Summary
We present a protocol to generate highly multiplexed spatial data at cellular and subcellular resolutions using iterative indirect immunofluorescence imaging (4i). We describe streamlined steps for using 4i across fixed cultured cells, formalin-fixed paraffin-embedded (FFPE) tissue sections, and metaphase chromosome spreads. We detail procedures for sample preparation, antibody and DNA staining, immunofluorescence imaging, antibody elution, and image processing. This protocol is adapted for high-throughput analysis of fixed cultured cells and addresses sample-specific challenges such as intrinsic tissue autofluorescence and chromosome fragility.
For complete details on the use and execution of this protocol for fixed cultured cells, please refer to Comandante-Lou et al.1
Subject areas: Cell Biology, Cancer, Microscopy, Systems biology
Graphical abstract

Highlights
-
•
Three variants of an iterative imaging protocol for cellular and subcellular analysis
-
•
Highly multiplexed immunofluorescence staining and antibody elution in diverse samples
-
•
Streamlined image processing steps for cultured cells, tissue sections, and chromosomes
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
We present a protocol to generate highly multiplexed spatial data at cellular and subcellular resolutions using iterative indirect immunofluorescence imaging (4i). We describe streamlined steps for using 4i across fixed cultured cells, formalin-fixed paraffin-embedded (FFPE) tissue sections, and metaphase chromosome spreads. We detail procedures for sample preparation, antibody and DNA staining, immunofluorescence imaging, antibody elution, and image processing. This protocol is adapted for high-throughput analysis of fixed cultured cells and addresses sample-specific challenges such as intrinsic tissue autofluorescence and chromosome fragility.
Before you begin
Protocol overview
Recent advances in highly-multiplexed imaging methods have enhanced our ability to study biology at cellular and subcellular resolutions. Among these methods, iterative indirect immunofluorescence imaging (4i) enables generation of multiplexed data on proteins, their posttranslational modifications, and their spatial context within samples via iterations of indirect immunostaining, imaging, and antibody elution.2 Here, we report streamlined 4i protocols for three sample types. First, we describe a detailed 4i protocol for cultured cells (cell culture-4i), which we recently applied to elucidate the heterogeneity in single-cell abundance of various proteins, including transcription factors, cell signaling, proliferation and differentiation state markers across genetically diverse melanoma cell lines.1 Then, we describe a 4i protocol for formalin-fixed paraffin embedded (FFPE) tissue sections (tissue-4i), which enables spatial mapping of protein localization at single-cell resolution from whole-tissue sections. Finally, we report a 4i protocol for metaphase chromosome spreads (chromosome spread-4i), which enables mapping of chromosomal proteins at single-chromosome resolution. Although the same chemical principles are used across three 4i protocols, there are sample-specific practical considerations and challenges, which we highlight in this paper.
Institutional permissions
All human specimens, including tonsil, melanoma, and breast carcinoma tissues, used in this study were obtained from the Biorepository and Tissue Research Facility (BTRF) at the University of Virginia School of Medicine and were collected in accordance with institutional and national guidelines and regulations.
4i-specific antibody considerations
A typical 4i experiment begins with assembling a panel of primary antibodies that are carefully chosen to address a specific biological question. In addition to general guidelines used widely for indirect immunostaining (such as validation of the antibody specificity, optimization of antibody concentrations to achieve maximal signal-to-background ratios, and consideration of host species when multiple primary antibodies are combined in a single round of immunostaining), there are a few 4i-specific guidelines that should be considered to achieve optimal results.
Optimizing the order of primary antibodies from one 4i round to the next
We recommend using antibodies against low-abundance epitopes in the early rounds of 4i. The rationale is based on our observation that later rounds are more prone to sample loss or epitope degradation. Furthermore, delaying the use of those antibodies until later 4i rounds minimizes the impact of residual fluorescence resulting from strong binding of some antibodies to highly abundant epitopes. However, immunofluorescence signal-to-background ratios for some protein targets increases in later rounds of 4i, likely due to epitope-specific antigen retrieval properties of the elution buffer. Thus, for the best performance, we recommend testing each new antibody across multiple rounds of 4i to determine its sensitivity to the 4i round.
Optimizing the combination of primary antibodies in each 4i round
Generally, there are no limitations in choosing protein targets within each 4i round. However, protein targets that are intended for high-resolution colocalization analysis (at a pixel level) should be stained within the same 4i round, if possible.
Optimizing the choice of fluorophores for secondary antibodies
In a typical 4i experiment, we use DAPI or Hoechst DNA stains in combination with secondary antibodies conjugated with Alexa Fluor dyes that are known for their brightness and photostability. We have successfully used both donkey and goat secondary antibodies. A combination of Alexa Fluor 488 (AF488), Alexa Fluor 568 (AF568), and Alexa Fluor 647 (AF647) enables immunofluorescence analysis of three protein targets in each 4i round, while minimizing spectral overlap among fluorescence channels. AF488 and AF568 have higher quantum yield and diffraction-limited resolution (in comparison with AF647), and are recommended for staining protein targets that have low abundance or require the highest resolution. However, AF647 offers the highest signal-to-noise ratio (in comparison with AF488 and AF568) due to its low background fluorescence and is ideal for immunostaining applications requiring high sensitivity. For example, AF647 is recommended for protein targets that are expected to be proximal to tissue sites with high intrinsic autofluorescence, e.g., collagen- or hemoglobin-rich regions. These considerations are weighed against one another as tradeoffs in the choice of fluorophore for each individual protein target.
Preparation for cell culture-4i
-
1.
If using the BioTek EL406 Washer Dispenser Protocol for high-throughput washing of 96-well plates, set up washing protocols as indicated in materials and equipment.
-
2.
Prepare filter-sterilized RPMI 1640 media for COLO858 cell culture (5% FBS, 1 mM sodium pyruvate, 1x penicillin-streptomycin, and 0.5 μg/mL Plasmocin Prophylactic).
-
3.
Prepare elution buffer and imaging buffer.
Preparation for tissue-4i
-
4.
Acquire and label FFPE tissue slides.
-
5.
Prepare elution buffer, imaging buffer, and sodium citrate buffer.
-
6.Prepare deparaffinization solutions.
-
a.In glass dishes or vertical staining jars within a chemical safety hood, dispense ∼200 mL of Sub-X (2 dishes), 1:1 Sub-X:ethanol, 100% ethanol, 95% ethanol, 70% ethanol, 50% ethanol, 30% ethanol, and PBS.
-
b.Arrange solutions in order of use.
-
i.Sub-X → Sub-X → 1:1 Sub-X:ethanol → 100% ethanol → 95% ethanol → 70% ethanol → 50% ethanol → 30% ethanol → PBS.
-
i.
-
a.
Note: 100% Sub-X can be left covered in the hood for future deparaffinization until it becomes depleted or noticeably cloudy (∼2 months). Ethanol solutions should not be reused, but 1 L stocks can be prepared beforehand and stored in a flammables cabinet to quicken preparation.
CRITICAL: Sub-X is a flammable liquid and should be handled under a chemical safety hood at all times.
Preparation for chromosome spread-4i
-
7.
A few days prior to the 4i experiment, culture HeLa cells in EMEM media (+ 10% FBS and 1% Penicillin-Streptomycin) in a 10-cm dish until cells reach 70%–80% confluency.
-
8.
Prepare elution buffer, imaging buffer, PBST, and 1% BSA-PBS solution.
-
9.
On the day of the 4i experiment, prepare fresh PBS-colcemid solution, hypotonic buffer, PFA-PBS solution, and 4i blocking buffer.
Key resources table
Note: All physical reagents or resources used are purchased from the US. US Catalog numbers are provided below.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 488 donkey anti-mouse IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-21202; RRID: AB_141607 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-21206; RRID: AB_2535792 |
| Alexa Fluor 568 donkey anti-goat IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-11057; RRID: AB_2534104 |
| Alexa Fluor 568 donkey anti-mouse IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A10037; RRID: AB_2534013 |
| Alexa Fluor 568 goat anti-human IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-21090; RRID: AB_2535746 |
| Alexa Fluor 647 donkey anti-mouse IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-31571; RRID: AB_162542 |
| Alexa Fluor 647 donkey anti-rabbit IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-31573; RRID: AB_2536183 |
| Alexa Fluor 647 goat anti-human IgG (H + L) secondary antibody (1:2,000 working dilution for cell culture- or tissue-4i; 1:1,000 working dilution for chromosome spread-4i) |
Thermo Fisher Scientific | CA# A-21445; RRID: AB_2535862 |
| Anti-ACA (human; 1:500 working dilution) | Antibodies Incorporated | CA# 15-234; RRID: AB_2939058 |
| Anti-CD3 (rabbit; 1:75 working dilution) | Abcam | CA# ab16669; RRID: AB_443425 |
| Anti-CD19 (rabbit; 1:250 working dilution) | Abcam | CA# ab134114; RRID: AB_2801636 |
| Anti-CD45 (mouse; 1:400 working dilution) | Agilent Dako | CA# M0701; RRID: AB_2314143 |
| Anti-H3pT3 (rabbit; 1:250 working dilution) | Abcam | CA# ab78351; RRID: AB_1566301 |
| Anti-INCENP (mouse; 1:1,000 working dilution) | Abcam | CA# ab23956; RRID: AB_447776 |
| Anti-Ki67 (mouse; 1:400 working dilution) | Cell Signaling Technology | CA# 9449S; RRID: AB_2715512 |
| Anti-MITF (mouse; 1:800 working dilution) | Abcam | CA# ab3201; RRID: AB_177350 |
| Anti-NGFR (rabbit; 1:400 working dilution) | Cell Signaling Technology | CA# CST8238; RRID: AB_10839265 |
| Anti-SOX10 (goat; 1:200 working dilution) | R&D Systems | CA# AF2864; RRID: AB_442208 |
| Biological samples | ||
| FFPE sections of tonsil tissue | Biorepository and Tissue Research Facility (BTRF) at the University of Virginia School of Medicine | N/A |
| FFPE sections of melanoma | Biorepository and Tissue Research Facility (BTRF) at the University of Virginia School of Medicine | N/A |
| FFPE sections of breast carcinoma with immune infiltration | Biorepository and Tissue Research Facility (BTRF) at the University of Virginia School of Medicine | N/A |
| Experimental models: Cell lines | ||
| COLO858 | MGH Cancer Center, primary source ECACC | CA# 93052613; RRID: CVCL_2005 |
| HeLa | American Type Culture Collection (ATCC) | CA# CCL-2; RRID: CVCL_0030 |
| Chemicals, peptides, and recombinant proteins | ||
| Bovine serum albumin (BSA) | Fisher BioReagents | CA# BP1600-1; CAS: 9048-46-8 |
| CellMask Green (1:5,000 working dilution) | Fisher Scientific | CA# H32714 |
| Colcemid solution in PBS | Gibco | CA# 15212012 |
| 4′, 6-diamidino-2-phenylindole (DAPI) (1:5,000 working dilution) | Sigma-Aldrich | CA# D9542 |
| EMEM medium | ATCC | CA# 30-2003 |
| Ethanol (pure, 100%) | Fisher Scientific | CA# BP2818-4; CAS: 64-17-5 |
| Fetal bovine serum (FBS) | Fisher Scientific | CA# 26140079 |
| Glycerol | Fisher Scientific | CA# BP229-1; CAS: 56-81-5 |
| Glycine | Sigma-Aldrich | CA# 50046-1KG; CAS: 56-40-6 |
| Guanidine hydrochloride | Sigma-Aldrich | CA# G3272-1KG; CAS: 50-01-1 |
| Hoechst 33342 (1:20,000 working dilution) | Invitrogen | CA# H3570 |
| Hydrochloric acid (1 N HCl) 37% | Sigma-Aldrich | CA# 258148-2.5L; CAS: 7647-01-0 |
| Hydrogen peroxide (H2O2) 30% | Sigma-Aldrich | CA# 216763-500ML; CAS: 7722-84-1 |
| LI-COR intercept (PBS) blocking buffer | Fisher Scientific | CA# NC1660556 |
| Maleimide | Sigma-Aldrich | CA# 129585-25G; CAS: 541-59-3 |
| N-acetyl cysteine | Sigma-Aldrich | CA# A7250-500G; CAS: 616-91-1 |
| 16% paraformaldehyde (PFA) | Fisher Scientific | CA# 50-980-489; CAS: 30525-89-4 |
| Penicillin-streptomycin solution 100x | Cytiva | CA# SV30010 |
| Phosphate-buffered saline (PBS) | Fisher Scientific | CA# BP665-1 |
| Dulbecco’s phosphate-buffered saline (DPBS) | Corning | CA# 21-031-CV |
| Plasmocin prophylactic | InvivoGen | CA# ant-mpp |
| Potassium chloride (KCl) | Acros Organics | CA# 193780010; CAS: 7447-40-7 |
| RPMI 1640 medium | Corning | CA# 10-040-CV |
| Sodium hydroxide (NaOH) 10 N | Fisher Scientific | CA# 18-606-545; CAS: 1310-73-2 |
| Sodium pyruvate 100 mM | Corning | CA# 25-000-Cl; CAS: 113-24-6 |
| Sub-X | Leica Biosystems | CA# 3803670 |
| Tri-sodium citrate dihydrate | Spectrum Chemical | CA# S1250; CAS: 6132-04-3 |
| Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) | Sigma-Aldrich | CA# C4706-10G; CAS: 51805-45-9 |
| Triton X-100 | ICN Biomedicals | CA# 807426; CAS: 9036-19-5x |
| Trypsin EDTA (0.25%), phenol red | Life Technologies | CA# 25200056 |
| Tween 20 | Fisher Scientific | CA# BP337100; CAS: 9005-64-5 |
| Urea | Sigma-Aldrich | CA# U5128-500G; CAS: 57-13-6 |
| Software and algorithms | ||
| ASHLAR, version 1.17.0+23.g387eff5 | Muhlich et al.3 | https://github.com/labsyspharm/ashlar; RRID: SCR_016266 |
| BaSiC | Peng et al.4 | https://sites.imagej.net/BaSiC/https://github.com/labsyspharm/basic-illumination/blob/master/imagej_basic_ashlar.py; RRID: SCR_016371 |
| CellProfiler, version 4.2.5 | Carpenter et al.5 | https://cellprofiler.org/; RRID: SCR_007358 |
| Fiji, version 2.14.0/1.54f | Schindelin et al.6 | https://imagej.net/software/fiji/; RRID: SCR_002285 |
| Fiji plugin: Find Focused Slices |
https://sites.google.com/site/qingzongtseng/find-focus https://raw.githubusercontent.com/qztseng/imagej_plugins/master/current/src/Find_focused_slices.java |
|
| Fiji plugin: SIFT | Lowe7 |
https://imagej.net/plugins/linear-stack-alignment-with-sift https://github.com/axtimwalde/mpicbg/blob/master/mpicbg_/src/main/java/SIFT_Align.java |
| MCMICRO | Schapiro et al.8 | https://github.com/labsyspharm/mcmicro; RRID: SCR_022832 |
| MATLAB, version 9.14.0.2206163 (R2023a) | https://www.mathworks.com/; RRID: SCR_001622 | |
| RStudio, version 2023.09.1 | RRID: SCR_000432 | |
| R, version 4.2.3 | https://www.r-project.org/; RRID: SCR_001905 | |
| Other | ||
| 96-well plates | Corning | CA# 3904 |
| 96-well plate foil seal | Agilent | CA# 5067-5154 |
| No. 1.5 glass coverslips | Electron Microscopy Sciences | CA# 71861-055-C |
| No. 1.5 glass coverslips (25 mm × 75 mm) | Ibidi | CA# 10812 |
| BioTek EL406 washer dispenser | Agilent | N/A |
| Cytology funnels | BMP Medical | CA# 80094-254 |
| Cytospin 4 centrifuge | Thermo Scientific | CA# TH-CYTO4 |
| Cytospin filter cards | Simport Scientific | CA# M965FW |
| Epitope retrieval steamer | IHC World | CA# NC1314441 |
| Fine point diamond scriber | Electron Microscopy Sciences | CA# 50-996-737 |
| Funnel clips | BMP Medical | CA# 80094-262 |
| Glass staining dish with glass rack | DWK Life Sciences | CA# 08-812 |
| LED lamp | Amazon | ASIN# B00OY4H6QE |
| Parafilm | Bemis | CA# PM992 |
| Manifold for microtest plates | Drummond Scientific | CA# 3-000-096 |
| Microscope slides (25 mm × 75 mm) | VWR | CA# 48311-703 |
| Silicone gasket | Invitrogen | CA# C24776 |
| StainTray slide staining system | Polysciences | CA# 25498-1 |
| Syringe (50 mL) | Fisher Scientific | CA# 13-689-8 |
| Syringe filter (5 μm) | VWR | CA# 28150-956 |
| Western blot box | Sigma-Aldrich | CA# Z742098-5EA |
| Operetta CLS 20X air objective | PerkinElmer/Revvity | CA# HH14000404 |
| Operetta CLS 63X water-immersion objective | PerkinElmer/Revvity | CA# HH14000423 |
| Operetta four slide holder | PerkinElmer/Revvity | CA# HH14000750 |
| Peristaltic pump 5 μL cassette | Agilent | CA# 7170011 |
| TC20 automated cell counter | Bio-Rad | CA# 1450102 |
Materials and equipment
Shared reagents
Elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| L-glycine (molar mass: 75.07 g/mol) | 0.64 M | 4.85 g |
| Urea (molar mass: 60.056 g/mol) | 3 M | 18.02 g |
| Guanidinum chloride (molar mass: 95.53 g/mol) | 3 M | 28.66 g |
| TCEP (molar mass: 286.65 g/mol) | 70 mM | 2 g |
| 3 N HCl | (to pH 2.5) | X mL |
| ddH2O | complete volume to 100 mL | |
| Total | 100 mL |
Note: Dissolve glycine, urea, and guanidinium chloride in water first before adding TCEP. After the addition of TCEP, avoid incorporating air into the buffer by stirring slowly. Adjust pH of the solution to 2.5 with HCl and add ddH2O to the final volume. “X” denotes the volume of 3 N HCl needed to bring the solution to pH 2.5. In our experience, 12–14.5 mL of 3 N HCl is typically sufficient to bring the pH to 2.5. Filter with 5-micron syringe filter. Store at 4°C for <1 month. TCEP can degrade and become ineffective over time.
CRITICAL: HCl is a powerful acid and can cause severe burns and irritation. Wear appropriate PPE.
-
•
Imaging buffer (cell culture-4i and chromosome spread-4i): Dissolve 11.42 g of N-acetyl cysteine (molar mass: 163.2 g/mol) in 80 mL ddH2O. pH solution to 7.4 with NaOH. Bring volume up to 100 mL with ddH2O and filter with 5-micron syringe filter. Prepare one-time use aliquots and store at −20°C.
CRITICAL: NaOH is a powerful base and can cause severe burns and irritation. Wear appropriate PPE.
-
•
4i Blocking Buffer (cell culture-4i and tissue-4i): Dissolve 0.219 g of maleimide (molar mass: 97.07 g/mol) in 15 mL LICOR Intercept (PBS) blocking buffer on a nutator.
Note: Prepare immediately before use. Maleimide takes approximately 10 min to dissolve on a nutator.
-
•
Hoechst solution (tissue-4i and chromosome spread-4i): Prepare 20,000-fold dilution of Hoechst 33342 (16.2 mM) in LICOR Intercept (PBS) blocking buffer.
Note: Store at 4°C for up to 1 week.
Reagents for cell culture-4i
-
•
4% PFA-PBS solution: Mix 10 mL of 16% PFA in 30 mL of 1x PBS to prepare 40 mL solution.
Note: Store at 4°C for up to 1 week.
-
•
0.5% Triton X-100-PBS solution: Dissolve 50 μL of 100% Triton X-100 in 10 mL of 1x PBS.
-
•
Hoechst solution: Dilute Hoechst 33342 (16.2 mM) 20,000-fold in 1x PBS.
Note: Store at 4°C for up to 1 week.
-
•
CellMask Green solution: Dilute CellMask Green 5,000-fold in 1x PBS.
BioTek EL406 washer dispenser settings
-
•
Although not essential, the use of an automated washer-dispenser is highly recommended to provide gentle washing and minimize cell loss or movement during 4i iterations. All steps, however, can be performed manually with a regular or multichannel pipette. Here, we provide optimized settings for BioTek EL406 washer dispenser.
-
•
Ensure there are carboys of water and PBS feeding into the EL406 as Buffer A and Buffer B.
-
•
Offset dimensions are optimized for Corning 96-well plates (CA# 3904). Settings may need to be adjusted for other products.
| Protocol name | Step details |
|---|---|
| AspirateTo4uL Aspirate liquid to approximately 4 μL of residual volume |
W- Aspirate Vacuum Filtration: False Travel Rate: 1 CW 4.1 mm/s Delay: 0 msec Z offset: 30 steps (3.1 mm above carrier) X offset: 42 steps (1.92 mm right of center) Y offset: 0 steps (center of well) Secondary Aspirate: Yes Secondary Z offset: 75 steps (9.53 mm above carrier) Secondary X offset: −45 steps (2.06 mm left of center) Secondary Y offset: 0 steps (center of well) |
| H2O_Prime PBS_Prime |
W- Prime Buffer: [A or B] Volume 45 mL Flow Rate: 9 Low Flow Path Volume: 25 mL Submerge tips: No |
| H2O_Wash_6X160uL PBS_Wash_6X160uL Aspirate liquid to approximately 40 μL, dispense 160 μL of PBS or ddH2O, and repeat for a total of 6 times |
W- Wash Pre-dispense before washing: No Bottom Wash: No Number of Wash Cycles: 6 Aspirate per cycle Travel Rate: 1 CW 4.1 mm/s Delay: 0 msec Z offset: 42 steps (5.34 mm above carrier) X offset: 42 steps (1.92 mm right of center) Y offset: 0 steps (center of well) Secondary Aspirate: Yes Secondary Z offset: 80 steps (10.16 mm above carrier) Secondary X offset: −45 steps (2.06 mm left of center) Secondary Y offset: 0 steps (center of well) Dispense per cycle Buffer: [A or B] Volume: 160 μL/well Flow Rate: 1 Z offset: 118 steps (14.99 mm above carrier) X offset: −36 steps (1.65 mm left of center) Y offset: 0 steps (center of well) Pre-dispense: not available Delay start of Vacuum until Volume dispensed: 160 μL/well Shake/Soak after dispense: No Pre-dispense between cycles No Final Aspirate: No |
| AspirateTo4uL_Peri40uL Aspirate liquid to approximately 4 μL of residual volume and dispense 40 μL of elution buffer using the peristaltic pump with a 5 μL cassette |
W- Aspirate Vacuum Filtration: False Travel Rate: 1 CW 4.1 mm/s Delay: 0 msec Z offset: 37 steps (4.7 mm above carrier) X offset: 42 steps (1.92 mm right of center) Y offset: 0 steps (center of well) Secondary Aspirate: Yes Secondary Z offset: 75 steps (9.53 mm above carrier) Secondary X offset: −45 steps (2.06 mm left of center) Secondary Y offset: 0 steps (center of well) P- Dispense Volume: 40 μL/tube Flow Rate: Low Required cassette: 5 μL Secondary Z offset: 336 steps (15.36 mm above carrier) Secondary X offset: −40 steps (1.83 mm left of center) Secondary Y offset: 0 steps (center of well) Pre-dispense: No Columns:111111111111 |
Reagents for tissue-4i
Sodium citrate buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tri-sodium citrate dihydrate (molar mass: 294.10 g/mol) | 10 mM | 2.94 g |
| Tween-20 | 0.05% | 500 μL |
| 3 N HCl | (to pH 6.0) | X mL |
| ddH2O | complete volume to 1 L | |
| Total | 1 L |
Note: Store at 25°C for up to 3 months. “X” denotes the volume of 3 N HCl needed to bring the solution to pH 6.0. In our experience, 1–1.5 mL of 3 N HCl is typically sufficient to bring the pH to 6.0.
CRITICAL: HCl is a powerful acid and can cause severe burns and irritation. Wear appropriate PPE.
Autofluorescence quenching solution
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O2 (30%) | 4.5% | 4.5 mL |
| 10 N NaOH | 24 mM | 72 μL |
| PBS | 24.428 mL | |
| Total | 30 mL |
Note: Prepare immediately before use.
10% glycerol imaging buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| N-acetyl cysteine (molar mass: 163.2 g/mol) | 700 mM | 11.42 g |
| Glycerol | 1.37 M or 10% | 10 mL |
| 10 N NaOH | (to pH 7.4) | X mL |
| ddH2O | complete volume to 100 mL | |
| Total | 100 mL |
Note: Add N-acetyl cysteine, 10 mL glycerol, and approximately 70 mL ddH2O to a beaker. Stir to dissolve. pH solution to 7.4 with NaOH. “X” denotes the volume of 10 N NaOH needed to bring the solution to pH 7.4. In our experience, approximately 5 mL–10 mL of 10 N NaOH is typically sufficient to bring the pH to 7.4. Filter with 5-μm syringe filter to remove particulate matter. Prepare 1–5 mL aliquots and store at −20°C indefinitely.
CRITICAL: NaOH is a powerful base and can cause severe burns and irritation. Wear appropriate PPE.
Reagents for chromosome spread-4i
-
•
PBST: Mix 50 μL of Tween-20 in 50 mL of 1x PBS for a final concentration of 0.1%.
Note: Store at 25°C indefinitely.
-
•
PBS-Colcemid Solution: Mix 70 μL of Colcemid (10 μg/mL) in 7 mL of 1x PBS for a final concentration of 100 ng/mL.
Note: Prepare immediately before use.
Hypotonic buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| KCl solution (75 mM) | 25 mM | 1.5 mL |
| 0.8% sodium citrate solution | 0.267% | 1.5 mL |
| ddH2O | 1.5 mL | |
| Total | 4.5 mL |
Note: Prior to making the hypotonic buffer, prepare a stock solution of 75 mM KCl (molar mass: 74.55 g/mol) by dissolving 0.2795 g of KCl in 50 mL in ddH2O and a stock solution of 0.8% sodium citrate solution by dissolving 0.4 g of tri-sodium citrate dihydrate in 50 mL ddH2O. Prepare the hypotonic buffer immediately before use.
-
•
2% PFA-PBS Solution: Mix 0.5 mL of 16% PFA in 3.5 mL of 1x PBS to prepare 4 mL solution.
Note: Store at 25°C for up to 1 week.
CRITICAL: PFA is a health hazard that can release corrosive formaldehyde gas when in aqueous solution. Wear appropriate PPE and handle under a fume hood at all times.
BSA-PBS 4i blocking buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 1% | 0.1 g |
| Maleimide (molar mass: 97.07 g/mol) | 150 mM | 0.146 g |
| PBS | 10 mL | |
| Total | 10 mL |
Note: Prepare immediately before use.
-
•
1% BSA-PBS Solution: Dissolve 0.1 g of BSA in 10 mL of 1x PBS.
Note: Store at 4°C for up to 1 week.
-
•
DAPI solution: Prepare 5,000-fold dilution of DAPI in 1x PBS.
Step-by-step method details
Cell culture-4i
Here, we describe a slightly modified protocol of the 4i method established by Gut et al., which we optimized for high-content imaging and used extensively in Comandante-Lou et al.1,2 Working in a 96-well format, various treated and fixed melanoma cell lines withstand at least 20 iterations of immunostaining and elution. The resulting highly multiplexed measurements of protein expression are then analyzed at a single-cell level.
Seeding of cultured cells into 96-well plates
Timing: 1 h
Sample preparation for fixed cell culture-4i involves seeding cells at an optimized density for imaging followed by fixation at a specific time point. Reproducible and accurate cell seeding helps to keep growth rate and confluency of cells consistent across replicates. If cells are too confluent, segmentation of nuclei during image analysis may be difficult. The following protocol uses the COLO858 melanoma cell line as an example of an adherent cell type.
-
1.Seeding of COLO858 cells in a 96-well plate.
-
a.Culture COLO858 cells in RPMI 1640 media supplemented with 5% FBS, 1 mM sodium pyruvate, 1x penicillin-streptomycin, and 0.5 μg/mL Plasmocin Prophylactic in a T75 flask until cells reach 70%–80% confluency. COLO858 cells are grown at 37°C with 5% CO2 in a humidified chamber.
-
b.In a biosafety cabinet, aspirate old media.
-
c.Wash cells with 10 mL of 1x DPBS.
-
d.Add 1 mL of 0.25% trypsin to the cells and incubate at 37°C for 1 min. Lightly tap the flask as necessary to detach the cells. Check under a microscope to ensure that cells are detached.
-
e.Add 10 mL of media to the flask to neutralize the trypsin. Pipette media up and down 1–3 times, dispensing media across the surface area of the flask to collect all of the cells in suspension.
-
f.Transfer cell suspension to a 15 mL tube.
-
g.Centrifuge cells at 200 × g for 3 min.
-
h.Aspirate the supernatant from the cell pellet.
-
i.Resuspend cells in 10 mL media.
-
j.Count cells using the TC20 automated cell counter or a hemocytometer. Take the average of two measurements.
-
k.Seed 5000 cells/well in 200 μL of media into the desired number of wells of a 96-well plate.
-
l.Incubate cells for 1–3 days at 37°C with 5% CO2 in a humidified chamber.
-
a.
Note: COLO858 cells will be relatively confluent when seeded at 5000 cells/well and incubated for 3 days. Seeding densities should be optimized for individual cell lines and experimental goals.
Fixation and permeabilization of cells
Timing: 1 h
Live cells are fixed with 4% PFA and permeabilized with Triton X-100 to allow for immunostaining of intracellular and intranuclear protein targets. Free aldehydes remaining from fixation are quenched with the acidic elution buffer. High-throughput processing of 96-well plates is enabled by the EL406 Washer Dispenser connected to water, PBS, and elution buffer.
-
2.Fixation and washing.
-
a.In a fume hood, aspirate media from 96-well plates with a vacuum manifold.
-
b.Add 100 μL 4% PFA-PBS solution per well.
-
c.Incubate the plate for 30 min at 25°C on a rocking platform.
-
d.Aspirate PFA and dispose as hazardous waste.
-
e.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
f.Aspirate 1x PBS from the wells leaving only a minimal residual volume using EL406 protocol AspirateTo4uL.
-
a.
Note: If you are processing many plates, wash all the plates first and then aspirate liquid at the end of washes to minimize the amount of time cells are without liquid.
Pause point: Seal plates containing 1x PBS with aluminum seals and store at 4°C for up to 3 months.
-
3.Permeabilization.
-
a.Add 100 μL 0.5% Triton X-100-PBS solution per well.
-
b.Incubate the plate for 15 min at 25°C on a rocking platform.
-
a.
-
4.Aldehyde quenching (troubleshooting).
-
a.Wash cells 6 times with ddH2O using EL406 protocols H2O_prime and H2O_Wash_6X160uL.
-
b.Prime peri-pump with elution buffer.
-
c.Aspirate ddH2O and add 40 μL elution buffer per well using EL406 protocol AspirateTo4uL_Peri40uL.Note: To be gentler to the cells and conserve resources, we opt to use the 5 μL peristaltic pump cassette on the EL406 to dispense elution buffer.
-
d.Incubate the plate for 12 min at 25°C on a rocking platform.
-
e.Repeat steps 4c–4d two more times.
-
f.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.Note: To reduce cell loss, AspirateTo4uL_Peri40uL can be modified to aspirate to 15 μL instead of 4 μL in the second and third rounds of elution.
-
a.
Antibody and DNA staining of fixed cells
Timing: 16 h
Fixed samples are prepared for 4i by using 4i blocking buffer. Maleimide reacts with free sulfhydryl groups to prevent disulfide bond formation between antibodies and proteins during imaging, thereby enabling elution when used alongside imaging and elution buffer. Nuclear and cytoplasmic stains are applied for tracking and segmentation purposes in post-processing.
-
5.Blocking.
-
a.Prepare fresh 4i blocking buffer immediately before blocking.
-
b.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
c.Add 50 μL of 4i blocking buffer per well.
-
d.Incubate the plate for 1 h at 25°C on a rocking platform.
-
e.Wash cells 6 times with 1x PBS to remove maleimide from the sample using EL406 protocol PBS_Wash_6X160uL.
-
a.
-
6.Primary antibody staining.
-
a.Prepare primary antibody solution at desired dilution in LICOR Intercept (PBS) blocking buffer. Keep on ice.
-
b.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
c.Add 40 μL primary antibody solution per well.
-
d.Seal plate with aluminum seal.
-
e.Incubate at 4°C for 12–18 h on a rocking platform.
-
a.
Note: Once the primary antibody solution is added to the sample, keep the plate covered with aluminum foil to minimize exposure of the samples to the light. Light can photo-crosslink antibodies to the sample leading to decreased antibody elution efficiency.
-
7.Secondary antibody staining.
-
a.Dilute secondary Alexa Fluor antibody 1:2000 in LICOR Intercept (PBS) blocking buffer. Vortex vigorously.
-
b.Remove aluminum seal from the plate.
-
c.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
d.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
e.Add 40 μL secondary antibody solution per well.
-
f.Incubate the plate for 1 h at 25°C on a rocking platform.
-
g.Wash cells 6 times with PBS using EL406 protocol PBS_Wash_6X160uL.
-
a.
-
8.Cytoplasmic staining.
-
a.Prepare CellMask Green solution.
-
b.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
c.Add 50 μL CellMask solution per well.
-
d.Incubate the plate for 20 min at 25°C on a rocking platform.
-
e.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
a.
Note: Staining with CellMask Green is needed only in the first round of 4i since it is not eluted during antibody elution.
-
9.Nuclear staining.
-
a.Prepare Hoechst solution.
-
b.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
c.Add 50 μL Hoechst solution per well.
-
d.Incubate the plate for 20 min at 25°C on a rocking platform.
-
e.Wash cells 6 times with ddH2O using EL406 protocols H2O_prime and H2O_Wash_6X160uL.
-
a.
Note: Hoechst staining needs to be repeated in every round of 4i, since it is eluted during antibody elution.
Immunofluorescence imaging of fixed cells
Timing: 30 min
Once immunostaining is completed, cells should be imaged in the presence of the imaging buffer. The N-acetyl cysteine in the imaging buffer scavenges free radicals formed from light excitation, helping to prevent the formation of photoreactive crosslinks.
-
10.Pre-imaging preparation.
-
a.Aspirate H2O from wells using EL406 protocol AspirateTo4uL.
-
b.Add 75 μL 25°C imaging buffer per well.
-
c.Seal plate with aluminum seal.
-
d.While holding the plate above eye-level and without jostling the imaging buffer, gently wipe the bottom of the plate with a Kimwipe soaked in 70% ethanol.
-
e.Use a compressed air duster to dry the ethanol and get rid of dust residue.
-
a.
Note: The sample and imaging buffer should be at 25°C to preserve the pH and efficacy of the imaging buffer.
-
11.Image cells using a 10X air objective on the Operetta CLS high content imaging system.
-
a.Use the minimal exposure time and light intensity to reduce photobleaching of samples while optimizing for a reasonably good signal intensity relative to the background.
-
b.Alternative microscopes and objectives can be used for imaging; however, it is imperative that the images are taken at the same positions in each well across all rounds.
-
a.
Antibody elution of fixed cells
Timing: 45 min
After imaging is completed, bound antibodies are removed with the elution buffer, which contains urea, guanidium chloride, TCEP-HCl, and L-glycine. Elution buffer disrupts hydrogen bonds and covalent disulfide bonds between antibodies and sample antigens. Once adequate elution of the bound antibodies is achieved, an additional round of 4i can be conducted starting with blocking (steps 5–11). Before eluting, the sample is washed with water to remove any PBS that may impact the acidity of the elution buffer. After elution, the plates are reimaged at one field of view per well to confirm removal of antibodies.
-
12.Antibody elution (troubleshooting).
-
a.Remove aluminum seal from the plate.
-
b.Wash cells 6 times with ddH2O using EL406 protocol H2O_Wash_6X160uL.
-
c.Prime peri-pump with elution buffer.
-
d.Add 40 μL elution buffer per well using EL406 protocol AspirateTo4uL_Peri40uL.
-
e.Incubate the plate for 12 min at 25°C on a rocking platform.
-
f.Repeat steps 12d–12e two more times.
-
g.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
a.
-
13.Reimage cells.
-
a.Wash cells 6 times with ddH2O using EL406 protocols H2O_prime and H2O_Wash_6X160uL.
-
b.Follow steps 10–11 to image the plate again in only one field of view.
-
c.Examine images to determine if fluorescence is diminished adequately. Ensure that fluorescence intensities are near background (i.e., unstained cell) intensity levels and ideally have an approximately 90% decrease in fluorescence intensities from true antibody signals under identical image acquisition parameters.
-
i.If the remaining signal is too bright, repeat the antibody elution in step 12.
-
i.
-
d.After antibody elution is completed, proceed to a subsequent round of 4i starting with blocking (steps 5–13). If an additional round of 4i is not needed immediately, but the samples are needed for further 4i analysis in the future, proceed to step 14.
-
a.
-
14.Refixation and storage.
-
a.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
b.Aspirate 1x PBS from wells using EL406 protocol AspirateTo4uL.
-
c.Add 100 μL 4% PFA-PBS solution per well.
-
d.Incubate the plate for 10 min at 25°C on a rocking platform.
-
e.Aspirate PFA and dispose as hazardous waste.
-
f.Wash cells 6 times with 1x PBS using EL406 protocols PBS_prime and PBS_Wash_6X160uL.
-
g.Seal plates containing 1x PBS with aluminum seals and store at 4°C.
-
a.
Note: When working with a cell line with relatively loose attachment, it is helpful to refix cells between every few rounds of 4i imaging to reduce cell loss. In our experience1 and as shown by others,9 refixation does not affect epitope availability or antibody performance in subsequent cycles. This optional step of refixation during a continuous 4i assay includes steps 14a–14f followed by aldehyde quenching (step 4), and should be performed after step 13 and before starting a new round of 4i.
Image processing for fixed cells
Timing: 1 day
Upon completion of a 4i experiment, raw images are processed for subsequent quantification and analysis. Image background subtraction is performed using ImageJ. To account for small shifts in image position across rounds, the Hoechst images are aligned and cropped with CellProfiler and MATLAB.5 CellProfiler is also used to segment and track the nuclei and cytoplasm using the Hoechst and CellMask Green stains, respectively. The resulting output of comma-separated text files contains single-cell intensity measurements of nuclei, cell, and cytoplasm with each cell containing a unique tracking number. Additional data organization and analysis can be performed using MATLAB, R, or Python.
-
15.Background subtraction.
-
a.Perform background subtraction using the rolling ball subtraction algorithm in ImageJ with a ball radius of 50.
-
a.
Note: Example ImageJ batch script: Fixed_Cell_Culture_BG_Subtraction.ijm (see data and code availability)
-
16.Alignment.
-
a.Align images from Hoechst channel across all rounds using normalized cross correlation method within the Align module in CellProfiler. Export X and Y shifts for each round from the alignment. The resulting comma-separated file will contain images as rows and X and Y shifts for each round as columns.Note: Example CellProfiler pipeline: Fixed_Cell_Culture_4i_Align.cpproj (see data and code availability).
-
b.Use the X and Y shifts saved from CellProfiler to align and crop images from all fluorescence channels using MATLAB. Save the final shifted and cropped images.Note: Example MATLAB script: Fixed_Cell_Culture_4i_Image_Shift_and_Crop.m (see data and code availability).
-
a.
-
17.Segmentation and tracking.
-
a.In CellProfiler, group images based on their location in the plate. The number of images in each group should be equal to the number of rounds.
-
b.Optimize segmentation of nuclei based on Hoechst staining using different parameters within the IdentifyPrimaryObjects module.
-
c.Optimize segmentation of whole cells based on CellMask Green staining with the Propagation method within the IdentifySecondaryObjects module. Cytoplasmic segmentation can be obtained using the IdentifyTertiaryObjects module.
-
d.Label individual nuclei across all rounds using the TrackObjects module with the Follow Neighbors method.
-
a.
Note: The resulting comma-separated text files for each image will contain cells as rows and intensity measurements for each fluorescence channel as columns. Additionally, there will be a column with a unique identifier for each nucleus. Additional organization and analysis of data can be performed with MATLAB, R, or Python. Example CellProfiler pipeline containing all steps from segmentation and tracking: Fixed_Cell_Culture_4i_Segmentation_and_Tracking.cpproj (see data and code availability)
Tissue-4i
The tissue-4i protocol follows the same principles as discussed for fixed cell culture, while including additional or modified steps that are unique to handling of FFPE tissue sections. These steps include: (i) deparaffinization and epitope retrieval, (ii) quenching of the intrinsic autofluorescence often seen in tissues with significant extracellular matrix, (iii) imaging in the presence of imaging buffer supplemented with 10% glycerol to reduce possible mechanical shearing of the tissue during coverslip removal in preparation for another round of immunostaining, and (iv) computational image background subtraction using post-quench images.
Deparaffinization and heat-induced epitope retrieval of FFPE tissue slides
Timing: 1 h
The first step of the tissue-4i protocol for FFPE sections is deparaffinization and antigen retrieval. This step prepares the FFPE sample for immunostaining and enables antibodies to access epitopes on target proteins within the tissue. Sub-X is used to dissolve paraffin, and increasingly diluted ethanol solutions are used to slowly rehydrate the tissue section. The application of sustained heat and buffered pH partially reverses formaldehyde crosslinks and unmask epitopes for immunostaining.
-
18.Chemical safety hood preparation.
-
a.In a chemical safety hood, prepare 9 glass staining dishes containing approximately 200 mL each of Sub-X (2 dishes), 1:1 Sub-X:ethanol, 100% ethanol, 95% ethanol, 70% ethanol, 50% ethanol, 30% ethanol, and 1X PBS. The volume should be sufficient to submerge the glass rack containing slides.
-
b.Place the slides in the glass rack and attach the wire handle.
-
a.
Note: The Epitope Retrieval Steamer can be prepared and heated to 100°C during the incubation intervals of deparaffinization.
-
19.Deparaffinization.
-
a.Perform the following incubations to deparaffinize the slides by quickly transferring the slide rack from one glass dish to the next, making sure to fully submerge the slides.
-
i.Sub-X for 5 min.
-
ii.Sub-X for 5 min.
-
iii.1:1 Sub-X:ethanol for 5 min.
-
iv.100% ethanol for 3 min.
-
v.95% ethanol for 3 min.
-
vi.70% ethanol for 3 min.
-
vii.50% ethanol for 3 min.
-
viii.30% ethanol for 3 min.
-
i.
-
a.
-
20.
Transfer the slide rack to 1x PBS until ready for antigen retrieval.
CRITICAL: Do not let the slides dry out between washes or at any time after deparaffinization, as this will cause non-specific antibody binding.
-
21.Steamer assembly.
-
a.Fill the water basin with deionized water.
-
b.Place plastic steaming compartment atop the water basin.
-
c.Place an empty glass staining dish in the steaming compartment.
-
d.Turn on steamer until steam is visible escaping from the vent. Keep the lid closed.
-
e.Using a hot plate or microwave, boil approximately 200 mL of sodium citrate buffer in a flask.
-
f.Using heat resistant gloves, carefully pour boiling sodium citrate buffer into the glass staining dish within the steaming compartment.
-
a.
-
22.Heat induced epitope retrieval.
-
a.Transfer slide rack from PBS to the heated sodium citrate buffer within the steaming compartment and close the lid.
-
b.Incubate for 20 min.
-
a.
-
23.
Transfer the slide rack to PBS and wash slides 3x by gently raising and submerging the slides.
Note: We have found that sodium citrate buffer (pH 6) works for antigen retrieval in several tissue types. However, using a buffer with a higher pH, such as Tris-EDTA buffer (pH 9.0), may be preferred if epitope availability is consistently poor. In general, the low-pH sodium citrate buffer is preferred for retrieving cytoplasmic or membrane-bound antigens, whereas the high-pH Tris-EDTA buffer is preferred for retrieving nuclear antigens.
Pause point: Slides can be kept submerged in 1x PBS for short term storage or coverslipped with 10% glycerol imaging buffer for storage up to 1–3 weeks. Store at 4°C.
Quenching of tissue autofluorescence
Timing: 1.5 h
Following deparaffinization and antigen retrieval, autofluorescence quenching is performed by oxidation with H2O2 and NaOH, combined with LED light excitation (Figure 1).10 By oxidizing endogenous fluorescent biomolecules in tissue, we reduce the background autofluorescence signals that are prominently detected in green (AF488) and yellow (AF568) fluorescent channels (Figure 2). In our hands, this method outperformed two other commonly used autofluorescence quenching methods (copper sulfate buffer and TrueBlack Lipofuscin Autofluorescence Quencher) in reducing the background autofluorescence while preserving antibody elution efficiency for iterative imaging. The performance of these quenching methods, however, could potentially vary depending on the tissue type, thickness of FFPE sections, or age of the archival specimens. Other autofluorescence methods may potentially be suitable for 4i, though it is important to ensure that the chosen method preserves antibody elution efficacy.
-
24.
Prepare fresh autofluorescence quenching solution and plug in the LED lamp.
CRITICAL: The autofluorescence quenching solution should be prepared fresh, because the reduction of H2O2 over time diminishes the oxidation power of the solution.
-
25.
Dispense autofluorescence quenching solution into a transparent western blot box or dish. Ensure there is enough solution to submerge all slides when slides are placed in a single, flat layer.
-
26.Autofluorescence quenching incubation.
-
a.Submerge slides in the H2O2/NaOH solution.
-
b.Turn on the LED light and lower the light source to be as close as possible to the box containing slides without touching the solution.
-
c.Incubate slides in autofluorescence quenching solution under the LED light for 1 h.
-
a.
Note: We have kept the LED light about 1–3 inches from the slide with effective results. If the light has to be raised higher to accommodate more slide surface area, use a larger LED lamp or multiple LED lamps.
-
27.
Transfer slides to a glass staining dish containing PBS and wash slides 3x by gently raising and submerging the slide rack. Keep slides in PBS until ready to proceed.
Optional: Following steps 31–33, take “Post-Quench” images of autofluorescence-quenched tissue stained with 1:20,000 dilution of Hoechst in LICOR Intercept (PBS) blocking buffer (0.5 μg/mL) for background subtraction.
Note: H2O2 and NaOH significantly reduce autofluorescence, but stubborn sections of tissue autofluorescence can be subtracted computationally to enhance the signal-to-background ratio. Imaging in the far-red (AF647) channel usually has little to no autofluorescence and does not require background subtraction. However, if using the green (AF488) or yellow (AF568) channels, we recommend taking “Post-Quench” images after autofluorescence quenching to use for background subtraction.
Pause point: Slides can be kept submerged in 1x PBS for short-term storage or coverslipped with 10% glycerol imaging buffer for storage up to 1–3 weeks. Store at 4°C.
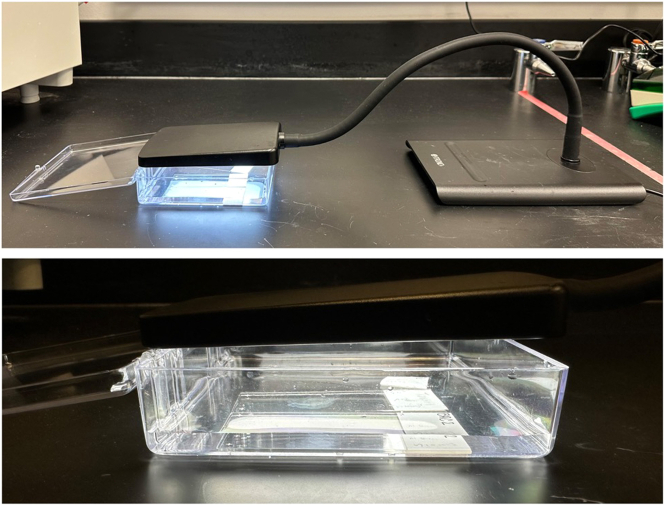
Figure 1.
H2O2, NaOH, and LED light set-up for autofluorescence quenching of tissue slides
(Bottom) Ensure that deparaffinized and antigen-retrieved tissue slides are fully submerged in H2O2-NaOH solution. (Top) Then, ensure that the LED lamp is set to its maximum brightness and brought as close as possible to the tissue sections for proper autofluorescence quenching.
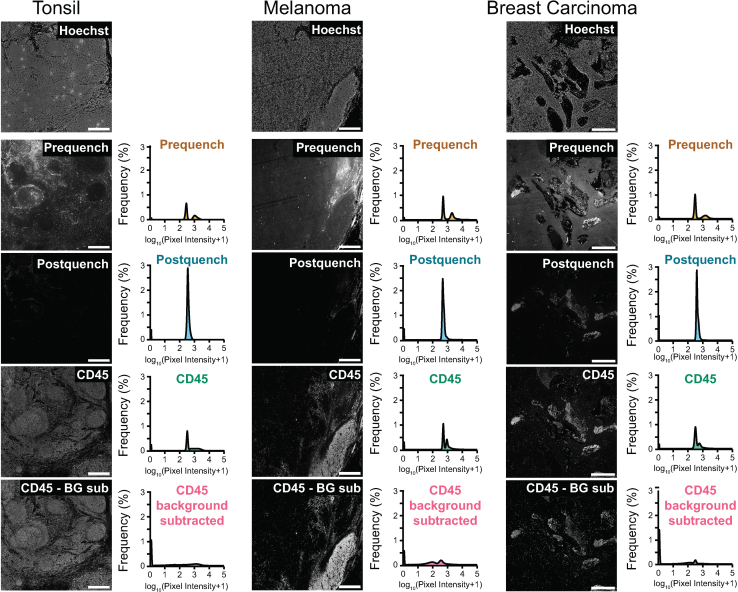
Figure 2.
Representative autofluorescence quenching results in tonsil, melanoma, and breast carcinoma
Representative cropped images and pixel intensity quantifications of Alexa Fluor 488 signal in human tonsil, melanoma, and breast carcinoma tissue before and after autofluorescence quenching, followed by staining for pan-immune cell marker CD45 in the same channel. To generate background-subtracted CD45 images, post-quench image signal intensities were subtracted from CD45 images using the background subtraction module in MCMICRO. The displayed pixel intensity quantifications represent 50% of the pixel intensities that were randomly sampled from each tissue type. Scale = 500 μm.
Antibody and DNA staining of FFPE tissue slides
Timing: 16 h
Following autofluorescence quenching, tissue sections are blocked with 4i blocking buffer, which prevents permanent disulfide bond formation between antibodies and the sample by reacting with free sulfhydryl groups. Samples are then stained with primary and secondary antibodies, followed by Hoechst to visualize the entire tissue and to serve as a consistent reference pattern of individual nuclei for downstream image analysis and processing.
CRITICAL: Performing a round of elution on unstained tissue enhances the immunofluorescence signal in the first round of 4i. For the first round in a 4i experiment, complete a preliminary elution step as detailed in step 35.
-
28.Blocking for immunostaining.
-
a.Prepare fresh 4i blocking buffer.
-
b.Place slides in the stain tray.
-
c.Pipette droplets (250–500 μL total) of 4i blocking buffer around the perimeter of the tissue until it fully covers the tissue section.
-
d.Close the stain tray with the lid and incubate slides in 4i blocking buffer for 1 h.
-
e.After 1 h, remove blocking buffer from each slide by resting the long edge of the slide on a paper towel. Immediately transfer slide to a slide rack submerged in PBS.
-
f.Wash slides 4x by gently raising and submerging the slide rack. Keep slides in PBS until ready to proceed.
-
a.
CRITICAL: When moving or drying slides, be careful not to damage the tissue section. When placing slides in the slide rack, be careful to avoid scraping slides on each other. Do not touch the pipette tip to the slide.
-
29.Primary antibody staining.
-
a.Prepare the primary antibody solution at the desired dilution in LICOR Intercept (PBS) blocking buffer and keep it on ice.
-
b.Minimize excess PBS on slides.
-
i.Remove slides, 1–2 at a time, from PBS. Rest the long edge of the slides on a towel to let liquid run off.
-
ii.Use a small Kimwipe to wipe parallel to the short edges of the slide, near the tissue section but allowing space to avoid contact with the tissue. There should be a rectangle of liquid left to avoid drying out the tissue section.
-
i.
-
c.Place slides back into the stain tray. Pipette droplets (250–500 μL total) of diluted primary antibody around the perimeter of the tissue section.
-
d.Use a small piece of parafilm to cover the tissue section and evenly spread the antibody solution.
-
e.Incubate slides at 4°C for 12–18 h.
-
f.The next day, carefully submerge slides in PBS to dislodge parafilm pieces from the slide.
-
g.Wash slides 4x in fresh PBS by gently raising and submerging the slide rack. Keep slides in PBS until ready to proceed.
-
a.
-
30.Secondary antibody staining.
-
a.Prepare secondary antibody dilution (1:2000 for Alexa Fluors) in LICOR Intercept (PBS) blocking buffer.
-
b.Place slides back into the stain tray. Pipette 250–500 μL of diluted secondary antibody around the perimeter of the tissue section.
-
c.Use a small piece of parafilm to cover the tissue section and evenly spread the antibody solution.
-
d.Incubate slides at 25°C for 1 h.
-
e.Carefully submerge slides in PBS to dislodge parafilm pieces from the slide.
-
f.Wash slides 4x in fresh PBS by gently raising and submerging the slide rack. Keep slides in PBS until ready to proceed.
-
a.
Optional: The 4i procedure also applies to direct immunostaining using fluorophore-conjugated primary antibodies. However, depending on the nature of protein target and antibody, the fluorescent signal resulting from direct immunostaining may be lower and masked by high tissue autofluorescence.
-
31.DNA staining.
-
a.Prepare a 1:20,000 dilution of Hoechst in LICOR Intercept (PBS) blocking buffer (0.5 μg/mL).
-
b.Place slides in the stain tray.
-
c.Pipette droplets (500–1000 μL total) of Hoechst solution around the perimeter of the tissue until it fully covers the tissue section.
-
d.Incubate slides at 25°C for 5 min.
-
e.Rest the long edge of each slide on a towel to let liquid run off. Immediately transfer slides to a slide rack in PBS.
-
f.Wash slides 4x in fresh PBS by gently raising and submerging the slide rack. Keep slides in PBS until ready to proceed.
-
a.
Note: We have found that 1:20,000 Hoechst solution is still effective when stored at 4°C for up to a week.
Pause point: Slides can be kept submerged in 1x PBS for short term storage or coverslipped with 10% glycerol imaging buffer for storage up to 1–3 weeks. Store at 4°C.
Immunofluorescence imaging of FFPE tissue slides
Timing: 1 h
In this section, we prepare tissue slides for imaging on the Operetta CLS High Content Analysis System by applying coverslips onto the slides. Glycerol-imaging buffer containing the antioxidant, N-acetyl cysteine, is used to prevent free radical-induced photocrosslinking between antibodies and antigens in the sample during imaging.
-
32.Apply coverslips to slides.
-
a.Minimize excess liquid on slides (See step 29b).
-
b.Place slides in the stain tray.
-
c.Pipette 50–100 μL of 25°C 10% glycerol imaging buffer onto the edge of the tissue within the remaining rectangle of liquid.
-
d.Gently apply the coverslip to spread the buffer evenly and without bubbles.
-
e.Sequentially rest the long and short edges of the coverslipped slides on a paper towel to draw out excess liquid.
-
f.Place slides with coverslips facing down into the Operetta slide holder.
-
a.
Note: The aim is not to dry out the slide, but to remove liquid that would leak out into the slide holder.
-
33.Image tissue sections using appropriate fluorescence channels. (troubleshooting).
-
a.Use the minimal exposure time and light intensity to reduce photobleaching of samples while looking for an optimized signal-to-background ratio.
-
b.Ensure that images are taken with at least 5% overlap if downstream stitching of individual fields of view is needed.
-
a.
Note: If images were taken of post-quench samples for background subtraction, keep light intensity consistent with the post-quench round of imaging. Exposure time can be varied and accounted for in post-processing.
Pause point: Slides can be kept submerged in 1x PBS for short term storage or coverslipped with 10% glycerol imaging buffer for storage up to 1–3 weeks. Store at 4°C.
-
34.Decoverslipping.
-
a.Decoverslip by placing the slides in a vertical staining jar filled with 1x PBS. Incubate slides for 3–5 min to loosen the coverslips from the slides with gravity.
-
b.Remove the slides from the vertical staining jar; the coverslips should slide off the slides.
-
a.
CRITICAL: Applying and removing coverslips is a major source of tissue loss. Do not remove coverslips manually and be careful when decoverslipping multiple slides in one staining jar. The coverslips can get jostled after they detach within the PBS.
Antibody elution of FFPE tissue slides
Timing: 40 min
Antibody elution in tissue sections follows the same general principles as described for fixed cell culture-4i. The step-by-step procedure for tissue sections is as follows.
-
35.Antibody Elution (troubleshooting).
-
a.Prepare a glass slide staining dish and stain tray filled with deionized water.
-
b.Wash slides 4x in H2O by gently raising and submerging the slide rack.
-
c.Minimize excess liquid on slides (See step 29b).
-
d.Place slides in the stain tray.
-
e.Pipette droplets (250–500 μL total) of 25°C elution buffer onto the tissue section until the section is fully covered.
-
f.Incubate for 10 min.
-
g.Gently remove the elution buffer from the slides.
-
h.Repeat steps 35e–35g 2x more.
-
i.Wash slides 3x in PBS. Keep slides in PBS until ready to proceed with another round of 4i starting with blocking and immunostaining (step 28).
-
a.
Optional: After every elution step, reimage the tissue to ensure fluorescence signals have diminished adequately in all appropriate channels. Stain the tissues with Hoechst but without any antibodies to visualize where the tissue is located on the slide. Adequate reduction in fluorescence signals entail fluorescence intensities that are near background or previous autofluorescence levels, and ideally have an approximately 90% decrease in fluorescence intensities from true antibody signals under identical image acquisition parameters. Furthermore, computational background subtraction using these post-elution images can be used to reveal true antibody signals in subsequent 4i rounds, as long as imaging acquisition parameters remain consistent.
Pause point: Slides can be kept submerged in 1x PBS for short term storage or coverslipped with 10% glycerol imaging buffer for storage up to 1–3 weeks. Store at 4°C.
Image processing for FFPE tissue slides
Timing: 1 h
To visualize the multiplexed protein localization attained through 4i at a whole-slide level, individual fields of view are stitched together and images from iterative rounds are registered using ASHLAR such that single cells are locatable across rounds.3 Uneven illumination artifacts, where each field of view has artificially low intensity at the borders, are corrected with BaSiC to create even whole-slide images with accurate representations of signal intensity.4
-
36.Generate profiles for illumination correction.
-
a.Generate dark-field and flat-field profiles for each round of imaging using the BaSiC ImageJ plugin.
-
a.
-
37.Stitching and registration after completion of all 4i rounds (troubleshooting).
-
a.Use ASHLAR to stitch fields of view and register iterative rounds simultaneously. Feed in the dark-field and flat-field profiles from BaSiC to correct illumination artifacts.
-
a.
Optional: If post-autofluorescence quenching images were taken, unquenched residual autofluorescence can be subtracted using the background subtraction module of MCMICRO found at https://mcmicro.org/parameters/other.html#backsub.
Note: We use a python script found at https://github.com/labsyspharm/basic-illumination/blob/master/imagej_basic_ashlar.py and run BaSiC on the command line to create dark-field and flat-field profiles.
Note: For best results after registering a complete 4i experiment, use the merge function on ImageJ with all rounds of nuclei stain to ensure correct registration after running ASHLAR. Inspect each round of nuclei stain for stitching errors as well. Overlaid nuclei should have no visual shift, though some loss of nuclei in later rounds is expected. For more details, see data and code availability.
Note: We provide step-by-step details on our tissue image processing and visualization workflow at https://github.com/fallahi-sichani-lab/4i-Protocols-Scripts/tree/main/Tissue-4i-Image-Processing-Workflow
Chromosome spread-4i
The metaphase chromosome spread-4i protocol follows the same principles as discussed for fixed cell culture-4i, while including additional or modified steps that are unique to handling of metaphase chromosome spreads and imaging with high-magnification objectives. Specifically, cells are cytospinned onto glass coverslips instead of microscope slides to facilitate using short working-distance, high-magnification water-immersion objectives. To facilitate imaging in the presence of imaging buffer, we create a reservoir on top of the glass coverslip by using a silicone gasket. Finally, to facilitate relocating the same metaphase spread across 4i rounds, the glass coverslip is etched with a diamond scriber.
Cell cycle arrest and harvesting
Timing: 4.5 h
HeLa cells in culture are arrested at metaphase with colcemid, a colchicine derivative that irreversibly binds onto soluble tubulin with high affinity to prevent microtubule assembly. Metaphase arrest enriches for cells with condensed chromosomes that are isolated for downstream visualization and processing.
-
38.HeLa cell cycle arrest.
-
a.Aspirate the old media and add 10 mL of fresh warm EMEM media with 100 μL of colcemid (100 ng/μL final concentration) to the dish.
-
b.Incubate HeLa cells at 37°C for 4 h.
-
a.
Note: Cells should be treated with colcemid for no more than 6 h, as longer incubation times may cause cell death.
-
39.Cell harvesting.
-
a.Pipette media up and down the dish to gently detach and collect arrested cells.
-
b.Transfer 8.5 mL of the EMEM-colcemid-cell suspension into a 15 mL conical tube.
-
c.Hit the dish against the benchtop and vigorously tap the sides of the dish to detach additional arrested cells.
-
d.Collect the remaining 1.5 mL EMEM-colcemid-cell suspension into the 15 mL conical tube.
-
e.Transfer 1.5 mL of the EMEM-colcemid-cell suspension from the 15 mL conical tube back onto the dish.
-
f.Repeat steps 39c–39d to collect a second round of arrested cells.
-
g.Centrifuge the 15 mL conical tube with arrested cells at 300 × g and 25°C for 5 min.
-
h.Aspirate most of the media and leave about 100 μL of media in the tube.
-
i.Resuspend cells by gently tapping on the 15 mL conical tube.
-
a.
CRITICAL: Do not resuspend the cell pellet by pipetting up and down.
-
40.Cell washing.
-
a.Add 5 mL of PBS-Colcemid solution to the resuspended cells and transfer the total contents to a 5 mL centrifuge tube.
-
b.Centrifuge cells at 300 × g at 25°C for 5 min.
-
c.Aspirate the solution and add 1 mL of PBS-Colcemid solution.
-
d.Resuspend cells by gently tapping on the 5 mL centrifuge tube.
-
e.Collect a small sample from the solution for cell counting.
-
f.Centrifuge cells at 300 × g and 25°C for 5 min and perform cell counting in the meantime.
-
g.Aspirate 900 μL of the solution and resuspend the cells in the remaining 100 μL by gently tapping on the 5 mL centrifuge tube.
-
a.
Spreading and fixation of metaphase chromosomes
Timing: 1.5 h
In this step, harvested cells are placed in a hypotonic solution to induce cell swelling and spun onto a glass coverslip using a cytospin to cause cytolysis.
-
41.Hypotonic buffer incubation.
-
a.Based on the cell counts, add the appropriate volume of hypotonic buffer for a cell concentration of approximately 100,000 cells per 300 μL and incubate at 25°C for 20 min, gently tapping the tube to mix every 2 min.
-
a.
-
42.Cytospin clip assembly.
-
a.Set up the clip from bottom to top in the following order: funnel clip → glass microscope slide → 25 × 75 mm glass coverslip → cytospin filter card → cytology funnel.Note: The glass microscope slide is used as a rigid support to prevent the glass coverslip from breaking under the centrifugal force of the cytospin.
-
b.Load the completed cytospin clip assemblies onto the cytospin.
-
c.Load 300 μL of the cell-hypotonic buffer solution into each clip funnel.Note: We recommend loading 75,000 to 150,000 total cells (with volumes between 300 μL and 400 μL) in hypotonic cell solution to the clip funnel to prevent overcrowding of the metaphase chromosome spreads.
-
a.
-
43.
Run cytospin at 164 × g (1000 RPM) for 5 min at 25°C.
-
44.
Disassemble the cytospin clip.
-
45.
Fix chromosome spreads with 350 μL of 2% PFA-PBS solution per coverslip for 20 min at 25°C. Ensure that droplets of the solution are added onto the sides of the chromosome spread to allow the solution to extend into the spreads without damaging or distorting them.
-
46.
Wash spreads 3x with 400 μL of PBS per coverslip for 5 min each by adding droplets of PBS to the sides of the chromosome spreads.
Antibody and DNA staining of metaphase chromosome spreads
Timing: 15 h
After fixation, chromosome spreads are blocked with BSA-PBS 4i blocking buffer. We then stain with primary and secondary antibodies followed by DAPI to visualize the chromosome spreads.
CRITICAL: To minimize chromosome spread distortion, all chromosome spread washing and staining steps should be performed delicately by adding droplets of solution distal to the sample and allowing the solution to smear into the chromosome spreads. Waste solution should be carefully tilted and drained into a beaker. The silicone gasket should be removed during washing and staining steps.
-
47.Primary antibody staining.
-
a.Block with 400 μL per coverslip of fresh BSA-PBS 4i blocking buffer for 1 h.
-
b.Prepare primary antibody solution at desired dilution in 1% BSA-PBS solution. Keep on ice.
-
c.Stain with 350 μL of primary antibodies diluted in 1% BSA-PBS solution.
-
d.Incubate at 4°C for 12–18 h.
-
a.
-
48.Secondary antibody staining.
-
a.Wash 2x with 400 μL of PBST per coverslip for 5 min each.
-
b.Prepare 1:1000 dilution of secondary antibody in 1% BSA-PBS solution.
-
c.Stain with 350 μL per coverslip of secondary antibody solution.
-
d.Incubate for 1 h at 25°C. Keep samples away from light.
-
a.
CRITICAL: Minimize sample exposure to light from this point onward.
-
49.DNA staining.
-
a.Wash 2x with 400 μL of PBST per coverslip for 5 min each.
-
b.Stain with 350 μL of DAPI solution at 25°C for 10 min.
-
c.Wash 2x with 400 μL of PBST per coverslip for 5 min each.
-
a.
Silicone gasket assembly and immunofluorescence imaging of metaphase chromosome spreads
Timing: 30 min
We avoid mechanical shearing of chromosome spreads by assembling a reservoir on top of the glass coverslip with a silicone gasket. The silicone gasket reservoir serves to hold the 4i imaging buffer that contains N-acetyl cysteine, an antioxidant that prevents free radical-induced photocrosslinking between antibodies and antigens in the sample during imaging. A microscope slide is sandwiched on top of the gasket to avoid spills. Imaging is then performed on the Operetta CLS High Content Analysis System using a 63X water-immersion objective.
-
50.Silicone gasket sandwich assembly.
-
a.Dry the sides of the coverslip with Kimwipes so that the silicone gasket can properly adhere onto the coverslip.
-
b.With a diamond scriber, etch 1 crosshair directly on the sample to create a reference for locating identical chromosome spreads in future staining rounds.Note: If there is residue on the coverslip from the previous steps, gently wipe the sides of the coverslip with 70% ethanol on a Kimwipe. Avoid touching the chromosome spreads. Then wipe the edges again with a dry Kimwipe.
-
c.On a flat surface, gently press the silicone gasket onto the coverslip without touching the chromosome spreads.
-
d.Add 500 μL of 25°C imaging buffer to the well with the chromosome spreads.
-
e.Gently press a glass microscope slide onto the open side of the silicone gasket to complete the assembly. (Figure 3).
-
f.Rest the long and short edges of the silicone gasket sandwich on a paper towel to draw out excess liquid.
-
g.Place the silicone gasket assembly with the coverslip facing down onto the Operetta slide holder.Note: Ensure there is no buffer leakage prior to imaging.
-
a.
-
51.Image a Z-stack of metaphase chromosome spreads by identifying the optimal signal-to-background ratio with minimal exposure time and light intensity to reduce photobleaching of samples. (troubleshooting).
-
a.To be able to locate identical chromosome spreads in future rounds of staining and imaging, use the reference crosshairs to identify the relative distance between the chromosome spread of interest and the crosshair.
-
a.
Optional: Store samples in the silicone gasket at 4°C for up to 1–2 weeks.
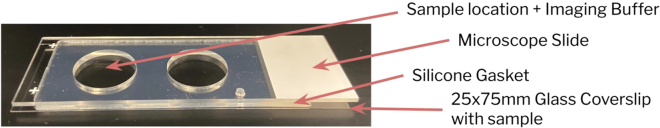
Figure 3.
Silicone gasket assembly for performing chromosome spread-4i
The silicone gasket assembly for chromosome spread-4i is composed of 3 major parts: a 25 × 75 mm glass coverslip, a silicone gasket, and a microscope slide. After chromosome spreads attach to the 25 × 75 glass coverslip, a silicone gasket is carefully attached to the coverslip, with the chromosome spreads at the center of one of the wells. Imaging Buffer is then added into the well containing the sample, followed by a microscope slide to: 1) prevent imaging buffer leakage, and 2) provide additional structural support for the coverslip during imaging.
Antibody elution of metaphase chromosome spreads
Timing: 1.5 h
Antibody elution for metaphase spreads follows the same general principles as described for fixed cell culture. The step-by-step procedure for metaphase spreads is as follows.
-
52.
Disassemble the silicone gasket sandwich, by slowly peeling the silicone gasket and microscope slide away from the coverslip.
-
53.
Wash the silicone gaskets in ddH2O and dry for future use.
-
54.Antibody elution (troubleshooting).
-
a.Wash chromosome spreads with ddH2O 2x for 5 min each.
-
b.Dry excess liquid from slides.
-
i.Remove excess liquid on the coverslip by resting the long edge of the coverslip on a paper towel to draw out excess liquid.
-
ii.Further dry the sides of the coverslip with Kimwipes,
-
i.
-
c.Add 400 μL elution buffer to spreads.
-
d.Incubate for 10 min.
-
e.Gently remove the elution buffer from the coverslip by resting the long edge of the coverslip on a paper towel to draw out excess liquid.
-
f.Repeat steps 54c–54e 2x more.
-
g.Wash coverslips 2x with PBST for 5 min each.
-
a.
Optional: Proceed with additional iterations of 4i (steps 47–54) with the same samples.
Image processing for metaphase chromosome spreads
Timing: 1 h
Chromosome spread images are processed in the following manner for future single-nucleus downstream analysis. A focused image slice from the z-stack for each channel is identified and identical chromosome spreads are manually cropped across all rounds of 4i imaging. Cropped images of chromosome spreads are then registered based on the DAPI channel across rounds.
-
55.
Identify the focused image slice from the image z-stack for each fluorescence channel either manually or with guidance using the Fiji plugin find focused slices.
Optional: Chromosome spreads can sometimes have multiple focal planes depending on the density and orientation of the chromosomes. In such cases, generating maximum intensity projection images from image Z-stacks (using Fiji) is an alternative. However, in our experience, image registration may be less successful for maximum intensity projection images compared to that of individual focused image slices.
-
56.
Using Fiji, create an image stack from the focused image slices for each channel of images within a round of 4i.
-
57.
From the stack, crop out identical chromosome spreads across each round of 4i.
-
58.
Using the Fiji plugin, SIFT (Linear Stack Alignment with SIFT Multichannel), perform image registration based on the DAPI channel.7
Note: Depending on the nature of chromosomal shifts across 4i rounds, different types of image registration may be required. In our experience, affine transformation has been frequently effective in registering images.
Expected outcomes
This protocol describes the process of 4i in three distinct sample types: fixed cell culture, FFPE tissue slides, and metaphase chromosome spreads. We use primary and secondary antibodies to stain one or more proteins in each round of imaging, and then elute antibodies to allow for continued multiplexing across iterative rounds. Subsequent image processing provides information on protein levels and their localization at single-cell or single-nucleus resolution. The main limits to multiplicity with iterative rounds are the availability of validated antibodies and sample integrity.
We have previously used fixed cell culture-4i to elucidate the heterogeneity in single-cell abundance of various proteins, including transcription factors, cell signaling, proliferation and differentiation state markers, across genetically diverse melanoma cell lines.1 Tissue-4i produces spatial maps of protein localization at single-cell resolution from whole-tissue sections. For example, to characterize the cellular heterogeneity across a tissue section, we stained a human melanoma tumor section with differentiation state markers (MITF, SOX10, NGFR), immune cell markers (CD3, CD45), and a proliferation marker (Ki67) (Figure 4). The differentiation state marker distinguishes melanocytic (MITFHigh/NGFRLow) versus neural-crest like (MITFLow/NGFRHigh) cells, and immune and proliferation markers characterize the tumor-immune microenvironment.
Figure 4.
Multiplexed 4i of 6 protein targets (SOX10, NGFR, MITF, CD45, CD3, Ki67) in a human melanoma tissue section
(A) Whole-slide merged image of melanoma tissue stained for 6 protein targets with Hoechst as a nuclear marker. The orange rectangle indicates a magnified region of interest for visualization in B. Scale = 2000 μm.
(B) Magnified region of interest from A, displaying the merged image of Hoechst and 6 protein targets. The yellow rectangle indicates a magnified region of interest for visualization in C and D. Scale = 200 μm.
(C) Representative antibody staining and elution images for each round of 4i. For each displayed 4i round/elution pair, minimum and maximum pixel intensities for Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 channels were matched. Scale = 100 μm.
(D) Magnified region of interest from B, displaying the merged image of Hoechst and 6 protein targets. Scale = 100 μm. Illumination correction and subsequent rolling-ball background subtraction were performed for all displayed images.
For chromosome spread-4i, we mapped multiple chromosomal proteins at single-chromosome resolution, providing an experimental path for interrogating recruitment of mitotic proteins by epigenetic marks. Specifically, we stained for kinetochores (ACA), histone modifications (H3pT3), and a subunit of the chromosomal passenger complex (INCENP) in a 2-round chromosome spread-4i experiment (Figure 5). Images of the chromosome spread after elution qualitatively confirm adequate elution of antibodies.
Figure 5.
A representative 2-round 4i experiment on metaphase chromosome spreads
The sample was stained iteratively for kinetochores using anti-centromere antibody (ACA), phosphohistone sites using phospho-histone H3 (H3pT3) antibody, and subunits of the chromosomal passenger complex using inner centromere protein (INCENP) antibody. Elution images of the chromosome spreads stained with DAPI qualitatively validated proper elution of antibodies. For each Round-Elution pair, minimum and maximum pixel intensities for Alexa Fluor 568 and Alexa Fluor 647 channels were matched; only the minimum pixel intensity was matched for the DAPI channel for visualization. Images across rounds were aligned based on the DAPI channel using affine transformation. Images were taken with a 63X water-immersion objective. Scale = 15 μm.
Limitations
Performing 4i for all three sample types is primarily constrained by sample damage from the iterations of elution, staining, and washing. Therefore, the number of 4i iterations relates to the fragility of samples and the potential sample loss or distortion over time. Sample disruptions hinder downstream image processing and analysis, but thus far we have achieved up to 22, 15, and 4 cycles of 4i in fixed cell lines, FFPE tissue sections, and metaphase chromosome spreads, respectively. We have not thoroughly explored the upper limit of 4i cycles.
The application of 4i protocol to fixed cell culture is optimized only for adherent cells, although performing 4i on suspension cell lines may be possible with additional modifications such as cytospinning. The elution buffer quenches fluorescence in cell lines engineered to express fluorescent reporters. If working with such cells, we recommend imaging right after permeabilization but before aldehyde quenching to record signals from the fluorescent proteins.
When applying 4i to FFPE tissue sections, we have observed that the overall immunofluorescence intensity from identical proteins may differ from one round to another. Thus, we do not recommend comparing absolute fluorescence intensities for the protein targets stained across distinct tissue-4i rounds. Nevertheless, the pixel-to-pixel signal correlation between iterative rounds of 4i for the same antibody remains high. We tested this rigorously across 8 rounds of 4i counterstaining for CD19 (a B cell marker) and CD3 (a T cell marker) in tonsil tissue. The correlation between any two rounds of CD19 staining and any two rounds of CD3 staining were >0.92 and >0.70, respectively (Figure 6). Despite the difference in pixel-to-pixel correlation strength for the two antibodies, both are substantially above the noise level (0.29–0.38). These results may also implicate that CD3 should be stained in earlier 4i rounds, due to potential decreases in epitope availability in later rounds. Tissue sections often undergo antigen retrieval in only one type of buffer (high pH or low pH), which may inadequately retrieve certain protein epitopes. In our experience, performing antigen retrieval sequentially in a low pH then high pH buffer could retrieve more protein epitopes. However, tissue specimens are prone to additional disintegration when using more than one type of antigen retrieval buffer. We also found that antigen retrieval methods that utilize both heat and pressure are more likely to damage tissue specimens in comparison with heat-only methods. We have currently validated Sub-X as the preferred paraffin clearing solution due to its lower toxicity. However, other paraffin clearing solutions (e.g., xylene and d-limonene) are in theory compatible with 4i. If these solutions are chosen for paraffin clearing, we recommend that additional optimization experiments and safety precautions be executed. When imaging large tissue sections, it is often possible that the sample is not uniformly flat. This leads to out-of-perfect-focus images of some fields of view that are detrimental to downstream computational image registration across rounds of 4i. This problem can be overcome largely by using automated wide-field microscopes (such as the Leica THUNDER Imager or EVOS M7000 Imaging System) with capability to create a so-called focus-map, an arbitrary number of focus points defined manually or automatically to interpolate the appropriate z position throughout the entire tissue section. A limitation of background subtraction with the post-quench image is that all subsequent rounds of imaging must have light intensity and gain levels matched to those used in the post-quench image. Exposure time, by contrast, does not need to be matched, because the MCMICRO pipeline compensates for variations in exposure time across different 4i rounds. However, empirical tests should be performed to ensure exposure times used during image acquisition is within ranges that maintain a linear relationship with signal intensities. Studies using background subtraction must determine a constant light intensity and gain level that is optimal for all antibodies. As an alternative, we recommend using periodically acquired post-elution images between rounds of 4i for background subtraction.
Figure 6.
Iterative staining of B cells and T cells in tonsil tissue using primary rabbit antibodies
(A) Representative images of a full tonsil tissue section counterstained for CD19 (B cells) or CD3 (T cells) across 8 rounds of tissue-4i. Alexa Fluor 647-conjugated anti-rabbit secondary antibodies were used to generate the fluorescent signal. All images shown are 11 by 14 field of views stitched together and aligned computationally using ASHLAR. Scale = 5000 μm.
(B) Pixel-to-pixel signal correlation (Pearson’s r) between eight rounds of 4i staining for either CD3+ T cells or CD19+ B cells. 5 × 5 pixel means were used for smoothing.
Finally, 4i of metaphase chromosome spreads requires a water immersion objective with high magnification (minimum 60X) and a numerical aperture of at least 1.20 to capture information on protein localization at the chromosome level. Oil immersion objectives may work, although a modified imaging buffer will be needed to better match the refractive index of the glass coverslip and imaging buffer; further optimizations to ensure minimal distortion of chromosome spreads, adequate removal of a modified highly viscous imaging buffer, and the silicone gasket stays on the coverslip, will be necessary. Furthermore, the Operetta CLS and its automated focus system is useful for high-content imaging applications but suboptimal for visualizing samples prepared with the silicone gasket assembly. As a result, we recommend performing chromosome-spread 4i on fluorescence microscopes with wide-field capabilities that allow for manual focus adjustment. Image processing and analysis of highly-multiplexed chromosome spread images is currently time-intensive and low-throughput. To our knowledge, the emerging field of highly-multiplexed imaging has not come to a consensus for processing or analyzing images at the resolution of chromosome spreads. Future efforts are needed towards developing high-throughput chromosome spread-4i image analysis and adapting chromosome spread-4i for use with confocal microscopy.
Troubleshooting
Problem 1: General
Antibodies from previous staining iterations are not adequately eluted (related to steps 12, 35, or 54).
Potential solution
-
•
Ensure that the reagents used for preparing the elution buffer (especially TCEP) are fresh. Store the elution buffer at 4°C and do not use it if it is older than 1 month.
-
•
Ensure that the elution buffer reaches 25°C before use.
-
•
Consider increasing the duration or frequency of sample incubation in the elution buffer.
-
•
Use a more dilute antibody concentration for immunostaining.
-
•
Minimize the exposure of the samples to light, especially after adding the antibody. Any photocrosslinks formed between the fluorophore, antibodies, and sample will be permanent.
-
•
For antibodies that are more difficult to remove, consider staining with fluorophores with longer fluorescence wavelength (e.g., AF647), since short-wavelength fluorophores (with high-energy excitation) are more prone to photo-induced crosslinking.
-
•
Optimize the order of primary antibodies from one 4i round to the next. We recommend using antibodies against low-abundance epitopes in the early rounds of 4i. Delaying the use of those antibodies until later 4i rounds minimizes the impact of residual fluorescence resulting from strong binding of antibodies to highly abundant epitopes.
Problem 2: Cell culture-4i
Significant amount of cell loss after each round of staining, washing, and elution (related to step 12).
Potential solution
-
•
Ensure washes are being done by pipetting against the wall of the wells instead of directly on the cells.
-
•
Check that the wash and aspiration protocols on the Washer Dispenser are set using manufacturer’s guidelines for cell-based assays (e.g., using a low-fluid path for dispensing fluid and using both a standard and crosswise aspiration). Also, consider increasing the volume of residual liquid with a corresponding increase in wash cycles to ensure enough liquid turnover.
-
•
Refix cells every two to three rounds of 4i.
-
•
For loosely adherent cell lines, consider coating plate wells with fibronectin, collagen, poly-D-lysine, or gelatin to improve cell attachment.
Problem 3: Cell culture-4i
If a cell line contains fluorescent protein reporters, most of the fluorescent signal will be lost after aldehyde quenching and antibody elution steps due to the low pH of the elution buffer (related to step 4).
Potential solution
-
•
Image fluorescent proteins after performing fixation, permeabilization, and nuclear staining, but before aldehyde quenching. During image analysis, treat these images as an individual cycle of 4i.
-
•
Consider using an antibody against the fluorescent protein in one of the 4i rounds.
Problem 4: Tissue-4i
Antibody does not stain consistently in tissue despite previous validation (related to step 33).
Potential solution
-
•
Adjust the imaging buffer to include 1 M HEPES buffer at pH 7.4.11 Alexa Fluor stains have a recommended pH range of 6.5–8.5 and pH fluctuations caused by changes in temperature or CO2 content may affect consistency.
Problem 5: Tissue-4i
Nuclei images across rounds do not stitch or register properly when using ASHLAR (related to step 37).
Potential solution
-
•
Try iterations of ASHLAR with combinations of different –maximum-shift and –filter-sigma parameters. We have successfully tried increasing maximum shift in intervals of 50 and filter sigma in intervals of 1 between 1–9.
-
•
Please see https://github.com/labsyspharm/ashlar/issues or https://forum.image.sc/tag/ashlar for other ASHLAR troubleshooting.
Problem 6: Chromosome spread-4i
Chromosome spreads come off the coverslip or become more distorted with each iteration of imaging (related to step 51).
Potential solution
-
•
Do not dip coverslips with chromosome spreads into buffers during washing steps. Instead, pipette buffers distal to the chromosome spreads and allow the buffer to seep into the chromosome spreads.
-
•
Chromosome spreads located in the center of the sample, rather than the periphery, will have less distortion.
-
•
Consider incubating coverslips in 0.02% gelatin for 2 h at 37°C before cytospinning.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mohammad Fallahi-Sichani (fallahi@virginia.edu).
Technical contact
All technical inquiries should be directed to and will be addressed by the technical contact, Mohammad Fallahi-Sichani (fallahi@virginia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw immunofluorescence microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
The original codes and image analysis pipelines used in this study are publicly available at GitHub: https://github.com/fallahi-sichani-lab/4i-Protocols-Scripts.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Doug Baumann, Mehwish Khaliq, P. Todd Stukenberg, Astrid Catalina Alvarez-Yela, B. Bishal Paudel, and members of the Fallahi-Sichani and Janes laboratories for their technical contributions, helpful suggestions, and discussion. We acknowledge the UVA School of Medicine Biorepository and Tissue Research Facility (RRID: SCR_022971) and Research Computing at the University of Virginia for providing computational resources and technical support. This work was supported by NIH grants U54-CA274499, R35-GM133404, R01-CA249229, T32-GM007267, T32-CA009109, and P30-CA044579.
Author contributions
J.H., K.T.N., M.B., K.A.J., and M.F.-S. conceived and designed the study and wrote the manuscript. J.H., K.T.N., and M.B. developed the protocols, performed the experiments, and analyzed the data. K.A.J. and M.F.-S. supervised the work.
Declaration of interests
The authors declare no competing interests.
References
- 1.Comandante-Lou N., Baumann D.G., Fallahi-Sichani M. AP-1 transcription factor network explains diverse patterns of cellular plasticity in melanoma cells. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gut G., Herrmann M.D., Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science. 2018;361 doi: 10.1126/science.aar7042. [DOI] [PubMed] [Google Scholar]
- 3.Muhlich J.L., Chen Y.-A., Yapp C., Russell D., Santagata S., Sorger P.K. Stitching and registering highly multiplexed whole-slide images of tissues and tumors using ASHLAR. Bioinformatics. 2022;38:4613–4621. doi: 10.1093/bioinformatics/btac544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng T., Thorn K., Schroeder T., Wang L., Theis F.J., Marr C., Navab N. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 2017;8 doi: 10.1038/ncomms14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004;60:91–110. doi: 10.1023/B:VISI.0000029664.99615.94. [DOI] [Google Scholar]
- 8.Schapiro D., Sokolov A., Yapp C., Chen Y.-A., Muhlich J.L., Hess J., Creason A.L., Nirmal A.J., Baker G.J., Nariya M.K., et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat. Methods. 2022;19:311–315. doi: 10.1038/s41592-021-01308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J.-R., Fallahi-Sichani M., Sorger P.K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 2015;6:8390. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J.-R., Izar B., Wang S., Yapp C., Mei S., Shah P.M., Santagata S., Sorger P.K. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife. 2018;7 doi: 10.7554/eLife.31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer B.A., Del Castillo J.S., Pelkmans L., Gut G. Iterative Indirect Immunofluorescence Imaging (4i) on Adherent Cells and Tissue Sections. Bio. Protoc. 2023;13 doi: 10.21769/BioProtoc.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Raw immunofluorescence microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
The original codes and image analysis pipelines used in this study are publicly available at GitHub: https://github.com/fallahi-sichani-lab/4i-Protocols-Scripts.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.