Abstract
Non-small cell lung cancer (NSCLC), the most prevalent form of lung cancer, accounts for approximately 85% of all lung cancer diagnoses. Circular RNAs (circRNAs) are non-coding RNAs that play an active role in gene expression regulation, influencing cell growth, migration, and apoptosis. Here, we aimed to investigate the function of circSNYJ1 in NSCLC. In the present study, we found that circSNYJ1 expression level was increased in NSCLC tissues and cell lines. Knockdown of circSNYJ1 suppressed NSCLC cell proliferation, colony formation and migration while promoting apoptosis. Mechanistically, we demonstrated that circSNYJ1 sponged miR-142-5p, thereby regulating the expression of CCND1, a well-known cell cycle regulator. In conclusion, this study uncovered a novel circSNYJ1/miR-142-5p/CCND1 axis involved in NSCLC progression, providing potential diagnostic and prognostic biomarkers for treating NSCLC.
Keywords: CircSNYJ1, NSCLC, miR-142-5p, CCND1, biomarkers
Introduction
Non-small cell lung cancer (NSCLC) is the most prevalent form of lung cancer, accounting for around 85% of annual cases [1]. Moreover, it is one of the most aggressive malignancies, acting as the leading cause of cancer-related deaths [1,2]. Despite advances in early detection and treatment, the prognosis for NSCLC patients remains poor [3,4]. Consequently, understanding the molecular pathways that drive NSCLC development and progression is essential for developing new, effective treatments [4].
Circular RNAs (circRNAs) are non-coding RNAs that actively regulate gene expression, affecting cell growth, migration, and apoptosis [5]. As a result, circRNAs have been implicated in various diseases, including cancer [6]. Consequently, circRNA has been proposed as novel biomarkers and pharmaceutical targets for treating cancer [7-9]. Their mode action in cancer is bimodal, functioning as either oncogenes or tumors suppressors [8-13]. Several circRNAs have been shown to regulate key signaling pathways involved in NSCLC progression, such as the Wnt/β-catenin and PI3K/Akt signaling [14,15]. Therefore, elucidating the function of circRNAs in NSCLC and the underlying molecular mechanisms is essential for developing new therapeutic approaches.
One circRNA, circSNYJ1, remains largely uncharacterized in NSCLC. In this study, we dissected the role of the circSNYJ1/miR-142-5p/CCND1 regulatory axis in NSCLC. We demonstrated that circSNYJ1 acts as a competing endogenous RNA (ceRNA), sponging miR-142-5p to regulate the expression of the downstream target gene, CCND1.
Methods
Tissue sample collection
NSCLC tissue samples along with paired adjacent non-tumor tissue samples, were collected from 68 patients who underwent tumor resection between 2018 and 2020. Written informed permission was obtained from each patient, and the experiments were conducted following the ethical protocol of our institution. The collected tissue samples were immediately flash-frozen in liquid nitrogen and stored at -80°C for further analysis.
Cell culture and transfection
The human NSCLC cell lines (SPCA1, A549, H1299 and CALU3) and human bronchial epithelial cells (16HBE) were acquired from the American Type Culture Collection (ATCC). Cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin-streptomycin (Gibco, USA), and incubated at 37°C in a 5% CO2 environment. The oligonucleotides for the miR-142-5p mimic, miR-142-5p inhibitor, and negative control (NC) were synthesized by GenePharma (China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, USA) following the guidelines provided by the manufacturer.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from tissues and cells was extracted using TRIzol reagent (Invitrogen, USA) following the manufacturer’s instruction. RNA purity and concentration were assessed by NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). Complementary DNA (cDNA) was synthesized using PrimeScript RT reagent kit (Takara, Japan). qRT-PCR was performed using a Takara SYBR Premix Ex Taq II kit (Takara, Japan) on an ABI 7500 Real-Time PCR System (Applied Biosystems, USA). Relative expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 small nuclear RNA (U6 snRNA) for circSNYJ1, miR-142-5p, and CCND1.
RNase R treatment assay
Three units of RNase (Epicentre, WI, USA) were added per gram of the total RNA and incubated at 37°C for 15 min. The reaction was terminated by heating to 80°C. Subsequently, circRNAs and linear RNAs were quantified by qPCR.
CircRNA and mRNA sequencing
To perform circRNA sequencing, ribosomal RNA (rRNA) was removed from total RNA using the RiboZero rRNA Elimination Kit (Epicentre, WI, USA) followed by the manufacturer’s protocol. Next, linear RNA was digested by RNase R, and the RNA samples were purified using Agencourt RNAClean XP magnetic particles. For mRNA sequencing, mRNA was enriched with Oligo (dT) magnetic beads. The RNA samples were fragmented for sequencing, and DNA libraries were prepared using the TruSeq® Stranded kit and the HiSeq2500 (Illumina, San Diego, USA).
Cell Counting Kit-8 (CCK8) assay
The Cell Counting Kit-8 (CCK-8) was used to evaluate cell proliferation. NSCLC cells were seeded in 96-well plates at a density of 2 × 10^3 cells per well and cultured for 24 hours. Afterward, si-circSNYJ1 or si-NC was transfected into the cells. At 0, 24, 48, and 72 hours post-transfection, 10 μl of CCK-8 solution was added to each well and incubated at 37°C for 2 hours. Absorbance at 450 nm was measured using a microplate reader (Thermo Fisher Scientific in Waltham, Massachusetts, USA).
Colony formation
For the colony formation assay, cells were seeded into 6-well plates at 5 × 10^2 cells per well and cultured for 7-10 days. After washing twice with PBS, the cells were treated with 0.1% crystal violet for 30 minutes, fixed in 4% paraformaldehyde for 30 minutes, and then stained. Colonies containing more than 50 cells were counted under a microscope.
Cell apoptosis and cell cycle analysis
To assess cell apoptosis, cells were infected with si-circSNYJ1 or si-NC lentiviruses for three days before treatment with FITC-tagged Annexin V and 7-amino-actinomycin D (7-AAD). Early and late apoptotic cells were then analyzed using a flow cytometer. Each experiment was repeated three times.
Flow cytometry was used for cell cycle analysis. Cells were first fixed in 70% ethanol at 4°C for two hours before staining with propidium iodide (PI) at 37°C in darkness for 30 minutes. After a PBS wash, the cell cycle was analyzed using a flow cytometer (Becton, Dickinson and Company, USA).
Migration and invasion assay
Cell migration and invasion were measured using 24-well transwell plates with 8-micron pores (Corning, USA). For migration assays, cells were seeded in the top chamber with serum-free medium, while the bottom chamber contained medium supplemented with 10% FBS. For invasion assays, the top chamber was coated with Matrigel (BD Biosciences, USA). After 24 hours of incubation, the cells were fixed with 4% paraformaldehyde, stained with crystal violet, and counted using an Olympus microscope (Japan).
Dual-luciferase reporter assay
PCR amplification was used to generate the circSNYJ1 sequence containing the potential miR-142-5p binding sites. The PCR product was gel-purified and cloned into the pmiR-Glo luciferase reporter vector (Promega, USA). Sanger sequencing was used to confirm the right sequences. The luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega, USA) following the instructions provided by the manufacturer.
RNA pull-down and RNA immunoprecipitation (RIP) assays
The RNA pull-down assay was performed following a previously published protocol [18]. Biotinylated probes for miR-218-5p probe and the NC probe were obtained from Sangon Biotech (Shanghai, China). After detachment, A549 and H1299 cells were lysed in RIP Lysis buffer. The lysate was incubated with magnetic Dynabeads M-280 Streptavidin beads (Invitrogen, USA) at 4°C overnight, and circSNYJ1 enrichment was determined via qRT-PCR.
RIP assay was performed using a Magna RIP kit (Millipore, Bedford, MA), following the protocol provided by the manufacturer. Cell lysates were incubated with Sepharose beads conjugated to an AgO2-specific antibody (Cell Signaling Technology, USA) at 4°C for four hours. The IgG antibody served as a control. RNA was isolated from the beads, and its enrichment was measured using quantitative reverse transcription-PCR [16].
Western blot
Cells were lysed in RIPA buffer (Beyotime, China) containing one millimeter of phenylmethylsulfonyl fluoride (PMSF) for 10 minutes on ice. The cell lysate was centrifuged at a speed of 14,000 × g and the supernatant was collected. Proteins were separated on a 10% SDS-PAGE gel and subsequently transferred onto a PVDF membrane (GE, USA). After blocking in 5% skimmed milk dissolved in TBST buffer (BD, USA), the membranes was incubated with primary antibody at 4°C overnight. Following five PBS washes, the membrane was incubated with near-infrared fluorescent anti-mouse (1:50,000, LI-COR, USA) or anti-rabbit (1:25,000, Jackson, USA) antibody for one hour at room temperature. The Odyssey Near-Infrared Fluorescence Imaging System (LI-COR, USA) was used to detect the fluorescent signal.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA). Data with normal distribution were presented as mean ± standard deviation (SD) from, while the non-normally distributed data were shown as median and interquartile range (IQR). Differ-ences between two groups were evaluated by Student’s t-test, while differences between more than two groups were evaluated using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Pearson correlation analysis was conducted to analyze normally distributed variables, while Spearman’s correlation analyzed the non-normally distributed variables. P values less than 0.05 were considered statistically significant. To analyze the expression patterns of circSYNJ1 in non-small cell lung cancer (NSCLC) tissues and cells, the limma R package was employed to compare circRNA expression in the GEO databases GSE112214 and GSE101586. The Kaplan-Meier analysis was performed using the KM-plotter to evaluate the relationship between variables and the patients’ prognosis. Each experiment was repeated at least three times.
Results
Expression of circSYNJ1 in NSCLC tissues and cells
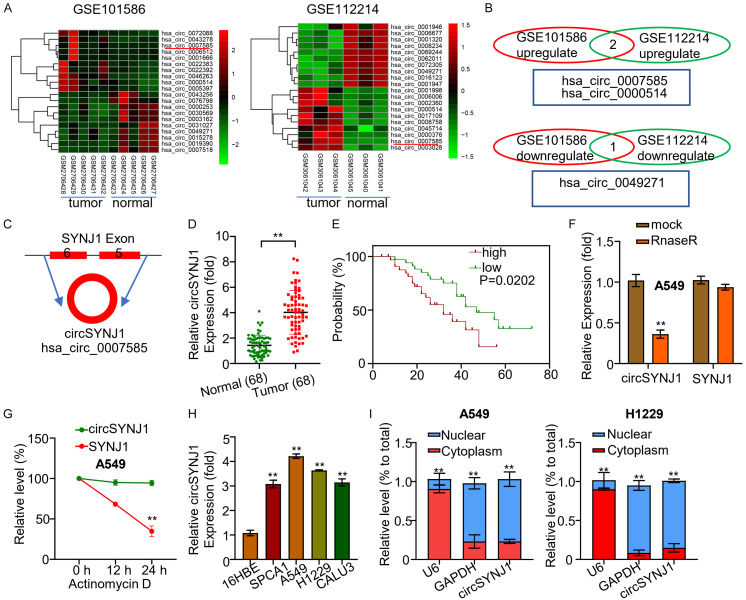
Circular RNAs (circRNAs) have been implicated in numerous biological processes, including the formation and progression of cancer. In our study, we observed that circSYNJ1 was highly expressed in both NSCLC tissues and cells compared to adjacent non-tumor tissues (Figure 1). To further investigate the differential expression of circRNAs between NSCLC cancer and surrounding tissues, we utilized the limma R package to analyze circRNA profiles in the GEO databases GSE112214 and GSE101586. In GSE112214, we found 16 upregulated and 100 downregulated circRNAs; whereas in GSE101586, 47 upregulated and 21 downregulated circRNAs were identified (Figure 1A). Bivariate analysis was employed to evaluate the expression of three circRNAs, hsa_circ_0007585, hsa_circ_0000514, and hsa_circ_0049271 in both datasets. The results highlighted that the expression of hsa_circ_0007585 and hsa_circ_0000514 was upregulated in cancer tissues, while hsa_circ_0049271 was downregulated (Figure 1B). Bioinformatic analysis revealed that circSYNJ1 was produced by reverse splicing of exon 5 and 6 of the Synaptojanin 1 (SYNJ1) gene (Figure 1C). Using qRT-PCR, we evaluated the expression of circSYNJ1 in 68 NSCLC patients, showing that circSYNJ1 was highly expressed in the cancer tissue compared to adjacent non-tumor tissue (Figure 1D). Furthermore, higher levels of circSYNJ1 expression were significantly correlated with poor prognosis in NSCLC patients, as demonstrated by the KM-plotter analysis (Figure 1E).
Figure 1.
CircSYNJ1 expression in NSCLC tissues and cells. A. Heatmaps illustrate the top upregulated and downregulated circRNAs in NSCLC and adjacent tissues from two databases (GSE112214 and GSE101586). B. A comparison of differentially expressed circRNAs between NSCLC and normal tissues. C. The diagram illustrates the circular architecture and origin of circSYNJ1. D. qRT-PCR analysis of circSYNJ1 expression in NSCLC and normal tissues from 68 patients. E. A Kaplan-Meier plot assessing the correlation between circSYNJ1 expression and the survival rates in NSCLC patients. F. qRT-PCR results showing RNase R digestion of SYNJ1 mRNA in A549 cells. G. The resistance of circSYNJ1 and SYNJ1 mRNA to RNase R treatment. H. Quantitative real-time PCR evaluation of circSYNJ1 expression in NSCLC cell lines and normal bronchial epithelial cells. I. Subcellular localization of circSYNJ1 in A549 and H1229 cells.
To evaluate the stability of circSYNJ1, we assessed the resistance of circSYNJ1 and the linear mRNA to RNase R digestion. The results indicated that RNase R readily digested the linear mRNA of SYNJ1, but not circSYNJ1 (Figure 1F). Additionally, circSYNJ1 had a longer half-life than its linear mRNA counterpart (Figure 1G). We also found that circSYNJ1 was more abundant in lung cancer cell lines compared to normal human bronchial epithelial cells (Figure 1H). Finally, to determine the subcellular localization of circSYNJ1, we performed qRT-PCR on RNA extracted from the cytoplasm and nucleus of A549 and H1229 cells, revealing a predominant cytoplasmic localization (Figure 1I).
Expression of circSNJY1 in NSCLC cell lines and its impact on proliferation and metastasis
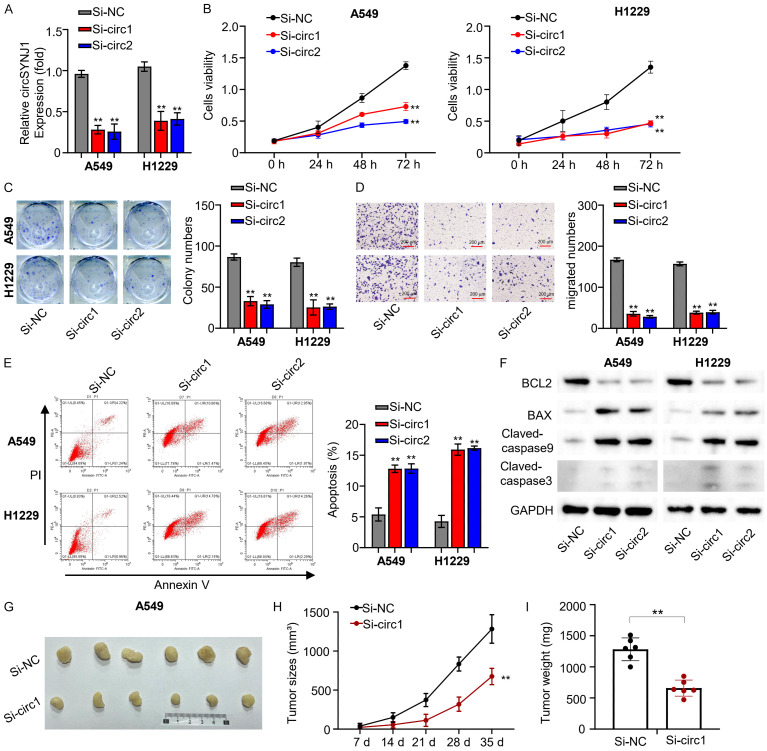
To investigate the role of circSNJY1 in NSCLC, we initially employed si-RNA to silence circSNJY1 expression in H1229 and A549 cells. QRT-PCR results showed that si-circ1 and si-circ2 efficiently reduced the circSNJY1 expression by more than 50% in both cell lines (Figure 2A). Subsequent CCK8 assays were conducted to examine cell viability after silencing circSNJY1 expression. The OD at 450 nm was measured at 0, 24, 48, and 72 hours. We observed a significant decrease cell viability in both H1229 and A549 cells following circSNJY1 silencing (Figure 2B). Colony formation assays were performed to evaluate the impact on colony-forming ability after circSNJY1 silencing, showing a substantial reduction in colony-formation ability of both cell types (Figure 2C). In addition, transwell experiment (without matrix gel) were employed to measure cell migration ability after circSNJY1 expression was silenced. The results indicated reduced cell migratory ability in both H1229 and A549 cells upon circSNJY1 silencing (Figure 2D). Flow cytometry was utilized to determine the effect of circSNJY1 silencing on apoptosis, revealing that circSNJY1 knockdown greatly enhanced apoptosis in H1229 and A549 cells (Figure 2E). Western blotting analysis supported these findings, as evidenced by an upregulation of key apoptotic factors including pro-apoptotic cytokines, BCL2, BAX, Claved-caspase9, Claved-caspase3, and GAPDH (Figure 2F). Lastly, we performed a tumor formation experiment in subcutaneous mice to investigate the tumorigenicity of A549 cells transfected with si-circ1. This experiment demonstrated that knocking down circSNJY1 notably reduced the tumorigenicity of A549 cells in mice (Figure 2G-I).
Figure 2.
CircSNJY1 expression and impact on proliferation and metastasis in NSCLC. A. qRT-PCR analysis of circSNJY1 expression in H1229 and A549 cells transfected with si-circ1 and si-circ2. B. The CCK-8 assay results indicating that suppression of circSNJY1 expression reduced cell viability in H1229 and A549 cells. C. Colony formation assay results demonstrating circSNJY1 knockdown impaired colony formation ability in H1229 and A549 cells. D. The transwell assay indicating that circSNJY1 knockdown impaired cell migration in H1229 and A549 cells. E. Flow cytometry analysis examining cell apoptosis in H1229 and A549 cells post circSNJY1 knockdown. F. Western blot analysis of apoptosis-related cytokines in H1229 and A549 cells following circSNJY1 knockdown. G-I. Tumor formation assay illustrating the reduced tumorigenic potential of A549 cells with circSNJY1 knockdown.
Regulation of miR-142-5p by circSNJY1 in NSCLC cell lines
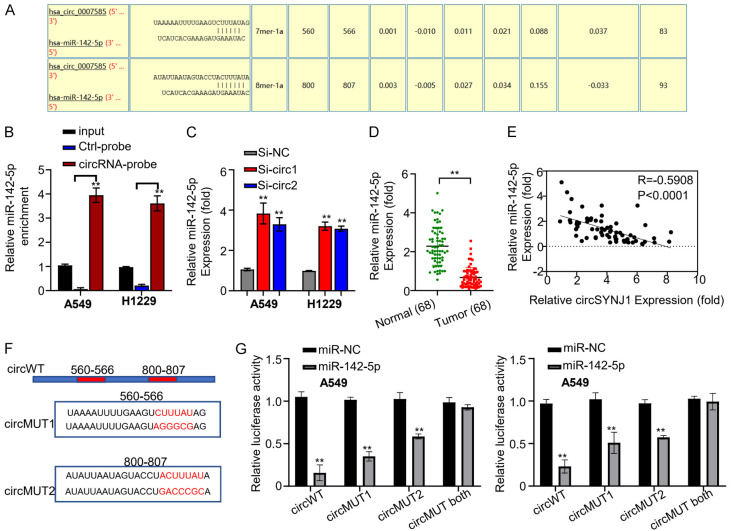
Circinteractome analysis predicted that circSNJY1 possesses two binding sites complementary to miR-142-5p (Figure 3A). An RNA pull-down assay was then conducted to examine the interaction between circSNJY1 and miR-142-5p, revealing a strong interaction (Figure 3B). Subsequent qRT-PCR was performed to assess the effect of circSNYJ1 knockdown on miR-142-5p expression. The results showed that silencing circSNYJ1 significantly increased miR-142-5p expression (Figure 3C). Additionally, qRT-PCR was used to measure miR-142-5p expression in tumor and adjacent non-tumor tissues of the 68 NSCLC patients, indicating significant downregulation of miR-142-5p in tumor tissues (Figure 3D).
Figure 3.
The interaction between circSNJY1 and miR-142-5p in NSCLC. A. Circinteractome analysis predicting miR-142-5p binding sites within circSNJY1. B. RNA Pulldown assay demonstrating the direct interaction between circSNJY1 and miR-142-5p. C. qRT-PCR analysis showing that dramatically increased miR-142-5p expression following circSNJY1 knockdown in H1229 and A549 cells. D. qRT-PCR results indicating lower expression levels of miR-142-5p in NSCLC tissues compared to adjacent normal tissues. E. Pearson correlation analysis revealing a significantly negative correlation between circSNJY1 and miR-142-5p expression in NSCLC patients. F. The fluorescent reporter construct containing wild-type and mutated miR-142-5p binding sites in circSNJY1. G. Transfection of H1229 and A549 cells with the reporter construct carrying wild-type, mutated, or partially mutated miR-143-5p binding sites, along with miR-142-5p or a control miRNA (miR-NC).
Pearson correlation analysis revealed that an inverse correlation between circSNJY1 and miR-142-5p expression among lung cancer patients (Figure 3E). To further elucidate the interaction between circSNJY1 and miR-142-5p, a fluorescent reporter assay was conducted. Reporter constructs containing either wild-type or mutated miR-142-5p binding sites of circSNJY1 were cloned into a vector with a luciferase reporter gene (Figure 3F). These constructs were transfected into H1229 and A549 cells with either mir-NC or miR-142-5p. Overexpression of miR-142-5p, but not miR-NC, significantly inhibited the expression of luciferase in the cells with the wild-type reporter construct, but this inhibitory effect was significantly reduced when the binding sites were mutated (Figure 3G).
Target gene of miR-142-5p
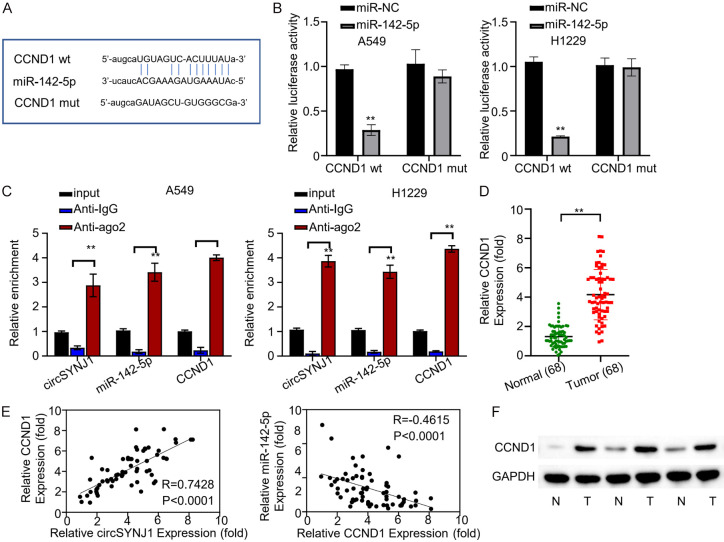
To identify the downstream target of miR-142-5p, we predicted CCND1 as a potential target using the Starbase database (Figure 4A). A fluorescent reporter assay was then employed to confirm this interaction. We constructed reporter plasmids containing either wild-type or mutated CCND1 binding sites within the miR-142-5p sequence. The expression of the fluorescent reporter gene was significantly reduced in the wild-type group compared to control, whereas the mutant group was unaffected (Figure 4B). These findings support that miR-142-5p can bind to CCND1 mRNA. Furthermore, we conducted RNA immunoprecipitation followed by RIP-qRT-PCR to examine the interaction between miR-142-5p, CCND1 mRNA, and the AGO2 protein. Both miR-142-5p and CCND1 mRNA were enriched by the AGO2 protein in H1229 and A549 cells (Figure 4C), confirming a direct interaction. Subsequent qRT-PCR analysis examined CCND1 expression in lung cancer and adjacent normal tissues from the 68 NSCLC patients. CCND1 was substantially upregulated in NSCLC compared to the normal tissues, suggesting an oncogenic function in lung cancer (Figure 4D). Additionally, pearson correlation analysis revealed that CCND1 expression was negatively correlated with miR-142-5p expression and positively correlated with circSNYJ1 expression (Figure 4E). These results imply that miR-142-5p targeted and downregulated CCND1 mRNA, a process regulated by circSNYJ1. Lastly, the expression of CCND1 protein in both cancer and surrounding samples was assessed by Western blot. The results confirmed that CCND1 protein was abundantly detected in lung cancer tissues (Figure 4F).
Figure 4.
CCND1 is the target gene of miR-142-5p. A. Prediction by Starbase identifying miR-142-5p as a regulator of the CCND1 gene. B. Effects of miR-142-5p on the expression of wild-type and mutant fluorescence reporter genes, which include miR-142-5p and CCND1 binding site sequences, in H1229 and A549 cells. C. RIP-QRT-PCR analysis revealed an enrichment of AGO2 protein on complex involving circSNYJ1, miR-142-5p, and CCND1 mRNA in H1229 and A549 cells. D. Comparative qRT-PCR analysis of CCND1 expression in malignant and adjacent normal tissues from lung cancer patients. E. Pearson correlation analysis examining the relationship between CCND1, circSNYJ1, and miR-142-5p expression across 68 lung cancer patients. F. WB study of EEF2 protein expression in in tumor and adjacent normal tissues from lung cancer patients.
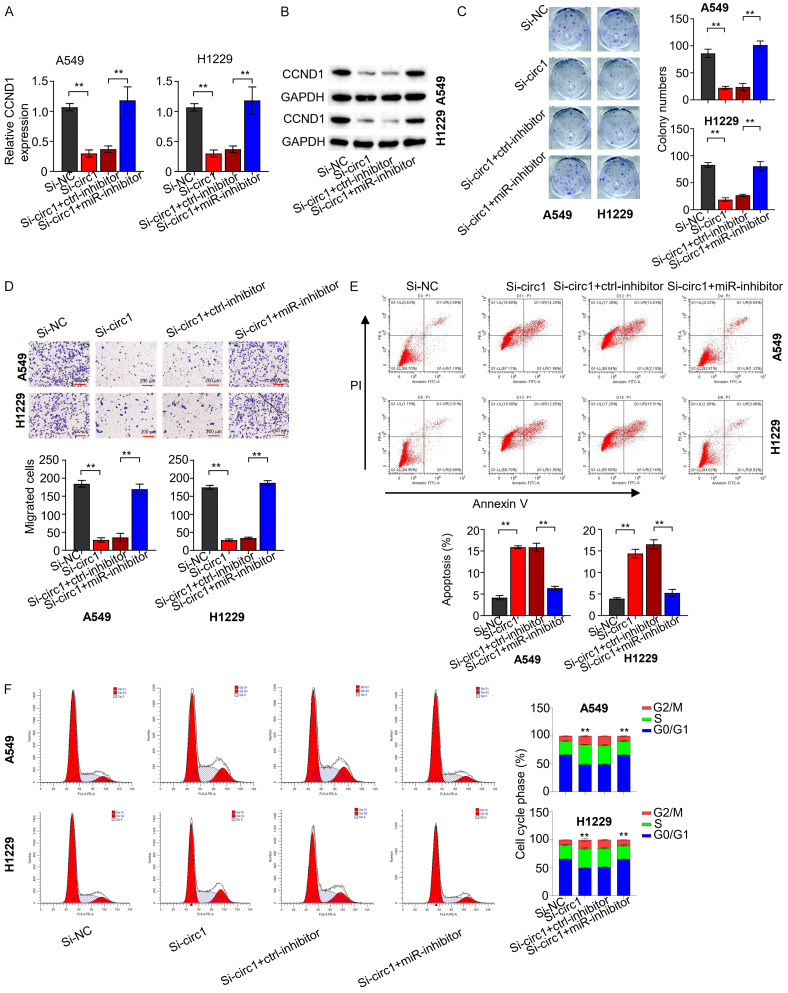
CircSNYJ1-mediated modulation of the miR-142-5p/CCND1 axis
To investigate the impact of circSNYJ1 on CCND1 expression, we performed qRT-PCR and Western blot analyses across different NSCLC cell groups (si-NC, si-circ1, si-circ1+NC inhibitor, si-circ1+miR-142-5p inhibitor) (Figure 5A, 5B). Our findings indicated that circSNYJ1 knockdown reduced CCND1 expression, which was restored by co-transfection with miR-142-5p inhibitor. A clonogenic assay was conducted with H1229 and A549 cells from different groups (Figure 5C), revealing that circSNYJ1 silencing significantly impaired clonogenic capacity, which was rescued by addition of the miR-142-5p inhibitor. We further conducted transwell assays using above cells to determine the effect of circSNYJ1 on the cell migration (Figure 5D). The results revealed that cell migration capacity was reduced following by circSNYJ1 knockdown, but this effect was reversed by miR-142-5p inhibition. To evaluate the influence of circSNYJ1 on NSCLC cell apoptosis, we performed flow cytometry experiments with H1229 and A549 cells from different groups (Figure 5E). Results showed that circSNYJ1 knockdown caused increased apoptosis rates, which was significantly diminished by the miR-142-5p inhibitor. Finally, we performed cell cycle tests using different cell groups (Figure 5F). Knockdown of CircSNYJ1 resulted in cell cycle arrest in the G2 phase, which was alleviated by co-transfection with the miR-142-5p inhibitor.
Figure 5.
CircSNYJ1-mediated modulation of the miR-142-5p/CCND1 axis. A, B. qRT-PCR and WB analyses assessing CCND1 expression in H1229 and A549 cells across different experimental groups. C. Clonogenic assay evaluating the clonogenic ability of H1229 and A549 cells in various groups. D. The Transwell assay (without matrix gel) examining the migratory capabilities of H1229 and A549 cells in different groups. E. Flow cytometry analysis determining the apoptotic rates of H1229 and A549 cells across the experimental groups. F. Cell cycle analysis assessing the cell cycle phases of H1229 and A549 cells from different groups.
Discussion
This study leveraged expression data from the GEO database to identified a highly expressed circRNA, circSYNJ1, in NSCLC tissues. We discovered that circSYNJ1 was significantly upregulated in cancer tissues from 68 lung cancer patients. Moreover, higher levels of circSYNJ1 expression were positively correlated with poor prognostic factors, including advanced staging, metastasis, and reduced survival. Functional experiments demonstrated that silencing circSYNJ1 inhibited NSCLC cell growth and migration, and promoted apoptosis. These findings suggest that circSYNJ1 may function as an oncogene in NSCLC progression, providing a potential therapeutic target for NSCLC diagnosis and treatment.
Lung cancer remains a major global health concern. In 2018, approximately 2.1 million new cases of lung cancer were reported, resulting in 1.8 million deaths directly related to this disease [17]. circRNAs have emerged as promising diagnostic biomarkers for NSCLC owing to their unique expression patterns in NSCLC tissues. Their stable circular structure confers the resistance to degradation, enabling their detection in various body fluids, including blood. This feature opens up the possibility of non-invasive diagnostics through liquid biopsies. Several circRNAs have been identified as differentially expressed in NSCLC patients, underscoring their potentials as high sensitivity and specificity diagnostic markers.
CircRNAs have garnered significant attention for their diverse roles in biological processes, including the development and progression of cancer [14,18,19]. Specifically, dysregulation of circRNAs have been frequently observed in various cancer, including lung cancer [20-22]. Our research study supports these findings, as we identified circSYNJ1 to be highly expressed in NSCLC tissues and cell lines. Notably, synaptojanin 2 (SYNJ2) has been implicated as a potential biomarker for lung squamous cell carcinoma [23]. In our study, the high expression of circSYNJ1 in NSCLC tissues was associated with poor prognosis. Additionally, we identified two miR-142-5p binding sites with circSYNJ1, aligning with the findings of Weixuman Yu et al., who reported elevated levels of miR-142-5p expression in breast cancer cells.
Our study confirmed Cyclin D1 (CCND1) as a target gene of miR-142-5p. CCND1 belongs to the highly conserved cyclin family and plays a crucial role in cell cycle regulation. Aberrant expression of CCND1 has been shown to disrupt the cell cycle control and promote tumor initiation and progression. A recent study highlighted the role of CCND1 in development of ulcerative colitis by dampening the function of miR-142-5p [24], thus supporting CCND1 as a downstream target gene of miR-142-5p.
Furthermore, our research demonstrated that high expression of circSNJY1 in NSCLC cell lines promotes cell proliferation and metastasis. This aligns with previous studies indicating the involvement of Protein-Tyrosine Phosphatase 4a2 in cancer progression [25,26], thus reinforcing the significance of our findings.
Conclusion
The study demonstrated that circSYNJ1 serves as a potential diagnostic and prognostic biomarker for NSCLC. Silencing circSYNJ1 expression inhibits NSCLC cell proliferation, colony formation, and migration, while simultaneously promoting apoptosis.
Disclosure of conflict of interest
None.
References
- 1.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med. 2011;32:703–740. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worley S. Lung cancer research is taking on new challenges: knowledge of tumors’ molecular diversity is opening new pathways to treatment. P T. 2014;39:698–714. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, Wang K, Li P. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017;13:1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao B, Liu G, Zhang W, Shi Y, Wei B. Role of circular RNAs and long non‑coding RNAs in the clinical translation of gastric cancer (Review) Int J Mol Med. 2021;47:77–91. doi: 10.3892/ijmm.2020.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren W, Yuan Y, Peng J, Mutti L, Jiang X. The function and clinical implication of circular RNAs in lung cancer. Front Oncol. 2022;12:862602. doi: 10.3389/fonc.2022.862602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Ao X, Yu W, Zhang Y, Wang J. Biogenesis, functions, and clinical implications of circular RNAs in non-small cell lung cancer. Mol Ther Nucleic Acids. 2021;27:50–72. doi: 10.1016/j.omtn.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sufianov A, Begliarzade S, Beilerli A, Liang Y, Ilyasova T, Beylerli O. Circular RNAs as biomarkers for lung cancer. Noncoding RNA Res. 2022;8:83–88. doi: 10.1016/j.ncrna.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Fraipont F, Gazzeri S, Cho WC, Eymin B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390. doi: 10.3389/fgene.2019.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussen BM, Abdullah SR, Hama Faraj GS, Rasul MF, Salihi A, Ghafouri-Fard S, Taheri M, Mokhtari M. Exosomal circular RNA: a signature for lung cancer progression. Cancer Cell Int. 2022;22:378. doi: 10.1186/s12935-022-02793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papatsirou M, Artemaki PI, Karousi P, Scorilas A, Kontos CK. Circular RNAs: emerging regulators of the major signaling pathways involved in cancer progression. Cancers (Basel) 2021;13:2744. doi: 10.3390/cancers13112744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HH, Zhang TN, Wu QJ, Huang XM, Zhao YH. Circular RNAs in lung cancer: recent advances and future perspectives. Front Oncol. 2021;11:664290. doi: 10.3389/fonc.2021.664290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, Xia Y, Tang W. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelidou K, Agelaki S, Mavridis K. Molecular markers related to immunosurveillance as predictive and monitoring tools in non-small cell lung cancer: recent accomplishments and future promises. Expert Rev Mol Diagn. 2020;20:335–344. doi: 10.1080/14737159.2020.1724785. [DOI] [PubMed] [Google Scholar]
- 18.Cheng D, Wang J, Dong Z, Li X. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int. 2021;21:11. doi: 10.1186/s12935-020-01703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Zheng L, Gao Y, Zhang W, Zhang Q, Xu Y. A comprehensive overview of circRNAs: emerging biomarkers and potential therapeutics in gynecological cancers. Front Cell Dev Biol. 2021;9:709512. doi: 10.3389/fcell.2021.709512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Yang Q, He AT, Yang BB. Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin Cancer Biol. 2021;75:49–61. doi: 10.1016/j.semcancer.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Zhu Q. Mechanisms regulating abnormal circular RNA biogenesis in cancer. Cancers (Basel) 2021;13:4185. doi: 10.3390/cancers13164185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou W, Li GS, Gao L, Lu HP, Zhou HF, Kong JL, Chen G, Xia S, Wei HY. SYNJ2 is a novel and potential biomarker for the prediction and treatment of cancers: from lung squamous cell carcinoma to pan-cancer. BMC Med Genomics. 2022;15:114. doi: 10.1186/s12920-022-01266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehman AU, Mohsin A, Cheema HA, Zahid A, Ebaad Ur Rehman M, Ameer MZ, Ayyan M, Ehsan M, Shahid A, Aemaz Ur Rehman M, Shah J, Khawaja A. Comparative efficacy and safety of tenecteplase and alteplase in acute ischemic stroke: a pairwise and network meta-analysis of randomized controlled trials. J Neurol Sci. 2023;445:120537. doi: 10.1016/j.jns.2022.120537. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Zhang M, Zheng Z, He Y, Zhu Y, Cheng Q, Rong J, Weng H, Chen C, Xu Y, Yun M, Zhang J, Ye S. Over-expression of protein tyrosine phosphatase 4A2 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:77527–77539. doi: 10.18632/oncotarget.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao D, Guo L, Neves H, Yuen HF, Zhang SD, McCrudden CM, Wen Q, Zhang J, Zeng Q, Kwok HF, Lin Y. The prognostic significance of protein tyrosine phosphatase 4A2 in breast cancer. Onco Targets Ther. 2015;8:1707–1717. doi: 10.2147/OTT.S85899. [DOI] [PMC free article] [PubMed] [Google Scholar]