Abstract
DNA of the avian hepadnavirus, duck hepatitis B virus, was found to be integrated at low abundance into the cellular DNA extracted from the livers of infected ducklings. The frequency of integration was estimated to be at least one viral genome per 103 to 104 cells by 6 days postinfection. The structures of virus-cell junctions determined by sequencing were compared with those of virus-virus junctions formed by nonhomologous recombination between the ends of linear viral DNA forms. This comparison allowed us to conclude that linear viral DNA was the preferential form used as an integration substrate. Potential factors promoting viral DNA integration during chronic infection are discussed.
Persistent infection of hepatocytes by hepatitis B virus (HBV) is not considered to be cytopathic. Rather, chronic liver disease is generally ascribed to consequences stemming from the action of the host immune system against virus-infected cells (for reviews see references 6, 8, and 21). In addition, variants of hepadnaviruses which have been shown or proposed to be directly cytotoxic have been described (1, 5, 14–16, 20, 23, 24). Hepadnaviruses have also been shown to cause genetic alterations in infected cells by integration into the host cell DNA, a process referred to as insertional mutagenesis (for a review see reference 3). Insertional mutations caused by hepadnaviruses have been implicated in the pathogenesis of hepatocellular carcinoma resulting from chronic HBV infection in humans and especially in chronic infections of woodchucks by the woodchuck hepatitis virus (WHV) (4, 7, 12). Integrated viral DNA sequences are often fragmentary or highly rearranged, indicating that specific mechanisms for the insertion of functional viral DNA into the chromosome are not encoded by the virus.
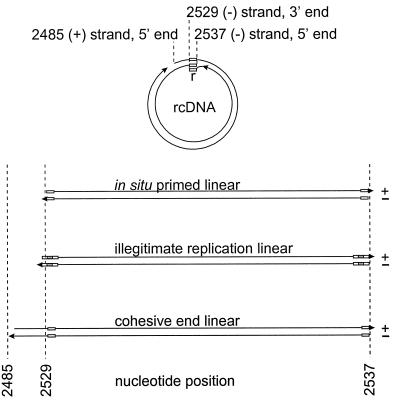
Circumstantial evidence suggests that various linear forms of viral DNA may recombine with cellular DNA to produce the variety of integrated structures seen in hepatocellular carcinomas (10, 31, 32). Evidence exists for at least three kinds of linear DNA produced during infection, as shown in Fig. 1. Linear viral double-stranded DNA is formed primarily as a minor product of abortive DNA replication due to failure of plus-strand priming to occur at a site on the genome that allows genome circularization (in situ-primed linear DNA) (17, 26). Linear forms can also be produced by the process of “illegitimate replication,” an inefficient replication pathway that bypasses the genome circularization step and results in a low sustained level of replication through double-stranded DNA in situ-primed linear intermediates (illegitimate linear DNA) (31, 33). Finally, evidence from chronically infected woodchuck livers suggests that about 20% of all linear-DNA forms involved in intramolecular recombination consist of molecules derived by denaturation of the cohesive 5′ ends of circular DNA followed by repair synthesis of the single-stranded ends (cohesive-end linear DNA) (32). These three forms of linear DNA have each been shown to participate in intramolecular blunt-end-joining reactions by nonhomologous recombination, and it has been suggested that linear DNAs might participate in intermolecular recombination with cellular DNA ends to produce integrated forms (9–11, 26, 31).
FIG. 1.
Viral double-stranded DNA forms in infected liver. rcDNA is shown at the top. The sites of the 9-nucleotide repeated sequence, r, positioned at either end of the viral minus strand, are indicated by small boxes. The three forms of linear double-stranded DNA shown at the bottom are produced by in situ priming of plus-strand synthesis (17, 26), illegitimate replication (31), or denaturation of the cohesive ends of rcDNA (32). The last two forms are hypothetical intermediates inferred from recombination joints found in cccDNA. The nucleotide numbering refers to the plus strand according to the method of Mandart et al. (18).
In the present study we have investigated the possibility that integration of viral linear DNA occurs even during short periods of viral DNA replication in the liver in the absence of any pathogenesis. Using ducklings infected with the avian hepadnavirus duck hepatitis B virus (DHBV) (19), we found that integrated DNAs could be detected at low levels at least as early as 6 days postinfection and that these integrated forms were probably derived from at least one of two types of linear precursor. Evidence for the production of abundant linear-DNA precursors to integrated DNA in the infected liver is presented.
MATERIALS AND METHODS
Viruses and infected ducklings.
The strain of DHBV used in these experiments was DHBV16 (18). A mutant genome of DHBV16, I2, containing an insertion of two bp in the r sequence, has been described (33). Ducklings infected with wild-type virus or with the I2 mutant were described in a previous study (33), and livers from these animals were used for this study.
Inverse-PCR assay for virus-cell junctions.
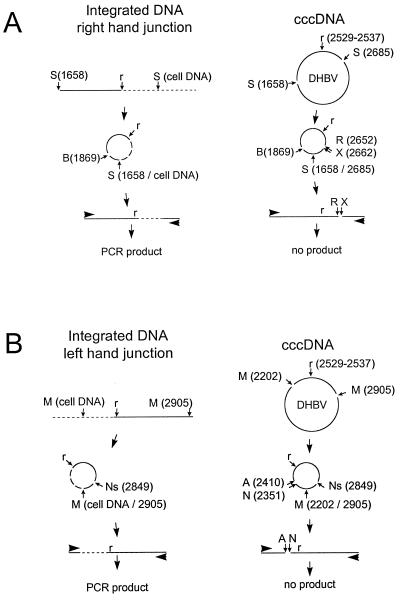
Measurements of the frequency of integration of viral DNA and the mapping of virus-cell junctions were performed by an inverse-PCR strategy similar to that described by Gong et al., as illustrated in Fig. 2 (11). DNA from infected liver was isolated by phenol extraction without any prior protease digestion. This procedure eliminates the viral-DNA replicative intermediates by extraction into the phenol phase or interface, producing a purified fraction containing high-molecular-weight chromosomal DNA as well as nonchromosomal forms, such as mitochondrial DNA and viral covalently closed circular DNA (cccDNA) (27). We refer to this fraction as “cellular DNA” even though it also contains viral cccDNA measured at approximately 1 part in 25,000 (data not shown). The inverse-PCR method we used was designed to amplify specific regions of viral DNA that were linked to nonviral sequences. Two reactions separately amplified regions covering the ends of in situ-primed linear DNA (26) that were linked either upstream or downstream to nonviral sequences. For convenience, we call the virus-cell junctions in which the cellular DNA is upstream of the small repeat sequence r the left-hand junctions (Fig. 1), while those in which the cellular sequences were downstream of r are called the right-hand junctions.
FIG. 2.
Inverse-nested-PCR amplification of virus-cell junctions. The strategies used for the detection of right-hand (A) and left-hand (B) virus-cell junctions are shown. The right and left ends refer to the two ends of linear DNAs, as defined in Fig. 1. The failure of the more abundant cccDNA species present in every infected cell to yield a PCR product according to these two strategies is illustrated on the right. (A) Cellular DNA was digested with Sau3AI (S) at position 1658 and at an adjacent position in covalently linked cellular DNA (left) or at 1658 and 2685 in cccDNA (right). The circular products produced by ligation were then digested with BspHI (B) at position 1869, and the linear products produced from cccDNA (right) were selectively cleaved with EcoRV (R) and XbaI (X) before PCR. (B) Cellular DNA was digested with MspI at position 2905 and at adjacent positions in covalently linked cellular DNA (left) or at 2202 in cccDNA (right). The circular products produced by ligation were then digested with NsiI (Ns) at position 2849, and the linear products produced from cccDNA (right) were selectively cleaved with NcoI (N) and AvaI (A) before PCR. A first round of PCR was followed by a second round with nested primers (not shown). The 9-nucleotide repeated sequence found at either end of minus strand DNA is indicated (r).
Cellular DNA was digested with the restriction enzyme Sau3AI, which recognizes a frequently occurring tetranucleotide sequence, GATC, or with MspI, whose recognition site is CCGG. Each digested DNA was diluted to approximately 1 μg/ml and treated with 40 U of T4 DNA ligase/ml overnight at 10°C. At low DNA concentration, this step resulted in circularization of the fragments through intramolecular ligation of their ends (2). Fragments containing a virus-cell junction derived from integrated viral DNAs would be expected to circularize through ligation of a Sau3AI or MspI end in the viral DNA with the corresponding end located in the adjacent cellular sequence, while all other viral sequences would circularize through two homologous sites located in the viral genome. Among all potential fragments containing virus-cell junctions, we selectively amplified those in which a recombination joint occurred in the vicinity of the small repeat sequence, r, which is found at either end of in situ-primed linear DNA.
To selectively amplify the right-hand joints in this region, we first digested the religated Sau3AI fragments with EcoRV and XbaI, which cleave at positions 2652 and 2662, respectively, on the viral genome, approximately 100 nucleotides downstream from r (ending at 2537), then with BspHI, which cleaves at position 1869. After cleavage, the DNA was diluted and subjected to PCR with primers RF1 (2093 to 2123) and RR1 (1778 to 1755). This primer set will only amplify template molecules that have the primer binding sites but have not been cleaved by either EcoRV or XbaI. This requirement prevented all cccDNA-derived Sau3AI fragments from acting as templates (Fig. 2A). A similar strategy was employed to selectively amplify the left-hand virus-cell junctions, starting with the MspI fragments, as illustrated in Fig. 2B. In this case, digestions with NcoI and AvaI were used to prevent cccDNA-derived MspI fragments from serving as templates for PCR. After an additional digestion with NsiI, intact linear DNAs were amplified with primers LF1 (2851 to 2873) and LR1 (2843 to 2820).
Using the inverse-PCR strategy, we performed amplification reactions on amounts of cellular DNA that yielded products in only a fraction of the individual reactions. To obtain visible products from such endpoint dilutions required a second set of nested primers to amplify 1 μl of the products of the first reactions (1/30 of the total) with an additional 35 cycles of amplification. For amplification of the left-hand junction fragments, we used primers LF2 (5′ biotin 2877 to 2900) and LR2 (2698 to 2675), and for the right-hand junction fragments the primers were RF2 (2217 to 2240) and RR2 (5′ biotin 1732 to 1709). All amplification reactions were carried out in a volume of 30 μl of reaction mixture containing 2.5 mM MgCl2, 15 pmol of each primer, and 1.5 U of Taq DNA polymerase (Promega catalog no. M1661) in a reaction buffer of the supplier (Promega). Each amplification cycle consisted of a 45-s denaturation at 94°C, a 60-s annealing at 55°C, and a 90 s elongation at 72°C. Direct sequencing of the biotinylated product from the second PCR was carried out as previously described (33).
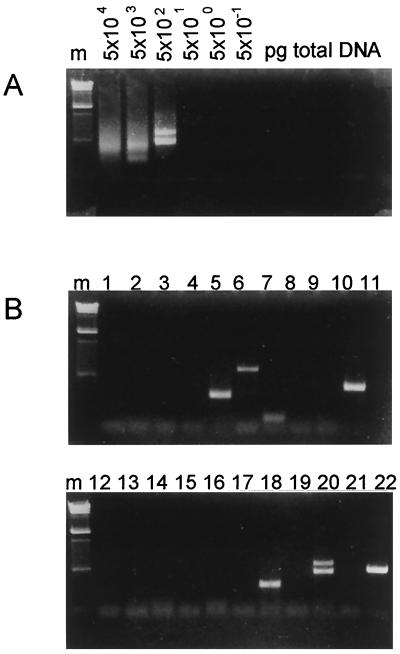
The advantages of the endpoint dilution method, which yielded products derived from a single template molecule in each positive reaction (see below), were threefold. First, since each product was amplified in the absence of any competing fragment, the collection of products obtained in individual reactions was unbiased with respect to their relative efficiencies of amplification. Only fragments that could not be amplified sufficiently to yield a detectable product, even in the absence of competitor, were not detected in the analysis. For example, virus-cell junction fragments containing long cellular sequences (>1,000 kb) would not be expected to be amplified. Second, since each product was clonal, PCR products could be sequenced directly to locate the position of the virus-cell junction. Finally, the amount of cellular DNA template in each reaction at the endpoint dilution, and the frequency of occurrence of virus-cell junctions in reactions performed on endpoint dilutions, allowed an estimate of the frequency of occurrence of amplifiable virus-cell junctions in the liver. An example of one such set of endpoint dilution PCRs for the detection of right-hand junctions is shown in Fig. 3.
FIG. 3.
Inverse nested PCR of right-hand virus-cell junctions. Determination of the limiting dilution for inverse nested PCR of cellular DNA (A) and the products from 22 individual limiting dilution amplifications (B) are shown. (A) Cellular DNA from wild-type-virus-infected duck 10 was processed as described in the legend to Fig. 2A and added to individual amplification reaction mixtures in the amounts indicated. After two rounds of PCR with nested primers, the products were analyzed by electrophoresis through a 1.5% agarose gel and stained with ethidium bromide. The absence of products in the reactions containing less than 500 pg of cellular DNA indicates that amplifiable junction fragments were not present in these dilutions. The presence of heterogeneous products in reactions containing more than 500 pg of cellular DNA indicates that multiple junction fragments were present. The two discrete products in the 500-pg reaction indicate that two junction fragments were present. (B) Twenty-two individual reactions (indicated above the lanes), each containing 100 pg of cellular DNA from the reaction shown in panel A, were assayed for the presence of discrete products. Nine unique products were produced in eight of the reactions. Therefore, the frequency of junction fragments was nine per 2.2 ng of cellular DNA. The PCR products were characterized by direct sequencing. The product from reaction 20 was unsuitable for sequencing because of the mixed product. Lane m, molecular weight markers of bacteriophage λ DNA digested with HindIII.
Validation of the single-molecule amplification.
In order to confirm that single template molecules were amplified as single sequence clones, we performed an experiment to see if nested PCR of appropriately diluted samples could produce the expected unique sequences known to be present in a mixture of two template sequences. Equal amounts of plasmids containing either cloned DHBV3 (25) or DHBV16 (18) sequences were diluted so that individual nested PCR produced single bands in only a fraction of the reactions. Forty reactions containing an amount of plasmid DNA calculated to be equivalent to 1.2 molecules per reaction (0.6 molecules for each strain) yielded eight reactions with products and 32 reactions with no product. Direct sequencing of each of the eight products generated unique sequence ladders characteristic of either DHBV3 (five of eight) or DHBV16 (three of eight). From this result we concluded that single templates were efficiently amplified to produce products and that products obtained by amplification of samples at endpoint dilution were derived from single templates.
Controls for artifacts.
Because the molecules we characterized as naturally occurring recombination joints could not be confirmed by any method with sensitivity comparable to PCR, we performed a series of control experiments to rule out artifacts that could produce PCR products with sequences resembling recombination joints. The potential known artifacts that we investigated included (i) T4-ligase-mediated in vitro joining of cellular DNA ends with ends of contaminating viral DNA and (ii) template switching during PCR. Both of these reactions would produce apparent recombination sites that were not located at a Sau3AI site.
Since in vitro ligation of cellular and viral DNA ends would require that one of the two ligation partners bear a 5′ phosphate end, we tested whether dephosphorylation of the sample prior to Sau3AI digestion influenced the frequency or type of products that appeared following nested PCR. Treatment of 10 μg of DNA from infected cells with 30 U of calf intestinal alkaline phosphatase at 37°C for 1 h (enzyme and reaction buffer from New England Biolabs [catalog no. 290S]) prior to Sau3AI digestion, dilution, and ligation did not result in any significant reduction in the frequency of PCR products derived from amplification reactions on 250 pg of treated or untreated samples (19 and 6 products per 20 reactions, respectively). Sequencing of the products from both the treated and untreated samples showed no pattern of differences in the structures of the recombination joints. From this result we concluded that the reaction products did not depend on templates produced during in vitro ligation.
In order to test for the possibility of template switching during the nested PCR, we used as a template a dimer-containing plasmid DNA that had been cut at multiple sites with Sau3AI and at the unique site with AlfII. After denaturation, this DNA sample contained a cleavage fragment with a structure similar to that of nascent virus minus strands, i.e., with the 5′ end located close to the AlfII site and the other end located at an upstream Sau3AI site. Intramolecular template switching on nascent viral minus strands during PCR could produce joints that resembled the recombination joints we detected in the infected-cell DNA. Using the nested primer set for inverse PCR (RF1-RR1 and RF2-RR2), we amplified various amounts of plasmid template and assayed for the expected-size PCR product. Only at high template concentrations, i.e., at 60,000 to 120,000 times that present in our reactions, did we observe any PCR product whatsoever. Although this experiment suggests that template switching may occur if the template molecule is discontinuous, the frequency of occurrence was at least 4 orders of magnitude below that required to contribute to generating the products that we characterized as recombination joints.
Finally, we tested whether nonintegrated viral sequences present in the sample could participate in an unforeseen way in generating the putative recombination joints. For this control, we added 1.5 ng of plasmid containing cloned DHBV3 DNA to 5 μg of a cellular-DNA sample from uninfected or infected birds prior to Sau3AI digestion and carried out the entire assay for right-hand joints as usual, using inverse PCR with nested primers. This ratio of plasmid to cellular DNA represents about seven times the abundance of viral cccDNA in the DNA from infected livers. The amplifiable plasmid-derived fragment in this assay would be expected to be cleaved by the EcoRV and XbaI after ligation and therefore to be unavailable for use as a template. We asked whether any other unforeseen reactions with the DHBV3 DNA produced apparent recombination joints. The data from this experiment are summarized in Table 1.
TABLE 1.
Artificial joints formed in vitro during inverse nested PCR
| Source of cell DNA | Cell DNA (pg)a | DHBV3 DNA (pg)a | No. of reactions | DHBV16 productsb | DHBV16 artifactsc | DHBV3 productsc | DHBV3 artifactsc |
|---|---|---|---|---|---|---|---|
| Uninfected liver | 200 | 0.06 | 30 | 0 | 0 | 15 | 15 |
| Infected liver | 50 | 0.015 | 96 | 4 | 1 | 22 | 22 |
Amount of cellular or plasmid DNA per reaction.
Total products sequenced that contained DHBV16 or DHBV3 DNA.
Joints due to ligation in vitro; see the text for detailed explanation.
Amplification of 124 samples containing uninfected or infected cellular DNA mixed with DHBV3 DNA (50 to 200 pg total DNA per reaction) yielded 37 single bands derived from DHBV3 DNA. Sequencing of these single bands revealed that two types of artifacts contributed to apparent recombinations joints, and both of these artifacts were easily recognized. One type of product was amplified due to a failure of either the EcoRV or XbaI sites to be cleaved (11 examples). Such molecules, when observed in the experiments, were considered as background and excluded from the analyses. A second type of product we observed was apparently due to a “star” activity of Sau3AI, i.e., cleavage at Sau3AI-like sites located at position 2567 (TATC; 12 examples), 2632 (GATT; 10 examples), or 2650 (GATA; 4 examples) and subsequent ligation of these ends to authentic Sau3AI ends in cellular or viral DNA in a way that allowed circularization and amplification. Of four DHBV16-containing sequences obtained from the infected-cell DNA sample, only one recombination joint was due to an apparent in vitro ligation, at the Sau3AI-like site located at position 2632, while the remaining three recombination joints were located at unrelated sites.
From this experiment, combined with the other controls, we concluded that artificial recombination joints were generated only at authentic Sau3AI or Sau3AI-like sites and that virus-cell junctions occurring at other positions on the DHBV genome were probably the results of in vivo recombination. Recombination joints occurring at Sau3AI-like sites were occasionally seen in the experiments and were excluded from the analysis as probable artifacts of in vitro ligation.
RESULTS
Infected ducks.
Two ducklings were infected at 4 days of age with 5 × 109 virus particles each and monitored for 6 days postinfection, at which time the ducklings were sacrificed and liver tissue was frozen. Peak viremia appeared at 4 days postinfection, and the liver was positive for viral DNA at 6 days postinfection. The virological assays of these two birds have been reported previously (see Fig. 5 in reference 33). Cellular DNA was extracted from the livers of the birds and assayed for virus-cell junctions by inverse nested PCR.
Frequency of occurrence of virus-cell junctions.
Left- and right-hand-junction fragments were amplified by similar strategies, as described in Materials and Methods. As outlined in Table 2 (WT rows), a total of 390 ng of cellular DNA from bird 10 and 55 ng from bird 14 was subjected to amplification reactions specific for right-hand junctions, and 6 ng from bird 10 was subjected to amplifications specific for the left-hand junctions. Right-hand-junction-specific reactions yielded a total of 60 unique products for bird 10 and 168 products for bird 14, while 140 products were produced from left-hand-junction-specific reactions in bird 10.
TABLE 2.
Inverse-nested-PCR products from infected livers
| Virusa | Bird no. | Total DNA assayed (μg) | Type of junctionb | Total no. of reactions | Total products | No. of virus-virus junctions | No. of virus-cell junctions | Frequency of integration (per 104 cells) |
|---|---|---|---|---|---|---|---|---|
| WT | 10 | 390 | R | 156 | 60 | 26 | 9 | 0.7 |
| WT | 14 | 55 | R | 300 | 168 | 45 | 11 | 2 |
| WT | 10 | 6 | L | 192 | 140 | 72 | 4 | 6 |
| I2 | 6 | 23 | R | 96 | 48 | 23 | 3 | 4 |
| I2 | 6 | 3 | L | 96 | 54 | 30 | 1 | 10 |
WT, wild type.
R, right-hand junction; L, left-hand junction.
Only a minority of viral recombination joints amplified in these reactions were apparently from virus-cell recombination events. Approximately 55% of all PCR products could not be positively identified by sequencing due to either (i) the presence of multiple bands in one reaction (42%), (ii) failure of the sequencing reaction (8%), or (iii) proximity of the recombination joint to the sequencing primer (4%). Sequencing information obtained from the remainder of these products to locate the position of the viral recombination joint and the nature of the joined sequences revealed that most recombination joints resulted from joining with other viral DNA sequences at various positions and either in direct or inverted orientation. Because some nonhomologous virus-virus recombinations resulted in the deletion of the EcoRV and XbaI sites (2652 and 2662, respectively), they were able to be amplified under the conditions employed for detecting the right-hand virus-cell junctions. Similarly, virus-virus junctions resulting from recombinations that deleted the AvaI and NcoI sites were able to be amplified under the conditions employed for detecting the left-hand virus-cell junctions. We have previously described the occurrence of such recombination joints in both DHBV-infected hepatocytes and in WHV-infected woodchuck liver (31, 32), concluding that such joints were formed by nonhomologous inter- and intramolecular recombination of the viral linear DNAs shown in Fig. 1. The abundance of these molecules in our assays suggests that linear-DNA molecules were present and available as recombination substrates during the short-term in vivo infection with wild-type DHBV.
Putative virus-cell junctions were readily detected (20 recombination joints) among the products of amplification of right-hand junctions; however, only four examples were found among the left-hand junctions. These junctions were identified by the nonviral nature of the sequence joined to viral DNA, and 23 of 24 tested from this and later experiments were confirmed to contain duck genomic DNA by hybridization to duck DNA on Southern blots or by PCR amplification of duck genomic sequences with specific primers designed from the known sequence (data not shown). The nature of the duck cellular sequences was not further investigated. The frequency of virus-cell junctions calculated from the number of sequenced right-hand joints was in the range of one in approximately 103 to 104 cells. A frequency of one joint in approximately 103 cells was calculated from left-hand virus-cell junctions, although this calculation was based on the detection of only four junctions. These numbers must represent a minimum frequency, since some junction fragments were probably not amplified due to their sizes and not all PCR products were identified in these experiments, e.g., reactions with multiple bands were not sequenced.
Distribution of virus-virus and virus-cell recombination joints.
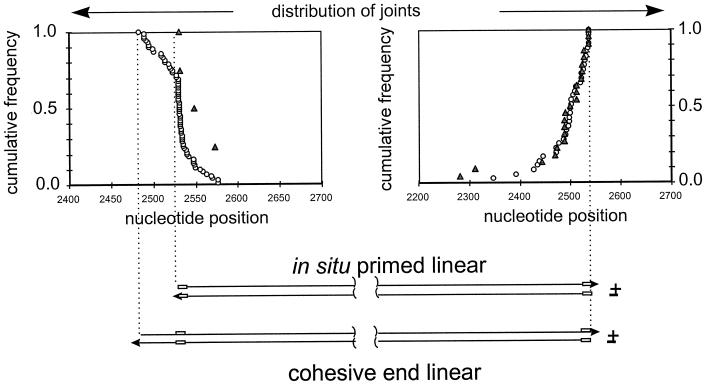
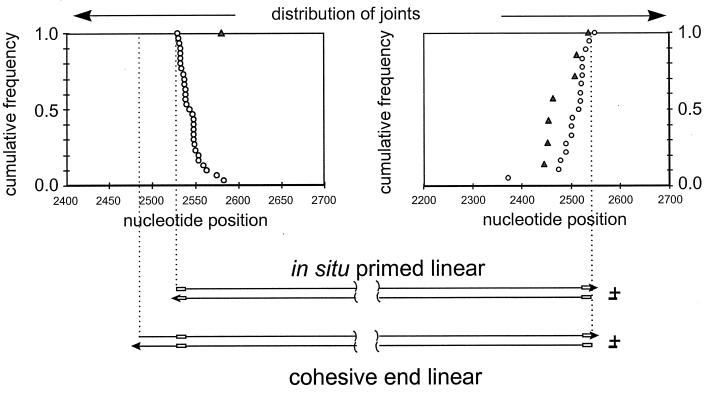
To examine whether virus-cell junctions appeared to be derived from linear viral-DNA precursors, we determined the cumulative distribution of virus-cell recombination joints in the vicinity of the two ends of the various forms of linear DNA. We compared these distributions to those seen in the virus-virus joints, presumed to be derived from linear DNA precursors. The results for both the right-hand and left-hand junctions are shown in Fig. 4. The distribution of 22 right-hand virus-cell junctions and a representative sample of 35 right-hand virus-virus junctions revealed that all recombinations (cumulative frequency = 1.0) within the viral sequences occurred to one side of a position corresponding to the right-hand end of the three linear DNAs (right-hand graph). Because the distribution of the virus-virus junctions was coincident with that of the virus-cell junctions (Fig. 4), we concluded that the two types of recombinants were derived from the same population of linear DNAs, at least with respect to the right-hand ends. This result suggests that linear DNA was the recombination substrate for integration into cellular DNA.
FIG. 4.
Distribution of virus-virus and virus-cell junctions in the livers of wild-type-virus-infected ducklings. The positions of the recombination joints derived from the two ends of viral DNA are shown. Joints in which viral sequences were colinear with either end of linear DNA were amplified by the reaction specific for that end, as shown in Fig. 2, and sequenced, and their cumulative distributions are shown relative to the ends of two types of linear DNA (bottom). A value of 1.0 indicates that 100% of all recombination joints were located to one side of the corresponding position on the genome. Symbols: ○, virus-virus junctions; ▵, virus-cell junctions. The r sequence is indicated by a box in the representations of linear DNAs.
The distribution of the 4 left-hand virus-cell junctions in comparison with that of 71 left-hand virus-virus junctions is shown in the left-hand graph of Fig. 4. As reported previously for WHV-infected liver, the distribution of left-hand virus-virus junctions suggested that two forms of linear DNA served as recombination substrates, corresponding to the in situ-primed and cohesive-end linear DNAs. The distinction between these two forms is seen as a break occurring at nucleotide 2526 in the cumulative distribution curve. This position corresponds to the left-hand end of in situ-primed linear DNA. The distribution ends at position 2489, the left-hand end of the hypothetical precursor, cohesive-end linear DNA. The yield of left-hand virus-cell junctions in our experiments was not sufficient to establish a high-resolution frequency distribution curve for comparison with that of the virus-virus junctions, but the positions of these four joints are consistent with either of the two linear DNA forms being the precursor for integration.
Effect of excess production of in situ-primed linear DNA.
Virus-cell junctions appeared to be derived from linear forms of viral DNA, which existed in at least two forms in DHBV-infected liver, judging from the distribution of virus-virus recombination joints. To determine the effect of production of excess linear DNA over relaxed circular DNA (rcDNA) on viral DNA integration, we used a mutant of DHBV, I2, that produced an amount of in situ-primed linear DNA estimated to be 5- to 10-fold higher than that produced by wild-type virus (50% of the total). Two ducklings were infected at 4 days of age with 5 × 109 I2 virus particles each and monitored for 6 days postinfection, at which time the ducklings were sacrificed and liver tissue was frozen. Peak viremia appeared at 4 days postinfection, and the liver was positive for viral DNA at 6 days postinfection. The virological assays of these two birds have also been reported previously (Fig. 5 in reference 33).
Cellular DNA was extracted from the liver of one of these birds, and limiting dilutions were subjected to PCR to isolate and sequence virus-virus and virus-cell junctions from both the left- and right-hand ends. The results are summarized in Table 2 (I2 rows). The frequency of amplification of right-hand virus-cell and virus-virus junctions was not significantly different in the I2 infection from that observed in the parallel wild-type DHBV infection, and at 6 days postinfection we estimated that right-hand joints could be detected in approximately 1 in 4 × 103 cell genome equivalents. Although 54 virus-virus junctions mapping to the left-hand end of the linear DNA were amplified, only 1 left-hand virus-cell junction was detected. This frequency is similar to that seen in the wild-type-virus-infected liver DNA, but the number of virus-cell junctions in both wild-type-virus- and I2-infected livers is too small for accurate comparison.
The distribution of virus-virus and virus-cell junctions mapping to both ends of the linear DNA in I2-infected birds is shown in Fig. 5. In general, the distribution of recombination joints mapping to the right-hand end is similar to that seen in wild-type-virus infection, supporting the conclusion that linear DNA was a significant integration substrate. The distribution of virus-virus junctions mapping to the left-hand end of linear DNA is dominated by the apparent contribution of the in situ-primed linear DNA end, consistent with the elevated production of this form of DNA and the reduced production of rcDNA, the precursor of the hypothetical cohesive-end linear DNA. The location of the single virus-cell junction mapping to this end of linear DNA is also consistent with a linear DNA precursor.
FIG. 5.
Distribution of virus-virus and virus-cell junctions in the liver of a duckling infected with the I2 virus. The positions of the recombination joints derived from the two ends of viral DNA are shown. Joints in which viral sequences were colinear with either end of linear DNA were amplified by the reaction specific for that end, as shown in Fig. 2, and sequenced, and their cumulative distributions are shown relative to the ends of two types of linear DNA (bottom). A value of 1.0 indicates that 100% of all recombination joints were located to one side of the corresponding position on the genome. The symbols are defined in the legend to Fig. 4.
DISCUSSION
The data we have presented support the conclusion that linear hepadnaviral DNA produced during the course of a short-term infection undergoes intermolecular recombination events resulting in the joining of viral sequences to the DNA of infected cells. We believe that these joining reactions represent integration events that result in the insertion of viral sequences into one or more chromosomes in the infected cell. On the basis of these experiments, we cannot exclude the possibility that some virus-cell junctions were ultimately derived from template switching to a cellular RNA during reverse transcription of viral RNA, as occurs commonly with retroviruses; however, the evidence favors the conclusion that the joints detected resulted from recombination at the DNA level, since (i) virus-cell joints were colinear with one of two forms of linear viral DNA and (ii) alterations in the ratio of linear to rcDNA forms correlated with an alteration of the sites of virus-virus recombination, which codistribute with the virus-cell junctions (compare Fig. 4 and 5). Moreover, stable integration of hepadnaviral DNA into the DNA of liver cells has been well documented (3, 4), while transduction of cellular sequences by hepadnaviruses has never been described. The frequency of putative viral integrations that resulted from inoculation and spread of infection throughout the liver during a period of 6 days was estimated to be, on average, at least one per 103 to 104 cells. We do not know whether these recombination events occurred in separate cells or if rare cells produced many recombination events.
The inference that linear DNAs were precursors to the integrated DNAs is supported by circumstantial evidence, namely, that the distributions of recombination sites on the viral genome were colinear with either end of the linear-DNA molecular map. This distribution corresponded closely with the distribution of virus-virus junctions that are known to be the result of nonhomologous recombination between the two ends of linear-DNA molecules (31–33). Moreover, no right-hand virus-cell junctions retained sequence continuity through position 2537 (the right-hand end of both linear DNAs) as might result if molecules other than the linear DNAs described were integration substrates. This finding suggests that linear DNA was the predominant integration substrate in this short-term infection.
The existence of two types of linear-DNA substrate for recombination, differing in the positions of the left-hand ends, was indicated by the distribution of the left-hand recombination joints from wild-type-virus-infected liver (Fig. 4). The left-hand end of one putative linear-DNA substrate corresponded to the 3′ end of the minus strand, consistent with a linear-DNA molecule formed by in situ priming of plus-strand synthesis. Seventy-four percent of all the left-hand virus-virus junctions were located to the right of this position. The remaining 26% of left-hand virus-virus junctions were located in the short region to the left referred to as the cohesive end region, between the 3′ end of the minus strand and the 5′ end of the plus strand at nucleotide 2485. These recombination joints can best be explained by a cohesive-end linear-DNA precursor formed by displacement of the cohesive ends of rcDNA by strand elongation. A strikingly similar distribution of left-hand recombination joints was observed in cccDNA isolated from livers chronically infected with WHV, indicating that this hypothetical precursor, cohesive-end linear DNA, may be a common product or intermediate in hepadnavirus replication (32).
In contrast, in DNA extracted from I2-infected livers, no virus-virus junctions that could be exclusively assigned to cohesive-end linear precursors were detected (Fig. 5). This result correlates well with the reduction of rcDNA in favor of in situ-primed linear DNA forms in livers infected with the I2 mutant, consistent with the assignment of these two forms of linear DNA as recombination substrates. Among all infected birds, only five left-hand virus-cell junctions were identified. Left-hand junctions were less easily detected than right-hand junctions because of a higher background of virus-virus joints and because the enzyme used for digestion of the integrated DNA, MspI, cut relatively infrequently in cellular DNA compared with Sau3AI, which was used for digestion of the right-hand junctions. Nevertheless, the positions of these recombination joints were consistent with either in situ-primed or cohesive-end linear precursors. None of the virus-cell recombination joints sequenced in any of the experiments appeared to be derived from linear DNA produced by illegitimate replication.
The frequency of nonhomologous right-hand virus-virus junctions in the wild-type-virus-infected livers exceeded that of virus-cell junctions by an average of 3.6-fold among the different sets of amplifications. These junctions are grossly underestimated, since such junctions were heavily selected against by the assay. Only virus-virus junctions in which intermolecular joining occurred in the antiparallel orientation, or in which joining resulted in a substantial deletion of sequences from one end of the linear precursor, would have been capable of being amplified in our assays. Intermolecular joining and large deletions were previously found to be infrequent events in nonhomologous recombination between the ends of linear DHBV DNA. In any case, the presence of a large excess of virus-virus recombination joints indicates that virus-cell recombination was a minor fraction of recombination events involving linear viral DNA. Moreover, enhancement of the production of in situ-primed linear DNA did not noticeably increase the frequency of virus-cell junctions, as has been reported to occur in DHBV-expressing chicken hepatoma cells (10). The results suggest that the availability of in situ-primed linear viral-DNA substrates is not essential for viral-DNA integration because rcDNA may provide additional linear substrates in the form of cohesive-end linear molecules with comparable efficiency. We speculate that the availability of cellular-DNA ends, resulting from DNA damage or replication, may be one of the major factors determining the frequency of integration, since DNA ends appear to be highly recombinogenic in hepatocytes. An effect of DNA damage on the integration frequency in a DHBV-expressing cell line, D2, has recently been reported (22).
Presumably, integration of linear DNA into cellular DNA requires that linear DNA be imported into the nucleus of the infected cell. Such nuclear import of viral DNA is known to occur in the phase of cccDNA amplification during the initiation of infection (29, 30). Moreover, conversion of linear DNA into cccDNA is regulated by the pre-S envelope protein, which prevents cccDNA synthesis by directing nucleocapsids containing mature linear or rcDNA into a pathway for assembly and secretion of enveloped virus particles (13, 28, 31). Thus, we might infer that the viral-DNA integration is regulated by the pre-S protein and may normally be limited to the initiation phase of infection. Moreover, because linear-DNA molecules efficiently undergo intramolecular recombination to produce cccDNA (31), linear-DNA substrates for integration are expected to exist only transiently in the nucleus. Therefore, opportunities for viral-DNA integration beyond the early phase of infection would require continued import of viral DNA into the nucleus and would depend on new rounds of infection or on loss and replacement of cccDNA by cell turnover or other mechanisms.
Although hepatocellular carcinoma does not appear to be a common result of infection with DHBV, other hepadnaviruses are strongly oncogenic in the liver (4). These include the human and the woodchuck hepadnaviruses, HBV and WHV, respectively. Hepatocellular carcinomas associated with these infections commonly contain integrated hepadnavirus DNA, indicating that viral-DNA integration occurred in the precursor cells that gave rise to the tumors. Moreover, 20 to 41% of WHV-induced hepatocellular carcinomas contain WHV DNA integrated in the vicinity of the N-myc1 or N-myc2 genes, resulting in activated transcription from this proto-oncogene (7, 12). Our studies suggest that integration of linear viral-DNA forms may occur preferentially during early phases of infection or during periods of extensive cell turnover and reinfection, when linear viral DNA is imported into the nucleus. In addition, viral-DNA integration may be enhanced during these periods by the availability of DNA ends resulting from DNA replication or from DNA damage (22). Thus, the production of conditions in the liver favoring viral-DNA integration and insertional mutagenesis may partially explain how chronic inflammation and regeneration increases the risk for hepatocellular carcinoma in chronic hepadnavirus infection.
ACKNOWLEDGMENTS
We thank C. J. Ramey for excellent technical assistance.
This work was supported by HHS grant CA-42542.
REFERENCES
- 1.Baumert T F, Marrone A, Vergalla J, Liang T J. Naturally occurring mutations define a novel function of the hepatitis B virus core promoter in core protein expression. J Virol. 1998;72:6785–6795. doi: 10.1128/jvi.72.8.6785-6795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckel-Mitchener A, Summers J. A novel transcriptional element in the duck hepatitis B virus that is involved in 3′ end formation of viral RNA. J Virol. 1997;71:7917–7922. doi: 10.1128/jvi.71.10.7917-7922.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buendia M A. Hepatitis B viruses and hepatocellular carcinoma. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 4.Buendia M A. Mammalian hepatitis B viruses and primary liver cancer. Semin Cancer Biol. 1992;3:309–320. [PubMed] [Google Scholar]
- 5.Bonino F, Rosina F, Rizzetto M, Rizzi R, Chiaberge E, Tardanico R, Callea F, Verme G. Chronic hepatitis in HBsAg carriers with serum HBV-DNA and anti-HBe. Gastroenterology. 1986;90:1268–1273. doi: 10.1016/0016-5085(86)90395-1. [DOI] [PubMed] [Google Scholar]
- 6.Chisari F V, Ferrari C. Hepatitis B virus immunopathology. Springer Semin Immunopathol. 1995;17:261–281. doi: 10.1007/BF00196169. [DOI] [PubMed] [Google Scholar]
- 7.Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia M A. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 8.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 9.Gong S S, Jensen A D, Rogler C E. Loss and acquisition of duck hepatitis B virus integrations in lineages of LMH-D2 chicken hepatoma cells. J Virol. 1996;70:2000–2007. doi: 10.1128/jvi.70.3.2000-2007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong S S, Jensen A D, Chang C J, Rogler C E. Double-stranded linear duck hepatitis B virus (DHBV) stably integrates at a higher frequency than wild-type DHBV in LMH chicken hepatoma cells. J Virol. 1999;73:1492–1502. doi: 10.1128/jvi.73.2.1492-1502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong S S, Jensen A D, Wang H, Rogler C E. Duck hepatitis B virus integrations in LMH chicken hepatoma cells: identification and characterization of new episomally derived integrations. J Virol. 1995;69:8102–8108. doi: 10.1128/jvi.69.12.8102-8108.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen L J, Tennant B C, Seeger C, Ganem D. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol. 1993;13:659–667. doi: 10.1128/mcb.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenhoff R, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenhoff R, Summers J. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol. 1994;68:5706–5713. doi: 10.1128/jvi.68.9.5706-5713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenhoff R, Luscombe C A, Summers J. Acute liver injury following infection with a cytopathic strain of duck hepatitis B virus. Hepatology. 1999;29:563–571. doi: 10.1002/hep.510290236. [DOI] [PubMed] [Google Scholar]
- 16.Lenhoff R L, Luscombe C A, Summers J. Competition in vivo between a cytopathic variant and a wild type duck hepatitis B virus. Virology. 1998;251:86–95. doi: 10.1006/viro.1998.9394. [DOI] [PubMed] [Google Scholar]
- 17.Loeb D, Hirsch R, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason W S, Seal G, Summers J. A virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriyama K, Okamoto H, Tsuda F, Mayumi M. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology. 1996;226:269–280. doi: 10.1006/viro.1996.0655. [DOI] [PubMed] [Google Scholar]
- 21.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 22.Petersen J, Dandri M, Burkle A, Zhang L, Rogler C E. Increase in the frequency of hepadnavirus DNA integrations by oxidative DNA damage and inhibition of DNA repair. J Virol. 1997;71:5455–5463. doi: 10.1128/jvi.71.7.5455-5463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 24.Scaglioni P P, Melegari M, Wands J R. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology. 1997;233:374–381. doi: 10.1006/viro.1997.8594. [DOI] [PubMed] [Google Scholar]
- 25.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 26.Staprans S, Loeb D, Ganem D. Mutations affecting hepadnavirus plus-strand synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers J, Smith P, Horwich A L. Hepadnaviral envelope proteins regulate amplification of covalently closed circular DNA. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summers J, Smith P, Huang M, Yu M. Regulatory and morphogenetic effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuttleman J, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 30.Wu T-T, Coates L, Aldrich C E, Summers J, Mason W S. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Summers J. Illegitimate replication of linear hepadnaviral DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Summers J. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J Virol. 1998;72:8710–8717. doi: 10.1128/jvi.72.11.8710-8717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]