Abstract
Background: An increasing number of studies demonstrate that abnormal miRNA expression contributes to the advancement of many tumors. Nonetheless, the potential role of miR-125b in multiple myeloma (MM) remains unknown. Objectives: To explore the potential effects and mechanism of miR-125b in MM. Methods: Real-time quantitative PCR was used to measure the expression levels of miR-125b and MKNK2 in a variety of MM samples. Colony formation and cell counting Kit-8 (CCK-8) assays were used to assess cell proliferation, the transwell assay was used to evaluate the cell invasion capability, and dual luciferase reporter gene assay and Western blot were used to examine the interaction between miR-125b and MKNK2. Results: The levels of miR-125b were higher in MM tissue samples, alongside increased expression of MKNK2. There was a negative correlation between MKNK2 and miR-125b expression in MM tissues. MKNK2 was identified as a direct target gene of miR-125b in MM cells. Overexpression of miR-125b suppressed MM cell growth, colony formation, and invasion. In addition, MKNK2 was found to mediate the effects of miR-125b on cell proliferation, colony formation, and invasion in MM. Conclusions: miR-125b acts as a suppressive factor in multiple myeloma and can affect the malignant behavior of MM by regulating the expression of MKNK2.
Keywords: miRNA, miR-125b, multiple myeloma, MKNK2, proliferation, target
Introduction
Multiple myeloma (MM) is an incurable plasma cell disease and the second most prevalent hematological malignancy after non-Hodgkin lymphoma [1,2]. Despite significant advancements in treatment over recent decades, MM remains incurable, often resulting in end-organ damage, with nearly all patients eventually relapsing and succumbing to these complications [3-5]. The high relapse and refractory nature of MM, as well as its severe final outcome, underscores the urgent need for identifying therapeutic targets in clinical research.
MAPK Interacting Serine/Threonine Kinases (MKNKs, also known as MNKs) are closely related to the MAPK signaling pathway [6]. Current research has identified two main subtypes of this family, namely MKNK1 and MKNK2, both of which play critical roles in cell protein synthesis, growth cycle regulation, differentiation, and response to environmental stress [7,8]. MKNKs have been shown to play a role in a variety of diseases including cancer [9,10] and inflammatory diseases [11,12]. In particular, MKNKs contribute to the malignancy of tumor cells by phosphorylating eIF4E, an important translation initiation factor [13,14]. Increasing evidence suggests that MKNKs also play a key role in tumor drug resistance [15,16]. Therefore, some therapeutic studies have targeted this kinase family, especially in hematological tumors [17,18]. Members of this family are recognized for their huge therapeutic potential [19,20]. Interestingly, some studies have found that MKNK2 has a higher basal activity compared to MKNK1 [21,22], highlighting the significance of regulating MKNK2 as a promising approach to developing therapeutic targets for tumors.
MicroRNAs are non-coding single-stranded RNA molecules produced by endogenous genes. MicroRNAs can regulate gene expression by binding to the 3’-untranslated region (3’-UTR) of particular target mRNAs, thereby inhibiting the post-transcriptional translation of target genes [23-25]. MicroRNAs influence the development and progression of various diseases by participating diverse biological processes, including cell death, development, proliferation, differentiation, lipid metabolism, and migration [26-29]. Cancers, including hepatocellular carcinoma, glioblastoma, colorectal cancer, ovarian cancer, renal cell carcinoma, and gastric cancer, have been linked to the dysregulation of microRNAs [30-35].
MiR-125b, the first mammalian homolog of miRNA lin-4 identified in Cryptomeria japonica, is one of the most important miRNAs regulating various physiological and pathological processes [36]. MiR-125b has been implicated in a variety of cancers as an oncogene or suppressor gene. miR-125b is known to operate primarily through multiple molecular targets that are involved in its signaling cascade. It has been demonstrated that miR-125b binds to the 3’ UTR of APC in triple-negative breast cancer and that inhibition of miR-125b activity results in reduced intracellular Wnt/β-catenin signaling and EMT activity [37]. MiR-125b has also been shown to regulate PI3K/Akt [38], STAT-3 [39], MAPK [40], NF-κB and p53 [41] signaling pathways. However, the mechanism by which miR-125b functions in MM is currently unclear. By exploring the role and mechanism of action of miR-125b in osteosarcoma, this study is conducive to the development of new targets for the treatment of osteosarcoma, and provides a new theoretical basis for the clinical targeted therapy of osteosarcoma. It also enriches the known miR-125b-related biological network, which can further reveal the pathological process of osteosarcoma and help find more clinical treatment options.
Materials and methods
Sample source
Twenty pairs of tissue samples from multiple myeloma (MM) patients, including MM tissue and adjacent non-tumor tissue, were obtained from our institution. The tissues were immediately frozen in liquid nitrogen until usage. Normal tissues adjacent to MM tissue served as the control. The Ethics Committee at the First Affiliated Hospital of Guangxi Medical University approved this study, and informed consent was obtained prior to the collection of human tissues. The expression of miR-125b and MKNK2 in MM tissues was assessed.
Cell culture and transfection
MM cancer cell lines (U266 myeloma) were provided by Shangen, Wuhan, China. Cells were cultured in RPMI-1640 medium (Beyotime, Shanghai, China) supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. miR-125b mimics, control miRNA (100 nM), miR-125b-inhibitor, inhibitor-NC or MKNK2 plasmid were transfected into the U266 cell line using Lipofectamine 2000 (Invitrogen), and the specific operating procedures were performed strictly in accordance with the manufacturer’s instructions. The sequences of the transfection plasmids are shown in Table 2.
Table 2.
Sequence of the transfection plasmid
| Genes | Sequences |
|---|---|
| miR-125b mimics | 5’-UCCCUGAGACCCUAACUUGUGA-3’ |
| Control miRNA | 5’-UCACAACCCUAGAAAGAGUAGA-3’ |
| miR-125b-inhibitor | 5’-UCA CAAGUUAGGGUCUCAGGGA-3’ |
| inhibitor-NC | 5’-UCUACUCUUUCUAGGAGGUUGUGA-3’ |
| MKNK2 | F: 5’-AAGCTTATGGTGCAGAAGAAACCAGCCGAACTT-3’ |
| R: 5’-TCTAGATCACTCATTCACAGTAACGGTTCTGA-3’ |
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using RNA TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. cDNA synthesis was performed using the PrimeScript RT Reagent Kit (Takara, Dalian, China). Quantification of miRNA and mRNA was performed by qRT-PCR experiments using CFX96 PCR system (BioRad), SYBR Premix ExTaqTM II (Takara, Dalian, China). The relative expression was normalized to the expression of U6 or GAPDH and calculated by the 2-ΔΔCT method. Each experiment was performed in triplicate. Primer sequences are shown in Table 1.
Table 1.
Sequences of the real-time PCR primers
| Gene | Forward Primer (5’ → 3’) | Reverse Primer (5’ → 3’) |
|---|---|---|
| MKNK2 | 5’-GTTCGAAGATGTCTATCAGC-3’ | 5’-TTCTAGAACATTCCTATGTCCC-3’ |
| GAPDH | 5’-GGTCTCCTCTGACTTCAACA-3’ | 5’-GTGAGGGTCTCTCTCTTCCT-3’ |
| miR-125b | 5’-TCCCTGAGACCCTAACTTGTGA-3’ | Universal primer |
| U6 | 5’-GCCAGCTCCTACATCTCAGC-3’ | 5’-AGCCTGACTTGCTAGTGGATTAT-3’ |
| Ki67 | 5’-TTACCGGGCGGAGGTATGAA-3’ | 5’-GCTGGCTCCTGTTCACGTAT-3’ |
Western blot analysis
The whole protein from the MM cell line was extracted using RIPA lysis buffer that pre-cooled (Beyotime, Shanghai, China). Protein samples were separated using 10% SDS-PAGE (Beyotime, Shanghai, China), transferred to 0.22 m PVDF membranes (Millipore, Temecula, CA), and blocked with 5% non-fat milk. Subsequently, the membranes were treated with specific primary antibodies. To identify immunoreactive bands, an ECL system (manufactured by Millipore and located in Temecula, California) was utilized, and GAPDH antibody was used as a control. Image Lab was utilized to quantitatively analyze the grayscale of protein bands. Antibodies against MKNK2, GAPDH, and secondary anti-rabbit antibodies were obtained from Cell Signaling Technology (Beverly, USA).
Detection of apoptosis by flow cytometry
Cell apoptosis rates were measured using Annexin-FITC/PI kit (C1062S, Beyotime, China). MM cells after intervention with miR-125b mimics or control miRNA were collected and washed with PBS, followed by incubation with 10 µl FITC and PI in the dark. Finally, the cell apoptosis rate was measured by flow cytometry.
Detection of cell proliferation
Kit-8 (CCK-8, Dojindo, Japan) was used to detect cell proliferation in accordance with the instructions. In brief, 1500 transfected MM cells were transplanted into a 96-well plate. The absorbance was measured at 450 nm after the CCK-8 solution was added at 0, 24, 48, 72, and 96 hours, and the cells were cultured in the dark for approximately 2 hours. Equal numbers of transfected MM cells were seeded in 6-well plates and counted every other day. After 12 days, colonies were stained and photographed.
Colony formation experiments
Following cell transfection, U266 cells were seeded into a 6-well plate at 500 cells/well and cultured at 37°C and 5% CO2 for 2 weeks. After removing the culture medium, the colonies were fixed with 4% paraformaldehyde for 15 minutes, stained with crystal violet for 10 minutes, and then photographed and counted using a microscope.
Transwell cell invasion assay
The experiment was performed using a Transwell chamber (8 µm, 6.5 mm, Merck, China). The transfected U266 cells (5 × 104) were seeded into the upper chamber, while the lower chamber was filled with culture medium containing 10% FBS. After incubation, the migrated cells were fixed in methanol and stained with 0.5% crystal violet for 15 minutes. Finally, a microscope was used for photography and cell counting.
Luciferase reporter assay
First, pG13 plasmid was introduced with MKNK2 fragments with or without anticipated miR-125b binding sites (Promega, USA). Next, the recombinant vectors miR-125b and scramble were co-transfected with Lipofectamine 2000 into U266 cells for 48 hours (Invitrogen). A dual-luciferase reporter gene assay was employed to determine the luciferase activity (Promega).
Statistical analysis
Each experiment was performed in triplicate. Data were analyzed using GraphPad Prism 8.0 (California, United States). The data were displayed as the mean ± standard deviation. Student’s t-test and ANOVA with Turkey’s test were utilized for comparisons between the two groups or among multiple groups as appropriate. Statistical significance was defined as P < 0.05.
Results
Expression of miR-125b and MKNK2 in MM tissues
The expression of miR-125b was significantly lower in MM tissues than that in non-tumor samples (Figure 1A), while the expression of MKNK2 was significantly higher in MM samples than that in non-tumor samples (Figure 1B). Additionally, the expression of MKNK2 in MM tissues was inversely correlated with the expression of miR-125b (Figure 1C).
Figure 1.
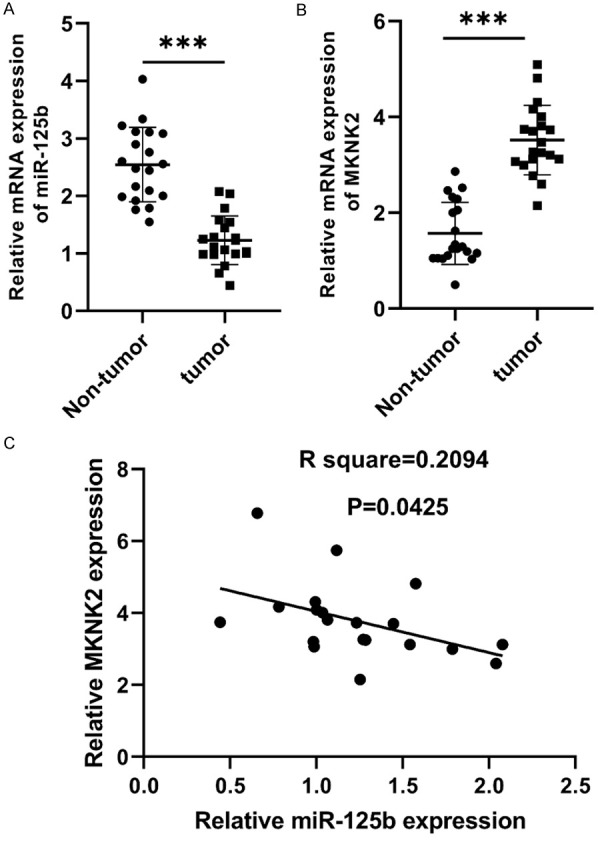
MiR-125b and MKNK2 expression in MM samples. A. miR-125b expression was down-regulated in comparison to non-tumor samples (***P < 0.001). B. MKNK2 expression was downregulated in non-tumor samples (***P < 0.001). C. The expression of MKNK2 was inversely correlated with the expression of miR-125b in MM samples (n=20).
miR-125b induced the malignant behaviors of MM cells and suppressed apoptosis
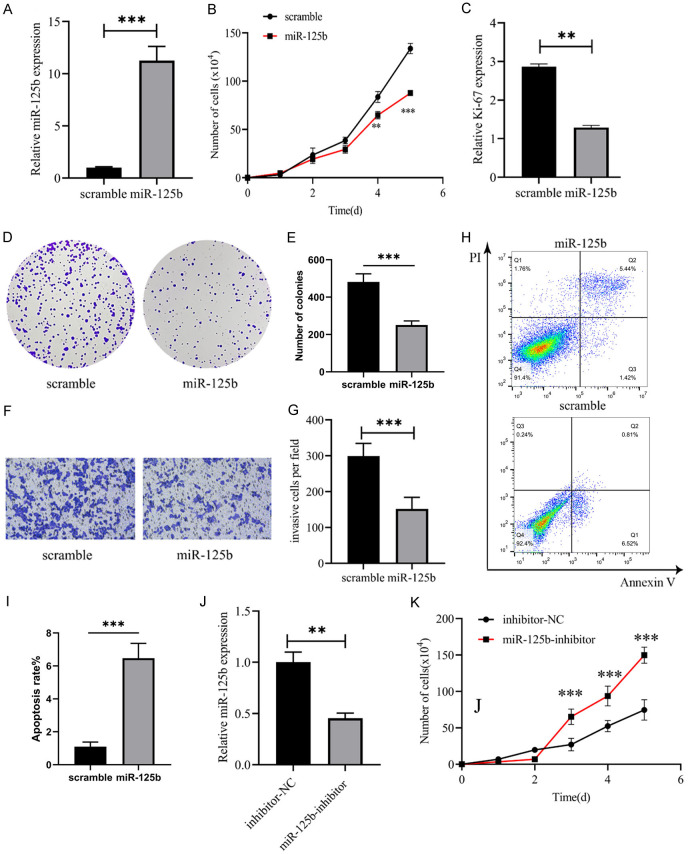
We transfected control miRNA (scramble) and miR-125b mimics into the U266 cell line, and RT-qPCR confirmed the upregulation of miR-125b expression by the mimic (Figure 2A). In addition, we also found that miR-125b overexpression inhibited the proliferation of U266 cells (Figure 2B). In contrast, miR-125b inhibitors promoted the proliferation of U266 cells (Figure 2J, 2K). Additionally, miR-125b mimic transfection led to a significant reduction in Ki-67 expression in U266 cells (Figure 2C) and decreased colony formation (Figure 2D, 2E). Furthermore, Transwell experiments revealed that the migration ability of U266 cells overexpressing miR-125b was significantly reduced (Figure 2F, 2G). Apoptosis experiments showed that miR-125b overexpression significantly promoted U266 cell apoptosis (Figure 2H, 2I). These results indicate that up-regulating miR-125b is beneficial to reducing the malignancy of MM cells and promoting apoptosis.
Figure 2.
Overexpression of miR-125b significantly reduced proliferation and migration of MM cells. A. Exogenous miR-125b mimic increased miR-125b expression in U266 cells. B. miR-125b mimics decreased U266 cell counts (**P < 0.01, ***P < 0.001, miR-125b group vs scramble group on the same day). C. miR-125b overexpression decreased ki-67 expression. D, E. miR-125b overexpression suppressed colony formation of U266 cells. F, G. miR-125b mimics suppressed migration of U266 cells (10×), Bar: 100 µm. H, I. U266 cell apoptosis rate was determined by the FACS analysis. J. miR-125b inhibitor significantly reduced miR-125b expression. K. miR-125b inhibitor increased U266 cell counts. **P < 0.01 and ***P < 0.001, n=3.
MKNK2 is a target gene of miR-125b in MM
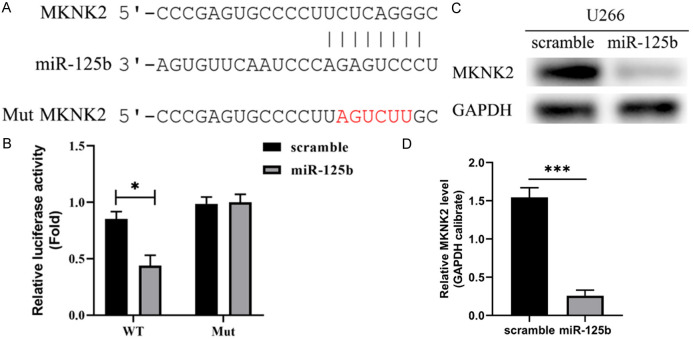
Using TargetScan, potential target genes for miR-125b were identified. A potential binding site for miR-125b was discovered in a conserved region of the 3’-UTR of the MKNK2 gene (Figure 3A). Both wild-type (WT) and mutant (MUT) MKNK2 luciferase reporter genes were co-transfected into U266 cells together with a miR-125b mimic or a scramble sequence. The luciferase activity of the WT reporter gene was lowered by miR-125b, but the Mut reporter gene remained unaffected (Figure 3B). Exogenous expression of miR-125b was shown to have a suppressive effect on MKNK2 expression in U266 cells (Figure 3C, 3D).
Figure 3.
Targeted relationship between miR-125b and MKNK2. A. The 3’-UTR of MKNK2 has a conserved area with putative miR-125b binding sites. B. Overexpression of miR-125b suppressed the luciferase activity of the WT reporter gene but not the Mut reporter gene. C. MKNK2 expression was down-regulated after exogenously transfecting miR-125b into U266 cells. D. Quantification of MKNK2 protein expression. *P < 0.05 and ***P < 0.001, n=3.
MKNK2 was involved in miR-125b-mediated cell behavior
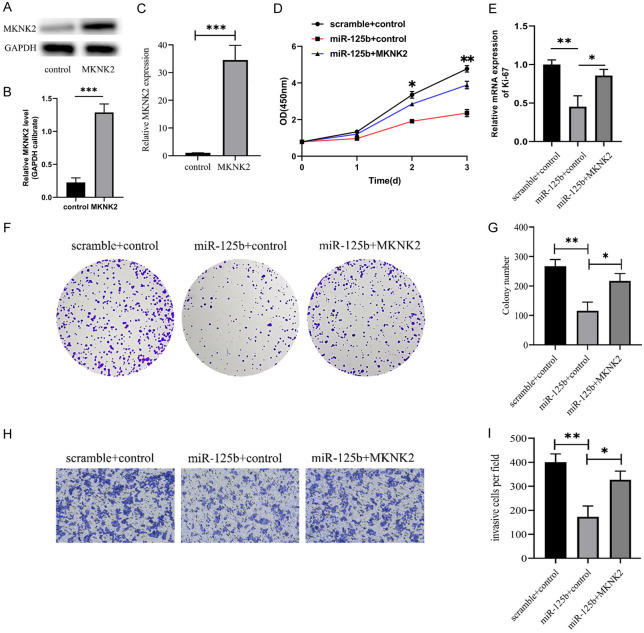
qRT-PCR and western blot analyses showed that transfection of MKNK2 plasmids up-regulated MKNK2 expression in U266 cells compared to the control vector (Figure 4A-C), and MKNK2 overexpression partly rescued the proliferation-suppressive effects induced by miR-125b mimics (Figure 4D). Additionally, MKNK2 overexpression improved miR-125b-induced ki-67 expression in U266 cells (Figure 4E). Reintroduction of MKNK2 dramatically reduced the suppression of cell colony formation in cells expressing miR-125b (Figure 4F, 4G). MKNK2 overexpression markedly mitigated the inhibition of cell invasion in miR-125b-expressing cells (Figure 4H, 4I).
Figure 4.
MKNK2 was engaged in the cellular activity regulated by miR-125b. A, B. MKNK2 protein expression detected by WB. C. Expression of MKNK2 mRNA was measured using qRT-PCR. D. The CCK-8 technique was used to detect the cell growth in each group. E. qRT-PCR was used to detect Ki-67 mRNA expression. F. Colony formation assays. G. The relative colony counts. H. Overexpression of MKNK2 significantly increased the invasion of cells expressing miR-125b (10×), Bar: 100 µm. I. The relative invasive cells were illustrated. *P < 0.05, **P < 0.01 and ***P < 0.001, n=3.
Discussion
Although it has been revealed that certain microRNAs are dysregulated in multiple myeloma (MM), the precise mechanism by which microRNAs influence carcinogenesis is not yet completely understood [42]. Our research found that the expression of miR-125b was decreased, while that of MKNK2 was up-regulated in MM. Additionally, there was an inverse relationship between the expression of MKNK2 and miR-125b in MM tissues. In MM cells, MKNK2 was proved to be a direct target gene of miR-125b. MiR-125b overexpression inhibited MM cell proliferation and invasion. Furthermore, we demonstrated that MKNK2 is involved in the proliferation and invasion of MM cells regulated by miR-125b. Our study demonstrates that miR-125b functions as a tumor suppressor in multiple myeloma by suppressing MKNK2 expression.
MicroRNAs are a type of gene that can either promote or restrict cell growth and are involved in a wide variety of cellular processes, including differentiation, proliferation, migration, and invasion [43]. In addition, an increasing body of evidence suggests that several miRNAs are responsible for regulating drug resistance [44]. MiR-125b is highly conserved throughout a wide range of organisms, including humans and nematodes, for example. It has been discovered that certain forms of cancer express miR-125b in an aberrant manner, which is strongly linked to resistance to treatment [45-47]. For instance, in anaplastic thyroid carcinoma, miR-125b inhibits cell migration and invasion by targeting PIK3CD [48], while in ovarian cancer, it suppresses epithelial-mesenchymal transition signaling to prevent tumor progression [49]. In addition, past research suggests that an aberrant expression of miR-125b is linked to chemotherapy resistance. Through the induction of DNA topoisomerase II deletion by doxorubicin and the regulation of miR-125b production by DNMT1, chemoresistance can be established in cells with positive anaplastic lymphoma kinase (ALK) [50]. In addition, miR-125b’s interaction with Foxp3 makes it possible to trigger autophagy in thyroid cancer [51]. However, very little is known about the function that miR-125b plays in MM. According to the results of our study, the expression of miR-125b was significantly decreased in MM tissue samples. In addition, overexpression of miR-125b inhibited MM cell proliferation, colony formation, and invasion, suggesting its tumor-suppressive role in MM.
Our molecular analysis revealed MKNK2 as a direct target of miR-125b in MM cells. MNKN2 is an oncogene that contributes to the progression of breast cancer as well as glioblastoma [52,53]. miR-125b is highly correlated with MKNK2 in MM and serves as an essential regulator of the proliferation of tumor cells. Recent studies have shown that MKNK2 is responsible for the increased drug resistance seen in prostate cancer patients [54]. According to our findings, MKNK2 is a direct target of miR-125b in MM. TargetScan identified a potential binding site for miR-125b in a conserved region of MKNK2’s 3’-UTR, critical for its protein activity. In luciferase reporter gene assay, ectopic production of miR-125b inhibited the luciferase activity of the WT reporter gene, while having no effect on the activity of the Mut reporter gene. Moreover, overexpression of miR-125b led to downregulation of MKNK2 expression in U266 cells. In addition to this, we discovered that the expression of MKNK2 was decreased in MM, inversely correlated with miR-125b expression. Additionally, it was found that MKNK2 is involved in the proliferation, colony formation, and invasion of MM cells regulated by miR-125b.
In exploring the role of miR-125b in MM, we found beneficial new targets for the treatment of MM and this can provide new directions for clinical treatment of MM. However, our study still has some limitations. This study is merely based on cellular experiments, without verifying its role in MM through animal experiments. Therefore, in-depth research on the precise mechanism by which miR-125b regulates the MKNK2 gene are required.
Conclusion
MiR-125b is down-regulated in multiple myeloma tissues. By reducing MKNK2 expression, miR-125b overexpression significantly impedes MM cell growth, colony formation, and invasion. Our findings indicate that miR-125b works as a tumor suppressor gene in multiple myeloma by inhibiting MKNK2 expression.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Kazandjian D. Multiple myeloma epidemiology and survival: a unique malignancy. Semin Oncol. 2016;43:676–681. doi: 10.1053/j.seminoncol.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95:548–567. doi: 10.1002/ajh.25791. [DOI] [PubMed] [Google Scholar]
- 4.Nijhof IS, van de Donk NWCJ, Zweegman S, Lokhorst HM. Current and new therapeutic strategies for relapsed and refractory multiple myeloma: an update. Drugs. 2018;78:19–37. doi: 10.1007/s40265-017-0841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Y, Mao X, Liu J, Fan H, Du C, Li Z, Yi S, Xu Y, Lv R, Liu W, Deng S, Sui W, Wang Q, Zou D, Wang J, Cheng T, Zhan F, Tai YT, Yuan C, Du X, Qiu L, Anderson KC, An G. The impact of response kinetics for multiple myeloma in the era of novel agents. Blood Adv. 2019;3:2895–2904. doi: 10.1182/bloodadvances.2019000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan Y, Guo J, Yang W, Goncalves C, Rzymski T, Dreas A, Żyłkiewicz E, Mikulski M, Brzózka K, Golas A, Kong Y, Ma M, Huang F, Huor B, Guo Q, da Silva SD, Torres J, Cai Y, Topisirovic I, Su J, Bijian K, Alaoui-Jamali MA, Huang S, Journe F, Ghanem GE, Miller WH Jr, Del Rincón SV. MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma. J Clin Invest. 2017;127:4179–4192. doi: 10.1172/JCI91258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzmil M, Huber RM, Hess D, Frank S, Hynx D, Moncayo G, Klein D, Merlo A, Hemmings BA. MNK1 pathway activity maintains protein synthesis in rapalog-treated gliomas. J Clin Invest. 2014;124:742–754. doi: 10.1172/JCI70198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Q, Yang C, Xiang Z, Huang G, Wu H, Chen T, Dou R, Song J, Han L, Song T, Wang S, Xiong B. LINC00924-induced fatty acid metabolic reprogramming facilitates gastric cancer peritoneal metastasis via hnRNPC-regulated alternative splicing of Mnk2. Cell Death Dis. 2022;13:987. doi: 10.1038/s41419-022-05436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie J, Shen K, Jones AT, Yang J, Tee AR, Shen MH, Yu M, Irani S, Wong D, Merrett JE, Lenchine RV, De Poi S, Jensen KB, Trim PJ, Snel MF, Kamei M, Martin SK, Fitter S, Tian S, Wang X, Butler LM, Zannettino ACW, Proud CG. Reciprocal signaling between mTORC1 and MNK2 controls cell growth and oncogenesis. Cell Mol Life Sci. 2021;78:249–270. doi: 10.1007/s00018-020-03491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartish M, Tong D, Pan Y, Wallerius M, Liu H, Ristau J, de Souza Ferreira S, Wallmann T, van Hoef V, Masvidal L, Kerzel T, Joly AL, Goncalves C, Preston SEJ, Ebrahimian T, Seitz C, Bergh J, Pietras K, Lehoux S, Naldini L, Andersson J, Squadrito ML, Del Rincón SV, Larsson O, Rolny C. MNK2 governs the macrophage antiinflammatory phenotype. Proc Natl Acad Sci U S A. 2020;117:27556–27565. doi: 10.1073/pnas.1920377117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiers S, Mwirigi J, Pradhan G, Kume M, Black B, Barragan-Iglesias P, Moy JK, Dussor G, Pancrazio JJ, Kroener S, Price TJ. Reversal of peripheral nerve injury-induced neuropathic pain and cognitive dysfunction via genetic and tomivosertib targeting of MNK. Neuropsychopharmacology. 2020;45:524–533. doi: 10.1038/s41386-019-0537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JRP, Alexandrou C, Skalka GL, Vlahov N, Pennel K, Officer L, Teodosio A, Kanellos G, Gay DM, May-Wilson S, Smith EM, Najumudeen AK, Gilroy K, Ridgway RA, Flanagan DJ, Smith RCL, McDonald L, MacKay C, Cheasty A, McArthur K, Stanway E, Leach JD, Jackstadt R, Waldron JA, Campbell AD, Vlachogiannis G, Valeri N, Haigis KM, Sonenberg N, Proud CG, Jones NP, Swarbrick ME, McKinnon HJ, Faller WJ, Le Quesne J, Edwards J, Willis AE, Bushell M, Sansom OJ. MNK inhibition sensitizes KRAS-mutant colorectal cancer to mTORC1 inhibition by reducing eIF4E phosphorylation and c-MYC expression. Cancer Discov. 2021;11:1228–1247. doi: 10.1158/2159-8290.CD-20-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brina D, Ponzoni A, Troiani M, Calì B, Pasquini E, Attanasio G, Mosole S, Mirenda M, D’Ambrosio M, Colucci M, Guccini I, Revandkar A, Alajati A, Tebaldi T, Donzel D, Lauria F, Parhizgari N, Valdata A, Maddalena M, Calcinotto A, Bolis M, Rinaldi A, Barry S, Rüschoff JH, Sabbadin M, Sumanasuriya S, Crespo M, Sharp A, Yuan W, Grinu M, Boyle A, Miller C, Trotman L, Delaleu N, Fassan M, Moch H, Viero G, de Bono J, Alimonti A. The Akt/mTOR and MNK/eIF4E pathways rewire the prostate cancer translatome to secrete HGF, SPP1 and BGN and recruit suppressive myeloid cells. Nat Cancer. 2023;4:1102–1121. doi: 10.1038/s43018-023-00594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun SY. mTOR-targeted cancer therapy: great target but disappointing clinical outcomes, why? Front Med. 2021;15:221–231. doi: 10.1007/s11684-020-0812-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, Wang C, Li M, Yang X. Targeting of MNK/eIF4E overcomes chemoresistance in cervical cancer. J Pharm Pharmacol. 2021;73:1418–1426. doi: 10.1093/jpp/rgab094. [DOI] [PubMed] [Google Scholar]
- 17.Xu W, Kannan S, Verma CS, Nacro K. Update on the development of MNK inhibitors as therapeutic agents. J Med Chem. 2022;65:983–1007. doi: 10.1021/acs.jmedchem.1c00368. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Chen JS, Li X, Bai X, Shi D. MNK, mTOR or eIF4E-selecting the best anti-tumor target for blocking translation initiation. Eur J Med Chem. 2023;260:115781. doi: 10.1016/j.ejmech.2023.115781. [DOI] [PubMed] [Google Scholar]
- 19.Mazewski C, Platanias LC. MNK proteins as therapeutic targets in leukemia. Onco Targets Ther. 2023;16:283–295. doi: 10.2147/OTT.S370874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed LM, Eltigani MM, Abdallah MH, Ghaboosh H, Bin Jardan YA, Yusuf O, Elsaman T, Mohamed MA, Alzain AA. Discovery of novel natural products as dual MNK/PIM inhibitors for acute myeloid leukemia treatment: pharmacophore modeling, molecular docking, and molecular dynamics studies. Front Chem. 2022;10:975191. doi: 10.3389/fchem.2022.975191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheper GC, Morrice NA, Kleijn M, Proud CG. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol Cell Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheper GC, Parra JL, Wilson M, Van Kollenburg B, Vertegaal AC, Han ZG, Proud CG. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol. 2003;23:5692–5705. doi: 10.1128/MCB.23.16.5692-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yazdani M, Beiki Z, Jahanian A. RNA secondary structured logic gates for profiling the microRNA cancer biomarkers. IET Nanobiotechnol. 2020;14:181–190. doi: 10.1049/iet-nbt.2019.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Z, An N, Lu BM, Zhou N, Yang SL, Zhang B, Li CY, Wang ZJ, Wang F, Wu CF, Bao JK. Identification of novel kinase inhibitors by targeting a kinase-related apoptotic protein-protein interaction network in HeLa cells. Cell Prolif. 2014;47:219–230. doi: 10.1111/cpr.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu D, Davis MP, Abreu-Goodger C, Wang W, Campos LS, Siede J, Vigorito E, Skarnes WC, Dunham I, Enright AJ, Liu P. MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming of mouse fibroblast cells to iPSCs. PLoS One. 2012;7:e40938. doi: 10.1371/journal.pone.0040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalman S, Garbett KA, Vereczkei A, Shelton RC, Korade Z, Mirnics K. Metabolic stress-induced microRNA and mRNA expression profiles of human fibroblasts. Exp Cell Res. 2014;320:343–353. doi: 10.1016/j.yexcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukerman P, Stern-Ginossar N, Gur C, Glasner A, Nachmani D, Bauman Y, Yamin R, Vitenshtein A, Stanietsky N, Bar-Mag T, Lankry D, Mandelboim O. MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012;72:5463–5472. doi: 10.1158/0008-5472.CAN-11-2671. [DOI] [PubMed] [Google Scholar]
- 30.Yu L, Ding GF, He C, Sun L, Jiang Y, Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661. doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Wang F, Xiao W, Sun J, Han D, Zhu Y. MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via suppressing Akt phosphorylation in glioblastoma. Tumour Biol. 2014;35:8653–8658. doi: 10.1007/s13277-014-2131-6. [DOI] [PubMed] [Google Scholar]
- 32.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu DD, Li XS, Meng XN, Yan J, Zong ZH. MicroRNA-873 mediates multidrug resistance in ovarian cancer cells by targeting ABCB1. Tumour Biol. 2016;37:10499–10506. doi: 10.1007/s13277-016-4944-y. [DOI] [PubMed] [Google Scholar]
- 34.Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL, Zhang MH, Liang HQ. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H, Zhang F, Lin X, Huang C, Zhang Y, Li Y, Lin J, Chen W, Lin X. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of beta-catenin signaling. Oncotarget. 2016;7:4647–4663. doi: 10.18632/oncotarget.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng B, Theng PY, Le MTN. Essential functions of miR-125b in cancer. Cell Prolif. 2021;54:e12913. doi: 10.1111/cpr.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie J, Jiang HC, Zhou YC, Jiang B, He WJ, Wang YF, Dong J. MiR-125b regulates the proliferation and metastasis of triple negative breast cancer cells via the Wnt/β-catenin pathway and EMT. Biosci Biotechnol Biochem. 2019;83:1062–1071. doi: 10.1080/09168451.2019.1584521. [DOI] [PubMed] [Google Scholar]
- 38.Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM, Araya JC, Roa JC. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr) 2016;39:23–33. doi: 10.1007/s13402-015-0247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY, Li S, Xue JS, Chen LL, Hu Y, Zhu H. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. 2019;234:6624–6632. doi: 10.1002/jcp.27403. [DOI] [PubMed] [Google Scholar]
- 40.Xiao T, Zhou Y, Li H, Xiong L, Wang J, Wang ZH, Liu LH. MiR-125b suppresses the carcinogenesis of osteosarcoma cells via the MAPK-STAT3 pathway. J Cell Biochem. 2019;120:2616–2626. doi: 10.1002/jcb.27568. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Liu Y, Huang WC, Zheng LC. MiR-125b-1-3p exerts antitumor functions in lung carcinoma cells by targeting S1PR1. Chin Med J (Engl) 2018;131:1909–1916. doi: 10.4103/0366-6999.238135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aass KR, Nedal TMV, Tryggestad SS, Haukas E, Slordahl TS, Waage A, Standal T, Mjelle R. Paired miRNA- and messenger RNA-sequencing identifies novel miRNA-mRNA interactions in multiple myeloma. Sci Rep. 2022;12:12147. doi: 10.1038/s41598-022-16448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alharbi M, Zuniga F, Elfeky O, Guanzon D, Lai A, Rice GE, Perrin L, Hooper J, Salomon C. The potential role of miRNAs and exosomes in chemotherapy in ovarian cancer. Endocr Relat Cancer. 2018;25:R663–R685. doi: 10.1530/ERC-18-0019. [DOI] [PubMed] [Google Scholar]
- 44.Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019;205:77–91. doi: 10.1016/j.trsl.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Zeng G, Jiang Y. The emerging roles of miR-125b in cancers. Cancer Manag Res. 2020;12:1079–1088. doi: 10.2147/CMAR.S232388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parayath NN, Gandham SK, Leslie F, Amiji MM. Improved anti-tumor efficacy of paclitaxel in combination with MicroRNA-125b-based tumor-associated macrophage repolarization in epithelial ovarian cancer. Cancer Lett. 2019;461:1–9. doi: 10.1016/j.canlet.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang HJ, Guo YQ, Tan G, Dong L, Cheng L, Li KJ, Wang ZY, Luo HF. miR-125b regulates side population in breast cancer and confers a chemoresistant phenotype. J Cell Biochem. 2013;114:2248–2257. doi: 10.1002/jcb.24574. [DOI] [PubMed] [Google Scholar]
- 48.Bu Q, You F, Pan G, Yuan Q, Cui T, Hao L, Zhang J. MiR-125b inhibits anaplastic thyroid cancer cell migration and invasion by targeting PIK3CD. Biomed Pharmacother. 2017;88:443–448. doi: 10.1016/j.biopha.2016.11.090. [DOI] [PubMed] [Google Scholar]
- 49.Ying X, Wei K, Lin Z, Cui Y, Ding J, Chen Y, Xu B. MicroRNA-125b suppresses ovarian cancer progression via suppression of the epithelial-mesenchymal transition pathway by targeting the SET protein. Cell Physiol Biochem. 2016;39:501–510. doi: 10.1159/000445642. [DOI] [PubMed] [Google Scholar]
- 50.Congras A, Caillet N, Torossian N, Quelen C, Daugrois C, Brousset P, Lamant L, Meggetto F, Hoareau-Aveilla C. Doxorubicin-induced loss of DNA topoisomerase II and DNMT1- dependent suppression of MiR-125b induces chemoresistance in ALK-positive cells. Oncotarget. 2018;9:14539–14551. doi: 10.18632/oncotarget.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, Wu J, Ren J, Vlantis AC, Li MY, Liu SYW, Ng EKW, Chan ABW, Luo DC, Liu Z, Guo W, Xue L, Ng SK, van Hasselt CA, Tong MCF, Chen GG. MicroRNA-125b interacts with Foxp3 to induce autophagy in thyroid cancer. Mol Ther. 2018;26:2295–2303. doi: 10.1016/j.ymthe.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogilevsky M, Shimshon O, Kumar S, Mogilevsky A, Keshet E, Yavin E, Heyd F, Karni R. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 2018;46:11396–11404. doi: 10.1093/nar/gky921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, Huang WL, Zeng YX, Shao JY. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–3562. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 54.Liu B, Sun Y, Tang M, Liang C, Huang CP, Niu Y, Wang Z, Chang C. The miR-361-3p increases enzalutamide (Enz) sensitivity via targeting the ARv7 and MKNK2 to better suppress the Enz-resistant prostate cancer. Cell Death Dis. 2020;11:807. doi: 10.1038/s41419-020-02932-w. [DOI] [PMC free article] [PubMed] [Google Scholar]