Abstract
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR associated nuclease (Cas) system has been proven to play an irreplaceable role in bacteria immunity activity against exogenous genetic elements. In recent years, this system has emerged as a valid gene engineering method and could be used to detect and treat various microorganisms such as bacteria and viruses, etc. Staphylococcus aureus, as a Gram-positive, opportunistic human and animal pathogen, can cause a variety of diseases greatly threatening human health. Here, we mainly reviewed the applications of the CRISPR-Cas system in Staphylococcus aureus infections in detail. Furthermore, the prospects and drawbacks of the CRISPR-Cas system were also discussed.
Keywords: Staphylococcus aureus, CRISPR-Cas, Gene editing, Detection, Treatment
1. Introduction
Staphylococcus aureus (S. aureus) is a member of the normal human bacterial flora, but when the host is in a state of low immunity, it can cause food poisoning by generating toxins and induce a variety of infectious diseases, ranging from minor skin infections (e.g., sores, boils, and abscesses) to even life-threatening diseases (e.g., pneumonia, meningitis, and endocarditis) [1,2]. Except for the common core genome and the specific core-variable genes, most natural S. aureus chromosome consists of different types of mobile genetic elements (MGEs) that can be transferred between cells [3,4]. This genetic constitution facilitates genetic variation, pathogenic evolution, and adaptation to new environments, which potentially promotes the highly widespread of S. aureus and the acquisition of antimicrobial-resistant (AMR) genes [5]. Consequently, the large propagation of AMR genes can induce the occurrence of AMR bacteria, which brings a huge burden on the proper antimicrobial prescription and public health. For example, methicillin-resistant S. aureus (MRSA) is highly prevalent in nosocomial infections and shows high morbidity and mortality rates [6,7]. A systematic analysis focused on the global burden of bacterial antimicrobial resistance showed that MRSA caused more than 100,000 deaths and 3.5 million disability-adjusted life-years (DALYs) attributable to AMR in 2019 [8].
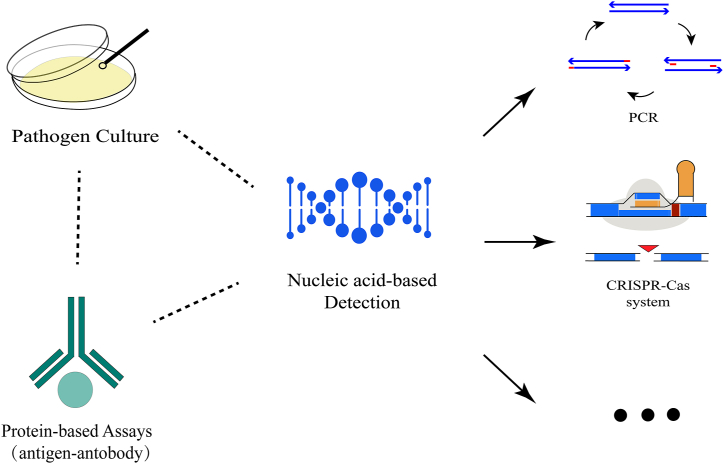
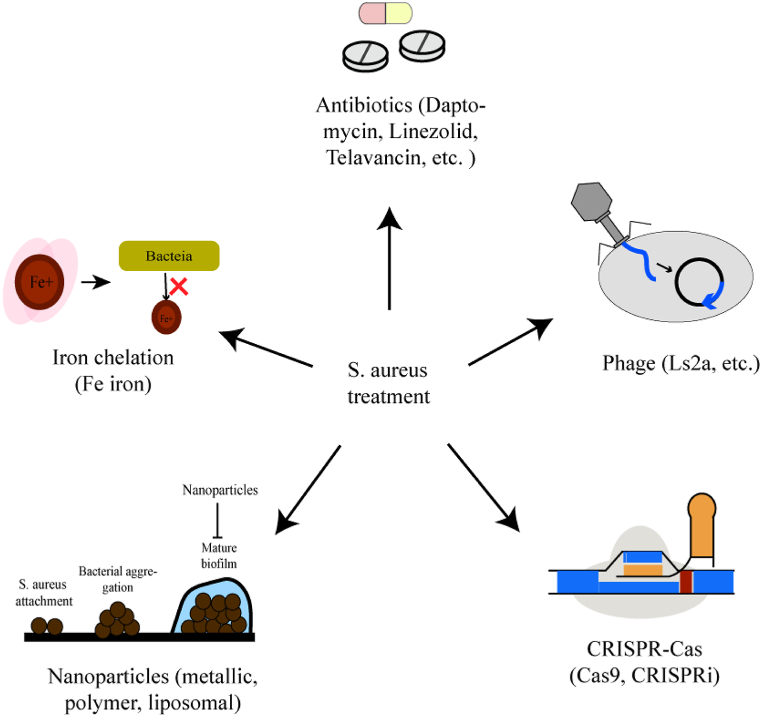
An accurate and rapid detection method for MRSA is deemed essential to guarantee the optimal administration of antibiotics and prevent the transmission and development of infectious diseases. Up to now, there are many methods to detect (pathogen culture, protein-based assays, and nucleic acid-based detection) [9,10] (Fig. 1) and treat S. aureus (antibiotics [11], iron chelation [12], phage [13,14], and nanoparticles [15]) (Fig. 2). However, most of them rely on expensive reagents, bulky and sophisticated equipment, and professional staff, rendering them unsuitable for use in resource-limited settings. As an emerging nucleic acid-based detection method, the CRISPR-Cas system recently caught more and more attention and was confirmed to have high sensitivity and specificity in pathogen detection [16]. What's more, a statistically significant relationship between the presence of the CRISPR-Cas system and the absence of the AMR genes in S. aureus has been proven [17], which indicates that the CRISPR-Cas system may inhibit the spread of the AMR genes and promote the development of the programable and sequence-specific antibiotics [[18], [19], [20], [21]].
Fig. 1.
The recent detection methods for S. aureus. To realize the early diagnosis, pathogen culture, protein-based assays, and nucleic acid-based detection (including PCR, CRISPR-Cas system, etc.) were developed.
Fig. 2.
The existing antimicrobial therapy in S. aureus. Recently, antibiotics, iron chelation, phage, and nanoparticles have been proven able to treat diseases caused by S. aureus.
The CRISPR-Cas system was first described in Escherichia coli in 1987 [22]. Later, researchers found this system exists in approximately half of the bacteria and almost all archaea [23]. It is reported that the CRISPR-Cas system is an adaptive immune system that defends against invasive genetic elements, such as viruses, bacteriophages, and plasmids [24]. Compared to traditional genetic manipulation methods, the CRISPR-Cas system provides a simple, sequence-specific platform to manipulate the gene of interest by generating a double-strand DNA break in the target genome, and consequently repairing the relevant break [25]. This process would enable the deeper investigation of uncharacterized genes responsible for bacterial virulence or resistance, allowing for more accurate diagnosis and targeted therapy for infectious diseases [26].
In this paper, we primarily summarized the characterizations of the CRISPR-Cas system and compared its different structure and function mechanisms. Then, its concrete applications in S. aureus were introduced from gene editing, nucleic acid detection, and antimicrobial therapy aspects. In addition, we generalized several current drawbacks and future research orientations.
1.1. Characterizations of the CRISPR-Cas system
The CRISPR-Cas system was proven to be a nucleic-acid-based immune system that comprises arrays of short, direct repetitive nucleotide sequences (repeats) and interspaced non-repetitive nucleotide sequences (spacers) [27], and uses RNA-guided nucleases to cleave invading mobile genetic elements (MGEs) [24,28]. In particular, the spacers, deriving from exogenous MGEs, were indispensable for specific and heritable immunity defense against ever-present invasion [[29], [30], [31]].
The function process of the CRISPR-Cas system can be summarized into three steps: adaption, CRISPR RNA (crRNA) maturation, and target interference [32]. Firstly, the invasive MGEs are cleaved by synthesizing corresponding proteins during the adaption stage [33]. Then, the snippets of exogenous MGEs, also termed new spacers, are often non-randomly captured and inserted into the genomic CRISPR array [[34], [35], [36]] under the participation of the proteins [37]. In the crRNA mature stage, the CRISPR array is first transcribed to a long precursor CRISPR RNA (pre-crRNA), which is further trimmed into individual and shorter mature crRNAs by enzymatic activity [27,38,39]. For instance, the Cas9 protein processes pre-crRNA acquiring the trans-activating crRNA (tracr-RNA) and RNase Ⅲ, while the Cas12 and Cas13 proteins process the pre-crRNA themselves [[40], [41], [42], [43], [44]]. Besides, the location of the spacer determines the ability of immunity, and the crRNA performs a stronger expression level and immunity activity when the spacer is closer to the leader region [45]. Consequently, during the interference stage, mature crRNAs interact with one or more Cas proteins to form an effector complex that recognizes the same or very similar sequences in the genome of the invasive MGEs and specifically cleaves and degrades exogenous MGEs [32,34,46]. Then, to maintain the cell's integrity, the lethal cleavage is repaired by the non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway [[47], [48], [49]].
According to differences in Cas protein composition and sequence diversity among the effector complexes, the CRISPR-Cas system can be categorized into two classes (class 1 and class 2) and further subdivided into six types (type Ⅰ-Ⅵ) and several subtypes [32,50]. The class 1 system includes type I, III, and IV, and owns a multi-subunit-protein complex consisting of crRNA and multiple Cas proteins [32,39,50]. The class 2 system includes type II, V, and VI, and possesses a large single-effector protein complex consisting of a Cas nuclease and a guide RNA [50]. Due to its relatively simple structure, the class 2 system is more easily investigated and widely applied [39,51]. What's more, type I, II, and V systems recognize and cleave DNA, type VI edits RNA, and type III edits both DNA and RNA. The concrete function of the type Ⅳ system is unknown yet [52].
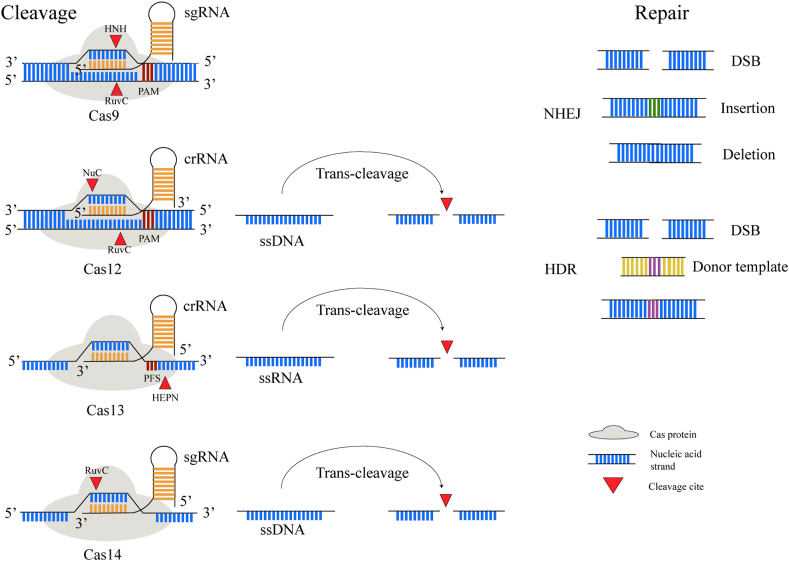
Every CRISPR-Cas system has a representative Cas protein and followingly a unique function process [53]. The detailed comparison is shown in Table 1 and the function process is illustrated in Fig. 3. Noticeably, recognizing a short protospacer adjacent motif (PAM) is indispensable for target binding and cleavage in some CRISPR-Cas systems such as type Ⅱ and type Ⅴ [54,55]. Likewise, the type Ⅵ system requires a relatively simple PAM-like sequence termed the protospacer flanking site (PFS) to direct specific cleavage of RNA [56,57]. Besides, Cas9 and Cas14 both require two RNAs: a mature crRNA and a partially complementary tracr-RNA [39,55,58]. The crRNA and tracr-RNA can also be artificially fused into a chimeric single guide RNA (sgRNA) which can recognize any target sequence of interest. Consequently, it is feasible to realize site-specific cleavage by re-programming the sgRNA sequence [59]. Furthermore, Cas12a, Cas13a, and Cas14a all perform cis cleavage to specifically cleave the target strand and trans cleavage to indiscriminately cleave the non-target strand, respectively [[60], [61], [62], [63], [64], [65], [66]].
Table 1.
The comparison of different Cas systems.
| Type |
Ⅱ |
Ⅴ |
Ⅵ |
|
|---|---|---|---|---|
| Cas protein | Cas9 | Cas12 | Cas14 | Cas13 |
| tracrRNA | Yes | No | Yes | No |
| Pre-crRNA processing | No | Yes | No | Yes |
| PAM/PFS | 3′, G-rich, NGG | 5′, T-rich, TTTV | TTTG | 3′, non-G (PFS) |
| Target substrate | dsDNA | dsDNA/ssDNA | dsDNA/ssDNA | ssRNA |
| Cleavage pattern | Blunt | Staggered | Nearly U or A | Nearly U or A |
| trans-cleavage activity | No | Yes (ssDNA) | Yes (ssDNA) | Yes (ssRNA) |
Note: N represents any nucleotide; V represents adenine, guanine or cytosine; PAM, protospacer adjacent motif; PFS, protospacer flanking site; dsDNA, double strand DNA; ssDNA, single strand DNA; ssRNA, single strand RNA.
Fig. 3.
The function process of the different CRISPR-Cas systems. The Cas9 system recognizes specific protospacer adjacent motif (PAM) sequences and performs cis cleavage to induce double-strand breaks (DSB). Particularly, Cas12, Cas13, and Cas14 also perform trans cleavage to accomplish indiscriminate nucleic acid strand cleavage. Then the resulting nucleic acid strand breaks are repaired through the non-homologous end-joining (NHEJ) pathway or homology-directed repair (HDR) pathway with a donor template.
1.2. CRISPR-cas system in S. aureus gene editing
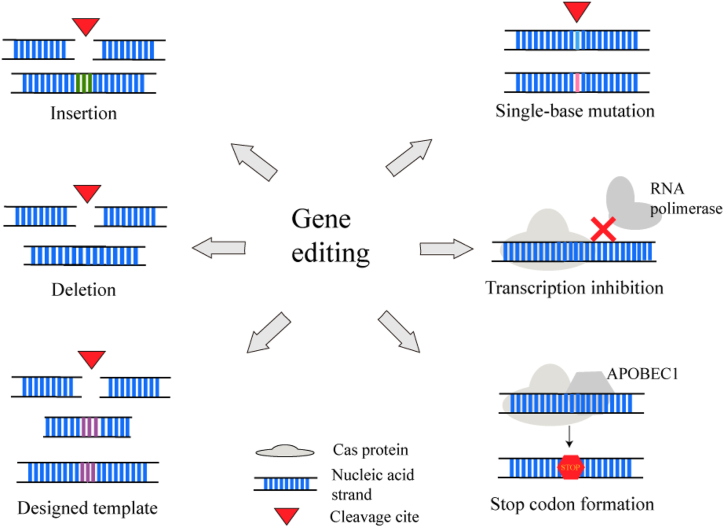
Efficient gene editing is essential for exploring and verifying the functional mechanisms of the uncharacterized genes or pathways responsible for pathogenicity and drug resistance. However, some conventional gene editing tools are labor-intensive, comparatively low-efficiency, and always leave a scar in the genome [9,10]. For that, a single-plasmid CRISPR-Cas9 genome editing tool in S. aureus was further developed and showed the ability of marker-free, scarless, and rapid genetic manipulation, which helps to study the gene function and pathogenicity molecular mechanism of S. aureus [7]. Noticeably, many gene applications, such as gene deletion or insertion, gene repression or inactivation, can be performed by using a programmed sgRNA/crRNA and combining the Cas protein's site-specific cleavage with the following repair process [67,68] (Fig. 4). Likewise, optimizing the gRNA and Cas protein can promote its gene editing efficiency [69].
Fig. 4.
The gene editing based on the CRISPR-Cas system in S. aureus. CRISPR-Cas system is capable of gene insertion by providing a designed template or not, gene deletion, single-base mutation, and transcription inhibition by restraining RNA polymerase connection or forming stop codon.
Based on the versatility of the CRISPR-Cas system, a CRISPR-Cas9 expression plasmid system (pCasSA) combined with the Streptococcus pyogenes Cas9 (SpCas9) was proven to be capable of gene indels, single-base substitutions, and showed high editing efficiencies and availability. Further research has proven that a highly efficient transcription inhibition system (pCasiSA) with mutation of the active sites of Cas9 protein can perform rapid and accurate screening of genes and pathways of interest in S. aureus, which may be helpful for gene characterization, enzymology, and drug development [70]. Besides, a CRISPR-Cpf1-mediated genome-editing (pCpfSA) system engineering Francisella novicida Cpf1 (FnCpf1) can perform multiplex gene editing and large-fragment DNA knockout by modifying the two crRNA expression cassettes and the corresponding donor templates. Surprisingly, this single-plasmid system provided more targetable sites and lower toxicity than pCasSA but with comparable editing efficiencies. However, given the multiplex-sites editing, some consideration should be taken: the length of the designed plasmids, and the repair process of multiplex editing [71].
Recently, the CRISPR interference (CRISPRi) system was found to be feasible for gene silencing, gene knockdown [5], and multiple genes repression simultaneously in S. aureus [58]. This system mostly uses a catalytically deactivated Cas9 (dCas9), in which amino acid mutations render two active sites completely inactive, but dCas9 still can bind the sgRNA [5,58,59,72]. As a consequence, the dCas9-sgRNA complex binds to the target gene and serves as a holdback of the elongating RNA polymerase, leading to obstruction of transcription initiation or elongation of target genes [73,74]. However, the unexpected toxicity of dCas9, also known as the “bad seed” effect, was observed at high dCas9 concentrations, which should be solved urgently [75,76].
Except above, there are also some Cas proteins or their combinations that can be involved in gene editing and exhibit high efficiencies and sensitivity. For example, a novel base-editing system, a CRISPR RNA-guided cytidine deaminase system (pnCasSA-BEC), was developed by engineering the fusion of a Cas9 nickase and a cytidine deaminase. This system can realize site-specific gene inactivation and point mutation in S. aureus via the guideline and cleavage of the Cas9 nickase, the conversion of C (cytidine) to U (uridine) through a deamination reaction without using repair templates or sacrificing transformation CFUs, and the occurrence of a premature stop codon. Given that, almost all the genes (98.81 %) of MRSA252 strains contain at least one PAM site and 68.8 % of the genes possess potential editable stop sites, the pnCasSA-BEC system can inactivate many genes in the S. aureus genome, thus promoting drug-target research in S. aureus or other microbes [[77], [78], [79]]. Next, a temperature-sensitive, two-vector system using single-stranded DNA (ssDNA) oligonucleotide recombineering with Cas9-mediated counterselection was developed to efficiently and precisely engineer point mutations and large single-gene deletions in S. aureus. Based on utilizing short, commercially synthesized synthetic DNA oligonucleotides as substrates, this system first transforms S. aureus through a recombinase to produce a recombinogenic strain. This system subsequently introduces the mutagenic oligonucleotide with the counterselection vector, and only cells realizing their successful recombineering are immune to lethal, double-stranded DNA breaks (DSBs). Furthermore, the system was proven to have excellent recombineering performance in multiple characterized strains (3 of 3 tested) and primary clinical isolates (6 of 6 tested). Given that the system proves a scalable, efficient, precise, and rapid tool, researchers will study the function mechanism of particular genes and specific mutations [[80], [81], [82]]. Overall, the abovementioned methods and previous research present the huge potential of the CRISPR-Cas9-based tool for gene editing in S. aureus [83].
1.3. CRISPR-cas system in S. aureus detection
Previous research has proven the high sensitivity and specificity of the CRISPR-Cas system in detecting S. aureus and MRSA. By combining the site-specific recognition and cleavage activity for the S. aureus representative genes with various signal output tools based on the indiscriminate trans cleavage, such as colorimetric signals [84], electrochemical signals [85], lateral flow strips signals [86], and fluorescence signals [87], visual and accurate detection results could be directly observed [88]. The detailed applications are shown in Table 2.
Table 2.
The application of the CRISPR-Cas system in S. aureus detection.
| Cas proteins | Target gene | Combining tools | Detection limitation | Detection time | Signal readouts | Practical application | References |
|---|---|---|---|---|---|---|---|
| Cas12a | nuc gene | ssDNA-FQ reporter | 5 copies/μL | 35 min | Fluorescence/strip | Water samples | [86] |
| Cas13a | nuc gene | PCR/T7 transcription | 1 CFU/mL | less than 240 min | Fluorescence | food samples | [89] |
| dCas9 | mecA gene (MARA) | SG I fluorescent probe | 10 CFU/mL | 30 min | Fluorescence | Clinical isolates | [87] |
| Cas14a | S. aureus cell | Specific aptamer, blocker DNA | 400 CFU/mL | 150 min | Fluorescence | Tilapia samples | [105] |
| Nucleic acid amplification | |||||||
| Cas12a | nuc/mecA gene | LAMP | 20 copies/μL | 60 min | Fluorescence | diabetic foot infectious patients samples | [93] |
| Cas12a | nuc gene | SRCA | 2.51 fg/μl for genomic DNA and 3 CFU/mL for S. aureus | 50 min | Electrochemical | Food samples | [85] |
| Cas12a | nuc gene | LAMP | 10 aM | 80 min | Fluorescence | clinical isolates | [92] |
| Cas12a | nuc gene | RPA | 102copies per reaction | 60 min | Strip | milk samples of the cow exhibiting clinical manifestations of mastitis | [94] |
| Cas12a | mecA gene | RPA | 8 CFU/mL | 15 min | Colorimetric | suspected MRSA isolates samples | [84] |
| Cas12a | mecA gene | RAA | 10 copies/μL | 60 min | Fluorescence | Clinical samples | [95] |
| Cas12a | Protein-A and PBP2a protein | RCA | 102 CFU/mL | 80 min | Fluorescence | sepsis blood samples | [97] |
| Cas12a | (sa)-16S rDNA | SDA | 0.473 fM | 80 min | Electrochemical luminescence | Human serum samples | [96] |
| Signal amplification | |||||||
| Cas12a | mecA gene | LAMP | 1aM | 85min | Strip | Bacterial suspension and clinical samples | [92] |
| Cas12a | mecA gene | silver metallization technology | 3.5 fM | 90min | Electrochemical | Human serum samples | [102] |
| Cas12a | mecA gene | magnetic relaxation switching sensor | 16 CFU/mL | 75 min | Transverse relaxation time | artificially contaminated food samples | [103] |
| Cas12a | femA gene | PCR and three logic gates | 103 CFU/mL | 120 min | Fluorescence | Milk samples | [107] |
| Cas12a | PBP2a protein | recycling signal amplification cascades | 102 CFU/mL | 45 min | Fluorescence | Skin and soft tissue infections samples | [98] |
| Cas12a | (sa)-16S rDNA | evanescent wave fluorescence enhancement | 13.2 CFU/mL | 90 min | Fluorescence | suspected clinical samples | [99] |
Note: ssDNA-FQ, single-stranded DNA-fluorophore-quencher; SG I fluorescent probe, SYBR Green I fluorescent probe; LAMP, loop-mediated isothermal amplification; SRCA, saltatory rolling circle amplification; RPA, recombinase polymerase amplification; RAA, recombinase-aided amplification; RCA, rolling circle amplification; SDA, strand displacement amplification; PBP2a protein, Penicillin-binding protein 2a.
The CCB-detection method (CRISPR-Cas13a-based bacterial detection) can detect the target genomic DNA (nuc gene) as low as 10° aM and showed a better linear range spanning from 10°-107 CFU/mL than the real-time quantitative PCR (105–109 CFU/mL) between the fluorescence intensity and S. aureus concentration. Its entire detection was completed within 4 h, which includes the extraction of genome DNA, specific gene amplification, in vitro transcription, the “collateral effect” cleavage, and the dequenching of fluorophores. It also demonstrated superior performance in real food samples including milk, juice, beer, and water with both known or unknown amounts of bacteria (spiked ones or non-spiked ones) [89]. Furthermore, the CRISPR-mediated DNA-FISH method, which combines CRISPR associated protein 9/single-guide RNA (dCas9/sgRNA) complex with SYBR Green I (SG I) fluorescent probe, can realize highly sensitive detection of MRSA with a detection limit of 10 CFU/mL within 30 min. This method also accurately distinguishes MRSA with the approximately 10–16 folds fluorescence intensity increase relative to that of MSSA. Besides, the target gene can be detected only by cell lysate without further gene separation and purification, which suggests this system may be applied in a relatively inexpensive point-of-care test (POCT) [87].
Due to its outstanding rapidity and simplicity, nucleic acid-based amplification technology caught more and more attention and researchers found that combining the amplification technology with CRISPR systems could largely magnify biosensing signals and promote its sensitivity [90,91]. The research that compared the sensitivity of three amplification methods including polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) found the CRISPR-Cas12a system coupled with LAMP showed 100 % specificity and 100 % sensitivity in 111 clinical isolates. Furthermore, the nuc-LAMP-Cas12a platform based on fluorescence readout and the mecA-LAMP-Cas12a platform based on strip readout respectively showed a limit of detection (LOD) of 10 aM (∼6 copies μL−1) and 1 aM (∼1 copy μL−1) [92]. Likewise, the another CRISPR-LAMP assay also showed 100 % specificity for the nuc gene and can accurately differentiate MRSA from 18 samples of diabetic foot infectious patients within 1 h, which suggests the CRISPR-LAMP method can be applied in more clinical diagnosis even in underprovided areas or at the POCT [93]. However, the CRISPR/Cas12a-LAMP system detecting the sea gene performed not well (a LOD of 104 copies of the plasmid containing the sea gene) in cow milk samples of bovine mastitis. In view of this, the CRISPR/Cas12a-RPA system coupled with the lateral flow assay was conducted and presented a LOD of 102 copies per reaction for the nuc gene and accurately identified S. aureus in 13 clinical isolates from cow milk exhibiting clinical manifestations of mastitis [94]. Then, a fluorescent biosensor integrating recombinase-aided amplification (RAA) and Cas12a system can detect S. aureus as low as 10 copies/μL in 1 h and distinguish MRSA from clinically common bacteria including Escherichia coli (E. coli), Staphylococcus epidermidis (S. epidermidis), Helicobacter pylori (H. pylori), Shigella sonnei (S. sonnei), Klebsiella pneumoniae (K. pneumoniae), and Salmonella typhimurium (S. typhimurium). Furthermore, its detection results in 83 clinical patient samples were coincident with that of antimicrobial susceptibility tests (AST) and PCR [95]. This suggests that the CRISPR-Cas system also performs excellently in clinical samples from different species and will play an irreplaceable role in the detection of pathogen and drug resistance genes, etc. Furthermore, a novel electrochemical biosensor combined saltatory rolling circle amplification (SRCA) with the CRISPR-Cas12a system has shown high sensitivity (the LOD was 2.51 fg/μL for genomic DNA and 3 CFU/mL for S. aureus). And this method also showed high specificity and reproducibility in distinguishing S. aureus from non-S. aureus bacteria and detecting S. aureus in food samples [85]. A large amount of substantial single-stranded DNA products (SP) was output after the cascade strand displacement amplification (SDA) and further repeated hybridization, cleavage, replacement, and other processes. Then the SP was combined with Cas12a/crRNA to form a Cas12a/crRNA/SP ternary complex, which activated its trans-cleavage ability and caused changes in the electrochemiluminescence (ECL) signal. The biosensor combining the two-stage amplification design demonstrated a wider linear range (1 fM to 10 nM), enhanced ECL luminescence efficiency, less false identification results, and a lower detection limit (0.473 fM). The high sensitivity and accuracy in the detection of real genome samples shows the CRISPR-Cas system may be applied in biomedical research [96].
Aptamers are a kind of single oligonucleotide fragments that can bind to targets such as proteins through specific interactions and then achieve the conversion from protein signals to nucleic acid signals. Integrating dual functionalized aptamer (PBP2a-specific aptamer and protein A-based aptamer) and CRISPR-Cas12a-assisted rolling circle amplification (RCA), this fluorescence detection tool can obtain the signal conversion and further dual signal amplification of the nucleic acid signals and demonstrated specific identification of MRSA and a linear correlation between the measured fluorescence intensity with MRSA concentration ranging from 102 to 106 CFU/mL [97]. With the specific aptamer, researchers have realized the conversion from nucleic acid detection to bacteria detection. In addition, a novel method comprising CRISPR-Cas12a-based cycling signal amplification cascades, including DNA polymerase-based target S. aureus release, ssDNA generation and the combination with CRISPR-Cas12a, showed accurate identification and sensitive quantitation of MRSA through PBP2a-specific aptamer in both clinical and experimental conditions and achieved a detection range from 102 to 106 CFU/mL [98]. Then, using triple sign amplification of RPA, CRISPR-Cas12a′s cleavage activity, and an aptamer-based (Ag+) colorimetric biosensor, a novel colorimetric detection method was shown to detect MRSA as low as 8 CFU/mL and represented high reliability, practicability, and results visualization in 12 suspected MRSA samples isolated from clinical patients [84]. Based on that, it's believed that the integrated methods will play a role in managing antimicrobial prescriptions and developing promising drug candidates in the future.
To further simplify the detection process and visually show detection results, some new methods combining the CRISPR-Cas system with several novel signal enhancement tools were developed and showed high sensitivity and specificity. For instance, a nucleic acid amplification-free quantitative detection method of pathogens, CRISPR-Cas12a-powered evanescent wave fluorescence nano-biosensing flatform (CREAT), consists of multiple signal enhancements, including nanophotonic structure-based evanescent wave fluorescence enhancement, Mg2+ or DNA-mediated fluorescence enhancement, air-displacement fluorescence enhancement, and the collateral cleavage activity of CRISPR-Cas system. The results demonstrated a LOD of 13.2 CFU/mL in 90 min, and a linear correlation between the fluorescence intensities and the S. aureus concentration measured by RT-PCR. However, the sample-to-answer time for this system was too long to achieve POCT and the clinical application was not performed [99]. A novel method based on RPA and CRISPR-Cas12a can acquire a shorter detection time (35 min, including 20 min genomic DNA amplification and 15 min trans-cleavage) and enhanced detection threshold (≥5 copies of pathogen DNA) by generating fluorescence signals with a single-stranded DNA-fluorophore-quencher (ssDNA-FQ) reporter or producing a naked-eye observed lateral flow strip with the destruction of a FITC and biotin-labeled ssDNA reporter. Taken together, this detection tool firstly was applied in the natural water environment [86]. Further, the cross-priming amplification (CPA) and CRISPR-Cas12a (CPA-Cas 12a) system integrating the paper-based strip with a microfluidic device can accurately detect S. aureus within 30 min with a LOD of 5 CFU/mL and realize portable, sensitive detection of S. aureus in bacterial suspension and 202 clinical samples. Given the high efficiency, portability and visualization, this system has great potential for POCT and clinical diagnostics [100]. Speaking of the electrochemical signals, a novel silver-enhanced E-CRISPR biosensor (E-Si-CRISPR) combining the silver metallization technology with the CRISPR-Cas12a was proven to achieve the amplification-free gene-based detection for the mecA gene in MRSA in 1.5 h. In the presence of the mecA gene, the cis- and trans-cleavage activity were performed, leading to degradation of the electrode's ssDNA surface layer, then the subsequent silver metallization and the measurement of final electrochemical signals via square ware voltammetry. As a consequence, the decreased electrochemical signal was positively proportional to the quantity of ssDNA remaining and thus the starting amount of the mecA gene, which could be inconvenient. In laboratory and practical applications (human serum samples), the E-Si-CRISPR methods can differentiate MRSA from other common bacteria even S. aureus with a LOD of 3.5 fM and linearity between 10 fM to 100 pM [101,102]. Furthermore, researchers developed a CRISPR-Cas12a-based magnetic relaxation switching (C-MRS) biosensor by synergistically combining the collateral activity of the CRISPR-Cas12a, on-particle rolling circle amplification, and ALP-triggered click chemistry into background-free MRS to achieve nucleic acid amplification-free and anti-contaminated detection for mecA gene of MRSA with a LOD of 16 CFU/mL. First, the crRNA specifically recognizes the mecA gene, leading to the cleavage for ssDNA of MNP-poly-alkaline phosphatase (MNP-poly-ALP) and the release of the fastened ALP. Then the freed ALP can engage in enzymatic activity, copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction and the formation of MNP30-MNP1000 complex, which has different saturation magnetization. Finally, the transverse relaxation time (T2) signal intensity by the MRS biosensor was proven to correspond to the unamplified mecA gene, and this excellent performance was still available in different food samples, such as eggs, milk, and pork. However, a highly simple and integrated strategy is still needed [103].
To reduce interference during the detection operation, researchers developed a contamination-free one-tube RPA-CRISPR/Cas12a system to detect MRSA, which firstly performed temporary separation of the two systems by adding RPA to the bottom of the tube and the CRISPR system to the cap, and then mixed by spinning after RPA reaction. The results demonstrated it could achieve specific MRSA detection in 20 min with a sensitivity of 10 copies for the fluorescence device and a range of 10–100 copies for the lateral flow strips. Then, the results in 23 clinical MRSA isolate samples also showed excellent consistency with qPCR (100 % and 95.7 % of the fluorescence and strips, respectively). All in all, the system is simple, non-polluting, inexpensive, rapid, and could potentially be applied to POCT [104]. Due to Cas14a′s small size, an aptamer-based Cas14a1 biosensor combining the aptamer that specifically binds to bacteria cells with the blocker for activation of Cas14a1/sgRNA was developed. When the live S. aureus is present, the blocker can be released and activate the Cas14a1 protein by binding with the sgRNA to generate a change of the fluorescent intensity. Thus, this method can distinguish live and dead bacteria accurately with a LOD of 400 CFU/mL for S. aureus. However, the comparatively higher LOD and longer reaction time (150 min) need to be solved rapidly and researchers aim to combine with other amplification methods and simplify detection procedure [105]. Different from the aforementioned methods, a signal-off Cas14a1 platform (SCOP) was established to efficiently detect MRSA by designing two specific primers that not only can induce the trans cleavage activity but also can be used for mecA gene amplification. In particular, those primers can be transformed into dsDNA without PAM site with PCR amplification in the presence of MRSA, resulting in the suppression of the trans-cleavage activity of Cas14a1 and thus the fluorescence signal turning off. Then, MRSA can be detected and the decreased fluorescent signal is proportional to the quantity of MRSA. Moreover, the SCOP showed high sensitivity (the calculated LOD of 1.23 ng/mL for genomic gene and accuracy for the mecA gene from infected biological samples and tilapia, which suggests the SCOP platform may be applied in broader fields [106]. Furthermore, the three 2-input elementary AND, OR, INHIBIT logic gates have been constructed to form a novel CRISPR-Cas12a-based tool, of which the LOD was 103 CFU/mL, and the dynamic range was 103–107 CFU/mL. Firstly, the genomic DNA is extracted and the femA gene of S. aureus is amplified through PCR amplification. Then, the amplified gene serving as input 1 and cognate crRNA serving as input 2 can initiate trans-cleavage of the reporter, leading to the cut of a fluorophore and a quencher modified ssDNA and further the emergence of fluorescent signals. This method also performs excellently in spiked milk samples and shows the possibility of developing intelligent bio-computer detection devices using the CRISPR-Cas system, which can be applied in larger areas such as food safety, disease diagnosis, and environment monitoring, etc [107].
1.4. CRISPR-cas system in S. aureus antimicrobial treatment
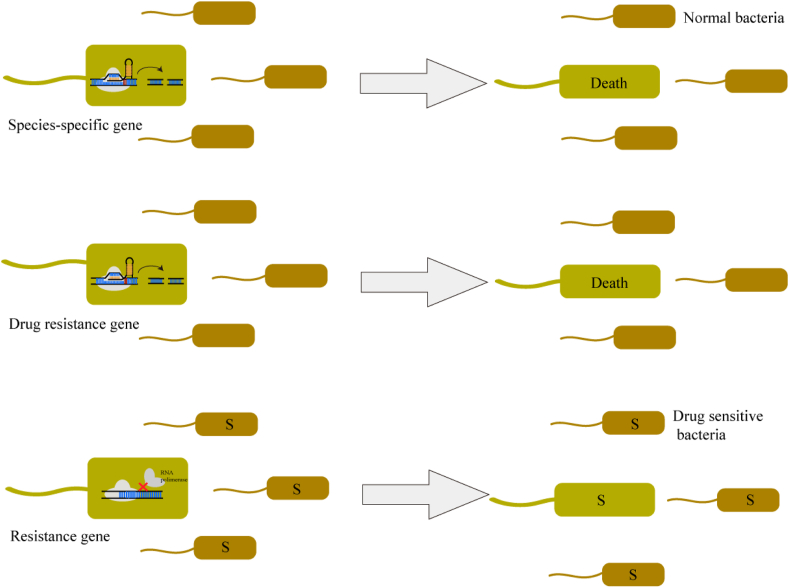
With the popularity of AMR, limiting broad-spectrum antibiotic abuse and selecting an individual antimicrobial treatment regimen are vital for shortening hospital time and impeding the spread of the resistance genes [98]. Due to its high gene editing efficiency and specificity, CRISPR-Cas plays an important role in anti-microbial therapy in several ways (Fig. 5). Firstly, it is feasible to cleave species-specific genes to result in the deployment of the target bacteria while maintaining the host's microbiome unimpressed [108]. Secondly, cleaving drug-resistant genes and eliminating relevant bacteria show high efficiency in decolonizing patients [109]. Thirdly, the CRISPR-Cas system can specifically modify or silence resistance genes to induce dysfunction in resistance genes while maintaining bacterial viability [37,110]. This process is defined as re-sensitization, which restores bacteria's susceptibility to antibiotics without damaging the patients' normal microbiota and can also be operated by curing plasmids carrying resistance genes [18,111,112].
Fig. 5.
The antimicrobial therapy based on the CRISPR-Cas system in S. aureus. Using the CRISPR-Cas system can cleave species-specific genes or drug resistance genes to induce bacteria death while keeping normal bacteria liveness. Besides, inhibiting resistance gene expression can recover bacteria's sensitivity to antimicrobials, termed re-sensitization.
Using Cas9 nuclease as a sequence-specific antimicrobial exhibited more efficiency in decolonizing patients of antibiotic-resistant bacteria including E. coli than other traditional therapies. However, the efficient delivery of Cas9 and its sgRNA into bacterial cells was a huge challenge. Given that the bacteriophages can naturally package their DNA into capsids and then host bacteria, researchers have chosen to deliver the Cas9 and sgRNA using a phagemid, which is designed to be packaged in phage capsids. A CRISPR-Cas9-based antimicrobial (pDB21mecA phagemid) programmed to target mecA genes was proven to selectively eradicate the clinical isolate USA300 strains in a mixed culture with RNΦ cells, of which the proportion dropped from 50 % before treatment to 0.4 % without cell death. Based on that, other CRISPR-Cas9 antimicrobials (pDB21aph phagemid) targeting aph-3 kanamycin resistance genes could lead to a decrease (from 50 % to 11.4 %) in proportion of fluorescence-labeled RNKΦ cells in a mouse skin colonization model. So, this novel, programmable, and sequence-specific antimicrobials based on the CRISPR system and bacteriophage provides new choices to manipulate bacterial populations in a sequence-specific manner. Although the phagemid in this technology provides an excellent delivery, the purity, large-scale production, potential transfer of virulence gene from the host chromosome, and narrow host range also need to be solved urgently [109]. Besides, it was proven that gene editing techniques based on the CRISPR-Cas system could expand the host range of temperate bacteriophage and promote bactericidal activity by modifying the tail fiber protein. Given that, a temperate bacteriophage with the CRISPR-Cas9 bactericidal activity and the modified tail fiber protein, was conducted to mitigate soft tissue infection caused by a biofilm-forming S. aureus strain. In vitro, the bacteriophage effectively killed 1 × 105 CFU S. aureus culture within 6 h, while the unmodified phage treatment increased to 1 × 109 CFU. In the biofilm-forming S. aureus induced dermal infection in vivo study, the bacteriophage mitigated almost dermal infections (∼1 log CFU/g tissue), while the control therapy showed a significantly higher bacterial load (∼3.5 log CFU/g tissue). What's more, the osteomyelitis and soft tissue infection models were used to compare the antimicrobial effects of bacteriophage, antibiotic (Fosfomycin), and combined therapies by analyzing histological, radiographic, and bacteriological performance. The results demonstrated the phage therapy performed as well as high dose Fosfomycin in mitigating soft tissue infection (the average bacterial counts: control: 4.713 ± 0.289 Log10(CFU); Fosfomycin: 4.146 ± 0.377 Log10(CFU); phage: 4.160 ± 0.516 Log10(CFU)) but not in bone infection. To sum up, further investigation of optimal dosing and infection type is still needed [113]. To further promote the efficiency of the phage therapy, a novel antimicrobial (ϕSaBov-Cas9-nuc phage), which integrated the CRISPR-Cas system into a temperate phage genome and removed virulence genes from the host chromosome preventing contamination of harmful bacterial products in the phage lysates and spread of virulent genes, has shown significantly enhanced efficiency in both in vivo and in vitro. S. aureus strain CTH96, an isolate susceptive to ϕSaBov phage, was treated with different multiplicities of infection (MOIs) and time of ϕSaBov-Cas9-nuc phage. Results showed that ϕSaBov-Cas9-nuc phage's corresponding number of viable bacteria significantly decreased after 8 h treatment with an MOI of 50 but ϕSaBov-Cas9-null phage's number was not decreased. In a mouse skin infection model, infected skin regions after the treatment of 24 h were excised and accessed, and results showed the ϕSaBov-Cas9-nuc phage's number of viable bacteria was significantly lower (0.647 ± 0.128 Log CFU/g of tissue, mean ± SEM) than the ϕSaBov-Cas9-null phage (3.333 ± 0.131 Log CFU/g of tissue, mean ± SEM). In conclusion, the ϕSaBov-Cas9-nuc phage can successfully decolonize S. aureus from the infected skin surface, which may be relevant to the CRISPR-Cas9 modified bacteriophage's dual killing mechanisms: direct lysis of target bacteria and CRISPR-Cas9 nuclease activity. The novel phage therapy coupled with CRISPR-Cas may provide a sequence-specific and safer antimicrobial platform for MRSA and other common pathogens treatment [114].
What's more, bacteriolytic enzymes are a promising alternative to antibiotics, which can eradicate bacterial pathogens by degrading bacterial cell wall peptidoglycan and inducing cell lysis. However, S. aureus slowly becomes resistant to various bacteriolytic enzymes in the presence of growth-supporting nutrients, which is due to the prevention of lysostaphin (Lst)-cell binding mediated by the wall teichoic acids (WTAs). For that, researchers have found that using the CRISPR-Cas system to downregulate genes encoding enzymes that anchor WTAs in the outer layer of cell wall peptidoglycan could produce lower drug resistance to bacteriolytic enzymes than antimicrobials and had great potential in eradicating bacterial pathogens in tryptic soy broth (TSB) within 24 h. For example, this paper demonstrated that inhibiting the expression of the tarO gene with CRISPR-Cas system could significantly sensitize S. aureus to Lst in TSB, as indicated by ∼4.7‐log reduction in cell viability compared with ∼1.3 log reduction in control cells. As a result, this may provide a potential treatment for AMR bacterial infection [[115], [116], [117], [118]]. Furthermore, the capability of multiplexing against different targets enables the CRISPR-Cas9 system to target different AMR genes simultaneously. However, studying how to design the appropriate temperate phages against multiple resistance genes and knowing the resistance genes carried by the bacteria is still needed [112]. For example, a study by Sato'o et al. constructed a novel CRISPRi-based vector, pBACi, which could silence various virulence and AMR genes in different types of clinical isolates from S. aureus. In detail, the pBAci was introduced into various clinical isolates, then decreased various targeted gene expressions, including four virulence and antibiotics resistant genes, and altered the knockdown strains' phenotypes with the sequence-specific activity of the dCas9 and crRNA. The results showed the silence of the icaA gene could significantly decrease the mass of the formed biofilm; the silence of the sec gene could reduce the amount of the encoded protein 50–100 folds compared to the control group; the silence of the coa gene could suppress coagulation of normal rabbit plasma; the silence of the blaZ gene encoding β-lactamase could reduce the β-lactamase activity by about 50 %. However, the designation of crRNA and the polar activity of pBACi still should be paid attention to Ref. [5]. Those experiments opened up a new era for sequence-specific antimicrobial therapy, but more possible therapeutic strategies still need to be further investigated.
1.5. Limitation and perspective
Every CRISPR-Cas system has a unique Cas protein constitution, recognition site, and cleavage function mechanism. Researchers have established some gene editing methods by combining the sequence-specific cleavage with the following repair processes to accomplish efficient gene manipulations, such as gene indels, gene silence, or gene repression. Based on these, a variety of detection methods were constructed to realize ultrasensitive, rapid, convenient, and precise early detection by combining the cis/trans cleavage of the Cas proteins with visual readouts, including fluorescence, colorimetric, and electrochemical signals. Furthermore, the novel CRISPR-Cas-based antimicrobials have attracted more and more attention recently. Some original therapeutic regimens by silencing or repressing resistance and virulence genes and cleaving relevant genes to eliminate the bacteria of interest were found to perform individual treatment and impede the spreading of the AMR genes.
However, some drawbacks need to be taken into consideration. Firstly, the off-target effect caused by non-specific nucleic acid-targeting was a matter of concern [[119], [120], [121]]. Then, to detect the off-target effect, some novel methods were conducted. For example, GOTI (genome-wide off-target analysis by two-embryo injection) can examine the off-target effects of various gene-editing tools by editing one blastomere of two-cell mouse embryos using either CRISPR-Cas9 or base editors [120]. To overcome the shortcomings, such as the need for purified DNA or cellular models and incapability of simple in vivo detection, of the current off-target discovery tools, DISCOVER-seq (discovery of in situ Cas off-targets and verification by sequencing) was developed to identify the unbiased off-target effect by leveraging the recruitment of DNA repair factors in cells and organisms. Further, the DISCOVER-seq can achieve characterization of new editing tools with various guide RNA formats and types of Cas enzymes [122]. Digenome-seq, in vitro Cas9-digested whole-genome sequencing, provides a robust, unbiased, and inexpensive tool to profile genome-wide Cas9 off-target effects in human cells, which detection limit is close to those of targeted deep sequencing. What's more, the methods verified that replacing 'promiscuous' single guide RNAs (sgRNAs) with modified sgRNAs could significantly reduce the off-target effects [123]. Given that, a comprehensive and high-density sgRNA activity map based on a genome-scale library was constructed to profile the association of sgRNA activity with Cas9 or its mutants. Based on that, an integrated algorithm was developed to accurately select the most suitable sgRNAs, leading to the reduction of the off-target effects and facilities of the CRISPR-Cas9-bases genome engineering [124]. What's more, there were some tools performing structural modifications in the Cas proteins to reduce the effect, including SaCas9's novel mutation (variant Mut268 harboring the single base-pair mismatches) which can effectively reduce off-target effects by approximately 2–90 folds compared to WT strains [125] and a modified version of Cas9 (Cas9 nickase) which can accurately edit bases up to 53 bp from the nicking site and show no off-target effects in yeast [126]. Secondly, how to deliver the CRISPR tool to the recipient was also taken into account. Although viral vector was proven efficient, there still were limitations in immunogenicity and duration of Cas expression genes in vivo [127]. Thirdly, some Cas proteins, such as dCas9, have shown unexpected toxicity, leading to host cell injury [[128], [129], [130]]. To avoid toxicity and preserve strong on-target repression activity, researchers optimized the expression level of dCas9 by using a specific vector, but how the toxicity is produced still needs to be further studied [76]. Fourthly, requiring recognizing specific PAM/PFS sequences, the applications of the CRISPR-Cas system were restricted [131]. To enlarge the scope of the target genes, novel Cas9 variants that recognized new PAM sequences were developed [132,133]. For example, the SaCas9 targeting range could be increased two to four times by modifying the PAM recognition sites [134]. Fifthly, more precise quantitation and field-deployable detection methods should be conducted to achieve ultra-accurate and convenient detection in source-poor areas [39].
Interestingly, with the fight between the CRISPR-Cas systems and invading MGEs, anti-CRISPR systems have been discovered in bacteriophages recently [135,136] and proven to inhibit many CRISPR-Cas systems, such as type I, type II, type III, and type V [[137], [138], [139]]. The system could escape the recognition of the CRISPR-Cas systems by point mutation, large-scale gene deletion, DNA modification, or specific encoded protein formation [140]. Although hosts could also successively acquire more new spacers derived from invasive MGEs to form new immunity memory in response to these escape processes [141,142], the interaction between the CRISPR-Cas defense system and adaptive escape might benefit the HGT of the AMR genes [143]. Furthermore, it is proven that S. aureus possesses many defense systems, including biofilm formation, persister cells, small colony variants, and efflux pump, bringing a huge burden on anti-MRSA treatment [144]. Therefore, how to reduce those defense systems and efficiently inhibit the dissemination of the AMR genes should be further investigated.
Nevertheless, the above methods were proven to have high sensitivity, specificity, and efficiency in S. aureus in both in vivo and in vitro experiments. Besides, the potential of CRISPR-Cas system was discovered in more and more microorganisms, especially infectious bacteria and viruses. Thus, further investigations focusing on the CRISPR-Cas system should be carried out to accomplish ultrarapid early detection of pathogens and highly efficient antimicrobial treatment, allowing inhibit the spread of drug resistance.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 82273696 and No. 81973105). The funders have no role in the data collection, data analysis, preparation of manuscript and decision to submission.
Data availability statement
The data supporting this study's findings are included in this article and available from the corresponding author upon reasonable requests.
CRediT authorship contribution statement
Jiamin Wang: Writing – review & editing, Writing – original draft, Conceptualization. Fang Liu: Data curation, Writing – review & editing. Jinzhao Long: Data curation, Writing – review & editing. Yuefei Jin: Data curation, Writing – review & editing. Shuaiyin Chen: Data curation, Writing – review & editing. Guangcai Duan: Conceptualization, Writing – review & editing. Haiyan Yang: Supervision, Funding acquisition, Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the support from the National Natural Science Foundation of China (No. 82273696 and No. 81973105).
References
- 1.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014;17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsay J.A., et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 2006;188(2):669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanza V.F., et al. The plasmidome of firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol. Spectr. 2015;3(2) doi: 10.1128/microbiolspec.PLAS-0039-2014. PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- 5.Sato'o Y., et al. Tailor-made gene silencing of Staphylococcus aureus clinical isolates by CRISPR interference. PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0185987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espedido B.A., Gosbell I.B. Chromosomal mutations involved in antibiotic resistance in Staphylococcus aureus. Front. Biosci. 2012;4(3):900–915. doi: 10.2741/s307. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q., et al. CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Acta Biochim. Biophys. Sin. 2017;49(9):764–770. doi: 10.1093/abbs/gmx074. [DOI] [PubMed] [Google Scholar]
- 8.Murray C.J.L., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/s0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prax M., Lee C.Y., Bertram R. An update on the molecular genetics toolbox for staphylococci. Microbiology (Read.) 2013;159(Pt 3):421–435. doi: 10.1099/mic.0.061705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato F., Sugai M. A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods. 2011;87(1):76–81. doi: 10.1016/j.mimet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Guo Y., et al. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020;10:107. doi: 10.3389/fcimb.2020.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgna-Pignatti C., Marsella M. Iron chelation in thalassemia major. Clin. Therapeut. 2015;37(12):2866–2877. doi: 10.1016/j.clinthera.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Krut O., Bekeredjian-Ding I. Contribution of the immune response to phage therapy. J. Immunol. 2018;200(9):3037–3044. doi: 10.4049/jimmunol.1701745. [DOI] [PubMed] [Google Scholar]
- 14.Wills Q.F., Kerrigan C., Soothill J.S. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob. Agents Chemother. 2005;49(3):1220–1221. doi: 10.1128/AAC.49.3.1220-1221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulme J. Application of nanomaterials in the prevention, detection, and treatment of methicillin-resistant Staphylococcus aureus (MRSA) Pharmaceutics. 2022;14(4) doi: 10.3390/pharmaceutics14040805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharyya R.P., Thakku S.G., Hung D.T. Harnessing CRISPR effectors for infectious disease diagnostics. ACS Infect. Dis. 2018;4(9):1278–1282. doi: 10.1021/acsinfecdis.8b00170. [DOI] [PubMed] [Google Scholar]
- 17.Mortensen K., Lam T.J., Ye Y. Comparison of CRISPR-cas immune systems in healthcare-related pathogens. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.758782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bikard D., Barrangou R. Using CRISPR-Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017;37:155–160. doi: 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Bikard D., et al. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12(2):177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian Y., et al. Application of CRISPR-Cas system in the diagnosis and therapy of ESKAPE infections. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1223696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishino Y., et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 24.Garneau J.E., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 25.Bhagwat A.C., Patil A.M., Saroj S.D. CRISPR/Cas 9-based editing in the production of bioactive molecules. Mol. Biotechnol. 2022;64(3):245–251. doi: 10.1007/s12033-021-00418-4. [DOI] [PubMed] [Google Scholar]
- 26.Cring M.R., Sheffield V.C. Gene therapy and gene correction: targets, progress, and challenges for treating human diseases. Gene Ther. 2022;29(1–2):3–12. doi: 10.1038/s41434-020-00197-8. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Huang C., Zhao W. Recent advances of the biological and biomedical applications of CRISPR/Cas systems. Mol. Biol. Rep. 2022;49(7):7087–7100. doi: 10.1007/s11033-022-07519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thakore P.I., et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun. 2018;9(1):1674. doi: 10.1038/s41467-018-04048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shipman S.L., et al. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353(6298):aaf1175. doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong T., et al. CRISPR-cas systems in streptococci. Curr. Issues Mol. Biol. 2019;32:1–38. doi: 10.21775/cimb.032.001. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko K.A., et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 32.Nidhi S., et al. Novel CRISPR-cas systems: an updated review of the current achievements, applications, and future research perspectives. Int. J. Mol. Sci. 2021;22(7) doi: 10.3390/ijms22073327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., et al. Application of CRISPR/cas systems in the nucleic acid detection of infectious diseases. Diagnostics. 2022;12(10) doi: 10.3390/diagnostics12102455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P., et al. Applications of the CRISPR-Cas system for infectious disease diagnostics. Expert Rev. Mol. Diagn. 2021;21(7):723–732. doi: 10.1080/14737159.2021.1922080. [DOI] [PubMed] [Google Scholar]
- 35.Mojica F.J.M., et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Read.) 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 36.Swarts D.C., et al. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez de Aledo M., et al. CRISPR-cas, a revolution in the treatment and study of ESKAPE infections: pre-clinical studies. Antibiotics. 2021;10(7) doi: 10.3390/antibiotics10070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deveau H., Garneau J.E., Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 39.Yuan B., et al. Application of the CRISPR/cas system in pathogen detection: a review. Molecules. 2022;27(20) doi: 10.3390/molecules27206999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deltcheva E., et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Z.Q., et al. Establishment of a highly efficient virus-inducible CRISPR/Cas9 system in insect cells. Antivir. Res. 2016;130:50–57. doi: 10.1016/j.antiviral.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Dong F., et al. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 2017;482(4):889–895. doi: 10.1016/j.bbrc.2016.11.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zetsche B., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fonfara I., et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., et al. Functional characterization of type III-A CRISPR-cas in a clinical human methicillin-R Staphylococcus aureus strain. CRISPR J. 2021;4(5):686–698. doi: 10.1089/crispr.2021.0046. [DOI] [PubMed] [Google Scholar]
- 46.Makarova K.S., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meliawati M., Schilling C., Schmid J. Recent advances of Cas12a applications in bacteria. Appl. Microbiol. Biotechnol. 2021;105(8):2981–2990. doi: 10.1007/s00253-021-11243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chayot R., et al. An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2010;107(5):2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szostak J.W., et al. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 50.Makarova K.S., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvam K., et al. CRISPR-Cas systems-based bacterial detection: a scoping review. Diagnostics. 2022;12(6) doi: 10.3390/diagnostics12061335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F., et al. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019;37(5):708–729. doi: 10.1016/j.biotechadv.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 53.Javed M.R., et al. CRISPR-cas system: history and prospects as a genome editing tool in microorganisms. Curr. Microbiol. 2018;75(12):1675–1683. doi: 10.1007/s00284-018-1547-4. [DOI] [PubMed] [Google Scholar]
- 54.Sternberg S.H., et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrington L.B., et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abudayyeh O.O., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindele P., Wolter F., Puchta H. Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett. 2018;592(12):1954–1967. doi: 10.1002/1873-3468.13073. [DOI] [PubMed] [Google Scholar]
- 58.Dong X., et al. CRISPR/dCas9-mediated inhibition of gene expression in Staphylococcus aureus. J. Microbiol. Methods. 2017;139:79–86. doi: 10.1016/j.mimet.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Jinek M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamano T., et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165(4):949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim H.K., et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017;14(2):153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 62.Swiat M.A., et al. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45(21):12585–12598. doi: 10.1093/nar/gkx1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S.Y., et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu L., et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168(1–2):121–134 e12. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 65.East-Seletsky A., et al. RNA targeting by functionally orthogonal type VI-A CRISPR-cas enzymes. Mol. Cell. 2017;66(3):373–383 e3. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gootenberg J.S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferrando J., et al. Barriers to simultaneous multilocus integration in Bacillus subtilis tumble down: development of a straightforward screening method for the colorimetric detection of one-step multiple gene insertion using the CRISPR-Cas9 system. Microb. Cell Factories. 2023;22(1):21. doi: 10.1186/s12934-023-02032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cong L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeussler M., Concordet J.P. Genome editing with CRISPR-cas9: can it get any better? J Genet Genomics. 2016;43(5):239–250. doi: 10.1016/j.jgg.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W., et al. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J. Am. Chem. Soc. 2017;139(10):3790–3795. doi: 10.1021/jacs.6b13317. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z., et al. CRISPR/Cpf1-Mediated multiplex and large-fragment gene editing in Staphylococcus aureus. ACS Synth. Biol. 2022;11(9):3049–3057. doi: 10.1021/acssynbio.2c00248. [DOI] [PubMed] [Google Scholar]
- 72.Ouellette S.P. Feasibility of a conditional knockout system for Chlamydia based on CRISPR interference. Front. Cell. Infect. Microbiol. 2018;8:59. doi: 10.3389/fcimb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi L.S., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang R., et al. Gene silencing through CRISPR interference in bacteria: current advances and future prospects. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.635227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui L., et al. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018;9(1):1912. doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Depardieu F., Bikard D. Gene silencing with CRISPRi in bacteria and optimization of dCas9 expression levels. Methods. 2020;172:61–75. doi: 10.1016/j.ymeth.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 77.Gu T., et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem. Sci. 2018;9(12):3248–3253. doi: 10.1039/c8sc00637g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji A.G.a.Q. Identification of editable sites, spacer oligonucleotide design, and generation of the gene-targeting CRISPR-nCas9 plasmid for gene disruption in Staphylococcus aureus using the CRISPR-nCas9 and cytidine deaminase system. Cold Spring Harb. Protoc. 2023 doi: 10.1101/pdb.prot107924. [DOI] [PubMed] [Google Scholar]
- 79.Ji A.G.a.Q. Introduction of a CRISPR-nCas9 gene-targeting plasmid into Staphylococcus aureus for gene disruption. Cold Spring Harb. Protoc. 2023 doi: 10.1101/pdb.prot107925. [DOI] [PubMed] [Google Scholar]
- 80.Penewit K., et al. Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-Mediated counterselection. mBio. 2018;9(1) doi: 10.1128/mBio.00067-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salipante A.G.a.S.J. Introduction of a recombineering oligonucleotide and a CRISPR-cas9 gene-targeting plasmid into Staphylococcus aureus for generating a gene-deletion strain. Cold Spring Harb. Protoc. 2023 doi: 10.1101/pdb.prot107921. [DOI] [PubMed] [Google Scholar]
- 82.Salipante A.G.a.S.J. Oligonucleotide design and construction of a gene-targeting CRISPR-cas9 plasmid in Escherichia coli for generating a gene-deletion strain in Staphylococcus aureus. Cold Spring Harb. Protoc. 2023 doi: 10.1101/pdb.prot107920. [DOI] [PubMed] [Google Scholar]
- 83.Angelika Gründling Q.J., Salipante Stephen J. Using CRISPR-cas9-based methods for genome editing in Staphylococcus aureus. Cold Spring Harb. Protoc. 2023 doi: 10.1101/pdb.top107919. [DOI] [PubMed] [Google Scholar]
- 84.Wei L., et al. Aptamer-based colorimetric detection of methicillin-resistant Staphylococcus aureus by using a CRISPR/Cas12a system and recombinase polymerase amplification. Anal. Chim. Acta. 2022;1230 doi: 10.1016/j.aca.2022.340357. [DOI] [PubMed] [Google Scholar]
- 85.Huang L., et al. An electrochemical biosensor for the highly sensitive detection of Staphylococcus aureus based on SRCA-CRISPR/Cas12a. Talanta. 2023;252 doi: 10.1016/j.talanta.2022.123821. [DOI] [PubMed] [Google Scholar]
- 86.Liu L., et al. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci. Total Environ. 2022;838(Pt 2) doi: 10.1016/j.scitotenv.2022.156048. [DOI] [PubMed] [Google Scholar]
- 87.Guk K., et al. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens. Bioelectron. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 88.Wang M., Zhang R., Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou J., et al. CRISPR-Cas13a based bacterial detection platform: sensing pathogen Staphylococcus aureus in food samples. Anal. Chim. Acta. 2020;1127:225–233. doi: 10.1016/j.aca.2020.06.041. [DOI] [PubMed] [Google Scholar]
- 90.Tomita N., et al. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 91.Xiong J., et al. A closed-tube loop-mediated isothermal amplification assay for the visual detection of Staphylococcus aureus. Appl. Biochem. Biotechnol. 2020;191(1):201–211. doi: 10.1007/s12010-020-03278-x. [DOI] [PubMed] [Google Scholar]
- 92.Cao X., et al. Cas12a/Guide RNA-based Platforms for Rapidly and accurately identifying Staphylococcus aureus and methicillin-resistant S. aureus. Microbiol. Spectr. 2023 doi: 10.1128/spectrum.04870-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Y., et al. Combining CRISPR-cas12a-based technology and metagenomics next generation sequencing: a new paradigm for rapid and full-scale detection of microbes in infectious diabetic foot samples. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.742040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amanzholova M., et al. Genetic identification of Staphylococcus aureus isolates from cultured milk samples of bovine mastitis using isothermal amplification with CRISPR/Cas12a-based molecular assay. Vet. Res. Commun. 2023 doi: 10.1007/s11259-023-10212-z. [DOI] [PubMed] [Google Scholar]
- 95.Wang Y., et al. Rapid and ultrasensitive detection of methicillin-resistant Staphylococcus aureus based on CRISPR-cas12a combined with recombinase-aided amplification. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.903298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y., et al. Programmable T-junction structure-assisted CRISPR/Cas12a electrochemiluminescence biosensor for detection of sa-16S rDNA. ACS Appl. Mater. Interfaces. 2023;15(1):617–625. doi: 10.1021/acsami.2c18930. [DOI] [PubMed] [Google Scholar]
- 97.Xu L., et al. Accurate MRSA identification through dual-functional aptamer and CRISPR-Cas12a assisted rolling circle amplification. J. Microbiol. Methods. 2020;173 doi: 10.1016/j.mimet.2020.105917. [DOI] [PubMed] [Google Scholar]
- 98.Wei J. Accurate and sensitive analysis of Staphylococcus aureus through CRISPR-Cas12a based recycling signal amplification cascades for early diagnosis of skin and soft tissue infections. J. Microbiol. Methods. 2021;183 doi: 10.1016/j.mimet.2021.106167. [DOI] [PubMed] [Google Scholar]
- 99.Song D., et al. CRISPR/Cas12a-powered evanescent wave fluorescence nanobiosensing platform for nucleic acid amplification-free detection of Staphylococcus aureus with multiple signal enhancements. Biosens. Bioelectron. 2023;225 doi: 10.1016/j.bios.2023.115109. [DOI] [PubMed] [Google Scholar]
- 100.Wu J., et al. CPA-Cas12a-based lateral flow strip for portable assay of Methicillin-resistant Staphylococcus aureus in clinical sample. J. Nanobiotechnol. 2023;21(1) doi: 10.1186/s12951-023-02002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chakraborty J., et al. CRISPR/Cas-Based biosensor as a new age detection method for pathogenic bacteria. ACS Omega. 2022;7(44):39562–39573. doi: 10.1021/acsomega.2c04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suea-Ngam A., Howes P.D., deMello A.J. An amplification-free ultra-sensitive electrochemical CRISPR/Cas biosensor for drug-resistant bacteria detection. Chem. Sci. 2021;12(38):12733–12743. doi: 10.1039/d1sc02197d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wei L., et al. CRISPR/Cas12a-based magnetic relaxation switching biosensor for nucleic acid amplification-free and ultrasensitive detection of methicillin-resistant Staphylococcus aureus. Biosens. Bioelectron. 2023;222 doi: 10.1016/j.bios.2022.114984. [DOI] [PubMed] [Google Scholar]
- 104.Li Y., et al. Rapid one-tube RPA-CRISPR/Cas12 detection platform for methicillin-resistant Staphylococcus aureus. Diagnostics. 2022;12(4) doi: 10.3390/diagnostics12040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei Y., et al. Aptamer-based Cas14a1 biosensor for amplification-free live pathogenic detection. Biosens. Bioelectron. 2022;211 doi: 10.1016/j.bios.2022.114282. [DOI] [PubMed] [Google Scholar]
- 106.Tao Z., et al. A signal-off Cas14a1-based platform for highly specific detection of methicillin-resistant Staphylococcus aureus. Anal. Chim. Acta. 2023;1256 doi: 10.1016/j.aca.2023.341154. [DOI] [PubMed] [Google Scholar]
- 107.Peng L., et al. Integration of logic gates to CRISPR/Cas12a system for rapid and sensitive detection of pathogenic bacterial genes. Anal. Chim. Acta. 2020;1125:162–168. doi: 10.1016/j.aca.2020.05.017. [DOI] [PubMed] [Google Scholar]
- 108.Gomaa A.A., et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5(1) doi: 10.1128/mBio.00928-13. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bikard D., et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014;32(11):1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y., et al. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 2018;84(23) doi: 10.1128/AEM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arcilla M.S., et al. Import and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect. Dis. 2017;17(1):78–85. doi: 10.1016/S1473-3099(16)30319-X. [DOI] [PubMed] [Google Scholar]
- 112.Zohra T., et al. Cracking the challenge of antimicrobial drug resistance with CRISPR/Cas9, nanotechnology and other strategies in ESKAPE pathogens. Microorganisms. 2021;9(5) doi: 10.3390/microorganisms9050954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cobb L.H., et al. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS One. 2019;14(11) doi: 10.1371/journal.pone.0220421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park J.Y., et al. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017;7 doi: 10.1038/srep44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu X., et al. Reducing Staphylococcus aureus resistance to lysostaphin using CRISPR-dCas9. Biotechnol. Bioeng. 2019;116(12):3149–3159. doi: 10.1002/bit.27143. [DOI] [PubMed] [Google Scholar]
- 116.Raz A., et al. Lysostaphin lysibody leads to effective opsonization and killing of methicillin-resistant Staphylococcus aureus in a murine model. Antimicrob. Agents Chemother. 2018;62(10) doi: 10.1128/AAC.01056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmelcher M., Loessner M.J. Bacteriophage endolysins: applications for food safety. Curr. Opin. Biotechnol. 2016;37:76–87. doi: 10.1016/j.copbio.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Wu X., et al. Flexible peptide linkers enhance the antimicrobial activity of surface-immobilized bacteriolytic enzymes. ACS Appl. Mater. Interfaces. 2018;10(43):36746–36756. doi: 10.1021/acsami.8b14411. [DOI] [PubMed] [Google Scholar]
- 119.Jin S., et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364(6437):292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 120.Zuo E., et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364(6437):289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cradick T.J., et al. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wienert B., et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364(6437):286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim D., et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. 1 pp. following 243. [DOI] [PubMed] [Google Scholar]
- 124.Guo J., et al. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res. 2018;46(14):7052–7069. doi: 10.1093/nar/gky572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xie H., et al. High-fidelity SaCas9 identified by directional screening in human cells. PLoS Biol. 2020;18(7) doi: 10.1371/journal.pbio.3000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Satomura A., et al. Precise genome-wide base editing by the CRISPR Nickase system in yeast. Sci. Rep. 2017;7(1):2095. doi: 10.1038/s41598-017-02013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kumar P., et al. CRISPR-cas system: an approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rock J.M., et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cho S., et al. High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018;7(4):1085–1094. doi: 10.1021/acssynbio.7b00462. [DOI] [PubMed] [Google Scholar]
- 130.Wurihan W., et al. Nonspecific toxicities of Streptococcus pyogenes and Staphylococcus aureus dCas9 in Chlamydia trachomatis. Pathog Dis. 2019;77(9) doi: 10.1093/femspd/ftaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moon S.B., et al. Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 2019;51(11):1–11. doi: 10.1038/s12276-019-0339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y., et al. Using Staphylococcus aureus Cas9 to expand the scope of potential gene targets for genome editing in soybean. Int. J. Mol. Sci. 2022;23(21) doi: 10.3390/ijms232112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kleinstiver B.P., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kleinstiver B.P., et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33(12):1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang F., Song G., Tian Y. Anti-CRISPRs: the natural inhibitors for CRISPR-Cas systems. Animal Model Exp Med. 2019;2(2):69–75. doi: 10.1002/ame2.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hwang S., Maxwell K.L. Meet the anti-CRISPRs: widespread protein inhibitors of CRISPR-cas systems. CRISPR J. 2019;2(1):23–30. doi: 10.1089/crispr.2018.0052. [DOI] [PubMed] [Google Scholar]
- 137.Marino N.D., et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362(6411):240–242. doi: 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rauch B.J., et al. Inhibition of CRISPR-cas9 with bacteriophage proteins. Cell. 2017;168(1–2):150–158 e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bhoobalan-Chitty Y., et al. Inhibition of type III CRISPR-cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell. 2019;179(2):448–458 e11. doi: 10.1016/j.cell.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 140.Hynes A.P., et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat Microbiol. 2017;2(10):1374–1380. doi: 10.1038/s41564-017-0004-7. [DOI] [PubMed] [Google Scholar]
- 141.Fineran P.C., et al. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U. S. A. 2014;111(16):E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jackson S.A., et al. CRISPR-Cas: adapting to change. Science. 2017;356(6333) doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 143.Malone L.M., Birkholz N., Fineran P.C. Conquering CRISPR: how phages overcome bacterial adaptive immunity. Curr. Opin. Biotechnol. 2021;68:30–36. doi: 10.1016/j.copbio.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 144.Hou H., et al. The crafty opponent: the defense systems of Staphylococcus aureus and response measures. Folia Microbiol. 2022;67(2):233–243. doi: 10.1007/s12223-022-00954-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this study's findings are included in this article and available from the corresponding author upon reasonable requests.