Abstract
Reliable and accurate laboratory assays to detect recent HIV-1 infection have potential as simple and practical methods of estimating HIV-1 incidence in cross-sectional surveys. This study describes validation of the limiting-antigen (LAg) avidity enzyme immunoassay (EIA) in a cross-sectional national survey, conducted in Swaziland, comparing it to prospective follow-up incidence. As part of the Swaziland HIV-1 Incidence Measurement Survey (SHIMS), 18,172 individuals underwent counseling and HIV rapid testing in a household-based, population survey conducted from December 2010 to June 2011. Plasma samples from HIV-positive persons were classified as recent infections using an incidence testing algorithm with LAg-Avidity EIA (normalized optical density ≤1.5) followed by viral load (VL ≥1,000 copies/mL). All HIV-seronegative samples were tested for acute HIV-1 infection by nucleic acid amplification test (NAAT) pooling. HIV-seronegative individuals who consented to follow-up were retested ~6 months later to detect observed HIV-1 seroconversion. HIV-1 incidence estimates based on LAg+VL and NAAT were calculated using assay-specific parameters and were compared with prospective incidence estimate. A total of 5,803 (31.9%) of 18,172 survey participants tested HIV seropositive; of these 5,683 (97.9%) were further tested with LAg+VL algorithm. The weighted annualized incidence from the longitudinal cohort study was 2.4% (95% confidence interval 2.0–2.7). Based on cross-sectional testing of HIV positives with LAg+VL algorithm, overall weighted annualized HIV-1 incidence was 2.5% (2.0–3.0), whereas NAAT-based incidence was of 2.6%. In addition, LAg-based incidence in men (1.8%; 1.2–2.5) and women (3.2%; 2.4–3.9) were similar to estimates based on observed incidence (men = 1.7%, women = 3.1%). Changes in HIV-1 incidence with age in men and women further validate plausibility of the algorithm. These results demonstrate that the LAg EIA, in a serial algorithm with VL, is a cost-effective tool to estimate HIV-1 incidence in cross-sectional surveys.
Keywords: HIV, LAg, viral load, incidence, SHIMS, Swaziland
Introduction
The past decade has seen a rapid scale-up of anti-retroviral (ARV) treatment and combination HIV-1 prevention programs, with a subsequent reduction in AIDS-related mortality and the number of new HIV-1 infections (UNAIDS).1,2 Further reductions in new infections are anticipated with the implementation of Test and Treat, the WHO recommendation to treat all HIV-positive people with ARVs regardless of CD4 cell count. To sustain these achievements, population-level data on the HIV-1 epidemic, mainly HIV-1 incidence and population VL as primary measures of program impact, will be increasingly important to assess progress, guide policy, and target resources to those who are still lacking access to HIV-1 prevention, care, and treatment services. The traditional gold standard method for estimating HIV-1 incidence relies on measuring observed HIV-1 seroconversion in a prospective cohort of HIV-negative individuals. Compared with cross-sectional studies, the follow-up visits inherent to prospective cohorts involve significantly higher costs, more time, and cohort biases related to recruitment and changes in behavior due to study enrollment. However, until recently, laboratory-based assays were inadequate in providing accurate cross-sectional incidence estimates due to: (1) the high level of misclassification among people with long-term infection and (2) the varying performance in different populations or subtypes.3-7 Recent developments in the accuracy of laboratory assays that distinguish between recent and long-term HIV-1 infections make it increasingly possible to estimate HIV-1 incidence in cross-sectional studies, promising less costly and timely results for decision making and resource allocation.4,8-18
We previously described the development, optimization, and characterization of the limiting antigen (“LAg”) avidity enzyme immunoassay (EIA) from Sedia BioSciences (Portland, OR) to detect recent HIV-1 infection in cross-sectional settings15,16; including the mean duration of recent infection (MDRI) of 130 days [95% confidence interval (CI) 118–142] that was optimized by balancing the length of MDRI while minimizing the proportion false recent (PFR).14-16 The LAg assay has been available from two commercial manufacturers since 2012 allowing global accessibility of the assay for the detection of recent HIV-1 infections.
The 2011 Swaziland HIV-1 Incidence Measurement Survey (SHIMS) was designed to measure the national HIV-1 incidence using prospective cohorts before and after expansion of national combination prevention programs. At the time, Swaziland had the highest known HIV-1 prevalence (31%, per 2007 Demographic Health Survey) and modeled 2011 HIV-1 annualized incidence of 2.9% (95% CI 2.7 – 3.2).19 As previously described, the baseline incidence was measured using a household-based, longitudinal cohort that observed seroconversions over the course of ~6 months among a nationally representative sample of men and women.20 SHIMS found a national HIV-1 prevalence of nearly 32% in Swaziland with observed annualized incidence of 2.4% (95% CI 2.0–2.7).20,21 With a directly observed cohort-based incidence estimate available for comparison, SHIMS offered an ideal opportunity for field validation of the LAg assay to estimate HIV-1 incidence from the cross-sectional survey that was conducted to identify eligible participants for the longitudinal cohort study. This study describes use of the LAg assay, in combination with VL results as part of an incidence testing algorithm, to classify recent and long-term infections and estimate the HIV-1 incidence estimate in the cross-sectional population. This field validation includes a comparison of incidence estimated from (1) the observed cohort, (2) the LAg+VL algorithm, and (3) nucleic acid amplification testing, which identified acute infections in the HIV-1 seronegative participants in the cross-sectional component of SHIMS.
Methods
SHIMS study design
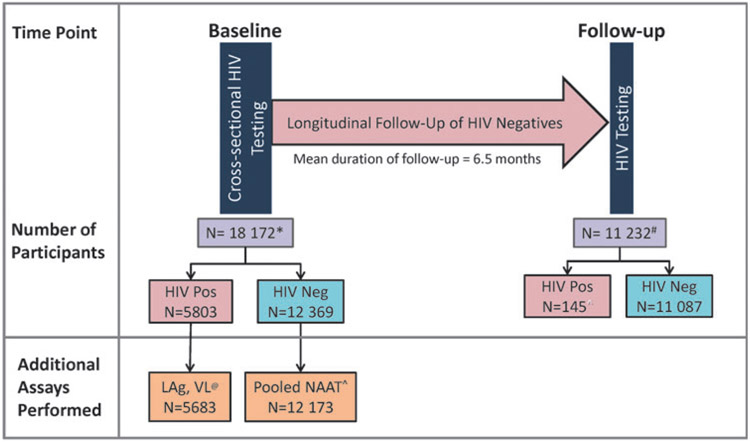
The SHIMS study design, sample size, eligibility criteria, and other survey details have been described elsewhere.20,21 Briefly, the cross-sectional component of SHIMS was conducted from December 2010 to June 2011 as a national survey of consenting adults, 18–49 years of age, who underwent counseling and HIV-1 rapid testing and provided demographic and HIV-1 risk behavior information in the household. A two-stage cluster sampling design was used to achieve a nationally representative sample of 14,891 households, of which 12,571 households participated. From these households, 18,172 individuals consented to participate in the initial cross-sectional study (Fig. 1), of which 5,803 were HIV-positive. Of the HIV-1-negative individuals, a total of 11,897 (96.2%) individuals consented to participate in the longitudinal cohort and 11,232 (94%) had a 6-month follow-up visit (mean follow-up time of 6.5 months) when they were retested for HIV-1 infection. A schematic of SHIMS study, with participant counts at each stage and tests performed, is shown in Figure 1.
FIG. 1.
A schematic representation of SHIMS study design, including various assays performed on study specimens. Number of individuals are indicated at each step. SHIMS, Swaziland HIV-1 Incidence Measurement Survey.
*131 Participants did not consent to additional testing and were excluded from LAg, VL and NAAT testing and laboratory database for analyses.
#Total participants who consented to participate and were retested in the cohort study.
@131 participants did not consent for LAg testing and were excluded; of the 5683 LAg-tested specimens, 5664 had VL results
^196 specimens were unavailable for pooled NAAT testing
Specimen collection and processing
Whole blood specimens were collected from all study participants at each household visit as previously described.22 Briefly, a phlebotomy-trained nurse collected whole blood from each participant in two separate EDTA tubes (Greiner Bio-One), one 9-mL and one 2-mL tube. Blood from the 2-mL tube was used for household HIV-1 rapid testing and for external quality assessment testing at the National Reference Laboratory (NRL) in Mbabane, whereas blood from the 9-mL tube was processed into plasma, aliquoted into 1.2 mL aliquots, and stored at −70°C at the NRL for further testing. Specimen integrity was maintained throughout the collection, transport, and processing using cold-chain methods to ensure specimen storage within 24 h of collection and specimens that were unable to meet these standards were either discarded or excluded from molecular testing.
HIV-1 testing algorithm
For SHIMS, HIV-1 diagnosis was determined using a serial rapid testing algorithm approved by the Swaziland Ministry of Health for use in this study as previously described.22 All HIV-1 rapid testing was performed in the household at the time of blood collection according to the manufacturer’s instructions. Teams participating in the household survey were well trained to perform the HIV-1 tests with strict adherence to protocol and quality assurance practices. Determine HIV-1/2 Ag/Ab Combo rapid test (Inverness Medical) was used as the screening test and all Determine Combo Ab+ samples were further tested using Uni-Gold Recombigen HIV-1/2 rapid test (Trinity Biotech). Those testing Ab+ on Uni-Gold rapid test were diagnosed as HIV positive as per national algorithm. Clients testing positive were provided posttest counseling in the household and referred to nearest clinic for care. Specimens identified as only Ag+ infection (Ag+/Ab−) by Determine Combo had a confirmatory VL test, whereas negative specimens (Ag−/Ab−) had additional pooled NAAT performed to detect acute infections22 (Fig. 1). Specimens with discordant Ab results between the Determine Combo and Uni-Gold rapid tests were resolved using a two HIV-1 EIA testing algorithm with Bio-Rad Genscreen HIV-1/2 V2 (Hercules, CA) as the screening EIA and Vironostika HIV-1 Uni-Form II Ag/Ab (bioMérieux) as the confirmatory EIA in accordance with Swaziland’s National HIV-1 testing algorithm. Specimens with low signal to cutoff values were further tested by HIV-1 western blot. All testing was performed at the National Institute of Communicable Disease (NICD), National Health Laboratory Services in Johannesburg, South Africa according to the manufacturer’s instructions.
Incidence testing
The LAg-Avidity EIA was performed according to the manufacturer’s instructions (Sedia BioSciences) and have been described in detailed elsewhere.14,15 For LAg, the raw optical density (OD) for each specimen was normalized using Calibration (CAL) OD on each plate as a ratio, such that normalized OD (ODn) = OD of specimen/median (OD of CAL). Plates were validated using acceptable values of OD and ODn for each control and CAL as determined for the kit. If one or more of the controls fell outside of the acceptable ranges defined in the kit insert, the run was rejected. Specimens were then retested and ODn values from only valid runs were used for analysis. To assist with data management and analysis, an Excel-based data management tool for each assay was used to autovalidate each plate, calculate ODn, and classify specimens as recent or long-term infections based on assay cutoff (1.5 ODn for LAg). As required, only confirmed antibody-positive specimens were subjected to testing with LAg EIA.
Molecular testing
Acute infections were detected among HIV-1-negative specimens by pooling 10 plasma samples (120 μL plasma/sample) per pool and testing for viral RNA using the Roche COBAS® AmpliPrep/COBAS TaqMan® System (CAP/CTM) HIV-1 Test, version 2.0 assay. The limit of detection (LOD) for the pooled testing was 200 copies/mL. Positive pools were then deconstructed and individual samples, diluted 1:2 with confirmed HIV-1 NAAT negative human plasma, were tested to identify the HIV-1 NAAT-positive sample with a LOD of 40 copies/mL. VL quantification was performed on all Determine Combo Ag-positive specimens and HIV-positive specimens using 1.2 mL of undiluted plasma on the CAP/CTM platform and the COBAS AmpliPrep/COBAS TaqMan HIV-1 Test, version 2.0 assay according to the manufacturer’s instructions. The LOD for the assay was 20 copies/mL.
HIV-1 incidence estimates using laboratory-based results and statistical considerations
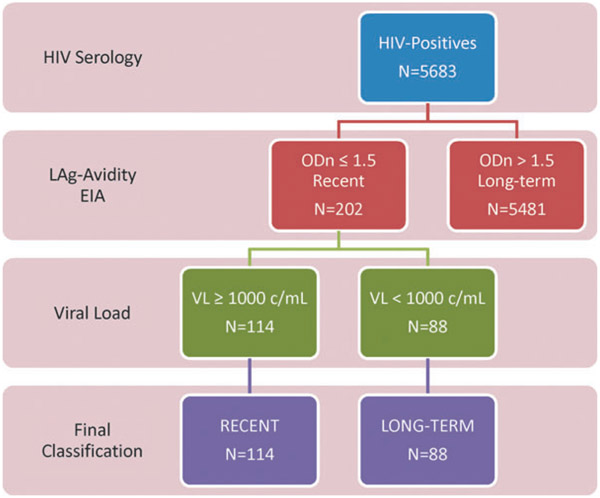
To improve accuracy of recent infection detection, LAg+VL algorithm (Fig. 2) was used to mitigate misclassification due to suppressed VL (<1,000 copies/mL) among elite controllers and those on ART. HIV-1 incidence estimates were calculated as annual instantaneous rate () for the LAg-Avidity and NAAT results using the UNAIDS/WHO-recommended incidence formula23 based on the Welte et al.24: , where R is the number of recent infections classified by the LAg+VL algorithm, is PFR, P is the number of HIV-1-positive people in the survey, is the MDRI, and N is the total number of HIV-1-negative people in the survey. Final input values for R, P, and were based on survey weighting to account for sampling methods and differences in nonresponse, whereas the MDRI for LAg+VL algorithm was 130 days (95% CI 118–142).14 The 95% CIs for incidence estimates were computed using a delta method approximation that included the error associated with the MDRI, which was assumed to be normally distributed. Since VL (>1,000 copies/mL) was included in the algorithm to correctly identify recent infections and remove misclassified cases, a PFR value of 0.0 was used to calculate incidence. Z-scores were calculated to compare similarity or differences between incidence estimates.
FIG. 2.
HIV incidence testing algorithm using LAg and viral load testing to determine recent infection, including SHIMS testing results at each step. LAg, limiting-antigen.
The MDRI for NAAT for detection of acute infection was calculated to be 15 days based on data from seven commercial seroconversion panels (Boston Biomedical, Inc., Boston, MA; data not shown). The panel data included preseroconversion period with NAAT results; 95% CI was not calculated due to lack of adequate data.
Prospective follow-up incidence
Prospectively observed incidence from the longitudinal cohort has been previously reported and was based on results of the number of individuals who seroconverted over the course of the cohort study.20 The results were weighted to adjust for sampling methods and differences in nonresponse to achieve nationally representative results. For comparison purposes, the reported follow-up instantaneous incidence estimate () was converted to an annualized incidence () value (shown in Table 1) using the following formula: .
Table 1.
HIV-1 Prevalence and Incidence Estimates Using Different Methods for Overall Population and by Gender
| Group | Prevalencea (%), 95% CI | Estimation of incidence by different method (%) | |||
|---|---|---|---|---|---|
| Observed (95% CI) | LAg+VL (95% CI) | Z-Statb | NAATc | ||
| Overall | 31.9 (31.0–32.9) | 2.4 (2.0–2.7) | 2.5 (2.0–3.0) | 0.41 | 2.6 |
| Men | 24.0 (22.8–25.3) | 1.6 (1.3–2.1) | 1.8 (1.2–2.5) | 0.64 | 1.3 |
| Women | 38.6 (37.5–39.7) | 3.1 (2.6–3.7) | 3.2 (2.4–3.9) | 0.23 | 3.6 |
Based on incidence dataset excluding 131 HIV-positive participants who did not consent for further testing.
Z statistics comparing observed incidence with LAg+VL algorithm.
95% CI not calculated due to uncertainty of MDRI.
95% VI, 95% confidence interval; LAg, limiting-antigen; MDRI, mean duration of recent infection; NAAT, nucleic acid amplification test; VL, viral load.
Ethics considerations
All study participants provided written informed consent before the collection of data and blood samples. The SHIMS protocol was approved by the Swaziland Ethics Committee and the Institutional Review Boards (IRBs) of Columbia University and the U.S. Centers for Disease Control and Prevention (CDC) before study initiation. All clinically relevant results were returned to clients or to the nearest clinic of their choice (such as VL) as soon as they were available.20,21 Recent infection testing using the LAg assay was for incidence surveillance only; therefore, these results were not returned to participants.
Results
Laboratory dataset
Previously reported SHIMS prevalence and observed incidence were based on a total of 18,172 participants that consented to household-based HIV-1 rapid testing. However, 131 participants did not consent for further laboratory testing, including LAg, VL, or NAAT and have been excluded from the dataset used for all analyses herein, including prevalence reported in Table 1. With the 131 participants excluded, a total of 5,730 participants were HIV-1 positive and 12,311 were HIV-1 negative for a total of 18,041 participants in laboratory dataset. Of the 5,730 HIV-1-positive results, 5,683 (99.2%) samples were tested on the LAg assay (Fig. 2), whereas 5,680 (99.1%) samples had VL results. A total of 5,664 (98.8%) samples had both LAg and VL results available for incidence analyses (Fig. 1).
Prevalence and prospectively observed HIV-1 incidence estimates
Using the SHIMS laboratory dataset, the resulting overall HIV-1 prevalence was of 31.9% (95% CI 31.0–32.9) in adults 18–49 years of age in Swaziland (Table 1), statistically equivalent to 32% as previously reported.21 Significant differences in prevalence were observed by gender where the prevalence for men was 24.0% (95% CI 22.8–25.3) and 38.6% (95% CI 37.5–39.7) for women. The observed incidence from the longitudinal cohort for the overall population was previously reported as 2.4 per 100 person years (95% CI 2.1–2.8) or 2.4% (95% CI 2.0–2.7) when converted to annual incidence. The annualized incidence for men was 1.6% (95% CI 1.3–2.1) and 3.1% (95% CI 2.6–3.7) for women (Table 1).
Incidence estimates using an incidence testing algorithm with LAg+VL
Applying the LAg+VL incidence testing algorithm (Fig. 2), 202 of the 5,683 HIV-positive specimens tested by the LAg EIA had ODn ≤1.5, and were classified as recent by the LAg assay. VL testing of these specimens indicated that 114 had VL ≥1,000 copies/mL and therefore given a final classification of recent HIV-1 infection. The other 88 samples had VL <1,000 copies and were reclassified as long-term infections. Of 114 samples with final recent classification, 35 were men and 79 were women. Applying these results to the incidence formula using the parameters described (see Methods section), incidence estimate was 2.5% (95% CI 2.0–3.0) for the overall population, 1.8% (95% CI 1.2–2.5) for men, and 3.2% (95% CI 2.4–3.9) for women (Table 1). The resulting incidence estimates do not differ significantly from the directly observed estimate (Z = 0.41, 0.64, 0.23, respectively).
Incidence estimation using NAAT
Incidence was also estimated using the NAAT-positive samples from the HIV-1 seronegative individuals at baseline. There were 13 acute infections identified (defined as NAAT positive and HIV seronegative), 3 were men and 10 were women, and resulted in an incidence estimate of 2.6% overall, 1.3% for men and 3.6% for women, using an MDRI of 15 days (Table 1). The 95% CIs were not calculated due to absence of sufficient available data around the MDRI (see Methods section). The overall estimate and estimates for men and women were very similar to the observed and LAg+VL-based HIV-1 incidence estimates (Table 1). Individuals with NAAT-positive results had 6-month follow-up visits and 12 of the 13 were confirmed for seroconversion, while 1 participant was lost to follow-up.22
Analysis of LAg-based HIV-1 incidence by gender and age in the context of HIV-1 prevalence
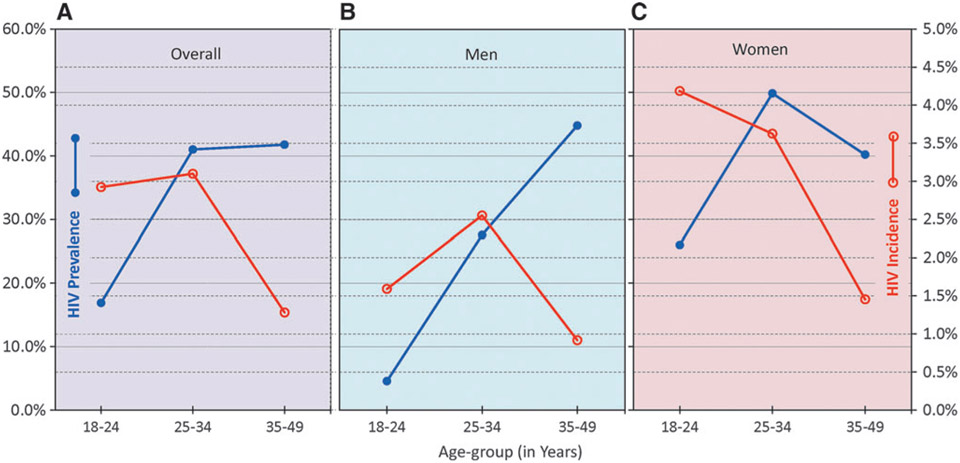
HIV-1 prevalence and incidence data were further analyzed by age groups. Overall, the prevalence was ~18% in the younger age group (18–24 years) which increased to more than 40% among adults ≥25 years of age (Fig. 3A). Among the younger age group, incidence was very high at 2.91%, which peaked among 25–34 years of age (3.1%) and then declined among older age group (1.28%). When further analyzed by gender (Fig. 3B and C), there were major differences among men and women. Men showed a steep increase in HIV-1 prevalence from 4.5% to almost 45% with age, which was accompanied by an increase in HIV-1 incidence from 1.6% (18–24 years) to 2.5% in men 25–34 years of age but then declined among older men (0.9%). Youngest women had the highest HIV-1 incidence (4.2%), which declined to about 3.6% in 25–34 years of age and about 1.3% among ≥35 years of age. High incidence was reflected in high prevalence in younger age group (26%), reaching almost to 50% among 25–34 years of age, followed by a decline among the oldest age group (40%).
FIG. 3.
HIV prevalence and incidence by age in Swaziland, overall (A), among men (B) and women (C). HIV-1 incidence was estimated using LAg+VL algorithm. Prevalence (closed blue circles), HIV-1 incidence (open red circles). VL, viral load.
Geolocation of recent infections
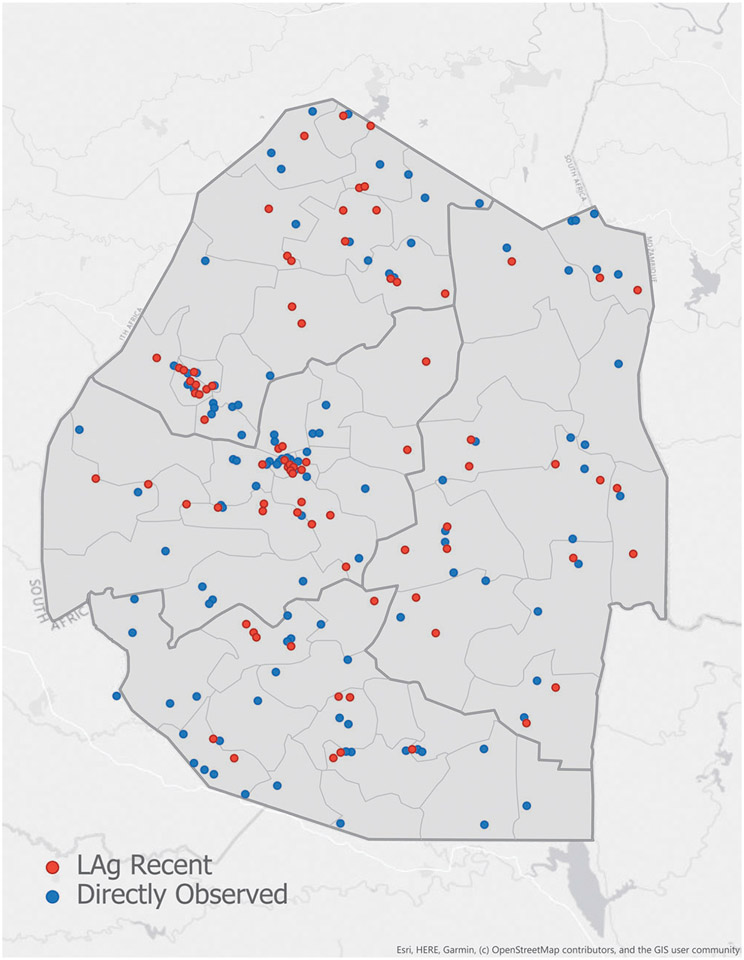
Based on collection of specimens from 575 enumeration areas, LAg-based recent infections and observed new infections were added to the map of Swaziland to examine the distributions and clusters of recent infections, if any. Both methods demonstrated two overlapping clusters coinciding with the location of Swaziland’s two main cities, Mbabane and Manzini (Fig. 4).
FIG. 4.
Geolocation of recent HIV infections, as detected by the LAg+VL algorithm (red dots) or by cohort follow-up seroconversion (blue dots), in 575 enumeration areas are shown on the map of Swaziland.
Discussion
This study describes the first field validation of the LAg assay as a laboratory method of estimating HIV-1 incidence in a cross-sectional survey by comparing the LAg-based estimate with observed HIV-1 seroconversion in SHIMS, a population-based, prospective cohort study conducted in Swaziland. The annualized HIV-1 incidence estimates for Swaziland in 2010–2011 based on LAg EIA (2.5%), the directly observed cohort (2.4%), NAAT (2.6%), and the 2010 UNAIDS estimate (2.6%) using the Spectrum model25 are all very similar. The convergence of HIV-1 incidence estimates in this population from multiple methods is remarkable and validates the use of LAg EIA, in combination with VL to identify misclassified individuals on ART and elite controllers and to calculate incidence in a cross-sectional survey.
The NAAT method produced comparable incidence estimates (2.6% overall, Table 1) to other methods, suggesting that in a high incidence setting and with a nationally representative large sample size, it can produce accurate incidence as others have noted.26 However, using NAAT for this purpose has its drawbacks: (1) the need to test large number of HIV-1 seronegatives to find few acute infections, (2) the high cost and complexity of performing NAAT, (3) the need to have highly skilled laboratory staff to perform the testing, (4) the potential for cross-contamination during the pooling and deconstruction of pools during NAAT testing, and (5) the inability to perform further risk factor analysis with few incident cases. In this population, with one of the highest national rates of HIV-1 incidence, only about 1 NAAT-positive case per 1,000 seronegatives tested was found, indicating that most national surveys will have even lower probability in finding such acute cases.
The comparability of incidence estimates based on the LAg+VL algorithm and observed incidence for the overall population and by gender (Table 1), along with the trends of incidence by age (Fig. 3) for both male and female, further validates use of this approach, demonstrating that the LAg+VL can be used in cross-sectional surveys to obtain reliable estimates of HIV-1 incidence. As expected, prevalence increases with age for both genders but incidence profile is unique but plausible for male and female. Highest incidence among women was in the youngest age group compared with men, where highest incidence was among middle age men; this is consistent with risk behavior and transmission dynamics between the two genders.20
Our results also demonstrate that the additional correction or adjustment with a PFR value is not needed. Although application of a residual PFR adjustment would reduce the overall number of recently classified individuals by a number that would be misclassified, this reduction is not targeted to specific individuals who may have been misclassified and thus does not improve positive predictive value (PPV) at the individual level which is important for further analysis. In contrast, the LAg+VL algorithm increases the overall PPV of detecting recent infection by removal of the individual most likely to be misclassified (elite controllers and those on ARV). Beyond a point estimate of incidence, additional risk factor analysis is important for public health officials and donor programs to identify the hot spots and understand drivers of transmission in these settings to make wise, targeted prevention investments to reduce the number of new infections.
This algorithm has the advantage of reducing or eliminating the need to determine a local, population-specific PFR to correct for misclassification as has been previously recommended by Consortium for Evaluation and Performance of HIV Incidence Assays (CEPHIA) and WHO/UNAIDS, 23 of which is not practical for each survey. Determination of a local PFR involves testing a large number of HIV-1-infected individuals with documented dates of infection greater than one or 2 years and determining the proportion of these individuals that are misclassified as recent infection by the incidence assay(s). PFR value for an assay is not constant in a given population but will vary over time, place, varying ART coverage, duration of ART, and accumulation of elite controllers.27 Therefore, the best strategy to remove misclassified individuals is to include meaningful biomarkers as part of the algorithm that will also identify misclassified elite controllers. Therefore, use of VL in an algorithm was considered as the best approach for correcting for misclassification and PFR was set at 0%. Data from a recent report from CEPHIA further confirm that LAg EIA, in combination with VL, had PFR of 0% and had the best performance among all the assays evaluated.28
The LAg assay has been applied in South Africa in 2012 and Kenya in 2012/2013 in large nationally representative household surveys to estimate HIV-1 incidence.29,30 Both studies used an incidence algorithm that also included ARV testing in addition to VL. For both studies, final recent classification required an ODn ≤1.5 on the LAg assay, negative for ART exposure (by ARV testing or self-report) and VL >1,000 copies/mL. Using these criteria, the national incidence estimates were 1.72% (95% CI 1.38–2.06) for adults 15–49 years of age in South Africa and 0.5% (95% CI 0.1–0.9) for adults 15–64 years of age in Kenya. Although this incidence algorithm includes additional information on ART exposure for recent classification, ARV testing may not be feasible in most countries as the technology is expensive and currently limited to a few laboratories worldwide that perform these analytical techniques. The validation of our LAg-based estimate of HIV-1 incidence in Swaziland with both seroconversion and NAAT-based estimates indicates that including the ARV testing in the algorithm may not be necessary to achieve an accurate incidence estimate. ARV testing was not done in this sample set, although self-reported ARV was available. Due to low coverage/duration of ARV and low VL suppression (~35%) in Swaziland in 2011, we do not see any change in incidence when self-reported ARV was factored in final incidence estimate. However, we are continuing to examine this issue in additional surveys in populations where ART coverage is high and Test and Start is widely implemented.
Use of LAg+VL algorithm was further validated by CEPHIA where they evaluated performance of multiple incidence assays.31 Their findings demonstrated that antibody-based assays do misclassify a proportion of people on treatment, which increases with duration of treatment. Therefore, the authors recommended including VL in the testing algorithm to improve the predictive value of detecting recent HIV-1 infection. Additional analysis suggested that when combined with VL in an algorithm, the LAg PFR was reduced to nearly zero.31 In spite of these data, the authors stated that a residual PFR of a finite number (not 0%) should be used when calculating incidence. Use of a residual PFR (e.g., 0.3%), although will lower the incidence, does not necessarily improve the accuracy. Additional complications result from occasional negative incidence estimates when further subcategory analysis is performed (data not shown) indicating that this same correction does not apply to all subcategories. This suggests that PFR should be replaced by the addition of appropriate biomarkers (e.g., VL, ARV detection) in the algorithm that help reduce misclassification close to zero.
Konikoff et al. have suggested two different multiassay algorithms, or MAAs, that incorporate the LAg assay.32 The first MAA combines the Bio-Rad Avidity assay at a cutoff of 40%, followed by the LAg assay with a cutoff ODn 2.8. The second MAA combines CD4 at cutoff of 50 cells/mm3, Bio-Rad Avidity at cutoff 85%, LAg at cutoff ODn 2.9, and VL at cutoff 400 copies/mL in a four-test MAA. These MAAs were derived from a combination of >500,000 possible MAAs using various cutoffs for each assay. The LAg cutoffs used were well outside the dynamic ranges of the assays and therefore are not appropriate. Moreover, randomly combining multiple assays in a large combination of algorithms can lead to artifacts that may not have biological relevance. Of note, Konikoff et al. show that using the incidence algorithm of LAg and VL, as we have used in this article, yields incidence estimates similar to estimates based on directly observed incidence estimates in three different cohorts from the United States.
Furthermore, our results (data not shown) and results from others28 show that low CD4 is not associated with LAg PFR; therefore, inclusion of CD4 will not improve accuracy of recent infection testing by the LAg assay. We believe the simplicity of our incidence testing algorithm using only LAg+VL testing will allow it to be widely applicable, easy to implement, cost effective, and most importantly, produce an accurate incidence estimate. It is to be noted that follow-up of about 12,000 seronegative individuals in SHIMS to derive cohort-based incidence was not only labor intensive, it was a very expensive exercise costing >$5 million. In comparison, cost of LAg-EIA reagents followed by VL testing on LAg-recent cases using cross-sectional specimens cost us less than $100,000, while reducing the time and eliminating the bias of recruitment and follow-up. If ARV testing is warranted, it should be used as a last step in serial algorithm, which will significantly reduce the numbers requiring ARV detection.
Furthermore, similar geolocation of recent infection clusters, indicate that LAg-based method can replace expensive cohort follow-up to get the same information (Fig. 4). Considering that a large number of samples were collected from these areas, this is not unexpected; however, this colocalization of hot-spots of transmission further validates LAg-based recent infection detection. As we strive to reduce incidence and interrupt further transmission, detection of recent infections in real time, with geolocation of recently infected individuals, may provide critical information for targeted HIV-1 prevention.
Limitation of this study include 472 (3.8%) of 12,389 seronegative individuals who did not consent to retesting as part of cohort prospective follow-up for incidence measurement, and of those consenting (11,897), 5.6% (665) did not retest. However, it is worthwhile noting that overall almost 92% of seronegative individuals retested for potential seroconversion.
In summary, we have validated the use of the LAg assay to estimate HIV-1 incidence in this high prevalence, high incidence setting in Swaziland. The LAg-based incidence estimates were highly comparable to the observed incidence derived from the longitudinal follow-up cohort. The use of the LAg+VL incidence algorithm provides timely, cost-effective, and accurate cross-sectional incidence estimates without the need for a lengthy, expensive prospective longitudinal cohort study that has the potential for cohort-based biases. The LAg-based incidence algorithm also provides important information on accurate demographic and risk behaviors associated with new HIV-1 infections that can be translated into targeted prevention to reduce the rate of HIV-1 transmission. With Swaziland having the highest prevalence and incidence in the world, it is a reminder that there is still a lot of work to do to achieve an AIDS-free generation, but through a scale-up of combinations of proven tools, we can not only achieve those goals, but also measure them with accuracy.
Acknowledgments
The authors would like to acknowledge and thank all the participants of SHIMS and the hard working SHIMS field staff, the laboratory staff at the NRL in Mbabane, Swaziland, and the invaluable contribution of the laboratory staff from the International Laboratory Branch in the Division of Global HIV & TB at CDC Atlanta who provided training and other technical support to the study. They would also like to thank the Ministry of Health for their support and acknowledge the efforts of many individuals from ICAP, Maromi/EpiCentre, and SCHARP who contributed to the success of the study. This research was supported by the PEPFAR through the CDC.
Footnotes
Disclaimer
President’s Emergency Plan for AIDS Relief (PEP-FAR)/CDC authorship disclaimer: the findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the funding agencies.
Author Disclosure Statement
As an inventor of LAg-Avidity EIA, BSP receives a portion of royalties from sale of LAg-Avidity EIA as per policy of the U.S. government. No other competing financial interests exist.
References
- 1.Brown JL, Sales JM, DiClemente RJ. Combination HIV prevention interventions: the potential of integrated behavioral and biomedical approaches. Curr HIV/AIDS Rep 2014;11:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson SJ, Cherutich P, Kilonzo N, et al. : Maximising the effect of combination HIV prevention through prioritisation of the people and places in greatest need: A modelling study. Lancet 2014;384:249–256. [DOI] [PubMed] [Google Scholar]
- 3.Parekh BS, Hu DJ, Vanichseni S, et al. : Evaluation of a sensitive/less-sensitive testing algorithm using the 3A11-LS assay for detecting recent HIV seroconversion, among individuals with HIV-1 subtype B or E infection in Thailand. AIDS Res Hum Retroviruses 2001;17:453–458. [DOI] [PubMed] [Google Scholar]
- 4.McDougal JS, Pilcher CD, Parekh BS, et al. : Surveillance for HIV-1 incidence using tests for recent infection in resource-constrained countries. AIDS 2005;19(Suppl 2):S25–S30. [DOI] [PubMed] [Google Scholar]
- 5.UNAIDS: UNAIDS Reference Group on estimates, modelling and projections—Statement on the use of the BED assay for the estimation of HIV-1 incidence for surveillance or epidemic monitoring. Wkly Epidemiol Rec 2006;81:40. [PubMed] [Google Scholar]
- 6.UNAIDS: WHO/UNAIDS technical update on HIV incidence assays for surveillance and epidemic monitoring. Available at www.who.int/diagnostics_laboratory/links/hiv_incidence_assay/en (2013), accessed May 30, 2014.
- 7.Young CL, Hu DJ, Byers R, et al. : Evaluation of a sensitive/less sensitive testing algorithm using the bioMerieux Vironostika-LS assay for detecting recent HIV-1 subtype B’ or E infection in Thailand. AIDS Res Hum Retroviruses 2003;19:481–486. [DOI] [PubMed] [Google Scholar]
- 8.Parekh B, Wei X, Dobbs T, Kuehl D, Hu D, Nkengasong J: Development of a limiting-antigen avidity-based enzyme immunoassay using a chimeric recombinant gp41 protein to detect recent HIV-1 infection. In: 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, February 25–28, 2007. [Google Scholar]
- 9.Parekh BS, McDougal JS: Application of laboratory methods for estimation of HIV-1 incidence. Indian J Med Res 2005;121:510–518. [PubMed] [Google Scholar]
- 10.Rawal BD, Degula A, Lebedeva L, et al. : Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J Acquir Immune Defic Syndr 2003;33:349–355. [DOI] [PubMed] [Google Scholar]
- 11.Parekh BS, Kennedy MS, Dobbs T, et al. : Quantitative detection of increasing HIV type 1 antibodies after seroconversion: A simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses 2002;18:295–307. [DOI] [PubMed] [Google Scholar]
- 12.Parekh BS, Pau CP, Kennedy MS, Dobbs TL, McDougal JS: Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res Hum Retroviruses 2001;17:137–146. [DOI] [PubMed] [Google Scholar]
- 13.Janssen RS, Satten GA, Stramer SL, et al. : New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. [Erratum appears in JAMA 1999 May 26;281(20):1893.] JAMA 1998;280:42–48. [DOI] [PubMed] [Google Scholar]
- 14.Duong YT, Kassanjee R, Welte A, et al. : Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 2015;10:e0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duong YT, Qiu M, De AK, et al. : Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: Potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012;7:e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X, Liu X, Dobbs T, et al. : Development of two avidity based assays to detect recent HIV-1 seroconversion using a multi-subtype gp41 recombinant protein. AIDS Res Hum Retroviruses 2010;26:61–71. [DOI] [PubMed] [Google Scholar]
- 17.Guy R, Gold J, Calleja JMG, et al. : Accuracy of serological assays for detection of recent infection with HIV and estimation of population incidence: A systematic review. Lancet Infect Dis 2009;9:747–759. [DOI] [PubMed] [Google Scholar]
- 18.Hargrove JW, Humphrey JH, Mutasa K, et al. : Improved HIV-1 incidence estimates using the BED capture enzyme immunoassay. AIDS 2008;22:511–518. [DOI] [PubMed] [Google Scholar]
- 19.http://aidsinfo.unaids.org. AIDSInfo. accessed July 1, 2013.
- 20.Justman J, Reed JB, Bicego G, et al. : Swaziland HIV Incidence Measurement Survey (SHIMS): A prospective national cohort study. Lancet HIV 2017;4:e83–e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bicego GT, Nkambule R, Peterson I, et al. : Recent patterns in population-based HIV prevalence in Swaziland. PLoS One 2013;8:e77101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duong YT, Mavengere Y, Patel H, et al. : Poor performance of the determine HIV-1/2 Ag/Ab combo fourth-generation rapid test for detection of acute infections in a National Household Survey in Swaziland. J Clin Microbiol 2014;52:3743–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UNAIDS/WHO Working Group on Global HIV/AIDS and STI Surveillance: When and how to use assays for recent infection to estimate HIV incidence at a population level. France: WHO Press; 2011. [Google Scholar]
- 24.Welte A, McWalter TA, Barnighausen T: A simplified formula for inferring HIV incidence from cross-sectional surveys using a test for recent infection [comment]. AIDS Res Hum Retroviruses 2009;25:125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaziland Country Report on Monitoring the Political Declaration on HIV and AIDS. Swaziland: Ministry of Health; 2014. [Google Scholar]
- 26.Cohen MS, Gay CL, Busch MP, Hecht FM; The detection of acute HIV infection. J Infect Dis 2010;202(Suppl 2):S270–S277. [DOI] [PubMed] [Google Scholar]
- 27.Hallett TB, Ghys P, Barnighausen T, Yan P, Garnett GP: Errors in “BED”-derived estimates of HIV incidence will vary by place, time and age. PLoS One 2009;4:e5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassanjee R, Pilcher CD, Keating SM, et al. : Independent assessment of candidate HIV incidence assays on specimens in the CEPHIA repository. AIDS 2014;28:2439–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(Kenya) Ministry of Health: Kenya AIDS Indicator Survey 2012 final report. Ministry of Health, National AIDS Control Program, Nairobi, 2014. pp. 43–46. [Google Scholar]
- 30.Shisana O, Rehle T, Simbayi LC, et al. : South African National HIV Prevalence, Incidence and Behaviour Survey, 2012. Cape Town: 2014. [Google Scholar]
- 31.Kassanjee R, Pilcher CD, Busch MP, et al. : Viral load criteria and threshold optimization to improve HIV incidence assay characteristics. AIDS 2016;30:2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konikoff J, Brookmeyer R, Longosz AF, et al. : Performance of a limiting-antigen avidity enzyme immunoassay for cross-sectional estimation of HIV incidence in the United States. PLoS One 2013;8:e82772. [DOI] [PMC free article] [PubMed] [Google Scholar]