Abstract
For many applications, specificity of gene expression by recombinant retroviral vectors is necessary. We wished to obtain transcriptional targeting in endothelial cells as part of an antivasculature approach to cancer treatment and have achieved specificity by using the promoter for human prepro-endothelin-1. In particular, we have inserted this heterologous promoter within the 3′ long terminal repeat (LTR), replacing all viral upstream transcriptional regulatory sequences, to generate a hybrid LTR with precise fusion at the TATA box for initiation of transcription at the viral start site. Reverse transcription and integration resulted in duplication of this hybrid promoter in the 5′ LTR of the provirus for transcription of the internal transgene. An important feature of our vectors is the absence of a selectable marker gene or additional promoters to avoid potential complications of silencing or interference and because selection will be inappropriate for clinical application. This vector design showed endothelial cell specificity of β-galactosidase expression when tested on a panel of human cell lines and primary breast microvascular endothelial cells, matching the specificity of expression of the endogenous promoter. Such simplified vectors exhibiting transcriptional specificity are likely to be useful for the development of a gene therapy approach to targeting tumor vasculature.
Recombinant retroviral vectors have found widespread use for gene delivery, both in vitro and in vivo. For human gene therapy it will be important to develop means of systemic application for in vivo transduction, requiring the ability to target gene expression at the levels of both restricted delivery and transcription. We are developing strategies for targeting tumor vasculature as an approach to cancer gene therapy, since tumor growth is dependent upon a blood supply and is associated with the switch to the angiogenic phenotype (20). Endothelial cells forming the tumor vasculature are suitable targets for cancer therapy due to the high ratio of dependent tumor cells to endothelial cells. Antiangiogenic drugs can slow tumor growth (24, 44), while angiostatin and endostatin, endogenous inhibitors of angiogenesis, can additionally cause tumor regression (6, 32, 33). The dependence on cell division for integration and expression when using murine retroviral vectors allows exploitation of the marked differential in the proliferation rates of tumor and uninvolved endothelial cells (11), providing one level of specificity. However, restriction of expression to the intended target cells is an essential consideration.
The U3 region of the murine leukemia virus (MLV) long terminal repeat (LTR) contains the enhancer and promoter for transcription of the integrated provirus from the 5′ LTR. MLV enhancer function maps to a 75-bp direct repeat in U3, within which binding sites for six different nuclear factors have been identified (36). The promoter contains a CAAT box and TATA box. Most of the transcription factors involved are ubiquitously expressed, accounting for the lack of any marked cell specificity of the LTR (37), although replacement of U3 with that derived from related murine retroviruses can modify specificity (3, 10). Transcriptional targeting, however, necessitates expression of the transgene under the control of heterologous sequences.
Vector design may influence the function of heterologous control sequences (41). The majority of vectors in use incorporate selectable marker genes, often the neomycin resistance gene, although this can act as a transcriptional silencer (2). Many such vectors retain a functional LTR, often in order to express the marker gene, and this can give rise to poor coexpression due to transcriptional interference (15, 16). Attempts to address this problem have involved deletion of LTR enhancer and/or promoter sequences in the 3′ LTR of the vector (8, 31, 45, 46), with this modification becoming duplicated at the 5′ end in the integrated provirus to leave a single transcriptional unit driven by an internal promoter. Alternatively, the entire transgene expression cassette can be shielded from these sequences by insertion upstream of the LTR enhancer, in the “double-copy” strategy (22). A reduced level of transcriptional specificity of the tyrosinase promoter has been obtained from a retroviral vector retaining an intact LTR (41). However, since selection will not be appropriate for clinical application of systemically administered vectors, we decided to omit a selectable marker gene from our vectors, thus avoiding such problems. Moreover, characterization of vectors in vitro without selection will be a more meaningful reflection of their behavior in vivo.
The transcriptional activity of the LTR itself can be modified by addition of, or replacement with, heterologous enhancer and/or promoter sequences. Thus, replacement of the viral enhancer with that of a polyomavirus mutant selected to grow in embryonal carcinoma cells (normally restrictive for MLV LTR function) yielded a hybrid LTR that was functional in these cells (39). The strength of the myeloproliferative sarcoma virus LTR could also be improved severalfold by addition of the cytomegalovirus IE enhancer upstream of the viral enhancer (40). However, addition of the muscle creatine kinase enhancer between the viral enhancer and promoter resulted in differentiation-specific expression in myogenic cells but failed to block constitutive LTR function in some cells (18), while incorporation of lymphoid cell-specific enhancers failed to modify specificity (28). In contrast, replacement of the viral enhancer with the tyrosinase enhancer and promoter did lead to specific expression in melanoma cells (12, 42). In this study, we explore the potential for deriving hybrid LTR retroviral vectors with endothelial cell transcriptional specificity, in the absence of selectable markers, by using the promoter for the human prepro-endothelin-1 (ppET1) gene.
MATERIALS AND METHODS
Cell lines.
NIH 3T3 (murine fibroblasts), CPAE (calf pulmonary artery endothelial cells), and PAE (porcine aorta endothelial cells) cells and the human cell lines TE671 (rhabdomyosarcoma), HT1080 (fibrosarcoma), ECV304 (human umbilical vein endothelial cell [HUVEC] derived), and EAhy926 (HUVEC derived, by fusion with A549 lung carcinoma cells) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. FLY-A13 (9) and TE-FLY-A8 (9a) amphotropic retroviral packaging cell lines were similarly maintained. HMEC-1 (simian virus 40 T-antigen-transformed human dermal microvascular endothelial cells) (1) were maintained in microvascular endothelial cell growth medium (Clonetics Corp., obtained from TCS Biologicals). Primary breast microvascular endothelial cells (a kind gift from C. Clarke and M. O’Hare, University College London) were obtained by selection with the antibody Q-BEND/40 attached to magnetic beads and were maintained as for HMEC-1; the cells were stained with CD31 for fluorescence-activated cell sorter analysis at passage 5, confirming the retention of this endothelial cell marker. HMEC-1 and EAhy926 were similarly confirmed to be positive for CD31 expression, while TE671, HT1080, and ECV304 were negative.
Cloning of the human ppET1 promoter region.
ppET1 promoter sequences were obtained following PCR amplification from genomic DNA prepared from a human lymphoblastoid cell line. Two regions were amplified: positions −177 to +33 and positions −177 to +158. Numbering is based on EMBL database entry HSETN1 (27). The forward primer (ETP5; see Fig. 1) was 5′-GCGAGATCTGCCTCTGAAGTTAGCAGTG, incorporating a BglII restriction site; the reverse primers were 5′-GCAGGATCCGTTCGCCTGGCGCAGATG (ETP3A) and 5′-AAAGGGATCCTTCAGCCCAAGTGCCC (ETP3B), incorporating BamHI restriction sites. PCR conditions were 30 cycles of 30 s at 94°C, 1 min at 60°C, and 30 s at 72°C. Amplified products were digested with BglII and BamHI before being cloned into these sites in pSP72 (Promega). DNA sequencing confirmed the nature of the cloned promoter regions, matching the reported sequence (25). To confirm the function of the cloned regions, the BglII-BamHI fragments were cloned into the BamHI site of the reporter vector pGCAT-C (21). Transient transfection verified orientation-dependent promoter function in CPAE cells but not 3T3 cells when expression of chloramphenicol acetyltransferase was assayed by an enzyme-linked immunosorbent assay (Boehringer Mannheim) (data not shown).
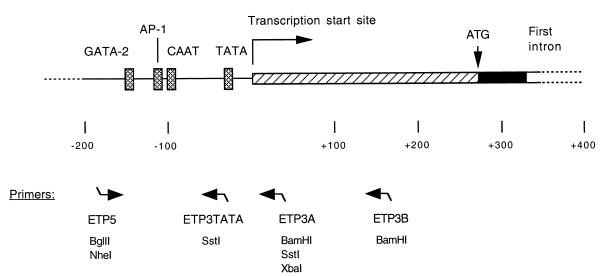
FIG. 1.
Schematic representation of DNA sequences around the transcription start site of the human ppET1 gene. Nucleotides are numbered with reference to the transcription start site at +1. Transcribed sequences are boxed: the untranslated 5′ leader (hatched box), the start of the coding sequence (solid) of exon 1, and the start of intron 1. The translation initiation codon is at +269. Deletion analysis of promoter activity showed that sequences between −141 and +145 gave endothelial cell specificity in transfected cell lines. Promoter function requires the presence of binding sites for GATA-2 and for AP-1. Both CAAT and TATA boxes are present. The positions of oligonucleotide primers for PCR are indicated below the diagram; these were designed with linker restriction sites, as indicated (refer to the text). Primer pair ETP5-ETP3A or ETP5-ETP3B generated promoter fragments −177 to +33 and −177 to +158, respectively. Primer ETP5 was used with ETP3A or ETP3TATA to generate ETP/LTR hybrids.
Retroviral vector construction with a hybrid ppET1/LTR promoter.
Plasmid pΔBN, containing a neomycin resistance (Neor) cassette and a single MLV LTR with a 3′-flanking NotI restriction site, was derived by partial deletion of sequences between the PstI sites in pBabeNeo (29). The multiple-cloning site was subsequently destroyed by digestion with BamHI and SalI, end repair with Klenow polymerase, and religation. The 5.5-kb NheI fragment from pMFGnlslacz (19) was inserted into the unique NheI site to create pNeoMFGnlslacz, equivalent to pMFGnlslacz but possessing a simian virus 40 Neor cassette on the same plasmid, outside of the retroviral transcription unit, and with the 3′-flanking NotI site to facilitate the exchange of 3′ LTR sequences.
ppET1/LTR hybrids were constructed by cloning ppET1 promoter sequences into pΔBN and verified by DNA sequencing. NheI-NotI fragments from the hybrids were used to replace the 3′ LTR in pNeoMFGnlslacz, following NotI and partial NheI digestion. Three hybrids were made, as follows. In the first two, the ppET1 promoter region from −177 to +33 was used to replace the NheI-XbaI or NheI-SstI fragment of pΔBN. This region was amplified from the cloned ppET1 promoter by using the forward primer 5′-ATAGCTAGCTCTGCCTCTGAAGTTAGCAGTG (ETP5Nhe) and the reverse primers 5′-ATATCTAGACCGTTCGCCTGGCGCAGATG (ETP3AXba) and 5′-ATAGAGCTCCGTTCGCCTGGCGCAGATG (ETP3ASst), incorporating NheI, XbaI, and SstI restriction sites, respectively. In the third construct, the reverse primer was 5′-ATAGAGCTCCCCTATTAGAGTGGGGGTAAAC (ETP3TATA), incorporating a SstI restriction site, yielding a product from −177 to −37, and placing the SstI site immediately before the TATA box to allow use of the LTR TATA box with equivalent spacing following replacement of the NheI-SstI fragment of pΔBN. PCR conditions were 25 cycles of 30 s at 94°C, 1 min at 60°C, and 1 min at 72°C.
Retroviral vector construction with an internal ppET1 promoter.
Plasmid pMBΔ, lacking the 3′ LTR enhancer, was derived following double digestion and religation to delete the 267-bp sequence between the NheI and XbaI sites of the 3′ LTR of pMB. The latter is a derivative of pBabePuro (29) that was obtained by replacing sequences between the BamHI site and the SstI site in the 3′ LTR (encompassing the puromycin selection cassette and the majority of U3) with myeloproliferative sarcoma virus U3 sequences from pMPSV (35).
ppET1 BglII-BamHI promoter fragments were cloned into the BamHI site of pMBΔ, re-creating a single BamHI site 3′ of the promoter. Vectors with an internal expression cassette in either orientation were constructed as follows. For ppET1 promoter expression in the same orientation relative to the viral transcription unit, a 3.5-kb BamHI fragment, encoding nucleus-localized β-galactosidase (nlslacz), was removed from pMFGnlslacz (19) and inserted into the BamHI site. For expression in the reverse orientation, a fragment containing polyadenylation sequences was first inserted. This was obtained by amplification of a 158-bp product surrounding the poly(A) site of pSV2cat, using the primers 5′-ACAGGATCCGAATGCAATTGTTGTTGTTAACTTG and 5′-CCTAGATCTCCAGACATGATAAGATACATTG, which incorporate BamHI and BglII restriction sites at the 5′ and 3′ ends, respectively. PCR conditions were 30 cycles of 30 s at 94°C, 1 min at 52°C, and 30 s at 72°C. The product was first cloned into pSP72 for sequence verification and then cloned into the BamHI site downstream of the ppET1 promoter in the “reverse” orientation in pMBΔ, re-creating a single BamHI site between the promoter and poly(A) sequences. The nlslacz fragment was subsequently inserted.
Recombinant retroviral vectors.
The packaging cell line FLY-A13 (9) was used to generate recombinant retroviral vectors following CaPO4-mediated transfection of plasmid DNA. For vectors constructed in the pMBΔ backbone, the plasmid pSV2neo was cotransfected at a molar ratio of 0.1. Stable transfectants were selected in 1.2 mg of G418 per ml. Virus was harvested following overnight incubation of confluent monolayers in fresh medium and filtered (0.45-μm-pore-size filter). Target cells were infected with serial dilutions of virus in the presence of 8 μg of Polybrene per ml for 4 to 6 h. The culture medium was changed and the cells were histochemically stained 2 to 3 days later with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). ELH5.10, a high-titer clone of the ppET1/LTR hybrid 5 construct, was obtained from TE-FLY-A8 packaging cells (a TE671-based equivalent of FLY-A13): following transfection and selection in 1 mg of G418 per ml, the clone with the highest titer on PAE cells was chosen. TELCeB6AF (9), a TE671-based MFGnlslacz virus producer clone, was used as a control. Since medium conditioned by virus producer cells can contain factors (such as chondroitin sulfate) that are inhibitory for retroviral infection (26), care was taken to ensure that titers were determined from the virus concentrations at which the infection efficiency titrated linearly.
Analysis of proviruses in transduced cells.
For confirmation of transduction, cells were lysed 5 days after infection in 50 mM Tris-HCl (pH 8.5)–1 mM EDTA–0.5% Tween 20–200 μg of proteinase K per ml. Lysates were subjected to PCR with primers 5′-GCACATGGCTGAATATCGACGG (beginning 78 bp 5′ of the EcoRI site within lacZ) and 5′-GCTTCAGCTGGTGATATTGTTGAG (spanning the PvuII site in the retrovirus backbone 3′ of the inserted lacZ gene). PCR conditions were 35 cycles of 30 s at 94°C, 1 min at 60°C, and 1 min at 72°C. Primers specific for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or porcine cytochrome oxidase (34) were used as controls for DNA quantification. Amplification with primer 5′-GGCAAGCTAGCTTAAGTAACGCC, matching the LTR downstream of the NheI site, or ETP5Nhe, matching the 5′ end of the ppET1 promoter, with the reverse primer 5′-GTTCCGAACTCGTCAGTTCCACC, matching part of the packaging sequence downstream of the 5′ LTR, was used to verify duplication of the enhancer deletion or hybrid construction for ETP-I and ETP/LTR hybrid 5, respectively.
Reverse transcription-PCR (RT-PCR) to determine endogenous expression of ppET-1.
Total RNA was extracted from cells at subconfluence by using the RNeasy kit (Qiagen) and digested with DNase I (5 U/30 μg of RNA; Promega). Following phenol-chloroform extraction and ethanol precipitation, 1 μg of RNA was reverse transcribed with oligo(dT) primer and 20 U of MLV reverse transcriptase for 1.5 h at 42°C. The resultant cDNA was heat denatured, and 5 μl (from an initial 30 μl reaction) was used for PCR. To determine endogenous expression from the endothelial cell-specific promoter, the forward and reverse primers were 5′-TACTTCTGCCACCTGGACATC and 5′-TGCTTGGCAAAAATTCCAGC, respectively (amplifying between positions +447 to +630). PCR conditions were 40 cycles of 1 min at 96°C, 1 min at 52°C, and 1 min at 72°C. To determine expression from a second upstream promoter, functional in some nonendothelial cells (4), the forward primer was 5′-TGTTTACCCCCACTCTAATA and the annealing temperature was 55°C (amplifying between positions −60 and +630). As a control, GAPDH sequences were amplified with primers 5′-TGGATATTGTTGCGATCAATGCC and 5′-GATGGCATGGACTGTGGTCATG, using a program of 1 min at 96°C, 30 s at 65°C, and 1 min at 68°C.
Determination of β-galactosidase expression.
β-Galactosidase activity in cell lysates was determined by a quantitative photometric assay (17). Cells in 24-well plates were infected with 50 μl of virus (ELH5.10 or the control virus without LTR modification) for 4 h, as above. At 3 days later, the cells were washed in phosphate-buffered saline and lysed in 350 μl of 250 mM Tris-HCl (pH 8.0)–0.1% Triton X-100. Lysates were stored at −80°C. To determine the amounts of β-galactosidase, 50-μl volumes of lysates and dilutions were combined with 50 μl of phosphate-buffered saline–0.5% bovine serum albumin and 150 μl of 60 mM Na2HPO4 (pH 8.0)–1 mM MgSO4–10 mM KCl–50 mM β-mercaptoethanol–0.1% CPRG (chlorophenol red galactopyranoside; Boehringer). After light-protected incubation at room temperature, absorption at 570 nm was measured and converted to picograms of β-galactosidase by using a standard curve obtained with purified enzyme (Sigma).
To estimate the (average) expression per cell transduced with ELH5.10, β-galactosidase activity was normalized as follows. For the control virus, the number of cells responsible for the observed enzyme activity was determined by histochemical staining from parallel experiments with all parameters, such as cell number and duration of exposure, kept constant. The number of cells transduced is dependent on the intrinsic infectability of the different target cells. The number of cells transduced with ELH5.10 could not be determined directly in this way due to the variable transcriptional activity of the hybrid promoter. It is, however, equivalent to the number transduced by the control virus multiplied by a factor that reflects the relative amounts of the two viruses, irrespective of their transcriptional activities. This factor is independent of the target cell but varies for different virus harvests, and it was determined from the relative LacZ titers of the two viruses for PAE cells. (An additional factor was applied for HMEC-1 cells to compensate for inhibition with undiluted virus, based on the observed nonlinearity of infection efficiency in this cell line.) This analysis assumes only that histochemical staining for the control virus on all cells, and for ELH5.10 on PAE cells, equates with transduction efficiency. Deviation from such a correlation for ELH5.10 would result in overestimation of the absolute values but would not undermine the relative values (see Results). Analysis of the data in this way is a consequence of the absence of selection in our vector design.
Confirmation of the transcription start site.
The start site for transcription from the wild-type and hybrid LTRs was determined by using RNA from PAE cells infected with ELH5.10 or control viruses. Total RNA was made with the RNeasy kit (Qiagen). A 2-μg sample of RNA was reverse transcribed with a primer specific for MLV bases 492 to 471, and the 5′ cDNA end was amplified with a 5′/3′ rapid amplification of cDNA ends (RACE) kit (Boehringer Mannheim). Two rounds of PCR were performed with the supplied anchor primer and primers specific for MLV bases 469 to 448 (first round) and 342 to 320 (second round). First-round PCR conditions were 40 cycles of 30 s at 94°C, 1 min at 60°C, and 1 min at 72°C; the second round of PCR used an annealing temperature of 65°C. The PCR product was purified and sequenced with the MLV-specific primer.
RESULTS
Cloning of ppET1 promoter sequences.
Two previous studies of sequences required for ppET1 promoter function in endothelial cells, using transient-transfection assays, reported specificity associated with sequences from positions −143 to +165 (27) and from −141 to +145 (43). A similar region (−177 to +158), as well as one with less downstream sequence (−177 to +33), was amplified by PCR from human genomic DNA (Fig. 1). Both promoter regions were used to generate retroviral vectors from pMBΔ (see below), with expression in the forward orientation (i.e., 5′ to 3′ with respect to the viral sequences). Viruses from bulk populations of stable packaging-cell transfectants led to expression in CPAE and PAE cells but not several other target cells, including TE671. Since both promoter regions functioned equivalently, only the shorter was used in the experiments discussed below. All constructs used expression of nucleus-localized β-galactosidase (nlslacz) as a reporter.
Retroviral vector design for use of hybrid ppET1/LTR promoters.
A series of vectors was made to examine the potential for incorporating ppET1 promoter sequences within the viral LTR. Modifications were made in the 3′ LTR so that they would be duplicated in the 5′ LTR of the provirus for expression of the reporter gene. The tyrosinase enhancer and promoter have previously been successfully inserted in the viral LTR, replacing the viral enhancer (42). We similarly inserted the ppET1 promoter between the NheI and XbaI sites flanking the viral enhancer (ETP/LTR hybrid 1; Fig. 2). However, we reasoned that this construct was potentially suboptimal in three respects. First, residual expression from the viral promoter due to incomplete removal of transcription regulatory sequences could compromise specificity. Second, when the transcriptional start site associated with the inserted promoter is placed a significant distance upstream of the viral polyadenylation sequences in the R region of the 5′ LTR, premature termination of transcription could result; this is usually silent due to the proximity of the viral start site and 5′ polyadenylation site and/or lack of upstream RNA sequences at the 5′ end of the viral transcript. Third, translation of a transcript with such a long leader may be inefficient.
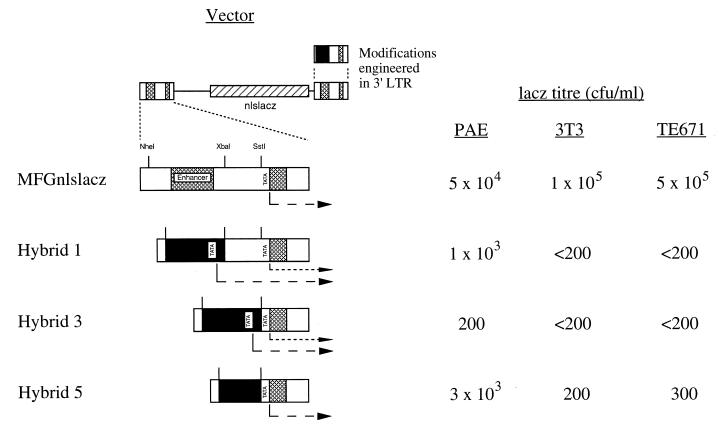
FIG. 2.
Vectors with hybrid ppET1/LTR promoters. The proviral forms, after transduction, of the vectors ppET1/LTR hybrids 1, 3, and 5 are illustrated, along with the control vector MFGnlslacz (derived from pNeoMFGnlslacz). In the three hybrids, variable amounts of the viral enhancer and promoter within U3 are replaced with ppET1 promoter sequence (solid). Cross-hatched boxes indicate enhancer and R regions of the viral LTR. The NheI, XbaI, and SstI restriction sites used during construction are indicated. Modifications were initially engineered in the 3′ LTR and duplicated at the 5′ LTR during RT. Also indicated are the starts of the expected transcripts, including potential transcripts from the residual viral promoter in hybrids 1 and 3. Average vector titers from four independent experiments, obtained following infection of PAE, 3T3, and TE671 cells and histochemical staining for β-galactosidase, are given as CFU per milliliter. The limit of detection in this assay was 200 CFU/ml for 3T3 and TE671 cells and 20 CFU/ml for PAE cells.
Two further ppET1/LTR hybrids were constructed. The majority of the viral promoter was removed by insertion between the NheI and SstI sites (ETP/LTR hybrid 3; Fig. 2). Also, the viral sequence upstream of the TATA box was replaced with ppET1 promoter sequence upstream of its TATA box (nucleotides −177 to −37), yielding a hybrid promoter in which the spacing between the ppET1 CAAT box and virus-derived TATA box was the same as in the ppET1 promoter (ETP/LTR hybrid 5; Fig. 2). This construct was expected to utilize the viral start site and so generate a spliced transcriptional leader identical to that generated by the intact LTR in the control vector. Sequences 3′ of the viral TATA box were not replaced since the terminal bases of U3 are necessary for recognition of the polyadenylation signal in the 3′ LTR, possibly due to their involvement in RNA secondary structure (5, 13).
The three hybrid LTRs were used to replace the 3′ LTR of the plasmid pNeoMFGnlslacz, which possesses a Neor selection cassette outside of the retroviral sequences. Virus derived from stable transfectants was used to infect target cells, in which marker gene expression was subsequently detected by histochemical staining. Bulk populations of transfected FLY-A13 cells were used to compare vector constructs without the potential complication of clone variability. The target cells used were 3T3, TE671, and PAE. Transfection experiments previously showed that the promoter is nonfunctional in 3T3 cells (27, 43). TE671 cells lack endogenous expression of ppET1 mRNA, by RT-PCR (see below), and were used as an additional negative control. Primers specific for porcine ppET1 confirmed expression in PAE cells (data not shown), which were used as a positive control. Consistent with our design rationale, the highest lacZ titer was shown by ETP/LTR hybrid 5; it was 6% of that of the unmodified LTR control vector on PAE cells but only 0.2% or less on 3T3 and TE671 cells (Fig. 2), suggesting specificity for PAE cells. Thus, replacement of the viral enhancer with the ppET1 promoter abolished LTR function in these negative control cell lines.
Retroviral vector design for use of the internal ppET1 promoter.
For comparison, vectors with an internal promoter in a vector with the 3′ LTR enhancer deleted were constructed in both forward (ETP-I) and reverse (ETPrev) orientations (Fig. 3). Both vectors gave lacZ titers for PAE cells, while the presence of an internal ppET1 promoter abolished residual LTR activity in 3T3 and TE671 cells. This is most probably because the ppET1 promoter, which is nonfunctional in these cells, overrides the disabled enhancerless LTR.
FIG. 3.
Vectors incorporating an internal ppET1 promoter. The proviral forms of the vectors ETP-I and ETPrev are illustrated, along with control vector MBΔ. All have LTR enhancer deletions, initially engineered in the 3′ LTR and duplicated at the 5′ LTR during RT. The ppET1 promoter is represented by the solid box. ETPrev additionally has an internal poly(A) site. Also indicated are the expected transcripts. Average vector titers from three independent experiments are given as CFU per milliliter. See also the legend to Fig. 2.
PCR analysis of transduced target cells.
Transduced target cells were lysed and used for PCR amplification of a specific lacZ/vector junction fragment to confirm transduction of both PAE and TE671 target cells (Fig. 4A), consistent with the cell type differences in functional titer (lacZ CFU per milliliter) being due to transcriptional specificity. Estimates based on a twofold serial dilution of the control vector indicated that the transduction efficiency for each of the modified vectors was reduced up to 10-fold on both PAE cells (Fig. 4B) and TE671 cells (data not shown). The reduced lacZ titer on PAE cells appears to be due in large part to reduced virus production, as a consequence of the LTR modification, although there may be an additional component due to reduced promoter function (e.g., hybrid 3).
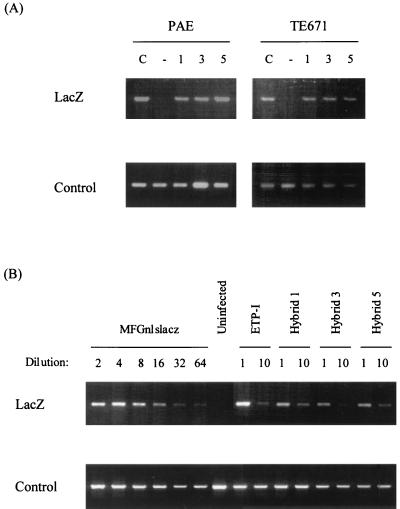
FIG. 4.
PCR analysis of transduced target PAE and TE671 cells. Cell lysates were used to amplify a ∼500-bp lacZ/vector junction region to detect the presence of integrated proviruses (LacZ). The product is slightly larger for ETP-I due to differences at the cloning site. The lower panels show controls for DNA quantification in the PCR, i.e., human GAPDH or porcine cytochrome oxidase, as appropriate. (A) PAE and TE671 cells were infected with MFGnlslacz control (lane C) or ETP/LTR hybrid 1, 3, and 5 viruses. Lane − contains uninfected controls. (B) PAE cells were infected with MFGnlslacz, ETP-I, and ETP/LTR hybrid viruses, diluted as shown. As is evident from the dilution series for MFGnlslacz, transduction is initially nonlinear, and so quantitation should be estimated from the 1:10-diluted viruses.
PCR of transduced cells was also used to analyze the nature of the provirus 5′ LTR and confirm duplication of the 3′ LTR deletions and modifications. Primers specific for sequences upstream or downstream of the NheI site or for the ppET1 promoter were used with primers specific for U5 or part of the packaging signal sequence downstream of the proviral 5′ LTR to verify duplication of the enhancer deletion for the vector ETP-I and of the hybrid construction for ETP/LTR hybrid 5 (data not shown).
Endothelial cell specificity of expression from ETP/LTR hybrid 5.
From the foregoing experiments, vector ETP/LTR hybrid 5 was chosen for further analysis. The producer cell clone with the highest lacZ titer on PAE cells was named ELH5.10 (for “ETP/LTR hybrid 5, clone 10”). Virus titers and β-galactosidase enzyme activities of transduced cells were measured for a panel of six human target cells. RT-PCR was used to determine the status of endogenous ppET-1 promoter function in these cells in order to assess how well this specificity was reproduced by the vector (Fig. 5). Specificity for ppET-1 and not ppET-2 or ppET-3 was also provided by the primers used. Moreover, recent studies have shown that a second, upstream promoter can lead to expression of ppET-1 sequences in nonendothelial cells (4). A second primer pair was used to assess expression from the upstream promoter to avoid misinterpreting the RT-PCR results. The endothelial cell-specific promoter was active in the cell line HMEC-1 and the primary breast microvascular endothelial cells but not in ECV304, HT1080, and TE671 cells.
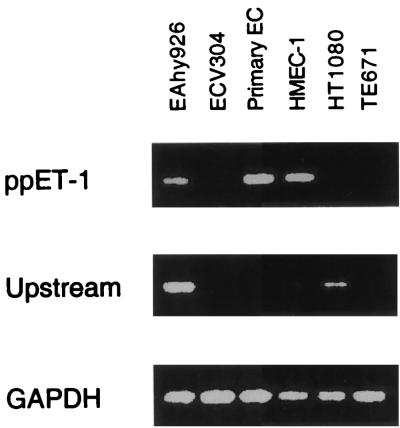
FIG. 5.
Endogenous expression from the ppET-1 promoter. RNA from human target cells, including primary endothelial cells (Primary EC), was isolated and used to amplify regions of the ppET-1 cDNA (+447 to +630), the cDNA derived from the upstream promoter (−60 to +630, based on the same numbering system relative to the endothelial cell-specific transcript [Fig. 1]), or the GAPDH cDNA as a control. Detection of −60 to +630 is more sensitive than of +447 to +630 (compare HT1080 products). Consequently, while it is evident that the ppET-1 promoter is active in HMEC-1 and primary endothelial cells, the +447 to +630 product for EAhy926 cannot be interpreted as such due to the activity of the upstream promoter in these cells.
Three independent virus harvests were subjected to titer determination on PAE, 3T3, and all six human target cells. The relative lacZ titer of ELH5.10 versus MFGnlslacz control virus was of the order of 1% for PAE, HMEC-1, and the primary endothelial cells and approximately 10-fold less for TE671 and all other target cells (Fig. 6A). Although the target cell infectabilities vary widely, the relative titers are directly comparable and indicate specificity consistent with the pattern of endogenous ppET-1 promoter activity. (The titer reduction on PAE cells is greater than that shown in Fig. 2 due to the nature of the particular clones used, as opposed to the bulk populations of producer cells used previously.)
FIG. 6.
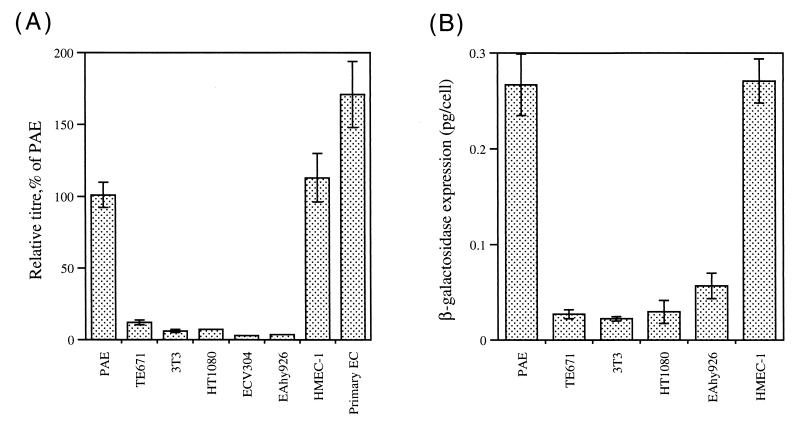
Endothelial cell specificity of expression from ETP/LTR hybrid 5. PAE, 3T3, and a panel of six human cell lines, including primary endothelial cells (Primary EC), were infected with virus ELH5.10 for titer determination by histochemical staining or for determination of β-galactosidase enzyme activities. The MFGnlslacz control virus was used in parallel. Three independent virus harvests of ELH5.10 were used. For each target cell, the titer of ELH5.10 relative to the control (A) or the β-galactosidase activity normalized to account for the variation in intrinsic infectability of the different target cells (B) is shown. (A) The relative titres are expressed as a percentage of that displayed on PAE cells in order to normalize variations in the absolute titers of the different harvests. Data shown are the mean and standard error of nine determinations. (B) Levels of expression per transduced cell for each cell target following infection with ELH5.10 (but see the text for a discussion of absolute values). Data shown are the mean and standard error of six (HT1080, EAhy926, and HMEC-1 cells) or nine (PAE, TE671, and 3T3 cells) determinations. The mean activity of the control virus in PAE cells was 0.8 pg/cell. Control virus titers were of the order of 107 CFU/ml for TE671 and 3T3 cells, 106 CFU/ml for HT1080, EAhy926, and PAE cells, 105 CFU/ml for ECV304 and HMEC-1 cells, and 104 CFU/ml for primary endothelial cells.
The same virus harvests were used for determination of β-galactosidase enzyme activities following transduction with either ELH5.10 or 1:100-diluted control virus. The poorly infectable primary endothelial cells and the ECV304 cell line did not give significant activity above background and so were not considered further for this analysis. β-Galactosidase expression for ELH5.10 was normalized in order to assess the relative expression per transduced cell (Fig. 6B). Normalization relies on the assumption that all PAE cells transduced with ELH5.10 stain positive in the histochemical assay. Although underestimation will lead to overestimation of the absolute values of enzyme expression, the relative magnitudes are reliable. Expression was approximately 10-fold greater in both PAE and HMEC-1 cells than in 3T3 and TE671 cells, consistent with the specificity indicated by the relative lacZ titers (Fig. 6A).
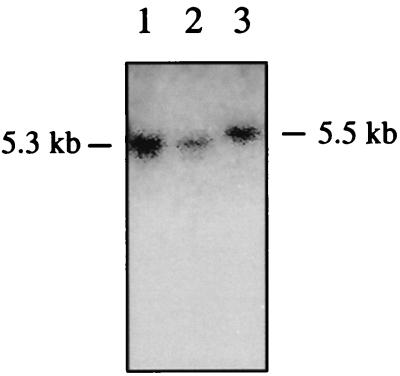
Southern blot analysis of TE671 cells infected with ELH5.10 virus indicated that the provirus was intact, confirming that reduced expression in these cells was not due to rearrangement (Fig. 7). The provirus was reduced in size relative to the unmodified vector due to replacement of the 5′ LTR with the (shorter) hybrid LTR. Finally, the performance of the hybrid LTR vector, designed to initiate transcription as for the unmodified LTR, was verified by using 5′-RACE to examine the transcripts. RNA was prepared from PAE cells infected with ELH5.10 and control viruses and also from the cells producing these viruses. The amplified RACE PCR products were sequenced, confirming that use of the +1 site was maintained for the initiation of transcription in both producer and transduced target cells (data not shown).
FIG. 7.
Detection of intact provirus. DNA from TE671 cells infected with two preparations of ELH5.10 (lanes 1 and 2) was digested with NheI. Southern blot analysis detected the 5.3-kb fragment expected for the intact provirus. The fragment is slightly smaller than that for the unmodified vector (lane 3) due to the nature of the LTR modification.
DISCUSSION
In considering retroviral vector designs for tissue-specific expression of therapeutic genes, we elected to avoid the presence of a functional LTR in the integrated provirus of the target cell, since this can lead to transcriptional interference. Similarly, optimal vectors will also avoid the use of an additional promoter for selectable-marker expression. Moreover, since selection is not applicable to a therapeutic protocol for in vivo gene therapy, we decided to omit any such gene. Although lack of a means of selection has attendant difficulties in terms of in vitro manipulation and evaluation, we reasoned that simplicity of design was likely to be valuable in terms of efficacy. Also, without the potential for selection to bias expression, the performance of such vectors in vivo will be more accurately reflected in vitro. In this important respect our study differs from previous studies in which regulatory sequences were incorporated into retroviral vectors.
The approach taken was to incorporate ppET1 promoter sequences into the LTR to generate hybrid promoters, as has been done previously with tyrosinase regulatory regions (12, 42). However, the construction most extensively studied in this work (hybrid 5) differs from any previously described. Sequences preceding the ppET1 TATA box were fused with the viral TATA box, eliminating all other viral transcription regulatory sequences and maintaining the spacing between CAAT and TATA boxes as in the ppET1 promoter. This resulted in initiation of transcription at the viral start site at the beginning of R identical to that obtained from the unmodified LTR. The MFG vector design employs a splicing mechanism to optimize translation efficiency (14), which is retained in the hybrid.
The hybrid promoter showed a 10-fold preferential expression in PAE and HMEC-1 cells compared to nonendothelial cells. Relative to a control vector used to correct for the widely varying infectabilities of the target cells, functional titers determined by measurement of β-galactosidase expression were similarly 10-fold higher for endothelial cells, including human primary breast microvascular endothelial cells. Vector specificity correlated with the endogenous activity of the ppET-1 promoter. However, it is apparent that the promoter activity of the hybrid was diminished compared to the unmodified LTR (Fig. 6). Additionally, the LTR modification resulted in some loss of vector titer.
We sought to incorporate sequences directing endothelial cell-specific expression within recombinant retroviral vectors because of our interest in developing a gene therapy approach to modify tumor vasculature. In the present study, we used the characterized promoter region of the human ppET1 gene (27, 43), which is small and of simple organization. ppET1 is expressed in large-vessel and microvascular endothelial cells (7). However, its expression is not entirely endothelial cell restricted: it is additionally expressed in renal and pulmonary epithelium, and, at least for the latter, expression in vitro is controlled by the same promoter region (38). Transgene expression controlled by the murine ppET1 promoter shows a similar pattern (23). More recently, a number of promoters for human genes whose expression is specifically upregulated during angiogenesis have been described and may afford greater potential. In particular, expression of KDR/flk-1, flt-1, and tie is induced on endothelial cells within human and experimental tumors (30). Such regulatory sequences may enable further improvements to give tightly restricted expression of genes in the tumor vasculature.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council.
We are grateful to C. Clarke and M. O’Hare, University College London, for the kind gift of primary breast microvascular endothelial cells; to the Centers for Disease Control and Prevention, Atlanta, Ga., for the HMEC-1 cell line; to F.-L. Cosset, University of Lyons, for the TE-FLY-A8 amphotropic retroviral packaging cell line; and to G. Mavria for Fig. 7. We thank M. K. L. Collins and Y. Takeuchi for critical discussion.
REFERENCES
- 1.Ades E W, Candal F J, Swerlick R A, George V G, Summers S, Bosse D C, Lawley T J. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Investig Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 2.Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H. The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene. 1991;99:249–254. doi: 10.1016/0378-1119(91)90134-w. [DOI] [PubMed] [Google Scholar]
- 3.Baum C, Hegewisch-Becker S, Eckert H G, Stocking C, Ostertag W. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benatti L, Bonecchi L, Cozzi L, Sarmientos P. Two preproendothelin 1 mRNAs transcribed by alternative promoters. J Clin Investig. 1993;91:1149–1156. doi: 10.1172/JCI116274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz E W, Wydro R M, Nadal-Ginard B, Dino D. Moloney murine sarcoma proviral DNA is a transcriptional unit. Nature. 1980;288:665–669. doi: 10.1038/288665a0. [DOI] [PubMed] [Google Scholar]
- 6.Boehm T, Folkman J, Browder T, O’Reilly M S. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature. 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 7.Bull H A, Bunker C B, Terenghi G, Springall D, Zhao Y, Polak J, Dowd P. Endothelin-1 in human skin: immunolocalisation, receptor binding, mRNA expression, and effects on cutaneous microvascular endothelial cells. J Investig Dermatol. 1991;97:618–623. doi: 10.1111/1523-1747.ep12483000. [DOI] [PubMed] [Google Scholar]
- 8.Cone R D, Weber-Benarous A, Baorto D, Mulligan R C. Regulated expression of a complete human beta-globin gene encoded by a transmissible retrovirus vector. Mol Cell Biol. 1987;7:887–897. doi: 10.1128/mcb.7.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosset F-L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High titre packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cosset, F.-L. Unpublished data.
- 10.Couture L A, Mullen C A, Morgan R A. Retroviral vector containing chimaeric promoter/enhancer elements exhibit cell-type-specific gene expression. Hum Gene Ther. 1994;5:667–677. doi: 10.1089/hum.1994.5.6-667. [DOI] [PubMed] [Google Scholar]
- 11.Denekamp J. Endothelial cell proliferation as a novel approach to targeting tumour therapy. Br J Cancer. 1982;45:136–139. doi: 10.1038/bjc.1982.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz R M, Eisen T, Hart I R, Vile R G. Exchange of viral promoter/enhancer elements with heterologous regulatory sequences generates targeted hybrid long terminal repeat vectors for gene therapy of melanoma. J Virol. 1998;72:789–795. doi: 10.1128/jvi.72.1.789-795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dougherty J P, Temin H M. A promoterless retroviral vector indicates that there are sequences in U3 required for 3′ RNA processing. Proc Natl Acad Sci USA. 1987;84:1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerman M, Temin H. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984;39:459–467. [PubMed] [Google Scholar]
- 16.Emerman M, Temin H M. Comparison of promoter suppression in avian and murine retrovirus vectors. Nucleic Acids Res. 1986;14:9381–9396. doi: 10.1093/nar/14.23.9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felgner J H, Kumar R, Sridhar C N, Wheeler C, Tsai Y J, Border R, Ramsey P, Martin M, Felgner P L. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]
- 18.Ferrari G, Salvatori G, Rossi C, Cossu G, Mavilio F. A retrovirus vector containing a muscle-specific enhancer drives gene expression only in differentiated muscle fibers. Hum Gene Ther. 1995;6:733–742. doi: 10.1089/hum.1995.6.6-733. [DOI] [PubMed] [Google Scholar]
- 19.Ferry N, Duplessis O, Houssin D, Danos O, Heard J M. Retroviral-mediated gene transfer into hepatocytes in vivo. Proc Natl Acad Sci USA. 1991;88:8377–8381. doi: 10.1073/pnas.88.19.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 21.Frebourg T, Brison O. Plasmid vectors with multiple cloning sites and cat-reporter gene for promoter cloning and analysis in animal cells. Gene. 1988;65:315–318. doi: 10.1016/0378-1119(88)90468-4. [DOI] [PubMed] [Google Scholar]
- 22.Hantzopoulos P A, Sullenger B A, Ungers G, Gilboa E. Improved gene expression upon transfer of the adenosine deaminase minigene outside the transcriptional unit of a retroviral vector. Proc Natl Acad Sci USA. 1989;86:3519–3523. doi: 10.1073/pnas.86.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harats D, Kurihara H, Belloni P, Oakley H, Ziober A, Ackley D, Cain G, Kurihara Y, Lawn R, Sigal E. Targeting gene expression to the vascular wall in transgenic mice using the murine preproendothelin-1 promoter. J Clin Investig. 1995;95:1335–1344. doi: 10.1172/JCI117784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingber D, Fujita T, Kishimoto S, Sudo K, Kanamaru T, Brem H, Folkman J. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 25.Inoue A, Yanagisawa M, Takuwa Y, Mitsui Y, Kobayashi M, Masaki T. The human preproendothelin-1 gene: complete nucleotide sequence and regulation of expression. J Biol Chem. 1989;264:14954–14959. [PubMed] [Google Scholar]
- 26.LeDoux J M, Morgan J R, Snow R G, Yarmush M L. Proteoglycans secreted by packaging cell lines inhibit retrovirus infection. J Virol. 1996;70:6468–6473. doi: 10.1128/jvi.70.9.6468-6473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M-E, Bloch K D, Clifford J A, Quertermous T. Functional analysis of the endothelin-1 gene promoter. J Biol Chem. 1990;265:10446–10450. [PubMed] [Google Scholar]
- 28.Moore K A, Scarpa M, Kooyer S, Utter A, Caskey C T, Belmont J W. Evaluation of lymphoid-specific enhancer addition or substitution in a basic retrovirus vector. Hum Gene Ther. 1991;2:307–315. doi: 10.1089/hum.1991.2.4-307. [DOI] [PubMed] [Google Scholar]
- 29.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustonen T, Alitalo K. Endothelial receptor tyrosine kinases involved in angiogenesis. J Cell Biol. 1995;129:895–898. doi: 10.1083/jcb.129.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima K, Ikenaka K, Nakahira K, Morita N, Mikoshiba K. An improved retroviral vector for assaying promoter activity. FEBS Lett. 1993;315:129–133. doi: 10.1016/0014-5793(93)81148-s. [DOI] [PubMed] [Google Scholar]
- 32.O’Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumour growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 33.O’Reilly M S, Holmgren L, Chen C, Folkman J. Angiostatin induces and sustains dormancy of human primary tumors in mice. Nat Med. 1996;2:689–692. doi: 10.1038/nm0696-689. [DOI] [PubMed] [Google Scholar]
- 34.Patience C, Patton G S, Takeuchi Y, Weiss R A, McClure M O, Rydberg L, Breimer M E. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet. 1998;352:699–701. doi: 10.1016/S0140-6736(98)04369-4. [DOI] [PubMed] [Google Scholar]
- 35.Porter C D, Parkar M H, Levinsky R J, Collins M K L, Kinnon C. X-linked chronic granulomatous disease: correction of NADPH oxidase defect by retrovirus-mediated expression of gp91-phox. Blood. 1993;82:2196–2202. [PubMed] [Google Scholar]
- 36.Speck N A, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speck N A, Renjifo B, Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun G, Angelis G D, Nucci F, Ackerman V, Bellini A, Mattoli S. Functional analysis of the preproendothein-1 gene promoter in pulmonary epithelial cells and monocytes. Biochem Biophys Res Commun. 1996;221:647–652. doi: 10.1006/bbrc.1996.0650. [DOI] [PubMed] [Google Scholar]
- 39.Valerio D, Einerhand M P W, Wamsley P M, Bakx T A, Li C L, Verma I M. Retrovirus-mediated gene transfer into embryonal carcinoma and haematopoietic stem cells: expression from a hybrid long terminal repeat. Gene. 1989;84:419–427. doi: 10.1016/0378-1119(89)90516-7. [DOI] [PubMed] [Google Scholar]
- 40.van den Wollenberg C L M, Hoeben R C, van Ormondt H, van der Eb A J. Insertion of the human cytomegalovirus enhancer into a myeloproliferative sarcoma virus long terminal repeat creates a high-expression retroviral vector. Gene. 1994;144:237–241. doi: 10.1016/0378-1119(94)90383-2. [DOI] [PubMed] [Google Scholar]
- 41.Vile R, Miller N, Chernajovsky Y, Hart I. A comparison of the properties of different retroviral vectors containing the murine tyrosinase promoter to achieve transcriptionally targeted expression of the HSVtk or IL-2 genes. Gene Ther. 1994;1:307–316. [PubMed] [Google Scholar]
- 42.Vile R G, Diaz R M, Miller N, Mitchell S, Tuszyanski A, Russell S J. Tissue-specific gene expression from Mo-MLV retroviral vectors with hybrid LTRs containing the murine tyrosinase enhancer/promoter. Virology. 1995;214:307–313. doi: 10.1006/viro.1995.9923. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D B, Dorfman D M, Orkin S H. A nonerythroid GATA-binding protein is required for function of the human preproendothelin-1 promoter in endothelial cells. Mol Cell Biol. 1990;10:4854–4862. doi: 10.1128/mcb.10.9.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaoka M, Yamamoto T, Masali T, Ikeyama S, Sudo K, Fujita T. Inhibition of tumour growth and metastasis of rodent tumors by the angiogenesis inhibitor O-(chloroacetyl-carbamoyl)fumagillol (TNP-470; AGM-1470) Cancer Res. 1993;53:4262–4267. [PubMed] [Google Scholar]
- 45.Yee J-K, Moores J C, Jolly D J, Wolff J A, Respess J G, Friedmann T. Gene expression from transcriptionally disabled retroviral vectors. Proc Natl Acad Sci USA. 1987;84:5197–5201. doi: 10.1073/pnas.84.15.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu S-F, von Ruden T, Kantoff P W, Garber C, Seiberg M, Ruther U, Anderson W F, Wagner E F, Gilboa E. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]