Abstract
How the KRAS oncogene drives cancer growth remains poorly understood. Therefore, we established a systemwide portrait of KRAS- and ERK-dependent gene transcription in KRAS-mutant cancer to delineate the molecular mechanisms of growth and of inhibitor resistance. Unexpectedly, our KRAS-dependent gene signature diverges significantly from the frequently cited Hallmark KRAS signaling gene signature, is driven predominantly through the ERK mitogen-activated protein kinase (MAPK) cascade, and accurately reflects KRAS- and ERK-regulated gene transcription in KRAS-mutant cancer patients. Integration with our ERK-regulated phospho- and total proteome highlights ERK deregulation of the anaphase promoting complex/cyclosome and other components of the cell cycle machinery as key processes driving PDAC growth. In summary, these findings elucidate mechanistically the critical role of ERK in driving KRAS-mutant tumor growth and in resistance to KRAS-ERK MAPK targeted therapies.
One-Sentence Summary:
Integration of multi-omics data reveals KRAS and ERK inhibitor targeting of a robust ERK-dependent cellular proliferation program in patients.
Main Text:
The recent approval of direct inhibitors for the KRASG12C mutation marks a milestone in cancer drug discovery and has stimulated intensive efforts to develop inhibitors for additional KRAS mutations (1). However, treatment-associated resistance mechanisms limit their long-term efficacy. Initially responsive patients who relapsed on KRAS inhibitor therapy displayed diverse genetic alterations that broadly drive reactivation of the ERK mitogen-activated protein kinase (MAPK) effector signaling network. To delineate the molecular mechanisms of growth and of inhibitor resistance, here we focused on the most KRAS-addicted cancer, pancreatic ductal adenocarcinoma (PDAC) (2, 3), to establish a systemwide portrait of KRAS- and ERK-dependent gene transcription.
KRAS drives PDAC growth primarily through sustained hyperactivation of the RAF-MEK-ERK MAPK cascade (4, 5). Whereas RAF and MEK each exhibit highly restricted substrate utilization, the two nearly identical ERK1 and ERK2 serine/threonine kinases (herein after referred to as ERK) phosphorylate and regulate the activities of a diverse spectrum of direct and indirect substrates (6, 7). Our recent determination of the ERK-regulated phosphoproteome in KRAS-mutant PDAC identified 2,295 direct/indirect ERK substrates that include 105 transcription factors (8). Thus, a key mechanism by which KRAS acts as a cancer driver involves systemwide deregulation of gene transcription. While analyses of candidate genes (e.g., MYC, FOSL1) have established important transcription factor drivers of KRAS-dependent PDAC growth (9, 10), a systemwide determination of the transcriptome that supports KRAS-driven oncogenesis remains to be completed.
Various approaches have been taken to define a KRAS gene expression signature and delineate a mechanistic basis for KRAS cancer driver function. Sweet-Cordero et al. determined an 89-gene expression signature of KRAS activation by cross-species gene set enrichment analyses (GSEA) of microarray data, profiling normal and tumor tissue from mouse models of mutant KrasG12D-induced lung adenocarcinoma together with a large set of KRAS-mutant and wild-type human lung adenocarcinoma tissue (11). Arena et al. used genetic knock-in of a mutant KrasG12D allele into mouse liver progenitor cells and established a 345-gene KRAS up/downregulated “leave-one-out” classification gene expression signature (12). Singh et al. utilized microarray profiles from mutant KRAS-dependent versus -independent human cancer cell lines to establish a KRAS addiction 196-gene (250 probe set) expression signature (13). Recently, East et al. utilized data from the Broad Institute Cancer Cell Line Encyclopedia to establish an 84-gene set to detect RAS pathway activation in lung cancer cell lines (14). However, these and other strategies to establish a mutant KRAS-regulated transcriptome identified signatures with limited overlap, likely reflecting the different experimental models and strategies taken to link gene transcription with KRAS function (9, 15, 16).
The Molecular Signatures Database (MSigDB) Hallmark KRAS Signaling gene set was developed to address the diversity of KRAS gene signatures identified from studies of different cell type models (17). This widely used signature, established in 2015, represents genes identified primarily from microarray profiling of a set of telomerase- and SV40 viral large T/small t antigen-immortalized human breast, kidney and lung epithelial cell lines ectopically overexpressing KRASG12V compared with control empty vector-matched cell lines (18), along with other correlated gene sets. The Hallmark signatures summarize information across multiple gene sets with the goal of reducing redundancy and providing diverse and broadly applicable transcriptional gene sets for GSEA. However, our recent comprehensive determination of an ERK-dependent phosphoproteome in KRAS-mutant PDAC cell lines revealed significant divergence in breadth and depth from the currently established ERK-regulated phosphoproteome generated from multiple cell types (8). We speculated that the Hallmark KRAS signature, which captured mutant-KRAS-correlated gene sets, may not accurately portray the mutant KRAS-dependent transcriptome in the context of endogenous mutant KRAS in genetically diverse cancer cells, thus providing the rationale for the present study.
Results
The KRAS-dependent transcriptome in PDAC diverges from the Hallmark signature
The Hallmark KRAS Signaling gene signatures are composed of genes differentially expressed within matched pairs of cell lines ectopically expressing control vector versus mutant KRAS (fig. S1A) (18). A potential limitation of the use of this and other mutant-KRAS-correlated signatures (11-13) is that they are also influenced by secondary transcriptional changes initiated by the oncogenic stress due to mutant KRAS-driven growth transformation. Therefore, for our KRAS gene signature, we determined the transcriptional changes caused by acute (24 hours) genetic suppression of KRAS. This approach also minimized the impact of the substantial genetic heterogeneity of KRAS-mutant PDAC, where, aside from the four major genetic alterations (fig. S1B), the majority of the remaining ~50 mutations occur at single digit frequencies (2, 19).
To determine our KRAS-dependent transcriptome, we used RNA-sequencing (RNA-Seq) to profile gene transcription changes in a panel of eight human KRAS-mutant PDAC cell lines subjected to 24 hours KRAS siRNA treatment (figs. S1A and S1B). Since KRAS protein half-life is approximately 24 hours (20), this time frame minimizes the onset of compensatory signaling (21). We verified KRAS protein knockdown (fig. S1C) concurrent with reduced KRAS mRNA expression (fig. S1D). Loss of KRAS signaling was supported by reductions in RAF, and, to a significantly lesser degree, in PI3K effector signal activation (phosphorylated and activated ERK and AKT, respectively), and by expression of previously well-established KRAS-ERK regulated genes (DUSP6 and SPRY4) (9, 11-13). Finally, recent RNA-Seq analysis has defined transcriptomic subtypes of PDAC (22). Seven of the eight PDAC lines in our panel comprise the basal subtype associated with poor prognosis, and short-term KRAS suppression did not significantly alter this subtype assignment (fig. S1E) (23).
We detected 14,799 protein coding genes represented in two or more samples (20 or more reads) and found that cell line had the largest impact on gene expression variability across samples (fig. S1F). We identified 677 KRAS-dependent (UP) and 1,051 KRAS-suppressed (DN) genes upon acute KRAS suppression (log2 fold-change/FC > 0.5, adj. p-val. < 0.05) shared across all eight cell lines (Fig. 1A and B, and data S1). Since the genes downregulated upon KRAS siRNA treatment represent putative KRAS-dependent genes, hereinafter we refer to this gene set as the PDAC KRAS upregulated (UP) signature (top 200, Fig. 1A). Conversely, since the genes upregulated upon KRAS siRNA treatment represent putative KRAS-suppressed genes, we refer to this gene set as the PDAC KRAS downregulated (DN) signature (top 200).
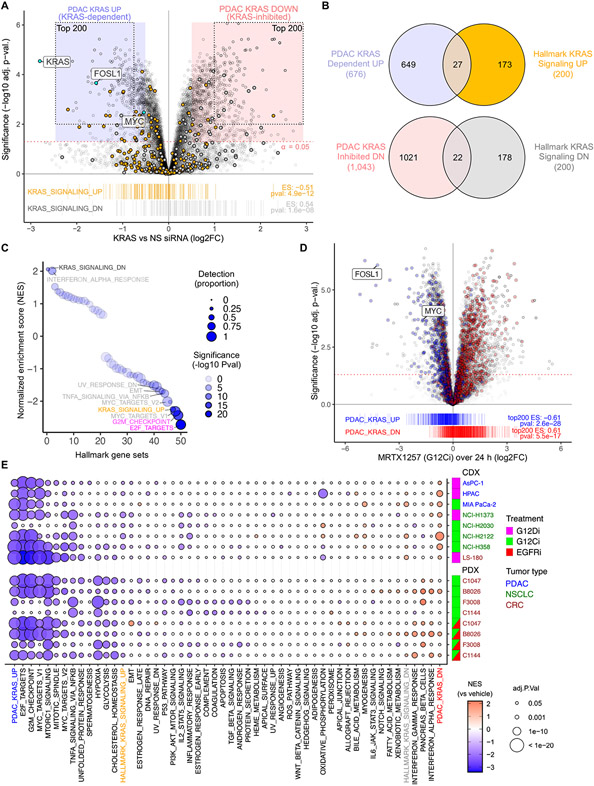
Fig. 1. Establishment and evaluation of a KRAS-dependent gene expression program in KRAS-mutant PDAC.
(A) KRAS-dependent gene expression changes upon acute (24 hours) KRAS suppression in eight KRAS-mutant PDAC cell lines transiently transfected with KRAS or control non-specific (NS) siRNA. Enrichment of Hallmark KRAS signaling gene sets is shown below. The 677 KRAS-dependent (UP) and 1,051 KRAS-suppressed (DN) genes (log2 fold-change/FC > 0.5, adj. p-val. < 0.05) are indicated by blue and red shaded circles, respectively. The top 200 KRAS-dependent (UP) and KRAS-inhibited (DN) genes comprising the PDAC KRAS UP/DN signatures are indicated by the dotted outlines. (B) Venn diagram indicates the overlap of differentially expressed genes (with unique Entrez gene IDs) upon KRAS siRNA treatment (refer to blue/red shading in (A)) compared to Hallmark KRAS signaling genes. N = 8 (cell lines as biological replicates) for each treatment and control. (C) GSEA for 50 Hallmark gene sets within DE genes following KRAS siRNA treatment. Detection is based on presence of a gene in all 8 PDAC cell lines at >5 reads. NES, normalized enrichment score. (D) KRAS-G12Ci (MRTX1257, 20 nM) induced expression changes summarized over 24 hours and three cell lines (PDAC, CRC, and NSCLC). Genes from siKRAS experiment shown as blue/red boxes in (A) are highlighted with blue/red and indicated by barcode plot below. Enrichment scores (ES) for top 200 UP/DN genes are indicated. (E) GSEA for 50 Hallmark gene sets within RNA-seq data from the indicated human cancer cell line-derived (CDX) or patient-derived (PDX) xenograft tumor samples. CDX mice were treated orally (24 hours) with either KRASG12C or KRASG12D selective inhibitors (G12Ci adagrasib or G12Di MRTX1133, respectively) or vehicle control. PDX mice were treated (21 days) with G12Ci sotorasib alone or together with the EGFRi panitumumab. The top 200 genes from the PDAC KRAS UP/DN gene sets (panel A) were also evaluated. N = 3 for each cell line/treatment combination except HPAC (control) and LS-180 (G12Di), where n = 6.
To assess the biological processes associated with our PDAC KRAS-dependent transcriptome, we conducted GSEA with the MSigDB 50 Hallmark gene sets (17). As expected, we found substantial enrichment of the E2F Targets, G2M Checkpoint, and MYC Targets gene sets (Fig. 1C). Enrichment of the Hallmark Interferon Alpha Response gene set is consistent with a recent study in a mouse model of PDAC showing that a KRAS- and MYC-dependent mechanism promotes immune evasion by suppressing the interferon pathway (24). Unexpectedly, we also found striking divergence between our PDAC KRAS and the Hallmark KRAS Signaling signatures. First, the Hallmark KRAS Signaling UP gene set was not the most significantly enriched among KRAS-dependent genes (Fig. 1C). Additionally, while the Hallmark KRAS Signaling DN signature was the most significantly enriched among the KRAS siRNA upregulated genes, the proportion of these genes detected across our eight PDAC cell lines was very low. The lack of concordance between our PDAC KRAS and the Hallmark KRAS Signaling genes was further explained upon examination of the individual founder gene sets used to derive the Hallmark KRAS Signaling gene sets, none of which accurately reflected the genes differentially expressed upon KRAS siRNA treatment of PDAC cells (fig. S1G).
The divergence of the PDAC KRAS and Hallmark KRAS Signaling signatures is even more evident at the level of specific genes (Fig. 1A). First, the most significantly altered PDAC KRAS UP and DN genes are not present in the Hallmark KRAS Signaling gene sets. In particular, two well-characterized genes with validated roles in supporting KRAS-driven cancer growth, MYC and FOSL1 (9, 10), are absent from the Hallmark KRAS Signaling gene set. Second, only 14% (27 genes) and 11% (22 genes) of the Hallmark KRAS Signaling UP and DN genes, respectively, are found among our 676 PDAC KRAS-dependent or 1,043 KRAS-suppressed genes (excluding genes without Entrez IDs, Fig. 1B). Third, within the PDAC KRAS-dependent genes (KRAS UP) are numerous Hallmark KRAS Signaling DN genes and vice versa.
Approximately half of the 200 genes that comprise the Hallmark KRAS Signaling UP signature were not detectable across our PDAC KRAS cell lines (Fig. 1C and data S1). They were also less frequently detected across RNA-seq data from the 41 PDAC cell lines in the Cancer Cell Line Encyclopedia (CCLE) (25) and from 66 laser capture microdissected (LCMD) PDAC patient tumor tissues (GSE93326) (fig. S1H) (26). Consistent with these observations, fewer Hallmark KRAS Signaling gene promoters exhibited signs of active transcription, including DNAse sensitivity, Pol II occupancy, and both H3K4me3 and H3K27me marks in PANC-1 and pancreas cells (fig. S1I).
The PDAC KRAS-dependent transcriptome accurately reflects KRAS-regulated genes in vivo
We next determined how accurately our cell culture-based PDAC KRAS-dependent transcriptome models KRAS-regulated gene expression in vivo. We first compared our human PDAC KRAS UP/DN signatures with a KrasG12D-dependent gene set determined in a doxycycline-inducible KrasG12D-driven mouse model of PDAC, designated iKras (KrasG12D;Trp53 null) (27). Ying et al. applied microarray profiling in orthotopic tumors generated from iKras-derived primary cell lines, where continued KrasG12D expression and tumor growth is dependent on doxycycline treatment (fig. S1A). To capture Kras-dependent gene transcription activities, microarray profiling of tumors was performed after 24 hours doxycycline withdrawal and loss of KrasG12D expression. The iKras differentially expressed (DE) genes include 847 KRAS-dependent (UP) and 684 KRAS-suppressed (DN) genes that have human homologs expressed in PDAC (log2FC > 0.25, adj. p-val. < 0.05) (fig. S1J and data S2). Our PDAC KRAS UP signature showed strong enrichment within iKras DE genes, though there was less enrichment of our PDAC KRAS DN signature (fig. S1J). Similarly, the top 200 iKras UP DE genes were enriched in the DE genes of our siKRAS experiment (fig. S1K).
We then extended our in vivo analyses to include RNA-Seq data sets of three KRASG12C/D-mutant human PDAC cell line-derived tumor xenografts (CDX) treated with pharmacologic inhibitors of KRASG12C (adagrasib/MRTX849; G12Ci) or KRASG12D (MRTX1133; G12Di) (28, 29) (fig. S1A, PRJNA578935, PRJNA831648). DE genes in tumors treated with G12C/Di ("KRASi") versus vehicle control comprised 697 KRAS-dependent (UP) and 704 KRAS-suppressed (DN) gene sets (log2FC > 0.25, adj. p-val. < 0.05) (data S3). PDAC KRAS UP and DN signatures were strongly enriched within KRASi DE genes (fig. S1L). Likewise, the top 200 KRASi UP/DN DE genes corresponded strongly with the DE genes of our siKRAS experiment (fig. S1M). The intersection between the human siKRAS and KRASi and the mouse iKras gene sets reveals a core set of 46 and 29 UP and DN genes, respectively, shared with all three KRAS-dependent gene signatures (fig. S1N and data S4). Approximately 29% and 18% of UP and DN DE genes, respectively, within the siKRAS cell line experiment were also DE within the KRASi CDX experiment (fig. S1N), compared with 10% for Hallmark KRAS UP/DN genes (fig. S1O), suggesting that this program may better reflect in vivo biology.
To evaluate whether PDAC KRAS UP/DN genes were relevant in other mutant KRAS cancer types, we applied RNA-Seq analyses in KRASG12C-mutant H353 non-small cell lung cancer (NSCLC), SW837 colorectal cancer (CRC) and MIA PaCa-2 (PDAC) cell lines, treated for 24 hours with the KRAS-G12C mutant selective inhibitor MRTX1257 (designated KRAS-G12Ci) (data S5). Detection of the PDAC KRAS gene signatures in an NSCLC and a CRC cell line (Fig. 1D) suggested broader tissue expression of this PDAC-derived gene signature. To address whether KRAS-regulated genes were consistent in PDAC and non-PDAC KRAS-mutant tumors, we conducted GSEA with the MSigDB 50 Hallmark gene sets and our PDAC KRAS UP/DN gene sets in human CDX models of PDAC, NSCLC, and CRC treated with KRASi versus vehicle control. We observed significant enrichment of both our PDAC KRAS UP and DN gene signatures in short-term (1-day) KRASi treated samples across cancer types, along with the Hallmark G2M Checkpoint, MYC Targets V1, and E2F Targets gene sets (Fig. 1E). In contrast, the Hallmark KRAS Signaling UP and DN gene sets were not consistently enriched across these KRASi treated CDX tumors. Unlike short-term treatment, PDAC KRAS signatures were not as prominent in some tumors that underwent long-term treatment for 5 or 7 days, likely due to onset of adaptive mechanisms as cancer cells attempt to offset the deleterious cellular consequences caused by the loss of KRAS activity (fig. S1P). Consistent with this possibility, apoptosis was detected upon short-term but not long-term KRASi treatment (28).
Finally, we conducted GSEA on RNA-Seq analyses of G12Ci-treated CRC patient-derived xenograft (PDX) models. Since combination treatment with EGFR inhibitor has shown significant enhancement of G12Ci response in CRC patients (30), we evaluated CRC PDX tumors treated with G12Ci (sotorasib) alone, or in combination with the EGFR inhibitor panitumumab, the response profiles of which we have described previously (31). We observed robust enrichment of the PDAC KRAS UP signatures in three of four PDX tumors treated with sotorasib alone, whereas Hallmark KRAS UP signature enrichment was only weakly detected (Fig. 1E). Upon concurrent treatment with panitumumab, we observed induction of the PDAC KRAS UP signature in the one CRC PDX that did not show a strong transcriptional response to G12Ci alone. Thus, our PDAC KRAS-dependent gene signature identified KRAS inhibition in vivo in pancreatic and other KRAS-mutant tumor types.
The PDAC KRAS-dependent transcriptome is driven through ERK
We next wanted to assess the specific KRAS effector signaling networks important for KRAS regulation of gene transcription. Recent studies have identified putative resistance mechanisms driving relapse in patients with NSCLC and CRC who initially responded to KRASG12C inhibitors (32-34). These studies identified genetic alterations that reactivate RAF or PI3K effector signaling networks, supporting their key roles in driving mutant KRAS-dependent growth of NSCLC and CRC.
To determine the degree to which RAF-MEK-ERK activation is sufficient to support KRAS-driven PDAC growth, we established cell lines stably expressing the constitutively activated phosphomimetic mutant MEK1S218/222D (MEK1-DD) in KRASG12C (MIA PaCa-2) and KRASG12D (Pa14C and Pa16C) mutant PDAC cell lines (Fig. 2A, and fig. S2A). Although MEK1-DD is insensitive to MEKi inhibition of RAF-mediated phosphorylation, it remains sensitive to MEKi inhibition of the homodimerization that is important for mediating ERK activation. We first determined the ability of activated MEK1-DD to drive resistance to pharmacologic inhibitors targeting components upstream (KRAS, RAF/LY3009120) or downstream (MEK/trametinib, ERK/SCH772984) of MEK. Immunoblot analyses showed that MEK1-DD alone was sufficient to prevent loss of phosphorylated and activated ERK1/2 (pERK) following treatment with inhibitors of mutant KRAS or RAF, while retaining sensitivity to inhibitors of MEK or ERK.
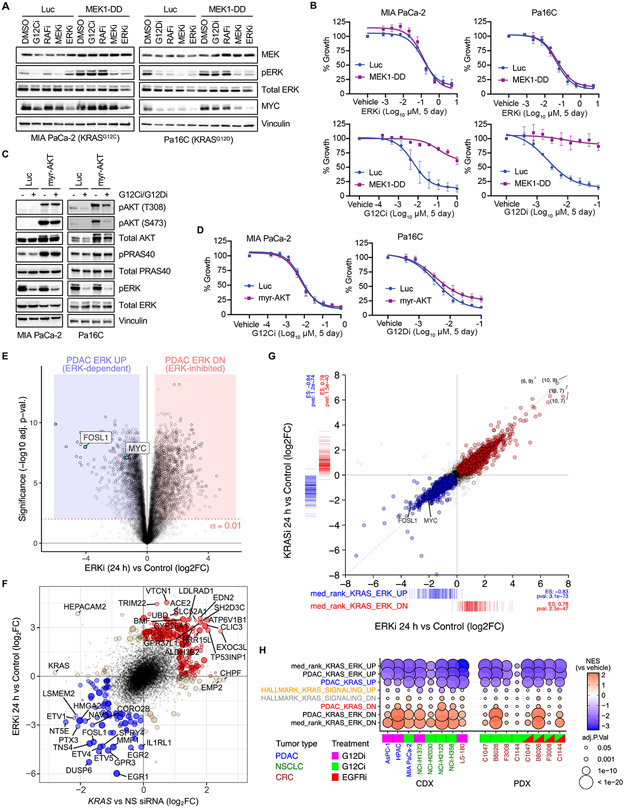
Fig. 2. Mutant KRAS is largely dependent on ERK for PDAC proliferation in vitro.
(A) Immunoblot analysis of ERK activity in PDAC cell lines stably infected with control vector (Luc) or constitutively activated MEK1 (MEK1-DD), treated with vehicle (DMSO) or indicated inhibitors of each level of the RAF-MEK-ERK cascade (G12Ci/G12Di, MRTX1257/MRTX1133 (20 nM); RAFi, LY3009120 (600 nM); MEKi, trametinib (6 nM); ERKi, SCH772984 (600 nM). Images are representative of 2-3 biological replicates. (B) Growth of PDAC cells stably infected with control vector (Luc) or activated MEK1-DD and treated with the indicated inhibitors. Error bars indicate SE of the mean with 3-4 biological replicates, each with three technical replicates. (C) Immunoblot analysis of ERK and AKT phosphorylation in PDAC cells stably infected with control vector (Luc) or constitutively activated AKT (myr-AKT), treated with G12Ci/G12Di (MRTX1257/MRTX1133, 20 nM) or ERKi (SCH772984, 600 nM). Representative of 2-3 biological replicates. (D) Growth of PDAC cells expressing activated AKT or control vector (Luc) and treated with KRAS G12C/D inhibitors. Error bars indicate SE of the mean with 3-4 biological replicates, each with 3 technical replicates. (E) Differential gene expression analysis for seven PDAC cell lines subjected to ERKi (SCH772984, 1000 nM) treatment for 24 hours versus paired untreated control cells. (F) Top 200 UP/DN genes from 24 hours KRAS siRNA and ERKi RNA-seq experiments are shown as filled blue (UP) or red (DN) circles, with a positive predictive value (agreement/total) for logFC = 0.84. Genes with strongest logFC values (n = 40) are labeled. (G) Evaluation of DE genes following 100 nM treatment of Pa16C PDAC cells with KRASi-G12Di (MRTX1133) or ERKi (SCH772984) for 24 hours (each condition, n = 2). Colored points are DE genes in either treatment (FDR < 0.05); red, log2FC > 0 with either treatment; blue, log2FC < 0 with either treatment; tan, log2FC opposite directions between treatments. Coordinate labels indicate points outside of plotting range. Barcode plots below and left represent log2FC for “median rank KRAS-ERK” signature genes within each experiment and ssGSEA enrichment statistics. (H) GSEA for PDAC KRAS UP/DN, PDAC KRAS-ERK UP/DN, and KRAS-ERK UP/DN based on median rank, along with Hallmark KRAS Signaling signatures in CDX and PDX models treated with G12Di, G12Ci or G12Ci+EGFRi with replicates as described in Fig. 1E. Inhibitors used in (H): G12Ci (CDX), adagrasib; G12Di, MRTX1133; G12Ci (PDX), sotorasib; EGFRi, panitumumab.
Next, we determined if activated MEK1-DD modulated sensitivity to inhibitor-mediated growth suppression. As expected, MEK1-DD expressing cells retained full sensitivity to ERKi and MEKi but not RAFi (Fig. 2B, and figs. S2B and C). Somewhat surprisingly, given that KRAS also utilizes non-RAF effectors to drive PDAC growth (3), MEK1-DD expressing cell lines showed complete to near-complete resistance to KRASG12C (G12Ci/MRTX1257) and KRASG12D (G12Di/MRTX1133) selective inhibitors.
PI3K is a second key effector supporting KRAS-driven cancer growth and does so primarily through activation of AKT (1). To assess the contribution of this effector in KRAS-dependent PDAC growth, we established cell lines stably expressing a constitutively activated, membrane-targeted variant of AKT1 (myr-AKT) (Fig. 2C). Myr-AKT expressing cells exhibited elevated phosphorylated and activated AKT (pAKT) and increased phosphorylation of the AKT substrate PRAS40 (pPRAS40). Surprisingly, no significant resistance to G12Ci or G12Di treatment was observed (Fig. 2D), indicating that PI3K-AKT signaling alone is not a significant driver of KRAS-dependent PDAC growth. Consistent with this lesser role, growth was strongly suppressed by inhibitors of RAF but not PI3K effector signaling (fig. S2D). Furthermore, treatment with a pharmacologic RAS(ON) multi-selective tri-complex inhibitor (RMC-7977) (35) inhibited growth that was associated primarily with a reduction in RAF but not PI3K effector signaling (fig. S2E). Based on these findings, we focused on the contribution of ERK MAPK signaling in driving KRAS-regulated gene transcription.
To assess the ERK-dependent transcriptome in KRAS-mutant PDAC, we performed additional analyses on our recently generated RNA-Seq profiling data set (PRJEB25806) (36) comprised of a panel of KRAS-mutant PDAC cell lines treated with the ERK1/2-selective inhibitor SCH772984 for 1, 4, 12, or 24 hours (ERKi). We identified 2,313 ERK-dependent (UP) and 2,648 ERK-suppressed (DN) genes during any time following ERKi treatment (logFC > 0.5, adj. p-val. < 0.01) (data S6). Most of these changes were observed at 24 hours, including 2,054 ERK-dependent (UP) and 2,519 ERK-suppressed (DN) genes (logFC > 0.5, adj. p-val < 0.01) (Fig. 2E and data S6). For consistency with our nomenclature for KRAS-dependent gene transcription, we refer to the gene set downregulated upon loss of ERK activity (i.e., upon ERKi treatment) as the PDAC ERK upregulated (UP) gene signature and the gene set upregulated upon ERKi treatment as the PDAC ERK downregulated (DN) gene signature.
To assess the role of ERK in KRAS-regulated gene transcription, we compared the top 200 DE UP/DN genes from ERKi (24 hours) and KRAS siRNA treatments versus control (Fig. 2F, colored points). In approximately 84% of cases, gene expression levels changed in the same direction under both treatments (Fig. 2F). We conclude that KRAS-regulated transcription is driven primarily through activation of the ERK MAPK effector signaling network. Consequently, we intersected siKRAS and ERKi UP/DN gene sets to derive PDAC KRAS-ERK UP/DN gene sets comprised of 278 and 307 genes, respectively (logFC > 0.5, adj. p-val. < 0.01, fig. S2F and data S4). Using over-representation analyses of KEGG, GO, and REACTOME gene sets, we found that the PDAC KRAS-ERK UP genes were predominantly enriched in cell cycle-related gene sets including chromosome segregation, RNA and DNA metabolism, and cell cycle processes (fig. S2G and H). Moreover, we recently observed that concurrent treatment of tumor xenografts with both G12Di (MRTX1133) and the anti-EGFR monoclonal antibody cetuximab impaired the compensatory ERK reactivation induced by G12Di alone and resulted in enhanced anti-tumor activity (28). The ability of this combination to block KRAS function more effectively was reflected in a more durable suppression of the KRAS-ERK gene signature (fig. S2I).
To further establish a KRAS-dependent gene signature that may be more broadly applicable to other tumor/tissue types, we next conducted differential expression (DE) analysis across eight KRASi-treated (24 hours) KRASG12C/D-mutant PDAC, CRC, and NSCLC CDX tumors (fig. S2J, data S7) (29, 37). We then compared DE genes (adj. p-val. < 0.01) among three experimental conditions for loss of KRAS or ERK function: siKRAS, ERKi, and KRASi, to find the top DE genes in at least two of the three conditions (median-rank of logFC). The top 200 negatively and positively changed genes from this analysis were used to identify the siKRAS-ERKi-KRASi UP and DN gene signatures, respectively (data S4), henceforth designated “median-rank” KRAS-ERK UP/DN. To validate that this generalized signature appropriately represented KRAS- and ERK-dependent expression, we evaluated cells of a single genetic background upon targeting different nodes of the KRAS-MAPK signaling pathway. We compared RNA-seq of a sensitive PDAC line (Pa16C) subjected to G12Di (MRTX1133), MEKi (trametinib), or ERKi (SCH772984) for 24 hours. The three inhibitors had largely the same effect on the transcriptome; (99.4% of 6,404 DE genes were similarly changed with G12Di versus ERKi, and 99.8% of 5,697 DE genes were similarly changed with KRASi versus MEKi (false discovery rate, FDR < 0.05 in at least one of two treatments). All treatments also showed strong enrichment of the median-rank KRAS-ERK UP/DN signature (Fig. 2G, fig. S2K, and data S8). Moreover, GSEA showed that these gene sets best recognized the gene transcription changes in G12C/D/EGFRi-treated CDX and PDX tumors in all cancer types examined (Fig. 2H).
Signaling networks are highly dynamic. A potential limitation of our median rank KRAS-ERK gene signatures is that we captured KRAS-ERK-dependent transcriptional changes at one time point. To first determine whether these KRAS-ERK transcriptional changes occurred acutely, we evaluated RNA-seq data (PRJNA947663) (38) of a series of 16 KRAS-mutant cancer cell lines treated for two hours with a pan-KRASi. While many KRAS-ERK-dependent genes exhibited expression changes at this early time, the degree of change was not as robust as upon 24 hours siKRAS or ERKi treatment (fig. S2L and data S9). To further assess temporally regulated gene transcription, we also evaluated changes in the median rank KRAS-ERK signatures across our aforementioned RNA-seq data sets upon various durations of treatment with ERKi or KRASi (fig. S2L). In both datasets we observed a largely monotonic increase in the magnitude of gene transcription changes, with the 24-hour time point for both showing strong overlap with the 24-hour median rank KRAS-ERK UP/DN gene sets. These observations support the relevance of median-rank KRAS-ERK signature genes to early responses to KRAS-ERK inhibition.
Integration of ERK-dependent transcriptomic, proteomic, and phosphosite activity
To delineate the mechanisms by which ERK regulates gene transcription, we first evaluated the gene promoters in ERK-regulated UP genes to identify putative transcription factor binding motifs. The predominant motifs in the top 50, 100, 500 and 1,000 genes were of the serum response factor (SRF; CArG DNA-binding motif), the FRA1 AP-1 subunit (TPA-responsive element), E2F (TTc/gGCGCg/c), and MYC (E-box motif CANNTG) transcription factors, respectively (Fig. 3A). Overall, for all 2,054 PDAC ERK-regulated UP genes, the E2F and MYC binding motifs were predominant (fig. S3A), consistent with the appearance of Hallmark E2F and MYC Targets among the top three KRAS-regulated gene sets (Fig. 1C).
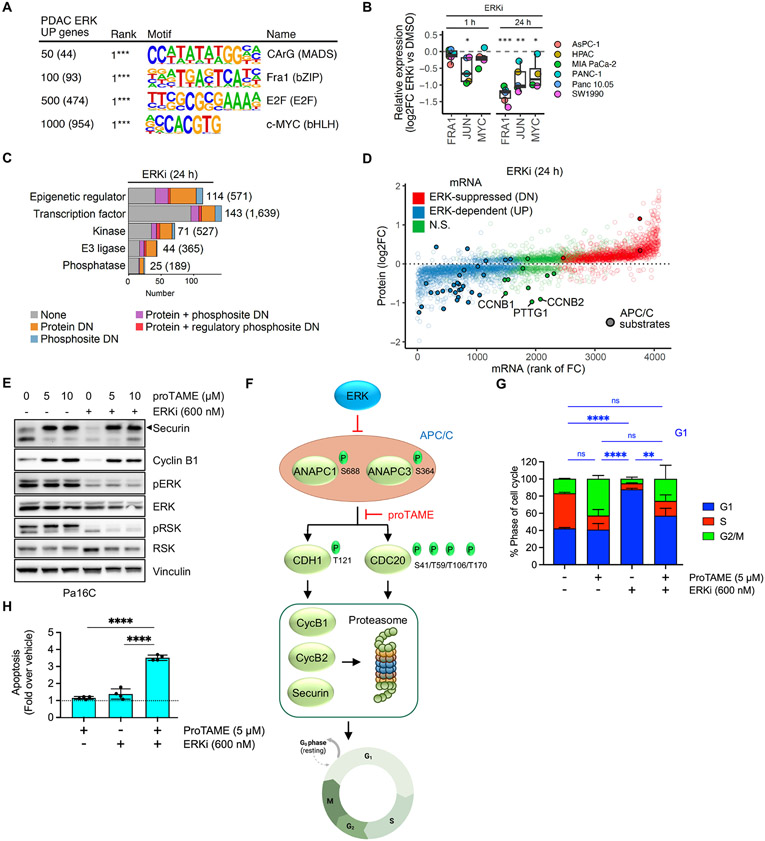
Fig. 3. Integration of transcriptomic, proteomic, and phosphosite activity data reveals multiple ERK dependent cell cycle control mechanisms.
(A) Supervised gene promoter motif enrichment analysis for different cutoffs of the top PDAC ERK UP genes. Numbers in parentheses indicate number of genes from each subset used in the analysis. (B) Protein abundance of AP-1 transcription factor components FRA1 and JUN, and of MYC, after 1 and 24 hours of ERKi, (SCH772984, 1000 nM) treatment across six PDAC cell lines, was determined by LC-MS2. (C) Categorization of proteins dependent on ERK for expression and/or phosphosite activity into five main functional groups. Numbers in parentheses indicate total number encoded in human genome. (D) Comparison of change in protein abundance (y-axis) with change in mRNA expression (x-axis) following ERKi treatment (24 hours) for proteins with significant changes (log2FC > 0.2 & adj. p-val. < 0.05). APC/C substrates are indicated by filled circles. (E) Immunoblot analysis of APC/C target proteins securin and cyclin B1 as well as ERK activity readouts pERK and pRSK following treatment with proTAME (APC/Ci) and/or ERKi in Pa16C cells. Images are representative of 2-3 biological replicates. (F) Schematic for ERK inhibition of APC/C E3 ligase function and degradation of mitotic regulators securin and cyclin B1/2. (G) Cell cycle distribution analysis of Pa16C cells treated with APC/Ci and/or ERKi for 24 hours (n = 3, each condition). (H) Apoptosis induction relative to vehicle control in Pa16C cells treated with APC/Ci and/or ERKi for 3 days (n = 4, each condition). (G-H), error bars represent +/− 1 S.D. All statistical comparisons, ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The expression/activity of these transcription factors can be driven by ERK through transcriptional and/or posttranscriptional mechanisms (39, 40). Therefore, we integrated the ERK-regulated phosphoproteome with proteomics from six KRAS-mutant PDAC cell lines treated for 1 or 24 hours with ERKi (fig. S3B and C). Among the earliest events (1 h) identified in the ERK-regulated phosphoproteome were decreased phosphorylation of RHO GTPase associated proteins and of 23 transcription factors that include regulatory sites of FRA1 and MYC (fig. S3C) (8). SRF is activated by RHO GTPase-stimulated F-actin polymerization (41), and our identification of direct ERK-regulated phosphosites of RHO-associated proteins is consistent with suppression of the SRF transcription factor motif.
Our evaluation of the ERK-regulated proteome found only three proteins rapidly downregulated at 1 hour ERKi, including members of the AP-1 transcription complex, JUN and JUNB. With 24 hours ERKi treatment we detected further downregulation of AP-1 proteins JUN and FRA1 as well as of MYC (Fig. 3B and data S10). The ERK-regulated phosphosites we identified (8) on these critical mediators of KRAS-dependent growth are known to regulate their protein stability (42, 43) (fig. S3C). Together, these data support an important role for ERK activity in maintenance of key transcriptional programs for PDAC growth.
We observed transcriptional downregulation of multiple E2F family transcription factors (fig. S3C). Though we were unable to detect E2F protein in the LC-MS2 proteomic assay, ERK regulation of E2F activity is likely mediated through loss of both mRNA transcript and protein levels of CCND1/3 (encoding cyclin D1/D3), CDK4, and CDK6, resulting in loss of RB1 transcripts and protein, and downregulation of RB hyperphosphorylation (fig. S3C).
We next assessed the impact of loss of ERK signaling on genes involved in aspects of protein regulation such as gene transcription, epigenetic modulation, protein phosphorylation and protein homeostasis (Fig. 3C). In the ERK-dependent (UP) gene set, we identified 114 non-kinase epigenetic regulators, 143 transcription factors, 71 protein/lipid kinases, 44 E3 ligases and 25 phosphatases. Additionally, in our ERK-dependent proteomics analyses (8), a significant fraction within each of these categories also corresponded to proteins that showed ERK-dependent changes in total protein or phosphorylation. Thus, the ERK-regulated transcriptome encodes proteins that can drive broad secondary changes in gene and protein activity/expression levels.
To further identify posttranscriptional regulatory mechanisms, we determined the relationship between the ERK-regulated transcriptome and proteome. Overall, 594/769 (77%) PDAC ERK UP proteins corresponded to PDAC ERK UP genes (adj. p-val. < 0.05) and 715/1019 (70%) PDAC ERK DN proteins corresponded to PDAC ERK DN genes (adj. p-val. < 0.05) (Fig. 3D, fig. S3D, data S6 and S10). Thus, while most ERK-regulated changes in protein expression are driven at the level of gene transcription, posttranscriptional regulatory mechanisms are also present.
Among ERK-dependent proteins that did not have corresponding changes in their mRNA transcripts, we investigated protein degradation as a potential mechanism of regulation. We observed a conspicuous loss of the anaphase promoting complex/cyclosome (APC/C) target proteins securin (PTTG1) and cyclin B1 and B2 (CCNB1 and CCNB2) (44) following ERKi treatment (Fig. 3D, fig. S3D). We also found the set of diminished proteins without mRNA loss (fig. S3D) was enriched in the KEN box (enrichment 1.6, p-val. 3.4E-3), a degron motif for the APC/C. Consistent with this, we identified many known APC/C substrates with decreased protein abundance (Fig. 3D). We verified that 24 hours ERKi reduced protein levels of securin and cyclin B1 and that concurrent treatment with the APC/C inhibitor ProTAME (45) (APC/Ci) prevented this reduction (Fig. 3E). These proteins are critical for cell cycle progression (44) and therefore give credence to a model where ERK activity suppresses APC/C function, leading to accumulation of the APC/C target proteins cyclin B1 and B2 to promote mitotic progression.
ERK regulation of APC/C is not well understood. APC/C activity is regulated by various complex posttranscriptional mechanisms that include phosphorylation. We leveraged our ERK-dependent phosphoproteome to determine if ERK regulates the phosphorylation state of components of APC/C. We identified rapid (1 hour ERKi) dephosphorylation of ANAPC1/APC1 (S688) and ANAPC3/APC3 (S364) (8), two structural components of the 19-subunit APC/C complex (Fig. 3F). Our kinase motif analysis predicted both phosphorylation sites to be direct ERK targets, thus potentially linking regulation of APC/C to ERK activity. By 24 hours of ERK inhibition, we found dephosphorylation of the two primary APC/C activators that regulate target associations: CDC20 (S41, T59, T106, and T170) and CDH1/FZR1 (T121).
The KRAS-ERK transcriptome regulates cell cycle progression and growth
We previously reported that ERKi potently inhibits cell cycle progression and causes G1 arrest in PDAC (5, 46). Deregulated APC/C function can likewise impact multiple cellular processes that contribute to cancer growth, including progression through G1 and mitosis. Treatment with the APC/Ci reduced the viability of PDAC cells (fig. S3E) and caused accumulation in G2/M (Fig. 3G and fig. S3F) but did not result in apoptosis (Fig. 3H). Concurrent APC/Ci and ERKi treatment partially reversed the cell cycle perturbations seen with each inhibitor alone and greatly enhanced apoptosis (Fig. 3H). Moreover, KRASi (G12Di) also resulted in degradation of securin and cyclin B1 (fig. S3G) and the combination with APC/Ci fully reversed the KRASi induced G1 arrest (fig. S3H). These observations support a role for KRAS-ERK suppression of APC/C function to facilitate cell cycle progression and survival.
Finally, we recently applied the multiplexed inhibitor bead (MIB)/mass spectroscopy (MS) chemical proteomics assay for kinome-wide profiling of activity and/or expression changes caused by ERK inhibition (24 hours) in a panel of six KRAS-mutant PDAC cell lines (21, 47). Although the original assay method was proposed to monitor kinase catalytic activity, comparison of our MIB/MS data with our total protein and gene transcription data indicate that, with limited exceptions (e.g., ERK1/2), the MIB/MS assay largely reflects changes in kinase protein abundance (fig. S3I). When we compared the KRAS-dependent changes observed in the MIB/MS assay to protein and transcript changes, we found a positive correlation with kinase mRNA and protein expression across the six cell lines (fig. S3J). Of the kinases downregulated in both assays, 11 are known to play driver roles in cell cycle progression and specifically in mitosis (e.g., Aurora A, PLK1, and WEE1). We also identified ERK-regulated phosphorylations on many of these kinases and a complete collapse of cell cycle regulatory components. Together, we found ERK-dependent regulation of nearly all aspects of cell cycle progression at the levels of gene transcription, protein expression, protein activity, protein stability, and protein phosphorylation. These mechanisms contribute crucially to the strong suppression of the Hallmark G2/M Checkpoint gene signature observed upon loss of KRAS (Fig. 1C).
We next evaluated which signaling pathways were enriched in KRAS and KRAS-ERK upregulated genes across three KRAS signatures: PDAC KRAS-ERK UP, PDAC KRASi UP, and PDAC iKras UP. Analysis of Reactome pathways revealed strikingly predominant regulation of cell cycle and mitotic processes in both the cell-based KRAS-ERK UP and in vivo PDX-based KRASi UP signatures (Fig. 4A, fig. S4A), consistent with the strong alignment of those gene sets (fig. S1L, M, N). In contrast, and despite otherwise significant overlap (fig. S1J, K, N), the in vivo mouse tumor-derived iKras UP signature was enriched in metabolic processes, consistent with the observed metabolic phenotype of these tumors (27). Further, as we anticipated given the limited overlap of the PDAC KRAS UP with the Hallmark KRAS Signaling UP signature (Fig. 1A), RTK signaling and immune processes predominated in the Hallmark gene set (fig. S4A). Together, these analyses revealed that, while the KRAS-ERK and KRASi signatures are dominated by ERK-dependent regulation of cell cycle progression, the frequently consulted Hallmark KRAS Signaling signatures are devoid of these essential cancer-driving genes.
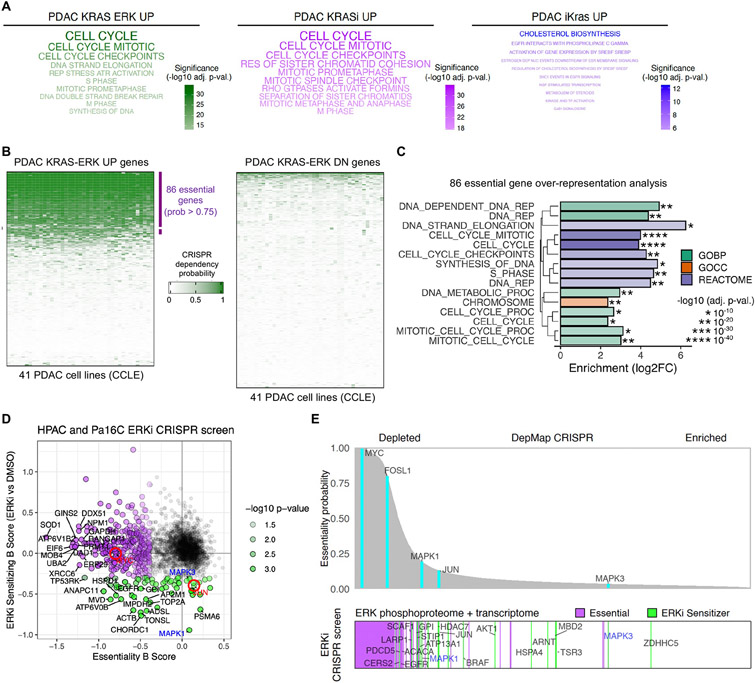
Fig. 4. KRAS-ERK dependent genes are essential for cell proliferation in PDAC.
(A) Over-representation analysis for REACTOME terms in three KRAS signatures: PDAC KRAS-ERK UP, PDAC KRASi UP, and PDAC iKras UP. Top 10 terms are shown. (B) Comparison of CRISPR dependency probabilities to gauge essentiality of genes in PDAC KRAS-ERK UP/DN signatures across 41 KRAS-mutant PDAC cell lines from CCLE. (C) Over-representation analysis for PDAC KRAS-ERK UP essential genes using KEGG, GO, and REACTOME. BP, biological process; CC, cellular component. (D) CRISPR drop-out screen using sgRNA library generated from ERK-dependent phosphoproteins and transcripts. Beta scores calculated with MAGeCK. Red circles highlight MYC and JUN. (E) Comparison of genes highlighted in (D) (bottom) with essentiality probabilities averaged across the 41 KRAS-mutant PDAC cell lines in DepMap (top).
We then examined the representation of genes essential for PDAC growth or survival (prob. >0.75) in the PDAC KRAS-ERK UP/DN signatures, using data available from the Cancer Dependency Map (DepMap) database. Across the 41 KRAS-mutant PDAC cell lines in DepMap, we found that essential genes were largely downregulated upon ERKi treatment (878/1016, 86.4%, data S11) and thus were more frequently observed within the PDAC KRAS-ERK UP than PDAC KRAS-ERK DN genes (31% vs 0.6%, respectively) (Fig. 4B). Cell cycle gene programming was a prominent feature within the top essential PDAC KRAS-ERK dependent genes (Fig. 4C, and fig. S4B).
We confirmed by pharmacological inhibition in three PDAC cell lines that expression of a subset of proteins from the KRAS-ERK UP gene signature was dependent on both KRAS and ERK (fig. S4C, data S1 and 6). These include proteins important for cell cycle control such as MYC, cyclin D1 (CCND1), Aurora B (AURKB), CHK1 (CHEK1), FRA1 (FOSL1), WEE1, and RAD51, along with chromosome separation such as SKA1, NUF2, and NCAPH1 (NCAPH). Importantly, expression of constitutively activated MEK (MEK1-DD) was unable to rescue cell growth following treatment with well-validated inhibitors of some of these ERK-dependent genes with high essentiality, such as CHK1, WEE1, or CDK4/6, confirming their essentiality downstream of ERK (figs. S4D and E).
DepMap analyses predict PDAC gene dependencies that may be associated with KRAS-ERK driver function. To determine genetic dependencies that sensitize cells to ERK inhibition and additionally to provide mechanistic analyses of ERK-dependent genes and phosphoproteins, we constructed a focused CRISPR-Cas9 library targeting 1,261 KRAS-ERK regulated UP genes and 1,930 ERK-dependent phosphoproteins (8). We applied this library to perform a loss-of-function screen in cells treated with a sublethal dose of ERKi (40 nM). We identified 354 genes with depleted sgRNAs during the duration of the experiment (Fig. 4D and data S12, beta score < −0.3, p-val. < 0.05). This essential gene set matched dependencies reported in DepMap CRISPR analyses across the 41 KRAS-mutant PDAC cell lines (Fig. 4E).
Consistent with previous analyses, 68 genes were solely or further depleted in ERKi treated cells (beta score > −0.3, p-val. < 0.05) included the AP-1 transcription factor subunit JUN, MAPK3/1 (ERK1/2) and other members of the RAS-MAPK signaling axis, such as EGFR and BRAF, as well as members of the PI3K signaling axis, such as AKT1, RPTOR (RAPTOR), and LARP1. Additional ERKi sensitizer genes encode proteins with previously identified roles in PDAC such as BCAR1 (p130Cas), TOP2A, SMARCA5, and WWTR1 (TAZ), as well as others with unknown roles in PDAC (Fig. 4E). These genes represent putative synthetic lethal interactions with KRAS or ERK inhibition.
Clinical validation of KRAS-ERK gene signatures
Having established that our PDAC KRAS-dependent transcriptome is regulated predominantly through the ERK MAPK signaling network, we then assessed whether this gene signature may have clinical relevance. Recently, we completed a Phase Ib clinical trial where PDAC patients were treated with the ERK1/2-selective inhibitor (ERKi) ulixertinib/BVD-523 together with nab-paclitaxel plus gemcitabine (NCT02608229) (48). Paired tumor biopsies were taken before and following the initial two-week treatment lead-in with ulixertinib alone, and immunohistochemistry analyses were done to assess ERK inhibition (as reported elsewhere (48)). No significant reductions in pERK or MYC protein levels were observed. Thus, it was not clear whether target inhibition was achieved in the treated patients.
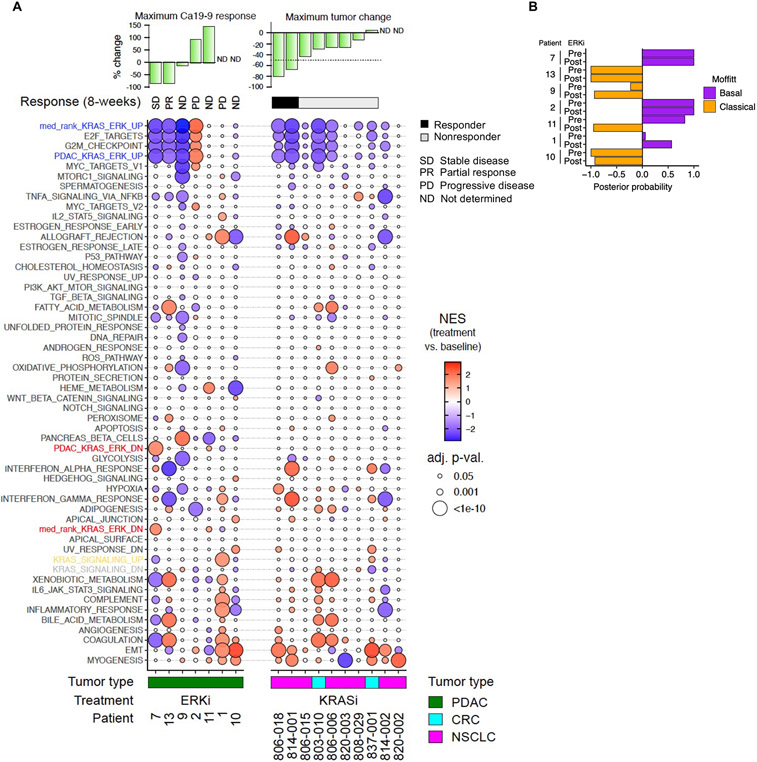
To provide a more sensitive analysis of target inhibition, RNA-Seq was also done on tumors pre- and 14 days post-treatment. We used GSEA to evaluate the KRAS-ERK median rank signature (Fig. 2G and data S4), the PDAC KRAS-ERK signature (Fig. S2F and data S4), and 50 Hallmark gene sets. RNA-Seq alignment data indicated that all seven patients evaluated were KRAS mutant. In three patients treated with ulixertinib (patients 7, 13 and 9), we noted a substantial loss of expression of genes in the KRAS-ERK signatures, as well as of the Hallmark cell cycle gene sets E2F Targets and G2M Checkpoint. This corresponded with reductions in CA 19-9, a serum biomarker used to monitor tumor response to therapy (49), suggesting that ulixertinib indeed diminished ERK-dependent transcriptional activity in those patients.
We did not observe a relationship between the KRAS-ERK signatures and the basal or classical subtype of PDAC, nor did we observe a change in PDAC subtype upon ERKi treatment in 6 of 7 patient tumors (Fig. 5B). Taken together, we did not observe a relationship between PDAC subtype and KRAS-ERK dependency in our analyses of cell lines in vitro (fig. S1E) and tumor xenografts in vivo (Fig. 5B). Thus, we did not find support for PDAC basal versus classical subtype as a predictive marker for response to KRAS or ERK inhibitor therapy.
Fig. 5. Changes in KRAS ERK dependent genes coincide with ERKi treatment response in patient tumors.
(A) GSEA evaluation of the median rank KRAS-ERK UP/DN, PDAC KRAS-ERK UP/DN and Hallmark signatures in patient biopsies, pre- and post-treatment. Patients with KRAS-mutant PDAC were treated with ERKi ulixertinib (14 days). Patients with KRASG12C-mutant NSCLC or CRC were treated with the KRASG12C-selective KRASi adagrasib (8 days). The serum biomarker CA 19-9 was used to monitor treatment response, and patient response was assessed according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1). NES, normalized enrichment score. (B) Evaluation of PDAC subtype in pre- and post-treatment patient biopsies using the Moffitt classifier (23).
To extend our findings beyond PDAC, we next evaluated our KRAS-ERK signatures in 10 KRASG12C-mutant lung or colorectal cancer patients treated with the G12Ci adagrasib, two of whom responded according to RECIST criteria (Fig. 5A and data S13). GSEA together with the 50 Hallmark signatures was conducted in targeted RNA-Seq datasets derived from needle core tumor biopsy samples pre- and 8 days post-treatment. The KRAS-ERK UP signatures, indicating KRAS dependence, were strongly altered in the two responders and also in two nonresponders whose tumors shrank but not enough to meet RECIST criteria. The remaining six nonresponders showed limited to no change in these signatures. Thus, there was a general association of the KRAS-ERK signature enrichment with tumor response. Surprisingly, enrichment in the Hallmark KRAS Signaling UP signature was largely absent from tumor expression changes, although Hallmark E2F Targets and G2/M Checkpoints signatures tracked with KRAS-ERK UP signatures, as we had observed in PDAC patients treated with ERKi (Fig. 5A). We are presently expanding these analyses to a larger patient population to determine if our KRAS-ERK signatures will have predictive value to define responsive versus nonresponsive patients.
Finally, unlike our findings in human CDX tumors treated with G12Ci adagrasib or G12Di MRTX1133 for 24 hours (Fig. 2H), we did not detect the KRAS-ERK DN signatures in patients treated with either ERKi ulixertinib or G12Ci adagrasib. These compensatory gene expression changes may be transient and not detected in the longer patient treatment windows (14- or 8-days, respectively).
Discussion
The mechanisms by which aberrant KRAS signaling drives cancer growth remain poorly understood. Here, we completed a systemwide determination of the KRAS-dependent transcriptome in KRAS-mutant PDAC and demonstrated that KRAS regulation of gene transcription and growth are driven predominantly through the ERK signaling network. We established consensus KRAS-ERK gene signatures that, unlike the widely applied Hallmark KRAS signature, accurately identified the molecular consequences of pharmacologic inhibition of mutant KRAS in KRAS-mutant PDAC, NSCLC and CRC tumor xenografts.
When evaluated in the same genetic background, we found that KRAS- and ERK-dependent gene transcription shared near 100% overlap in KRAS-mutant PDAC. Similarly, the KRAS- and ERK-phosphoproteomes were also near-identical (8). Although a previous study determined that the inability of mutant KrasG12D to engage PI3Kα caused a near complete loss of lung carcinogenesis (50) , this may not be relevant for PDAC. By contrast, we recently determined that KRASG12R is impaired in binding PI3Kα, yet is the third most common KRAS mutation in PDAC (15%; cBioPortal GENIE Cohort v15.0). That KRASG12R is found rarely in KRAS-mutant NSCLC (0.2%) and CRC (0.6%) suggests the possibility that the minor role of PI3K in KRAS-mutant PDAC may be specific for this tissue (51).
To fully elucidate the molecular mechanisms by which aberrant KRAS-ERK signaling drives deregulated cell cycle regulation and growth, we found it essential to integrate our determinations of the ERK-dependent transcriptome together with the ERK-dependent phospho- and total proteome. We identified mechanisms that impinged on multiple stages of cell cycle progression that included ERK-dependent disruption of APC/C function. Since direct inhibition of ERK as a therapeutic approach to block KRAS function has been hampered by normal cell toxicity, we further identified ERK-regulated genes that displayed synthetic lethality with ERK inhibition.
Finally, we found that enrichment of our KRAS-ERK gene signatures was consistent with tumor responses, including regressions, in a limited set of patients treated with an ERK inhibitor or a KRASG12C-specific inhibitor, suggesting possible correlation with target inhibition. That less than half of KRASG12C mutant patients respond to KRASG12C inhibitors (52, 53) may reflect insufficient target inhibition rather than loss of KRAS dependency, supporting the need for molecular signatures that accurately monitor pharmacologic disruption of mutant KRAS signaling. In summary, our determination of the KRAS-ERK regulated transcriptome together with our concurrent determination of the ERK-regulated phosphoproteome (8) provide the most comprehensive profile of the highly complex molecular events that drive KRAS- and ERK-dependent cancer growth.
Materials and Methods
Cell lines
Patient-derived xenograft (PDX) human KRAS-mutant PDAC cell lines (Pa01C, Pa02C, Pa04C, Pa14C, and Pa16C) were provided by A. Maitra (MD Anderson Cancer Center) and were maintained in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Established PDAC cell lines (AsPC-1, HPAC, MIA PaCa-2, PANC-1, and SW1990) were obtained from the American Type Culture Collection (ATCC) and maintained in RPMI 1640 supplemented with 10% FBS (AsPC-1 and SW1990) or DMEM supplemented with 10% FBS. PDAC line identity was verified by short tandem repeat analysis and were evaluated and determined negative for mycoplasma.
Plasmid constructs and viral transduction
Stable cell lines were generated using lentivirus expression vectors encoding untagged MEK1DD (MEK1S218D/S222D-pcw107), Myr-AKT (myr-FLAG-AKT1-pcw107), or LUC (luciferase-pcw107), all of which were provided by K. Wood (Duke) and described previously (57). Lentivirus was generated using HEK293T cells plated in T25 flasks (0.9 X 106 cells/flask) in DMEM supplemented with FBS (10%). The following day, cells were transfected with the target construct (4 μg), psPax (3 μg), and pMD2.G (1 μg) diluted in 500 μl of OptiMEM and combined with 25 μl of Fugene 6 (Promega). The medium was replaced 24 hours post transfection with DMEM supplemented with 20% FBS and incubated for 48 hours. The supernatant was removed and cleared using 0.45 μm PES syringe filter (Nalgene) and either used directly to infect target cells or frozen in aliquots at −80°C for later use. Cells were incubated for 12 hours with viral supernatant diluted with DMEM supplemented with 10% FBS and polybrene (8 μg/ml). Cells were then given fresh media and puromycin selection was initiated 24-48 hours post transduction.
Inhibitors
The ERK1/2-selective inhibitor SCH772984 (58) was provided by Merck, the pan-RAF inhibitor LY3009120 (59) was provided by Eli Lilly, and the MEK1/2-selective inhibitor trametinib was obtained from Selleckchem (S2673). The KRASG12C-selective inhibitor MRTX1257 and the KRASG12D-selective inhibitor MRTX1133 were synthesized at WuXi AppTec (Wuhan, China) or purchased from Selleckchem (E1051). Sotorasib was obtained from MedChemExpress (HY-114277) and panitumumab from McKesson . ProTAME was purchased from MedChem Express (HY-124955). The CHK1 inhibitor prexasertib (LY2606368, S7178), the WEE1 inhibitor adavosertib (S1525), and the CDK4/6 inhibitor palbociclib (S4482) were obtained from Selleckchem. The RAS(ON) multi-selective tri-complex inhibitor RMC-7977 was obtained from Revolution Medicines.
Antibodies
The following primary antibodies were used for immunoblotting. From Cell Signaling Technology: p44/42 MAPK (ERK1/2) (L34F12), cat. no. 4696 [RRID:AB_390780]; phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (D13.14.4E), cat. no. 4370 [RRID:AB_2315112]; phospho-p90RSK (Thr359/Ser363), cat. no. 9344 [RRID:AB_331650]; RSK1/2/3 (32D7), cat. no. 9355 [RRID:AB_659900]; c-MYC (D84C12), cat. no. 5605 [RRID:AB_1903938]; MEK1/2 (L38C12). cat. no. 4694 [RRID:AB_10695868]; phospho-AKT (Ser473), cat. no. 9271 [RRID:AB_329825]; phospho-AKT (Thr308) (D25E6), cat. no. 13038 [RRID:AB_2629447]; AKT (pan) (40D4), cat. no. 2920 [RRID:AB_1147620]; phospho-PRAS40 (Thr246) (C77D7), cat. no. 2997 [RRID:AB_2258110]; phospho-S6 (Ser235/236), cat. no. 2211 [RRID:AB_331679]; S6 (54D2), cat. no. 2317 [RRID:AB_22238583]; phospho-histone H2A.X (Ser139) (20E3), cat. no. 9718 [RRID:AB_2118009]; cyclin D1 (E3P5S) XP, cat. no. 55506 [RRID:AB_2827374]; DDX21, cat. no. 75762 [RRID:NA]; PRAS40 (D23C7), cat. no. 2691 [RRID:AB_2225033]; WEE1 (D10D2), cat. no. 13084 [RRID:AB_2713924]; FRA1 (D80B4), cat. no. 5281 [RRID:AB_10557418]; Aurora B/AIM1, cat. no. 3094 [RRID:AB_10695307]; CHK1 (2G1D5), cat. no. 2360 [RRID:AB_2080320]; phospho-cdc2 (Tyr15), cat. no. 4539 [RRID:AB_560953]; Securin (D2B6O), cat. no. 13445 [RRID:AB_2798220]; and cyclin B1, cat. no. 4138 [RRID:AB_2072132]. From Sigma-Aldrich: vinculin, cat. no. V9131 [RRID:AB_477629]; KRAS (3B10-2F2), cat. no. WH0003845M1 [RRID:AB_1842235]. From Invitrogen: RAD51, cat. no. PA5-27195 [RRID:AB_2544671]; SKA1, cat. no. PA5-61019 [RRID:AB_2647324], and NUF2, cat. no. MA5-31536 [RRID:AB_2787164]. From Thermo Fisher: phospho-RSK1 (Thr359/Ser363), cat. no. PA5-38309 [RRID:AB_2554910]; and from Proteintech: NCAPH (3D2F11), cat. no. 67655-1-IG [RRID:AB_2918495]. Secondary antibodies used for immunoblotting included IgG (H+L) cross-adsorbed goat anti-mouse, HRP, Invitrogen cat. no. PI31432, and IgG (H+L) cross-adsorbed goat anti-rabbit, HRP, Invitrogen cat. no. PI31462, both from Fisher Scientific.
siRNA transfection
KRAS siRNA (5’ GCCUUGACGAUACAGCUAAtt 3’) (Thermo Fisher) cat. No. 4390824 or NS siRNA (5’ XXXXXXXXXX 3’) (Thermo Fisher) cat. no. 4390843 ID s7940 were performed using lipofectamine RNAiMax (ThermoFisher), cat. no. 13778150, and 10 nM of siRNA in Opti-MEM. Opti-MEM was combined with RNAiMax and incubated for 5 minutes to equilibrate. The siRNA(s) were then added and incubated for another 20 minutes. All incubations were done at room temperature.
Viral transduction
Following plating in T25 flasks, HEK293T cells (0.8 X 106) were incubated overnight in DMEM supplemented with 10% FBS. Cells were transfected using Fugene 6 according to the manufacturer's recommendation (500 μl of OptiMEM was combined with 25 μl of Fugene 6, 3 μg of psPax, 1 μg of pMD2.G, and 4 μg of target construct). Cells were incubated for 12 hours and then the medium was replaced with DMEM supplemented with 20% FBS. The cells were incubated for a further 48 hours, at which time the viral supernatant was collected and filtered through a 0.45 μm PES syringe filter (Nalgene). Polybrene (8 μg/ml) plus the filtered viral supernatant was used to infect target cells. Fresh medium was provided 12 hours post infection. Selection was then initiated 24 hours after transduction.
PDAC cell line RNA-seq analyses
To determine the KRAS-dependent transcriptome, total RNA for RNA-seq analyses was isolated from samples of eight KRAS-mutant PDAC cell lines (HPAC, MIA PaCa-2, Pa01C, Pa02C, Pa04C, Pa14C, Pa16C, and PANC-1) subjected to 24 hours of KRAS or NS siRNA transfection. To evaluate the timing, tissue type specificity, and comparison to pharmacological inhibition of KRAS, total RNA was then also isolated from NCI-H358 (NSCLC), SW837 (CRC), and MIA PaCa-2 (PDAC) cells, all subjected to KRAS-G12Ci MRTX1257 (20 nM) for 0, 2, 6, 12, and 24 hours. Illumina TruSeq library preparation kits were used to generate cDNA libraries with poly(A) mRNA selection using oligo(dT) magnetic beads. Agilent Bioanalyzer and Invitrogen Qubit were used for cleaning and quantitation of PCR amplified products. Sequencing was conducted on the Illumina HiSeq 2500 generating 150 bp paired-end (PE) reads. RNA-seq for Pa16C drug treatment study was conducted via isolation of total RNA from Pa16C cells with 24 hours of treatment followed by generation of poly(A) mRNA stranded cDNA libraries at Novogene (Sacramento, CA) and sequencing on an Illumina NovaSeq 2000. RNA-seq analyses on KRAS-mutant PDAC cell lines (HPAC, HPAFII, Pa01C, Pa04C, Pa14C and PANC-1) treated with ERKi SCH772984 for 24 hours were done as described previously (36).
CRC PDX RNA-seq analyses
Establishment and drug treatment of CRC PDX models were done as we described previously (31, 60). Treatment was discontinued and the mice were sacrificed after 3 weeks (21 days). Tumors of three mice per arm were harvested 2 hours post treatment. The tumors were segmented and immediately flash frozen in liquid nitrogen for subsequent RNA-seq analysis. Snap-frozen CRC PDX tumors were submitted to Admera Health for RNA sequencing. Raw sequence reads in the form of FASTQ files were pre-processed using Xenome (v1.0.1) (61) to classify human and mouse reads. Human reads were aligned to the human reference genome (GRCh38) using STAR aligner (v2.7.2b) (62), with duplicated reads marked using biobambam (v0.0.191) (63). Reads were annotated using GENCODE (v22) (64). Un-normalized genes counts were generated using HTseq (v0.11.0) (65) and subsequently filtered to keep only protein-coding genes and then processed through the R software package, DESeq2 (v1.41.4) (66), to generate differentially expressed genes for pathway analysis. Pathway enrichment statistics were estimated with fGSEA (v1.26.0) (67) to evaluate the PDAC KRAS UP/DN, median rank KRAS-ERK UP/DN, KRAS-ERK UP/DN, and the 50 Hallmark signatures in sotorasib or sotorasib plus panitumumab treated KRASG12C metastatic CRC PDX pre- and post-treatment samples.
Patient RNA-seq sample collection
Imaging-guided biopsies were performed before and after two weeks of ulixertinib monotherapy patient treatment (NCT02608229) as we have described (48). Biopsied samples were snap-frozen for RNA-seq. Treatment response was assessed by measurement of serum CA19-9 levels and assessed according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1), time to progression (TTP). Phase II evaluation of adagrasib was completed as we have described (53), with RNA-seq conducted on biopsies performed before and after 8 days of treatment. For RNA-seq analyses of adagrasib-treated KRASG12C-mutant NSCLC and CRC patients, tumor samples were evaluated with the HTG EdgeSeq Transcriptome Panel. Library size was used to normalize raw counts, resulting in log2CPM expression values for each sample (edgeR v3.32.1, R v4.0.2). Lowly expressed genes, with mean log2CPM ≤ −1, were removed. Filtering was done based on mean expression values of a larger cohort of 111 NSCLC and CRC patient samples. Treatment response was assessed according to RECIST v1.1 and custom visualization of progression-free survival (PFS) was generated using ggplot2 (v3.3.5) in R.
Growth assays
Cells were plated in 96-well plates for growth assays. Approximately 1,000-2,000 cells were added per well, depending on the cell line, and grown for 24 hours, with an additional day 0 plate. Pharmacological inhibitors were added to cells 24 hours later using a Tecan D300e Digital Dispenser. The day 0 plate was labeled with calcein AM and counted live. Treated cells were then incubated for 5 days and then labeled with calcein AM. Cells were imaged and counted using a SpectraMax i3X Multi-Mode Detection Platform (Molecular Devices). Growth percentages for each biological replicate were calculated as normalized treated values with respective control samples set to 100% growth. GraphPad Prism (v9.3.1) was used to construct three-parameter drug response curves. Growth (%) represents the mean of three-four biological replicates and error bars depict the standard deviation between biological replicates. For each biological replicate, fold growth was calculated using the day 5 control wells divided by the mean value calculated from the day 0 plate. Fold growth was used to verify that cells were growing as expected in biological replicates.
Generation of CRISPR/Cas9 ERK phosphoprotein/transcript library
Genes for establishing an ERK-focused CRISPR library were selected as follows. Genes with significant phosphoproteome hits (∣log2FC∣ > 0.25, adj. p-val. < 0.05) were selected first. Positive control genes are those defined in DepMap PDAC (21Q4, Achilles) data with beta scores less than −1. A subset of RAS-ERK pathway members was included. Twentyfive negative control genes were selected from protein-coding genes (autosomal) not detected in PDAC RNA-seq data in this study or in the CCLE. Finally, additional genes that had transcript changes in 24 hours ERKi RNA-seq (∣logFC∣ > 0.5, adj. p-val. < 0.01) were also selected, for a total of 2497 unique ensembl gene IDs. For each gene, five sgRNAs were selected using CRISPick online (Broad) with Human genome GRCh38 Ensembl v.103 and SpyoCas9.
For library generation, oligos were ordered from TWIST Bioscience in the following format:
5’-GGAAAGGACGAAACACCGXXXXXXXXXXXXXXXXXXXXGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC – 3’
X* Denotes unique 20mer sgRNA sequence; total 73 bp ultramer
The guides were amplified using the following primers and conditions:
ArrayF: 5’ – TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG – 3’
ArrayR: 5’ – ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC – 3’
PCR was performed as follows: 1. Initial denaturation 98° (3 minutes); 2. Denaturation 98°C (20 seconds); 3. Annealing 63°C (15 seconds); 4. Extension 72°C (15 seconds) (steps 2-4 X8 cycles); 5. Final extension 72°C (2 minutes). The resulting PCR product was gel purified and ligated into lentiGuide-Puro (Addgene, 52963) using Esp3I. The ligated product was transformed into electrocompetent Endura cells (Lucigen, Cat # 60242-1) and serial plating was used to ensure the colony forming units exceeded 200x fold of the library size. Purified plasmid DNA was used to generate lentivirus as described above. Prior to infection, the virus was validated using Illumina NextSeq 500 with 75 bp single end reads. Multiplicity of infection (MOI) was titrated in the Cas9 stable expressing cells as described in (46). Stable Cas9 expressing PDAC cell lines were generated using the lentiviral transduction of pKLV2-EF1a-Cas9Bsd-W (Addgene, 68343) as described above.
CRISPR/Cas9 screening protocol
PDAC cells were infected at an MOI of 0.2 and then maintained in growth medium supplemented with puromycin. At 9 days post infection, genomic DNA was harvested and sequenced to determine library coverage. Cells were divided into growth medium supplemented with a sublethal concentration of SCH772984 (40 nM) or vehicle (DMSO) control. Each group contained four replicates of 107 cells. Cells were continuously cultured for 20 days in the presence of vehicle or drug, refreshed every 4 days. Each replicate was maintained and split independently and not allowed to reach confluency. For all replicates, 107 cells each were maintained to achieve >1000x coverage of the library. Following 20 days of vehicle/treatment, genomic DNA was harvested. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN) and analyzed using next-generation sequencing. Specifics on virus titering and infection, as well as sample preparation including DNA extraction, amplification, purification, and sequencing, has been described previously (46). To evaluate sgRNA abundance/enrichment for each gene, the prioritization algorithm in MAGeCK (v0.5.9.5) was used (68).
Flow cytometry assays: cell cycle
To analyze cell cycle, adherent cells were fixed, resuspended in 40 μg/ml propidium iodide (PI) and 100 μg/ml RNase A in PBS, and analyzed on a BD LSRFortessa flow cytometer. At least 20,000 cells per sample were exported and analyzed using FCS Express 7 (De Novo Software, Pasadena, CA). Cells were gated on SSC-A and SSC-H to discriminate singlet cells and SSC-A vs FSC-A to select cells from debris. Cell cycle phases were assigned using the Multicycle DNA fit function for cells based on PI height, with the three categories G0/G1, S, and G2/M defined from lower to higher PI intensity. Three biological replicates for each condition were performed and the mean values were graphed.
Immunoblot analyses
Cells were washed in cold PBS, then scraped into lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40 (Igepal, CA-630), 0.5% sodium deoxycholate, 0.1% SDS) supplemented with phosphatase (Sigma-Aldrich) and protease (Roche) inhibitors. Cells were incubated in lysis buffer for 10 minutes on ice and then centrifuged at 8,000 rpm for 10 minutes at 4°C. Cleared lysates were transferred to new tubes and protein concentrations were measured using Bio-Rad Protein Assay Dye Reagent cat. no. 5000006 according to the manufacturer’s protocol. Equal amounts of protein from each sample were combined with 4X Laemmli buffer and 2.5% 2-mercaptoethanol, boiled for 5 minutes, loaded at equal volume into SDS-PAGE gels and subjected to standard immunoblot procedures.
Drug Sensitivity Resistance Testing (DSRT) chemical library screen
Drugs used, their sources, concentration ranges, and more extensive method descriptions have been previously described (69). In brief, DMSO was used to dissolve compounds which were pre-printed on 384-well tissue culture plates (Corning) using the Echo 550 acoustic liquid handling device (Labcyte Inc.). Each compound was assayed across five concentrations in a 10,000-fold concentration range, without technical replicates. Single-cell suspensions were added to each plate, followed by incubation at 37°C for 72 hours. Cell viability was measured in a Paradigm plate reader (Molecular Devices) using the Realtime-Glo luminescent assay (Promega) according to the manufacturer’s protocol. Vehicle (DMSO) and positive (100 μM benzethonium chloride) controls were used to noromalize response readout and obtain relative growth inhibition percent. Dose-response data were first processed using Dotmatics Browser/Studies software (Dotmatics Ltd.) and then analyzed using the DSS pipeline (70).
RNA-seq data preprocessing
QC and adapter trimming of RNA-seq data were performed with TrimGalore (v0.4.5) (71), a wrapper for the tools FastQC (72) and Cutadapt (73), retaining paired end reads with 35 or more high quality (>28 Sanger/Illumina 1.9) bases. High quality reads were then mapped to the hg38 (GRCh38.p12) human genome using the STAR aligner (v2.7.8a) (62) and Gencode (v30) (64) basic transcriptome. Transcript quantitation was performed using Salmon (v0.11.3) (74) and summarized at gene level using the R package tximport (v1.22.0) (75). Gene annotations were assigned using biomaRt (v2.50.3) (76). RNA-seq experiment-wise data were managed and normalized using edgeR (v3.36.0 and 3.32.1) (77). Only protein coding genes and genes not encoded by Y-chromosome or mitochondrial DNA were considered prior to TMM (78) or counts-per-million (CPM) normalization and differential expression comparisons. Principal components analyses were conducted using the stats package of base R.
RNA-seq data analyses
The glmQLFTest function (EdgeR) was used to perform differential expression comparisons either across groups or using specific experimental contrasts. Gene set enrichment analyses were conducted using fGSEA (v1.26.0) (67) for custom gene sets as well as Hallmark KRAS Signaling Founder, and other gene sets of the Molecular Signatures Database (MSigDB) (17, 79)) accessed with the R package msigdbr (v7.5.1) (80). Gene expression subtyping was performed using nearest shrunken centroid estimation with the pamr (v1.56.1) (81) R package along with the Moffitt basal/classical signature gene set (23) trained on 181 PDAC samples of The Cancer Genome Atlas (TCGA). APC/C substrates were identified from UbiBrowser 2.0 (82) and Zhou et al. 2016 (83).
ChIP-seq data analyses
PANC-1 and pancreas histone and transcription factor ChIP-Seq and DNase-Seq data were downloaded from ENCODE (84, 85). Positions within +/− 3 kb of the transcription start site (TSS) of PDAC_KRAS_UP/DN and Hallmark_KRAS_Signaling_UP/DN signatures were evaluated for the proportion of genes with a detected IDR (irreproducible discovery rate) thresholded peak at each nucleotide position.
Gene expression microarray data analyses
Microarray data from dox-inducible KrasG12D mouse tumors were analyzed using the R package affy (v1.72.0) (86) with updated probeset ensembl gene mappings “mouse4302mmensgcdf (v25.0.0)” provided by Brainarray (87), and normalized using the robust multichip average (RMA) (88) approach. Limma (v3.50.3) (89) was used for differential expression comparisons and biomaRt (v2.50.3) for annotations and mouse-human homology assignments. Multiple testing was accounted for with false discovery rate (FDR) p-value adjustments by Benjamini-Hochberg method.
Overrepresentation analyses
KEGG, REACTOME, and GO gene sets were obtained using the R package msigdbr (v7.5.1). Overrepresentation analyses were conducted using the base R hypergeometric test. Clustering of gene sets based on Jaccard index similarity was accomplished with the R package vegan (v.2.5-4) (90) and hierarchical clustering with complete linkage. Exemplar gene sets were chosen as those with the largest number of genes within one of the eight main gene set clusters.
Liquid chromatography mass spectrophotometry (LC-MS2)
Six KRAS-mutant PDAC cell lines (AsPC-1, HPAC, MIA PaCa-2, PANC-1, Pa16C/Panc 10.05, and SW1990) were treated with SCH772984 (1 μM) or vehicle (DMSO) for 1 and 24 hours. Four biological replicates were used. Proteins were digested using Trypsin/Lys-C (Wako, 1:75 w/w) and tandem mass tags were added with the TMTpro 16plex Label Reagent Set (Thermo Fisher). Reverse phase separation at high pH was conducted offline to generate 24 fractions for LC-MS2. LC-MS2 was conducted with the Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher).
Proteomics integration and protein classification
Global proteomics and phosphoproteomics were performed as described in Klomp et al. (2023) (8). Proteins/genes were annotated as human kinases from KinHub. Human transcription factors were annotated from Supplemental Data S1 of Lambert et al. (2018) (91). Protein phosphatases were annotated from Supplemental Data S1 from Chen et al. (2017) and filtered for proteins within the evolutionarily conserved human protein phosphatome (92). E3 ubiquitin ligases were annotated from UbiNet 2.0 (93). Epigenetic regulators were annotated as published in EpiFactors (94, 95) and filtered out if they were already annotated to be kinases, transcription factors, phosphatases, or E3 ubiquitin ligases. Except for epigenetic regulators, other protein classifications were found to be mutually exclusive. The membership of each gene, protein, and phosphoprotein within each of these datasets was annotated and used for further analysis.
Promoter motif enrichment analysis
For enrichment of transcription factor promoter motifs, gene promoters were defined within −2000 to +1000 nucleotides around the TSS. The HOMER (v4.11) (96) command line tool for hypergeometric optimization of motif enrichment was used for supervised evaluation of promoter sequence motif enrichment in gene lists. Promoter motifs are defined by those in the JASPAR database (97) as well as several model organism motif collections. Background genes were randomly sampled from genes that were expressed in PDAC cells but did not vary with ERKi treatment (n = 1000, adj. p-val. > 0.5 and ∣log2FC∣ < 0.2).
Proteomics analyses
LC-MS2 protein data were analyzed using MaxQuant (v2.0.3.0) (98) with the UniProt Human Reference Proteome (99). Raw intensity values were log2 transformed and a variance stabilizing normalization was applied with limma (v3.50.3). BiomaRt (v2.50.3) was used for annotations and protein alias identification. Proteins that were not detected in at least two cell lines were removed from evaluation. Differential protein expression was evaluated with limma (v3.50.3) and adjusted for multiple testing using the FDR method (Benjamini-Hochberg).
Supervised enrichment for protein motifs in gene lists was conducted using SEA (100) (simple enrichment analysis) of the MEME Suite (101). Specifically for the evaluation of protein motifs enriched in proteins that were down-regulated with ERKi without mRNA changes, a list of down-regulated proteins was generated (prot: log2FC < −0.25 AND adj. p-val. < 0.05; mRNA: log2FC > 0 OR adj. p-val. > 0.05) and compared to a list of the top 100 (log2FC) up-regulated proteins (adj. p-val. < 0.05). Peptide sequences were obtained with BiomaRt (v2.50.3) and the longest sequence was chosen for each gene. Functional motifs were identified from the functional Eukaryotic Linear Motif (ELM2018) set (102) (n = 164) and evaluated for potential degrons.
DepMap CRISPR analysis
Essential gene probabilities, gene effect scores, and KRAS mutation status were obtained from CRISPR screens and analyses provided by the Cancer Dependency Map (55, 56) (DepMap v22Q2). CCLE cell lines with common KRAS mutations (at codons 12, 13, or 61; n = 41) were evaluated for dependency on KRAS and ERK-related gene sets.
General statistical tools
The statistical computing environment R (103) (various versions) and Bioconductor (v3.14) (104, 105) were used for analyses and graphics as well as software management. Heatmaps were produced using ComplexHeatmap (v2.10.0) (106) and general graphics with ggplot2 (v3.4.2 and 3.3.5) (107). Unless otherwise noted, multiple testing was controlled with FDR (Benjamini-Hochberg method) to generate adjusted p-values, and t-tests and Wilcoxon rank-sum tests were two-tailed.
Animal research
We have complied with the relevant ethical guidelines and regulations regarding care and use of animals in research. Our work with PDX mouse models subjected to sotorasib or panitumumab treatments were reviewed and approved by the Animal Care and Use Committee to meet all guidelines established by the University of Texas MD Anderson Cancer Center.
Human research
Please refer to clinical trial KRYSTAL-1 (NCT03785249), published protocol (https://www.nejm.org/doi/suppl/10.1056/NEJMoa2204619/suppl_file/nejmoa2204619_protocol.pdf), and referencing publications at ClinicalTrials.gov (https://classic.clinicaltrials.gov/ct2/show/NCT03785249). The study has complied with all relevant ethical regulations regarding the use of human research participants overseen by institutional review boards and has established informed consent from participating patients.
Supplementary Material
Fig. S1. Establishment and evaluation of a KRAS-dependent gene expression program in KRAS-mutant PDAC. (A) Flow chart for the experimental approaches applied to generate signatures for Hallmark KRAS Signaling, KRAS siRNA-based “PDAC KRAS” (this study), inducible Kras iKrasG12D/Trp53−/− “iKras” and KRASG12C/D inhibitor-treated tumor xenograft “KRASi” models (upper panel). The characteristics of the three KRAS-dependent models are summarized (lower panel). (B) Genetic and patient profiles of PDAC cell lines used in this study. (C-D) Immunoblot analysis of KRAS-mutant PDAC cell lines treated with control non-specific (“NS”) siRNA or targeting human KRAS (“K”) to monitor suppression of KRAS protein expression. (C) pERK and pAKT are also shown with quantitation below. Vinculin was a loading control to verify equivalent total cellular protein, and images are representative of 2-3 biological replicates. (D) Evaluation of relative KRAS expression in RNA-seq samples of PDAC cell lines treated with KRAS versus NS siRNA. Increased expression of negative feedback regulatory genes DUSP6 and SPRY4 indicates functional validation of KRAS activity inhibition. (E) Determination of basal versus classical subtype status using the gene signature from Moffitt et al., 2015 (23), for PDAC cells treated with KRAS or NS siRNA. (F) Principal component analysis of the 500 most variable genes (estimated by largest interquartile ranges) across the PDAC KRAS and NS siRNA datasets. (G) Barcode plot showing gene ranks for the Hallmark KRAS Signaling UP/DN gene sets along with founder gene sets as compared to ranked expression changes in KRAS versus NS siRNA samples. (H) Comparison of gene detection across RNA-seq data from NS siRNA treated control cells, 41 KRAS-mutant PDAC cell lines in the Cancer Cell Line Encyclopedia (CCLE), and laser-capture microdissection (LCMD)-purified epithelium from human PDAC tumor tissue (PRJNA360619). Gene sets evaluated included the top 200 KRAS siRNA genes (from Fig. 1A), Hallmark KRAS Signaling gene sets, and genes from the Hallmark KRAS Signaling founder gene sets. (I) Proportion of genes with the indicated epigenetic marks/characteristics from either PDAC KRAS UP/DN top 200 gene sets (see Fig. 1A) or the Hallmark KRAS Signaling UP/DN gene sets (data from Encode). (J) Ranked expression changes for genes in iKras doxycycline-off vs control mouse PDAC tumor model microarray dataset (GSE32277) (top). Enrichment for PDAC KRAS UP/DN signatures (bottom). (K) Comparison of DE genes in iKras OFF experiment showing UP (orange) and DN (grey) genes within PDAC siKRAS experiment of Fig. 1A and enrichment statistics for top 200 DE genes. (L) Ranked expression changes for genes in human PDAC CDX RNA-seq samples from mice treated with G12Ci/G12Di (KRASi) for one day versus control (top). Enrichment for PDAC KRAS UP/DN signatures (bottom). (M) Comparison of DE genes in PDAC KRASi CDX experiment showing UP (orange) and DN (grey) genes within PDAC siKRAS experiment of Fig. 1A and enrichment statistics for top 200 DE genes. (N) Intersection of top differentially expressed genes among the three KRAS modulation experiments described in Fig. A. (J, L, N) siKRAS genes are from Fig 1A-B and genes of iKras and KRASi experiments are filtered by those with unique Entrez gene IDs with ∣logFC∣ > 0.25 and adj. p-val. < 0.05. (O) Intersection of DE genes from iKras and KRASi experiment with Hallmark KRAS signaling gene sets. (P) Enrichment for PDAC KRAS UP/DN and Hallmark signatures in CDX models treated with G12Ci/G12DI (KRASi) for 5 or 7 days.
Fig. S2. Mutant KRAS is largely dependent on ERK for its proliferation programming. (A) Immunoblot analysis of ERK activity in Pa14C cells stably infected with control vector (Luc) or constitutively activated MEK1 (MEK1-DD), treated with vehicle (DMSO) or indicated inhibitors of each level of the RAF-MEK-ERK cascade: (G12Di, MRTX1133 (20 nM); RAFi, LY3009120 (600 nM); MEKi, trametinib (6 nM); ERKi, SCH772984 (600 nM). Images are representative of 2-3 biological replicates. (B) Cell proliferation assays for Pa14C cells stably expressing activated MEK1-DD versus Luc control when treated with ERKi SCH772984 or G12Di MRTX1133. Error bars represent standard error (SE) of the mean for three to four biological replicates each with three technical replicates. (C) Cell proliferation assays for PDAC cell lines treated with RAFi LY3009120 or MEKi trametinib. Error bars represent SE of the mean for three to four biological replicates each with 3 technical replicates. (D) Antiproliferative Drug Sensitivity Score (DSS) for indicated MAPK and PI3K inhibitors in a panel of PDAC cell lines. Higher numbers indicate greater sensitivity to target inhibition. (E) Immunoblot analysis of ERK and/or AKT activity in a panel of PDAC cell lines treated for 24 hours with RMC-7977, a broad-spectrum active-state RAS inhibitor (35), at the indicated dose. 0 nM refers to vehicle treatment (DMSO). (F) Venn diagram indicating intersection of siKRAS and ERKi UP/DN gene sets to derive PDAC KRAS ERK UP/DN gene sets (logFC > 0.5, adj. p-val. < 0.01). (G) Similarity clustering (Jaccard index) for the top 100 gene sets from REACTOME, GO, and KEGG databases that were over-represented in PDAC KRAS-ERK UP genes. Eight main gene set clusters are indicated with labels and separated by blue dashed lines. Red boxes highlight the largest gene sets within a cluster as exemplars for that cluster. (H) Comparison of gene expression for time series ERKi treatment and 24 hours KRAS siRNA experiments, highlighting genes in common from D and enrichment of exemplar gene sets from E. (I) GSEA for the PDAC KRAS-ERK UP signature in AsPC-1 tumor xenografts treated with G12Di (MRTX1133) and/or EGFRi (cetuximab) for 7 days. Enrichment statistics shown as circle size/intensity. Sample t-test significance shown above box plots. (J) Schematic for identification of the top 200 most consistent KRAS-ERK dependent genes (UP and DN) based on median rank across three independent RNA-seq datasets (all adj. p-val. < 0.01). (K) Evaluation of DE genes following 100 nM treatment with KRASi-G12Di (MRTX1133) or MEKi (trametinib) treatment for 24 hours in Pa16C PDAC cells (each condition n = 2). Colored points are DE genes in either treatment (FDR < 0.05); red, log2FC > 0 with either treatment; blue, log2FC < 0 with either treatment; tan, log2FC opposite directions between treatments. Coordinate labels indicate points outside of plotting range. Barcode plots below and left represent log2FC for “median-rank KRAS-ERK” signature genes within each experiment and ssGSEA enrichment statistics. (L) Comparison of median-rank KRAS-ERK signatures in RNA-seq data from multiple cell lines representing three cancer types: PDAC, LUAD, and COAD/CRC with various treatments and durations. Genes highlighted on right are a subset with strong changes at the 24-hour time points.
Fig. S3. Integration of transcriptomic, proteomic, and phosphosite activity data reveals multiple ERK dependent cell cycle control mechanisms. (A) Supervised promoter motif enrichment analysis for all 2,054 genes downregulated (UP) upon 24 hour ERKi. (B) Schematic for LC-MS2 proteomics workflow. (C) Schematic for integration of gene and protein expression with phosphosite activity data for transcription factors relevant to motifs identified in Fig 3A. (D) Differential expression analysis of proteomics data across six PDAC lines. Corresponding mRNA was downregulated (blue), upregulated (red), or not significantly changed (yellow) upon 24 hour ERKi treatment. Labels indicate subset of proteins with disagreement in protein and mRNA expression change. (E) Viability and apoptosis assays in PDAC cells treated with ERKi with or without APC/Ci for three days. Error bars represent SE of the mean for three biological replicates, each with three technical replicates. (F) Example gating strategy for flow sorting experiments. SSC, side scatter; FSC, forward scatter; A, area; H, height. (G) Immunoblot analysis of APC/C target proteins securin and cyclin B1 as well as ERK activity readouts pERK and pRSK following treatment with ProTAME (APC/Ci) and/or KRASi (G12Di, MRTX1133) in Pa16C cells. Images are representative of two to three biological replicates. (H) Cell cycle distribution analysis of Pa16C cells treated with APC/Ci and/or KRASi (G12Di, MRTX1133) for 24 hours. Error bars are +/− 1 S.D. (I) Comparison of protein kinase changes using MIB/MS versus MS in PDAC cells treated with ERKi for 24 hours. (J) MIB/MS proteomics analysis compared to RNA-seq or MS analysis of kinases in cells as in (I).
Fig. S4. KRAS-ERK dependent genes are essential for cell proliferation in PDAC. (A) Over-representation analysis for REACTOME terms in Hallmark KRAS Signaling UP and PDAC KRAS UP signatures. (B) Heatmap displaying CRISPR dependency probabilities for PDAC KRAS-ERK UP signature genes in 41 KRAS-mutant PDAC cell lines (DepMap). Genes with strongest probabilities (>0.75) are labeled. (C) Immunoblot evaluation of KRAS and ERK dependent protein expression changes for a subset of PDAC KRAS-ERK UP signature genes across three PDAC cell lines, treated with G12Ci/G12Di, MRTX1257/MRTX1133 (20 nM) or ERKi, SCH772984 (600 nM). Images are representative of two to three biological replicates. (D) Immunoblot evaluation of target inhibition for KRAS, ERK, and downstream kinases (CHK1, WEE1, and CDK4/6), treated with G12Di (MRTX1133, 20 nM); ERKi (SCH772984, 500 nM); CHK1i (prexasertib, 50 nM); WEE1i (adavosertib, 100 nM); or CDK4/6i (palbociclib, 500 nM). Images are representative of two to three biological replicates. (E) Proliferation assays for PDAC cells stably expressing control vector (Luc) or MEK1-DD treated with CHK1i, WEE1i, or CDK4/6i. Error bars represent SE of the mean for three to four biological replicates, each with three technical replicates.
Data S1. Differential expression statistics for genes in KRAS versus NS siRNA treated PDAC cells, using RNA-sequencing.
Data S2. Differential expression statistics and human homolog assignments for genes in dox-on versus dox-off iKras mouse orthotopic tumors, using gene expression microarrays.
Data S4. Top differentially expressed genes within KRAS modulation experiments (from data S1-S3).
Data S3. Differential expression statistics for genes in PDAC CDX tumors in animals, 24 hours after administration of KRASi versus vehicle.
Data S5. Differential expression statistics for genes in KRAS-G12Ci treated PDAC, CRC, and NSCLC cell lines over time, using RNA-sequencing.
Data S6. Differential expression statistics for genes in ERKi treated PDAC cell lines at various time points, using RNA-sequencing.
Data S7. Differential expression statistics for genes across PDAC, CRC, and NSCLC CDX tumors in animals, 24 hours after administration of KRASi versus vehicle.
Data S8. Differential expression statistics for genes in the PDAC cell line Pa16C treated with KRASi, MEKi, or ERKi versus control for 24 hours, using RNA-sequencing.
Data S10. Differential expression statistics for proteins across PDAC cell lines treated with ERKi for 1 or 24 hours, summarized by gene names.
Data S11. Median DepMap essentiality probability scores across KRAS mutant PDAC cell lines and 24-hour ERKi differential expression statistics for top differentially expressed genes.
Data S12. CRISPR drop-out screen statistics (MAGeCK) across two PDAC cell lines.
Data S13. HTG EdgeSeq gene expression counts for CRC and NSCLC patient tumors before and after KRAS-G12Ci treatment.
Data S9. Differential expression statistics for genes in KRASi treated PDAC, CRC, and NSCLC cell lines versus control for 2 hours, using RNA-sequencing.
ACKNOWLEDGMENTS
We thank A. Maitra (MD Anderson Cancer Center) for PDAC cell lines and K. Wood (Duke University) for the MEKDD and Myr-AKT expression vectors. We thank Revolution Medicines for their RAS(ON) multi-selective tri-complex inhibitor, RMC-7977.
Funding:
National Cancer Institute grants R01CA42978, P50CA196510, P50CA257911, U01CA199235, P01CA203657 and R35CA232113 (A.D.C. and/or C.J.D.)
Pancreatic Cancer Action Network 22-WG-DERB (C.J.D.)
Department of Defense grant W81XWH2110692 (C.J.D.)
Lustgarten Foundation grant 388222 (C.J.D.)
Pancreatic Cancer Action Network/AACR grant 15-70-25-BRYA (K.L.B.)
National Cancer Institute grants P50CA196510 and R37CA251877 (K.L.B.)
Department of Defense grant W81XWH2110693 (K.L.B.)
Sky Foundation grant (K.L.B.)
National Institute of General Medical Sciences grant R01GM138520 (L.M.B)
National Cancer Institute grant P50CA196510 (K.H.L. and A.W.G.)
National Cancer Institute grants T32CA009156, F32CA239328, and K99CA276700 (J.E.K.)
American Cancer Society grant PF-20-069 (J.E.K.)
National Cancer Institute grants T32CA009156 and F32CA232529 (C.A.S.)
National Institute of General Medical Sciences grant T32GM119999 (A.C.E.)
National Cancer Institute grant F31CA275260 (A.C.E.)
National Cancer Institute grant T32CA009156 (K.D.M.)
American Cancer Society grant PF-22-066-01-TBE (K.D.M.)
Slomo and Cindy Silvian Foundation grant (P.S.H. and J.N.D.)
National Cancer Institute grant T32CA071341 (P.S.H. and J.N.D.)
National Cancer Institute grant F30CA243253 (J.N.D.)
National Institute of General Medical Sciences grant T32GM007040 (Y.S.L.)
This research is based in part upon work conducted using the UNC Proteomics Core Facility, which is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Competing interests: C.J.D. was a consultant/advisory board member for Cullgen, Deciphera Pharmaceuticals, Kestrel Therapeutics, Mirati Therapeutics, Reactive Biosciences, Revere Pharmaceuticals, Revolution Medicines, Ribometrics, Sanofi and SHY Therapeutics. C.J.D. has received research funding support from Deciphera Pharmaceuticals, Mirati Therapeutics, Reactive Biosciences, Revolution Medicines, and SpringWorks Therapeutics. C.A.S. has consulted for Reactive Biosciences. A.D.C. has consulted for Eli Lilly and Mirati Therapeutics. J.H., P.O. and J.G.C. are employees and shareholders of Mirati Therapeutics, Inc. S.K. has ownership interest in Lutris, Iylon, Frontier Medicines, Xilis, Navire and is a consultant for Genentech, EMD Serono, Merck, Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, AstraZeneca/MedImmune, Bayer Health, Redx Pharma, Ipsen, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, Abbvie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics, Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical, Cardiff Oncology, Ono Pharmaceutical, Bristol-Myers Squibb-Medarex, Amgen, Tempus, Foundation Medicine, Harbinger Oncology, Inc, Takeda, CureTeq, Zentalis, Black Stone Therapeutics, NeoGenomics Laboratories, Accademia Nazionale Di Medicina, and received research funding from Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, Daiichi Sankyo; J.P.S. is an advisor for Engine Biosciences and receives research funding from Celsius Therapeutics. The other authors declare no competing interests.
Data and materials availability:
Primary and public data sets used in this manuscript are indicated below.
PDAC cell line with KRAS siRNA RNA-seq (NCBI SRA PRJNA980201)
PDAC LCMD tumor epithelium RNA-seq (54) (NCBI SRA PRJNA360619)
PDAC and NSCLC CDX with G12Ci RNA-seq (37) (NCBI SRA PRJNA578935)
PDAC, NSCLC, and CRC CDX with G12Di RNA-seq (28) (NCBI SRA PRJNA831648)
CRC PDX with G12Ci or G12Ci/EGFRi RNA-seq (NCBI SRA PRJNA996641)
KRAS mutant cell lines with 2 hours pan-KRASi RNA-seq (NCBI SRA PRJNA947663)
PDAC, CRC, NSCLC cells with KRAS-G12Ci RNA-seq (NCBI SRA PRJNA1054293)
PDAC cell line with ERKi RNA-seq (36) (NCBI SRA PRJEB25806)
PDAC cell line Pa16C with KRASi, MEKi, or ERKi RNA-seq (NCBI SRA PRJNA1063706)
PDAC cell lines with ERKi LC-MS2 (ProteomeXchange Consortium, PXD048531 and 10.6019/PXD048531)
PDAC cell line Depmap essentiality probabilities and gene effect scores (55, 56) (DepMap v22Q2)
PDAC cell line with ERKi CRISPR screen (data S11)
PDAC paired tumor biopsies from patients treated with ERKi RNA-seq (48) (NCBI GEO GSE213797, clinical trial NCT02608229)
CRC and NSCLC paired tumor biopsies from patients treated with KRASi HTG EdgeSeq (data S13)
PDAC cell line with ERKi MIB/MS (46)
REFERENCES AND NOTES
- 1.Punekar SR, Velcheti V, Neel BG, Wong K-K, The current state of the art and future trends in RAS-targeted cancer therapies. Nat Rev Clin Oncol, 1–19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi A, Hong J, Iacobuzio-Donahue CA, The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol 18, 469–481 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Waters AM, Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med 8, a031435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles R-P, Rabinovich BA, Hann B, Dankort D, Spellman PT, Phillips WA, Gray JW, McMahon M, A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2, 685–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, Aziz M, Kassner M, Bryant KL, Pierobon M, Marayati R, Kher S, George SD, Xu M, Wang-Gillam A, Samatar AA, Maitra A, Wennerberg K, Petricoin EF, Yin HH, Nelkin B, Cox AD, Yeh JJ, Der CJ, Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 29, 75–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavoie H, Gagnon J, Therrien M, ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 21, 607–632 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Klomp JE, Klomp JA, Der CJ, The ERK mitogen-activated protein kinase signaling network: the final frontier in RAS signal transduction. Biochem Soc Trans 49, 253–267 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Klomp JE, Diehl JN, Klomp JA, Edwards AC, Yang R, Morales AJ, Taylor KE, Drizyte-Miller K, Bryant KL, Schaefer A, Johnson JL, Huntsman EM, Yaron TM, Pierobon M, Baldelli E, Prevatte AW, Barker NK, Herring LE, Petricoin EF III, Graves LM, Cantley LC, Cox AD, Der CJ, Stalnecker CA, Determining the ERK-regulated phosphoproteome driving KRAS-mutant cancer. Science (In Review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, Valencia K, Arricibita A, Gwinn D, Sayles LC, Chuang C-H, Guembe L, Bailey P, Chang DK, Biankin A, Ponz-Sarvise M, Andersen JB, Khatri P, Bozec A, Sweet-Cordero EA, Sage J, Lecanda F, Vicent S, An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun 8, 14294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, Ozkan-Dagliyan I, Gautam P, Bryant KL, Pearce KH, Herring LE, Han H, Graves LM, Witkiewicz AK, Knudsen ES, Pecot CV, Rashid N, Houghton PJ, Wennerberg K, Cox AD, Der CJ, KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell 34, 807–822.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T, An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 37, 48–55 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A, Knock-in of oncogenic Kras does not transform mouse somatic cells but triggers a transcriptional response that classifies human cancers. Cancer Res 67, 8468–8476 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J, A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.East P, Kelly GP, Biswas D, Marani M, Hancock DC, Creasy T, Sachsenmeier K, Swanton C, Downward J, de Carné Trécesson S, RAS oncogenic activity predicts response to chemotherapy and outcome in lung adenocarcinoma. Nat Commun 13, 5632 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Wang T, Zhang K, Li M, Shen Q, Lu S, Zhang J, Pan-KRAS inhibitors suppress proliferation through feedback regulation in pancreatic ductal adenocarcinoma. Acta Pharmacol Sin 43, 2696–2708 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muzumdar MD, Chen P-Y, Dorans KJ, Chung KM, Bhutkar A, Hong E, Noll EM, Sprick MR, Trumpp A, Jacks T, Survival of pancreatic cancer cells lacking KRAS function. Nat Commun 8, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC, Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW, Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K, Rumschlag M, Shankar S, Whitehead C, Schipper M, Basrur V, Southworth DR, Chinnaiyan AM, Rehemtulla A, Beer DG, Lawrence TS, Nyati MK, Ray D, KRAS Protein Stability Is Regulated through SMURF2: UBCH5 Complex-Mediated β-TrCP1 Degradation. Neoplasia 16, 115–W5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan P-F, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL, Dynamic Reprogramming of the Kinome In Response to Targeted MEK Inhibition In Triple Negative Breast Cancer. Cell 149, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collisson EA, Bailey P, Chang DK, Biankin AV, Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ, Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthalagu N, Monteverde T, Raffo-Iraolagoitia X, Wiesheu R, Whyte D, Hedley A, Laing S, Kruspig B, Upstill-Goddard R, Shaw R, Neidler S, Rink C, Karim SA, Gyuraszova K, Nixon C, Clark W, Biankin AV, Carlin LM, Coffelt SB, Sansom OJ, Morton JP, Murphy DJ, Repression of the Type I Interferon Pathway Underlies MYC- and KRAS-Dependent Evasion of NK and B Cells in Pancreatic Ductal Adenocarcinoma. Cancer Discov 10, 872–887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, Barretina J, Gelfand ET, Bielski CM, Li H, Hu K, Andreev-Drakhlin AY, Kim J, Hess JM, Haas BJ, Aguet F, Weir BA, Rothberg MV, Paolella BR, Lawrence MS, Akbani R, Lu Y, Tiv HL, Gokhale PC, de Weck A, Mansour AA, Oh C, Shih J, Hadi K, Rosen Y, Bistline J, Venkatesan K, Reddy A, Sonkin D, Liu M, Lehar J, Korn JM, Porter DA, Jones MD, Golji J, Caponigro G, Taylor JE, Dunning CM, Creech AL, Warren AC, McFarland JM, Zamanighomi M, Kauffmann A, Stransky N, Imielinski M, Maruvka YE, Cherniack AD, Tsherniak A, Vazquez F, Jaffe JD, Lane AA, Weinstock DM, Johannessen CM, Morrissey MP, Stegmeier F, Schlegel R, Hahn WC, Getz G, Mills GB, Boehm JS, Golub TR, Garraway LA, Sellers WR, Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnes L, Liu Z, Wang J, Maurer C, Sagalovskiy I, Sanchez-Martin M, Bommakanti N, Garofalo DC, Balderes DA, Sussel L, Olive KP, Rabadan R, Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma. Gut 68, 499–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik J, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA, Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallin J, Bowcut V, Calinisan A, Briere DM, Hargis L, Engstrom LD, Laguer J, Medwid J, Vanderpool D, Lifset E, Trinh D, Hoffman N, Wang X, David Lawson J, Gunn RJ, Smith CR, Thomas NC, Martinson M, Bergstrom A, Sullivan F, Bouhana K, Winski S, He L, Fernandez-Banet J, Pavlicek A, Haling JR, Rahbaek L, Marx MA, Olson P, Christensen JG, Anti-tumor efficacy of a potent and selective non-covalent KRASG12D inhibitor. Nat Med 28, 2171–2182 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Kemp SB, Cheng N, Markosyan N, Sor R, Kim I-K, Hallin J, Shoush J, Quinones L, Brown NV, Bassett JB, Joshi N, Yuan S, Smith M, Vostrejs WP, Perez-Vale KZ, Kahn B, Mo F, Donahue TR, Radu CG, Clendenin C, Christensen JG, Vonderheide RH, Stanger BZ, Efficacy of a Small-Molecule Inhibitor of KrasG12D in Immunocompetent Models of Pancreatic Cancer. Cancer Discov 13, 298–311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, Whaley A, Pinnelli M, Murciano-Goroff YR, Vakiani E, Valeri N, Liao W-L, Bhalkikar A, Thyparambil S, Zhao H-Y, de Stanchina E, Marsoni S, Siena S, Bertotti A, Trusolino L, Li BT, Rosen N, Di Nicolantonio F, Bardelli A, Misale S, EGFR Blockade Reverts Resistance to KRASG12C Inhibition in Colorectal Cancer. Cancer Discov 10, 1129–1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan MB, Coker O, Sorokin A, Fella K, Barnes H, Wong E, Kanikarla P, Gao F, Zhang Y, Zhou L, Kopetz S, Corcoran RB, KRASG12C-independent feedback activation of wild-type RAS constrains KRASG12C inhibitor efficacy. Cell Reports 39, 110993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, Michel AG, Syed MU, Fella KA, Sakhi M, Baiev I, Juric D, Gainor JF, Klempner SJ, Lennerz JK, Siravegna G, Bar-Peled L, Hata AN, Heist RS, Corcoran RB, Clinical Acquired Resistance to KRASG12C Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS-MAPK Reactivation. Cancer Discov 11, 1913–1922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, Johnson ML, Heist RS, Patil T, Riely GJ, Jacobson JO, Yang X, Persky NS, Root DE, Lowder KE, Feng H, Zhang SS, Haigis KM, Hung YP, Sholl LM, Wolpin BM, Wiese J, Christiansen J, Lee J, Schrock AB, Lim LP, Garg K, Li M, Engstrom LD, Waters L, Lawson JD, Olson P, Lito P, Ou S-HI, Christensen JG, Jänne PA, Aguirre AJ, Acquired Resistance to KRASG12C Inhibition in Cancer. N Engl J Med 384, 2382–2393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Murciano-Goroff YR, Xue JY, Ang A, Lucas J, Mai TT, Da Cruz Paula AF, Saiki AY, Mohn D, Achanta P, Sisk AE, Arora KS, Roy RS, Kim D, Li C, Lim LP, Li M, Bahr A, Loomis BR, de Stanchina E, Reis-Filho JS, Weigelt B, Berger M, Riely G, Arbour KC, Lipford JR, Li BT, Lito P, Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 599, 679–683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holderfield M, Lee BJ, Jiang J, Tomlinson A, Seamon KJ, Mira A, Patrucco E, Goodhart G, Dilly J, Gindin Y, Dinglasan N, Wang Y, Lai LP, Cai S, Jiang L, Nasholm N, Shifrin N, Blaj C, Shah H, Evans JW, Montazer N, Lai O, Shi J, Ahler E, Quintana E, Chang S, Salvador A, Marquez A, Cregg J, Liu Y, Milin A, Chen A, Ziv TB, Parsons D, Knox JE, Klomp JE, Roth J, Rees M, Ronan M, Cuevas-Navarro A, Hu F, Lito P, Santamaria D, Aguirre AJ, Waters AM, Der CJ, Ambrogio C, Wang Z, Gill AL, Koltun ES, Smith JAM, Wildes D, Singh M, Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature, 1–8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang G-F, Witkiewicz AK, Knudsen ES, Petricoin EF, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ, Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 25, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA, Burkard MR, Fell JB, Fischer JP, Vigers GP, Xue Y, Gatto S, Fernandez-Banet J, Pavlicek A, Velastagui K, Chao RC, Barton J, Pierobon M, Baldelli E, Patricoin EF, Cassidy DP, Marx MA, Rybkin II, Johnson ML, Ou S-HI, Lito P, Papadopoulos KP, Jänne PA, Olson P, Christensen JG, The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Herdeis L, Rudolph D, Zhao Y, Böttcher J, Vides A, Ayala-Santos CI, Pourfarjam Y, Cuevas-Navarro A, Xue JY, Mantoulidis A, Bröker J, Wunberg T, Schaaf O, Popow J, Wolkerstorfer B, Kropatsch KG, Qu R, de Stanchina E, Sang B, Li C, McConnell DB, Kraut N, Lito P, Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature 619, 160–166 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talotta F, Casalino L, Verde P, The nuclear oncoprotein Fra-1: a transcription factor knocking on therapeutic applications’ door. Oncogene 39, 4491–4506 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Das SK, Lewis BA, Levens D, MYC: a complex problem. Trends Cell Biol 33, 235–246 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill CS, Wynne J, Treisman R, The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81, 1159–1170 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M, Ubiquitin-Independent Proteasomal Degradation of Fra-1 Is Antagonized by Erk1/2 Pathway-Mediated Phosphorylation of a Unique C-Terminal Destabilizer. Mol Cell Biol 27, 3936–3950 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M, Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 354, 494–496 (1991). [DOI] [PubMed] [Google Scholar]
- 44.Zhou Z, He M, Shah AA, Wan Y, Insights into APC/C: from cellular function to diseases and therapeutics. Cell Div 11, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh D-C, Hathaway N, Dimova N, Cuny GD, King RW, Pharmacologic Inhibition of the Anaphase-Promoting Complex Induces A Spindle Checkpoint-Dependent Mitotic Arrest in the Absence of Spindle Damage. Cancer Cell 18, 382–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klomp JE, Lee YS, Goodwin CM, Papke B, Klomp JA, Waters AM, Stalnecker CA, DeLiberty JM, Drizyte-Miller K, Yang R, Diehl JN, Yin HH, Pierobon M, Baldelli E, Ryan MB, Li S, Peterson J, Smith AR, Neal JT, McCormick AK, Kuo CJ, Counter CM, Petricoin EF, Cox AD, Bryant KL, Der CJ, CHK1 protects oncogenic KRAS-expressing cells from DNA damage and is a target for pancreatic cancer treatment. Cell Rep 37, 110060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diehl JN, Klomp JE, Snare KR, Hibshman PS, Blake DR, Kaiser ZD, Gilbert TSK, Baldelli E, Pierobon M, Papke B, Yang R, Hodge RG, Rashid NU, Petricoin EF, Herring LE, Graves LM, Cox AD, Der CJ, The KRAS-regulated kinome identifies WEE1 and ERK coinhibition as a potential therapeutic strategy in KRAS-mutant pancreatic cancer. J Biol Chem 297, 101335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grierson PM, Tan B, Pedersen KS, Park H, Suresh R, Amin MA, Trikalinos NA, Knoerzer D, Kreider B, Reddy A, Liu J, Der CJ, Wang-Gillam A, Lim K-H, Phase Ib Study of Ulixertinib Plus Gemcitabine and Nab-Paclitaxel in Patients with Metastatic Pancreatic Adenocarcinoma. Oncologist 28, e115–e123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, Mulvihill SJ, The clinical utility of CA 19–9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med 13, 340–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J, Binding of Ras to Phosphoinositide 3-Kinase p110α Is Required for Ras- Driven Tumorigenesis in Mice. Cell 129, 957–968 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Hobbs GA, Baker NM, Miermont AM, Thurman RD, Pierobon M, Tran TH, Anderson AO, Waters AM, Diehl JN, Papke B, Hodge RG, Klomp JE, Goodwin CM, DeLiberty JM, Wang J, Ng RWS, Gautam P, Bryant KL, Esposito D, Campbell SL, Petricoin EF, Simanshu DK, Aguirre AJ, Wolpin BM, Wennerberg K, Rudloff U, Cox AD, Der CJ, Atypical KRASG12R Mutant Is Impaired in PI3K Signaling and Macropinocytosis in Pancreatic Cancer. Cancer Discov 10, 104–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H, Barlesi F, Kato T, Curioni-Fontecedro A, Sacher A, Spira A, Ramalingam SS, Takahashi T, Besse B, Anderson A, Ang A, Tran Q, Mather O, Henary H, Ngarmchamnanrith G, Friberg G, Velcheti V, Govindan R, Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 384, 2371–2381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jänne PA, Riely GJ, Gadgeel SM, Heist RS, Ou S-HI, Pacheco JM, Johnson ML, Sabari JK, Leventakos K, Yau E, Bazhenova L, Negrao MV, Pennell NA, Zhang J, Anderes K, Der-Torossian H, Kheoh T, Velastegui K, Yan X, Christensen JG, Chao RC, Spira AI, Adagrasib in Non–Small-Cell Lung Cancer Harboring a KRASG12C Mutation. N Engl J Med 387, 120–131 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Maurer C, Holmstrom SR, He J, Laise P, Su T, Ahmed A, Hibshoosh H, Chabot JA, Oberstein PE, Sepulveda AR, Genkinger JM, Zhang J, Iuga AC, Bansal M, Califano A, Olive KP, Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68, 1034–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, Ansari R, Harper S, Jackson DA, McRae R, Pooley R, Wilkinson P, van der Meer D, Dow D, Buser-Doepner C, Bertotti A, Trusolino L, Stronach EA, Saez-Rodriguez J, Yusa K, Garnett MJ, Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, Anderson GR, Winter PS, Wang T, Alley HM, Kwong LN, Cooper ZA, Tetzlaff M, Chen P-L, Rathmell JC, Flaherty KT, Wargo JA, McDonnell DP, Sabatini DM, Wood KC, Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal 7, ra121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, DiNunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, Xiao L, Shipps G, Hruza A, Wang J, Kelly J, Paliwal S, Gao X, Babu BS, Zhu L, Daublain P, Zhang L, Lutterbach BA, Pelletier MR, Philippar U, Siliphaivanh P, Witter D, Kirschmeier P, Bishop WR, Hicklin D, Gilliland DG, Jayaraman L, Zawel L, Fawell S, Samatar AA, Discovery of a Novel ERK Inhibitor with Activity in Models of Acquired Resistance to BRAF and MEK Inhibitors. Cancer Discov 3, 742–750 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Peng S-B, Henry JR, Kaufman MD, Lu W-P, Smith BD, Vogeti S, Rutkoski TJ, Wise S, Chun L, Zhang Y, Van Horn RD, Yin T, Zhang X, Yadav V, Chen S-H, Gong X, Ma X, Webster Y, Buchanan S, Mochalkin I, Huber L, Kays L, Donoho GP, Walgren J, McCann D, Patel P, Conti I, Plowman GD, Starling JJ, Flynn DL, Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell 28, 384–398 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Katsiampoura A, Raghav K, Jiang Z-Q, Menter DG, Varkaris A, Morelli MP, Manuel S, Wu J, Sorokin AV, Rizi BS, Bristow C, Tian F, Airhart S, Cheng M, Broom BM, Morris J, Overman MJ, Powis G, Kopetz S, Modeling of Patient-Derived Xenografts in Colorectal Cancer. Mol Cancer Ther 16, 1435–1442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, Beresford-Smith B, Xenome—a tool for classifying reads from xenograft samples. Bioinformatics 28, i172–i178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tischler G, Leonard S, biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol Med 9, 13 (2014). [Google Scholar]
- 64.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A, Fast gene set enrichment analysis. bioRxiv, 060012 (2021). [Google Scholar]
- 68.Wang B, Wang M, Zhang W, Xiao T, Chen C-H, Wu A, Wu F, Traugh N, Wang X, Li Z, Mei S, Cui Y, Shi S, Lipp JJ, Hinterndorfer M, Zuber J, Brown M, Li W, Liu XS, Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 14, 756–780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozkan-Dagliyan I, Diehl JN, George SD, Schaefer A, Papke B, Klotz-Noack K, Waters AM, Goodwin CM, Gautam P, Pierobon M, Peng S, Gilbert TSK, Lin KH, Dagliyan O, Wennerberg K, Petricoin EF, Tran NL, Bhagwat SV, Tiu RV, Peng S-B, Herring LE, Graves LM, Sers C, Wood KC, Cox AD, Der CJ, Low-Dose Vertical Inhibition of the RAF-MEK-ERK Cascade Causes Apoptotic Death of KRAS Mutant Cancers. Cell Rep 31, 107764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yadav B, Pemovska T, Szwajda A, Kulesskiy E, Kontro M, Karjalainen R, Majumder MM, Malani D, Murumägi A, Knowles J, Porkka K, Heckman C, Kallioniemi O, Wennerberg K, Aittokallio T, Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep 4, 5193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krueger F, Trim Galore!, version 0.4.1, Babraham Bioinformatics (2012); http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- 72.Andrews S, FastQC, version 0.11.5, Babraham Bioinformatics (2010); http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- 73.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12 (2011). [Google Scholar]
- 74.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soneson C, Love MI, Robinson MD, Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W, BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson MD, Oshlack A, A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dolgalev I, msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format, (2022); https://CRAN.R-project.org/package=msigdbr. [Google Scholar]
- 81.Hastie T, Tibshirani R, Narasimhan B, Chu G, pamr: Pam: Prediction Analysis for Microarrays, (2019); https://CRAN.R-project.org/package=pamr. [Google Scholar]
- 82.Wang X, Li Y, He M, Kong X, Jiang P, Liu X, Diao L, Zhang X, Li H, Ling X, Xia S, Liu Z, Liu Y, Cui C-P, Wang Y, Tang L, Zhang L, He F, Li D, UbiBrowser 2.0: a comprehensive resource for proteome-wide known and predicted ubiquitin ligase/deubiquitinase–substrate interactions in eukaryotic species. Nucleic Acids Res 50, D719–D728 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Z, He M, Shah AA, Wan Y, Insights into APC/C: from cellular function to diseases and therapeutics. Cell Div 11, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, Baymuradov UK, Graham K, Litton C, Miyasato SR, Strattan JS, Jolanki O, Lee J-W, Tanaka FY, Adenekan P, O’Neill E, Cherry JM, New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 48, D882–D889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, Rowe LD, Dreszer TR, Roe G, Podduturi NR, Tanaka F, Hong EL, Cherry JM, ENCODE data at the ENCODE portal. Nucleic Acids Res 44, D726–D732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gautier L, Cope L, Bolstad BM, Irizarry RA, affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F, Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33, e175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP, Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Solymos P, Stevens MHH, Szoecs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, Caceres MD, Durand S, Evangelista HBA, FitzJohn R, Friendly M, Furneaux B, Hannigan G, Hill MO, Lahti L, McGlinn D, Ouellette M-H, Cunha ER, Smith T, Stier A, Braak CJFT, Weedon J, vegan: Community Ecology Package, (2022); https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 91.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, The Human Transcription Factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Chen MJ, Dixon JE, Manning G, Genomics and evolution of protein phosphatases. Sci Signal 10, eaag1796 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Li Z, Chen S, Jhong J-H, Pang Y, Huang K-Y, Li S, Lee T-Y, UbiNet 2.0: a verified, classified, annotated and updated database of E3 ubiquitin ligase–substrate interactions. Database 2021, baab010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marakulina D, Vorontsov IE, Kulakovskiy IV, Lennartsson A, Drabløs F, Medvedeva YA, EpiFactors 2022: expansion and enhancement of a curated database of human epigenetic factors and complexes. Nucleic Acids Res 51, D564–D570 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Medvedeva YA, Lennartsson A, Ehsani R, Kulakovskiy IV, Vorontsov IE, Panahandeh P, Khimulya G, Kasukawa T, The FANTOM Consortium, Drabløs F, EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database 2015, bav067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, Fornes O, Leung TY, Aguirre A, Hammal F, Schmelter D, Baranasic D, Ballester B, Sandelin A, Lenhard B, Vandepoele K, Wasserman WW, Parcy F, Mathelier A, JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 50, D165–D173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LL, UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32, D115–D119 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bailey TL, Grant CE, SEA: Simple Enrichment Analysis of motifs. bioRxiv [Preprint] (2021). 10.1101/2021.08.23.457422. [DOI] [Google Scholar]
- 101.Bailey TL, Johnson J, Grant CE, Noble WS, The MEME Suite. Nucleic Acids Res 43, W39–W49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kumar M, Gouw M, Michael S, Sámano-Sánchez H, Pancsa R, Glavina J, Diakogianni A, Valverde JA, Bukirova D, Čalyševa J, Palopoli N, Davey NE, Chemes LB, Gibson TJ, ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res 48, D296–D306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2021; https://www.R-project.org/). [Google Scholar]
- 104.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M, Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J, Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gu Z, Eils R, Schlesner M, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Wickham H, Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; New York, 2016; https://ggplot2.tidyverse.org) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Establishment and evaluation of a KRAS-dependent gene expression program in KRAS-mutant PDAC. (A) Flow chart for the experimental approaches applied to generate signatures for Hallmark KRAS Signaling, KRAS siRNA-based “PDAC KRAS” (this study), inducible Kras iKrasG12D/Trp53−/− “iKras” and KRASG12C/D inhibitor-treated tumor xenograft “KRASi” models (upper panel). The characteristics of the three KRAS-dependent models are summarized (lower panel). (B) Genetic and patient profiles of PDAC cell lines used in this study. (C-D) Immunoblot analysis of KRAS-mutant PDAC cell lines treated with control non-specific (“NS”) siRNA or targeting human KRAS (“K”) to monitor suppression of KRAS protein expression. (C) pERK and pAKT are also shown with quantitation below. Vinculin was a loading control to verify equivalent total cellular protein, and images are representative of 2-3 biological replicates. (D) Evaluation of relative KRAS expression in RNA-seq samples of PDAC cell lines treated with KRAS versus NS siRNA. Increased expression of negative feedback regulatory genes DUSP6 and SPRY4 indicates functional validation of KRAS activity inhibition. (E) Determination of basal versus classical subtype status using the gene signature from Moffitt et al., 2015 (23), for PDAC cells treated with KRAS or NS siRNA. (F) Principal component analysis of the 500 most variable genes (estimated by largest interquartile ranges) across the PDAC KRAS and NS siRNA datasets. (G) Barcode plot showing gene ranks for the Hallmark KRAS Signaling UP/DN gene sets along with founder gene sets as compared to ranked expression changes in KRAS versus NS siRNA samples. (H) Comparison of gene detection across RNA-seq data from NS siRNA treated control cells, 41 KRAS-mutant PDAC cell lines in the Cancer Cell Line Encyclopedia (CCLE), and laser-capture microdissection (LCMD)-purified epithelium from human PDAC tumor tissue (PRJNA360619). Gene sets evaluated included the top 200 KRAS siRNA genes (from Fig. 1A), Hallmark KRAS Signaling gene sets, and genes from the Hallmark KRAS Signaling founder gene sets. (I) Proportion of genes with the indicated epigenetic marks/characteristics from either PDAC KRAS UP/DN top 200 gene sets (see Fig. 1A) or the Hallmark KRAS Signaling UP/DN gene sets (data from Encode). (J) Ranked expression changes for genes in iKras doxycycline-off vs control mouse PDAC tumor model microarray dataset (GSE32277) (top). Enrichment for PDAC KRAS UP/DN signatures (bottom). (K) Comparison of DE genes in iKras OFF experiment showing UP (orange) and DN (grey) genes within PDAC siKRAS experiment of Fig. 1A and enrichment statistics for top 200 DE genes. (L) Ranked expression changes for genes in human PDAC CDX RNA-seq samples from mice treated with G12Ci/G12Di (KRASi) for one day versus control (top). Enrichment for PDAC KRAS UP/DN signatures (bottom). (M) Comparison of DE genes in PDAC KRASi CDX experiment showing UP (orange) and DN (grey) genes within PDAC siKRAS experiment of Fig. 1A and enrichment statistics for top 200 DE genes. (N) Intersection of top differentially expressed genes among the three KRAS modulation experiments described in Fig. A. (J, L, N) siKRAS genes are from Fig 1A-B and genes of iKras and KRASi experiments are filtered by those with unique Entrez gene IDs with ∣logFC∣ > 0.25 and adj. p-val. < 0.05. (O) Intersection of DE genes from iKras and KRASi experiment with Hallmark KRAS signaling gene sets. (P) Enrichment for PDAC KRAS UP/DN and Hallmark signatures in CDX models treated with G12Ci/G12DI (KRASi) for 5 or 7 days.
Fig. S2. Mutant KRAS is largely dependent on ERK for its proliferation programming. (A) Immunoblot analysis of ERK activity in Pa14C cells stably infected with control vector (Luc) or constitutively activated MEK1 (MEK1-DD), treated with vehicle (DMSO) or indicated inhibitors of each level of the RAF-MEK-ERK cascade: (G12Di, MRTX1133 (20 nM); RAFi, LY3009120 (600 nM); MEKi, trametinib (6 nM); ERKi, SCH772984 (600 nM). Images are representative of 2-3 biological replicates. (B) Cell proliferation assays for Pa14C cells stably expressing activated MEK1-DD versus Luc control when treated with ERKi SCH772984 or G12Di MRTX1133. Error bars represent standard error (SE) of the mean for three to four biological replicates each with three technical replicates. (C) Cell proliferation assays for PDAC cell lines treated with RAFi LY3009120 or MEKi trametinib. Error bars represent SE of the mean for three to four biological replicates each with 3 technical replicates. (D) Antiproliferative Drug Sensitivity Score (DSS) for indicated MAPK and PI3K inhibitors in a panel of PDAC cell lines. Higher numbers indicate greater sensitivity to target inhibition. (E) Immunoblot analysis of ERK and/or AKT activity in a panel of PDAC cell lines treated for 24 hours with RMC-7977, a broad-spectrum active-state RAS inhibitor (35), at the indicated dose. 0 nM refers to vehicle treatment (DMSO). (F) Venn diagram indicating intersection of siKRAS and ERKi UP/DN gene sets to derive PDAC KRAS ERK UP/DN gene sets (logFC > 0.5, adj. p-val. < 0.01). (G) Similarity clustering (Jaccard index) for the top 100 gene sets from REACTOME, GO, and KEGG databases that were over-represented in PDAC KRAS-ERK UP genes. Eight main gene set clusters are indicated with labels and separated by blue dashed lines. Red boxes highlight the largest gene sets within a cluster as exemplars for that cluster. (H) Comparison of gene expression for time series ERKi treatment and 24 hours KRAS siRNA experiments, highlighting genes in common from D and enrichment of exemplar gene sets from E. (I) GSEA for the PDAC KRAS-ERK UP signature in AsPC-1 tumor xenografts treated with G12Di (MRTX1133) and/or EGFRi (cetuximab) for 7 days. Enrichment statistics shown as circle size/intensity. Sample t-test significance shown above box plots. (J) Schematic for identification of the top 200 most consistent KRAS-ERK dependent genes (UP and DN) based on median rank across three independent RNA-seq datasets (all adj. p-val. < 0.01). (K) Evaluation of DE genes following 100 nM treatment with KRASi-G12Di (MRTX1133) or MEKi (trametinib) treatment for 24 hours in Pa16C PDAC cells (each condition n = 2). Colored points are DE genes in either treatment (FDR < 0.05); red, log2FC > 0 with either treatment; blue, log2FC < 0 with either treatment; tan, log2FC opposite directions between treatments. Coordinate labels indicate points outside of plotting range. Barcode plots below and left represent log2FC for “median-rank KRAS-ERK” signature genes within each experiment and ssGSEA enrichment statistics. (L) Comparison of median-rank KRAS-ERK signatures in RNA-seq data from multiple cell lines representing three cancer types: PDAC, LUAD, and COAD/CRC with various treatments and durations. Genes highlighted on right are a subset with strong changes at the 24-hour time points.
Fig. S3. Integration of transcriptomic, proteomic, and phosphosite activity data reveals multiple ERK dependent cell cycle control mechanisms. (A) Supervised promoter motif enrichment analysis for all 2,054 genes downregulated (UP) upon 24 hour ERKi. (B) Schematic for LC-MS2 proteomics workflow. (C) Schematic for integration of gene and protein expression with phosphosite activity data for transcription factors relevant to motifs identified in Fig 3A. (D) Differential expression analysis of proteomics data across six PDAC lines. Corresponding mRNA was downregulated (blue), upregulated (red), or not significantly changed (yellow) upon 24 hour ERKi treatment. Labels indicate subset of proteins with disagreement in protein and mRNA expression change. (E) Viability and apoptosis assays in PDAC cells treated with ERKi with or without APC/Ci for three days. Error bars represent SE of the mean for three biological replicates, each with three technical replicates. (F) Example gating strategy for flow sorting experiments. SSC, side scatter; FSC, forward scatter; A, area; H, height. (G) Immunoblot analysis of APC/C target proteins securin and cyclin B1 as well as ERK activity readouts pERK and pRSK following treatment with ProTAME (APC/Ci) and/or KRASi (G12Di, MRTX1133) in Pa16C cells. Images are representative of two to three biological replicates. (H) Cell cycle distribution analysis of Pa16C cells treated with APC/Ci and/or KRASi (G12Di, MRTX1133) for 24 hours. Error bars are +/− 1 S.D. (I) Comparison of protein kinase changes using MIB/MS versus MS in PDAC cells treated with ERKi for 24 hours. (J) MIB/MS proteomics analysis compared to RNA-seq or MS analysis of kinases in cells as in (I).
Fig. S4. KRAS-ERK dependent genes are essential for cell proliferation in PDAC. (A) Over-representation analysis for REACTOME terms in Hallmark KRAS Signaling UP and PDAC KRAS UP signatures. (B) Heatmap displaying CRISPR dependency probabilities for PDAC KRAS-ERK UP signature genes in 41 KRAS-mutant PDAC cell lines (DepMap). Genes with strongest probabilities (>0.75) are labeled. (C) Immunoblot evaluation of KRAS and ERK dependent protein expression changes for a subset of PDAC KRAS-ERK UP signature genes across three PDAC cell lines, treated with G12Ci/G12Di, MRTX1257/MRTX1133 (20 nM) or ERKi, SCH772984 (600 nM). Images are representative of two to three biological replicates. (D) Immunoblot evaluation of target inhibition for KRAS, ERK, and downstream kinases (CHK1, WEE1, and CDK4/6), treated with G12Di (MRTX1133, 20 nM); ERKi (SCH772984, 500 nM); CHK1i (prexasertib, 50 nM); WEE1i (adavosertib, 100 nM); or CDK4/6i (palbociclib, 500 nM). Images are representative of two to three biological replicates. (E) Proliferation assays for PDAC cells stably expressing control vector (Luc) or MEK1-DD treated with CHK1i, WEE1i, or CDK4/6i. Error bars represent SE of the mean for three to four biological replicates, each with three technical replicates.
Data S1. Differential expression statistics for genes in KRAS versus NS siRNA treated PDAC cells, using RNA-sequencing.
Data S2. Differential expression statistics and human homolog assignments for genes in dox-on versus dox-off iKras mouse orthotopic tumors, using gene expression microarrays.
Data S4. Top differentially expressed genes within KRAS modulation experiments (from data S1-S3).
Data S3. Differential expression statistics for genes in PDAC CDX tumors in animals, 24 hours after administration of KRASi versus vehicle.
Data S5. Differential expression statistics for genes in KRAS-G12Ci treated PDAC, CRC, and NSCLC cell lines over time, using RNA-sequencing.
Data S6. Differential expression statistics for genes in ERKi treated PDAC cell lines at various time points, using RNA-sequencing.
Data S7. Differential expression statistics for genes across PDAC, CRC, and NSCLC CDX tumors in animals, 24 hours after administration of KRASi versus vehicle.
Data S8. Differential expression statistics for genes in the PDAC cell line Pa16C treated with KRASi, MEKi, or ERKi versus control for 24 hours, using RNA-sequencing.
Data S10. Differential expression statistics for proteins across PDAC cell lines treated with ERKi for 1 or 24 hours, summarized by gene names.
Data S11. Median DepMap essentiality probability scores across KRAS mutant PDAC cell lines and 24-hour ERKi differential expression statistics for top differentially expressed genes.
Data S12. CRISPR drop-out screen statistics (MAGeCK) across two PDAC cell lines.
Data S13. HTG EdgeSeq gene expression counts for CRC and NSCLC patient tumors before and after KRAS-G12Ci treatment.
Data S9. Differential expression statistics for genes in KRASi treated PDAC, CRC, and NSCLC cell lines versus control for 2 hours, using RNA-sequencing.