Abstract
The M2 protein of influenza A virus forms a proton channel that is required for viral replication. The M2 ion channel is a homotetramer and has a 24-residue N-terminal extracellular domain, a 19-residue transmembrane domain, and a 54-residue cytoplasmic tail. We show here that the N-terminal methionine residue is cleaved from the mature protein. Translational stop codons were introduced into the M2 cDNA at residues 46, 52, 62, 72, 77, 82, 87, and 92. The deletion mutants were designated truncx, according to the amino acid position that was changed to a stop codon. We studied the role of the cytoplasmic tail by measuring the ion channel activity (the current sensitive to the M2-specific inhibitor amantadine) of the cytoplasmic tail truncation mutants expressed in oocytes of Xenopus laevis. When their conductance was measured over time, mutants trunc72, trunc77, and trunc92 behaved comparably to wild-type M2 protein (a decrease of only 4% over 30 min). In contrast, conductance decreased by 28% for trunc82, 27% for trunc62, and 81% for trunc52 channels. Complete closure of the channel could be observed in some cells for trunc62 and trunc52 within 30 min. These data suggest that a role of the cytoplasmic tail region of the M2 ion channel is to stabilize the pore against premature closure while the ectodomain is exposed to low pH.
The M2 integral membrane protein of influenza A virus is a minor but essential component of virions (48). However, it is abundantly expressed at the cell surface of virus-infected cells (27). The M2 mRNA encodes a polypeptide of 97 amino acids (26), and the protein forms a homotetramer either consisting of a pair of disulfide-linked dimers or completely disulfide linked as a tetramer (16, 29, 40). The protein is orientated in membranes such that it has 24 N-terminal extracellular (or lumenal) residues, a 19-residue transmembrane (TM) domain, and a 54 residue cytoplasmic tail (27, 48).
The M2 protein has an ion channel activity that functions during uncoating of virions in endosomes, permitting protons to enter the virion and bringing about dissociation of protein-protein interactions, principally weakening those between the matrix (M1) and nucleocapsid protein (for reviews, see references 14 and 25). In addition, the M2 protein functions during its transport through the exocytic pathway. There the M2 protein ion channel activity causes equilibration of pH of the lumen of the trans Golgi network with the cytoplasm (4, 13, 33). Evidence that the M2 protein has ion channel activity has been obtained by expressing the M2 protein in oocytes of Xenopus laevis or in mammalian cells and measuring cell surface currents (3, 17, 18, 32, 37, 43–45). The M2 protein ion channel activity is specifically blocked by the anti-influenza virus drug amantadine, and the M2 ion channel is activated at the lowered pH found intralumenally in endosomes and the trans Golgi network (3, 32, 37, 45). Measurement of the ion channel activity of mixed oligomers of M2 containing both amantadine-sensitive and amantadine-resistant subunits indicated that the active oligomeric form of the M2 ion channel is the homotetramer (34). In addition to the in vivo measurements, the M2 protein ion channel activity has also been shown by reconstitution of purified M2 protein into planar lipid bilayers and by incorporation of a synthetic peptide corresponding to the TM domain into planar bilayers. In both cases, amantadine-sensitive currents were measured (5, 36, 41).
As the M2 protein contains only a single hydrophobic domain, the four TM domains must encompass the pore region of the ion channel. If the TM domain is modeled as an α-helical bundle, then mutations in the M2 TM domain which confer amantadine resistance (predominantly residues 27, 30, 31, and 34) map to one face of the presumptive α-helix (14, 15, 40). Furthermore, histidine 37, which is believed to be directly involved in proton conduction (31, 43), maps to the same face of the α-helix. The inhibition of the M2 ion channel activity by Cu2+ is also consistent with histidine 37 facing the pore (11). The model that M2 residues 27, 30, 34, and 37 (heptad repeat residues a and d) would face the interior pore region of the four-helix bundle has been proposed. Several lines of evidence support this model: (i) circular dichroism studies of the TM domain peptide in membranes indicates it is α-helical in structure (7, 23); (ii) when a series of mutants with successive cysteine substitutions in the TM domain were expressed and properties of the altered M2 ion channels (reversal potential, ion currents, and amantadine resistance) were measured, Fourier analysis indicated a periodicity of 3.4 amino acid residues per turn of the helix, consistent with that expected for a four-stranded coiled coil or helical bundle (31); (iii) when the ability of the TM domain cysteine substitution mutants to form disulfide bonds under oxidizing conditions was measured, the most rapid cross-linking was at residues 27, 30, 34, 37, and 41, positions which correspond with the predicted interior of the pore (1); (iv) molecular modeling and simulation studies predict a left-handed four-helix bundle (9, 24, 31, 35, 49); and (v) solid-state nuclear magnetic resonance data, using a synthetic peptide corresponding to the TM domain region of M2, indicate that the helices are packed together in a left-handed four-helix bundle with a 33° tilt angle with respect to the membrane bilayer (23).
Although it has been suggested that the M2 protein ectodomain may be involved in incorporation of the M2 protein into virions (30) and that the M2 cytoplasmic tail may be important for interactions with other influenza virus proteins, in particular the matrix (M1) protein (2, 46), the role of these domains, if any, in the ion channel activity has not been determined. Here we describe properties of the M2 protein containing truncations to its cytoplasmic tail and show that the M2 protein cytoplasmic tail is important for maintaining the ion channel activity.
MATERIALS AND METHODS
Construction of plasmids.
The M2 cDNA of influenza A virus/Udorn/72 (48) was cloned into plasmid pGEM3 such that M2-specific RNA could be transcribed using the bacteriophage T7 promoter and T7 RNA polymerase. The USE (unique site elimination) mutagenesis system (Pharmacia Biotech, Piscataway, N.J.) was used to generate C-terminal truncation mutants of the M2 protein. The nucleotide sequences of the mutants were confirmed by using an ABI Prism 310 genetic analyzer (Applied Biosystems Inc., Foster City, Calif.).
Cell culture and transient expression of proteins.
HeLa T4 cells were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. The M2 proteins were expressed in HeLa T4 cells by using the vaccinia virus-T7 RNA polymerase (vac-T7)-mediated transient expression system as described previously (17). In brief, cells at 70 to 80% confluency were infected with recombinant vaccinia virus vTF7.3, which expresses the bacteriophage T7 RNA polymerase gene. Fourty-five minutes postinfection, cells were transfected with pGEM3 containing the cDNA for the wild-type (wt) M2 protein or the C-terminal deletion mutants.
Flow cytometry.
The M2 proteins were expressed in HeLa T4 cells by using the vac-T7-mediated transient expression system. Six hours posttransfection, cells were washed with ice-cold phosphate-buffered saline (PBS) containing 0.02% sodium azide. All further incubations were performed at 4°C. The monolayer was incubated for 30 min with PBS supplemented with 0.02% sodium azide, 0.1% bovine serum albumin, and 2% fetal calf serum (PBS-A), incubated for 1 h with M2-specific monoclonal antibody (MAb) 14C2 (47) at 1:1,000 dilution in PBS-A, washed four times with PBS, incubated for 30 min with fluorescein-conjugated goat anti-mouse immunoglobulin G diluted 1:1,000, and washed four times more with PBS to remove unbound secondary antibody. To detach the cells from the dish, cells were incubated with 0.5 ml of PBS containing 5 mM EDTA. The cells were transferred to tubes containing 0.5 ml of PBS and supplemented with 2% formaldehyde. Fluorescence intensity of 10,000 cells was measured by a FACScalibur flow cytometer (Becton Dickinson & Co., Mountain View, Calif.).
Immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Proteins were radiolabeled with [35S]methionine and/or [35S]cysteine (50 μCi/ml for 1 h at 5 h posttransfection. Cells were lysed, and M2 proteins were immunoprecipitated with MAb 14C2 (47). Polypeptides were analyzed on 17.5% acrylamide gels containing 4 M urea (17). 14C-labeled molecular weight standards (Amersham Life Sciences, Arlington Heights, Ill.) were used. The gels were subjected to autoradiography.
In vitro transcription.
pGEM3 plasmid DNA encoding the M2 proteins was linearized downstream of the M2-encoding region with HindIII, and capped RNA was transcribed in vitro by using an mMessage mMachine T7 transcription kit as instructed by the manufacturer (Ambion, Austin, Tex.).
Microinjection and culture of oocytes.
Ovarian lobules from X. laevis females were surgically removed and treated with collagenase B (2 mg/ml; Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Defolliculated oocytes were injected via a 20-μm-diameter glass pipette with mRNA transcribed in vitro, and the oocytes were maintained in ND96 at 17°C (32).
Recording of ion channel activity.
Whole-cell current was measured with a two-electrode voltage-clamp apparatus 2 to 3 days after mRNA injection. The electrodes were filled with 3 M KCl, and the oocytes were bathed in ND96. The traces were recorded with pCLAMP (version 7.1; Axon Instruments, Foster City, Calif.). Whole-cell currents were measured while gradually increasing the membrane potential from −120 to +60 mV. The holding voltage was −20 mV. For both wt and mutant channels, the current was first measured in Barth’s solution at pH 7.5. The pH of the medium was then decreased to pH 6.2; after a 2-min incubation in solution of pH 6.2, the current was remeasured. The conductance, measured as the slope of the current-voltage relationship, was determined between −80 and −40 mV. The amount of endogenous current and leakage current was determined by measuring the current-voltage relationship while the cell was bathed in Barth’s solution containing amantadine (pH 6.2). The current attributable to the M2 ion channel was calculated by subtracting the current measured in the presence of amantadine from the current measured without amantadine (pH 6.2). We did not use the data from cells in which significant endogenous currents or leakage currents were observed.
RESULTS
Expression of M2 proteins containing cytoplasmic tail deletions.
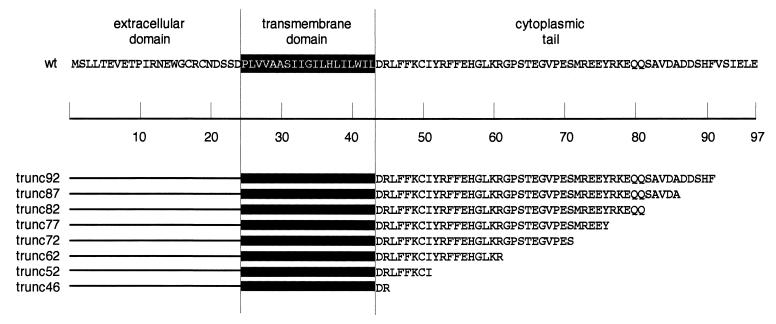
To test whether the cytoplasmic tail of the M2 protein is involved in regulating its ion channel activity, truncation mutants were constructed. Translational stop codons were introduced into the M2 cDNA at amino acid positions 46, 52, 62, 72, 77, 82, 87, and 92 (Fig. 1). The deletion mutants were designated truncx, according to the amino acid position that was changed to a stop codon.
FIG. 1.
Amino acid sequence of the influenza virus M2 protein (strain A/Udorn/72) and the C-terminal truncation mutants. The presumptive transmembrane domain residues are shaded.
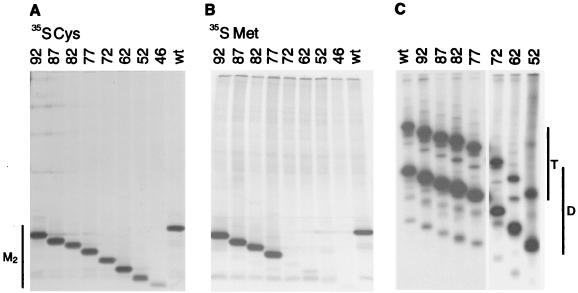
To determine whether the M2 protein expression was affected by deletion of the cytoplasmic tail, the proteins were expressed in HeLa T4 cells by using the vac-T7-mediated transient expression system. The transfected cells were metabolically labeled with [35S]methionine or [35S]cysteine, and M2 proteins were immunoprecipitated from cell lysates by using MAb 14C2, which is specific to the N-terminal ectodomain, and polypeptides were analyzed by SDS-PAGE. As shown in Fig. 2A, when the proteins were labeled with [35S]cysteine, the mutant M2 proteins exhibited a cascade of mobilities consistent with the extent of the truncation. The M2 protein contains three cysteine residues (17, 19, and 50), and trunc46 lacks cysteine residues 50. Thus, the observation that M2 trunc46 accumulated less [35S]cysteine-labeled polypeptide than the other truncation mutants is consistent with the protein sequence. When the M2 truncation mutants were labeled with [35S]methionine, mutants trunc92, trunc87, trunc82, and trunc77 were readily detected and the other truncation mutants were essentially undetectable (Fig. 2B). The M2 protein sequence predicts methionine residues at residue 1 (the initiation methionine residue) and at residue 72. Therefore, the data shown in Fig. 2B are consistent with cleavage of the initiation methionine residue from the mature protein, and thus M2 protein contains 96 residues. This experimental finding is consistent with the finding that for cytosolic proteins, small, uncharged residues next to the initiation codon (Ala, Gly, Pro, Ser, and Thr) promote N-terminal processing of the initiation methionine residue (8).
FIG. 2.
Expression of the M2 protein truncation mutants from cDNAs. HeLa T4 cells were infected with vac-T7 and transfected with plasmids encoding the wt M2 protein or the truncation mutants. At 5 h posttransfection, the cells were labeled with [35S]methionine (A), [35S]cysteine (B), or [35S]cysteine (C). The cells were lysed, proteins were immunoprecipitated with MAb 14C2, and polypeptides were analyzed by SDS-PAGE on 17.5% acrylamide gels containing 4 M urea. (A and B) Electrophoresis under reducing conditions; (C) electrophoresis under nonreducing conditions. T, tetramers; D, disulfide-linked dimers.
Disulfide bond formation of M2 proteins containing cytoplasmic tail truncations.
As homotetramer formation is believed to be obligatory for forming a functional M2 ion channel, the ability of the deletion mutants to form disulfide-linked dimers and tetramers was examined. The wt M2 protein and the deletion mutants were expressed in HeLa T4 cells and radiolabeled with [35S]cysteine. The M2 proteins were immunoprecipitated from cell lysates, and proteins were analyzed by SDS-PAGE under nonreducing conditions. As shown in Fig. 2C, all of the truncation mutants formed a mixture of disulfide-linked dimers and tetramers on SDS-PAGE, much like wt M2 protein, and thus the data suggest that all of the truncation mutants formed homotetrameric species that were either a dimer of disulfide-linked dimers or a completely disulfide-linked tetramer.
Cell surface expression of M2 proteins containing cytoplasmic tail truncations.
The wt M2 protein is abundantly expressed on the surface of influenza virus-infected cells (27). As the length of the C-terminal cytoplasmic tails might influence the transport of the truncation mutants to the plasma membrane, surface expression of the truncation mutants in mammalian cells was quantified by flow cytometry. The wt M2 protein and the truncation mutant proteins were expressed by using the vac-T7-mediated transient expression system; at 6 h posttransfection, cells were analyzed by flow cytometry using MAb 14C2. The percentage of cells expressing M2 proteins and the mean fluorescence intensity were similar for trunc92, trunc82, trunc77, and trunc72, although for all of these mutants the number of cells expressing M2 protein was less than for wt M2 protein (Table 1). For trunc62, both the percentage of cells expressing M2 protein and the mean fluorescent intensity were reduced, and this lower-level surface expression was more pronounced for trunc52. M2 trunc46 was not detected at the cell surface. We have not investigated further the reason for the reduced surface expression of trunc62 and trunc52 or the lack of surface expression of trunc46.
TABLE 1.
Surface expression of C-terminal truncation mutants
| Construct | Mean ± SD (n = 3)
|
|
|---|---|---|
| % Gated cells | Mean fluorescence intensity | |
| wt | 46.08 ± 2.11 | 197.86 ± 4.44 |
| trunc92 | 34.84 ± 3.61 | 239.76 ± 11.15 |
| trunc87 | 41.61 ± 3.59 | 307.95 ± 10.49 |
| trunc82 | 33.86 ± 5.27 | 231.29 ± 9.62 |
| trunc77 | 36.51 ± 4.11 | 258.91 ± 8.35 |
| trunc72 | 32.17 ± 2.96 | 256.69 ± 13.99 |
| trunc62 | 26.26 ± 0.70 | 162.25 ± 8.07 |
| trunc52 | 19.75 ± 1.10 | 64.60 ± 0.71 |
| trunc46 | 1.93 ± 0.28 | 56.23 ± 3.91 |
| Mock | 1.87 ± 0.29 | 50.43 ± 2.39 |
Ion channel activities of M2 truncation mutant proteins.
We compared the ion channel activity of each of the M2 truncation proteins with that of the wt M2 protein by expressing the proteins in oocytes of X. laevis and recording the membrane currents with a two-electrode voltage clamp apparatus (32). The wt M2 protein has very low ion channel activity at neutral pH, but its activity increases after the pH of the bathing medium is lowered and is rapidly eliminated by application of amantadine (100 μM) to the bathing medium (32, 45). We quantified the ion channel activity of the M2 proteins by measuring the current induced by a range of applied voltages (−120 to +60 mV). This was done first in high-pH medium and then for each of seven time points over the 30-min interval in which the oocytes were bathed in medium of low pH. The measure of ion channel activity that we used was the slope of the relationship between current and voltage, or the conductance (Fig. 3 and 4).
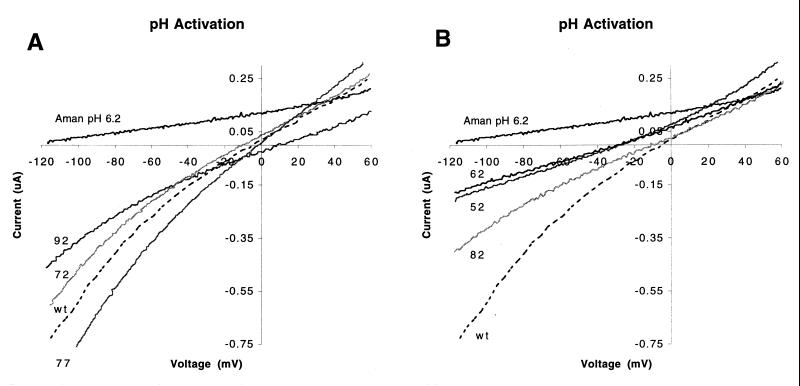
FIG. 3.
Comparison of the ion channel activity of M2 truncation mutants with that of the wt M2 protein ion channel. The transmembrane voltage was varied from −120 to +60 mV while membrane current was measured after incubation of the oocyte for 2 min in medium of pH 6.2. (A) Truncation mutants with ion channel properties similar to those of wt M2 protein (trunc92, trunc77, and trunc72); (B) truncation mutants with activity less than that of wt M2 protein (trunc82, trunc62, and trunc52). The interrupted lines show the currents of wt M2 protein in pH 6.2 medium. The inhibition of the wt ion channel activity by amantadine is shown in the top traces. Each trace represents the average total current of at least two cells.
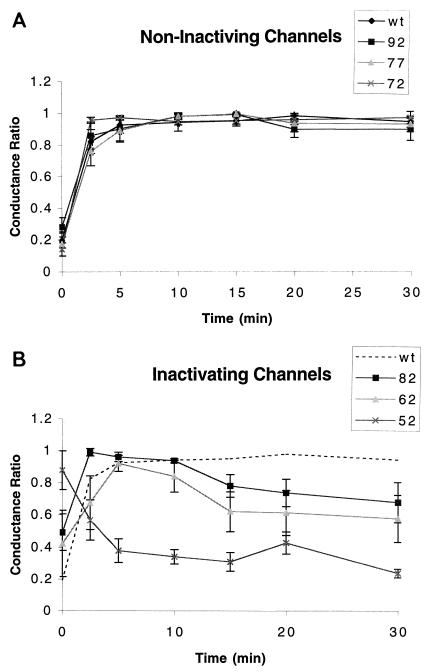
FIG. 4.
Comparison of abilities of the wt M2 protein and truncation mutant proteins to sustain currents. The ability to sustain was evaluated by measuring the amantadine-sensitive conductance. Conductances for the mutant proteins were normalized individually (conductance ratio) and plotted against time during which the oocytes were bathed in a medium of pH 6.2 (−20 mV holding voltage). (A) Mutants trunc92, trunc77, and trunc72 sustained currents during the 30-min incubation in solution of pH 6.2. (B) Conductances of mutants trunc82, trunc62, and trunc52 declined by more than 25% during the incubation in pH 6.2 medium.
We measured the current-voltage relationship of oocytes expressing the M2 truncation mutant proteins and compared them to oocytes expressing the wt M2 protein. Only cells exhibiting currents that could be eliminated by the application of amantadine and that were able to be recorded for the full time of the experiment are reported. Three of the truncation mutant proteins (trunc92, trunc77, and trunc72) had ion channel activity similar to that of the wt M2 protein: they had low conductance at neutral pH, their conductance increased when the pH of the bathing solution was lowered, and their conductance was decreased by amantadine (Fig. 3A). In contrast, the ion channel activity of oocytes expressing the remaining truncation mutant proteins (trunc82, trunc62, and trunc52) was less than that of oocytes expressing the wt M2 protein (Fig. 3B). Recordings of trunc87 were not performed. Despite the lower ion channel activity, the ion selectivity of these proteins did not differ greatly from that of the wt M2 protein. However, the reversal potentials of lower conducting channels such as trunc62 and trunc52 were shifted. This is because the lower conducting channels were more adversely affected by endogenous currents than the higher conducting channels (Fig. 3B). Nonetheless, when the amantadine-sensitive current was determined, it was found that the lower conducting mutants had a reversal potential comparable to that of wt M2 (data not shown). Thus, the truncation mutant proteins exhibited ion channel activities that could be quantitatively compared to that of the wt M2 protein.
We compared the ability of each of the truncation mutant proteins with that of the wt M2 protein to sustain ion channel activity while bathed in medium of low pH. The ion channel activity of the wt M2 protein was constant for as long as 30 min, while the oocyte was bathed medium of pH 6.2 (Fig. 4). Ion channel activity was also sustained in oocytes that expressed truncation mutant proteins trunc92, trunc77, and trunc72 (Fig. 4A). However, the same truncation mutant proteins associated with lower ion channel activity (trunc82, trunc62, and trunc52) also failed to maintain sustained ion channel activity when expressed in oocytes that were bathed in a solution of pH 6.2 (Fig. 4B).
One of the truncation mutant proteins that was similar to wt M2 protein in most respects, trunc92, differed from wt M2 protein in one physiological property. After injection of mRNA encoding the wt M2 protein, it is usually satisfactory to incubate the oocytes in a medium of pH 7.5 while the protein is being expressed. At this pH, the ion channel activity is minimal, and the proton flux across the plasma membrane does not result in oocyte death; however, when bathed in medium of lower pH, oocytes die rapidly (12, 25). We noticed that oocytes expressing the trunc92 mutant protein had a higher rate of cell death than oocytes expressing either the wt M2 protein or the other truncation mutant proteins incubated in medium of pH 7.5. However, cell death of oocytes expressing the trunc92 mutant protein was decreased when these oocytes were incubated in a medium of pH 8.5 starting from the time of injection of mRNA. This suggests that the trunc92 mutant protein has greater ion channel activity than the wt M2 protein at neutral pH.
DISCUSSION
Oocytes expressing three truncation mutant proteins, trunc82, trunc62, and trunc52, had lower ion channel activity than oocytes expressing the wt M2 protein (Fig. 3B). In principle, this lower ion channel activity can result from either reduced surface expression of the protein or reduced activity of the ion channel macromolecular complex. It is likely that reduced ion channel activity of oocytes expressing the trunc62 and trunc52 mutant proteins is the result of reduced protein surface expression (Table 1). However, it is unlikely that reduced surface expression can explain the reduced ion channel activity of oocytes expressing the trunc82 protein (Table 1). Instead, the reduced ion channel activity probably results from an alteration in the structure of the trunc82 mutant ion channel protein. Truncation of the cytoplasmic N and C termini of several other ion channels has been demonstrated to alter their activity. In most cases, the result is an increase in the activity of the channel (22, 28). One possible basis for the increase in activity of these other ion channels is the removal of a peptide that is capable of blocking the channel pore and thus causing inactivation (20). However, the decrease in activity of the trunc82 mutant protein must have a mechanism different from the increase in activity seen for truncation mutations of other ion channels.
Oocytes expressing the three truncation mutant proteins trunc82, trunc62, and trunc52 were also unable to sustain ion channel activity while bathed in medium of low pH (Fig. 4). Two fundamental properties of ion channels are activation and inactivation. Activation is the increase in ion channel activity that occurs upon presentation of the appropriate stimulus. In the case of the M2 protein, this stimulus is lowered pH. Inactivation is the decrease in activity that occurs after prolonged application of the activating stimulus. Oocytes expressing these three truncation mutant proteins displayed a decrease in inward current during prolonged exposure to low-pH medium. However, a decrease in inward current does not necessarily signal that inactivation has occurred, especially in oocytes. The reason is that normally silent endogenous currents can be up-regulated by expression of exogenous proteins, among them variants of the M2 protein (37). Two lines of evidence indicate that the decrease of inward current that we observed does reflect the process of inactivation for these mutant proteins. The currents of the M2 protein are up-regulated by low pH of the bathing medium and are sensitive to the inhibitor amantadine. Application of amantadine eliminated the currents of oocytes that expressed the truncation mutant proteins. The second argument against endogenous currents being responsible for the observed decrease in current is based on the observation that the amplitude of endogenous currents varies widely from frog to frog (38). If endogenous currents were responsible for the measured decrease in inward current, then the decrease should have varied from frog to frog. However, the variability that we observed in the decrease in inward current was from cell to cell from the same frog and not from frog to frog. A second potential cause for a decrease in current while the oocyte was bathed in a medium of low pH is acidification of the intracellular medium of the oocyte (37). However, this is not a likely explanation, as the decrease in current was not greater for oocytes expressing greater currents.
Posttranslational modifications are capable of modifying the function of many proteins, including ion channel proteins. The cytoplasmic domain of the M2 protein is modified in several ways. Cysteine residue 50 is palmitylated (39, 42), and serine residue 64 is the major site for phosphorylation, but serine residues, 82, 89, and 93 are also minor phosphorylation sites (18). However, site-altered forms of the protein in which all of these modifications are ablated show ion channel activity that is indistinguishable from that of the wt M2 protein (18). Thus, it seems unlikely that the mechanism by which the truncations alter ion channel activity is due to lack of a site for a posttranslational modification.
What is responsible for the observed decrease in ion channel activity that results from truncation of the C terminus of the M2 protein? It is unlikely that the alterations are a sole consequence of the number of residues truncated from the C terminus of the protein. If this were so, then oocytes expressing the trunc82 protein would have conductances larger than those of oocytes expressing the trunc77 protein. A second possibility is that the C terminus of each M2 monomer has a secondary structure that is essential for the stability of the M2 ion channel protein and that disruption of the secondary structure of the channel protein results in a decrease in channel activity, perhaps by destabilizing the protein. However, if such a secondary structure exists, it cannot be one whose stability is directly proportional to its length, or the decrease in current would be proportional to the length of the truncated segment of the C terminus. An example of truncation affecting the state of an ion channel can be found in the amiloride-sensitive Na+ channel, the activity of which is decreased by truncation of the C terminus. The result of this truncation is to increase the probability of the channel to exist in a low-conductance state at the expense of a high-conductance state (10). Although the conducting states of the M2 ion channel protein are not yet known, it is possible that this mechanism also serves to explain the present results.
Three truncation mutant channels (trunc82, trunc62, and trunc52) did not sustain ion channel activity in low pH; i.e., they partially inactivated. Alterations of C-type inactivation also occur as a result of deletions in the cytoplasmic C-terminal domain of Shaker-type K+ channels. C-type inactivation (21) has slow onset and can be studied in the absence of the more rapid N-type inactivation that results from occlusion of the channel pore by amino acids in the N terminus of the channel protein. Small deletions of the C terminus of the Kv1.1 channel result in a slower onset of inactivation, while large deletions in the region result in a more rapid onset of inactivation (19). The alteration that results in slower inactivation was mapped to a region containing five amino acids. Thus, it is possible that certain C-terminal truncations of the M2 protein result in the initiation of a process similar to C-terminal inactivation of Shaker-type K+ channels, although with a slower time course.
These results have important implications for the study of the structure and function of the M2 protein. Functional studies of the ion channel activity (5) and structural studies using circular dichroism (7, 23), neutron diffraction (6), and solid-state nuclear magnetic resonance (23) using the isolated M2 transmembrane peptide have been reported. Our findings indicate that truncation of C terminus of the protein leads to modification of the function of the ion channel activity which is not directly proportional to the length of the truncated segment. This implies that measurements made on the isolated transmembrane peptide may not precisely reflect the properties of the intact M2 ion channel protein.
The M2 cytoplasmic tail may be multifunctional. In addition to being important for sustaining ion channel activity, the cytoplasmic tail may be important for interactions with other viral proteins, in particular the M1 protein. In a reverse genetics experiment, it was found that deletion of 5 or 10 residues from the M2 cytoplasmic tail abrogated the ability to rescue influenza virus (2). However, the failure to rescue the virus does not permit distinction between alteration of ion channel activity or a loss of protein-protein interactions. However, influenza virus variants that escaped the growth restriction of MAb 14C2 contained changes in the M2 cytoplasmic tail or in the M1 protein, suggesting an interaction between the M2 cytoplasmic tail and the M1 protein (46, 47).
ACKNOWLEDGMENTS
This research was supported by Public Health Service research grants R37 AI-20201 (R.A.L.) and AI-31882 (L.H.P.) from the National Institute of Allergy and Infectious Diseases. K.T. was the recipient of a fellowship from the Swiss National Science Foundation. R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bauer C M, Pinto L H, Cross T A, Lamb R A. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 2.Castrucci M R, Kawaoka Y. Reverse genetics system for generation of an influenza A virus mutant containing a deletion of the carboxyl-terminal residue of M2 protein. J Virol. 1995;69:2725–2728. doi: 10.1128/jvi.69.5.2725-2728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chizhmakov I V, Geraghty F M, Ogden D C, Hayhurst A, Antoniou M, Hay A J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciampor F, Bayley P M, Nermut M V, Hirst E M, Sugrue R J, Hay A J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 5.Duff K C, Ashley R H. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 6.Duff K C, Gilchrist P J, Saxena A M, Bradshaw J P. Neutron diffraction reveals the site of amantadine blockade in the influenza A M2 ion channel. Virology. 1994;202:287–293. doi: 10.1006/viro.1994.1345. [DOI] [PubMed] [Google Scholar]
- 7.Duff K C, Kelly S M, Price N C, Bradshaw J P. The secondary structure of influenza A M2 transmembrane domain. FEBS Lett. 1992;311:256–258. doi: 10.1016/0014-5793(92)81114-2. [DOI] [PubMed] [Google Scholar]
- 8.Flinta C, Persson B, Jornvall H, von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986;154:193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 9.Forrest L R, DeGrado W F, Dieckmann G R, Sansom M S. Two models of the influenza A M2 channel domain: verification by comparison. Fold Design. 1998;3:443–448. doi: 10.1016/S1359-0278(98)00061-3. [DOI] [PubMed] [Google Scholar]
- 10.Fuller C M, Ismailov I I, Berdiev B K, Shlyonsky V G, Benos D J. Kinetic interconversion of rat and bovine homologs of the alpha subunit of an amiloride-sensitive Na+ channel by C-terminal truncation of the bovine subunit. J Biol Chem. 1996;271:26602–26608. doi: 10.1074/jbc.271.43.26602. [DOI] [PubMed] [Google Scholar]
- 11.Gandhi C S, Shuck K, Lear J D, Dieckmann G R, DeGrado W F, Lamb R A, Pinto L H. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J Biol Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 12.Giffin K, Rader R K, Marino M H, Forgey R W. Novel assay for the influenza virus M2 channel activity. FEBS Lett. 1995;357:269–274. doi: 10.1016/0014-5793(94)01369-c. [DOI] [PubMed] [Google Scholar]
- 13.Grambas S, Hay A J. Maturation of influenza A virus hemagglutinin—estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 14.Hay A J. The action of adamantanamines against influenza A viruses: inhibition of the M2 ion channel protein. Semin Virol. 1992;3:21–30. [Google Scholar]
- 15.Hay A J, Wolstenholme A J, Skehel J J, Smith M H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holsinger L J, Lamb R A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 17.Holsinger L J, Nichani D, Pinto L H, Lamb R A. Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holsinger L J, Shaughnessy M A, Micko A, Pinto L H, Lamb R A. Analysis of the posttranslational modifications of the influenza virus M2 protein. J Virol. 1995;69:1219–1225. doi: 10.1128/jvi.69.2.1219-1225.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopkins W F, Demas V, Tempel B L. Both N- and C-terminal regions contribute to the assembly and functional expression of homo- and heteromultimeric voltage-gated K+ channels. J Neurosci. 1994;14:1385–1393. doi: 10.1523/JNEUROSCI.14-03-01385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshi T, Zagotta W N, Aldrich R W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 21.Hoshi T, Zagotta W N, Aldrich R W. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–556. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- 22.Jerng H H, Covarrubias M. K+ channel inactivation mediated by the concerted action of the cytoplasmic N- and C-terminal domains. Biophys J. 1997;72:163–174. doi: 10.1016/S0006-3495(97)78655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs F A, Cross T A. Transmembrane four-helix bundle of influenza A M2 protein channel: structural implications from helix tilt and orientation. Biophys J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kukol A, Adams P D, Rice L M, Brunger A T, Arkin T I. Experimentally based orientational refinement of membrane protein models: a structure for the influenza A M2 H+ channel. J Mol Biol. 1999;286:951–962. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- 25.Lamb R A, Holsinger L J, Pinto L H. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Receptor-mediated virus entry into cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1994. pp. 303–321. [Google Scholar]
- 26.Lamb R A, Lai C-J, Choppin P W. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci USA. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb R A, Zebedee S L, Richardson C D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Taffet S, Stoner L, Delmar M, Vallano M L, Jalife J. A structural basis for the unequal sensitivity of the major cardiac and liver gap junctions to intracellular acidification: the carboxyl tail length. Biophys J. 1993;64:1422–1433. doi: 10.1016/S0006-3495(93)81508-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panayotov P P, Schlesinger R W. Oligomeric organization and strain-specific proteolytic modification of the virion M2 protein of influenza A H1N1 viruses. Virology. 1992;186:352–355. doi: 10.1016/0042-6822(92)90096-8. [DOI] [PubMed] [Google Scholar]
- 30.Park E K, Castrucci M R, Portner A, Kawaoka Y. The M2 ectodomain is important for its incorporation into influenza A virions. J Virol. 1998;72:2449–2455. doi: 10.1128/jvi.72.3.2449-2455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto L H, Dieckmann G R, Gandhi C S, Papworth C G, Braman J, Shaughnessy M A, Lear J D, Lamb R A, DeGrado W F. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi T, Leser G P, Lamb R A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakaguchi T, Tu Q, Pinto L H, Lamb R A. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc Natl Acad Sci USA. 1997;94:5000–5004. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sansom M S P, Kerr I D, Smith G R, Son H S. The influenza A virus M2 channel: a molecular modeling and simulation study. Virology. 1997;233:162–173. doi: 10.1006/viro.1997.8578. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder C, Ford C M, Wharton S A, Hay A J. Functional reconstitution in lipid vesicles of influenza virus M2 protein expressed by baculovirus: evidence for proton transfer activity. J Gen Virol. 1994;75:3477–3484. doi: 10.1099/0022-1317-75-12-3477. [DOI] [PubMed] [Google Scholar]
- 37.Shimbo K, Brassard D L, Lamb R A, Pinto L H. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimbo K, Brassard D L, Lamb R A, Pinto L H. Viral and cellular small integral membrane proteins can modify ion channels endogenous to Xenopus oocytes. Biophys J. 1995;69:1819–1829. doi: 10.1016/S0006-3495(95)80052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugrue R J, Belshe R B, Hay A J. Palmitoylation of the influenza A virus M2 protein. Virology. 1990;179:51–56. doi: 10.1016/0042-6822(90)90272-s. [DOI] [PubMed] [Google Scholar]
- 40.Sugrue R J, Hay A J. Structural characteristics of the M2 protein of the influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tosteson M T, Pinto L H, Holsinger L J, Lamb R A. Reconstitution of the influenza virus M2 ion channel in lipid bilayers. J Membr Biol. 1994;142:117–126. doi: 10.1007/BF00233389. [DOI] [PubMed] [Google Scholar]
- 42.Veit M, Klenk H-D, Kendal A, Rott R. The M2 protein of influenza A virus is acylated. Virology. 1991;184:227–234. doi: 10.1099/0022-1317-72-6-1461. [DOI] [PubMed] [Google Scholar]
- 43.Wang C, Lamb R A, Pinto L H. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Lamb R A, Pinto L H. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Takeuchi K, Pinto L H, Lamb R A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zebedee S L, Lamb R A. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci USA. 1989;86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zebedee S L, Lamb R A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zebedee S L, Richardson C D, Lamb R A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong Q, Husslein T, Moore P B, Newns D M, Pattnaik P, Klein M L. The M2 channel of influenza A virus: a molecular dynamics study. FEBS Lett. 1998;434:265–271. doi: 10.1016/s0014-5793(98)00988-0. [DOI] [PubMed] [Google Scholar]