cisplatin resistance in SCLC
(A) Boxplots showing PCDHB4 expression (left panel) and methylation level of CpG sites within the PCDHB4 promoter (middle panel) in cisplatin-sensitive and cisplatin-resistant SCLC cell lines. Correlation plot (right panel) showing the negative correlation between the CpG methylation level and the expression of PCDHB4 in SCLC cell lines. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(B) Correlation plot (left panel) showing the positive correlation between the PCDHB4 expression and the cisplatin IC50 in SCLC cell lines. Correlation plot (right panel) showing the negative correlation between the PCDHB4 CpG methylation level and the cisplatin IC50 in SCLC cell lines.
(C) Kaplan-Meier curves of SCLC patients treated with platinum-based chemotherapy using the George dataset.
In 54 SCLC cell lines, the expression of PCDHB4 significantly correlated with the IC50 of cisplatin (Figure 4B). In addition, the methylation levels of the 2 CpG sites (cg24918705 and cg07129523) were negatively correlated with the IC50 of cisplatin in SCLC cells (Figure 4B). Notably, we found that in SCLC patients treated with chemotherapy, the OS of patients with low PCDHB4 expression was significantly longer, and the progression-free survival (PFS) of SCLC patients with high PCDHB4 expression was significantly shorter (Figure 4C). Therefore, the hypomethylation of CpG sites in the PCDHB4 gene correlates with its high expression and cisplatin resistance, and high expression of PCDHB4 in SCLC patients who received chemotherapy is associated with fast disease progression.
Hypomethylation of the PCDHB4 gene in small cell lung cancer was validated by Methylated DNA immuno-precipitation sequencing and bisulfite sequencing PCR
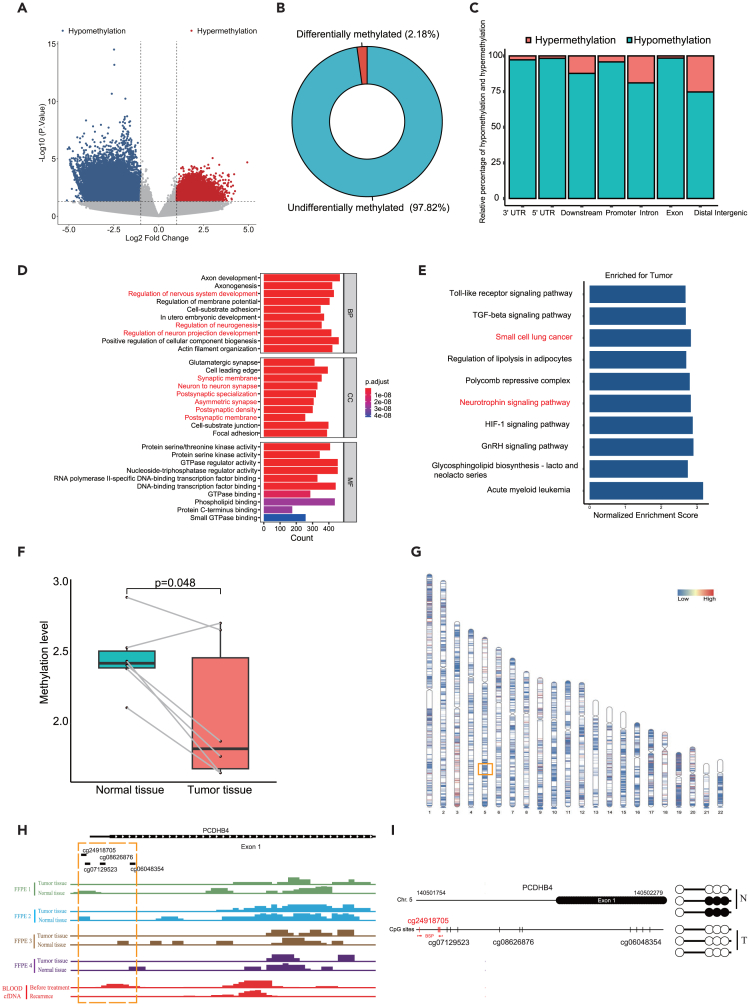
We further detected the methylation status of the PCDHB4 gene in our collected SCLC clinical samples. First, we used MeDIP-seq to detect the whole genome methylation level in SCLC tumors and adjacent normal tissues. According to the cut-off of p < 0.05 and |logFC|≥1, 179,468 (2.18%) differentially methylated sites were detected between SCLC tumors and adjacent normal tissues (Figures 5A and 5B). 83.56% (149,959/179,468) of these differentially methylated sites were hypomethylated. The 149,959 hypomethylated sites were located in different regions of genes, including the Exon (98.45%), 3′UTR (97.28%), 5′UTR (98.30%), Downstream (87.77%), Distal Intergenic (74.72%), Intron (81.09%) and Promoter (95.83%) (Figure 5C). To understand the biological functions of the identified differentially methylated genes, we performed GO annotation analysis. The result indicated that the differentially methylated genes were significantly enriched in the regulation of neurogenesis, regulation of neuron projection development, and neuron to neuron synapse (Figure 5D). GSEA indicated that genes were significantly enriched in small cell lung cancer and neurotrophin signaling pathways (Figure 5E).
Figure 5.
The validation of hypomethylation of the PCDHB4 gene in SCLC specimens by MeDIP-seq and BSP
(A) Volcano plot of differentially methylated sites between SCLC tumors and adjacent normal tissues by MeDIP-seq.
(B) The proportion of differentially methylated sites in all detected methylated sites.
(C) The category of genomic locations for hypermethylated or hypomethylated sites.
(D) GO analysis for the differentially methylation genes. Top 10 terms are shown. Neuron-associated pathways are marked in red.
(E) Bar plot of GSEA based on the differentially methylation genes.
(F) Boxplot showing the methylation level within the PCDHB4 promoter in SCLC tumors and adjacent normal tissues.
(G) Distribution of differentially methylated sites on chromosomes. The red area represents the hypermethylated region, and the blue area represents the hypomethylated region. The PCDHB4 gene locates in the orange box.
(H) The peak maps of methylation with PCDHB4 promoter by MeDIP-seq. DNA methylation was detected in SCLC tumors and adjacent normal tissues (FFPE 1, FFPE 2, FFPE 3, and FFPE 4) by MeDIP-seq. cfDNA methylation of an SCLC patient (before treatment and after recurrence) was detected by MeDIP-seq.
(I) BSP analysis of the CpG site cg24918705 in SCLC tumor and adjacent normal tissue.
We then examined the methylation level of PCDHB4 with the promoter region between SCLC tumors and normal tissues. We found that the methylation level of the PCDHB4 promoter was significantly lower in SCLC tumors, compared with normal tissues (Figure 5F). Furthermore, we mapped the differentially methylated sites to the 22 autosomal chromosomes and visualized the site locations across the chromosomes, and PCDHB4 gene was located in the hypomethylation region of chromosome 5 (Figure 5G). As indicated in Figure 5H, the methylation level in the PCDHB4 promoter region including the 4 CpG sites (cg06048354, cg24918705, cg08626876, and cg07129523) was lower in SCLC tumors than in adjacent normal tissues. Interestingly, by MeDIP-seq, we detected the methylation level of the PCDHB4 gene of blood cfDNA in an SCLC patient who was treated with carboplatin and etoposide. We observed that the methylation level of cfDNA in the CpG sites was lower when the patient relapsed than before treatment. Finally, by BSP, the CpG site (cg24918705) was validated to be de-methylated in SCLC tumor, compared with normal tissue (Figure 5I). Therefore, our results validate that the PCDHB4 gene is lowly methylated in SCLC.
The expression of PCDHB4 in small cell lung cancer is high, which promotes cellular proliferation and chemo-resistance
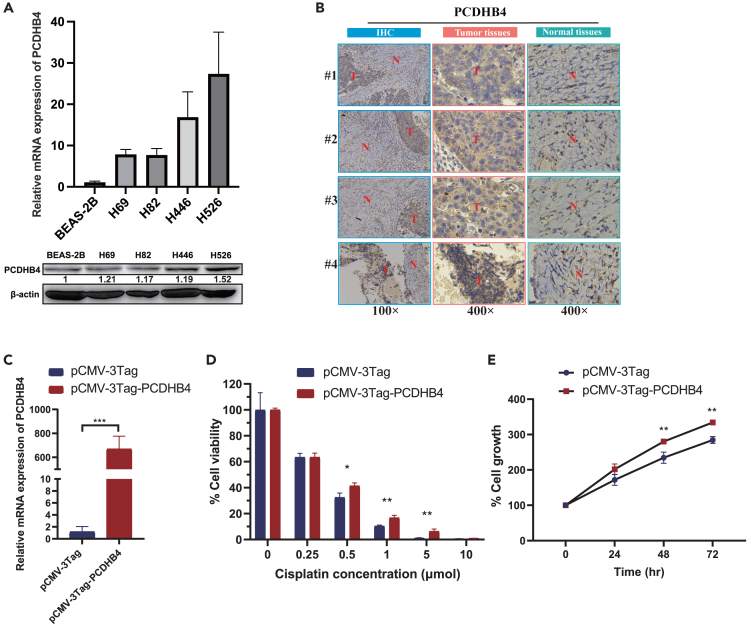
Next, we performed RT-PCR, Western blot, and IHC experiments to validate whether the expression of PCDHB4 was higher in SCLC. The result of RT-PCR showed higher mRNA levels of PCDHB4 in four SCLC cell lines compared with normal cells (BEAS-2B). Meanwhile, western blot showed that the protein expressions were also higher in SCLC cells (Figure 6A). To further verify the expression of PCDHB4 in SCLC clinical samples, IHC staining demonstrated that PCDHB4 expression was higher in SCLC tumor tissues compared to normal tissues (Figure 6B).
Figure 6.
The expression of PCDHB4 and in vitro functional analysis in SCLC
(A) qRT-PCR and western blot showing the higher expression of PCDHB4 in SCLC cell lines (H69, H82, H446 and H526) compared with human lung epithelial cells (BEAS-2B).
(B) Representative IHC images of PCDHB4 expression in SCLC clinical specimens. T: Tumor tissues, N: Normal tissues.
(C) PCDHB4 overexpression in SBC2 cells confirmed by qRT-PCR. The differences between groups were analyzed by unpaired t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(D) PCDHB4 overexpression reduces the cell death of SBC2 cells induced by different concentrations of cisplatin. The differences between groups were analyzed by unpaired t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(E) Cell proliferation assay of SBC2 cells transfected with control vector (pCMV-3Tag) and PCDHB4 overexpression vector (pCMV-3Tag-PCDHB4). The differences between groups were analyzed by unpaired t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
To further elucidate the function of PCDHB4 in SCLC, in vitro functional experiments were conducted. PCDHB4 overexpression plasmid was transfected in SBC2 SCLC cells (Figure 6C). Cell proliferation assay demonstrated that the cisplatin-induced cell death was reduced when SBC2 cells were transfected with PCDHB4 overexpression plasmid, compared with empty vector transfection (Figure 6D). Furthermore, overexpression of PCDHB4 enhanced cell proliferation (Figure 6E). Therefore, these data indicate that PCDHB4 is highly expressed in SCLC, which promotes cellular proliferation and chemo-resistance.
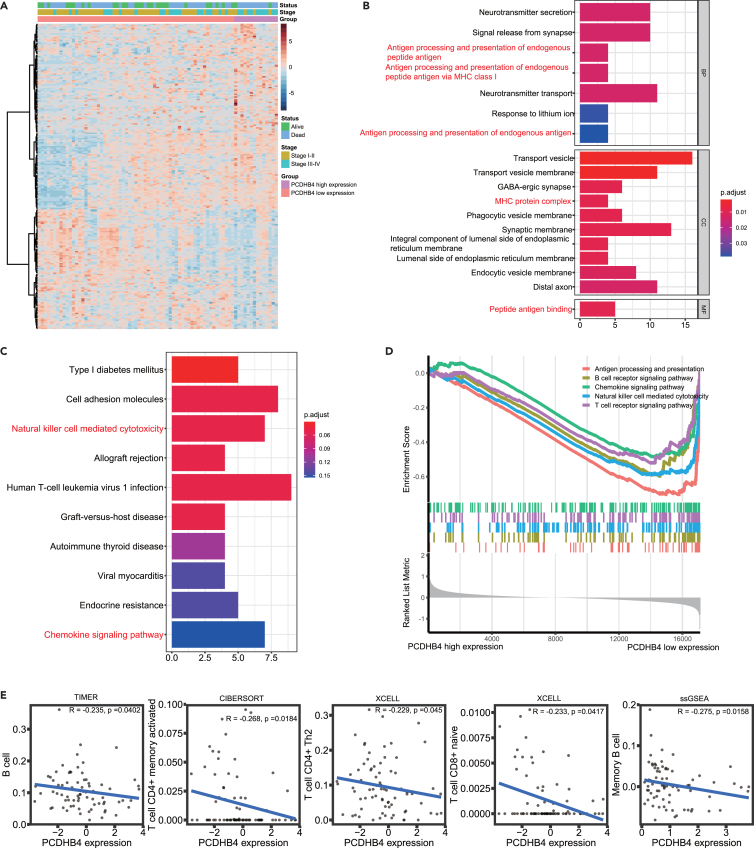
Higher PCDHB4 expression in small cell lung cancer tumors is associated with a lower infiltration level of immune cells
To explore the mechanism by which PCDHB4 affects the prognosis of SCLC patients, functional analyses were used to identify potential biological processes and pathways enriched in SCLC tumors with high or low PCDHB4 expression. In the George dataset, we identified the differentially expressed genes between the low and high PCDHB4 expression groups. As indicated in the heatmap, 226 differentially expressed genes were identified (Figure 7A). Using the differentially expressed genes, the GO and KEGG analysis indicated significant enrichment of immune-associated signaling pathways, such as antigen processing and presentation, MHC protein complex, and natural killer cell-mediated cytotoxicity (Figures 7B and 7C). GSEA indicated that genes associated with low PCDHB4 expression were significantly enriched in immune-related biological processes, such as antigen processing and presentation, B and T cell receptor signaling, and natural killer cell-mediated cytotoxicity (Figure 7D). Different algorithms, including TIMER, CIBERSORT, and XCELL, were used to evaluate immune cell infiltration in SCLC tumors (George cohort). The TIMER algorithm indicated that SCLC tumors with low PCDHB4 expression had higher infiltration of B cells. The CIBERSORT and XCELL algorithms revealed that higher infiltration of T cells was present in SCLC tumors with low PCDHB4 expression (Figure 7E). The ssGSEA analysis showed that PCDHB4 expression was negatively correlated with the content of memory B cells (Figure 7E). These data suggest that SCLC tumors with high PCDHB4 expression exhibit low immune infiltration, which may lead to poor survival of SCLC patients.
Figure 7.
Functional enrichment analysis of SCLC tumors with low or high PCDHB4 expression
(A) Heatmap of the differentially expressed genes between SCLC tumors with high and low PCDHB4 expression.
(B) GO analysis for the differentially expressed genes. Immune-associated terms are marked in red.
(C) KEGG analysis for the differentially expressed genes. Immune-associated pathways are marked in red.
(D) GSEA between PCDHB4-high and PCDHB4-low expression group. Multiple immune-related gene sets are enriched in the PCDHB4-low expression group.
(E) The significant correlation of immune cell infiltration and PCDHB4 expression in SCLC tumors obtained by different algorithms (TIMER, CIBERSORT, XCELL and ssGSEA).
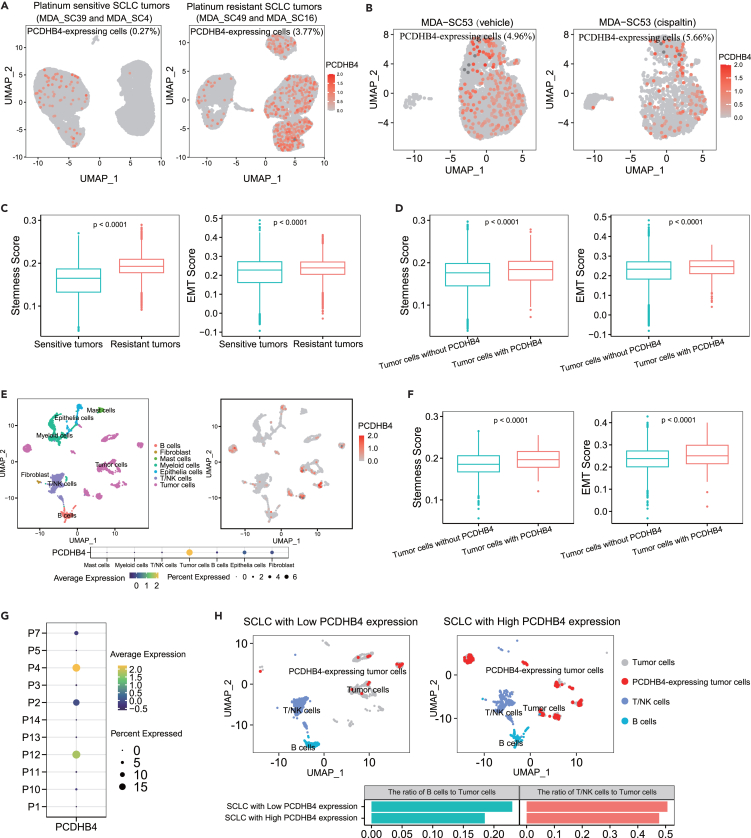
Single-cell dissection of small cell lung cancer tumors with PCDHB4 expression
Next, we included two scRNA-seq datasets (GEO: GSE138474 and publicly available dataset by Tian et al.13) to further dissect the expression characteristics of PCDHB4 in SCLC tumors at single-cell resolution. In the GEO: GSE138474 dataset containing scRNA-seq data of SCLC CDXs,6 we screened out SCLC cells by neuroendocrine markers (UCHL1, NCAM1, SYP, and CHGA) and showed the dissimilarity and distribution of these cells by UMAP dimensionality reduction. We then dissected the PCDHB4-expressing cells at the single-cell level. In platinum-sensitive SCLC tumors (MDA-SC39 and MDA-SC4), PCDHB4 was only expressed in 0.27% of SCLC cells. However, in platinum-resistant SCLC tumors (MDA-SC16 and MDA-SC49), PCDHB4-expressing cells were more abundant (3.77%) (Figure 8A). MDA-SC53 (vehicle) and MDA-SC53 (ciaplatin) were SCLC CDXs (MDA-SC53) treated with vehicle control and cisplatin, until cisplatin-treated SCLC tumor relapsed. The UMAP plot showed that cisplatin-relapsed SCLC cells had a higher proportion of PCDHB4-expressing cells (5.66%) than vehicle-treated SCLC cells (4.96%) (Figure 8B). Furthermore, we found that both epithelial-mesenchymal transition (EMT) and stemness scores of SCLC cells were significantly higher in the platinum-resistant group, compared to the sensitive group (Figure 8C). We also assessed EMT and stemness scores in SCLC cells with PCDHB4 expression and without PCDHB4 expression. SCLC cells with PCDHB4 expression had higher EMT and stemness scores, compared to SCLC cells without PCDHB4 expression (Figure 8D).
Figure 8.
Single-cell dissection of SCLC tumors with PCDHB4 expression
(A) UMAP plot of all SCLC tumor cells indicates the expression status of PCDHB4 in platinum-sensitive and platinum-resistant SCLC tumors.
(B) UMAP plot of all SCLC tumor cells indicates the expression status of PCDHB4 in SCLC tumors receiving vehicle and cisplatin treatment.
(C) Scoring of stemness and EMT gene signatures in platinum-sensitive and resistant SCLC tumors. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(D) Scoring of stemness and EMT gene signatures in SCLC cells with or without PCDHB4 expression. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(E) UMAP plots indicate the clustering of all cell types and the expression of PCDHB4 in all cell types. Bubble plot indicates that PCDHB4 is predominantly expressed in tumor cells.
(F) Scoring of stemness and EMT gene signatures in SCLC tumor cells with or without PCDHB4 expression. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(G) Expression of PCDHB4 in tumor cells of each SCLC patient. The SCLC patients (P2, P4, P7 and P12) are divided into the group with high PCDHB4 expression. The SCLC patients (P1, P3, P5, P10, P11, P13, and P14) are divided into the group with low PCDHB4 expression.
(H) UMAP plot of all B cells, T/NK cells, and tumor cells indicates that the ratio of B cells or T/NK cells to tumor cells is higher in the group with low PCDHB4 expression.
The dataset by Tian et al. contained scRNA-seq data of surgically resected SCLC tumors from 11 SCLC patients, including the single-cell transcriptome data of 7 distinct cell types (B cells, fibroblast, mast cells, myeloid cells, epithelial cells, T/NK cells, and tumor cells).13 The clustering of all cells revealed that PCDHB4 was predominantly expressed in tumor cells (Figure 8E). Similarly, tumor cells expressing PCDHB4 exhibited higher stemness and EMT scores, compared to tumor cells without PCDHB4 expression (Figure 8F). Furthermore, according to the expression of PCDHB4 in tumor cells, SCLC patients were divided into the group of SCLC with high PCDHB4 expression (P2, P4, P7, and P12) and the group of SCLC with low PCDHB4 expression (P1, P3, P5, P10, P11, P13, and P14) (Figure 8G). The clustering of T/NK cells, B cells, and tumor cells indicated that the ratio of T/NK cells or B cells to tumor cells was higher in the group with low PCDHB4 expression, compared with the group with high PCDHB4 expression (Figure 8H).
In summary, the single-cell sequencing data indicate that PCDHB4-expressing SCLC cells are associated with the “stemness” and “EMT” characteristics, as well as low immune infiltration.
Evaluation of PCDHB4 as a prognostic marker in other subtypes of lung cancer
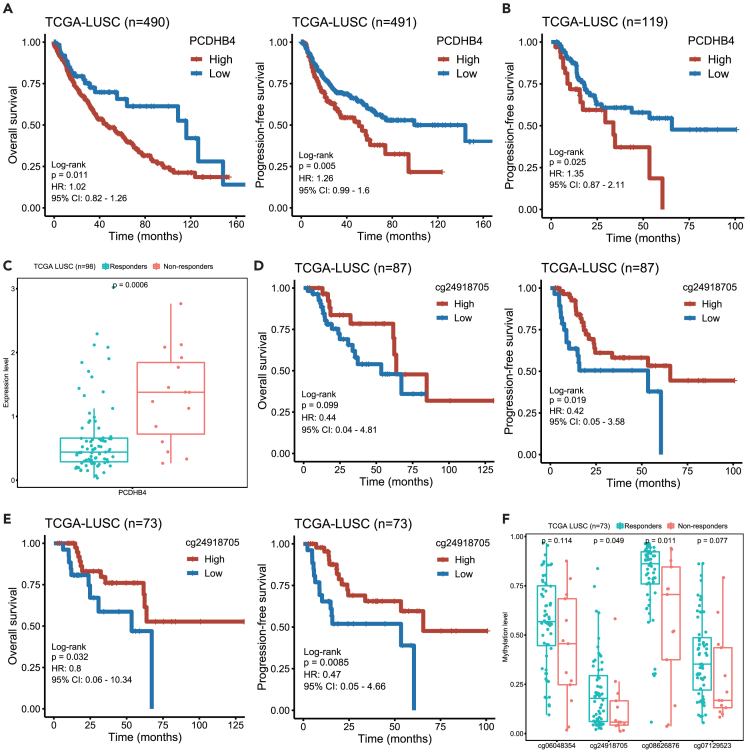
We further assessed whether the expression and CpG methylation of the PCDHB4 gene were able to predict prognosis in other subtypes of lung cancer, lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC). By mining TCGA data, shorter progression-free survival (PFS) and OS were observed in LUSC patients with higher PCDHB4 expression (Figure 9A). In LUSC patients who had received platinum-based chemotherapy, we also found that the FPS was shorter in patients with higher PCDHB4 expression (Figure 9B). According to the response to platinum-based chemotherapy, the LUSC patients were further divided into two groups: responders (patients who had a partial or complete response) and non-responders (patients who had clinical progressive or stable disease). As shown in Figure 9C, the expression of PCDHB4 was significantly higher in the non-responder group.
Figure 9.
The expression and CpG methylation of the PCDHB4 as prognostic markers in LUSC
(A) Kaplan-Meier curves of OS (left panel) and PFS (right panel) of LUSC patients with low and high PCDHB4 expression.
(B) Kaplan-Meier curves of the PFS of LUSC patients with low and high PCDHB4 expression, who were treated with platinum-based chemotherapy.
(C) Boxplot showing the expression of PCDHB4 in LUSC patients who were responsive or non-responsive to platinum-based chemotherapy. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
(D) Kaplan-Meier curves of OS (left panel) and PFS (right panel) of LUSC patients with low and high methylation levels of the PCDHB4 CpG site (cg24918705).
(E) Kaplan-Meier curves of OS (left panel) and PFS (right panel) of LUSC patients with low and high methylation levels of PCDHB4 CpG site (cg24918705), who were treated with platinum-based chemotherapy.
(F) Boxplot showing the methylation level of PCDHB4 CpG sites in LUSC patients who were responsive or non-responsive to platinum-based chemotherapy. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
In addition, shorter PFS and OS were also observed in LUSC patients with lower methylation levels of the CpG site (cg24918705) in the PCDHB4 promoter (Figure 9D). Similarly, in LUSC patients receiving platinum-based chemotherapy, the PFS and OS of the patients with lower methylation levels of the CpG site (cg24918705) were shorter (Figure 9E). As shown in Figure 9F, in the non-responder group, the methylation level of the CpG site (cg24918705) was significantly lower.
However, we did not observe the correlation of the PCDHB4 expression or CpG methylation with the survival in LUAD patients (data not shown). Therefore, the data suggest that the expression and CpG methylation (cg24918705) of the PCDHB4 gene are prognostic markers in LUSC.
Discussion
In this study, by integrating DNA methylation, gene expression, drug response, and clinical survival data, we found that the PCDHB4 gene was poorly methylated and highly expressed in SCLC tumors, which was associated with the poor survival of SCLC patients.
PCDHB4 is a family member of protocadherin (PCDH) transmembrane proteins, which are predominantly expressed in the nervous system, and participate in cell-cell adhesion.14 Previous studies have demonstrated that PCDH expression is controlled by DNA methylation and its dysregulation is common in different types of cancer. Novak et al. reported DNA hypermethylation and transcriptional down-regulation of PCDH family genes in breast cancer.15 Dallosso et al. found DNA hypermethylation of the PCDHA, PCDHB and PCDHG genes in both adenomas and colorectal carcinomas. Particularly, PCDHGC3 has been found to be highly methylated only in carcinomas and not in adenomas, thereby being proposed as a driver for the progression from adenoma to carcinoma.16 Abe et al. showed that the high methylation of PCDHB16 was associated with poor survival in neuroblastoma.17
However, our study found that PCDHB4 was hypomethylated and up-regulated in SCLC tumors. Some studies have demonstrated that hypomethylation and high expression of specific PCDHs correlated with cancer progression. Vega-Benedetti et al. indicated that hypomethylation of PCDHGC5 correlated with its high expression in low-grade glioma (LGG), which was a poor survival predictive biomarker for LGG patients.18 PCDH10 has been shown to be required for the proliferation and tumorigenicity of glioblastoma.19 Mukai et al. indicated that PCDHB9 was over-expressed in gastric cancer, which correlated with poor survival.20 Zhou et al. demonstrated that the over-expression of PCDH7 was associated with poor clinical outcome in non-small cell lung cancer.21
The molecular mechanisms of PCDH proteins in cancer progression and drug resistance remain largely unknown. Previous studies have demonstrated that PCDH proteins regulate cancer cell proliferation and apoptosis via Wnt/β-catenin, Pi3K/AKT, and NF-κB signalings.14 Our single-cell analysis found that PCDHB4-expressing SCLC cells exhibited more characteristics of stemness and EMT. Previous study has demonstrated that the activation of stemness and EMT program promotes the development of treatment resistance in cancer cells.22 A previous study indicated that the expression of PCDHB family genes was high in SCLC with high SOX4 expression. The SOX4 transcription factor was recruited to the promoter of PCDHB family genes to induce their expression.23 Sox family proteins have been shown to play an important role in the maintenance of cancer stem cells.24 A previous study reported that SOX4 expression was up-regulated in glioblastoma and particularly high in malignant glioma initiating cells.25 On the other hand, our study indicated that PCDHB4-expressing cells correlated with a low level of immune infiltration in SCLC tumors. Previous studies have indicated that protocadherin-18 (PCDH18) is an inhibitory signaling receptor that regulates tumor-infiltrating CD8+ memory T cells.26 Therefore, our study suggests that PCDHB4-expressing SCLC cells exhibit the characteristics of stemness and EMT, which could lead to chemo-resistance and progression. Moreover, PCDHB4-expressing SCLC cells may have an inhibitory effect on tumor immune infiltration in SCLC.
In conclusion, the integrated analysis of DNA methylation and gene expression identified a novel prognostic marker, PCDHB4, in SCLC. The low CpG methylation and high expression of PCDHB4 were also poor prognostic markers in LUSC. SCLC tumors with high PCDHB4 expression were characterized by the cancer stem cell phenotype and low immune infiltration. The PCDHB4 gene as a potential prognostic biomarker warrants further investigation in large SCLC and LUSC clinical samples.
Limitations of the study
Although this study already performed multi-omics analyses combined with clinical data and the validation at different levels, there were still several limitations. Firstly, the prognostic value of PCDHB4 expression and methylation needs be validated in a larger population of SCLC patients. Secondly, the detailed mechanism that PCDHB4 expression leads to chemo-resistance and SCLC progression needs to be studied furtherly.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-PCDHB4 | ThermoFisher Scientific | Cat# PA5-120577; RRID: AB_2914149 |
| Anti-β-actin antibody | TransGen Biotech | Cat# HC201; RRID: AB_2860007 |
| Bacterial and virus strains | ||
| DH5α | TOLOBIO | Cat# CC96102-02 |
| pClone007 Simple Vector | TsingKe Biotech | Cat# TSV-007S |
| Biological samples | ||
| FFPE samples of human SCLC and adjacent non-tumor tissues | The First Affiliated Hospital of USTC (Hefei, China) | N/A |
| Blood samples of SCLC patients | The First Affiliated Hospital of USTC (Hefei, China) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 | Gibco | Cat# 11875500BT |
| Fetal bovine serum | ExCell Bio | Cat# FSP500 |
| Penicillin-streptomycin solution | Hyclone | Cat# SV30010 |
| Cisplatin | MCE | Cat# HY-17394 |
| Critical commercial assays | ||
| GeneRead DNA FFPE Kit | Qiagen | Cat# 180134 |
| Circulating Nucleic Acids Kit | Qiagen | Cat# 55114 |
| DNA Methylation kit | Zymo Research | Cat# D5001 |
| RNA extraction kit | Tiangen | Cat# DP451 |
| Reverse transcription kit | TransGen | Cat# AT301-03 |
| BCA Protein Assay Kit | Beyotime | Cat# P0009 |
| NEBNext Multiplex Oligos for Illumina | New England BioLabs | Cat# E7335L |
| KAPA HiFi Hotstart ReadyMix | KAPA Biosystems | Cat# KK2602 |
| KAPA HyperPrep Kit | KAPA Biosystems | Cat# KK8504 |
| MagMeDIP Kit | Diagenode | Cat# C02010021 |
| HighGene transfection reagent | ABclonal Technology | Cat# RM09014 |
| CellTiter-Glo Luminescent Cell Viability assay | Promega | Cat# G7572 |
| Deposited data | ||
| Gene expression data and clinical information of SCLC | George et al.12 | https://doi.org/10.1038/nature14664 |
| GEO: GSE60052 | Gene Expression Omnibus (GEO) database | https://www.ncbi.nlm.nih.gov/geo/ |
| GEO: GSE40275 | Gene Expression Omnibus (GEO) database | https://www.ncbi.nlm.nih.gov/geo/ |
| GEO: GSE138474 | Gene Expression Omnibus (GEO) database | https://www.ncbi.nlm.nih.gov/geo/ |
| SCLC scRNA-seq dataset | Tian et al.13 | https://doi.org/10.1038/s41392-022-01150-4 |
| Gene expression data and clinical information of LUSC | TCGA | https://xenabrowser.net/datapages/ |
| Genomics of Drug Sensitivity in Cancer (GDSC) database | GDSC | https://www.cancerrxgene.org/ |
| MeDIP-seq data of SCLC | This study | GSA-Human: HRA007706 |
| Experimental models: cell lines | ||
| BEAS-2B | Anhui Medical University | N/A |
| H69 | ATCC | Cat# HTB-119 |
| H82 | ATCC | Cat# HTB-175 |
| H446 | Chinese Academy of Sciences | N/A |
| H526 | ATCC | Cat# CRL-5811 |
| SBC-2 | HyCyte | Cat# TCH-C432 |
| Oligonucleotides | ||
| See Table S4 for primer sequence used in this study | This study | N/A |
| Recombinant DNA | ||
| pCMV-3Tag | Agilent Technologies | Cat# 240195 |
| pCMV-3Tag-PCDHB4 | This study | N/A |
| Software and algorithms | ||
| R | The R Project for Statistical Computing | www.r-project.org/ |
| Survival | R CRAN | https://cran.r-project.org/web/packages/survival/index.html |
| survminer | R CRAN | https://cran.r-project.org/web/packages/survminer/index.html |
| limma | Bioconductor | https://bioconductor.org/packages/release/bioc/html/limma.html |
| ChAMP | Bioconductor | https://bioconductor.org/packages/release/bioc/html/ChAMP.html |
| clusterProfiler | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Seurat | satijalab | https://satijalab.org/seurat/ |
| GSVA | R package | https://github.com/rcastelo/GSVA |
| TIMER2.0 | X Shirley Liu Lab 2020 | Dana Farber Cancer Institute | http://timer.cistrome.org/ |
| Macs2 | Github | https://github.com/taoliu/MACS |
| deeptools | Github | https://github.com/deeptools/deepTools/ |
| Integrative Genomics Viewer (IGV) | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ | ImageJ Software | https://imagej.nih.gov/ij |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bo Hong (bhong@hmfl.ac.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
MeDIP-seq raw data of SCLC samples associated with this study have been deposited in the Genome Sequence Archive (GSA) in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA007706) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Ethics approval and consent to participate
This study was approved by the Clinical Research Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China, with the approval number 2023-KY-136. SCLC clinical samples were obtained with written informed consent of the patients.
Method details
Data collection
We obtained the data on cisplatin IC50 values, gene expression and DNA CpG methylation of SCLC cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database.27 The gene expression of SCLC tumors and corresponding survival data were obtained from the datasets by George et al.12 and GEO: GSE60052.28 Among 77 SCLC patients in George dataset, 50 SCLC patients received chemotherapy. In GEO: GSE60052 dataset, all 48 SCLC patients received chemotherapy. The gene expression of SCLC tumors and adjacent normal tissues were obtained from GEO: GSE60052 and GEO: GSE40275. Two SCLC scRNA-seq datasets (GEO: GSE138474 and the dataset by Tian et al.13) were included in this study. The data of LUSC, including gene expression, CpG methylation and clinical information, were obtained from TCGA.
DNA extraction
For formalin-fixed and paraffin-embedded (FFPE) samples, genomic DNAs of SCLC tumors and paired normal tissues were extracted using the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany). For cell-free DNA (cfDNA) of blood samples, cfDNAs of SCLC patients were isolated from 3 to 4 mL of plasma using the Circulating Nucleic Acids Kit (Qiagen, Hilden, Germany).
Cell culture and in vitro functional experiment
SCLC cell lines H69, H82, H446, H526, SBC-2 and human lung epithelial cells (BEAS-2B) were cultured in RPMI-1640 medium (Gibco, Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (ExCell Bio, Shanghai, China) and 1% penicillin and streptomycin (Hyclone, GE Healthcare Life Sciences, Logan, UT, USA) in a wet incubator with 37°C in a 5% CO2 atmosphere.
The coding region of PCDHB4 gene was cloned into pCMV-3Tag expression vector to construct PCDHB4 expression vector (pCMV-3Tag-PCDHB4). SBC2 cells (3×105) were seeded onto 6-well plates and incubated for 24 h. Cells were then transfected with 1 μg PCDHB4 expression vector and empty vector using HighGene (ABclonal Technology, Wuhan, China). Two days after transfection, one part of transfectants was collected for RT-PCR to check whether PCDHB4 was over-expressed. Other part of transfectants was replated onto 96-well plate and was treated with different doses of cisplatin and vehicle control for 72 h, and then cell viability was measured by CellTiter-Glo Luminescent assay (Promega, WI, USA).
To detect the growth of SCLC cells overexpressing PCDHB4, transfected SBC2 cells were replated onto 96-well plate. After 1, 2 and 3 days, cell viability was measured by CellTiter-Glo Luminescent assay (Promega, WI, USA).
Methylated DNA immuno-precipitation sequencing (MeDIP-seq) and data processing
MeDIP-seq was performed using previously published methods.29 In brief, DNA samples were first subjected to fragmentation, end repair, A-tailing and adaptor ligation. Then, methylated DNA was immunoprecipitated with a 5-mC monoclonal antibody. The immunoprecipitated methylated-DNA was sequenced on the Illumina Nova 6000 platform to generate 150-bp paired-end reads. The sequencing statistics for each sample were summarized in Table S3. After sequencing, Macs2 was used to identify the methylation peaks. Reads per kilobase per million mapped reads (RPKM) was used to normalize the number of reads by deeptools and visualized using IGV tools.
Bisulfite sequencing PCR (BSP)
Genomic DNA was chemically modified with sodium bisulfite using the DNA Methylation kit (Zymo Research, CA, USA). Bisulfite-treated genomic DNA was amplified with BSP primers, specific for the cg24918705 site. Nested PCR was performed for 35 cycles with an annealing temperature of 55°C. The “outer” and “inner” primers were shown in Table S4. The PCR products were cloned into the T-vector (TsingKe Biotech, Beijing, China). The colonies were randomly chosen for Sanger sequencing.
Real-time quantitative PCR (RT-PCR)
Total RNA from cells was extracted using total RNA extraction kit (Tiangen, Beijing, China) and first strand cDNA was synthesized with a reverse transcription kit (TransGen Biotech, Beijing, China). The primers were shown in Table S4. The quantification of mRNA expression was obtained with the 2−ΔΔCt method.
Western blot
Lysis buffer was used to extract cellular proteins, and BCA Protein Assay Kit (Beyotime, Shanghai, China) was used to quantify protein concentration. The same amount of protein was run for SDS-polyacrylamide gel electrophoresis. The primary antibody used was anti-PCDHB4 (1:1000 dilution, PA5-120577, ThermoFisher Scientific, MA, USA). The β-actin (1:5000 dilution, HC201, TransGen Biotech, Beijing, China) was used as loading control. The density of the band was quantified by ImageJ (NIH, USA).
Immunohistochemistry (IHC)
Immunostaining of PCDHB4 protein was performed using a rabbit polyclonal anti-PCDHB4 antibody (1:400 dilution, PA5-120577, ThermoFisher Scientific, MA, USA). For the IHC assay, paraffin sections were incubated with primary antibody against PCDHB4 at room temperature for 60 min, secondary antibody at 37°C for 30 min then stained with DAB and haematoxylin.
Functional enrichment and immune infiltration analyses
The clusterProfiler was used to determine Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Gene set enrichment analysis (GSEA) was conducted to explore the underlying pathway variations between two groups.
We estimated immune infiltration using the CIBERSORT, TIMER and XCELL algorithms through the TIMER2 resource. We also evaluated immune cell infiltration in each sample using single-sample GSEA (ssGSEA) with the GSVA.
The process of scRNA-seq data analysis
The scRNA-seq expression matrix was processed with the Seurat. The gene expression data were log-normalized by the “NormalizeData” function. The top 2,000 highly variable genes were used to aggregate samples into a merged dataset and then scale. The main cell clusters were reduced and visualized using uniform manifold approximation and projection (UMAP). “FeaturePlot” was used to visualize gene expression. The scores of gene expression signatures were evaluated with the AddModuleScore function provided by the Seurat package. The stemness score was calculated with a geneset by Boyer et al.30 and the EMT score was evaluated using a geneset by Schliekelman et al.31
Quantification and statistical analysis
Univariate Cox model was conducted to identify genes that were significantly correlated with the survival. Multivariate Cox analysis was conducted to identify genes that were independent prognostic factors. Kaplan-Meier analysis was conducted to evaluate the survival rate for each group using the “survimer” R package, and the patients were assigned into two groups according to the optimal cut-off value. Differentially expressed genes were analyzed by the limma package. Differentially methylated CpGs were analyzed by the ChAMP package. The differences between groups were analyzed by Wilcoxon test and p < 0.05 was considered statistically significant.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (52072360 and 81872438), the Program of Research and Development of Key Common Technologies and Engineering of Major Scientific and Technological Achievements in Hefei (2021YL007), the Collaborative Innovation Program of Hefei Science Center, CAS (2022HSC-CIP015), and the Program of Clinical Medical Translational Research in Anhui Province (202304295107020092).
Author contributions
B.H. and W.X. designed the study. Q.Z., J.Q., and J.N. performed the bioinformatic analysis. M.F. and R.D. performed the MeDIP-seq, BSP, RT-PCR, Western blot, and IHC experiments. Z.X., D.W., J.L., X.W., and H.W. collected the SCLC clinical samples. Y.W and Z.W. performed the in vitro functional experiments. Q.Z. and B.H. wrote the manuscript. All authors reviewed the manuscript.
Declaration of interests
The authors have no financial/commercial conflicts of interest regarding the study.
Published: June 28, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110413.
Contributor Information
Bo Hong, Email: bhong@hmfl.ac.cn.
Weiping Xu, Email: weipingx@ustc.edu.cn.
Supplemental information
References
- 1.Gazdar A.F., Bunn P.A., Minna J.D. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 2017;17:765. doi: 10.1038/nrc.2017.106. [DOI] [PubMed] [Google Scholar]
- 2.Oronsky B., Reid T.R., Oronsky A., Carter C.A. What's New in SCLC? A Review. Neoplasia. 2017;19:842–847. doi: 10.1016/j.neo.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabari J.K., Lok B.H., Laird J.H., Poirier J.T., Rudin C.M. Unravelling the biology of SCLC: implications for therapy. Nat. Rev. Clin. Oncol. 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudin C.M., Poirier J.T., Byers L.A., Dive C., Dowlati A., George J., Heymach J.V., Johnson J.E., Lehman J.M., MacPherson D., et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gay C.M., Stewart C.A., Park E.M., Diao L., Groves S.M., Heeke S., Nabet B.Y., Fujimoto J., Solis L.M., Lu W., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart C.A., Gay C.M., Xi Y., Sivajothi S., Sivakamasundari V., Fujimoto J., Bolisetty M., Hartsfield P.M., Balasubramaniyan V., Chalishazar M.D., et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat. Cancer. 2020;1:423–436. doi: 10.1038/s43018-019-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.M., Quintanal-Villalonga Á., Gao V.R., Xie Y., Allaj V., Chaudhary O., Masilionis I., Egger J., Chow A., Walle T., et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39:1479–1496.e18. doi: 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner E.E., Lok B.H., Schneeberger V.E., Desmeules P., Miles L.A., Arnold P.K., Ni A., Khodos I., de Stanchina E., Nguyen T., et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laird P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 10.Kalari S., Jung M., Kernstine K.H., Takahashi T., Pfeifer G.P. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32:3559–3568. doi: 10.1038/onc.2012.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krushkal J., Silvers T., Reinhold W.C., Sonkin D., Vural S., Connelly J., Varma S., Meltzer P.S., Kunkel M., Rapisarda A., et al. Epigenome-wide DNA methylation analysis of small cell lung cancer cell lines suggests potential chemotherapy targets. Clin. Epigenetics. 2020;12:93. doi: 10.1186/s13148-020-00876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George J., Lim J.S., Jang S.J., Cun Y., Ozretić L., Kong G., Leenders F., Lu X., Fernández-Cuesta L., Bosco G., et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y., Li Q., Yang Z., Zhang S., Xu J., Wang Z., Bai H., Duan J., Zheng B., Li W., et al. Single-cell transcriptomic profiling reveals the tumor heterogeneity of small-cell lung cancer. Signal Transduct. Target. Ther. 2022;7:346. doi: 10.1038/s41392-022-01150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pancho A., Aerts T., Mitsogiannis M.D., Seuntjens E. Protocadherins at the Crossroad of Signaling Pathways. Front. Mol. Neurosci. 2020;13:117. doi: 10.3389/fnmol.2020.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak P., Jensen T., Oshiro M.M., Watts G.S., Kim C.J., Futscher B.W. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68:8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dallosso A.R., Øster B., Greenhough A., Thorsen K., Curry T.J., Owen C., Hancock A.L., Szemes M., Paraskeva C., Frank M., et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31:4409–4419. doi: 10.1038/onc.2011.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe M., Ohira M., Kaneda A., Yagi Y., Yamamoto S., Kitano Y., Takato T., Nakagawara A., Ushijima T. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–834. [PubMed] [Google Scholar]
- 18.Vega-Benedetti A.F., Loi E., Moi L., Blois S., Fadda A., Antonelli M., Arcella A., Badiali M., Giangaspero F., Morra I., et al. Clustered protocadherins methylation alterations in cancer. Clin. Epigenetics. 2019;11:100. doi: 10.1186/s13148-019-0695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echizen K., Nakada M., Hayashi T., Sabit H., Furuta T., Nakai M., Koyama-Nasu R., Nishimura Y., Taniue K., Morishita Y., et al. PCDH10 is required for the tumorigenicity of glioblastoma cells. Biochem. Biophys. Res. Commun. 2014;444:13–18. doi: 10.1016/j.bbrc.2013.12.138. [DOI] [PubMed] [Google Scholar]
- 20.Mukai S., Oue N., Oshima T., Imai T., Sekino Y., Honma R., Sakamoto N., Sentani K., Kuniyasu H., Egi H., et al. Overexpression of PCDHB9 promotes peritoneal metastasis and correlates with poor prognosis in patients with gastric cancer. J. Pathol. 2017;243:100–110. doi: 10.1002/path.4931. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X., Updegraff B.L., Guo Y., Peyton M., Girard L., Larsen J.E., Xie X.J., Zhou Y., Hwang T.H., Xie Y., et al. PROTOCADHERIN 7 Acts through SET and PP2A to Potentiate MAPK Signaling by EGFR and KRAS during Lung Tumorigenesis. Cancer Res. 2017;77:187–197. doi: 10.1158/0008-5472.CAN-16-1267-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillo S.D., Matheu A., Mariani N., Carretero J., Lopez-Rios F., Lovell-Badge R., Sanchez-Cespedes M. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 2012;72:176–186. doi: 10.1158/0008-5472.CAN-11-3506. [DOI] [PubMed] [Google Scholar]
- 24.Pouremamali F., Vahedian V., Hassani N., Mirzaei S., Pouremamali A., Kazemzadeh H., Faridvand Y., Jafari-Gharabaghlou D., Nouri M., Maroufi N.F. The role of SOX family in cancer stem cell maintenance: With a focus on SOX2. Pathol. Res. Pract. 2022;231 doi: 10.1016/j.prp.2022.153783. [DOI] [PubMed] [Google Scholar]
- 25.Vervoort S.J., van Boxtel R., Coffer P.J. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 26.Vazquez-Cintron E.J., Monu N.R., Burns J.C., Blum R., Chen G., Lopez P., Ma J., Radoja S., Frey A.B. Protocadherin-18 is a novel differentiation marker and an inhibitory signaling receptor for CD8+ effector memory T cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L., Huang J., Higgs B.W., Hu Z., Xiao Z., Yao X., Conley S., Zhong H., Liu Z., Brohawn P., et al. Genomic Landscape Survey Identifies SRSF1 as a Key Oncodriver in Small Cell Lung Cancer. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi J., Hong B., Tao R., Sun R., Zhang H., Zhang X., Ji J., Wang S., Liu Y., Deng Q., et al. Prediction model for malignant pulmonary nodules based on cfMeDIP-seq and machine learning. Cancer Sci. 2021;112:3918–3923. doi: 10.1111/cas.15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schliekelman M.J., Taguchi A., Zhu J., Dai X., Rodriguez J., Celiktas M., Zhang Q., Chin A., Wong C.H., Wang H., et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015;75:1789–1800. doi: 10.1158/0008-5472.CAN-14-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
MeDIP-seq raw data of SCLC samples associated with this study have been deposited in the Genome Sequence Archive (GSA) in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA007706) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.