Abstract
The herpes simplex virus (HSV) proteins VP16 and ICP0 play key roles in stimulating the onset of the viral lytic cycle. We sought to explore the regulatory links between these proteins by studying the phenotypes of viral mutants in which the activation functions of both were simultaneously inactivated. This analysis unexpectedly revealed that truncation of the C-terminal transcriptional activation domain of VP16 (allele V422) in an ICP0-deficient background almost completely eliminated immediate-early gene expression and virus replication in Vero and HEL cells. The doubly mutant viral genome persisted in a quiescent state for at least 10 days in HEL cells infected at high multiplicity and could be reactivated by superinfection with wild-type HSV. In contrast, the in1814 VP16 mutation produced a markedly less severe phenotype in the same ICP0-deficient background. These data demonstrate that expression of the immediate-early genes requires ICP0 when the C-terminal activation domain of VP16 is deleted and raise the possibility that the in1814 form of VP16 retains a residual ability to stimulate gene expression during virus infection.
Herpes simplex virus type 1 (HSV-1) is a large nuclear DNA virus that induces both lytic and latent infections in its natural human host (60). HSV-1 gene expression occurs during lytic infection in three phases, termed immediate-early (IE), early, and late. The IE genes are the first to be transcribed, and the resulting IE polypeptides are essential for early- and late-gene expression (31). Transcription of the IE genes is coordinately activated by the virion protein VP16, which is targeted to IE promoters through the TAATGARAT (R = purine) element (10, 44, 55, 70, 71). VP16 is a 65-kDa phosphoprotein that is synthesized late in infection and packaged into the tegument of HSV-1 virions (41, 45, 49). It contains an extremely potent C-terminal transcriptional activation domain (62, 71, 72) and forms a complex with the cellular factors Oct-1 and HCF that binds TAATGARAT with high affinity (reviewed in references 51 and 60). VP16-induced activation of IE gene expression plays an important role in triggering the onset of the HSV-1 lytic cycle, as illustrated by the phenotypes of viral mutants encoding transactivation-deficient forms of VP16. For example, in1814, a mutant in which the ability of VP16 to form a complex with Oct1, HCF, and DNA is disrupted by an in-frame linker insertion, displays a greatly increased particle-to-PFU ratio and substantially reduced IE gene expression during infection (2). Truncation of the C-terminal acidic transcriptional activation domain produces a similar phenotype (66). These data demonstrate that VP16 greatly increases the probability that cells infected with a single HSV-1 virion enter the lytic cycle.
Four of the IE genes induced by VP16 encode nuclear phosphoproteins (ICP0, ICP4, ICP22, and ICP27) that act at a variety of levels to regulate IE-, early-, and late-gene expression (reviewed in reference 60). The IE protein ICP0 occupies an unusual position in this regulatory cascade, because it is required for efficient expression of the IE genes: ICP0 mutants display a greatly increased particle-to-PFU ratio and reduced levels of IE gene expression during infection (4, 5, 42, 43, 67, 68, 76), and ICP0 activates the expression of IE, early, and late genes in transient-transfection assays (7, 14, 24, 42, 52, 53, 59). Taken in combination, these observations indicate that VP16 is unable to fully activate IE gene expression in the absence ICP0. In this sense, the regulatory function of ICP0 appears to lie “upstream” of those of the other IE gene products.
The mechanism of action of ICP0 has yet to be precisely defined. ICP0 behaves as a promiscuous activator in transient-cotransfection assays, stimulating expression from a variety of HSV and heterologous promoters (reviewed in reference 22). Nuclear runoff transcription assays indicate that it acts at the transcriptional or pretranscriptional level (35, 64), and Lium et al. have shown that it contains a promoter-specific amino-terminal acidic transcriptional activation domain (42). ICP0 localizes to nuclear ND10 domains and disperses their constituent proteins (17, 47), an activity that correlates with activation function in mutational studies (20, 46). ICP0 also directly interacts with a variety of cellular proteins including the G1-phase cell cycle regulator cyclin D3 (37), translation elongation factor 1δ (36), and a ubiquitin-specific protease, HAUSP (19, 50). The association between ICP0 and HAUSP correlates with ICP0 function (18), suggesting a link to ubiquitin-mediated protein turnover pathways. Consistent with this view, ICP0 induces proteasome-dependent degradation of the catalytic subunit of the DNA-dependent protein kinase (39, 54), some isoforms of the ND10-associated PML protein (16), and the kinetochore binding protein CENP-C (15). Moreover, ICP0 activation function is blocked by proteasome inhibitors (21). These data have led to the emerging hypothesis that ICP0 acts by altering the stability of specific cellular proteins, thus enhancing the environment for HSV-1 replication (3, 16, 18). This hypothesis may explain how ICP0 plays a key role in the establishment and reactivation phases of latency (4, 8, 28, 40, 73, 77), as well as the lytic replication cycle.
Although VP16 and ICP0 appear to stimulate gene expression through very different mechanisms, viral mutants lacking VP16 or ICP0 activation function display a number of striking similarities, including (i) severely impaired replication at low multiplicities of infection (2, 61, 69), (ii) increased particle-to-PFU ratio (2, 61, 69), (iii) efficient growth on U2OS osteosarcoma cells (66, 76), and (iv) cell cycle-dependent variation in the severity of the mutant phenotype (6, 9). In addition, Ace et al. reported that the expression of ICP0 in trans at least partially complements the defect of a VP16 mutation (2). These findings suggest that the functions of VP16 and ICP0 are interlinked and/or overlap. One interpretation is that the primary physiological role of the transactivation function of VP16 is to stimulate expression of ICP0, which then suffices to activate the other IE genes. We sought to explore the regulatory links between VP16 and ICP0 by studying the phenotypes of viral mutants in which the activation functions of both proteins were simultaneously inactivated. This analysis unexpectedly revealed a major phenotypic difference between the in1814 and V422 VP16 alleles when these mutations were placed in an ICP0-deficient background and demonstrated that accumulation of IE RNAs is rendered almost completely dependent on ICP0 when the C-terminal activation domain of VP16 is deleted.
MATERIALS AND METHODS
Cells and viruses.
Vero and U2OS cells, obtained from the American Type Culture Collection, were maintained in Dulbecco’s minimal essential medium (DMEM) supplemented with 5 and 10% fetal bovine serum, respectively. Human embryonic lung fibroblasts (HEL cells) were generously provided by C. Spencer and maintained in DMEM–10% fetal bovine serum. HSV-1 KOS and 17syn+ were propagated on Vero cells. HSV-1 in1814 (2), V422 (38), n212 (7), KM100, and KM110 (see below) were propagated on U2OS cells in the presence of 3 mM hexamethylene bisacetamide (HMBA; Sigma).
Construction of recombinant viruses.
Two ICP0/VP16 double mutants that combine the n212 ICP0 mutation with either the in1814 or V422 VP16 mutation (KM100 and KM110, respectively) were constructed. U2OS cells were coinfected with each parental virus at a multiplicity of infection (MOI) of 5, and plaque-purified progeny were screened by Southern blot hybridization for the SpeI, BamHI, and NheI linkers that define the n212, in1814, and V422 mutations respectively. Doubly mutant recombinants were then plaque purified three times to obtain a final working stock.
Southern blot analysis.
Monolayers growing in wells of a six-well plate were infected with the indicated virus at an MOI of 10, and harvested 24 h postinfection directly into 1× lysis buffer (0.6% sodium dodecyl sulfate [SDS], 10 mM Tris [pH 7.5], 10 mM EDTA, 100 μg of proteinase K per ml). Following incubation at 37°C for 4 h, DNA was precipitated with 95% ethanol and resuspended in 10 mM Tris (pH 7.6)–1 mM EDTA. DNA was cleaved with the indicated restriction endonuclease, separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized to a 32P-labeled probe generated by random priming in ExpressHyb (Clontech) buffer as specified by the manufacturer. The VP16 probe was a 2.9-kb BamHI fragment spanning the entire VP16 open reading frame, and the ICP0 probe was a 0.5-kb XhoI-BamHI fragment internal to the ICP0 gene.
Northern blot analysis.
Cells growing in 100-mm dishes were infected at the indicated MOI. Where indicated, cycloheximide (100 μg/ml) was added 1 h prior to infection and maintained continuously. Total cellular RNA was extracted from infected monolayers by using Trizol (Gibco-BRL). Aliquots (5 μg) of RNA were prepared in MOPS buffer (20 mM morpholinepropanesulfonic acid [MOPS], 5 mM sodium acetate, 1 mM EDTA) containing 50% formamide and 20% formaldehyde, incubated at 55°C for 15 min, cooled on ice, and then loaded onto a 1% agarose gel containing 1× MOPS buffer, 2% formaldehyde, and 0.5 μg of ethidium bromide per ml. Following electrophoresis, RNA was transferred to a nylon membrane and hybridized as described above. Probes for ICP22 and ICP8 were 1.2- and 1.9-kb fragments, respectively, derived from the 5′ portions of these genes.
Western blot analysis.
Cellular lysates harvested directly into 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer were separated on 9% polyacrylamide gels, transferred to nitrocellulose membranes, and blocked with 5% skim milk in TBS-T (1× Tris-buffered saline, 0.2% Tween 20). The membranes were washed extensively in TBS-T and incubated with both primary and secondary antibodies for 30 min each in TBS-T. Protein was visualized by enhanced chemiluminescence (Gibco-BRL). VP16 was detected with a 1:5,000 dilution of LP1 (kindly provided by A. Minson), and VP5 was detected with a 1:20,000 dilution of NC-1 (kindly provided by G. H. Cohen and R. J. Eisenberg).
Purification of virions.
Roller bottles of U2OS cells were infected at an MOI of 1 in the presence of 3 mM HMBA. Cells were harvested 2 days postinfection, pelleted at 1,700 × g for 10 min, and resuspended in 1 mM sodium phosphate buffer (pH 7.4). The cells were Dounce homogenized on ice, and nuclei were pelleted at 3,000 × g for 5 min at 4°C. Supernatants were loaded onto dextran gradients and centrifuged at 50,000 × g for 1 h at 4°C. Linear dextran gradients were made by mixing solutions of dextran (Sigma) prepared in 1 mM phosphate buffer (pH 7.5) in a gradient maker (18.4 ml of the lighter solution [ρ = 1.04] was mixed with 17.6 ml of the denser solution [ρ = 1.09]). Banded virus was removed from the gradient, resuspended in serum-free DMEM, and pelleted by centrifugation at 78,000 × g for 2 h at 4°C.
RESULTS
Construction of HSV-1 ICP0/VP16 double mutants.
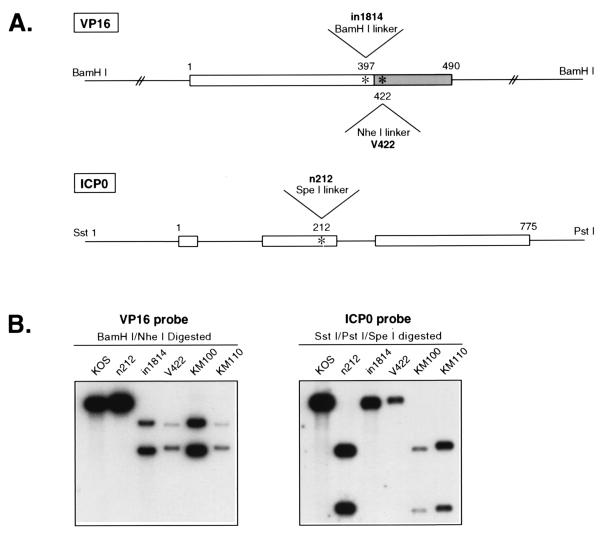
HSV-1 mutants bearing lesions in ICP0 and VP16 display similar multiplicity- and cell cycle-dependent defects in viral gene expression and are “complemented” to approximately the same extent by growth on U2OS osteosarcoma cells. Our initial objective was to explore the degree to which the functions of these two activators overlap. To this end, we constructed two ICP0/VP16 double mutants (Fig. 1). These isolates harbor the n212 ICP0 mutation (7) and the in1814 (2) or V422 (38) VP16 mutations (isolates KM100 and KM110, respectively). KM100 and KM110 were produced by in vivo recombination between n212 and in1814 or V422 in coinfected U2OS cells in the presence of 3 mM HMBA (see Materials and Methods). Following plaque purification, recombinants were identified by Southern blot analysis of the ICP0 and VP16 loci. As diagrammed in Fig. 1A, all three parental mutations are marked by a diagnostic restriction endonuclease cleavage site: n212 was derived by inserting a synthetic SpeI linker bearing an in-frame termination codon into the second exon of the ICP0 gene (truncating the protein after amino acid residue 212), in1814 bears an in-frame BamHI linker that inserts four extra amino acids into VP16 following residue 397, and V422 is marked by a chain-terminating NheI linker that truncates VP16 after residue 422 (removing the majority of the C-terminal acidic transcriptional activation domain). Southern blot analysis confirmed the status of the ICP0 and VP16 alleles in recombinants KM100 and KM110 (Fig. 1B). We also generated derivatives of KM100 and KM110 in which the VP16 and ICP0 mutations were individually rescued to the wild type (data not shown).
FIG. 1.
Construction of ICP0/VP16 double mutants KM100 and KM110. (A) Schematic diagram of the VP16 and ICP0 loci, indicating the locations of the linkers corresponding to the in1814, V422, and n212 mutations. The C-terminal acidic activation domain of VP16 is displayed as a shaded box. Numbers refer to amino acid residues. Diagrams are not to scale. (B) Southern blot analysis of viral DNA from wild-type and mutant viruses. Viral DNA was digested with the indicated restriction endonucleases and subjected to Southern blot hybridization as described in Materials and Methods.
Plaquing efficiency of mutant viruses.
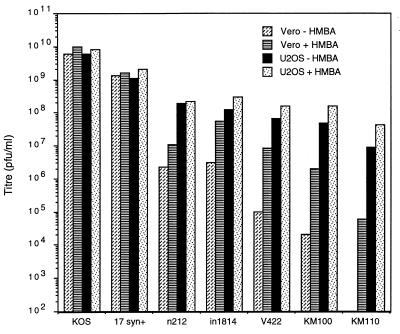
Mutations that inactivate the trans-inducing activity of VP16 or ICP0 greatly reduce the probability that cells infected with a single HSV virion will enter the lytic cycle (2, 69). As a result, these mutations lead to an increased particle-to-PFU ratio (reduced titer) in plaque assays conducted under noncomplementing conditions. The defect of VP16 mutants can be largely overcome by adding HMBA to the culture medium (48), and ICP0 and VP16 mutants are efficiently “complemented” on U2OS cells (66, 76). We subjected stocks of our VP16/ICP0 double mutants to titer determination on Vero and U2OS cells in the presence or absence of 3 mM HMBA, to determine if simultaneous inactivation of ICP0 and VP16 produces a more severe defect than loss of only one of these activators (Fig. 2). As previously described (48, 66, 76), wild-type strains KOS and 17syn+ plaqued with similar efficiency under all four conditions, while n212, in1814, and V422 displayed a titer that was ca. 2 log units (n212 and in1814) to 3 log units (V422) lower on Vero cells than on U2OS cells (Fig. 2). HMBA markedly stimulated the VP16 mutants (and had a marginal effect on n212) in Vero cells but had a smaller effect in U2OS cells. The KM100 and KM110 double mutants displayed a much more severe defect under “noncomplementing” conditions than did either of their singly mutated parents (the titer was 4 and >5 log units lower on Vero cells minus HMBA than on U2OS cells plus HMBA, respectively). Indeed, KM110 was essentially incapable of forming plaques on Vero cells in the absence of HMBA. Derivatives of KM100 and KM110 in which the VP16 and ICP0 mutations were individually rescued to wild type could not be distinguished from their respective single-mutant parental counterparts in this assay (data not shown), indicating that the severe defect displayed by the double mutants stems from simultaneous inactivation of ICP0 and VP16.
FIG. 2.
Plaquing efficiency of mutant viruses. Virus stocks were subjected to titer determination on Vero and U2OS cells in the presence or absence of 3 mM HMBA, as indicated.
These data provide genetic evidence that ICP0 and VP16 make largely independent contributions to plaquing efficiency on Vero cells and that U2OS cells are able to bypass the requirement for both proteins. Moreover, they demonstrate that the V422 VP16 allele produces a more severe phenotype than does the in1814 allele, especially in an ICP0-deficient background.
KM110 is severely defective in IE gene expression.
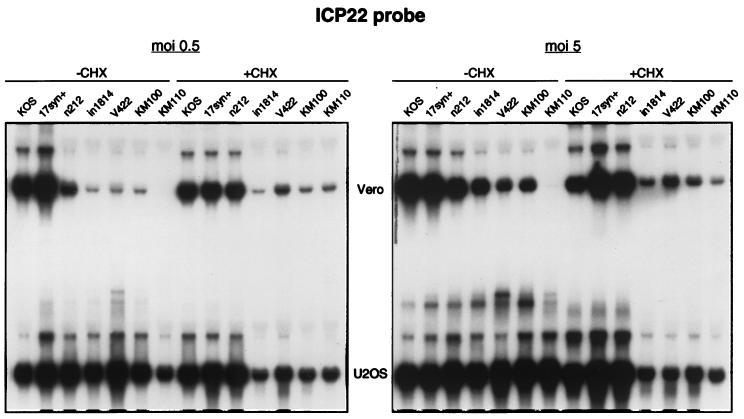
The remarkably severe defect exhibited by KM110 in plaque assays on Vero cells prompted us to examine viral IE gene expression during infection at higher input MOIs (Fig. 3). Vero and U2OS cells were infected at 0.5 and 5 PFU/cell in the presence or absence of cycloheximide, and total RNA harvested at 6 h postinfection was scored for ICP22 mRNA by Northern blot hybridization. Input MOIs were based on the titers obtained in U2OS cells in the presence of HMBA. Consistent with previous work, Vero cells infected with n212, in1814 and V422 showed reduced levels of ICP22 RNA relative to the wild-type strains KOS and 17syn+, particularly at the lower input MOI (Fig. 3). In contrast, these mutants displayed little if any defect in U2OS cells. Perhaps surprisingly, KM100 was not obviously impaired in this assay relative to its single-mutant parents (in1814 and n212), a result which may reflect the higher MOIs used in this experiment. In striking contrast, KM110 produced virtually no ICP22 mRNA during infection of Vero cells at either MOI. However, the same aliquot of KM110 induced high levels of ICP22 RNA in U2OS cells, confirming that biologically active virus was present. Moreover, cycloheximide increased the level of ICP22 RNA in Vero cells infected with KM110 to roughly that observed with in1814, V422, and KM100. Preston et al. (58) have shown that cycloheximide actively stimulates HSV IE gene transcription under conditions where the major viral transactivators are absent. Our data support this conclusion and argue that the KM110 genome is delivered to infected Vero cells in a potentially expressible state.
FIG. 3.
Northern blot analysis of ICP22 RNA levels. Vero and U2OS cells were infected with either 0.5 or 5 PFU of the indicated virus per cell in the presence or absence of 100 μg of cycloheximide (CHX) per ml. The same inoculum was used to infect both cell types. At 6 h postinfection, RNA was extracted and analyzed for ICP22 RNA levels by Northern blot hybridization.
Entirely analogous results were obtained when probes for ICP4 and ICP27 RNA were used (data not shown). Taken in combination, these data indicate that KM110 displays a severe defect in IE gene expression in Vero cells, even at high MOIs.
KM110 does not inhibit IE gene expression from KM100.
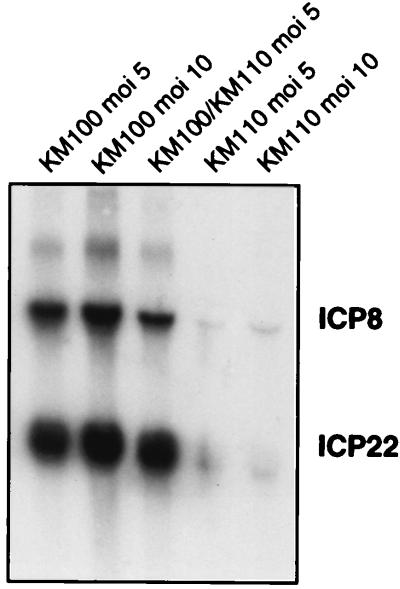
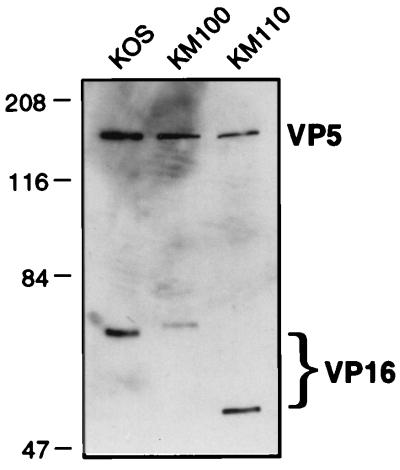
The data presented in Fig. 2 and 3 demonstrate that the V422 and in1814 VP16 mutations produced readily distinguishable phenotypes when combined with the n212 ICP0 mutation, with V422 displaying a more severe defect. This observation was somewhat surprising, because the in1814 mutation is thought to completely eliminate the transactivation function of VP16 (by preventing the assembly of VP16 into the complex with Oct1, HCF, and DNA [1, 2, 27, 30]). We therefore considered the possibility that the V422 protein actively represses the VP16-independent basal activity of IE promoters and thus produces a more severe phenotype than that encoded by a simple loss-of-function mutation. Consistent with this hypothesis, the V422 mutation truncates the C-terminal acidic activation domain of VP16 but leaves the region of the protein required for promoter recognition intact (25, 26). Indeed, Greaves and O’Hare have shown that VP16 truncated at residue 422 (as in V422) retains the ability to assemble into a complex with Oct1, HCF, and DNA but is incapable of activating IE promoters (26). Moreover, McKnight and colleagues have shown that a similarly truncated VP16 derivative blocks transactivation mediated by wild-type VP16 (71) and serves as a trans-dominant inhibitor of HSV-1 replication (23). Alternatively, it was conceivable that the in1814 form of VP16 retains one or more residual functions that marginally stimulate IE gene expression in the context of a viral infection. As one approach to distinguishing between these two scenarios, we asked if KM110 inhibits viral gene expression in cells coinfected with KM100 (Fig. 4). The results demonstrated that cells coinfected with KM100 and KM110 accumulated approximately the same amount of ICP22 and ICP8 RNA as did cells singly infected with KM100. Similar results were obtained for ICP4 and ICP27 transcripts (data not shown). Inasmuch as purified KM110 virions appear to contain roughly the same amount of VP16 as wild-type HSV-1 KOS does (Fig. 5), these results suggest that the V422 VP16 protein present in KM110 virions does not act as a strong trans-acting repressor of VP16-independent IE gene expression. This interpretation is further supported by our finding that KM110 expresses easily detectable levels of IE RNAs during infection of U2OS cells and in Vero cells treated with cycloheximide (Fig. 3).
FIG. 4.
ICP22 and ICP8 RNA levels in cells coinfected with KM100 and KM110. Vero cells were singly infected or coinfected with KM100 or KM110 at the indicated MOI. RNA extracted 6 h postinfection was analyzed for ICP22 and ICP8 RNA by Northern blot hybridization.
FIG. 5.
VP16 levels in purified virions. KOS, KM100, and KM110 virions were purified by centrifugation through dextran gradients (see Materials and Methods), pelleted, resuspended in SDS-PAGE sample buffer, and subjected to electrophoresis through an SDS–9% polyacrylamide gel. The relative amounts of the capsid protein (VP5) and VP16 were then visualized by Western blot analysis with antibodies NC-1 and LP1, respectively. Note the differences in the electrophoretic mobilities of the wild-type, in1814, and V422 forms of VP16.
KM110 is markedly less cytotoxic than KM100 and persists in a quiescent state in restrictive cells.
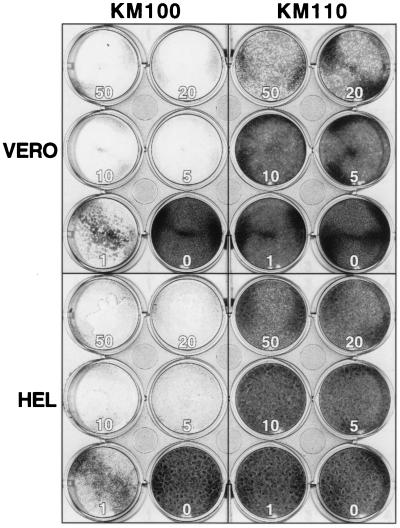
Early studies by Johnson et al. established that expression of HSV-1 IE proteins is cytotoxic to cultured cells (33, 34). More recently, the groups of Preston and DeLuca have shown that the cytotoxicity of HSV-1 can be reduced or eliminated by introducing multiple mutations into the viral genome that prevent synthesis of the IE proteins of HSV (29, 32, 56, 57, 63). The substantial defect in IE gene expression exhibited by KM110 suggested that this isolate might display a similar reduction in cytotoxicity. To assess this possibility, we infected Vero cells and HEL cells with varying input MOIs of KM110 and KM100 and examined the cultures 3 days postinfection (Fig. 6). The results demonstrated that KM110 is much less toxic than KM100, particularly on HEL cells. Vero cell monolayers tolerated infection with 1 and 5 PFU of KM110 and produced only small isolated foci of cytopathic effect, while HEL cells could be infected with 10 and 20 PFU/cell and showed no detectable cytotoxicity. In contrast, KM100 induced virtually complete destruction of monolayers of both cell types at 1 PFU/cell. These data provide additional evidence that KM110 is substantially more impaired than KM100 and indicate that KM110 is essentially incapable of entering the lytic cycle in HEL cells.
FIG. 6.
KM110 is less cytotoxic than KM100. Monolayers of Vero and HEL cells were infected at the indicated MOIs, fixed, and stained 72 h postinfection.
Previous studies have demonstrated that similarly compromised viral mutants bearing multiple lesions in several viral transactivators persist for extended periods in nonproductively infected cells, in a quiescent form that can be induced into the lytic cycle by superinfection with wild-type HSV (29, 32, 56, 57, 63). To clarify the nature of the defect exhibited by KM110, we investigated whether potentially expressible copies of the KM110 genome persist in infected HEL cells. These experiments used the altered electrophoretic mobility of VP16 encoded by KM110 (Fig. 5) as a means of specifically monitoring expression from the KM110 genome in the presence of superinfecting wild-type HSV-1.
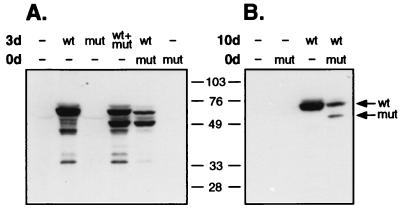
Confluent monolayers of HEL cells were infected with 10 PFU of KM110 per cell or left untreated. Three days later the cells were either superinfected with wild-type HSV-1 KOS or mock infected. At the same time, control monolayers were infected with KOS, KM110, or both viruses. The cells were then harvested 24 h later (i.e., 4 days after the initial infection with KM110), and expression of VP16 was examined by Western blot analysis (Fig. 7A). HEL cells singly infected with KM110 expressed little if any VP16. However, high levels of VP16 arising from the KM110 genome were detected in cells that were either coinfected or superinfected with KOS. Further analysis revealed that the resident KM110 genome remained susceptible to activation by superinfecting KOS for at least 10 days (Fig. 7B). Taken in combination, these data indicate that the KM110 genome persists in a quiescent but inducible form in infected HEL cells. These results provide additional evidence that KM110 displays a severe defect in launching the viral lytic cycle and argue that the defect operates at the level of gene expression (as opposed to a defect in adsorption or penetration). Thus, this recombinant will probably provide a useful tool for analyzing the quiescent state adopted by HSV genomes in the absence of IE gene expression.
FIG. 7.
Persistence of quiescent KM110 genomes in infected HEL cells. Confluent monolayers of HEL cells were either left untreated (0d, −) or infected with KM110 at an MOI of 10 (0d, mut). At 3 days (A) or 10 days (B) later, the cells were superinfected with 10 PFU of KOS (wt) or KM110 (mut) per cell or mock infected (−). Monolayers were harvested 24 h later, and VP16 was detected by Western blot analysis. Arrows indicate the mobility of VP16 arising from either the KOS (wt) or KM110 (mut) genome.
DISCUSSION
Several lines of evidence indicate that VP16 and ICP0 play interrelated roles in stimulating the onset of the HSV lytic cycle. First, mutations that eliminate the activation functions of ICP0 and VP16 produce similar multiplicity- and cell cycle-dependent defects in viral gene expression (2, 6, 9, 13, 61, 69). Second, expression of ICP0 in trans at least partially alleviates the effects of a VP16 mutation (2). Similarly, ICP0 stimulates the production of virus from transfected protein-free viral DNA (7). Third, ICP0 and VP16 mutants are both “complemented” on U2OS cells (66, 76), which have been proposed to express a cellular ICP0-like function (76). Fourth, ICP0 contains an amino-terminal transcriptional activation domain that selectively activates IE promoters (42). Taken in combination, these data suggest that, once expressed, ICP0 can at least partially substitute for VP16 activation function. This in turn raises the possibility that the primary role of the transactivation function of VP16 is to stimulate expression of ICP0, which then suffices to activate the other IE genes. If activation of ICP0 was the only physiologically relevant function of VP16, VP16/ICP0 double mutants should display a phenotype similar to that of an ICP0 mutant. However, we found that such double mutants exhibit a 4- to >5-log-unit reduction in titer under noncomplementing conditions while their singly mutant parents display only a 2- to 3-log-unit reduction relative to wild-type virus. These data indicate that VP16 and ICP0 make largely independent contributions to plaquing efficiency on Vero cells. Preston et al. (56) have reported similar results with another in1814-based VP16/ICP0 double mutant (in1820), in which ICP0 expression was reduced by placing the gene under the control of the murine leukemia virus long terminal repeat. The simplest interpretation of these findings is that although ICP0 can partially substitute for VP16 function, VP16-induced transactivation of other IE genes plays a major role in triggering the onset of the lytic cycle when ICP0 is inactivated.
We found that the VP16/ICP0 double mutant bearing the V422 VP16 allele (KM110) displayed a markedly more severe phenotype than did the double mutant harboring the in1814 allele (KM100). Thus, KM110 was essentially incapable of forming plaques on Vero cells and displayed a much more severe defect in IE gene expression during high-MOI infection. In addition, HEL cell monolayers survived infection with 20 PFU of KM110 per cell without obvious cytopathic effect, while KM100 induced extensive cell death at 1 PFU/cell. The large difference between the two double mutants was surprising, because the in1814 mutation is thought to eliminate the transactivation function of VP16. One possible explanation was that the V422 protein actively represses IE expression, thereby reducing expression below the level obtained with a simple loss-of-function mutant (in1814). However, although similarly truncated derivatives of VP16 can block transactivation mediated by wild-type VP16 (23, 71), KM110 did not interfere with IE gene expression in cells coinfected with KM100. This result argues that the V422 form of VP16 does not serve as a strong trans-acting inhibitor of the VP16-independent activity of IE promoters. Consistent with this view, KM110 expressed readily detectable levels of IE transcripts during infection of U2OS cells and in Vero cells treated with cycloheximide. In our view, the simplest interpretation of our data is that the V422 mutation eliminates the ability of VP16 to stimulate IE gene expression in the context of an HSV infection whereas the in1814 protein retains residual function. How can one reconcile this hypothesis with previous data that clearly demonstrate that the in1814 mutation abolishes the ability of VP16 to stimulate IE transcription in the absence of other HSV proteins, by preventing the assembly of VP16 into the complex with Oct1, HCF, and DNA (1, 2, 27, 30)? One possibility is that VP16 influences the packaging or activity of other tegument proteins that facilitate IE gene expression, in addition to directly stimulating IE transcription. According to this hypothesis, the V422 lesion inactivates both of these stimulatory functions whereas the in1814 mutation affects only direct transactivation. In this context, it is interesting that VP16 directly binds to at least two tegument proteins, the virion host shutoff (vhs) protein (65) and VP22 (12). Intriguingly, the interaction with VP22 occurs through the C-terminal transcriptional activation domain (12).
KM110 exhibited a much more severe defect in IE gene expression than did either KM100 or V422 at high MOI (Fig. 3). This finding indicates that IE gene expression becomes almost completely dependent on ICP0 when the activation domain of VP16 is deleted. Inasmuch as ICP0 is itself an IE gene product, an interesting question arises: what is the source of the ICP0 protein that allows V422 to express its IE genes more efficiently than KM110 does? One possibility is that sufficient quantities of ICP0 are produced in the newly infected cell to launch the infection. However, an alternative explanation is suggested by the finding that small amounts of ICP0 are packaged into the tegument of HSV virions produced in Vero and HEp-2 cells (74, 75). In this context, Dargan et al. have provided evidence that ICP0 delivered into cells by noninfectious L particles is biologically active in that it can enhance the infectivity of transfected viral DNA (11). Thus, it is possible that ICP0 delivered by the infecting virion plays a key role in initiating the V422 infection. Further experiments are required to test this hypothesis.
Although KM110 is effectively unable to replicate in Vero and HEL cells, it can be readily propagated on U2OS osteosarcoma cells (Fig. 2), which have been previously shown to “complement” ICP0 and VP16 mutants (66, 76). This result demonstrates that U2OS cells can compensate for the simultaneous loss of both ICP0 and VP16 activation functions. The molecular basis for this “complementation” is unknown. Yao and Schaffer have suggested that U2OS cells express a functional homologue of ICP0, and our data are consistent with this hypothesis. However, an equally plausible explanation is that these cells lack one or more inhibitors of IE gene expression that are present in most other cell types. In this context, it is interesting that current evidence suggests that ICP0 stimulates viral gene expression by inducing proteosome-dependent degradation of one or more cellular proteins (15, 16, 18, 21, 54). Defining the mechanism of “complementation” by U2OS cells will probably enhance our understanding of the regulation of HSV IE gene expression. It will also be important to determine if other human tumor-derived cell lines exhibit similar complementing properties. If so, KM110 may have considerable potential for viral antitumor therapy.
Vero and HEL cells tolerated infection with KM110 at quite high MOIs without showing major cytopathic effects, and the KM110 genome persisted in a quiescent form in infected HEL cells for at least 10 days. These findings demonstrate that it is possible to effectively prevent the onset of the HSV lytic cycle by mutating two viral regulatory proteins: VP16 and ICP0. KM110 differs from previously described HSV constructs displaying similar properties (56, 63), in that it retains functional copies of four of the viral IE genes. We therefore believe that KM110 and derivatives will serve as useful tools for investigating the quiescent state adopted by HSV genomes in the absence of IE gene expression and the process of reactivation from this state. KM110 or related constructs may also prove to be useful platforms for the development of HSV vectors for gene therapy.
ACKNOWLEDGMENTS
We thank Rob Maranchuk and Holly Saffran for excellent technical assistance and Stephen A. Rice for generously providing laboratory space to K.L.M. during the early stages of this study.
This work was supported by a grant from the Medical Research Council of Canada (MT-121720). K.L.M. holds postdoctoral fellowships from the MRC and the Alberta Heritage Foundation for Medical Research. J.R.S. was a Terry Fox Senior Scientist of the National Cancer Institute of Canada until that National Program of ongoing career support was terminated by the NCI(C).
REFERENCES
- 1.Ace C I, Dalrymple M A, Ramsay F H, Preston V G, Preston C M. Mutational analysis of the herpes simplex virus type 1 transinducing factor Vmw65. J Gen Virol. 1988;69:2595–2605. doi: 10.1099/0022-1317-69-10-2595. [DOI] [PubMed] [Google Scholar]
- 2.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkham J, Coen D, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Astor T L, Lipak L M, Cho C, Coen D M, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Schaffer P. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Schaffer P A. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J Virol. 1991;63:4578–4590. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4570–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clements G B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latent-competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 9.Daksis J I, Preston C M. Herpes simplex virus immediate early gene expression in the absence of transinduction by Vmw65 varies during the cell cycle. J Gen Virol. 1992;189:196–202. doi: 10.1016/0042-6822(92)90695-l. [DOI] [PubMed] [Google Scholar]
- 10.Dalrymple M A, McGeoch D J, Davison A J, Preston C M. DNA sequence of the herpes simplex virus type 1 gene whose product is responsible for transcriptional activation of immediate-early promoters. Nucleic Acids Res. 1985;13:7865–7879. doi: 10.1093/nar/13.21.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dargan D J, Subak-Sharpe J H. The effects of herpes simplex virus type 1 L-particles on virus entry, replication, and the infectivity of naked herpesvirus DNA. Virology. 1997;239:378–388. doi: 10.1006/viro.1997.8893. [DOI] [PubMed] [Google Scholar]
- 12.Elliott G, Mouzakitis G, O’Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 14.Everett R D. Trans-activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 ring finger helix affect activation of gene expression, viral growth, and interaction with PML-containing structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes virus gene expression. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 49–76. [Google Scholar]
- 23.Friedman A D, Triezenberg S J, McKnight S L. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature. 1988;333:452–454. doi: 10.1038/335452a0. [DOI] [PubMed] [Google Scholar]
- 24.Gelman I H, Silverstein S. Identification of immediate-early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goding C R, O’Hare P. Herpes simplex virus Vmw65-octamer binding protein interaction: a paradigm for combinatorial control of transcription. Virology. 1989;173:363–367. doi: 10.1016/0042-6822(89)90548-5. [DOI] [PubMed] [Google Scholar]
- 26.Greaves R, O’Hare P. Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J Virol. 1989;63:1641–1650. doi: 10.1128/jvi.63.4.1641-1650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greaves R F, O’Hare P. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J Virol. 1990;64:2716–2724. doi: 10.1128/jvi.64.6.2716-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris R A, Preston C M. Establishment of latency in vitro by the herpes simplex virus type 1 mutant in1814. J Gen Virol. 1991;72:907–913. doi: 10.1099/0022-1317-72-4-907. [DOI] [PubMed] [Google Scholar]
- 30.Hayes S, O’Hare P. Mapping of a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J Virol. 1993;67:852–862. doi: 10.1128/jvi.67.2.852-862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamieson D R S, Robinson L H, Daksis J I, Nicholl M J, Preston C M. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus Vmw65 mutants. J Gen Virol. 1995;76:1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 33.Johnson P A, Miyanohara A, Levine F, Cahill T, Friedmann T. Cytotoxicity of a replication-defective mutant of herpes simplex virus type 1. J Virol. 1992;66:2952–2965. doi: 10.1128/jvi.66.5.2952-2965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson P A, Wang M J, Friedmann T. Improved cell survival by the reduction of immediate-early gene expression in replication-defective mutants of herpes simplex virus type 1 but not by mutation of the virion host shutoff function. J Virol. 1994;68:6347–6362. doi: 10.1128/jvi.68.10.6347-6362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawaguchi Y, Sant C V, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 39.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type I transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemaster S, Roizman B. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J Virol. 1979;35:798–811. doi: 10.1128/jvi.35.3.798-811.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lium E K, Panagiotidis C A, Wen X, Silverstein S J. The NH2 terminus of the herpes simplex virus type 1 regulatory protein ICP0 contains a promoter-specific transcription activation domain. J Virol. 1998;72:7785–7795. doi: 10.1128/jvi.72.10.7785-7795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lium E K, Silverstein S. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol. 1997;71:8602–8614. doi: 10.1128/jvi.71.11.8602-8614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackem S, Roizman B. Structural features of the herpes simplex virus α gene 4, 0, and 27 promoter-regulatory sequences which confer α regulation on chimeric thymidine kinase genes. J Virol. 1982;44:939–949. doi: 10.1128/jvi.44.3.939-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsden H S, Stow N D, Preston V G, Timbury M C, Wilkie N M. Physical mapping of herpes simplex virus-induced polypeptide. J Virol. 1978;28:624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 47.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 48.McFarlane M, Daksis J I, Preston C M. Hexamethylene bisacetamide stimulates herpes simplex virus immediate-early gene expression in the absence of trans-induction by Vmw65. J Gen Virol. 1992;73:285–292. doi: 10.1099/0022-1317-73-2-285. [DOI] [PubMed] [Google Scholar]
- 49.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 50.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 51.O’Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 52.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoter. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Hare P, Hayward G S. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J Virol. 1985;56:723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pellett P E, McKnight J L C, Jenkins F J, Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing α genes. Proc Natl Acad Sci USA. 1985;82:5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preston C M, Mabbs R, Nicholl M J. Construction and characterization of herpes simplex virus type 1 mutants with conditional deletions in immediate early gene expression. Virology. 1997;229:228–239. doi: 10.1006/viro.1996.8424. [DOI] [PubMed] [Google Scholar]
- 57.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preston C M, Rinaldi A, Nicholl M J. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J Gen Virol. 1998;79:117–124. doi: 10.1099/0022-1317-79-1-117. [DOI] [PubMed] [Google Scholar]
- 59.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 61.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadowski I, Ma J, Triezenberg S J, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 63.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smiley J R, Duncan J. 1814 linker insertion mutation. J. Virol. 71:6191–6193. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stow E C, Stow N D. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J Gen Virol. 1989;70:695–704. doi: 10.1099/0022-1317-70-3-695. [DOI] [PubMed] [Google Scholar]
- 68.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 69.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 70.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 71.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 72.Triezenberg S J, LaMarco K L, McKnight S L. Evidence of DNA:protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 73.Wilcox C L, Smith R L, Everett R, Mysofski D. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J Virol. 1997;71:6777–6785. doi: 10.1128/jvi.71.9.6777-6785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang T Y, Courtney R J. Influence of the host cell on the association of ICP4 and ICP0 with herpes simplex virus type 1. Virology. 1995;211:209–217. doi: 10.1006/viro.1995.1393. [DOI] [PubMed] [Google Scholar]
- 75.Yao F, Courtney R. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66:2709–2716. doi: 10.1128/jvi.66.5.2709-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu X, Chen J, Young C, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]