Abstract
Glypican-3 (GPC3) is overexpressed in hepatocellular carcinomas and hepatoblastomas and represents an important therapeutic target but the biologic importance of GPC3 in liver cancer is unclear. To date, there are limited data characterizing the biological implications of GPC3 knockout (KO) in liver cancers that intrinsically express this target. Here, we report on the development and characterization of GPC3-KO liver cancer cell lines and compare to them to parental lines. GPC3-KO variants were established in HepG2 and Hep3B liver cancer cell lines using a lentivirus-mediated CRISPR/Cas9 system. We assessed the effects of GPC3 deficiency on oncogenic properties in vitro and in murine xenograft models. Downstream cellular signaling pathway changes induced by GPC3 deficiency were examined by RNAseq and western blot. To confirm the usefulness of the models for GPC3-targeted drug development, we evaluated the target engagement of a GPC3-selective antibody, GC33, conjugated to the positron-emitting zirconium-89 (89Zr) in subcutaneous murine xenografts of wild type (WT) and KO liver cancer cell lines. Deletion of GPC3 significantly reduced liver cancer cell proliferation, migration, and invasion compared to the parental cell lines. Additionally, the tumor growth of GPC3-KO liver cancer xenografts was significantly slower compared with control xenografts. RNA sequencing analysis also showed GPC3-KO resulted in a reduction in the expression of genes associated with cell cycle regulation, invasion, and migration. Specifically, we observed the downregulation of components in the AKT/NFκB/WNT signaling pathways and of molecules related to cell cycle regulation with GPC3-KO. In contrast, pMAPK/ERK1/2 was upregulated, suggesting an adaptive compensatory response. KO lines demonstrated increased sensitivity to ERK (GDC09994), while AKT (MK2206) inhibition was more effective in WT lines. Using antibody-based positron emission tomography (immunoPET) imaging, we confirmed that 89Zr-GC33 accumulated exclusively in GPC3-expression xenografts but not in GPC3-KO xenografts with high tumor uptake and tumor-to-liver signal ratio. We show that GPC3-KO liver cancer cell lines exhibit decreased tumorigenicity and altered signaling pathways, including upregulated pMAPK/ERK1/2, compared to parental lines. Furthermore, we successfully distinguished between GPC3+ and GPC3- tumors using the GPC3-targeted immunoPET imaging agent, demonstrating the potential utility of these cell lines in facilitating GPC3-selective drug development.
Keywords: Hepatocellular carcinoma, hepatoblastoma, glypican-3, proliferation, migration, invasion, codrituzumab, MAPK/ERK pathway, radiopharmaceutical therapy, molecular imaging
Introduction
Liver cancer remains a global health challenge and is the third leading cause of cancer-related deaths worldwide in 2020, having a very high prevalence in Eastern Asia and sub-Saharan Africa [1,2]. In the United States, it is estimated that the 5-year relative survival rate of liver cancer is 21%, which remains low despite recent improvements in systemic therapies [3]. Hepatocellular carcinoma (HCC) is the predominant type of liver cancer, accounting for approximately 85% of all cases. Most patients with HCC are diagnosed with advanced-stage disease and have limited curative treatment options.
Glypicans, a family of heparan sulfate proteoglycans, consist of a core protein, heparan sulfate chains, and a glycosylphosphatidylinositol linkage [4]. They play essential roles in biological processes, including cell proliferation, cell motility, morphogenesis, and inflammation. The structure of glypicans is highly conserved across the family with 14 cysteine residues. There are six known glypican family members (GPC-1 to GPC-6) in mammals, primarily found in the cell membrane and the extracellular matrix due to glycosylphosphatidylinositol anchors [5]. Furthermore, glypicans have been reported to be involved in angiogenesis, immune cell migration, inflammation, and metastasis in various cancers [6,7]. Glypican-3 (GPC3) regulates several signaling pathways, including WNT, Hedgehog, fibroblast growth factor, and bone morphogenic protein [8-11]. GPC3 is highly expressed in approximately 80-90% of HCC and hepatoblastoma but is not found in normal hepatocytes, hepatocellular adenoma, focal nodular hyperplasia, or large regenerative nodules [12-14]. Previous studies have demonstrated the superior sensitivity of serum GPC3 protein compared to alpha-fetoprotein (AFP), the most commonly used in detection of HCC [15,16]. In addition, GPC3 has been used as a reliable immunohistochemistry marker for HCC diagnosis in surgical pathology [17]. GPC3 also serves as an independent negative prognostic factor for survival in HCC patients [18]. Expression of GPC3 has also been reported in squamous cell lung carcinoma, lung small cell carcinoma, Merkel cell carcinoma, melanoma, head and neck squamous cell cancer, and neuroendocrine prostate cancer, among others [19-22]. Thus, various GPC3 targeted strategies have been developed and evaluated in HCC and other cancers [23,24].
Radiopharmaceutical Therapy (RPT), an emerging cancer treatment delivering radiation selectively to cancer cells, can improve patient survival while preserving quality of life [25,26]. GPC3-target radiopharmaceuticals have been studied for antibody-based positron emission tomography (immunoPET) imaging in preclinical [27-29] and clinical [30] studies in HCC. Our recent work demonstrated enhanced tumor uptake and tumor-to-liver signal ratio with an engineered single-domain antibody (ssHN3) compared to the conventionally modified antibody (nHN3) in xenograft models [31]. Tumor-selective PET imaging of liver cancer would be incredibly valuable clinically, given that assessing viability, especially following locoregional therapy, remains challenging with conventional imaging techniques. Therapeutically, GPC3-selective RPT studies using full-length antibodies have shown promise, though optimization is needed prior to clinical translation [32-34]. Given the important role of GPC3 as a therapeutic target, it is critical to better characterize its oncogenic role in liver cancer. One facile way to assess its role is to engineer knockout lines from parental liver cancer lines that intrinsically express GPC3. Notably, beyond allowing characterization of the role of GPC3, this approach yields isogenic liver cancer cell lines, which theoretically differ only in GPC3 expression and represent a critical tool in assessing target specificity for drug development [35].
The aim of this study is to explore the role of GPC3 expression in HepG2 and Hep3B, two of the most widely used liver cancer cell lines. Furthermore, we clarify the underlying functional mechanisms responsible for the depletion of GPC3 in liver cancer cells, while assessing the target engagement of GC33, a GPC3-specific antibody, by PET imaging.
Materials and methods
Cell culture
HepG2 (a human hepatoblastoma cell line) and Hep3B (a human hepatocellular carcinoma cell line) cells were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Carlsbad, CA, USA) or Eagle’s minimal essential medium (EMEM, ATCC) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) at 37°C under 5% CO2. HepG2-GPC3 knockout (HepG2-KO) and Hep3B-GPC3 knockout (Hep3B-KO) cells were maintained in the same media used for parental cells. All cell lines were confirmed to be mycoplasma-negative using a MycoAlert Mycoplasma Detection kit (Lonza, Basel, Switzerland).
GPC3-KO cell lines construction
GPC3 gene knockout was performed using CRISPR/Cas9 gene editing system as previously described [36]. The pLentiCRISPR-v2 plasmid containing the small guide RNA (sgRNA) targeting exon 3 of the human GPC3 gene (Target DNA sequence: GACATCAATGAGTGCCTCCGAGG, PAM sequence: AGG) (pLentiCRISPR-v2-GPC3) was purchased from GenScript (Piscataway, NJ, USA). To produce lentiviral particles, HEK293T cells were transfected with pLentiCRISPR-v2-GPC3 and a ViraPower lentiviral packaging mix (Thermo Fisher Scientific) using Lipofectamine 3000 (Thermo Fisher Scientific). After 6 hours, the medium was replaced with complete growth medium. Lentivirus-containing supernatant was harvested at 24 h and 48 h, filtered through a 0.45 μM syringe filter (EMD Millipore, Billerica, MA, USA), and concentrated using Lenti-X Concentrator (Takara, Shiga, Japan). The titer of the concentrated lentiviruses was determined using Lenti-X GoStix Plus kit (Takara). Cells were transduced using the lentiviral supernatant and 5 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA) in complete media. GPC3-KO stable cells were achieved via puromycin selection (2 µg/mL; Thermo Fisher Scientific). Single-cell clones were expanded and validated as GPC3 knockout by reverse-transcription polymerase chain reaction (RT-PCR), Western blotting, flow cytometry, and immunofluorescence analyses as well as genomic DNA sequencing.
RT-PCR
Total RNA was extracted from 1 × 106 cells using the RNeasy Micro Kit (Qiagen, Valencia, CA, USA). Subsequently, 1 µg of RNA was used with the QuantiTect reverse transcription kit (Qiagen). PCR amplification was conducted with an initial denaturation of 5 min at 94°C, comprising 25 cycles of 30 s at 94°C, 30 s at 56°C, and 45 s at 72°C, followed by a final elongation at 72°C for 5 min. PCR primers were purchased from Eurofins Genomics (Louisville, KY, USA): HuGPC3, 5’-GTTACTGCAATGTGGTCATGC-3’ (forward), 5’-ACATGTGCTGGGCACCAG-3’ (reverse) and Huβ-actin, 5’-ACCATGGATGATGATATCGC-3’ (forward), 5’-ACATGGCTGGGGTGTTGAAG-3’ (reverse). The PCR was carried out on a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) in a total volume of 20 µL, consisting of a mixture of 2 µL of cDNA solution, 2 pmol of each primer, and PCR master mix (Takara). The amplification products were analyzed by electrophoresis on 1% agarose gel (Thermo Fisher Scientific) in TAE running buffer (20 mM Tris-acetate, 0.5 mM EDTA) containing SYBR green (Thermo Fisher Scientific). PCR amplicons were detected using a ChemiDoc MP Imaging system (Bio-Rad).
Western blotting
Cell lysates were extracted with RIPA lysis and extraction buffer (Thermo Fisher Scientific) containing Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and PMSF (Sigma-Aldrich). Lysates were vortexed and cleared by centrifugation at 13,200 rpm for 20 min at 4°C. Supernatants were transferred to fresh microcentrifuge tubes and protein concentrations were measured using Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific). To detect the cellular localization of β-catenin, four different cell lines (HepG2, Hep3B, HepG2-KO, and Hep3B-KO) were fractionated with the NE-PER nuclear and cytoplasmic extraction kit according to the manufacturer’s protocol (Thermo Fisher Scientific). Proteins (20-50 μg) were subjected to western blot analysis, which were probed with the following primary antibodies. GPC3 antibody was purchased from Cell Marque (261M-96, Rocklin, CA, USA). Non-phospho (Active) β-catenin (#8814), β-catenin (#8480), AKT (#9272), phospho-AKT (#4060), PTEN (#9559), ERK1/2 (#9102), phsopho-ERK1/2 (#4370), NFκB (#8242), phospho-NFκB (#3033), mTOR (#2972), phospho-mTOR (#2976), CDK4 (#2906), CDK6 (#3136), Cyclin D1 (#55506), Cyclin D3 (#2936), p27 Kip1 (#2552), N-myc (#84406), Survivin (#2808), and Lamin B1 (#12586) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Calnexin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were purchased from BD Transduction Lab (#610523, San Jose, CA, USA) and Sigma-Aldrich (#CB1001), respectively. The membrane was incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (HRP) for 1 h at room temperature (RT). The immunoreactive bands were developed with the Clarity or Clarity Max ECL substrate (Bio-Rad), and signals were detected using a ChemiDoc MP imaging system (Bio-Rad). ImageJ software (NIH, Bethesda, MD, USA) was used to evaluate the relative expression of each molecule normalized to GAPDH.
gDNA sequencing
Genomic DNA extraction from cells was performed using the QIAamp DNA kit (Qiagen). The DNA fragment encompassing the guide RNA region at exon 3 of GPC3 gene was amplified using a Takara PCR amplification kit (Takara) and the following primers (Eurofins Genomics): GP1F, 5’-TGCCAAGAACTACACCAATGCC-3’ and GP1R, 5’-ATCTCCACCACACCTGCCATAC-3’. The reactions were incubated for 2 min at 95°C for initial denaturing, followed by 30 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec; and a final extension for 5 min at 72°C. Amplified DNA fragments were purified using the QIAquick gel extraction kit (Qiagen) from agarose gel, ligated into pCR2.1-TOPO vector (Thermo Fisher Scientific), and subsequently transformed into Escherichia coli DH5a (Thermo Fisher Scientific). The plasmid DNAs were extracted using QIAprep Spin Miniprep kit (Qiagen), and were submitted for Sanger sequencing by Eurofins Genomics.
Flow cytometry
Parental liver cancer cells and GPC3-KO cells were harvested by trypsinization and washed with ice-cold phosphate-buffered saline (PBS) three times. Cell pellets were resuspended in FcR block (1:5 final dilution) (Miltenyi Biotec, Bergisch Gladbach, Germany) and incubated for 30 minutes on ice. The cells were incubated with mouse anti-GPC3 monoclonal antibody (Cell Marque), mouse isotype control IgG1 (ab37355; Abcam, Cambridge, MA, USA), or incubated without antibody for 45 min at 4°C. After incubation, cells were washed with ice-cold staining buffer (PBS, 1% BSA (w/v), 0.5 mM ethylenediaminetetraacetic acid (EDTA)), and incubated with 1 μg of APC-labeled goat-anti-mouse IgG1 secondary antibody (Thermo Fisher Scientific) on ice for 30 min in the dark. Stained cells were diluted using the staining buffer and immediately analyzed by a flow cytometer (CytoFLEX Flow Cytometer, Beckman Coulter, Brea, CA, USA). Data analysis was performed using FlowJo software (FlowJo LLC, Ashland, OR, USA).
Immunohistochemistry and immunofluorescence
Formalin-fixed paraffin-embedded (FFPE) xenograft tissue sections (5 µm thickness) were deparaffinized and rehydrated through xylenes and decreasing alcohol gradient. Antigen retrieval was performed using an antigen retrieval buffer (pH 6.0; Dako, Carpinteria, CA, USA) within a pressure cooker. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 15 min. After non-specific binding blocking using protein block (Dako) for 20 min, the sections were incubated with rabbit anti-GPC3 monoclonal antibody (clone no. SP86; dilution 1:3000; Abcam) or rabbit monoclonal anti-Ki-67 antibodies (clone no. SP6; dilution 1:100; Abcam) for 1 h at room temperature. The reaction of antigen and antibody was visualized by the Dako Envision+ peroxidase kit with 3,3-diaminobenzadine (Dako). Negative controls (IgG and omission of primary antibody) were concurrently performed. Thyroid and placenta tissues were used as positive controls. The stained slides were scanned using the NanoZomer XR digital pathology (NDP) system (Hamamatsu, Hamamatsu City, Japan). Digitized immunohistochemical staining images were assessed using Visiopharm software version 6.9.1 (Visiopharm, Hørsholm, Denmark). GPC3 and Ki-67 expression were calculated according to the percentage of positively stained cells and the percentage of cell nuclei stained (potential range of 0-100%), respectively.
Four different cell lines (HepG2, Hep3B, HepG2-KO, and Hep3B-KO) were seeded and cultured on Labtek II chamber slides (Nalgene Nunc, Naperville, IL, USA), rinsed with PBS, fixed with 4% paraformaldehyde (Sigma-Aldrich), and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich). Subsequently, the cells were washed with PBS and incubated with 5% BSA in PBS for 30 min at room temperature. Mouse anti-GPC3 antibodies (clone no. 1G12; Cell Marque) were diluted to 1:200 in 2% BSA in PBS and incubated overnight at 4°C, followed by 1 h incubation at 1:500 dilution with Alexa Fluor 555 goat anti-mouse IgG (Thermo Fisher Scientific). Mouse IgG1 (ab37355; Abcam) was used as the isotype (negative) control. After nucleus visualization with DAPI, slides were mounted with Prolong Diamond antifade (Thermo Fisher Scientific) and imaged on a Zeiss Axio Imager 2 (Carl Zeiss Microscopy, Jena, Germany).
Cell proliferation assay
Cell proliferation of each cell line was measured using cell proliferation enzyme-linked immunosorbent assay (ELISA), BrdU (colorimetric) kit (Roche, Indianapolis, IN, USA) according to the manufacturer’s instructions. 5 × 103 cells were seeded per well into 96-well culture plates. The absorbance in each well was measured at 450 nm using a BioTek Epoch 2 microplate spectrometer (Agilent, Palo Alto, CA, USA). Each experiment was repeated three times.
Wound healing migration assay
HepG2 and HepG2-KO (1 × 106 cells per well) and Hep3B and Hep3B-KO cells (5 × 105 cells per well) were seeded into 6-well culture plates and allowed to grow to a confluent monolayer state. The cell monolayers were wounded with a pipette tip and the medium was changed to fresh media after washing the free-floating cells with PBS. Photos were captured at 0 and 24 h after wounding. The gap distance was measured using ImageJ software (NIH). Wound healing migration rate was represented as % of control.
Transwell matrigel invasion assay
Cell invasion assay was performed using 24-well Corning Biocoat Matrigel invasion chambers with 8.0 μm pore membrane (Corning, Inc., Corning, NY, USA). Cells were seeded into the upper chamber at a density of 2 × 104 in 200 μL serum-free medium. Cell culture media (DMEM and EMEM) supplemented with 10% FBS was added to the lower chamber. Following incubation for 48 h, the non-migrated cells on the upper surface of the filters were removed with cotton swabs, and migrated cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature. Subsequently, migrated cells were stained with 0.5% crystal violet for 20 min. The number of migrated cells was counted under a light microscope by randomly selecting ten fields. Three independent experiments were performed.
Xenograft tumor models
All animal procedures were performed in accordance with appropriate standards under a protocol approved by the Institutional Animal Care and Use Committee at the National Institutes of Health (protocol ROB-104). HepG2, HepG2-KO, Hep3B, and Hep3B-KO cells (5 × 106 cells per mouse) were subcutaneously inoculated into the right flank of female athymic nu/nu 8- to 10-wk-old mice (Charles River Laboratories, Wilmington, MA, USA). In order to validate how GPC3 deficiency affects tumor growth, the tumor volume was measured by a digital caliper 2-3 times a week from 21 days after tumor inoculation. Tumor volume was calculated from digital caliper raw data using the formula V (mm3) = (L × w2)/2. The value w (width) was the smaller of the two perpendicular tumor axes, and the value L (length) was the larger of the two perpendicular axes. The tumor endpoint volume was set at 2000 mm3, and tumors were monitored until the animals were euthanized either due to tumors reaching 2000 mm3 or animals reaching the study endpoint at 60 days for HepG2 and HepG2-KO xenografts and 50 days for Hep3B and Hep3B-KO xenografts after tumor inoculation. The excised tumors from euthanized mice were weighed. Validation of GPC3-KO with GPC3-selective immunoPET agent (89Zr-DFO-GC33) for PET imaging and biodistribution studies was performed once tumors were grown to approximately 200 mm3.
ELISA for serum alpha-fetoprotein
The alpha-fetoprotein (AFP) level (ng/mL) in the serum of HepG2 xenografts (WT and KO; each n=7) and Hep3B xenografts (WT and KO; each n=7) was analyzed using a Mouse alpha-fetoprotein (AFP) Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA), following the manufacturer’s instructions. Serum samples were diluted (1:20) in calibrator diluents, respectively before the assay according to the recommendation of the manufacturer. The microplate was read at 450 nm in BioTek Epoch 2 Microplate Spectrophotometer (Agilent).
Antibody array
Proteins extracted from four different liver cancer cell lines were analyzed using a Human Phosphorylation Pathway Profiling Array C55 kit (Ray Biotech Inc., Norcross, GA, USA), following the manufacturer’s instructions. Briefly, after blocking, phosphorylation profiling array membranes were incubated with 1 mL 10-fold dilution of cell lysates overnight at 4°C. After washing, membranes were incubated with a detection antibody cocktail at RT for 2 h, followed by incubation with HRP-labeled secondary antibody for an additional 2 h. Subsequently, membranes were washed and exposed to a peroxidase substrate. Chemiluminescent signals were detected using a ChemiDoc MP Imaging System (Bio-Rad), and spot intensities were quantified using ImageQuant version 5.2 (Molecular Dynamics; Sunnyvale, CA, USA).
RNASeq data analysis
RNA extracted from parental liver cancer cells and GPC3-KO cells were analyzed by RNA sequencing. RNA seq samples were pooled and sequenced on NovaSeq 6000 S1 using Illumina Stranded Total RNA Prep, ligation with RiboZero Plus, and paired-end sequencing. Initial raw sequencing data were demultiplexed using Bcl2fastq (v2.20). Reads of the samples were trimmed for adapters and low-quality bases using Cutadapt before alignment with the reference genome (hg38) and the annotated transcripts using STAR. Picard (v2.18.26) was employed for RNA statistics, and duplication removal and quantification of gene and isoform abundances were conducted using RSEM (v1.3.1) for accurate expression level estimation. Differential gene expression analysis was carried out using DESeq2 (v1.34.0). Genes having a false-discovery corrected p-value (padj) <0.05 and a fold change >1.0 were considered as differentially expressed. The R package ComplexHeatmap (v2.14.0) was used to generate gene expression heatmaps. The R package clusterProfiler (v4.6.2) was used to examine gene ontology, molecular function, and cell composition signatures. Gene set enrichment analysis (GSEA) was performed using the GSEA tool (v4.1.0) (https://www.gsea-msigdb.org/gsea/index.jsp). RNA sequencing raw gene counts derived from STAR were first normalized using DESeq2 (v1.34.0). The table of normalized gene counts generated was used for GSEA analysis, using the log2 ratio as the enrichment metric. 1,000 gene set permutations were run to generate an empirical distribution of enrichment scores, and the p-value was calculated based on score distribution.
Radiolabeling of GC33 with Zr-89
Codrituzumab (GC33), a humanized anti-GPC3 IgG1, was obtained from Chugai Pharmaceutical Co. Ltd. (Tokyo, Japan). Zirconium-89 oxalate was obtained from 3D Imaging (Little Rock, AR, USA). The GPC3-selective immunoPET agent was prepared using a previously published method [37,38]. Briefly, GC33 was reacted with a 3-fold molar excess of p-isothiocyanatobenzyl-desferrioxamine (DFO-Bz-NCS, Macrocyclics, Inc., Plano, TX, USA) to prepare DFO conjugate (DFO-GC33) and the concentrations of the conjugates (3.3 mg/mL) were measured using bicinchoninic acid assay (BCA, Thermo Fisher Scientific). For radiolabeling, 150 MBq of Zr-89 oxalate in HEPES buffer (0.5 M, pH 7.1-7.3, Thermo Fisher Scientific) was mixed with DFO-GC33 (0.15 mg, 3.3 mg/mL, 45 µL) and the reaction mixture was incubated for 1 h at room temperature and the reaction was quenched by the addition of DTPA (0.1 M, 5 µL, pH 7). The radiolabeled conjugate was purified by PD-10 column (GE Healthcare Biosciences, Pittsburgh, PA, USA) using 0.9% NaCl (pH 7, Sigma Aldrich). The molar activity and purity of the conjugate were determined by HPLC (Agilent 1200 Series) using a size exclusion column (tR=8.0 min, 4.6 mm ID × 30 cm, 4 µm, TSKgel SuperSW3000, Tosoh Bioscience LLC; Montgomeryville, PA, USA). High-performance liquid chromatography (HPLC) condition: eluent, 0.1 M sodium phosphate, 0.1 M sodium sulfate, 0.05% sodium azide, 10% isopropyl alcohol (pH 6.8), flow rate: 0.3 mL/min.
ImmunoPET and biodistribution
HepG2 xenografts (WT and KO; each n=3) and Hep3B xenografts (WT and KO; each n=3) were injected through a tail vein with 3,696 ± 63 kBq (5 μg) of 89Zr-DFO-GC33 and at 72 h after injection, PET/CT images were acquired using PET/CT (BioPET/CT; Sedecal, Madrid, Spain) as follows: the mice were anesthetized using 2% isoflurane, and static PET scans were acquired over 10 min. Whole-body computed tomography (CT) scans (8.5 min, 50 kV, 180 μA) were obtained immediately after PET images and were used to provide attenuation correction and anatomic co-registration for the PET scans. PET data were reconstructed using 3-dimensional ordered-subsets expectation maximization (OSEM) and were normalized, decay-corrected, and dead-time-corrected before analysis using MIM software (MIM Software Inc., Cleveland, OH, USA). After PET/CT imaging, the mice were euthanized by CO2 asphyxiation, and 12 tissues, including tumor, were collected for the ex vivo biodistribution (BioD). All samples were weighed and counted on a γ-counter (2480 Wizard3; Perkin Elmer Inc., Waltham, MA, USA). The counts were converted to percentage injected activity (%IA) using a standard solution of known activity prepared from the injection solution. %IA/g was calculated by dividing the activity in each organ by its weight.
Statistical analysis
Statistical analyses were conducted using Prism 10.0 (GraphPad Software Inc., San Diego, CA, USA). The data are expressed as the mean ± standard deviation (SD) or standard error (SEM). Unpaired two-tailed t-tests were employed for statistical comparisons between the two groups. For datasets with more than two groups, a one-way ANOVA was performed, followed by Tukey’s post hoc test. Statistical significance was established at P<0.05 and is indicated in the figure legends.
Results
CRISPR/Cas9-mediated knockout of GPC3 effectively disrupts its expression in liver cancer cells
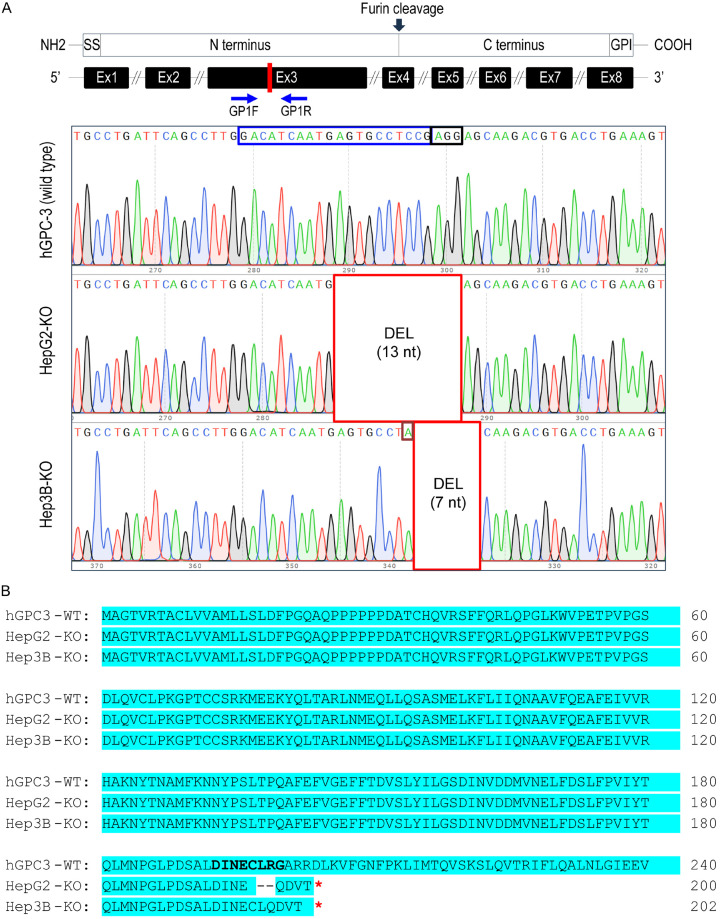
We used CRISPR/Cas9 to generate GPC3 knockout (KO) liver cancer cells (HepG2 and Hep3B) and assessed the knockout status through genomic DNA sequencing. After single colony selection, surviving clones (10 clones per cell line) were expanded and passaged, and half of the total cells were harvested for genomic DNA isolation. The rest were expanded and cryopreserved. To analyze the consequences of sgRNA-mediated GPC3-KO at the genomic level, we conducted PCR amplification of the genomic region encompassing the targeted exon 3. Subsequently, the resulting PCR amplicons were cloned into a TA-cloning plasmid and subjected to sequencing. The sequencing results confirmed the presence of a 13-bp and 7-bp nonsense deletion in HepG2-KO and Hep3B-KO, respectively (Figure 1A). A stop codon (TGA) was identified at positions 601-bp and 607-bp of the GPC3 open reading frame (ORF) in both HepG2-KO and Hep3B-KO clones, respectively (data not shown). Additionally, a point mutation (C>A) was detected at position 554 bp of the GPC3 ORF in Hep3B-KO (Figure 1A). Translation of the ORF yielded a 200-amino acid polypeptide for HepG2-KO and 202-amino acid residues for Hep3B-KO, respectively (Figure 1B), whereas GPC3-WT has a length of 603 amino acids. These results suggest that the sgRNA-mediated gene editing system effectively disrupted the GPC3 gene, thereby hindering the synthesis of functional protein.
Figure 1.
Generation of GPC3 knockout (KO) liver cancer cell lines using CRISPR-Cas9. A. Schematic representation of the genomic structure of GPC3 with the engineered exon site highlighted in red (Exon 3). The gRNA region (blue box) and PAM region (black box) were amplified using the GP1F and GP1R primer set, and the resulting DNA was sequenced. In HepG2-KO, a 13-nt deletion was observed, while in Hep3B-KO, a 7-nt deletion accompanied by a single nucleotide mutation (burgundy box) was identified compared to the wild type. B. Alignment of the amino acid sequences of partial human GPC3 and the two GPC3-KO proteins, presented in the one-letter code. Protein synthesis in the KO cells was terminated by a stop codon (indicated by a red-colored asterisk in the box). Open reading frames are highlighted in cyan, and amino acid sequences corresponding to the sgRNA-targeted regions are shown in bold letters.
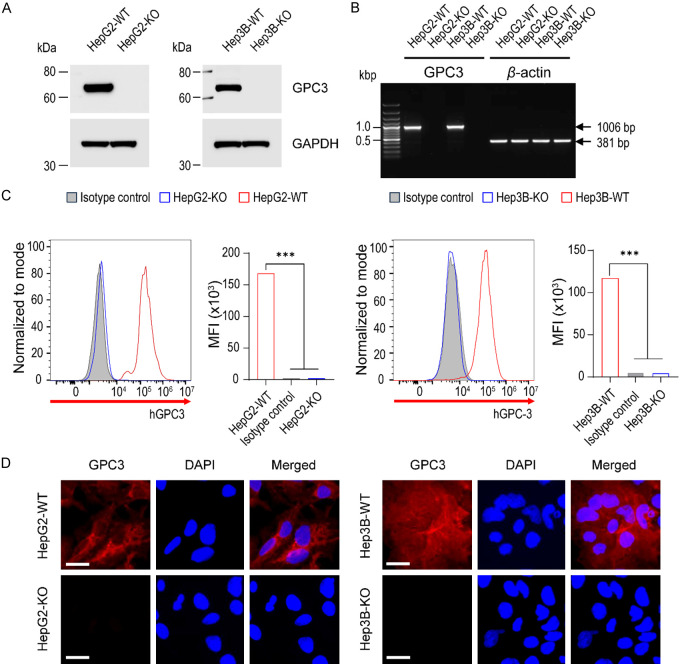
Next, we assessed the knockout efficiency through Western blotting. It was evident that GPC3 protein expression was undetectable in the majority of clones (8/10 clones, 80%). Two single-cell clones (KO-LG3 and KO-VG5 for HepG2 and Hep3B, respectively) were chosen for all subsequent analyses (Supplementary Figure 1 and Figure 2A). Subsequently, we checked the transcriptional expressional level of GPC3 by RT-PCR. As shown in Figure 2B, the PCR resulted in a 1006 bp target band exclusively in wild-type liver cancer cells.
Figure 2.
Confirmation of GPC3 knockout at the transcriptional and translational level. A. Western blots of cell lysates extracted from parental and GPC3-KO cells. Endogenous GPC3 protein was detected by the protein-specific antibody in parental cells, whereas protein loss was confirmed in the KO cells. GAPDH was used as the internal control. B. RT-PCR analysis of parental and GPC3 knockout HepG2 and Hep3B cell lines. Successful knockout of the GPC3 gene was confirmed in the KO cell lines. Beta-actin was used as the internal control. C. Analytical flow cytometry of surface GPC3 expression on parental and GPC3-KO cells. Parental HepG2 and Hep3B cells (red line) expressed GPC3, while KO cells (blue line) lacked intact GPC3 expression. Data are presented as the mean fluorescence intensity (MFI). Mouse immunoglobulin G1 (IgG1, grey-filled area) served as the isotype control. Differences in MFI were statistically tested using ANOVA. ***, P<0.001. D. Immunofluorescence staining with GPC3 (in red) and DAPI (in blue) on parental and GPC3 knockout HepG2 and Hep3B cells. Highly expressed GPC3 in parental cells was completely knocked out in HepG2 and HepG2-KO clones. The bottom panel shows immunofluorescence with the isotype negative control. Scale bars are shown for 20 µm.
In the context of radiopharmaceutical applications, it is crucial to identify the expression of potential target molecules on cancer cell membranes. We confirmed the membrane expression of KO cell lines by flow cytometry. The targeted HepG2 and Hep3B cells exhibited a complete lack of GPC3 surface expression, in contrast to wild-type (WT) HepG2 and Hep3B cells (Figure 2C). The mean fluorescence intensity (MFI) of HepG2-WT and Hep3B-WT was 66-fold and 25-fold higher, respectively, compared to HepG2 GPC3-KO and Hep3B GPC3-KO. Meanwhile, the MFI of Hep3B-WT was 1.4-fold lower than that of HepG2-WT. Furthermore, we validated the GPC3-KO using immunofluorescence. Fixed cells were immunolabeled with primary antibodies against GPC3 from both parental liver cancer cells and GPC3-KO cell cultures. GPC3 expression was notably absent in GPC3-KO liver cancer cells, while clear expression was observed in HepG2-WT and Hep3B-WT cells (Figure 2D and Supplementary Figure 2). These results confirm the complete knockout of GPC3 expression at both transcriptional and translational levels.
GPC3 deletion impairs cell proliferation, migration, and invasion
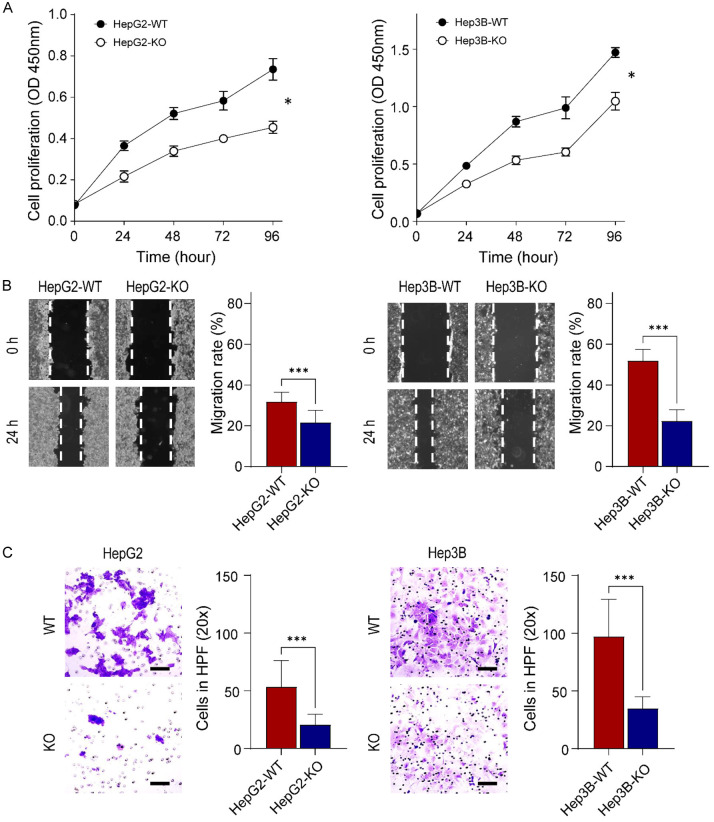
Next, we investigated whether GPC3-KO affects the oncogenic properties in liver cancer cells. A BrdU cell proliferation assay, which detects actively proliferating cells, showed that the depletion of GPC3 resulted in the inhibition of cell proliferation. At 96 h, the proliferation rate of HepG2-KO and Hep3B-KO was significantly reduced by 61.8 and 71.1%, respectively, compared to their parental cells (both P<0.05, Figure 3A). This reduction suggests that GPC3-KO in liver cancer cells led to a marked decrease in cell numbers and an extended cell doubling time.
Figure 3.
Effect of GPC3 ablation on liver cancer cell growth, migration, and cell invasion. A. Cell growth rate of parental and GPC3-KO cells. Cell proliferation was measured using the BrdU cell proliferation assay kit after 24, 48, 72, and 96 h. The left panel represents HepG2, and the right panel represents Hep3B. The cell growth rate was significantly reduced by GPC3 deficiency in both HepG2 and Hep3B. B. Scratch wound healing assay demonstrates a significant decrease in the rate of wound healing in GPC3-KO cells compared to the wild-type liver cancer cells. C. Cell invasion was significantly inhibited after the knockout of GPC3 expression. Invading cells were assessed by counting cells in five random high-power fields (HPFs). Scale bars are shown for 100 µm. All experiments were performed in triplicate, and error bars represent standard deviations from the mean. Differences in cell growth and migration rates were statistically tested using the t-test. *, P<0.05; ***, P<0.001.
Subsequently, we assessed the cell migration of GPC3-KO cells using the wound healing assay. The cell monolayer was scraped, and the width of the wound gap was measured after 24 h. This scratch assay revealed that GPC3-KO significantly inhibited the wound healing ability of HepG2 (67.6% diminution compared with the parental cell, P<0.001, Figure 3B left panel). Similar inhibitory effects were also observed in Hep3B-KO cells (43.2% less than Hep3B-WT decrease, P<0.001, Figure 3B right panel). Through the transwell matrigel invasion assay, GPC3 depletion showed significantly reduced HepG2 (0.4-fold decrease, P<0.001) and Hep3B (0.4-fold decrease, P<0.001) invasion ability (Figure 3C). These results collectively suggest that GPC3 levels correlate with cell proliferation, migration, and invasion ability of HepG2 and Hep3B cells.
GPC3 deficiency suppresses tumor growth in mice
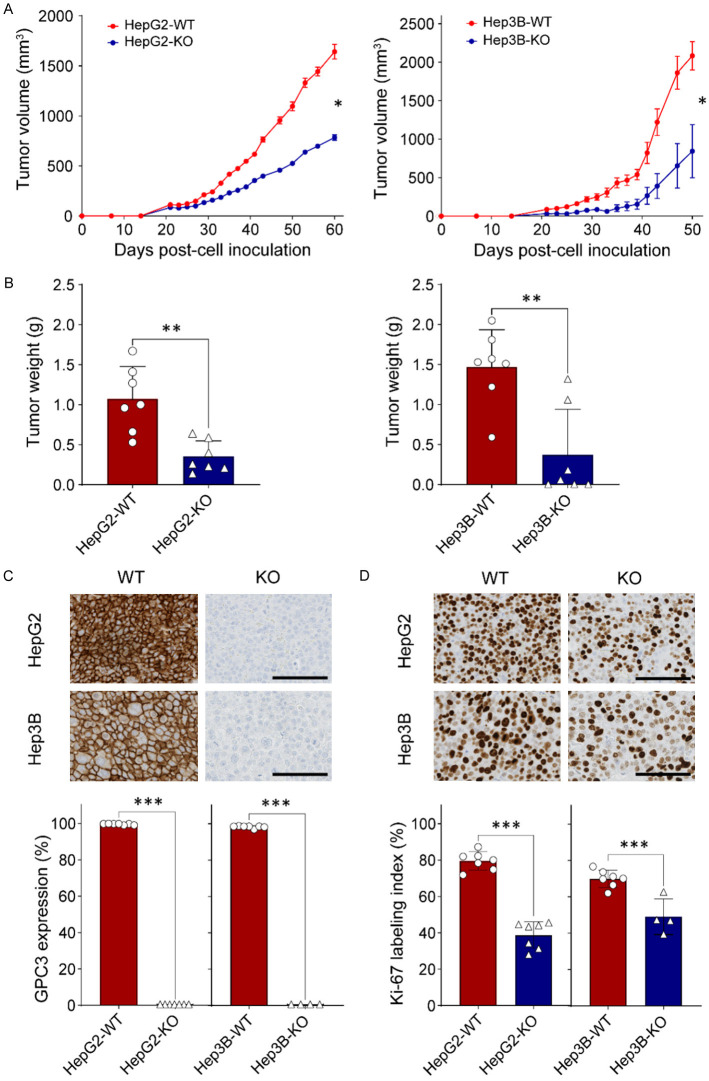
Cell lines harboring GPC3-KO were injected subcutaneously into nude mice to further validate the functional consequences of GPC3-KO and examine tumor growth. We observed significantly delayed growth of HepG2-KO and Hep3B-KO tumors compared to wildtype tumors. While the average tumor volume in the HepG2-WT group on day 60 was 1641.8 ± 192.0 mm3, HepG2-KO tumor volume was 784.2 ± 73.5 mm3 (52.3% less than HepG2-WT, P<0.05, Figure 4A left panel). Similarly, while the average tumor volume in Hep3B-WT group on day 50 was 2082.8 ± 183.8 mm3, Hep3B-KO tumor volume was 842.9 ± 343.6 mm3 (59.5% less than Hep3B-WT, P<0.05, Figure 4A right panel). Tumors in all mice of the HepG2-WT group were larger than those in the HepG2-KO group (Supplementary Figure 3A). In the Hep3B-KO tumor group, tumors did not develop in three mice, and in two mice, the tumor volumes were significantly smaller compared to those in the Hep3B-WT group, measuring 124.2 mm3 and 400.9 mm3, respectively, at day 50 (Supplementary Figure 3B).
Figure 4.
Impact of GPC3 deficiency in liver cancer cells in a xenograft mouse model. A. Comparison of in vivo tumor growth between parental and GPC3 knockout (KO) cells. Five million parental (HepG2 & Hep3B) and GPC3-KO cells were subcutaneously inoculated into BALB/c athymic nu/nu mice, with each group comprising n=7 mice. The values represent the mean ± standard error (SE). B. Tumor weights of xenograft tumors derived from wild-type HepG2, GPC3-KO HepG2, wild-type Hep3B, and GPC3-KO Hep3B cells. The data represents the mean ± standard deviation (SD). C. Immunohistochemical staining for GPC3 in xenograft tumor tissues. Digital analysis using Visiopharm software categorized staining intensity as negative or positive based on intensity. The final score was calculated as the mean positive ratio from six randomly selected different regions of the tumor section. D. Immunohistochemical staining for Ki-67 in xenograft tumor tissues. The Ki-67 expression index represents the mean percentage of positively stained cells from six representative regions of the xenograft tumor section. The data represent the mean ± SD. Statistical significance in experimental values was determined using the t-test. *, P<0.05; ***, P<0.001.
The weight of tumors excised from the four groups at the study endpoint was directly compared (Figure 4B). The average weight of HepG2-KO tumors (0.35 ± 0.20 g) was 3 times lower than that of tumors from the HepG2-WT group (1.07 ± 0.41 g) (P<0.01, Figure 4B left panel). Similarly, the average weight of Hep3B-KO tumors (0.37 ± 0.21 g) was 4 times lower than that of tumors from Hep3B-WT group (1.47 ± 0.17 g) (P<0.01, Figure 4B right panel). Together, these results suggest that GPC3 deficiency suppressed tumor growth in mice.
To investigate whether GPC3-KO in mouse tumors would result in any differences in AFP levels, a widely used tumor marker for HCC, we measured serum AFP levels from xenografts. Even though differences in serum AFP levels were observed based on tumor type (HepG2 and Hep3B), GPC3-KO did not affect serum AFP levels between HepG2-WT and HepG2-KO (48.0 ± 3 0.2 ng/mL and 55.8 ± 35.9 ng/mL, respectively, P=0.644) as well as Hep3B-WT and Hep3B-KO (19.5 ± 8.9 ng/mL and 14.6 ± 3.8 ng/mL, respectively, P=0.171) (Supplementary Figure 4A). Moreover, we also analyzed the correlation between serum AFP levels and tumor volume, but the results failed to demonstrate any correlation (Supplementary Figure 4B).
The formalin-fixed paraffin-embedded sections were immunohistochemically stained with antibodies for GPC3 and Ki-67 proliferation marker. The results revealed membranous expression exclusively in parental liver cancer cells for GPC3 (Figure 4C). The Ki-67 labeling index in HepG2-KO and Hep3B-KO cells were significantly decreased by 51.2 and 25.3% (P<0.01, Figure 4D), respectively.
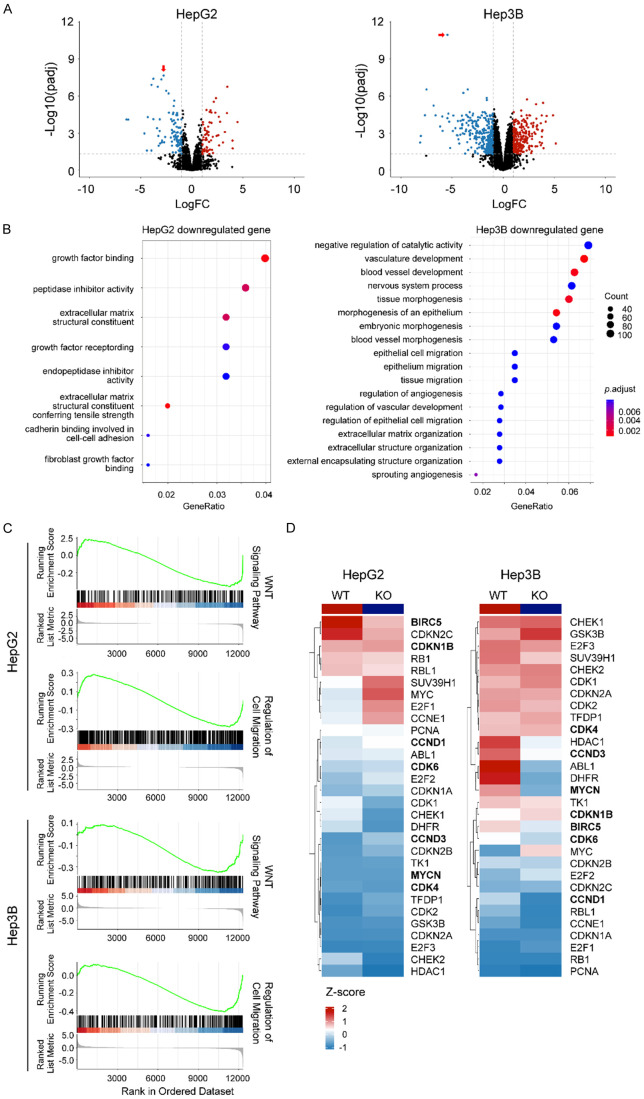
GPC3 deficiency alters the expression of genes involved in WNT signaling and cell cycle and migration at the transcriptomic level
We observed a statistically significant downregulation of GPC3 (P=2.18 × 10-8 for HepG2 and P=1.09 × 10-11 for Hep3B) in KO cell lines (Figure 5A). Using an adjusted p-value threshold <0.05 and fold change >1.0 to define differentially expressed genes (DEGs), we noted that GPC3-KO showed an effect on gene expression patterns with 98 genes upregulated and 64 downregulated for HepG2 and 234 genes upregulated and 299 downregulated for Hep3B. In enrichment analyses of DE genes in Gene Ontology Biological Process (GOBP) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, downregulated genes in HepG2 were enriched for growth factor binding and cell invasion and migration for Hep3B (Figure 5B). The effect of GPC3-KO on cell invasion and migration was consistent with the observation from the wound healing assay. We also confirmed these findings using Gene Set Enrichment Analysis (GSEA), a threshold-independent approach to examine biological processes enriched. Top GSEA-enriched terms indicated a strong effect of GPC3-KO on the WNT signaling pathway and protein kinase B (AKT) pathway (Figure 5C), both of which are critical regulators of cell proliferation, survival, and differentiation. Furthermore, other enriched pathways cell migration and the cell cycle in both HepG2 and Hep3B. We observed that specific genes related to the cell cycle were less expressed in GPC3-KO cell lines compared to WT, implying the potential effect of GPC3 in cell cycle regulation (Figure 5D). In addition to these shared pathways, the Hep3B cell line with GPC3-KO was downregulated in for genes involved in related to cell-cell adhesion, signal transduction, response to growth factor, and signaling receptor activity (Supplementary Figure 5), suggesting distinct pathways that GPC3 may contribute to in different liver cancer subtypes/disease/cell line. In the comparison of DEGs between the HepG2 and Hep3B cell lines following GPC3-KO, we observed a minimal overlap between DEGs in HepG2 and Hep3B cell lines (8.02% of HepG2 DEGs and 2.44% of Hep3B DEGs), suggesting different compensatory responses to the loss of GPC3.
Figure 5.
Transcriptional effects of GPC3 knockout in HepG2 and Hep3B cells. A. Volcano plot showing differential gene expression upon GPC3 knockout using CRISPR/Cas9 in HepG2 and Hep3B cell lines. Plot depicts log2 fold change vs. -log10 False discovery-corrected p-value (FDR) for individual genes. Downregulated genes are represented in blue and upregulated genes are represented in red. GPC3 is represented with red arrow. B. Plot showing enriched KEGG pathway and gene ontology, biological process (GO BP) terms in genes downregulated upon GPC3 knockout, relative to parental liver cancer cell lines. Gene ratio indicates proportion of the GO/KEGG term containing query genes, dot size indicates number of query genes in the GO term, and color indicates the false-discovery rate corrected/adjusted p-value. C. Gene set enrichment analysis (GSEA) of RNA sequencing data from HepG2 and Hep3B cells with GPC3-KO. Results show representative GSEA plots and tables of top enriched GO BP gene sets downregulated in GPC3-KO cells. D. Heatmap representation of cell cycle related genes upon GPC3-KO.
GPC3 deficiency modulates critical signaling pathways related to cell proliferation, survival and cell cycle regulation in liver cancer cells
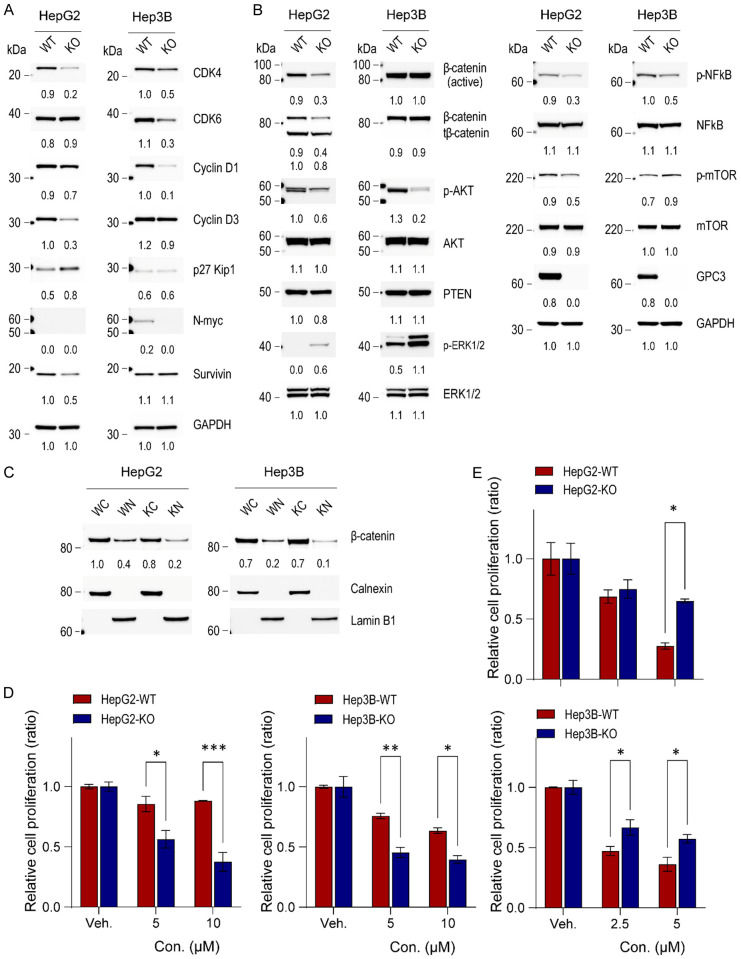
Because genes related to the cell cycle were downregulated in GPC3-KO liver cancer cells according to RNASeq data analysis, we investigated multiple components related to cell cycle regulation in liver cancer cells harboring GPC3-KO vs. wild type. Among cell-cycle regulators, the expression of CDK4, CDK6, Cyclin D1, Cyclin D3, survivin, and N-myc was decreased significantly in GPC3 deficient HepG2 and Hep3B cancer cell lines. Additionally, the expression of p27 Kip1, a negative regulator of cell proliferation, was increased in HepG2 GPC3-KO cells (Figure 6A), aligning with the observed reduction in the cell proliferation rate in GPC3-KO liver cancer cells.
Figure 6.
Impact of GPC3 knockout on cellular signaling in liver cancer cells. (A) Western blot analysis of cell-cycle related molecules, (B) signaling pathways, and (C) cytoplasmic and nuclear protein fractions in HepG2 and Hep3B parental and GPC3 knockout cells. The numerical values beneath the blot images represent the expression levels measured as fold-change. The cytoplasmic proteins were normalized using anti-calnexin antibody, while the nuclear proteins were normalized using anti-lamin B1 as a loading control. WC, wildtype cytoplasmic fraction; WN, wildtype nuclear fraction; KC, GPC3 knockout cytoplasmic fraction; KN, GPC3 knockout nuclear fraction. Additionally, the impacts of ERK and AKT inhibitors on HepG2 and Hep3B parental and GPC3 knockout cells was investigated. Cells were seeded at 2 × 105 cells and then treated with GDC0994 (ERK inhibitor, 5 and 10 µM) (D) or MK2206 (AKT inhibitor, 2.5 and 5 µM) (E) after 24 h. Viable cell numbers were counted on day 2. The data are presented as the mean ± SD. Differences in cell viability were statistically assessed using the t-test. *, P<0.05; **, P<0.01; ***, P<0.001.
To elucidate the critical signaling pathways associated with GPC3 ablation in HepG2 and Hep3B cells, we performed an antibody array using a Human Phosphorylation Pathway Profiling Array kit. Our findings revealed a significant decrease in phosphorylated AKT (p-AKT) and an increase in phosphorylated mitogen-activated protein kinase (p-MAPK/ERK1/2) in GPC3-KO liver cancer cells (Supplementary Figure 6). We further validated quantitative changes using an antibody array and Western blot. In addition, we investigated the expression level and nuclear translocation of β-catenin due to the pivotal role of the WNT/β-catenin signaling pathway in HCC and the well-described modulation of WNT/β-catenin by GPC3 [8]. Ablation of GPC3 expression in HepG2 cells led to a significant reduction in p-AKT, phosphorylated nuclear factor-κB (p-NFκB), phosphorylated mammalian target of rapamycin (p-mTOR), full-length β-catenin and truncated β-catenin (tβ-catenin), but slight upregulation of p-MAPK/ERK1/2. Similar molecular alterations were observed in GPC3-KO Hep3B cell. However, full-length β-catenin expression was not changed (Figure 6B). We further investigated the nuclear translocation of β-catenin in both liver cancer cell lines. Nuclear translocation of full-length β-catenin was reduced by half in both HepG2-KO and Hep3B-KO compared to their respective wild-type lines, while the cytoplasmic β-catenin level was slightly reduced in HepG2-KO cells (Figure 6C). These data suggest divergent roles of GPC3 in the two liver cancer cell lines.
To further assess whether the compensatory activation of MAPK/ERK, in response to decreased AKT signaling, we treated both parental and GPC3-KO liver cancer cells with either ERK inhibitor (GDC09994) or AKT inhibitor (MK2206). We then assessed the efficacy of each inhibitor on cell proliferation. When compared with their respective wild-type cell lines, HepG2-KO (1.5- and 2.3-fold at low and high concentrations, respectively) and Hep3B-KO (1.7- and 1.6-fold at low and high concentrations, respectively) exhibited higher sensitivity to MAPK/ERK inhibitor GDC09994 (Figure 6D). However, the AKT inhibitor MK2206 was less effective on HepG2-KO and Hep3B-KO lines than on their respective parental lines (Figure 6E). Taken together, these data demonstrate that GPC3 deficiency results in compensatory upregulation of MAPK/ERK in response to AKT downregulation. This can be inhibited pharmacologically with subsequent suppression of cellular proliferation.
GPC3-targeted immunoPET agent demonstrates that GPC3 deficiency was persistently sustained in GPC3-KO xenografts
GC33 is a humanized anti-GPC3 IgG1 that binds to human GPC3 with high affinity (Kd of 0.673 nM) [39]. The radiochemical yields were over 95% with radiochemical purity >98% and the molar activities of the radio-conjugates were 85 GBq/µmol.
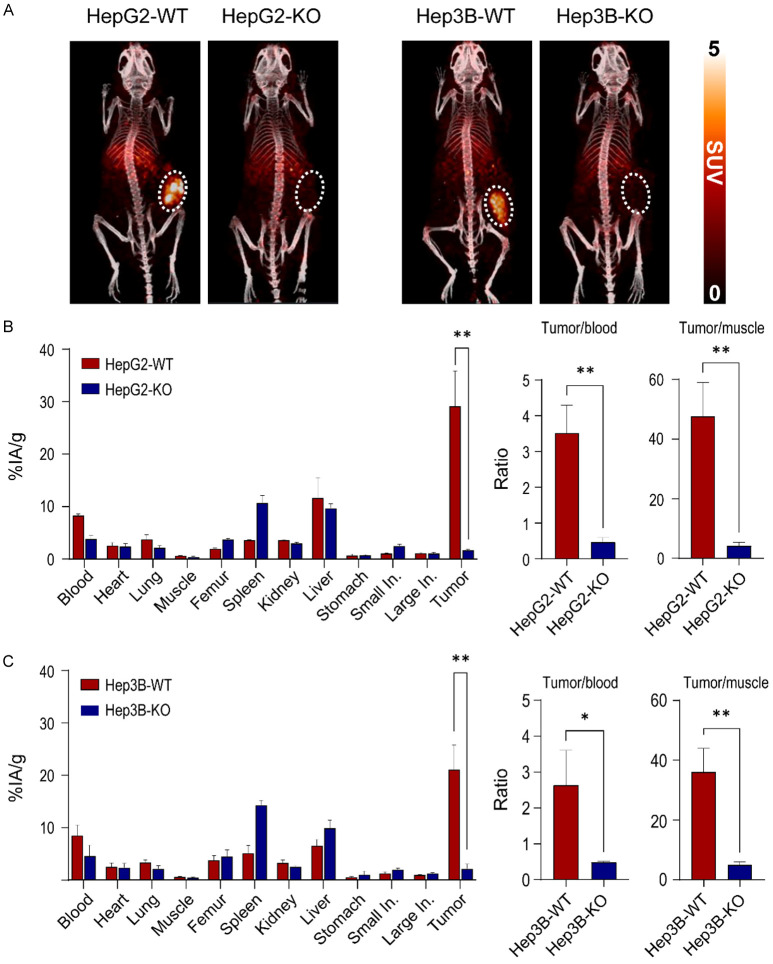
89Zr-DFO-GC33 was injected in HepG2 and Hep3B xenografts (WT and KO), and the tumor uptake was evaluated at 72 h post-injection. HepG2-WT tumors were clearly visible, whereas the HepG2-KO tumors showed minimal signal (Figure 7A left panel). Because of the lower GPC3 expression in Hep3B-WT vs. HepG2-WT cells, uptake of 89Zr-DFO-GC33 in Hep3B-WT tumors was lower but still evident. However, almost no signal was observed in Hep3B-KO tumors (Figure 7A right panel).
Figure 7.
Validation of GPC3 knockout by GPC3-targeted immunoPET imaging and biodistribution in xenograft models. A. Representative MIP-PET/CT images of 89Zr-DFO-GC33 in HepG2 xenografts (WT and KO) and Hep3B xenografts (WT and KO) at 72 h post-injection (n=3, respectively). The tumor is indicated by white dotted circles. PET images display radioactivity calibrated in standardized uptake values (SUV). B. Biodistribution data of 89Zr-DFO-GC33 in the major organs of HepG2-WT and HepG2-KO xenografts at 72 h post-injection. Comparison of tumor-to-blood ratio and tumor-to-muscle ratio of 89Zr-DFO-GC33 at 72 h post-injection between HepG2-WT and HepG2-KO xenografts. Data are present as the mean ± SD (n=3, respectively). C. Biodistribution data of 89Zr-DFO-GC33 in the major organs of Hep3B-WT and Hep3B-KO xenografts at 72 h post-injection. Comparison of tumor-to-blood ratio and tumor-to-muscle ratio of 89Zr-DFO-GC33 at 72 h post-injection between Hep3B-WT and Hep3B-KO xenografts. Data are presented as the mean ± SD (n=3, respectively). Radioactivity uptakes in biodistribution were calculated as the percentage of the injected activity per gram of tissue (%IA/g). Statistical significance in biodistribution data and tumor-to-organ ratio calculation was determined by the t-test. A p-values of <0.05 was considered statistically significant: *, P<0.05; **, P<0.01.
These results were further confirmed by ex vivo biodistribution right after PET imaging at 72 h time point. The highest uptake was found in the tumor of HepG2-WT xenografts (29.2 ± 6.7% IA/g), whereas HepG2-KO tumor uptake (1.7 ± 0.2% IA/g) was 17.2 times lower than that of tumors from HepG2-WT xenografts (P<0.01, Figure 7B left panel). The tumor-to-blood and tumor-to-muscle ratios in HepG2-WT xenografts were calculated to be 3.5 and 47.6, whereas HepG2-KO xenografts showed 7.0 and 11.6 times lower compared to that of HepG2-WT xenografts (Figure 7B right panel). Similarly, the highest uptake was found in the tumor of Hep3B-WT xenografts (21.1 ± 4.7% IA/g), whereas Hep3B-KO tumor uptake (2.2 ± 0.9% IA/g) was 9.8 times lower than that of tumors from Hep3B-WT xenografts (P<0.01, Figure 7C left panel). The tumor-to-blood and tumor-to-muscle ratios in Hep3B-WT xenografts were calculated to be 2.6 and 35.9, whereas Hep3B-KO xenografts showed 5.2 and 7.5 times lower compared to that of Hep3B-WT xenografts (Figure 7C right panel). These results align with the PET imaging results, indicating that immunoPET using 89Zr-DFO-GC33 accurately visualizes the expression levels of GPC3 in mice. Moreover, the above PET imaging and ex vivo biodistribution data strongly demonstrate that GPC3 deficiency was persistently sustained in HepG2-KO and Hep3B-KO xenografts.
Discussion
GPC3 is a biomarker of hepatocellular carcinoma, but its role in pathogenesis is unclear. To the best of our knowledge, the present study is the first to engineer and fully characterize GPC3-KO liver cancer cell lines to facilitate development of a GPC3 targeted immunoPET agent (89Zr-DFO-GC33). In this study, we generated stable GPC3-KO cell lines in HepG2 and Hep3B via the CRISPR/Cas9 gene editing system and confirmed deletion at both gene and protein levels. The results of this study confirmed previous findings: GPC3 is an oncogenic protein, and its deficiency resulted in reduced cell proliferation, migration, invasion, and in vivo tumor growth compared to their wildtype counterparts. In addition, GPC3 is linked to several signaling pathways, including WNT pathway. We also revealed unprecedented evidence highlighting the effect of GPC3 deficiency on the MAPK/ERK pathway in liver cancer cells. Furthermore, using a previously described GPC3-targeted immunoPET agent, we confirmed the ability to discern GPC3-positive from -negative xenograft models [37]. Moreover, positive uptake on GPC3 immunoPET suggests increased aggressiveness. As disease models, these isogenic cell lines can serve a critical role in developing GPC3-directed therapies for cancers with GPC3 expression, especially liver cancer.
Although it is known that GPC3 plays a crucial role in liver cancer tumorigenesis and progression, its impact on cell proliferation remains controversial. A previous study reported that overexpression of GPC3 in Huh7, a liver cancer cell line with low intrinsic GPC3 expression, inhibited cell proliferation and invasion through induction of apoptosis [40]. Additionally, Lin et al. reported hepatocyte growth inhibition of GPC3 using hepatocyte-targeted GPC3 transgenic mice [41]. Conversely, prior studies showed that the knockdown of GPC3 expression using GPC3-targeted small interfering RNA (siRNA) effectively inhibited liver cancer cell proliferation, migration, and invasion [42-45]. Cheng et al. also observed excessive cell proliferation in NIH3T3 cells following transfection with GPC3 [46]. These data suggest that the role of GPC3 in cell proliferation may hinge on its environmental context.
In the present study, we confirmed that knockout of GPC3 resulted in reduced liver cancer cell proliferation, migration, and invasion compared to parental cells. Furthermore, we demonstrated that CDK4, Cyclin D3, and survivin were significantly reduced in GPC3 deficient HepG2 cells, while expression of p27 Kip1, a negative regulator of cell proliferation, was increased in GPC3-KO HepG2 cells. Similarly, the expression of CDK4/6, Cyclin D1, and N-myc was downregulated in GPC3 deficient Hep3B cells (Figure 6A). These data demonstrate that GPC3 plays an important role in the regulation of liver cancer cell proliferation, but the molecular mechanism underlying GPC3 mediated cell proliferation differs between these two liver cancer cell lines. This is unsurprising, given that HepG2 is a hepatoblastoma while Hep3B is a hepatocellular carcinoma line. GPC3 may play different roles in these two liver tumor subtypes.
Investigating the specific pathogenic role of GPC3 in liver cancer provides an opportunity to identify additional, potentially synergistic, therapeutic targets. Previous studies have shown that GPC3 is involved in the modulation of various signaling pathways, including WNT/β-catenin [47], Hippo/Yes-associated protein (Hippo/YAP) [44], fibroblast growth factor/FGF receptor (FGF/FGFR) [48,49], Insulin-like growth factor/IGF receptor (IFG/IGFR) [46], and hepatocyte growth factor/tyrosine kinase MET (HGF/Met) [48,50]. Interestingly, GPC3 has been shown to interact with some of these growth factors and modulate their activity through its heparin sulfate chains [51]. This finding was underscored by the discovery that mRNA and protein levels of these growth factors were downregulated by GPC3 knockdown using siRNA in HepG2 [52]. It has also been shown that GPC3 binds to FGF2 via its heparin sulfate side chains [48], and sulfatase 2 enhances its expression, promoting FGF signaling [49]. However, Kwack et al. demonstrated that GPC3 inhibits FGF2-mediated cell proliferation in HCC cells [53]. In our work, we show down-regulation of p-AKT, p-NFκB, and p-mTOR in GPC3 knockout liver cancer cells, which could, in part, be explained by the activation of the phosphoinositide 3-kinase (PI3K)/AKT pathway via FGF2/FGFR binding.
A previous study demonstrated that mTOR, a downstream activator of AKT signaling, controls NF-κB activity in PTEN-null/inactive prostate cancer cells [54]. This suggests that the PI3K/AKT pathway is closely linked to the NFκB and mTOR pathways. Wu et al. also reported a positive correlation between GPC3 expression and p-MAPK/ERK in 45 HCC patients. In addition, authors showed that administration of exogenous soluble GPC3 increased p-MAPK/ERK levels in a dose-dependent manner in liver cancer cells, leading to cell migration and invasion [42]. By contrast, we found that the MAPK/ERK pathway is upregulated in GPC3 knockout liver cancer cells, which made these cells more susceptible to ERK inhibition with GDC09994 than parental lines. This discordance may be explained through possible binding between excess soluble GPC3 and receptors upstream to MAPK/ERK, such as G-protein coupled receptors, receptor tyrosine kinases, or integrins. Collectively, GPC3 is involved in the crosstalk among various oncogenic pathways, suggesting that targeting GPC3 could be potential therapeutic strategy against liver cancers.
Among oncogenic signaling pathways, the activation of the canonical WNT/β-catenin signaling pathway is a common feature in liver cancer. GPC3 promotes cell proliferation by acting as a coreceptor for WNT proteins [55,56]. The GPC3 core protein interacts with the WNT/Frizzled (FZD) complex to promote WNT/β-catenin signaling in HCC cells [57]. Furthermore, GPC3 expression significantly correlated with the nuclear/cytoplasmic localization of β-catenin, suggesting that GPC3 is involved in the activation of the WNT signaling pathway [52,55]. Corroborating previous studies, we found that nuclear translocation of β-catenin was significantly inhibited in GPC3 deficient HepG2 and Hep3B cells compared to their respective wild type lines, while the cytoplasmic β-catenin level was slightly reduced in GPC3 deficient HepG2 cells (Figure 6C). Loss of β-catenin transcriptional activity lead to decreased tumor growth, reduced tumor invasion, and inhibited metastasis formation in vivo using a metastatic breast mouse model [58]. These data suggest that GPC3 is involved in nuclear/cytoplasmic translocation of β-catenin and interfering with this process may be a potential therapeutic strategy. Further studies are needed to clarify the precise mechanism of GPC3 underlying β-catenin translocation and activation in liver cancer.
The role of GPC3 in tumor growth has been explored using xenografts from both HepG2 and Hep3B, demonstrating that the tumor growth of GPC3 knockdown xenografts was significantly slower than that of wild-type xenografts [59]. Li et al. found that tumors did not develop when inoculated mice with GPC3 knockout Hep3B cells [57]. The authors established a stable GPC3 knockout cell line using CRISPR/Cas9, targeting the promoter region with gRNA. In a recent report, delayed tumor growth was noted in GPC3 knockout Hep3B xenografts compared to the wild-type group [60]. Disparate findings may be attributed to differences in tumor cell clonal heterogeneity or variation and differences in the disrupted regions within the GPC3 gene. Consistent with some previous reports, we found that the GPC3 deficient HepG2 group exhibited significantly slower tumor growth and reduced tumor weight compared to the control group. Although GPC3 knockout in liver cancer cells did not completely inhibit tumorigenesis, it is clear that GPC3 plays an important role in liver cancer tumorigenesis and progression in these cell lines.
The correlation between GPC3 and AFP expression levels in liver cancer has been previously studied, but its relation has remained undetermined. Previous studies indicated that serum GPC3 levels did not exhibit correlations with serum AFP level, tumor size, stage, or metastasis in liver cancer patients [52,61,62]. Xie et al. reported a positive correlation trend between serum GPC3 and AFP levels using an Affirm-Mab chemiluminescence immunoassay [63]. Recently, Li et al. also demonstrated a notable difference in the serum AFP levels between the GPC3-KO group and the Hep3B WT group. Serum AFP levels were relatively high in the Hep3B WT group, while they were barely detectable in GPC3-KO Hep3B xenografts that failed to form tumors. Interestingly, the re-expression of WT or mutant GPC3 in GPC3-KO cells showed a similar pattern to the tumor size formed, suggesting a potential correlation between GPC3 and serum AFP levels [57]. In contrast, we observed that there is no meaningful association between serum AFP levels of GPC3 deficient and parental xenografts. Further studies are needed to elucidate the potential link between GPC3 and AFP in liver cancer.
Despite the conflicting data regarding the biological role of GPC3 in liver cancer, its presence in the majority of HCC and some hepatoblastomas underscores its potential role as a diagnostic and therapeutic target. Early detection and diagnosis of HCC is a critical first step in improving patient survival. Because it is challenging to identify residual or recurrent disease using conventional imaging, such as CT or MRI, several imaging and therapeutic agents looking at different targets are being studied to address this unmet clinical need [64-67]. 18F-Fluorodeoxyglucose (FDG) PET, which is commonly used in other cancer histologies, is not routinely employed for HCC because it lacks both sensitivity and specificity. In this context, GPC3 has emerged as a favored target for HCC diagnosis and therapy.
GPC3-selective imaging was first described by Sham et al. using a monoclonal antibody PET probe in HepG2 orthotopic xenografts [27]. PET imaging using novel GPC3-specific probes, such as 18F- or 68Ga-labeled GPC3 peptides [68-70] and 124I-labeled GC33 [30], showed good sensitivity for the detection of GPC3 expression in tumors. Here, we demonstrated that 89Zr-DFO-GC33 accumulated exclusively in GPC3+ xenografts. These data suggest that GPC3-targeted imaging can detect GPC3-expressing liver cancer cells and, perhaps, be used to assess therapeutic response to liver-directed therapies, such as ablation, embolization, and stereotactic radiotherapy, as well as novel GPC3-targeted therapy, including chimeric antigen receptor (CAR) T cell therapy [71] and radiopharmaceutical therapy [32,33,70].
In conclusion, we engineered GPC3-KO liver cancer cell lines and show that they exhibit lower proliferation, invasion, migration, and suppressed tumor growth kinetics compared to parental lines. We confirmed that GPC3 activates the AKT/NFκB/WNT signaling pathway and observed compensatory upregulation of pMAPK/ERK1/2 in GPC3-KO cells. Notably, the inhibition of MAPK/ERK1/2 using GDC09994 exhibited greater efficacy in GPC3-KO cells than their WT counterparts. Conversely, AKT inhibition using MK2206 showed a higher response in WT cells than in GPC3-KO liver cancer cells, suggesting that therapeutic strategies for liver cancer patients should be tailored based on GPC3 expression. Furthermore, we demonstrate exclusive target engagement in GPC3+ xenografts using our GPC3-selective immunoPET agent. The GPC3-KO cell lines engineered in this study, when paired with the parental lines, could be a valuable tool for developing diagnostic or therapeutic agents for patients with liver cancer.
Acknowledgements
This work was supported by the Intramural Cancer Institute of the National Institutes of Health, including the National Cancer Institute, Center for Cancer Research, and the Molecular Imaging Branch. The projects funded under this support are ZIA BC011800 and ZIA BC010891. We thank Chugai Pharmaceutical Co. (Japan) for providing GC33 for this study.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73(Suppl 1):4–13. doi: 10.1002/hep.31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 4.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 5.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knelson EH, Nee JC, Blobe GC. Heparan sulfate signaling in cancer. Trends Biochem Sci. 2014;39:277–288. doi: 10.1016/j.tibs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagarajan A, Malvi P, Wajapeyee N. Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front Endocrinol (Lausanne) 2018;9:483. doi: 10.3389/fendo.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolluri A, Ho M. The role of glypican-3 in regulating Wnt, YAP, and Hedgehog in liver cancer. Front Oncol. 2019;9:708. doi: 10.3389/fonc.2019.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho M, Kim H. Glypican-3: a new target for cancer immunotherapy. Eur J Cancer. 2011;47:333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha YL, Liu S, Yan WW, Dong B. Wnt/beta-catenin signaling as a useful therapeutic target in hepatoblastoma. Biosci Rep. 2019;39:BSR20192466. doi: 10.1042/BSR20192466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell D, Ranganathan S, Tao J, Monga SP. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: a Wnt/beta-catenin perspective. Gene Expr. 2017;17:141–154. doi: 10.3727/105221616X693639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi N, Watanabe A, Hishinuma M, Ohashi K, Midorikawa Y, Morishita Y, Niki T, Shibahara J, Mori M, Makuuchi M, Hippo Y, Kodama T, Iwanari H, Aburatani H, Fukayama M. The glypican 3 oncofetal protein is a promising diagnostic marker for hepatocellular carcinoma. Mod Pathol. 2005;18:1591–1598. doi: 10.1038/modpathol.3800436. [DOI] [PubMed] [Google Scholar]
- 14.Zynger DL, Gupta A, Luan C, Chou PM, Yang GY, Yang XJ. Expression of glypican 3 in hepatoblastoma: an immunohistochemical study of 65 cases. Hum Pathol. 2008;39:224–230. doi: 10.1016/j.humpath.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 16.Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, Tokita S, Iwanari H, Ito Y, Nakano K, Nezu J, Tsunoda H, Yoshino T, Ohizumi I, Tsuchiya M, Ohnishi S, Makuuchi M, Hamakubo T, Kodama T, Aburatani H. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res. 2004;64:2418–2423. doi: 10.1158/0008-5472.can-03-2191. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi Y, Dungubat E, Kusano H, Ganbat D, Tomita Y, Odgerel S, Fukusato T. Application of immunohistochemistry in the pathological diagnosis of liver tumors. Int J Mol Sci. 2021;22:5780. doi: 10.3390/ijms22115780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, Nakagohri T, Konishi M, Kobayashi N, Kinoshita T, Nakatsura T. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100:1403–1407. doi: 10.1111/j.1349-7006.2009.01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Gao W, Zhang YF, Ho M. Glypicans as cancer therapeutic targets. Trends Cancer. 2018;4:741–754. doi: 10.1016/j.trecan.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler W, Xu L, Zhou Y, Cheng Q, Hauck JS, He Y, Marek R, Hartman Z, Cheng L, Yang Q, Wang ME, Chen M, Zhang H, Armstrong AJ, Huang J. Oncofetal protein glypican-3 is a biomarker and critical regulator of function for neuroendocrine cells in prostate cancer. J Pathol. 2023;260:43–55. doi: 10.1002/path.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moek KL, Fehrmann RSN, van der Vegt B, de Vries EGE, de Groot DJA. Glypican 3 overexpression across a broad spectrum of tumor types discovered with functional genomic mRNA profiling of a large cancer database. Am J Pathol. 2018;188:1973–1981. doi: 10.1016/j.ajpath.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Ishiguro T, Sano Y, Komatsu SI, Kamata-Sakurai M, Kaneko A, Kinoshita Y, Shiraiwa H, Azuma Y, Tsunenari T, Kayukawa Y, Sonobe Y, Ono N, Sakata K, Fujii T, Miyazaki Y, Noguchi M, Endo M, Harada A, Frings W, Fujii E, Nanba E, Narita A, Sakamoto A, Wakabayashi T, Konishi H, Segawa H, Igawa T, Tsushima T, Mutoh H, Nishito Y, Takahashi M, Stewart L, ElGabry E, Kawabe Y, Ishigai M, Chiba S, Aoki M, Hattori K, Nezu J. An anti-glypican 3/CD3 bispecific T cell-redirecting antibody for treatment of solid tumors. Sci Transl Med. 2017;9:eaal4291. doi: 10.1126/scitranslmed.aal4291. [DOI] [PubMed] [Google Scholar]
- 23.Filmus J, Capurro M. Glypican-3: a marker and a therapeutic target in hepatocellular carcinoma. FEBS J. 2013;280:2471–2476. doi: 10.1111/febs.12126. [DOI] [PubMed] [Google Scholar]
- 24.Feng M, Ho M. Glypican-3 antibodies: a new therapeutic target for liver cancer. FEBS Lett. 2014;588:377–382. doi: 10.1016/j.febslet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strosberg JR, Caplin ME, Kunz PL, Ruszniewski PB, Bodei L, Hendifar A, Mittra E, Wolin EM, Yao JC, Pavel ME, Grande E, Van Cutsem E, Seregni E, Duarte H, Gericke G, Bartalotta A, Mariani MF, Demange A, Mutevelic S, Krenning EP NETTER-1 investigators. 177Lu-Dotatate plus long-acting octreotide versus high-dose long-acting octreotide in patients with midgut neuroendocrine tumours (NETTER-1): final overall survival and long-term safety results from an open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:1752–1763. doi: 10.1016/S1470-2045(21)00572-6. [DOI] [PubMed] [Google Scholar]
- 26.Sartor O, de Bono J, Chi KN, Fizazi K, Herrmann K, Rahbar K, Tagawa ST, Nordquist LT, Vaishampayan N, El-Haddad G, Park CH, Beer TM, Armour A, Perez-Contreras WJ, DeSilvio M, Kpamegan E, Gericke G, Messmann RA, Morris MJ, Krause BJ VISION Investigators. Lutetium-177-PSMA-617 for metastatic castration-resistant prostate cancer. N Engl J Med. 2021;385:1091–1103. doi: 10.1056/NEJMoa2107322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sham JG, Kievit FM, Grierson JR, Miyaoka RS, Yeh MM, Zhang M, Yeung RS, Minoshima S, Park JO. Glypican-3-targeted 89Zr PET imaging of hepatocellular carcinoma. J Nucl Med. 2014;55:799–804. doi: 10.2967/jnumed.113.132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An S, Zhang D, Zhang Y, Wang C, Shi L, Wei W, Huang G, Liu J. GPC3-targeted immunoPET imaging of hepatocellular carcinomas. Eur J Nucl Med Mol Imaging. 2022;49:2682–2692. doi: 10.1007/s00259-022-05723-x. [DOI] [PubMed] [Google Scholar]
- 29.Labadie KP, Ludwig AD, Lehnert AL, Hamlin DK, Kenoyer AL, Sullivan KM, Daniel SK, Mihailovic TN, Sham JG, Orozco JJ, Yeung RS, Chen DL, Wilbur DS, Miyaoka RS, Park JO. Glypican-3 targeted delivery of 89Zr and 90Y as a theranostic radionuclide platform for hepatocellular carcinoma. Sci Rep. 2021;11:3731. doi: 10.1038/s41598-021-82172-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrasquillo JA, O’Donoghue JA, Beylergil V, Ruan S, Pandit-Taskar N, Larson SM, Smith-Jones PM, Lyashchenko SK, Ohishi N, Ohtomo T, Abou-Alfa GK. I-124 codrituzumab imaging and biodistribution in patients with hepatocellular carcinoma. EJNMMI Res. 2018;8:20. doi: 10.1186/s13550-018-0374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fayn S, King AP, Gutsche NT, Duan Z, Buffington J, Olkowski CP, Fu Y, Hong J, Sail D, Baidoo KE, Swenson RE, Cheloha RW, Ho M, Choyke PL, Escorcia FE. Site-specifically conjugated single-domain antibody successfully identifies glypican-3-expressing liver cancer by immuno-PET. J Nucl Med. 2023;64:1017–1023. doi: 10.2967/jnumed.122.265171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell MM, Gutsche NT, King AP, Baidoo KE, Kelada OJ, Choyke PL, Escorcia FE. Glypican-3-targeted alpha particle therapy for hepatocellular carcinoma. Molecules. 2020;26:4. doi: 10.3390/molecules26010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labadie KP, Hamlin DK, Kenoyer A, Daniel SK, Utria AF, Ludwig AD, Kenerson HL, Li L, Sham JG, Chen DL, Orozco JJ, Yeung RS, Orvig C, Li Y, Wilbur DS, Park JO. Glypican-3-targeted 227Th alpha-therapy reduces tumor burden in an orthotopic xenograft murine model of hepatocellular carcinoma. J Nucl Med. 2022;63:1033–1038. doi: 10.2967/jnumed.121.262562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo M, Zhang H, Zheng J, Liu Y. Glypican-3: a new target for diagnosis and treatment of hepatocellular carcinoma. J Cancer. 2020;11:2008–2021. doi: 10.7150/jca.39972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman RM, Kelada OJ, Gutsche NT, Natarajan R, Swenson RE, Fu Y, Hong J, Ho M, Choyke PL, Escorcia FE. In vitro performance of published glypican 3-targeting peptides TJ12P1 and L5 indicates lack of specificity and potency. Cancer Biother Radiopharm. 2019;34:498–503. doi: 10.1089/cbr.2019.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelada OJ, Gutsche NT, Bell M, Berman RM, Baidoo KE, Warner BM, Szajek LP, Hong J, Ho M, Choyke P, Escorcia FE. ImmunoPET as stoichiometric sensor for glypican-3 in models of hepatocellular carcinoma. Int J Radiat Oncol Biol Phys. 2020;108:S180–181. [Google Scholar]
- 38.Jagoda EM, Basuli F, Olkowski C, Weiss I, Phelps TE, Wong KR, Ton AT, Lane KC, Adler S, Butcher D, Edmondson EF, Langermann S, Choyke PL. Immuno-PET imaging of siglec-15 using the zirconium-89-labeled therapeutic antibody, NC318. Mol Imaging. 2023;2023:3499655. [Google Scholar]
- 39.Nakano K, Ishiguro T, Konishi H, Tanaka M, Sugimoto M, Sugo I, Igawa T, Tsunoda H, Kinoshita Y, Habu K, Orita T, Tsuchiya M, Hattori K, Yamada-Okabe H. Generation of a humanized anti-glypican 3 antibody by CDR grafting and stability optimization. Anticancer Drugs. 2010;21:907–916. doi: 10.1097/CAD.0b013e32833f5d68. [DOI] [PubMed] [Google Scholar]
- 40.Pan Z, Chen C, Long H, Lei C, Tang G, Li L, Feng J, Chen F. Overexpression of GPC3 inhibits hepatocellular carcinoma cell proliferation and invasion through induction of apoptosis. Mol Med Rep. 2013;7:969–974. doi: 10.3892/mmr.2013.1279. [DOI] [PubMed] [Google Scholar]
- 41.Lin CW, Mars WM, Paranjpe S, Donthamsetty S, Bhave VS, Kang LI, Orr A, Bowen WC, Bell AW, Michalopoulos GK. Hepatocyte proliferation and hepatomegaly induced by phenobarbital and 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene is suppressed in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2011;54:620–630. doi: 10.1002/hep.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Liu H, Weng H, Zhang X, Li P, Fan CL, Li B, Dong PL, Li L, Dooley S, Ding HG. Glypican-3 promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells through ERK signaling pathway. Int J Oncol. 2015;46:1275–1285. doi: 10.3892/ijo.2015.2827. [DOI] [PubMed] [Google Scholar]
- 43.Sun CK, Chua MS, He J, So SK. Suppression of glypican 3 inhibits growth of hepatocellular carcinoma cells through up-regulation of TGF-beta2. Neoplasia. 2011;13:735–747. doi: 10.1593/neo.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao HL, Pan ZJ, Lei CJ, Wen JY, Li MY, Liu ZK, Qiu ZD, Lin MZ, Chen NP, Chen M. Knockdown of GPC3 inhibits the proliferation of Huh7 hepatocellular carcinoma cells through down-regulation of YAP. J Cell Biochem. 2013;114:625–631. doi: 10.1002/jcb.24404. [DOI] [PubMed] [Google Scholar]
- 45.Schepers EJ, Lake C, Glaser K, Bondoc AJ. Inhibition of glypican-3 cleavage results in reduced cell proliferation in a liver cancer cell line. J Surg Res. 2023;282:118–128. doi: 10.1016/j.jss.2022.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng W, Tseng CJ, Lin TT, Cheng I, Pan HW, Hsu HC, Lee YM. Glypican-3-mediated oncogenesis involves the Insulin-like growth factor-signaling pathway. Carcinogenesis. 2008;29:1319–1326. doi: 10.1093/carcin/bgn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Cat B, Muyldermans SY, Coomans C, Degeest G, Vanderschueren B, Creemers J, Biemar F, Peers B, David G. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol. 2003;163:625–635. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song HH, Shi W, Filmus J. OCI-5/rat glypican-3 binds to fibroblast growth factor-2 but not to insulin-like growth factor-2. J Biol Chem. 1997;272:7574–7577. doi: 10.1074/jbc.272.12.7574. [DOI] [PubMed] [Google Scholar]
- 49.Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao W, Kim H, Ho M. Human monoclonal antibody targeting the heparan sulfate chains of glypican-3 inhibits HGF-mediated migration and motility of hepatocellular carcinoma cells. PLoS One. 2015;10:e0137664. doi: 10.1371/journal.pone.0137664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Midorikawa Y, Ishikawa S, Iwanari H, Imamura T, Sakamoto H, Miyazono K, Kodama T, Makuuchi M, Aburatani H. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer. 2003;103:455–465. doi: 10.1002/ijc.10856. [DOI] [PubMed] [Google Scholar]
- 52.Akutsu N, Yamamoto H, Sasaki S, Taniguchi H, Arimura Y, Imai K, Shinomura Y. Association of glypican-3 expression with growth signaling molecules in hepatocellular carcinoma. World J Gastroenterol. 2010;16:3521–3528. doi: 10.3748/wjg.v16.i28.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwack MH, Choi BY, Sung YK. Cellular changes resulting from forced expression of glypican-3 in hepatocellular carcinoma cells. Mol Cells. 2006;21:224–228. [PubMed] [Google Scholar]
- 54.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 56.Capurro M, Martin T, Shi W, Filmus J. Glypican-3 binds to Frizzled and plays a direct role in the stimulation of canonical Wnt signaling. J Cell Sci. 2014;127:1565–1575. doi: 10.1242/jcs.140871. [DOI] [PubMed] [Google Scholar]
- 57.Li N, Wei L, Liu X, Bai H, Ye Y, Li D, Li N, Baxa U, Wang Q, Lv L, Chen Y, Feng M, Lee B, Gao W, Ho M. A frizzled-like cysteine-rich domain in glypican-3 mediates wnt binding and regulates hepatocellular carcinoma tumor growth in mice. Hepatology. 2019;70:1231–1245. doi: 10.1002/hep.30646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buechel D, Sugiyama N, Rubinstein N, Saxena M, Kalathur RKR, Luond F, Vafaizadeh V, Valenta T, Hausmann G, Cantu C, Basler K, Christofori G. Parsing beta-catenin’s cell adhesion and Wnt signaling functions in malignant mammary tumor progression. Proc Natl Acad Sci U S A. 2021;118:e2020227118. doi: 10.1073/pnas.2020227118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao W, Kim H, Feng M, Phung Y, Xavier CP, Rubin JS, Ho M. Inactivation of Wnt signaling by a human antibody that recognizes the heparan sulfate chains of glypican-3 for liver cancer therapy. Hepatology. 2014;60:576–587. doi: 10.1002/hep.26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun L, Gao F, Gao Z, Ao L, Li N, Ma S, Jia M, Li N, Lu P, Sun B, Ho M, Jia S, Ding T, Gao W. Shed antigen-induced blocking effect on CAR-T cells targeting glypican-3 in hepatocellular carcinoma. J Immunother Cancer. 2021;9:e001875. doi: 10.1136/jitc-2020-001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou S, O’Gorman MR, Yang F, Andresen K, Wang L. Glypican 3 as a serum marker for hepatoblastoma. Sci Rep. 2017;7:45932. doi: 10.1038/srep45932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tangkijvanich P, Chanmee T, Komtong S, Mahachai V, Wisedopas N, Pothacharoen P, Kongtawelert P. Diagnostic role of serum glypican-3 in differentiating hepatocellular carcinoma from non-malignant chronic liver disease and other liver cancers. J Gastroenterol Hepatol. 2010;25:129–137. doi: 10.1111/j.1440-1746.2009.05988.x. [DOI] [PubMed] [Google Scholar]
- 63.Xie C, Tiede C, Zhang X, Wang C, Li Z, Xu X, McPherson MJ, Tomlinson DC, Xu W. Development of an Affimer-antibody combined immunological diagnosis kit for glypican-3. Sci Rep. 2017;7:9608. doi: 10.1038/s41598-017-10083-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell M, Turkbey EB, Escorcia FE. Radiomics, radiogenomics, and next-generation molecular imaging to augment diagnosis of hepatocellular carcinoma. Cancer J. 2020;26:108–115. doi: 10.1097/PPO.0000000000000435. [DOI] [PubMed] [Google Scholar]
- 65.Marker SC, Espinoza AF, King AP, Woodfield SE, Patel RH, Baidoo K, Nix MN, Ciaramicoli LM, Chang YT, Escorcia FE, Vasudevan SA, Schnermann MJ. Development of Iodinated indocyanine green analogs as a strategy for targeted therapy of liver cancer. ACS Med Chem Lett. 2023;14:1208–1215. doi: 10.1021/acsmedchemlett.3c00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mena E, Shih J, Chung JY, Jones J, Rabiee A, Monge C, Turkbey B, Lindenberg L, Salerno KE, Kassin M, Wood B, Hernandez J, Maass-Moreno R, Saboury B, Jakhete N, Molitoris JK, Unger KR, Choyke PL, Escorcia FE. Functional Imaging of Liver Cancer (FLIC): study protocol of a phase 2 trial of 18F-DCFPyL PET/CT imaging for patients with hepatocellular carcinoma. PLoS One. 2022;17:e0277407. doi: 10.1371/journal.pone.0277407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takao S, Fukushima H, King AP, Kato T, Furusawa A, Okuyama S, Kano M, Choyke PL, Escorcia FE, Kobayashi H. Near-infrared photoimmunotherapy in the models of hepatocellular carcinomas using cetuximab-IR700. Cancer Sci. 2023;114:4654–4663. doi: 10.1111/cas.15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Han YJ, Huang S, Wang M, Zhou WL, Li HS, Wang QS, Wu HB. Imaging the expression of glypican-3 in hepatocellular carcinoma by PET. Amino Acids. 2018;50:309–320. doi: 10.1007/s00726-017-2517-z. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Zhang J, Gu J, Hu K, Huang S, Conti PS, Wu H, Chen K. Radiofluorinated GPC3-binding peptides for PET imaging of hepatocellular carcinoma. Mol Imaging Biol. 2020;22:134–143. doi: 10.1007/s11307-019-01356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin F, Clift R, Ehara T, Yanagida H, Horton S, Noncovich A, Guest M, Kim D, Salvador K, Richardson S, Miller T, Han G, Bhat A, Song K, Li G. Peptide binder to glypican-3 as a theranostic agent for hepatocellular carcinoma. J Nucl Med. 2024;65:586–592. doi: 10.2967/jnumed.123.266766. [DOI] [PubMed] [Google Scholar]
- 71.Xie C, Monge B MC, Mabry-Hrones D, Coffman KL, Hicks S, Redd B, Wood B, Highfill S, Ho M, Greten TF. A phase I study of GPC3 targeted CAR-T cell therapy in advanced GPC3-expressing hepatocellular carcinoma (HCC) J. Clin. Oncol. 2023;41 Suppl TPS624. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.