Abstract
The commitment of stem cells to differentiate into osteoblasts is a highly regulated and complex process that involves the coordination of extrinsic signals and intrinsic transcriptional machinery. While rodent osteoblastic differentiation has been extensively studied, research on human osteogenesis has been limited by cell sources and existing models. Here, we systematically dissect human pluripotent stem cell-derived osteoblasts to identify functional membrane proteins and their downstream transcriptional networks involved in human osteogenesis. Our results reveal an enrichment of type II transmembrane serine protease CORIN in humans but not rodent osteoblasts. Functional analyses demonstrated that CORIN depletion significantly impairs osteogenesis. Genome-wide chromatin immunoprecipitation enrichment and mechanistic studies show that p38 MAPK-mediated CCAAT enhancer binding protein delta (CEBPD) upregulation is required for CORIN-modulated osteogenesis. Contrastingly, the type I transmembrane heparan sulfate proteoglycan SDC1 enriched in mesenchymal stem cells exerts a negative regulatory effect on osteogenesis through a similar mechanism. Chromatin immunoprecipitation-seq, bulk and single-cell transcriptomes, and functional validations indicated that CEBPD plays a critical role in controlling osteogenesis. In summary, our findings uncover previously unrecognized CORIN-mediated CEBPD transcriptomic networks in driving human osteoblast lineage commitment.
Keywords: p38 MAPK, induced pluripotent stem cells (iPSCs), osteoblasts, mesenchymal stem cells (MSCs), ChIP-sequencing (ChIP-seq)
The human skeletal system plays a crucial role in providing both static and dynamic stability to the body. The development of skeletal elements during embryogenesis, as well as the ongoing remodeling of bone in the adult, require a delicate interplay of developmental cues, signaling proteins, transcription factors, and their associated coregulatory proteins. This process supports the differentiation of osteogenic lineage cells from the initial mesenchymal stem cells (MSCs) to mature osteocytes in mineralized connective tissues (1). The synthesis, deposition, and mineralization of extracellular matrix are key functions of specialized cells called osteoblasts, which are responsible for the formation and maintenance of skeletal architecture. Osteoblasts, as the primary cells responsible for bone formation and maintenance, play a crucial role in extracellular matrix protein production, including collagen and osteocalcin, matrix mineralization, and differentiation of osteoclasts (2). Dysregulation of osteoblast functions can result in a variety of bone-related disorders such as osteoporosis, osteopenia, osteogenesis imperfecta, and osteosarcoma (3, 4), suggesting a crucial role of osteoblasts in maintaining bone homeostasis.
The osteogenic process involved in osteoblast differentiation from MSCs remains incompletely understood. Osteoblast commitment and differentiation are regulated by a complex interplay between signal transduction and transcriptional gene regulation. Recent advances in molecular and genetic studies have revealed several signaling cascades that play critical roles in osteoblast development, such as the Wnt, transforming growth factor-β (TGF-β), Hedgehog, fibroblast growth factor, ephrin, and sympathetic β2-adrenergic receptor signaling pathways (1). These intracellular signaling pathways are essential for transducing external signals into internal processes to promote lineage differentiation, such as the activation of intercellular transcriptional profiles. Several osteoblastic transcription factors functioning downstream of these signaling systems have been identified in mouse models for their critical functions in bone development. They included Runx2, Osterix/Sp7, Atf4, Satb2, and β-Catenin (1). However, it remains unclear how upstream signals, such as membrane proteins and cascades, fine-tune the activities of these transcriptional factors and transmit extracellular signals to intracellular transcriptional machinery, ultimately leading to osteoblast differentiation and maturation in humans.
While mouse models are commonly utilized to study osteogenesis, it is important to recognize that the genetic and molecular processes involved in bone development vary between species. Critical genes involved in human bone development may be absent in mice, and vice versa. Unfortunately, access to human bone tissue is often limited, which has hindered our ability to study human osteogenesis. This makes validating findings from rodent model systems a challenging task. Additionally, osteoblast-specific cell surface markers used to define functional human osteoblasts, such as membrane proteins, are not well-characterized. Therefore, a deeper understanding of the mechanisms by which membrane proteins and developmental signals regulate gene expression through transcription factors is needed in order to bridge this knowledge gap and develop more effective treatments for bone-related disorders.
Type II transmembrane serine protease CORIN, also known as atrial natriuretic peptide-converting enzyme, plays a crucial role in regulating cardiovascular physiology (5). The main function of CORIN is to activate natriuretic peptides, specifically atrial natriuretic peptide (ANP) and brain natriuretic peptide by cleaving them from their inactive precursor forms (6, 7). This cleavage is a crucial step in the physiological response to cardiac stress and is essential for maintaining cardiovascular homeostasis. These natriuretic peptides are key regulators of blood pressure and fluid balance. When released in response to increased atrial pressure or stretching of the heart walls, ANP and brain natriuretic peptide promote vasodilation, increase sodium excretion by the kidneys, and suppress the release of renin and aldosterone—actions collectively aimed at reducing blood volume and pressure. Therefore, CORIN plays a pivotal role in the regulation of blood pressure and fluid balance, contributing to overall cardiovascular health. Dysregulation of CORIN activity has been implicated in various cardiovascular disorders, emphasizing its importance in maintaining proper cardiovascular function. While CORIN is notably expressed primarily in the atria, it remains unclear whether CORIN could also be expressed and play a role in bone development, particularly in osteoblasts.
Type I transmembrane heparan sulfate proteoglycan syndecan-1 (SDC1), has been found to be a critical modulator of bone-cell communication through the nuclear factor kappa-B ligand (RANKL)/osteoprotegerin axis (8), which is one of the major regulators of bone resorption and osteoclastogenesis. Knockdown of SDC1 in human MSCs has been shown to result in a proadipogenic phenotype, followed by enhanced maturation of osteoblasts during subsequent osteogenic differentiation (9). This implicates SDC1 as a facilitator of human MSC osteo-adipogenic balance during early induction of lineage differentiation. Furthermore, in the context of multiple myeloma (MM)-associated bone disease, which is characterized by severely impaired osteoblast activity resulting from disrupted osteogenesis, MM cell-derived SDC1+ circulating extracellular vesicles have been found to inhibit osteogenesis of bone marrow-derived MSCs (10), implying that SDC1 and/or its regulated signaling in MM cell-derived SDC1+ circulating extracellular vesicles can directly impair normal osteogenesis. However, it remains unclear whether SDC1 plays a role in regulating osteogenesis.
Therapies using engineered bone tissue derived from human-induced pluripotent stem cell (hiPSC)-derived osteoblasts have been proposed as a potential treatment for osteoporosis and related disorders of bone mineral density (11). Studies have shown that hiPSC-derived osteoblasts share many characteristics with bone marrow-MSC-derived osteoblasts and primary osteoblasts, including the secretion of organic matrix substances and the production of bone minerals (12, 13, 14, 15, 16). In this study, we aimed to identify osteoblastic membrane proteins and their regulated transcriptome networks in functional osteoblasts derived from human pluripotent stem cells (hPSCs), including both human embryonic stem cells (hESCs) and hiPSCs. We found that CORIN positively modulates human osteogenesis and osteoblast identity, whereas SDC1 negatively regulates these processes. Further analysis of the CORIN-modulated transcriptome revealed that upregulation of CCAAT enhancer binding protein delta (CEBPD), activated by the p38 mitogen-activated protein kinase (MAPK), is critical to the downstream signaling cascade that governs osteoblastic differentiation. In contrast, the type I transmembrane heparan sulfate proteoglycan SDC1 negatively regulates these processes. In summary, our study highlights the important role of cell membrane proteins as potential osteoblastic markers governing osteoblast function in human osteogenesis.
Results
hPSC-derived osteoblasts recapitulate functional osteoblast characteristics
To gain insight into the crucial signals regulating osteogenesis and osteoblast lineage commitment in humans, we used a cellular model based on osteoblasts derived from hPSCs. These cells share many features with osteoblasts derived from bone marrow MSCs and primary osteoblasts, including the secretion of organic matrix and the production of bone minerals (11, 17). Using a previously described two-stage differentiation process (14, 16), three hESC lines (H1, H9, and HES2) and five WT hiPSC lines (WT-1H, WT-1G, WT-1J, WT-F6, and WT-F37) were differentiated to multipotent MSCs and then to osteoblasts (Fig. 1A and S1A). The three hESC lines used in this study were commercial cell lines, whereas the five WT hiPSC lines were derived from healthy fibroblasts in our laboratory (14, 16, 18). The high purity (>95%) of hPSC-derived MSCs was validated by the expression of MSC markers CD73, CD105, and CD166 (Fig. S1A). Osteoblast identity and maturity were verified by the identification of matrix mineralization on Alizarin Red S (ARS) staining (Fig. 1A). We then investigated and compared global transcripts among hPSC-derived MSCs and osteoblasts. The extracted RNAs were isolated at four time points during the course of differentiation (day 0, MSCs; day 15–18, preosteoblasts; day 21–24, maturing osteoblasts; and day 27, mature osteoblasts). Expression profiles of time-course samples analyzed by Spearman’s correlation and principal component analysis demonstrated that gene expression profiles of preosteoblast, maturing osteoblast, and mature osteoblast samples clustered together and distinctly from MSC samples (Fig. 1, B and C), indicating significant transcriptome changes during differentiation from MSCs to osteoblasts. We observed a distinct separation of transcriptome profiles between MSCs and osteoblasts at various stages of differentiation (Fig. 1C). Mouse Gene Atlas and MGI Mammalian Phenotype analyses demonstrated that genes upregulated in preosteoblasts, maturing osteoblasts, and mature osteoblasts are also enriched in differentiating mouse osteoblast gene profiles (day 14 and 21); these same genes are also involved in abnormal skeletal development (Fig. 1, D and E). On the other hand, genes downregulated in preosteoblasts, maturing osteoblasts, and mature osteoblasts were most highly expressed in the early stages of differentiating mouse osteoblasts (day 5), bone marrow cells, and MSC-like cells; these genes were also involved in cell cycle dysregulation of MGI mammalian phenotypes (Fig. 1, D and E). Gene Ontology biological process (GO_BP) analysis showed that genes related to extracellular structure organization, bone mineralization, regulation of ossification, and osteoblast differentiation development are enriched in osteoblasts; in contrast, genes related to the regulation of the mitotic cell cycle, DNA replication, and DNA metabolic process are enriched in undifferentiated MSCs (Fig. 1F and S1, B and C). The enriched expression of genes involved in bone development (ANKH, CLEC3B, IGF2, JUNB, PDGFRB, and PRRX1) (19, 20, 21, 22, 23), osteogenesis imperfecta genes (COL1A1, COL1A2, SERPINF1, and CRTAP) (24), osteoporosis (CLCN7) (25), and Paget’s disease of bone (SQSTM1) (26) (Fig. S1, D and E) were detected in hPSC-derived osteoblasts compared to MSCs. Collectively, these findings provide compelling evidence that osteoblast-like characteristics are recapitulated by osteoblasts derived from hPSCs.
Figure 1.
hPSC-derived osteoblasts recapitulate functional osteoblast characteristics.A, the morphology of hESCs (H9) and hiPSCs (WT1-G, WT-1H, and WT-F6) at different stages: MSCs, preosteoblasts (Pre-OBs), maturing osteoblasts (Mat OBs), and osteoblasts (OBs). ARS staining revealed matrix mineralization in MSCs, preosteoblasts, maturing osteoblasts, and osteoblasts. The scale bar represents 100 mm. B, Spearman’s correlation of the RNA-seq results of hESC and hiPSC-derived MSCs, preosteoblasts, maturing osteoblasts, and mature osteoblasts. The pairwise correlation ranges from 0.91 (blue) to 1 (orange). C, principal component analysis (PCA) was used to calculate and compare the gene expression data of MSCs (light blue), preosteoblasts (Pre; green), maturing osteoblasts (Mat; orange), and osteoblasts (OBs; dark red). The two axes, PC1 and PC2, represent the first two principal components identified by the analysis. Squares indicate hESC-derived cells, while circles indicate hiPSC-derived cells. The grouping of MSCs, as well as Pre, Mat, and OBs, is outlined. D and E, Mouse Gene Atlas (D) and MGI Mammalian Phenotype (E) analyses indicate that the genes enriched in preosteoblasts (Pre-OBs), maturing osteoblasts (Mat OBs), and osteoblasts (OBs) are most significantly expressed in late-stage mouse osteoblasts (day 14 and 21) and associated with abnormal skeletal development, while genes enriched in MSCs are most highly expressed in early-stage mouse osteoblasts (day 5) and associated with cell cycle dysregulation. F, upper left graph: GSEA analyses identified Gene Ontology biological processes (GO_BP) from a collection of 7525 gene sets enriched in MSCs or osteoblasts. GO_BP gene sets enriched (orange, corresponding to a positive normalized enrichment score (NES)) or depleted (green, corresponding to a negative NES) in the transcriptome of osteoblasts compared to MSCs are shown. Enriched gene sets are selected based on statistical significance (normalized p-value < 0.05 and FDR q-value < 0.25). Upper center-right panels: heatmap of significantly altered GO_BPs in preosteoblasts (Pre-OBs), maturing osteoblasts (Mat OBs), and mature osteoblasts (OBs). Lower panel: GSEA leading edge analysis demonstrates the overlap between gene sets enriched in osteoblasts or MSCs. ARS, Alizarin Red S; FDR, false discovery rate; GO_BP, Gene Ontology biological process; GSEA, gene set enrichment analysis; hESC, human embryonic stem cell; hiPSC, human-induced pluripotent stem cell; hPSC, human pluripotent stem cell; MGI, Mouse Genome Informatics; MSC, mesenchymal stem cell.
Identification of enriched membrane proteins in osteoblasts through systematic analysis of osteoblast-specific gene clusters
To identify genes contributing to functional osteoblasts, we analyzed the gene expression patterns of hPSC-derived MSCs and osteoblasts at key time points during osteogenic differentiation, reflecting progressive differentiation from MSCs through mature osteoblasts, and grouped them into 12 gene expression clusters (Fig. 2A, left panel). Enrichr analysis of these 12 clusters by GO_BP revealed that genes enriched in Cluster 6 (C6, genes gradually upregulated during osteogenesis) are highly associated with bone development processes including skeletal system development, extracellular matrix organization, regulation of osteoblast differentiation, positive regulation of ossification, and bone morphogenesis (Fig. 2A, right panel). Conversely, Cluster 12 genes (C12, genes gradually downregulated during osteogenesis) are highly associated with cell cycle events including DNA replication, sister chromatid segregation, and the G2/M transition of the mitotic cell cycle (Fig. 2A, right panel). These findings point to the critical role of C6 and C12 genes in regulating osteogenesis and maintaining bone homeostasis.
Figure 2.
Systematic transcriptome analyses of hPSC-derived cells reveal enriched membrane proteins in human osteoblasts.A, left panel: Cluster analysis of the gene expression patterns during osteogenesis. The horizontal axis represents the stages of osteoblast development: MSCs, preosteoblasts (Pre), maturing osteoblasts (Mat), and osteoblasts (OBs). The vertical axis represents FPKM values using 2 as the logarithmic base of a gene at different developmental stages. The red lines indicate the change in gene expression level between samples. Right panel: GO_BP pathway analysis of 12 gene expression clusters showed the enrichment of genes involved in different biological processes as indicated. B, left panel: diagrammatic depiction of stages of osteogenesis. Right panel: RNA-seq analysis of hPSC-derived osteoblasts revealed the enriched expression of AMIGO2, CDH13, CDH2, PVR, and SDC1 in MSCs and the enriched expression of CORIN, IL1R1, CD82, APOD, TGFBR2, ADAMTSL4, SERPING1, MME1, VCAM1, and LEPR in osteoblasts. C, heatmap showing the differential expression of identified membrane proteins during osteogenesis. The enriched membrane proteins in osteoblasts are in Clusters 3 and 6. The enriched membrane proteins in MSCs are in Cluster 12. D, RT-qPCR validated the upregulation/downregulation of the identified membrane proteins in multiple hPSC-derived osteoblasts as biological repeats (WT-1H, WT-1G, WT-1J, WT-F6, and WT-F37) compared to MSCs. MSCs were derived from hPSCs using the bFGF/PDGF-AB method. E, RT-qPCR demonstrated the upregulation/downregulation of the identified membrane proteins in H1, WT-F6, and WT-F37 hPSC-derived osteoblasts compared to MSCs. MSCs were derived from hPSCs using the SB-431542/7.5%CO2 method. (n = 3, mean ± S.D.). F, RT-qPCR demonstrated the upregulation/downregulation of the identified membrane proteins in osteoblasts differentiated from ad-HMSCs and bm-HMSCs. (n = 3, mean ± S.D.). G, RT-qPCR demonstrated the upregulation/downregulation of the identified membrane proteins in hFOB-differentiated osteoblasts. (n = 3, mean ± S.D.). ad-HMSC, adipose tissue-derived human MSC; CD, clusters of differentiation; FPKM, Fragments Per Kilobase per Million mapped fragments; GO_BP, Gene Ontology biological process; HMSC, human MSC; hPSC, human pluripotent stem cells; MSC, mesenchymal stem cell; RT-qPCR, real-time quantitative PCR; SDC1, syndecan-1; TGF-β transforming growth factor-β.
Cell membrane protein and surface receptor-mediated signaling play a critical role in regulating gene expression as part of lineage determination. We hypothesized that during our differentiation process, osteoblasts and their precursors would express a set of unique membrane proteins corresponding to osteoblastic gene signatures that would distinguish them from other cell populations differentiating into other lineages and thereby permit quantification of osteoblastic development and maturation during this process. With this in mind, we focused on the differential expression of 389 clusters of differentiation (CD) molecules and cell membrane proteins in C1-C12. Among these CD molecules and membrane proteins, CORIN, IL1R1, APOD, TGFBR2, ADAMTSL4, SERPING1, MME (also known as CD92), VCAM1, and LEPR were enriched in osteoblasts, while AMIGO2, CDH13, PVR, and SDC1 were enriched in MSCs (Fig. 2B). Hierarchical clustering of their expression profiles in distinct osteogenic stages demonstrated that the expressions of C6 genes CORIN, IL1R1, TGFBR2, ADAMTSL4, SERPING1, VCAM1, and LEPR are initiated at the very beginning of osteogenesis and increase through the entire osteogenic process; the expressions of C3 genes CD82 and MME are highly enriched in preosteoblasts and maintain the same level during osteogenesis; and the expressions of C12 genes SDC1, PVR, CDH2, CDH13, and AMIGO2 are particularly enriched in MSCs and dramatically decrease in early osteogenesis (Fig. 2C). The enriched expression of these cell membrane proteins in osteoblasts was confirmed by real-time quantitative PCR (RT-qPCR) in an independent experimental setting (Fig. 2D). These significant differences were further validated in multiple MSC types differentiated to mature osteoblasts using SB-431542/7.5%-induced MSCs (16), adipose tissue-derived human MSCs (ad-HMSCs), bone marrow-derived human MSCs (bm-HMSCs), and preosteoblast line hFOB (Fig. 2, E–G). Consistent with our results, a number of membrane proteins that are known to regulate osteogenesis, such as IL1R1 (27), TGFBR2 (28), and MME (29), have previously been shown to be involved in sustaining osteoblast identity and facilitating bone development. In summary, our findings indicate that hPSC-derived osteoblasts offer a promising platform to elucidate osteoblast-specific gene clusters and identify key osteogenic molecules and signaling pathways in humans.
Functional validation of membrane proteins CORIN and SDC1 in regulating osteogenesis and bone formation
Upon analyzing the expression of these membrane proteins identified in hPSC-derived osteoblasts in mouse tissues, we observed that most of the C3 and C6 genes (IL1R1, TGFBR2, ADAMTSL4, SERPING1, VCAM1, LEPR, and MME) are highly expressed in bone, bone marrow, and osteoblasts. CD82, on the other hand, is enriched in bone and bone marrow (Fig. S2A), suggesting the evolutional conservation of their roles in regulating osteogenesis. Interestingly, while the C6 gene CORIN, a transmembrane serine protease acting as a pro-ANP-converting enzyme, is enriched in differentiated human osteoblasts, it does not appear to be expressed in mouse bone tissues (Fig. S2A), implying that CORIN may have a unique role in regulating osteogenesis in humans that is not present in rodents. Moreover, SDC1 displays decreased expression during human osteogenesis, but not in rodent models (Fig. S2A). Due to the unclear understanding of the roles of CORIN and SDC1 in regulating osteogenesis, we have intentionally chosen to conduct a more in-depth investigation into these proteins and the cellular signaling pathways they regulate in the context of osteogenesis.
We first evaluated the protein expression of CORIN and SDC1 using flow cytometry. Our analysis revealed a marked increase in CORIN expression and a significant reduction in SDC1 expression in osteoblasts compared to MSCs (Figs. 3A and S2B), which is consistent with our RNA-seq and RT-qPCR findings. Immunohistochemistry (IHC) staining for CORIN and SDC1 proteins in the human femur bone tissues revealed expression of CORIN but not SDC1 in trabecular bone (Fig. 3B). Interestingly, CORIN was not expressed in mouse tibial bone tissue (Fig. S2C), suggesting that CORIN may have functions specific to human bone development. Notably, SDC1 protein was not detected in mouse tibia bone (Fig. S2C), which is consistent with transcriptome data from Mouse Gene Atlas MOE430 (Fig. S2A), implying that posttranslational regulation of SDC1 mRNA may be involved in rodent bone formation. To determine the precise role of CORIN and SDC1 in human osteoblast differentiation, we depleted CORIN or ectopically expressed tetracycline (Tet)-inducible SDC1 in hPSC-derived osteoblasts. Both CORIN depletion as well as ectopic SDC1 expression led to impaired expression of the osteoblastic genes COL1A1, PTH1R, and BGLAP, and transcription factor RUNX2 upon osteogenesis (Fig. 3C). ARS staining validated impaired osteoblastic differentiation and decreased mineral deposition in CORIN-depleted and SDC1-overexpressed osteoblasts (Fig. 3D). To examine the role of CORIN and SDC1 in three-dimensional (3D) bone formation, we used gelatin methacrylate (GelMA) scaffolds, because their biomechanical properties can be tuned to closely resemble tissues of interest and their extracellular matrix, including trabecular bone matrix (30). We cultured CORIN-depleted or SDC1-overexpressed MSCs in GelMA discs (Fig. 3, E–G) and then differentiated them into osteoblasts. Control MSC (shCtrl (control shRNA) and Vector)-seeded matrices demonstrated the progressive formation of calcified tissue after 6 weeks of culture in osteogenic differentiation conditions by 3D microcomputed tomography (Fig. 3, H and I, left panels). In contrast, CORIN knockdown or SDC1 overexpression in MSCs was associated with impaired osteoblastic calcification (Fig. 3, H and I, left panels), lower bone mineral density, and lower bone volume/total tissue volume ratios in 3D GelMAs (Fig. 3, H and I, middle and right panels). Together, our functional studies suggest the essential roles of CORIN and SDC1 in regulating human osteoblast function and bone formation.
Figure 3.
Functional studies on CORIN and SDC1 in osteogenesis.A, flow cytometry analysis of CORIN and SDC1 expression in hESC H9-derived MSCs and osteoblasts (OBs). B, IHC staining of CORIN and SDC1 expression in human femur bone tissue using anti-human CORIN and anti-human SDC1 antibodies. Goat and mouse IgG were utilized as a negative control. The scale bar represents 100 μm. C, RT-qPCR confirmed the impairment of osteoblast lineage genes COL1A1, PTH1R1, BGLAP, and RUNX2 in CORIN-depleted or SDC1-overexpressed hESC H1-derived osteoblasts. (n = 3, mean ± S.D.). D, ARS staining indicated that depletion of CORIN as well as overexpression of SDC1 in osteoblasts led to impaired bone mineral production. The scale bar represents 100 μm. E, diagrammatic depiction of preparation of GelMA discs. Hybridization of gelatin and methacrylic anhydride was followed by UV cross-linking to make the GelMA discs. F, morphology of GelMA discs, which had a diameter of 4 mm and a thickness of 1 mm. G, scanning electron microscope (SEM) image showing the surface and hollow structures of the GelMA discs. The scale bar represents 400 μm. H and I, left: 3D μCT images of GelMA discs loaded with CORIN-depleted or SDC1-overexpressed hESC H1-derived osteoblasts compared to control osteoblasts at 3 weeks and 6 weeks culture in osteoblastic differentiation media. Right: bone mineral density and BV/TV (%) analyses of GelMA discs loaded with CORIN-depleted (upper), or SDC1-overexpressed (lower) hESC H1-derived osteoblasts. The scale bar represents 1 mm. The difference between two groups was compared by the two-tailed unpaired Student’s t test or ANOVA to calculate the p-value. (n = 3, mean ± S.D.). ARS, Alizarin Red S; BV/TV, bone volume/total tissue volume; GelMA, gelatin methacrylate; hESC, human embryonic stem cell; IHC, immunohistochemistry; MSC, mesenchymal stem cell; RT-qPCR, real-time quantitative PCR; SDC1, syndecan-1; μCT, microcomputed tomography.
Transcriptome analysis dissecting the impact of CORIN and SDC1 on the gene signature associated with osteoblasts
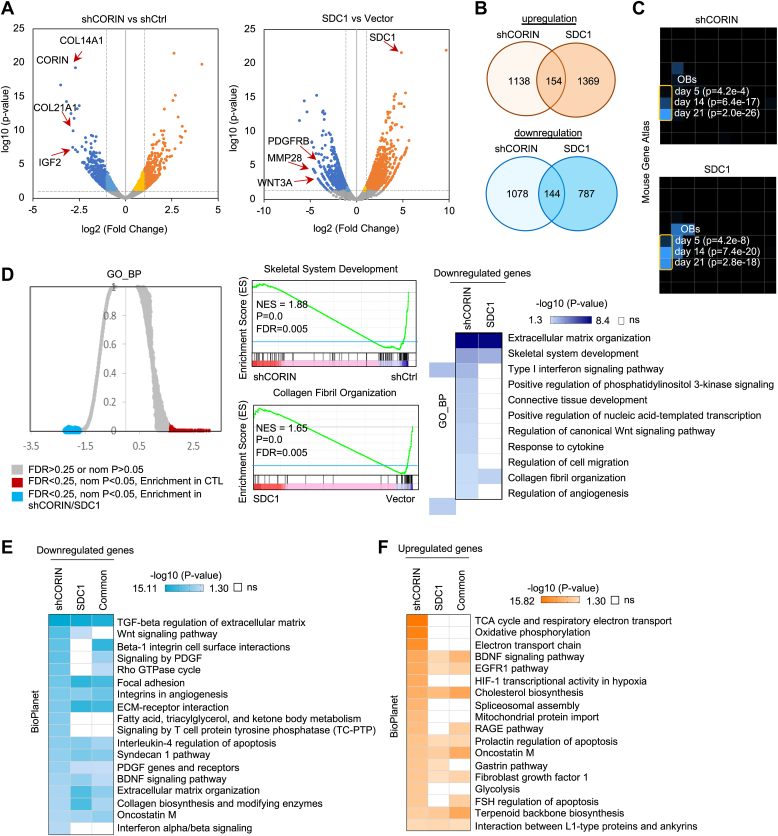
Next, we sought to understand the role of CORIN and SDC1 in osteogenesis. To achieve this, we examined the altered transcriptomes of osteoblasts that were either depleted of CORIN or overexpressed with SDC1 and subjected them to RNA-seq. Analysis of differentially expressed genes using volcano plots revealed that several genes involved in cellular matrix formation and bone development were significantly downregulated upon CORIN knockdown or SDC1 overexpression. These genes included COL14A1, COL21A1, and IGF2 in CORIN-depleted osteoblasts, and PDGFRB, MMP28, and WNT3A in SDC1-overexpressed osteoblasts (Fig. 4A). By investigating these changes, we were able to gain insight into the mechanisms by which CORIN and SDC1 regulate osteogenesis. Screening for overlapping differentially expressed genes in CORIN-knockdown and SDC1-overexpression samples, we identified 144 genes that were codownregulated and 154 genes that were coupregulated (Fig. 4B). Mouse Gene Atlas analyses by Enrichr indicated that downregulated genes in CORIN-depleted and SDC1-overexpressed osteoblasts are significantly enriched in the later stages of mouse osteoblast differentiation (day 14 and day 21) (Fig. 4C). Gene set enrichment analysis (GSEA) of GO_BP indicated that genes significantly downregulated in CORIN-depleted and SDC1-overexpressed osteoblasts are involved in extracellular matrix organization, skeletal system development, and collagen fibril organization (Fig. 4D), supporting the critical role of CORIN and SDC1 in regulating human osteogenesis. BioPlanet pathway analysis further confirmed that CORIN-upregulated and SDC1-downregulated genes are related to TGF-β-mediated regulation of extracellular matrix, extracellular matrix organization, and extracellular matrix receptor interactions (Fig. 4E), which were consistent with the enriched gene signatures in osteoblasts compared to MSCs. In contrast, CORIN-downregulated and SDC1-upregulated genes shared fewer similarities, mainly in epidermal growth factor receptor and cholesterol biosynthesis (Fig. 4F). These findings provide a global view of osteogenesis regulated by CORIN and SDC1.
Figure 4.
Systematic analyses of the CORIN/SDC1-regulated transcriptome.A, volcano plots for DEGs in CORIN-depleted (left panel) or SDC1-overexpressed (right panel) hESC H9-derived osteoblasts compared with control osteoblasts. DEGs were selected with the parameters p < 0.05 and log2 (fold-change) > 1. Vertical dashed lines demarcate the 2-fold change cutoff. Blue and dark blue dots represent genes significantly downregulated ≤2-fold or >2-fold, respectively; yellow and orange dots represent genes significantly upregulated ≤2-fold or >2-fold, respectively; gray dots represent genes without significantly differential expression following depletion or CORIN or overexpression of SDC1. B, Venn diagram depicting the number of genes considered to be DEGs in CORIN-knockdown and SDC1-overexpressed hESC H9-derived osteoblasts. The intersection represents commonly upregulated (upper panel) or downregulated (lower panel) genes in both the shCORIN and SDC1 groups. C, Mouse Gene Atlas analyses by Enrichr indicated that downregulated genes in CORIN-depleted and SDC1-overexpressed osteoblasts are significantly enriched in the later stages of mouse osteoblast differentiation (days 14 and 21). D, GSEA GO_BP analyses (left panel) demonstrated that genes significantly downregulated in CORIN-depleted and SDC1-overexpressed hESC H9-derived osteoblasts were involved in skeletal system development (right upper panel) and collagen fibril organization (right lower panel). Gene sets enriched in control cells are shown in dark blue (FDR q value < 0.25; normalized p value < 0.05), while white indicates a nonsignificant difference. E, BioPlanet pathway analyses demonstrate that genes significantly downregulated in CORIN-depleted and SDC1-overexpressed hESC H9-derived osteoblasts are involved in TGF-β regulation of extracellular matrix, extracellular matrix organization, and ECM receptor interactions. Gene sets enriched in control cells are shown in blue (FDR q value < 0.25; normalized p value < 0.05), while white indicates a nonsignificant difference. F, BioPlanet pathway analyses demonstrate that genes significantly upregulated in CORIN-depleted and SDC1-overexpressed hESC H9-derived osteoblasts are overrepresented for genes in cellular functions such as EGFR1 and cholesterol biosynthesis. ECM, extracellular matrix; EGFR, epidermal growth factor receptor; FDR, false discovery rate; hESC, human embryonic stem cell; GO_BP, Gene Ontology biological process; GSEA, gene set enrichment analysis; SDC1, syndecan-1; TGF-β transforming growth factor-β.
CEBPD is a key mediator of CORIN and SDC1 regulation of osteoblast identity
To identify the critical signaling regulated by CORIN and SDC1-mediated osteogenesis, we focused on transcription factors, which are key molecules controlling osteoblast identity. Chromatin immunoprecipitation (ChIP) enrichment analysis revealed that CORIN-upregulated and SDC1-downregulated genes are the main targets of the transcription factors CEBPD (mouse, m), POU3F2 (human, h), ZNF217 (h), CTNNB1 (h), YAP1 (m), WT1 (h), and FOXA2 (h) (Fig. 5A). Among these transcription factors, CEBPD targets were most commonly downregulated upon CORIN depletion as well as SDC1 overexpression (Fig. 5A), indicating that CEBPD is a potential key downstream transcription factor that links CORIN and SDC1 to characteristic osteoblast gene signatures. Conversely, genes upregulated by CORIN depletion and downregulated by SDC1 were mainly targeted by ATF3 (h), E2F1 (m), EGR1 (h), ELK3 (h), KMD2B (h), REL1A (h), and p53 (m) (Fig. 5A), suggesting that these transcription factors may play an inhibitory role in regulating osteogenesis. Immunoblotting and RT-qPCR demonstrated reduced CEBPD expression in CORIN-knockdown or SDC1-overexpressed osteoblasts compared to control osteoblasts (Fig. 5, B and C), indicating that CORIN and SDC1 are capable of regulating CEBPD expression.
Figure 5.
CORIN/SDC1 governs the determination of osteoblast identities via CEBPD.A, ChEA analysis of DEGs in hESC H9-derived osteoblasts upon CORIN depletion as well as SDC1 overexpression. Genes enriched in control cells are shown in blue, and genes enriched in osteoblasts with CORIN depletion or SDC1 overexpression are shown in orange. The color intensity of the circle indicates the number of targets by a given transcription factor in the dataset, while the circle size represents the significance of the interaction. B and C, immunoblotting of CEBPD in CORIN-depleted (B) and SDC-overexpressed (C) hESC H9-derived osteoblasts. RT-qPCR confirmed the efficacy of shCORIN and SDC1 in modifying gene expression. (n = 3, mean ± S.D.). D, immunoblotting showed an increase in p38 MAPK phosphorylation and subsequent CORIN and CEBPD expression in hESC H9-derived osteoblasts compared to MSCs. E, immunoblotting showed inhibition of p38 MAPK by p38 MAPK inhibitor SB202190 (200 nM) and an associated downregulation of CEBPD expression. F, ARS staining of hESC H9-derived differentiated osteoblasts treated with p38 MAPK inhibitor SB202190 (200 nM) compared with control osteoblasts and MSCs. The scale bar represents 5 mm. G, immunoblotting showed a decrease in p38 MAPK phosphorylation upon CORIN knockdown (upper panel) or SDC1 overexpression (lower panel) in H1 hESC-derived osteoblasts. H, IHC staining showed the expression of CEBPD in human and mouse femur bone tissues. Rabbit IgG served as a control. I, scRNA-seq analysis of human osteoblasts reveals the expression of CEBPD in a distinct osteoblast population. Left panel: three osteoblast clusters (preosteoblasts [Pre-OBs], osteoblasts [OBs], and undetermined osteoblasts [Undetermined OBs]) are defined, colored, and visualized by UMAP using 5329 osteoblasts. Middle panel: logarithm-normalized expression showed CEBPD is highly expressed in the three osteoblast clusters. Right panel: distributions of CEBPD expression within the three osteoblast clusters showed the highest CEBPD expression in the undetermined osteoblast cluster. J, ARS staining confirmed that depletion of CEBPD impairs bone mineralization in hESC H9-derived osteoblasts. The scale bar represents 100 μm. K, RT-qPCR reveals the impairment of osteoblast lineage genes COL1A1, PTH1R1, BGLAP, and RUNX2 in CEBPD-depleted hESC H9-derived osteoblasts. (n = 3, mean ± S.D.). L, left panels: 3D μCT images of GelMA discs loaded with CEBPD-depleted hESC H9-derived osteoblasts (sgCEBPD-1) at 3 weeks and 6 weeks culture in osteoblast differentiation media. Right panels: bone mineral density and BV/TV (%) analyses of GelMA discs loaded with CEBPD-depleted hESC H9-derived osteoblasts (sgCEBPD). The scale bar represents 1 mm. The difference between two groups was compared by the two-tailed unpaired Student’s t test or ANOVA to calculate the p-value. (n = 3, mean ± S.D.). ARS, Alizarin Red S; BV/TV, bone volume/total tissue volume; CEBPD, CCAAT enhancer binding protein delta; ChEA, ChIP enrichment analysis; DEG, differentially expressed gene; GelMA, gelatin methacrylate; hESC, human embryonic stem cell; IHC, immunohistochemistry; MAPK, mitogen-activated protein kinase; MSC, mesenchymal stem cell; RT-qPCR, real-time quantitative PCR; scRNA-seq, single-cell RNA-seq; SDC1, syndecan-1; μCT, microcomputed tomography; UMAP, uniform manifold approximation and projection.
We next investigated the signaling cascade by which CORIN and SDC1 regulate CEBPD expression. High-throughput screening has identified p38 MAPK’s activator DIPQUO as a promoter of human osteoblast differentiation (31). We are exploring whether CORIN positively regulates, while SDC1 negatively regulates, CEBPD expression through the p38 MAPK signaling pathway. Indeed, we observed upregulation of CORIN, p38 MAPK phosphorylation, and CEBPD in hPSC-derived osteoblasts compared to MSCs (Fig. 5D). Inhibition of p38 MAPK by SB202190 led to decreased CEBPD and impaired osteogenesis (Fig. 5, E and F), confirming the essential role of p38 MAPK in regulating CEBPD expression. Notably, the knockdown of CORIN or ectopic expression of SDC1 led to decreased p38 MAPK phosphorylation and CEBPD expression (Fig. 5G). Taken together, our findings suggest that CORIN and SDC1 may regulate osteogenesis through the p38 MAPK/CEBPD signaling pathway.
IHC staining of human femur and mouse bone tissues demonstrated high expression of CEBPD in trabecular bone (Fig. 5H). Single-cell RNA-seq (scRNA-seq) analysis of human ALPLhigh/CD45low osteoblasts isolated from human femur heads (32) revealed that CEBPD is highly expressed in all preosteoblasts (COL1A1high/VCAM1high/LEPRhigh), osteoblasts (BGLAPhigh/IBSPhigh/SPP1high/COL1A1high), and undetermined osteoblasts (COL1A1high/VCAM1high/LEPRhigh/NR4A1high/NR4A2high) (Fig. 5I), suggesting that CEBPD is a critical transcription factor that participates in all stages of osteogenesis. ARS staining demonstrated impaired osteoblastic differentiation and decreased mineral deposition in CEBPD-depleted osteoblasts (Fig. 5J). CEBPD depletion led to reduced expression of RUNX2, PTH1R, BGLAP, and COL1A1 (Fig. 5K). Furthermore, CEBPD KO was associated with impaired osteoblast-mediated calcification in GelMA discs (Fig. 5L, left panel), lower bone mineral density, and lower BV/TV, bone volume/total tissue volume ratios (Fig. 5L, middle and right panels). Together, our functional studies suggest that CEBPD functions as a downstream mediator of CORIN/SDC1-controlled bone formation and osteoblast function.
Multiomics studies indicate that CEBPD serves as a crucial transcription factor in maintaining osteoblast function
To elucidate how CORIN/SDC1-CEBPD axis regulates osteoblastic lineage-associated gene expression, we carried out CEBPD ChIP followed by next-generation sequencing (ChIP-seq) and assay for transposase-accessible chromatin sequencing (ATAC-seq) to assess CEBPD genome occupancies and genome-wide chromatin accessibility in osteoblasts. Integration of CEBPD ChIP-seq and ATAC-seq data revealed that the gene-proximal regions of transcription start sites bound by CEBPD showed no preference for open or closed chromatin (Fig. 6A) and exhibited similar genome binding affinities (Fig. 6B). A significant proportion of CEBPD+/ATAC+ cotargeted regions (29.01%) were promoter regions (Fig. 6C), suggesting that CEBPD functions as a transcriptional regulator in modulating gene expression. Homer known motif and de novo motif analyses revealed that the top motif enriched in CEBPD+/ATAC+ cooccupied motifs is identical to the CEBPD family binding motif and osteoblast/bone formation-associated transcription factor motifs associated with Fos1 (33), Fra1 (33), JUNB (34, 35), RUNX (36) and STAT3 (37) (Figs. 6D and S3A).
Figure 6.
ChIP-seq, ATAC-seq, and RNA-seq studies suggest CEBPD functions as an osteoblastic transcription factor.A, heatmaps depicting the commonalities and differences in the 3 kb genomic loci surrounding each of the identified ChIP-seq peaks and ATAC peaks, grouped by cluster. B, composite plots showing the average binding intensities of CEBPD+/ATAC− and CEBPD+/ATAC+ peak regions. C, pie chart showing the genomic positional distribution of CEBPD/ATAC cotargeted binding sites for known and de novo RefSeq genes. D, motif analyses plot of the top-scoring Homer known binding motifs for CEBPD/ATAC cotargeted genomic binding peaks in osteoblasts. E, Venn diagram of genes identified via ChIP-seq, ATAC-seq, and RNA-seq studies reveal 179 genes transcriptionally regulated by CEBPD in osteoblasts. F, upper panel: heatmaps of CEBPD peak intensities in CEBPD/ATAC cotargeted gene regions (±3 kb from TSS). lower panel: heatmaps of changes in gene expression in CEBPD-depleted hESC H9-derived osteoblasts as examined by RNA-seq. G, GO_BP analyses showed that the 179 enriched genes regulated by CEBPD are involved in extracellular matrix organization, regulation of ossification, regulation of bone mineralization, and other bone physiology-related functions. H, IGV snapshot of CEBPD/ATAC occupancy over the promoter regions of COL12A1, COL21A1, SMAD6, and ADAMTS5. I, ChIP-qPCR at CEBPD TSS peak sites assess CEBPD or IgG enrichment (ChIP/input). ChIP-qPCR confirms specific enrichment of COL12A1, COL21A1, SMAD6, and ADAMTS5 at peak regions in hESC H9-derived osteoblasts. (n = 3, mean ± S.D.). J, scRNA-seq analysis demonstrated logarithm-normalized expression of the CEBPD transcriptional targets COL12A1, SMAD6, C1RL, SMIM14, and FBXO32, and are displayed according to the three osteoblast clusters. The difference between two groups was compared by the two-tailed unpaired Student’s t test to calculate the p-value. ATAC, assay for transposase-accessible chromatin; CEBPD, CCAAT enhancer binding protein delta; ChIP, chromatin immunoprecipitation; GO_BP, Gene Ontology biological process; hESC, human embryonic stem cell; IgG, immunoglobulin G; IGV, Integrative Genomics Viewer; scRNA-seq, single-cell RNA-seq; TSS, transcription start site.
To further characterize the roles (activator or repressor) of CEBPD in transcriptional regulation, we performed RNA-seq to assess alterations to the osteoblast transcriptome upon CEBPD depletion. By integrating ChIP-seq, ATAC-seq, and RNA-seq studies, we pinpointed 179 downregulated genes in CEBPD-depleted osteoblasts which were CEBPD+/ATAC+ cotargeted genes (Fig. 6E). This intersection represents genes containing promoters with open chromatin architecture that were also positively regulated by CEBPD binding during osteogenesis. A representative subset of the 179 genes is shown in Figure 6F. GO_BP analysis revealed that these genes are mainly involved in osteoblast differentiation and bone formation, supporting biological functions including extracellular matrix organization, regulation of ossification, collagen fibril organization, and Wnt signaling pathways (Fig. 6G). This further supports the idea that CEBPD functions as a transcriptional activator and actively regulates osteoblastic determination. The Integrative Genomics Viewer revealed CEBPD+/ATAC+ cooccupancy over promoter regions of the osteoblast lineage-associated or upregulated genes COL12A1 (38), COL21A1 (39), SMAD6 (40), ADAMTS5 (41), ESCO1, MED30, HSD17B1, VCAM1, C1RL, and APOL6 (Figs. 6H and S3B). ChIP-qPCR validated the binding of CEBPD to COL12A1, COL21A1, SMAD6, and ADAMTS5 promoter regions (Fig. 6I). Furthermore, the expressions of COL12A1, COL21A1, SMAD6, and ADAMTS5 were reduced in CEBPD-depleted osteoblasts (Fig. S3C). scRNA-seq analysis of human ALPLhigh/CD45low osteoblasts revealed the enrichment of many CEBPD-regulated genes that are expressed in osteoblasts at different maturation stages (Figs. 6J and S3D). Among them, COL12A1 was enriched in osteoblasts, SMAD6 and C1RL were enriched in preosteoblasts, and SMIM14 and FBXO32 were enriched in all types of osteoblasts (Figs. 6J and S3D). Taken together, these results strongly connect CORIN and SDC1 expression to CEBPD activity, which plays crucial roles in modulating human osteoblast differentiation and physiology.
Discussion
Bone diseases resulting from dysregulation of bone homeostasis, whether congenital or acquired, represent a significant public health issue. These diseases, particularly those associated with bone fractures and large bone defects, are a leading cause of disability, and effective bone regeneration is critical to recovery. While decades of applied research in bone biology have resulted in improved preventive and therapeutic interventions, many of which are pharmacologic, the incidence of illness, disability, and mortality caused by bone diseases remain high. One of the main factors contributing to this challenge is the limited foundational understanding of the cells that regulate human bone homeostasis. Osteoblasts are bone-forming cells that are one of the key cell types in regulating skeletal bone development by constructing mineralized bone matrix. Although mouse and human osteogenesis share many similarities, there are important differences between human and mouse bone development. The genetic and molecular mechanisms involved in bone development differ between species, with critical genes playing a role in human bone development that are not present in mice, and vice versa. Limited access to human bone tissue further complicates our understanding of human osteoblasts, making it challenging to validate findings from in vivo rodent model systems. Currently, with well-established hPSC culture and osteogenic lineage differentiation methods, functional human osteoblasts can be generated for in vitro studies. This powerful platform enables the identification of critical osteoblast membrane proteins and their regulated transcription factors. Our research has revealed that both CORIN/p38 MAPK/CEBPD and SDC1/p38 MAPK/CEBPD axis integrate extrinsic and intrinsic signaling to determine osteoblast cell fate and govern human osteoblast identity. These findings provide important insights into the fundamental mechanisms underlying human osteoblast regulation, which may lead to the development of novel preventive and therapeutic interventions for bone diseases.
CORIN is known to convert ANP precursors to mature ANP and is crucial to maintaining electrolyte balance and blood pressure (5). Our transcriptome profile and IHC analyses interestingly revealed that CORIN is highly expressed in human osteoblasts/bone but not mouse osteoblasts/bone. Clinical studies have shown that reduced serum CORIN levels are associated with osteoporosis (42), suggesting that dysregulation of CORIN could lead to impaired bone homeostasis and bone disease development. However, the precise roles of CORIN in bone formation have yet to be fully elucidated. In this study, we investigated the expression and function of CORIN in hPSC-derived osteoblasts. Our results showed that CORIN is highly expressed in these cells and its depletion leads to impaired osteogenic differentiation and bone mineralization. Further analysis revealed that p38 MAPK-mediated CEBPD upregulation was responsible for the observed effects of CORIN on osteoblast function. Previous studies have shown that ANP/natribretic peptide receptor 3 can activate p38 MAPK through cGMP-regulated protein kinase G and its downstream regulator peroxisome proliferator-activated receptor gamma coactivator 1-alpha, which plays a role in human brown adipocyte-associated thermogenicity (43). These findings provide one potential mechanism explaining the association between ANP, the natribretic peptide receptor 3 G-protein-linked receptor, and p38 MAPK, and explain how CORIN is able to activate p38 MAPK. Further studies are necessary to comprehensively dissect the role of CORIN in bone development, including the downstream transcription factors and CORIN-mediated signaling pathways involved. Such further work is essential to the identification of potential therapeutic targets to treat bone diseases.
CEBPD is a transcription factor that belongs to the CEBP family and plays a crucial role in regulating various cellular processes such as cell differentiation, proliferation, growth arrest, cell death, metabolism, and immune response (44). Previous research has indicated that CEBPD is upregulated in osteoblasts through α1B-adrenoceptor signaling, which can modulate bone formation (45). Moreover, CEBPB and CEBPD have been found to not only synergize with Runx2 to upregulate bone-specific genes such as osteocalcin (46), but also regulate the expression of IGF1, a critical factor involved in skeletal growth, osteoblast proliferation, and differentiation (47). However, the upstream signals regulating CEBPD and how the CEBPD transcriptome profile contributes to osteogenesis remain poorly understood. In this study, we have identified CORIN and SDC1-mediated p38 MAPK signaling as key regulators of CEPBD expression during osteogenesis. We found that depletion of CORIN, overexpression of SDC1, or inhibition of p38 MAPK impaired osteoblast differentiation and bone mineralization, likely due to the downregulation of CEBPD. scRNA- sequencing analysis of human osteoblastic lineage cells (preosteoblasts, osteoblasts, and undifferentiated osteoblasts) revealed enriched expression of CEBPD in all three cell types, indicating its involvement in all stages of bone development. RNA-seq and ChIP-seq analysis of CEBPD regulatory targets showed that many CEBPD-regulated genes are involved in extracellular matrix organization, regulation of ossification, and BMP/Wnt signaling. Our results suggest that CEBPD plays a versatile role in regulating osteogenesis through the modulation of multiple signaling molecules associated with bone development, providing valuable insights for reevaluating CEBPD's potential in human osteogenesis.
In summary, we used an hPSC-based culture and differentiation platform to generate functional human osteoblasts, define their cell membrane proteins CORIN and SDC1 and the p38 MAPK signaling pathway regulating their activity, and identify their downstream effectors: a transcription factor CEBPD that controls human osteogenesis. In particular, CORIN is a membrane protein that specifically regulates osteogenesis in humans. These findings are a monumental addition to our foundational understanding of the cellular signaling networks that govern the determination of human osteoblast identity, paving the way for novel developments in bone regeneration technology and clinical therapies for osteoblast-mediated diseases in humans.
Experimental procedures
hESC/hiPSC culture
The hESC H1 and H9 lines were obtained from WiCell, while the HES2 line was generously gifted by Dr Ihor Lemischka from the Icahn School of Medicine at Mount Sinai. The hiPSC lines WT-1H, WT-1G, WT-1J, WT-F6, and WT-F37 were generated by reprogramming health donor fibroblasts using the CytoTune-iPS 2.0 reprogramming kit (Invitrogen, A16517) with four reprogramming factors (OCT4, SOX2, KLF4, and MYC), as detailed previously (14, 16, 18). All hESCs and hiPSCs were cultured and maintained in mTeSR1 medium, following established protocols (48, 49). hPSC-derived MSCs and hFOB cells (American Type Culture Collection, CRL-11372) were cultured in the MSC medium (α-minimal essential medium supplemented with 10% fetal bovine serum and L-glutamine). Adipose-derived human mesenchymal stem cells (ad-HMSCs; ScienCell Research Laboratories) and bone marrow-derived human mesenchymal stem cells (bm-HMSCs; ScienCell Research Laboratories) were maintained in mesenchymal stem cell medium (ScienCell Research Laboratories).
Differentiation of hESCs/hiPSCs to MSCs and then osteoblasts
The in vitro differentiation of hESCs/hiPSCs into MSCs using bFGF/PDGF-AB for hESCs and SB-431542/7.5% for hiPSCs was previously described (14, 16). The differentiated MSCs were characterized using CD73, CD105, and CD166, which are established markers for MSCs. To induce osteogenic differentiation, hPSC-derived MSCs, ad-HMSCs, bm-HMSCs, and hFOB cells were seeded in a 6-well plate at a density of 2 × 104 cells per well and cultured in the osteogenic differentiation medium (α-minimal essential medium supplemented with 10% fetal bovine serum, 10 mM β-glycerol phosphate, 200 μM ascorbic acid, and 0.1 μM dexamethasone). Osteogenic differentiation was assessed at various time points (day 0 to day 30) to monitor the progression of differentiation.
Alizarin Red S staining
Alizarin Red S staining was used to detect bone mineralization by mature osteoblasts. Differentiated cells were washed twice with 1×Dulbecco's phosphate-buffered saline and then fixed with ice-cold 70% ethanol at room temperature for 15 min. After fixation, cells were washed twice with tap water and then stained with 2% (w/v) ARS Solution (Sigma-Aldrich, A5533; pH 4.2) for 30 min with gentle rotation on a rotor. Following the staining process, the cells were washed with tap water five times. The stained cells were then visualized under a microscope, using the H-filter in color mode to capture images.
Flow cytometry analysis
In total, 14.7 × 104 MSCs were seeded onto 10 cm culture dishes and cultured in the osteogenic differentiation medium for 18 days. At day 18 (D18), the differentiated osteoblasts and MSCs were incubated with 2 ml of 0.1% Collagenase Type II (Invitrogen, 17101015) at 37 °C for 30 min, followed by an additional 5-min incubation with 1 ml of 0.25% Trypsin-EDTA (GenDEPOT, CA014-010). The cells were then washed with 1×PBS and filtered with a 40 μm cell strainer (Thermo Fisher Scientific 22-363-547). To analyze the expression of surface receptors CORIN and SDC1, the detached cells were stained with human CORIN antibody (R&D System, AF2209) and PE-conjugated human SDC1 (BD Bioscience, 552026). Alexa fluor 488 AffiniPure donkey anti-goat immunoglobulin G (IgG) (H+L) (Jackson ImmunoResearch, 705-545-003) was used to detect CORIN primary antibodies. Nonimmune isotype controls were used as negative controls. The flow cytometry results were analyzed with FlowJo software (https://www.flowjo.com).
Plasmids, shRNAs, and sgRNAs
The design of lentiviral sgRNAs and shRNAs was performed using the Benchling (https://benchling.com) and TCR library databases (https://portals.broadinstitute.org/gpp/public/). These sequences were then inserted into lentiCRISPR v2 (Addgene, 52961) and pLKO.pig vectors (50, 51), respectively. The primer sequences used for generating sgCEBPD and shCORIN are provided in Table S1. To create the TetO-SDC1 construct, pTT5-hSDC1 (Addgene, 52326) was first digested with PmeI and BamHI. The resulting fragment was then inserted into an AfeI- and BamHI-digested TetO-8MCS vector (52). Lentiviral sgRNAs and shRNAs were produced in HEK-293T cells using pMD2.G (Addgene, 12259) and psPAX2 (Addgene, 12260) packaging vectors.
Gene KO, knockdown, and ectopic expression
For gene KO and knockdown, sgRNA and shRNA lentiviral particles were generated in HEK-293T cells by transfecting pMD2.G (Addgene, 12259) and psPAX2 (Addgene, 12260) packaging vectors along with lentiCRISPR v2 sgCEBPD or pLKO.pig shCORIN using Lipofectamine 3000 reagent (Thermo Fisher Scientific). After 48 h of posttransfection, viral particles were harvested in polypropylene microfuge tubes and used to infect/transduce the differentiated osteoblasts in the presence of 8 μg/ml polybrene. Infected osteoblasts were then selected in the culture medium containing 2 μg/ml puromycin for 3 days to generate indicated KO or knockdown cells. To ectopically express SDC1 in osteoblasts, TetO-SDC1 and M2rTA (Addgene, 20342) lentiviruses were packaged using pMD2.G and psPAX2, as described above. Inducible SDC1 osteoblasts were generated by coinfecting osteoblasts with TetO-SDC1 and M2rTA lentiviruses at a multiplicity of infection of 10, treating them with 1 mg/ml doxycycline, and selecting with 2 μg/ml puromycin for 3 days. The expression of CEBPB, CORIN, and SDC1 was confirmed through immunoblotting using their respective antibodies.
RT-qPCR
Total mRNA was isolated using the TRIzol reagent (Invitrogen, 15596026) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from the isolated mRNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, 1708891). For each PCR reaction, 1 μl of cDNA, 1 μl of each 10 μM forward and reverse qPCR primers, 10 μl of SYBR Green PCR Master Mix (Bio-Rad Laboratories, 1725124), and 7 μl of RT-PCR grade water were mixed to make a total volume of 20 μl. RT-qPCR was carried out using a CFX96 machine (Bio-Rad Laboratories). All reactions were performed in triplicate and normalized to GAPDH expression. The primer sequences are provided in Table S2.
GelMA disc preparation
Gelatin Type A, methacrylic anhydride, PBS, and 2-hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959) were obtained from Sigma-Aldrich. Dialysis tubing (SnakeSkin, 10K MWCO) was purchased from Thermo Fisher Scientific. To synthesize GelMA, 3 g of gelatin Type A was dissolved in 30 ml of PBS solution at 50 °C, followed by dropwise addition of 1.739 ml of methacrylic anhydride to initiate the methacrylation of gelatin, and the reaction was allowed to proceed for 1 h. The reaction was terminated by adding an additional 30 ml of PBS solution, and the mixture was dialyzed at 50 °C for 48 h. After dialysis, the pH value was adjusted to 7.0 to 7.4, and the solution was freeze-dried by a lyophilizer (FreeZone 2.5, Labconco) for at least 72 h. For the fabrication of GelMA discs, 100 mg of freeze-dried GelMA was added to 1 ml of PBS containing 1% (w/v) of Irgacure 2959 as the photoinitiator at 50 °C. The solution was dispensed into a polydimethylsiloxane mold for casting the GelMA discs with micropatterns designed by SolidWorks. GelMA was cross-linked by UV-light (365 nm) for 300 s, and the mold was then transferred to an ice bath for 5 min to further stabilize the discs. The cross-linked discs were removed from the mold and stored in PBS, then placed in an incubator at 37 °C for 24 h to ensure stability. The resulting GelMA discs were stored at 4 °C for further cell culture experiments. The porous structure of each GelMA disc was observed by scanning electron microscopy (Vega 3, TESCAN). The hydrated GelMA discs were lyophilized and attached to a specimen stage with carbon tapes, followed by gold sputtering with a 20 nm gold layer. The operating voltage was 20 kV with a beam intensity of 10.0 and the working distance was 15 mm. The size distribution was calculated from a population of 50 holes with ImageJ software (https://imagej.net/ij/).
Microcomputed tomography scanning and analysis
To investigate bone formation in vitro, 5 × 105 MSCs were seeded onto each GelMA disc. After a 2-h incubation period at 37 °C to allow for cell infiltration, the MSC-laden GelMA discs were cultured in 1 ml of osteogenic differentiation medium. The medium was replenished every 3 days. After 3 or 6 weeks of culture, the GelMA discs were fixed in 4% paraformaldehyde at 4 °C for 3 days and subsequently washed with PBS. The fixed samples were kept hydrated by placing them into PCR tubes containing 200 μl PBS. Viva CT 40 (Scanco Medical) was used to scan the samples, and the scanning resolution (15 μm) and protocol were consistent for all samples in accordance with previous publications (53). The parameters used for GelMA disc analysis were Gauss = 0.8, Sigma = 1, and threshold = 163. 3D evaluation software BoneJ (https://bonej.org/) was used to generate all bone parameters.
Human tibia tissues
The specimens of normal tibia adjacent to osteosarcoma were collected from patients diagnosed with osteosarcoma of the femur who underwent femoral amputation as part of their clinical treatment. The samples were obtained with patient or parental informed consent and were surgically removed during the amputation procedure. These samples were acquired from the Department of Musculoskeletal Oncology at the First Affiliated Hospital of Sun Yat-sen University. Prior to their use in this study, the samples were deidentified.
Immunohistochemistry staining
Bone tissue, obtained from C57BL/10J mice sacrificed at 4 months of age or human tibia as described above, was decalcified with 10% EDTA and 1% sodium hydroxide (PH 7.0) for 4 weeks, and then paraffin-embedded as previously described (53). Sections were cut at 5 μm thickness. Prior to staining, the sections were deparaffinized with xylene and hydrated with a gradient of ethanol and water. IHC was performed following the previously reported protocol (54). Briefly, after deparaffinization, the slides were washed with PBS and blocked with 5% donkey serum. Primary antibodies, including anti-human CORIN (1:1000) (R&D Systems, AF2209), anti-human SDC1 (DL-101) (1:50) (Santa Cruz Biotechnology, sc-12765), anti-mouse CORIN (Abnova, PAB12762), and anti-mouse SDC1 (MilliporeSigma, ZRB1360), as well as anti-CEBPD (1:500) (Santa Cruz Biotechnology, sc-365546x), were applied and incubated overnight at 4 °C. The following day, sections were incubated with 0.5% H2O2 in PBS for 30 min at room temperature and then washed with PBS. The corresponding secondary biotinylated horse anti-goat IgG (1:300) (Vector Laboratories, BA9500), goat anti-mouse IgG (1:300) (Vector Laboratories, BA9200), and goat anti-rabbit IgG (H+L)-horseradish peroxidase (HRP) (1:300) (Bio-rad, #1706515) were applied and incubated. After washing with PBS, the slides were incubated with ABC reagents (PK 7200, Elite ABC kits, Vector Laboratories) for 30 min at room temperature. Then, the DAB staining kit (SK-4100, Vector Laboratories) was used to reveal positive staining. Hematoxylin QS (Vector Laboratories, H3404) was used to counterstain the nucleus. The slides were dehydrated through an ethanol gradient, cleared with xylene, and sealed with Cytoseal. Images were captured using a NIKON CI microscope (Nikon).
Immunoblotting
Whole-cell extracts were collected from MSC-differentiated osteoblasts (D18) in complete 1× radioimmunoprecipitation assay buffer (150 mM NaCl, 20 mM Tris pH 7.4, 1% NP-40, and 0.1% SDS) with HALT protease inhibitor cocktail (Thermo Fisher Scientific). Proteins were separated by electrophoresis on a 6 to 12% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 5% milk in TBS with 0.5% Tween-20 for 30 min and then incubated overnight at 4 °C with primary antibodies, including anti-CEBPD Antibody (C-6) (Santa Cruz Biotechnology, sc-365546), anti-β-ACTIN (AC-15) (Sigma-Aldrich, A5441), human CORIN antibody (R&D Systems, AF2209), phospho-p38 MAPK antibody (Thr180/Tyr182) (D3F9) (Cell Signaling Technology, 4511T), and p38 MAPK (D13E1) (Cell Signaling Technology, 8690T). The secondary antibodies used were goat anti-mouse IgG & IgM HRP conjugated antibody (EMD Millipore, AP130P), goat anti-rabbit IgG HRP Conjugate antibody (Bio-Rad, 1706515), and rabbit anti-goat IgG HRP conjugated antibody (EMD Millipore, AP107P). The images were obtained using enhanced chemiluminescence Western blotting detection reagents (GE HealthCare, RPN2209).
Inhibition of p38 MAPK pathway by SB202190 treatment
MSC-derived osteoblasts were treated with the p38 MAPK inhibitor SB202190 (Selleckchem, S1077) at a concentration of 200 nM/ml for 24 h. Cells were then collected for immunoblotting analysis. For ARS staining, MSC-derived osteoblasts were cultured in the osteogenic differentiation medium with or without SB202190 for 4 weeks.
RNA-seq
Time course samples of hPSC-derived MSCs and osteoblasts were collected at the indicated days of differentiation. Cell samples were lysed in TRIzol reagent. The RNA sample preparations and RNA-seq data analyses were performed as previously described (48).
Single-cell RNA-seq data processing
The raw read count data from GSE147390 was imported and processed with the Seurat package according to their standard pipeline (55). The SCTransform function was used to perform regularized negative binomial regression and normalize UMI count data utilizing the top 2000 variable genes, while the cells with <200 genes or >5000 genes were removed. RunPCA was used to perform principal component analysis. The first 18 principal components were used for uniform manifold approximation and projection dimension reduction. The cells with a resolution of 0.1 were clustered. Based on the feature plot of preosteoblasts, osteoblasts, and undetermined osteoblasts, Clusters 3, 4, and 6 (all representing nonosteoblast cells) were excluded.
ChIP-seq
The CEBPD ChIP assay was performed in accordance with previously published methods albeit with some modifications (16, 48). Briefly, cells were pretreated with 1.5 mM disuccinimidyl glutarate (DSG; Thermo Fisher Scientific, 20593) for 30 min and then cross-linked with 4% paraformaldehyde (Thermo Fisher Scientific, 28906) for 10 min at room temperature. Subsequently, the cells were cross-linked in 1% formaldehyde for an additional 10 min. After glycine quenching, cell pellets were lysed and collected, and the DNA was sonicated using a Branson Sonifier 450 under the following conditions: 200 cycles, power 10%, 10 s on, and 10 s off. The supernatant was diluted with the same buffer and subjected to immunoprecipitation using CEBPD antibodies (Santa Cruz Biotechnology, sc-365546x) or IgG at 4 °C overnight. The beads were washed, and the DNA was reverse cross-linked and purified.
ChIP-seq analysis and identification of promoter regions
The ChIP-seq analysis method was previously reported (16, 48). The peak-calling for the ChIP-seq data was conducted using MACS2 with default parameters. The genomic annotations, such as the labeling of promoter regions, and motif analyses were performed using Homer.
Enrichr and GSEA analyses
GO_BP pathway analysis, BioPlanet pathway analysis, Mouse Gene Atlas, and MGI mammalian phenotype analysis were performed using Enrichr (https://maayanlab.cloud/Enrichr/). GSEA analysis was performed using default the GO_BP gene set. The analyzed results were considered significant with the settings of false discovery rate (FDR) q-value less than 0.25 and nominal (NOM) p-value less than 0.05.
Quantification and statistical analysis
Statistical analysis was performed using Prism 8.0 (https://www.graphpad.com/), and the difference between two groups was compared by two-tailed unpaired Student’s t test or ANOVA to calculate the p-value. The results were expressed as the mean ± SD.
Data availability
The data supporting the findings of this study are included in the article and its supporting information. In addition, the RNA-seq and ChIP-seq data can be accessed in the Gene Expression Omnibus (GEO) repository under accession number GSE202147.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to express our gratitude to Drs Jie Su and Brendan Lee, Brian C. Dawson, and members of the Lee laboratory for their valuable technical assistance and insightful discussions. We also thank Dr Zhen Sun from the Department of Medicine at Baylor College of Medicine for providing liver tissue. lentiCRISPR v2 was a gift from Feng Zhang (Addgene, 52961). pTT5-hSDC1 was a gift from Gordon Laurie (Addgene, 52326). pMD2.G was a gift from Didier Trono (Addgene, 12259). psPAX2 was a gift from Didier Trono (Addgene, 12260).
Author contributions
D. Z., M.-F. H., A. X., X. G., Y.-W. H., T. T. T. P., L. L., T.-Y. C., L. K. P., J. T., Z. H., R. S., J. K., L. L. W., and R. Z. investigation; D. Z., M.-F. H., A. X., X. G., Y.-W. H., T. T. T. P., L. L., L. K. P., J. T., Z. H., L. L. W., R. Z. and D.-F. L. conceptualization; D. Z., M.-F. H., A. X., X. G., J. T., R. S., J. W., C. G. A., J. S., methodology; D. Z., M.-F. H., T. T. T. P., J. A. G., D. A. B., and D.-F. L. visualization; D. Z., M.-F. H., A. X., and X. G., validation; D. Z. L. K. P., J. A. G., R. Z., and D.-F. L. writing–original draft; M.-F. H., A. X., L. K. P., Y. Z., Y. D., and Z. Z. formal analysis; M.-F. H., J. A. G., D. A. B., R. Z., and D.-F. L. writing–review and editing; A. X., R. Z., and D.-F. L. project administration; A. X. funding acquisition; T.-Y. C., J. W., C. G. A., J. S., and J. K., resources; Y. D. and Z. Z. data curation; Y. Z., R. Z., and D.-F. L. supervision.
Funding and additional information
This work was mainly supported by UTHealth Houston start-up funds (D.-F. L. (37516-11998) and R. Z. (37516-11999)). D. Z. was supported by DoD Horizon Award (W81XWH-20-1-0389). A. X. was a CPRIT Postdoctoral Fellow in the Biomedical Informatics, Genomics and Translational Cancer Research Training Program (BIG-TCR, CPRIT grant RP210045). J. W. was supported by R01 HL14270. Z. Z. was partially supported by NIH/NLM (R01LM012806) and CPRIT (RP180734 and RP210045). L. L. W. was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development [HD42136], the Doris Duke Charitable Foundation Clinician Scientist Development Program, The Rolanette and Berdon Lawrence Bone Disease Program of Texas, the BCM Curtis and Doris K. Hankamer Foundation Collaborative Research Grant, the Amschwand Sarcoma Cancer Foundation, the Kurt Groten Family Research Scholar’s Program, and the Gillson Longenbaugh Foundation. Y. Z. was supported by the Guangdong Basic and Applied Basic Research Foundation (2021A1515220115). D.-F. L. was supported by R01 CA246130, R01 HL14270, and the Rolanette and Berdon Lawrence bone disease program of Texas, and the Pablove Foundation childhood cancer research grant (690785). D.-F. L. is a CPRIT Scholar in Cancer Research.
Reviewed by members of the JBC Editorial Board. Edited by Paul Shapiro
Contributor Information
Yang Zhang, Email: zhangyang07@hit.edu.cn.
Ruiying Zhao, Email: ruiying.zhao@uth.tmc.edu.
Dung-Fang Lee, Email: dung-fang.lee@uth.tmc.edu.
Supporting information
References
- 1.Huang W., Yang S., Shao J., Li Y.P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. 2007;12:3068–3092. doi: 10.2741/2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathews S., Bhonde R., Gupta P.K., Totey S. Extracellular matrix protein mediated regulation of the osteoblast differentiation of bone marrow derived human mesenchymal stem cells. Differentiation. 2012;84:185–192. doi: 10.1016/j.diff.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y.H., Jewell B.E., Gingold J., Lu L., Zhao R., Wang L.L., et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol. Med. 2017;23:737–755. doi: 10.1016/j.molmed.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neve A., Corrado A., Cantatore F.P. Osteoblast physiology in normal and pathological conditions. Cell Tissue Res. 2011;343:289–302. doi: 10.1007/s00441-010-1086-1. [DOI] [PubMed] [Google Scholar]
- 5.Dong N., Niu Y., Chen Y., Sun S., Wu Q. Function and regulation of corin in physiology and disease. Biochem. Soc. Trans. 2020;48:1905–1916. doi: 10.1042/BST20190760. [DOI] [PubMed] [Google Scholar]
- 6.Ichiki T., Huntley B.K., Burnett J.C., Jr. BNP molecular forms and processing by the cardiac serine protease corin. Adv. Clin. Chem. 2013;61:1–31. doi: 10.1016/b978-0-12-407680-8.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan W., Wu F., Morser J., Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmen M., Hidding H., Gotte M., Khassawna T.E., Kronenberg D., Stange R. The heparan sulfate proteoglycan syndecan-1 influences local bone cell communication via the RANKL/OPG axis. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-77510-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C., Peall I.W., Pham S.H., Okolicsanyi R.K., Griffiths L.R., Haupt L.M. Syndecan-1 facilitates the human mesenchymal stem cell osteo-adipogenic balance. Int. J. Mol. Sci. 2020;21:3884. doi: 10.3390/ijms21113884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Lei Q., Wang H., Xu C., Liu T., Kong F., et al. Tumor-derived extracellular vesicles inhibit osteogenesis and exacerbate myeloma bone disease. Theranostics. 2019;9:196–209. doi: 10.7150/thno.27550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez J.R., Kouroupis D., Li D.J., Best T.M., Kaplan L., Correa D. Tissue engineering and cell-based therapies for fractures and bone defects. Front. Bioeng. Biotechnol. 2018;6:105. doi: 10.3389/fbioe.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marie P.J., Fromigue O. Osteogenic differentiation of human marrow-derived mesenchymal stem cells. Regen. Med. 2006;1:539–548. doi: 10.2217/17460751.1.4.539. [DOI] [PubMed] [Google Scholar]
- 13.Ozasa R., Matsugaki A., Matsuzaka T., Ishimoto T., Yun H.S., Nakano T. Superior alignment of human iPSC-osteoblasts associated with focal adhesion formation stimulated by oriented collagen scaffold. Int. J. Mol. Sci. 2021;22:6232. doi: 10.3390/ijms22126232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D.F., Su J., Kim H.S., Chang B., Papatsenko D., Zhao R., et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewell B.E., Xu A., Zhu D., Huang M.F., Lu L., Liu M., et al. Patient-derived iPSCs link elevated mitochondrial respiratory complex I function to osteosarcoma in Rothmund-Thomson syndrome. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu J., Huo Z., Yu Y., Zhu D., Xu A., Huang M.F., et al. Hereditary retinoblastoma iPSC model reveals aberrant spliceosome function driving bone malignancies. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2117857119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H., Kimura T., Swami S., Wu J.Y. Pluripotent stem cells as a source of osteoblasts for bone tissue regeneration. Biomaterials. 2019;196:31–45. doi: 10.1016/j.biomaterials.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., Yoo S., Zhou R., Xu A., Bernitz J.M., Yuan Y., et al. Oncogenic role of SFRP2 in p53-mutant osteosarcoma development via autocrine and paracrine mechanism. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11128–E11137. doi: 10.1073/pnas.1814044115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szeri F., Lundkvist S., Donnelly S., Engelke U.F.H., Rhee K., Williams C.J., et al. The membrane protein ANKH is crucial for bone mechanical performance by mediating cellular export of citrate and ATP. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen K.H., Frederiksen C.M., Burns J.S., Abdallah B.M., Kassem M. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J. Bone Miner Res. 2010;25:796–808. doi: 10.1359/jbmr.091018. [DOI] [PubMed] [Google Scholar]
- 21.Uchimura T., Hollander J.M., Nakamura D.S., Liu Z., Rosen C.J., Georgakoudi I., et al. An essential role for IGF2 in cartilage development and glucose metabolism during postnatal long bone growth. Development. 2017;144:3533–3546. doi: 10.1242/dev.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meixner A., Zenz R., Schonthaler H.B., Kenner L., Scheuch H., Penninger J.M., et al. Epidermal JunB represses G-CSF transcription and affects haematopoiesis and bone formation. Nat. Cell Biol. 2008;10:1003–1011. doi: 10.1038/ncb1761. [DOI] [PubMed] [Google Scholar]
- 23.Lu X., Beck G.R., Jr., Gilbert L.C., Camalier C.E., Bateman N.W., Hood B.L., et al. Identification of the homeobox protein Prx1 (MHox, Prrx-1) as a regulator of osterix expression and mediator of tumor necrosis factor alpha action in osteoblast differentiation. J. Bone Miner Res. 2011;26:209–219. doi: 10.1002/jbmr.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valadares E.R., Carneiro T.B., Santos P.M., Oliveira A.C., Zabel B. What is new in genetics and osteogenesis imperfecta classification? J. Pediatr. (Rio J.) 2014;90:536–541. doi: 10.1016/j.jped.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Li L., Lv S.S., Wang C., Yue H., Zhang Z.L. Novel CLCN7 mutations cause autosomal dominant osteopetrosis type II and intermediate autosomal recessive osteopetrosis. Mol. Med. Rep. 2019;19:5030–5038. doi: 10.3892/mmr.2019.10123. [DOI] [PubMed] [Google Scholar]
- 26.Numan M.S., Jean S., Dessay M., Gagnon E., Amiable N., Brown J.P., et al. Gene-environment interactions in Paget's disease of bone. Joint Bone Spine. 2019;86:373–380. doi: 10.1016/j.jbspin.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Simsa-Maziel S., Zaretsky J., Reich A., Koren Y., Shahar R., Monsonego-Ornan E. IL-1RI participates in normal growth plate development and bone modeling. Am. J. Physiol. Endocrinol. Metab. 2013;305:E15–E21. doi: 10.1152/ajpendo.00335.2012. [DOI] [PubMed] [Google Scholar]
- 28.Wu M., Chen G., Li Y.P. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4 doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graneli C., Thorfve A., Ruetschi U., Brisby H., Thomsen P., Lindahl A., et al. Novel markers of osteogenic and adipogenic differentiation of human bone marrow stromal cells identified using a quantitative proteomics approach. Stem Cell Res. 2014;12:153–165. doi: 10.1016/j.scr.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Nichol J.W., Koshy S.T., Bae H., Hwang C.M., Yamanlar S., Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook B., Rafiq R., Lee H., Banks K.M., El-Debs M., Chiaravalli J., et al. Discovery of a small molecule promoting mouse and human osteoblast differentiation via activation of p38 MAPK-beta. Cell Chem Biol. 2019;26:926–935.e6. doi: 10.1016/j.chembiol.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong Y., Yang J., Li X., Zhou C., Chen Y., Wang Z., et al. A systematic dissection of human primary osteoblasts in vivo at single-cell resolution. Aging (Albany NY) 2021;13:20629–20650. doi: 10.18632/aging.203452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eferl R., Hoebertz A., Schilling A.F., Rath M., Karreth F., Kenner L., et al. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004;23:2789–2799. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenner L., Hoebertz A., Beil F.T., Keon N., Karreth F., Eferl R., et al. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J. Cell Biol. 2004;164:613–623. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner E.F. Functions of AP1 (Fos/Jun) in bone development. Ann. Rheum. Dis. 2002;61(Suppl 2):ii40–ii42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komori T. Cbfa1/Runx2, an essential transcription factor for the regulation of osteoblast differentiationNihon Rinsho. 2002;60(Suppl 3):91–97. [PubMed] [Google Scholar]
- 37.Sims N.A. The JAK1/STAT3/SOCS3 axis in bone development, physiology, and pathology. Exp. Mol. Med. 2020;52:1185–1197. doi: 10.1038/s12276-020-0445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izu Y., Sun M., Zwolanek D., Veit G., Williams V., Cha B., et al. Type XII collagen regulates osteoblast polarity and communication during bone formation. J. Cell Biol. 2011;193:1115–1130. doi: 10.1083/jcb.201010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Hao X., Yin M., Xu T., Guo F. Long non-coding RNA in osteogenesis: a new world to be explored. Bone Joint Res. 2019;8:73–80. doi: 10.1302/2046-3758.82.BJR-2018-0074.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q., Wei X., Zhu T., Zhang M., Shen R., Xing L., et al. Bone morphogenetic protein 2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J. Biol. Chem. 2007;282:10742–10748. doi: 10.1074/jbc.M610997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang L., Lin J., Zhao S., Wu J., Jin Y., Yu L., et al. ADAMTS5 in osteoarthritis: biological functions, regulatory network, and potential targeting therapies. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H., Liu W., Zhu J., Liu M., Fang C., Wu Q., et al. Reduced serum corin levels in patients with osteoporosis. Clin. Chim. Acta. 2013;426:152–156. doi: 10.1016/j.cca.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Bordicchia M., Liu D., Amri E.Z., Ailhaud G., Dessi-Fulgheri P., Zhang C., et al. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J. Clin. Invest. 2012;122:1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko C.Y., Chang W.C., Wang J.M. Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J. Biomed. Sci. 2015;22:6. doi: 10.1186/s12929-014-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K., Hirai T., Kodama D., Kondo H., Hamamura K., Togari A. alpha1B -Adrenoceptor signalling regulates bone formation through the up-regulation of CCAAT/enhancer-binding protein delta expression in osteoblasts. Br. J. Pharmacol. 2016;173:1058–1069. doi: 10.1111/bph.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez S., Javed A., Tennant D.K., van Rees M., Montecino M., Stein G.S., et al. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 47.Umayahara Y., Ji C., Centrella M., Rotwein P., McCarthy T.L. CCAAT/enhancer-binding protein delta activates insulin-like growth factor-I gene transcription in osteoblasts. Identification of a novel cyclic AMP signaling pathway in bone. J. Biol. Chem. 1997;272:31793–31800. doi: 10.1074/jbc.272.50.31793. [DOI] [PubMed] [Google Scholar]
- 48.Xu A., Liu M., Huang M.F., Zhang Y., Hu R., Gingold J.A., et al. Rewired m(6)A epitranscriptomic networks link mutant p53 to neoplastic transformation. Nat. Commun. 2023;14:1694. doi: 10.1038/s41467-023-37398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou R., Xu A., Tu J., Liu M., Gingold J.A., Zhao R., et al. Modeling osteosarcoma using Li-fraumeni syndrome patient-derived induced pluripotent stem cells. J. Vis. Exp. 2018:57664. doi: 10.3791/57664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee D.F., Su J., Sevilla A., Gingold J., Schaniel C., Lemischka I.R. Combining competition assays with genetic complementation strategies to dissect mouse embryonic stem cell self-renewal and pluripotency. Nat. Protoc. 2012;7:729–748. doi: 10.1038/nprot.2012.018. [DOI] [PubMed] [Google Scholar]
- 51.Lee D.F., Su J., Ang Y.S., Carvajal-Vergara X., Mulero-Navarro S., Pereira C.F., et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu A., Huang M.F., Zhu D., Gingold J.A., Bazer D.A., Chang B., et al. LncRNA H19 suppresses osteosarcomagenesis by regulating snoRNAs and DNA repair protein complexes. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.611823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao X., Usas A., Tang Y., Lu A., Tan J., Schneppendahl J., et al. A comparison of bone regeneration with human mesenchymal stem cells and muscle-derived stem cells and the critical role of BMP. Biomaterials. 2014;35:6859–6870. doi: 10.1016/j.biomaterials.2014.04.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao X., Tang Y., Amra S., Sun X., Cui Y., Cheng H., et al. Systemic investigation of bone and muscle abnormalities in dystrophin/utrophin double knockout mice during postnatal development and the mechanisms. Hum. Mol. Genet. 2019;28:1738–1751. doi: 10.1093/hmg/ddz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are included in the article and its supporting information. In addition, the RNA-seq and ChIP-seq data can be accessed in the Gene Expression Omnibus (GEO) repository under accession number GSE202147.