Abstract
Background
Cancer stem-like cells (CSCs) play an important role in initiation and progression of aggressive cancers, including esophageal cancer. Natural killer (NK) cells are key effector lymphocytes of innate immunity that directly attack a wide variety of cancer cells. NK cell-based therapy may provide a new treatment option for targeting CSCs. In this study, we aimed to investigate the sensitivity of human esophageal CSCs to NK cell-mediated cytotoxicity.
Methods
CSCs were enriched from human esophageal squamous cell carcinoma cell lines via sphere formation culture. Human NK cells were selectively expanded from the peripheral blood of healthy donors. qRT-PCR, flow cytometry and ELISA assays were performed to examine RNA expression and protein levels, respectively. CFSE-labeled target cells were co-cultured with human activated NK cells to detect the cytotoxicity of NK cells by flow cytometry.
Results
We observed that esophageal CSCs were more resistant to NK cell-mediated cytotoxicity compared with adherent counterparts. Consistently, esophageal CSCs showed down-regulated expression of ULBP-1, a ligand for NK cells stimulatory receptor NKG2D. Knockdown of ULBP-1 resulted in significant inhibition of NK cell cytotoxicity against esophageal CSCs, whereas ULBP-1 overexpression led to the opposite effect. Finally, the pro-differentiation agent all-trans retinoic acid was found to enhance the sensitivity of esophageal CSCs to NK cell cytotoxicity.
Conclusions
This study reveals that esophageal CSCs are more resistant to NK cells through down-regulation of ULBP-1 and provides a promising approach to promote the activity of NK cells targeting esophageal CSCs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05549-1.
Keywords: All-trans retinoic acid, Cancer stem-like cells, Esophageal cancer, NK cells, ULBP-1
Background
Esophageal cancer has a high incidence, and ranks as the sixth cause of cancer-related deaths worldwide largely due to failure of early diagnosis and high potential of metastases [1]. Esophageal squamous cell carcinoma (ESCC) is the predominant subtype in certain Asian countries, especially in China [2]. Although substantial progress has been made in cancer treatments, the 5-year survival rate for ESCC remains low [3]. Cancer stem-like cells (CSCs) or tumor-initiating cells have been considered as the key mechanism for cancer formation, progression, metastasis, and relapse [4, 5]. Increasing evidence suggests that CSCs contribute to poor prognosis and treatment failure for ESCC [6]. Thus, targeting CSCs may be a promising strategy for the treatment of ESCC [7].
Natural killer (NK) cells are cellular components of the innate immune system that contribute to the immune surveillance of tumors. NK cells can recognize and directly eliminate target cells without prior immune sensitization [8]. NK cell-based adoptive therapy showed promising results in some types of blood cancers [9]. Recently, NK cells exhibit killing activity against CSCs derived from a variety of solid tumors via the decreased MHC class I molecules and the altered ligands for NK cells [10, 11]. However, little is known about the susceptibility of human esophageal CSCs to NK cells.
NK cells can distinguish self from non-self through the engagement of various surface receptors that trigger activating or inhibitory signals [12]. The natural cytotoxicity receptors (NCR) are an important family of activating receptors which include NKp46, NKp44 and NKp30. However, the tumor-associated ligands for NCR have yet to be completely defined. NKG2D is another major activating receptor exclusively on NK cells and some T cells [13]. NKG2D binds to several families of ligands, including MHC I Chain-related molecules A and B (MICA/B) and the UL16-binding proteins (ULBPs). NKG2D ligands are rarely expressed on normal cells, but could be induced in malignant or virus-infected cells. On the other hand, NK cells also express multiple inhibitory receptors, such as the killer cell immunoglobulin-like receptor (KIR) for MHC class I molecules and recently identified TIGIT for PVR [14].
The effector functions of NK cells are regulated by the dynamic balance between activating and inhibitory signals [12]. Many strategies have been proposed to enhance the susceptibility of cancer cells to NK cell cytotoxicity. In this study, we demonstrate that CSCs derived from ESCC spheres are more resistant to NK cell killing through down-regulation of ULBP-1. All-trans retinoic acid (ATRA) improved the anti-tumor activity of NK cells against ESCC spheres by inducing sphere differentiation. These findings reveal the immune escape of esophageal CSCs from NK cell surveillance, providing a rationale for combining ATRA treatment to overcome the resistance of esophageal CSCs in response to NK cells.
Materials and methods
Cell lines and patient samples
Human ESCC cell lines KYSE510, TE1, KYSE450 and KYSE70 were obtained from Cell Bank, Chinese Academy of Sciences (China). ESCC cell lines and human myelogenous leukemia K562 cells were cultured in RPMI-1640 medium, and human embryonic kidney 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), in the presence of 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 µg/mL of streptomycin. All cells were maintained at 37ºC in a humid atmosphere with 5% CO2. Peripheral blood and tumor tissues were obtained from healthy donors or ESCC patients in the First Affiliated Hospital of Zhengzhou University, respectively.
Tumor sphere formation assay
KYSE510 and TE1 cells were incubated in serum-free DMEM/F12 medium supplemented with 4 µg/mL heparin (Sigma, USA), B27 (1: 50, Gibco, USA), 20 ng/mL human recombinant basic fibroblast growth factor (Peprotech, USA), 20 ng/mL human recombinant epidermal growth factor (Peprotech, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin in ultralow attachment culture plates (Corning, USA) at a density of 4000 cells/mL. After 5–7 days of culture, numbers of tumor spheres (diameter > 75 μm) were counted under microscope. For further analysis, the spheres were collected and enzymatically dissociated into single cells with the addition of trypsin.
RNA extraction, cDNA synthesis, and quantitative PCR (qPCR)
Total RNA was extracted from tumor spheres or adherent cells using TRIzol reagent (TaKaRa, Japan). cDNA was synthesized by reverse transcription from total RNA using PrimerScript TM RT reagent Kit with gDNA Eraser (TaKaRa, Japan). qPCR reactions were performed using SYBR Green qPCR Master Mix (Roche, Germany). The primers used in this study were synthesized by Sangong Biotech (China) and listed in Additional file 1: Table S1. GAPDH was used as a control to determine the relative gene expression of target genes.
Preparation of human activated NK cells
Peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood of 20 healthy donors by density gradient centrifugation using Ficoll-Paque (Haoyang, China). The PBMCs were cultured with irradiated K562 cells as feeder cells in GT-T551 H3 medium (TaKaRa, Japan), supplemented with 5% heat-inactivated autologous plasma, IL-2 (1000 IU/mL) (SL PHARM, Beijing, China), penicillin (100 U/mL), and streptomycin (100 µg/mL). The fresh medium was replaced every 2–3 days during the amplification. NK cells were harvested on day 14 and analyzed by flow cytometry. To verify direct NK cell-mediated cytotoxicity, NK cells were purified from expanded or fresh NK cells by positive selection using CD56 microbeads (Miltenyi Biotec, Germany).
Flow cytometry
For surface staining, cells were washed in FACS buffer (PBS supplemented with 2% FBS) and then incubated with the indicated fluorescein-conjugated antibodies: APC-conjugated anti-CD3, APC-Cy7-conjugated anti-CD56, and PE-conjugated anti-CD107a. For intracellular staining, cells were first stimulated with phorbol myristate acetate (PMA, 50 ng/mL) and ionomycin (250 ng/mL) for 1 h, followed by the addition of protein transport inhibitor brefeldin A (BFA) for 5 h. After surface staining, the cells were then fixed with 4% paraformaldehyde and permeabilized with the permeabilization buffer (Biolegend) before incubation with PE-Cy7-conjugated anti-IFN-γ, FITC-conjugated anti-granzyme-B antibodies. Isotype control antibodies were used for non-specific staining. Flow cytometric analysis was performed on FACS Canto II flow cytometer. Data were analyzed with the FlowJo software. Information of the antibodies were listed in Additional file 2: Table S2.
Cytotoxicity assay
A flow cytometric assay was used to determine NK cell activity. Target cells were stained with carboxyfluorescein succinimidyl ester (CFSE) in serum-free medium at 37℃ for 15 min. Human activated NK cells were incubated with CFSE-labeled target cells at different effector: target (E: T) ratios for 6 h in 96-well U-bottom plates at 37℃ in a humidified 5% CO2 incubator. Cell culture supernatant was collected for cytokines detection. Cells were then collected and stained with propidium iodide (PI). The percentages for dead target cells were determined by FACS Canto II flow cytometry. Specific lysis was calculated as the percentage of dead target cells with NK cells – the percentage of dead target cells with medium alone.
In vivo xenograft model
Female nude mice aged 5 weeks were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd. (China) and housed in the SPF Laboratory Animal Center of Zhengzhou University (China). KYSE510 cells (5 × 106) were subcutaneously injected into the flank of each nude mouse (n = 5 per group). On day 5 and 8, mice were injected with human activated NK cells (5 × 107) or PBS in the control group via tail vein. Tumor growth was monitored twice a week. After 3 weeks of xenografting, the mice were euthanized and the tumors were excised. The animal study was performed under the guidelines of Zhengzhou University’s Animal Care and Use Committee.
siRNA transfection
Human ULBP-1 siRNA and control siRNA were purchased from Genepharma (China). The sequences of siRNA targeting human ULBP-1 were shown in Additional file 3: Table S3. ESCC cell lines were transfected with siRNAs using Lipofectamine 3000 (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Lentiviral production and transduction
Lentiviral vectors containing human ULBP-1 or a non-target control shRNA were purchased from Genepharma Company (China). To generate ULBP-1 overexpressing cells, full-length cDNA of human ULBP-1 was amplified by PCR and cloned into the lentiviral vector pCDH-EF1a-GFP. DNA sequencing was performed to verify the cloning quality. Lentivirus was generated by transfecting 293T cells with lentiviral vectors, packaging plasmid pxPAX2 and pMD2.G using Lipofectamine 3000. Lentiviral supernatants were collected 2 days after transfection. KYSE510 and TE1 cells were transduced with lentivirus in the presence of polybrene (10 µg/mL). Transduced target cells were selected by puromycin or by flow cytometry. The efficacy of silencing or overexpressing ULBP-1 was determined by qPCR analysis and flow cytometry.
Enzyme-linked immunosorbent assay (ELISA)
Interferon-γ (IFN-γ) levels were determined by an ELISA kit (Biolegend, USA) according to the manufacturer’s instructions. The data analysis was performed using the Star Station software.
Statistical analysis
All data are shown as the means ± standard deviation (SD) from at least 2–3 independent experiments. Student’s t-test was performed using GraphPad Prism 8 software. A p value of less than 0.05 was defined as statistically significant.
Results
Characterization of cancer stem-like cells from ESCC cell lines
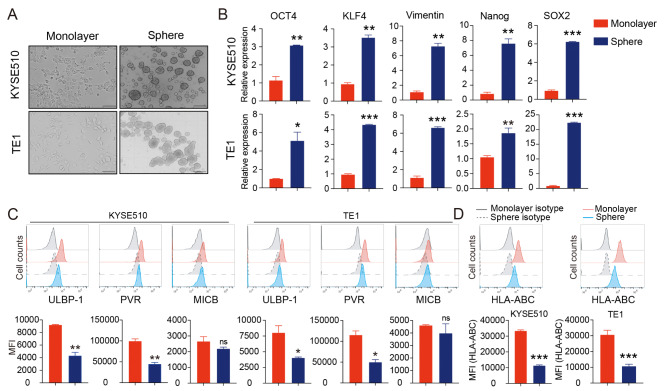
To enrich CSCs, two human ESCC cell lines KYSE510 and TE1 were cultured with serum-free media in ultra-low plates for 5–7 days to form tumor spheres (Fig. 1A). We first assessed the stemness-related gene expression of tumor spheres using qPCR assay. Both KYSE510 spheres and TE1 spheres expressed higher levels of OCT4, KLF4, Vimentin, Nanog and SOX2 than that of monolayer cells, indicating the stemness properties of ESCC cells-derived spheres (Fig. 1B).
Fig. 1.
Esophageal CSCs enriched from tumor spheres exhibit distinct NK cell ligands. (A) Two ESCC cell lines KYSE510 cells and TE1 cells were used in tumor sphere formation assays to enrich CSCs. Scale bar: 500 μm. (B) qPCR was carried out to validate the expression of stemness-associated genes in tumor spheres. (C) Flow cytometry analysis of selected NK cell ligands on tumor spheres. (D) Surface expression of MHC class I molecules were detected by flow cytometry on tumor spheres. Data are shown as the mean ± SD. MFI, mean fluorescence intensity; ns, no significance. *p < 0.01, **p < 0.01, ***p < 0.001
The sensitivity of tumor cells to NK cell lysis is largely related to the expression of ligands specific for NK cells [15]. We next examined the expression of specific ligands for NK activating or inhibitory receptors. Compared with the monolayer counterparts, the expression of MICB and ULBP-1 significantly decreased in both KYSE510 spheres and TE1 spheres, and PVR remarkably decreased in TE1 spheres (Additional file 4: Figure S1). The three different ligands were further examined by flow cytometry, confirming the consistent decrease of ULBP-1 and PVR at protein level (Fig. 1C). In addition, MHC class I expression was dramatically down-regulated following tumor sphere formation (Fig. 1D). The results suggest that CSCs derived from KYSE510 and TE1 spheres may exhibit altered sensitivity to NK cell cytotoxicity.
Generation of activated human NK cells from peripheral blood of healthy donors
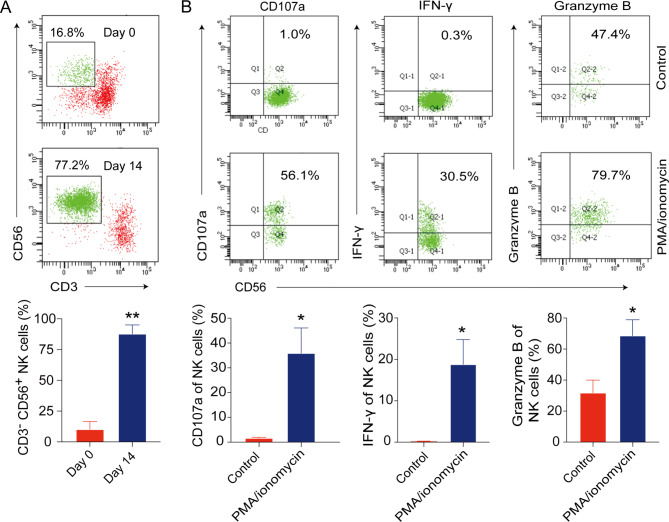
Next, we generated the activated human NK cells from the peripheral blood of healthy donors. After 14-day culture, the majority of expanded NK cells developed a CD56brihgt phenotype (Fig. 2A). The expression of activating receptors NCR and NKG2D were significantly increased after ex vivo expansion (Additional file 5: Figure S2). Additionally, the expanded NK cells expressed high levels of CD107a and produced significant amounts of IFN-γ and granzyme B under PMA and ionomycin stimulation (Fig. 2B). These data indicate that the expanded human NK cells are fully functional.
Fig. 2.
Ex vivo expansion of NK cells from peripheral blood of healthy donors. (A) The percentages of NK cells were analyzed by flow cytometry after 14 days of ex vivo expansion. (B) CD107a, IFN-γ and granzyme B were analyzed in expanded NK cells upon stimulation with PMA and ionomycin. Data are shown as the mean ± SD. *p < 0.05, **p < 0.01
Tumor spheres derived from ESCC cells exhibit significant resistance to NK cell-mediated cytotoxicity
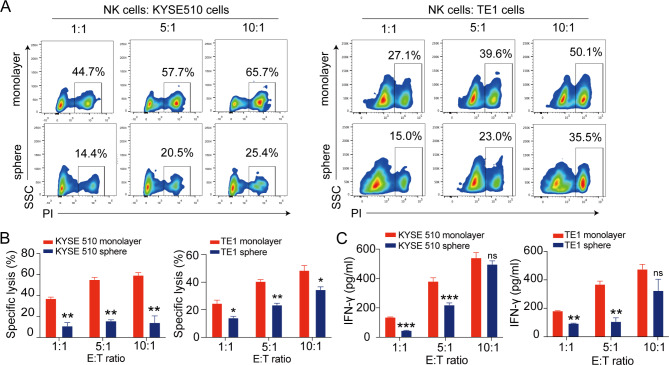
We then evaluated the susceptibility of tumor spheres to allogeneic NK cells using a flow cytometry-based cytotoxicity assay. Both KYSE510 spheres and TE1 spheres were less susceptible to NK cell-mediated lysis compared with their adherent counterparts at various E: T ratios (Fig. 3A and B). Thus, ESCC-derived CSCs may possess the potential to escape NK cell surveillance. This resistance to NK cell-mediated killing is consistent with the pronounced decrease of ligands for activating NK receptors on esophageal tumor spheres, such as ULBP-1 (Fig. 1C). To further validate the sensitivity of ESCC spheres to NK cells, we cultured NK cells with either monolayer or sphere ESCC cells and assayed for cytokine production. We observed a remarkable decrease of IFN-γ in NK cells cultured with tumor spheres at E: T ratios of 1:1 and 5:1, relative to monolayer ESCC cells (Fig. 3C). Together, these results provide direct evidence that CSCs derived from ESCC cells are resistant to NK cell-mediated cytotoxicity.
Fig. 3.
Esophageal CSCs derived from tumor spheres are more resistant to NK cell cytotoxicity. (A) Representative flow plots of apoptotic CSFE-labeled KYSE510 cells (left panel) and TE1 cells (right panel) in a 6 h cytotoxicity assay with NK cells at different effector: target (E: T) ratios. (B) Summarized data showing the cytotoxicity of NK cells against tumor sphere cells derived from KYSE510 and TE1 cells. (C) The release of IFN-γ was assessed by ELISA assays in culture supernatants of NK cells stimulated by tumor spheres derived from KYSE510 and TE1 cells, respectively. Data are shown as the mean ± SD; ns, no significance. *p < 0.05, **p < 0.01, ***p < 0.001
It should be noted that a small fraction of T cells still exists in our feeder cell expansion system of NK cells. Typically, T cells recognize and kill target cells through MHC class I-restricted presentation of specific antigens. To verify the cytotoxicity of expanded NK cells, we purified the expanded NK cells using anti-CD56 magnetic beads (Additional file 6: Figure S3A). Similarly, tumor spheres were resistant to the cytotoxicity of expanded and highly purified NK cells (Additional file 6: Figure S3B). Finally, we further confirm the resistance of ESCC-derived tumor spheres using isolated fresh NK cells (Additional file 6: Figure S3C and D).
Considering the alteration of ligands for NK cell receptors, the pronounced reduction of activating NK cell receptor ligand ULBP-1 but not inhibitory NK cell receptor ligands PVR and HLA class could explain the resistance of CSCs to NK cell cytotoxicity. We further confirmed the down-regulation of ULBP-1 in tumor spheres derived from another two ESCC cell lines (Additional file 7: Figure S4A) and CSCs isolated from tumor tissues of four ESCC patients (Additional file 7: Figure S4B). Thus, we speculate that ULBP-1 is more likely a possible mechanism responsible for the resistance of esophageal CSCs to NK cells.
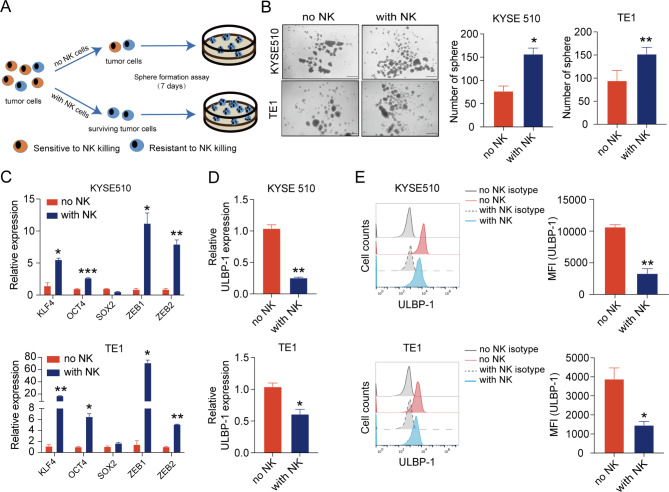
ESCC cells survived from co-incubation with NK cells show the stemness features and down-regulation of ULBP-1
To characterize ESCC cells resistant to NK cytotoxicity, we developed a transient co-culture system of NK cells with ESCC cells. ESCC cells were plated and on the next day activated NK cells were added at a E: T ratio of 1:1. After 6 h of co-culture, NK cells were removed and the remaining live ESCC cells were washed and then subjected to sphere formation assays (Fig. 4A). Compared to control ESCC cells, ESCC cells survived from NK cells possess a higher ability to form tumor spheres (Fig. 4B). Additionally, higher levels of stemness-associated markers (KLF4, OCT4) and EMT markers (ZEB1, ZEB2) were observed in both KYSE510 cells and TE1 cells that survived from NK cells (Fig. 4C). Notably, tumor cells derived from surviving ESCC cells also expressed lower levels of ULBP-1 mRNA (Fig. 4D) and ULBP-1 protein (Fig. 4E). The results further support the hypothesis that ESCC cells with stemness properties have the potential to escape NK-mediated killing through down-regulation of ULBP-1, a ligand for NK cell activating receptor NKG2D.
Fig. 4.
ESCC cells survived from co-incubation with NK cells show cancer stem-like and low ULBP-1 phenotypes. (A) ESCC cells were incubated with or without NK cells for 6 h. After removal of NK cells by washing, the remaining tumor cells were subjected to sphere formation assays. (B) Representative images (left panel) and summarized results (right panel) showing sphere formation efficiency of NK cells-treated ESCC cell lines (KYSE510 and TE1). Scale bar: 500 μm. In KYSE510 cells (upper panel) and TE1 cells (lower panel) survived from NK cells, the expression of stemness-associated genes (C), ULBP-1 mRNA (D) and ULBP-1 protein (E) were detected by qPCR and flow cytometry, respectively. Data are shown as the mean ± SD. MFI, mean fluorescence intensity. *p < 0.05, **p < 0.01, ***p < 0.001
To reinforce clinical relevance, nude mice were subcutaneously injected with KYSE510 cells and then intravenously treated with human activated NK cells twice. Adoptive transfer of NK cells significantly inhibited KYSE510 tumor growth in vivo (Additional file 8: Figure S5A and B). The qPCR analysis of tumor tissues confirmed higher expression of stemness-related genes in tumor tissues from NK cell treatment group compared to untreated group (Additional file 8: Figure S5C).
ULBP1 plays an important role in NK cell cytotoxicity against ESCC cells
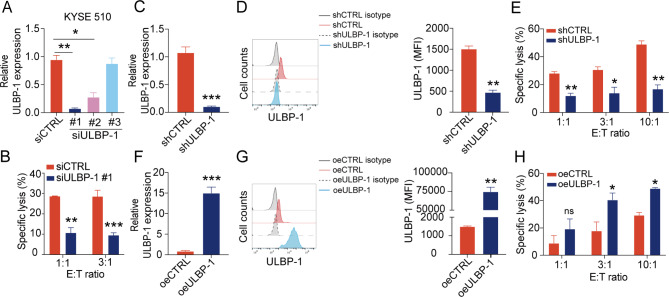
To investigate whether ULBP-1 is essential for lysis of ESCC cells by NK cells, we transfected synthetic siRNAs against ULBP-1 (siULBP-1) into monolayer culture of KYSE510 cells. The mRNA level of ULBP-1 was significantly decreased in the cells transfected with si-ULBP-1 #1 in comparison with those transfected with control siRNA (siCTRL) (Fig. 5A). The cytotoxic activity of NK cells was substantially suppressed by knockdown of ULBP-1 in KYSE510 cells (Fig. 5B), indicating that loss of ULBP-1 protects KYSE510 cells from lysis by NK cells. To further validate the functional role of ULBP-1, we silenced ULBP-1 expression with lentivirus carrying ULBP-1 shRNA (shULBP-1) in KYSE510 cells, verified by qPCR and flow cytometry (Fig. 5C and D). Consistently, stable knockdown of ULBP-1 significantly decreased NK cell-mediated cytotoxicity against KYSE510 cells (Fig. 5E). In addition, we established KYSE510 cells stably overexpressing ULBP-1 (oeULBP-1). The increased expression of surface ULBP-1 was confirmed in KYSE510 cells by qPCR and flow cytometry (Fig. 5F and G). As expected, NK cells exhibited more efficient cytotoxicity against KYSE510 cells overexpressing ULBP-1 at E: T ratios of 3:1 and 10:1 (Fig. 5H). We repeated the above experiments using another ESCC cell line TE1 and observed similar results (Additional file 9: Figure S6). These results strongly suggest that ULBP-1 plays an important role in the killing of ESCC cell lines by NK cells.
Fig. 5.
ULBP-1 plays an important role in NK cell-mediated cytotoxicity against KYSE510 cells. (A) qPCR analysis of ULBP-1 knockdown in KYSE510 cells transfected with control siRNA (siCTRL) or ULBP-1 siRNA (siULBP-1). (B) Cytolytic activity of NK cells against KYSE510 transfected with siCTRL or siULBP #1 at different E: T ratios. qPCR (C) and flow cytometry (D) analysis of stable ULBP-1 knockdown on KYSE510 cells transduced with control shRNA (shCTRL) or ULBP-1 shRNA (shULBP-1). E. Cytolytic activity of NK cells against KYSE510 cells transduced with shCTRL or shULBP-1 at different E: T ratios. qPCR (F) and flow cytometry (G) analysis of stable ULBP-1 over-expression on KYSE510 cells with control (oeCTRL) and ULBP-1 (oeULBP-1) vectors. H. Cytolytic activity of NK cells against KYSE510 cells with oeCTRL or oeULBP-1 vectors at different E: T ratios. Data are shown as the mean ± SD. MFI, mean fluorescence intensity. *p < 0.05, **p < 0.01, ***p < 0.001
ULBP-1 is involved in the resistance of tumor spheres derived from ESCC to NK cell cytotoxicity
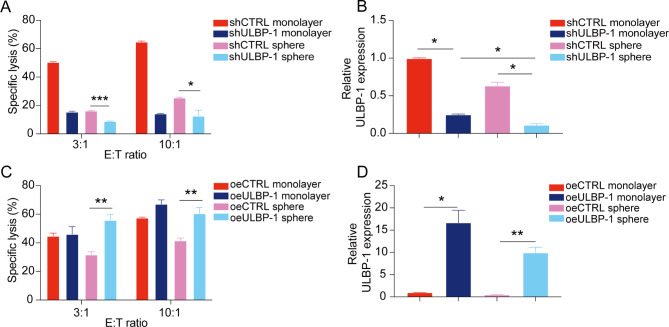
The possible role of ULBP-1 in protecting tumor spheres from NK cell lysis was explored in functional assays. Indeed, ULBP-1 knockdown further decreased the sensitivity of TE1 spheres to NK cell cytotoxicity (Fig. 6A). The lowest level of ULBP-1 was identified in spheres derived from TE1 shULBP1 cells (Fig. 6B). On the other hand, ULBP-1 overexpression restored the sensitivity of TE1 spheres to NK cell cytotoxicity (Fig. 6C). This is likely due to the increase of ULBP-1 expression in TE1 spheres (Fig. 6D). Thus, ULBP-1 might be a key mechanism for ESCC tumor spheres to escape NK cell lysis.
Fig. 6.
ULBP-1 is involved in the resistance of esophageal tumor spheres to NK cell-mediated cytotoxicity. (A) Cytolytic activity of NK cells against monolayer or sphere-derived TE1 cells in the presence or absence of ULBP-1 knockdown by shRNA. (B) qPCR analysis of ULBP-1 expression in monolayer or sphere-derived TE1 cells in the presence of absence of ULBP-1 knockdown by shRNA. (C) Cytolytic activity of NK cells against monolayer or sphere-derived TE1 cells in the presence or absence of ULBP-1 overexpression. (D) qPCR analysis of ULBP-1 expression in monolayer or sphere-derived TE1 cells in the presence of absence of ULBP-1 overexpression. Data are shown as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001
It has been reported that celecoxib (a selective COX-2 inhibitor) upregulates ULBP-1 expression in cancer cells, suggesting the role of COX-2 in inhibition of ULBP-1 expression [16, 17]. Interestingly, COX-2 is involved in promoting CSCs-like activity [18]. Therefore, we investigated whether ULBP-1 down-regulation is mediated by COX2 signaling in ESCC CSCs. As expected, ESCC spheres expressed higher level of COX-2, as compared to monolayer controls (Additional file 10: Figure S7A). More importantly, the treatment of a COX-2 inhibitor efficiently recovered the ULBP-1 expression in ESCC CSCs (Additional file 10: Figure S7B). The results indicate that increased COX-2 expression is likely related to the down-regulation of ULBP-1 in ESCC CSCs.
All-trans retinoic acid (ATRA) sensitizes ESCC-derived tumor spheres to NK cell killing
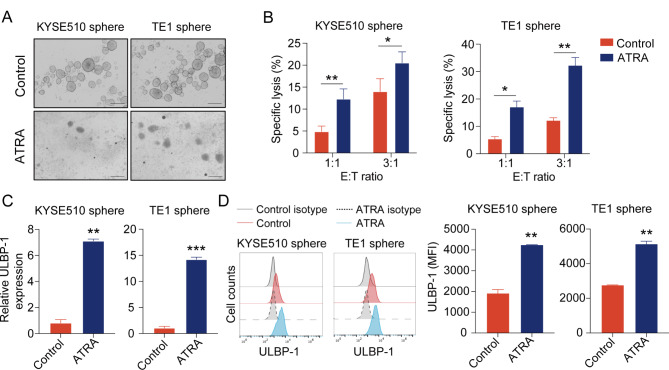
Finally, we further validate the sensitivity of ESCC tumor spheres to NK cell cytotoxicity through re-differentiation assays. ATRA is a powerful agent to induce differentiation of cancer stem cells [19]. To re-differentiate cancer stem cells, KYSE510 or TE1 spheres were treated with 10 µM ATRA for 48 h. The addition of ATRA led to morphological disassociation of tumor spheres (Fig. 7A) but had little impact on cell apoptosis (Additional file 11: Figure S8A). The re-differentiation of KYSE510 or TE1 spheres were further confirmed by decreased expression of stemness-related genes at the mRNA level (Additional file 11: Figure S8B). Of note, ATRA treatment caused KYSE510 or TE1 spheres to be more sensitive to NK cell-mediated lysis (Fig. 7B), suggesting the susceptibility of re-differentiated tumor spheres to NK cells. Furthermore, increased expression of ULBP-1 was observed in KYSE510 or TE1 spheres after ATRA treatment, by qPCR and flow cytometry (Fig. 7C and D). These results indicate that ATRA-treated ESCC spheres are more sensitive to NK cell cytotoxicity likely through up-regulation of ULBP-1.
Fig. 7.
ATRA promotes the sensitivity of esophageal tumor spheres to NK cell-mediated cytotoxicity. (A) Images showing the effect of ATRA on tumor spheres derived from KYSE510 and TE1. Scale bar: 500 μm. (B) ATRA-treated tumor spheres derived KYSE510 cells (left panel) and TE1 cells (right panel) at different E: T ratios. qPCR (C) and flow cytometry (D) analysis of ULBP-1 expression on ATRA-treated tumor spheres derived KYSE510 cells (left panel) and TE1 cells (right panel). Data are shown as the mean ± SD. MFI, mean fluorescence intensity. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
There is accumulating evidence that cancer stem cells play a crucial role in esophageal cancer progression and treatment resistance [20]. NK cells are key innate effector lymphocytes and represent an attractive strategy for cancer immunotherapy. In this study, we investigated the sensitivity of human esophageal CSCs to NK cell cytotoxicity. Sphere-forming cultures have been widely used in the enrichment of cancer stem cells [21, 22]. We have shown that tumor spheres derived from two ESCC cell lines are less susceptible to NK-mediated killing. Loss of ULBP-1 expression is likely involved in the evasion of esophageal CSCs from NK cell surveillance. ESCC-derived tumor spheres restored the sensitivity to NK cell cytotoxicity through ATRA-induced re-differentiation, providing a novel therapeutic potential of targeting esophageal CSCs.
Although the concept of cancer stem cells has been widely accepted, the model for study of esophageal CSCs remains to be established. Several specific markers have been used to identify and isolate CSCs from various cancers, including CD133, CD44, CD90, CD34, and ALDH1 [23–25]. In addition, isolation of side population (SP) cells via exclusion of Hoechst 33,342 dye was discovered in 1996 [26]. From then on, SP cells are regarded as a phenotype of stem cells and have been isolated from both blood and solid tumors [27, 28]. However, the phenotypes of CSCs are variable and often affected by the tumor microenvironment. The culture system of tumor spheres provides a convenient and efficient method to enrich cancer cells with stem-cell properties in many types of solid tumors, including esophageal carcinoma [29]. It remains poorly understood how the immune system recognizes and attacks cancer stem cells. We examined several stress-related ligands on the surface of ESCC-derived tumor spheres. Our data indicate that down-regulation of ULBP-1 may affect the sensitivity of ESCC CSCs to NK cell surveillance.
Recently NK cell-based therapies have become the new frontier of cancer treatment. However, the susceptibility of cancer stem cells to NK cells varies considerably in different types of cancers. It has been reported that melanoma stem cells defined by CD133 expression are more sensitive to NK cell-mediated cytotoxicity than non-stem cells [30]. Similarly, CSCs derived from bladder cancer [31], and liver cancer [32] also were more susceptible to NK cells. In contrast, breast cancer stem cells [33] and leukemic stem cells [34] are more resistant to NK cell-mediated cytotoxicity ex vivo. In this study, we demonstrated that ESCC-derived spheres exhibit reduced responsiveness to NK cells. We also showed that ESCC cells surviving from NK cell killing display CSCs properties. Our results further support the possibility of a tissue-specific context to NK cell killing. Indeed, dissecting the sensitivity of CSCs derived from different tissues to NK cells in the tumor microenvironment is of great scientific importance and translational potential.
The cytotoxic function of NK cells depends on the balance of signals delivered by activating and inhibitory cell surface receptors. MHC class I molecules are the most important ligands for inhibitory receptors, whereas NKG2D and NCR are the major activating receptor [35, 36]. Previous studies revealed that the higher susceptibility of CSCs to NK cell-mediated killing is likely due to lower expression of MHC class I and higher expression of NCR ligands in colorectal cancer [37] or up-regulated NKG2D ligands in ovarian cancer [38]. Similarly, different mechanisms contribute to the resistance of CSCs to NK cell cytotoxicity. For example, two NKG2D ligands, MICA and MICB, were down-regulated in breast CSCs [33]. We identified a significant loss of NKG2D ligand ULBP-1 in esophageal CSCs. These reports highlight the significant role of NKG2D ligands in anti-tumor immunity.
Induction of differentiation represents an attractive strategy for eradication of cancer stem cells. ATRA is a natural compound with pro-differentiating properties, and has achieved great success in the treatment of acute promyelocytic leukemia by inducing the differentiation of abnormal blasts to normal cells [39]. Recent studies have shown that ATRA also has anti-tumor activities for solid tumors such as breast cancer [40]. Similar to the previous reports [41], we found that ATRA can induce differentiation of ESCC-derived tumor spheres. More importantly, the re-differentiated tumor spheres became more sensitive to NK cell lysis. These results suggest that the combination of ATRA and NK cell therapy may provide a potential approach to eliminate esophageal CSCs. Further pre-clinical studies are needed to validate the combination strategy for the treatment of esophageal cancer.
Conclusions
In this study, we demonstrate that ESCC-derived CSCs were resistant to NK cell cytotoxicity, likely due to down-regulation of ULBP-1 expression. In addition, ATRA treatment can restore the sensitivity of ESCC-derived CSCs to NK cells through induction of CSCs re-differentiation. This study not only reveals the escape of ESCC-derived CSCs from NK cell killing, but also provides a combinatorial strategy for NK cell-based targeting of esophageal CSCs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We appreciate our laboratory members for their technical assistance and fruitful discussion.
Abbreviations
- ATRA
All-trans retinoic acid
- CFSE
Carboxyfluorescein succinimidyl ester
- CSC
Cancer stem-like cell
- ELISA
Enzyme-linked immunosorbent assay
- ESCC
Esophageal squamous cell carcinoma
- E
T ratio: Effector: target ratio
- FBS
Fetal bovine serum
- IL-2
Interleukin-2
- IFN-γ
Interferon-γ
- KIR
Killer cell immunoglobulin-like receptor
- MFI
Mean fluorescence intensity
- MIC
MHC I Chain-related
- NCR
Natural cytotoxicity receptors
- NK
Natural killer
- PBMC
Peripheral blood mononuclear cells
- PI
Propidium iodide
- qPCR
Quantitative PCR
- SD
Standard deviation
- SP
Side population
- ULBP
UL16-binding proteins
Author contributions
BT, MG, YZ, KZ and KN conducted the experiments and performed the data analysis. BT, MG and LH wrote and revised the manuscript. LH and YZ designed and supervised the project. All authors approved the final draft of the paper.
Funding
This study was supported by the National Natural Science Foundation of China (82172704, 81773045) and Medical Science and Technology Key Project of Henan Province, China (SBGJ202302038).
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study has been approved by the Ethical Committee of the First Affiliated Hospital of Zhengzhou University, China (Protocol No. 2019-KY-255).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bo Tang and Mengxing Guo contributed equally to this work.
Contributor Information
Yi Zhang, Email: yizhang@zzu.edu.cn.
Lan Huang, Email: lanhuang@zzu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021;134:783–91. 10.1097/CM9.0000000000001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555–67. 10.1016/S2214-109X(18)30127-X [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. 10.1016/j.stem.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 5.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34. 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 6.Lv M, Gong Y, Liu X, Wang Y, Wu Q, Chen J, Min Q, Zhao D, Li X, Chen D, et al. CDK7-YAP-LDHD axis promotes D-lactate elimination and ferroptosis defense to support cancer stem cell-like properties. Signal Transduct Target Ther. 2023;8:302. 10.1038/s41392-023-01555-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke MF. Clinical and therapeutic implications of cancer stem cells. N Engl J Med. 2019;380:2237–45. 10.1056/NEJMra1804280 [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 9.Bednarski JJ, Zimmerman C, Berrien-Elliott MM, Foltz JA, Becker-Hapak M, Neal CC, Foster M, Schappe T, McClain E, Pence PP, et al. Donor memory-like NK cells persist and induce remissions in pediatric patients with relapsed AML after transplant. Blood. 2022;139:1670–83. 10.1182/blood.2021013972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castriconi R, Daga A, Dondero A, Zona G, Poliani PL, Melotti A, Griffero F, Marubbi D, Spaziante R, Bellora F, et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J Immunol. 2009;182:3530–9. 10.4049/jimmunol.0802845 [DOI] [PubMed] [Google Scholar]
- 11.Ames E, Canter RJ, Grossenbacher SK, Mac S, Chen M, Smith RC, Hagino T, Perez-Cunningham J, Sckisel GD, Urayama S, et al. NK cells preferentially target tumor cells with a cancer stem cell phenotype. J Immunol. 2015;195:4010–9. 10.4049/jimmunol.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol. 2019;16:430–41. 10.1038/s41423-019-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyrysyuk O, Wucherpfennig KW. Designing Cancer immunotherapies that Engage T cells and NK cells. Annu Rev Immunol. 2023;41:17–38. 10.1146/annurev-immunol-101921-044122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupo KB, Matosevic S. CD155 immunoregulation as a target for natural killer cell immunotherapy in glioblastoma. J Hematol Oncol. 2020;13:76. 10.1186/s13045-020-00913-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raulet DH, Marcus A, Coscoy L. Dysregulated cellular functions and cell stress pathways provide critical cues for activating and targeting natural killer cells to transformed and infected cells. Immunol Rev. 2017;280:93–101. 10.1111/imr.12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Noh M, Hur D, Kim B, Kim Y, Lee H-K. Celecoxib upregulates ULBP–1 expression in lung cancer cells via the JNK/PI3K signaling pathway and increases susceptibility to natural killer cell cytotoxicity. Oncol Lett. 2020;20:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S-J, Ha G-H, Bae J-H, Kim GR, Son C-H, Park Y-S, Yang K, Oh S-O, Kim S-H, Kang C-D. COX-2- and endoplasmic reticulum stress-independent induction of ULBP-1 and enhancement of sensitivity to NK cell-mediated cytotoxicity by celecoxib in colon cancer cells. Exp Cell Res. 2015;330:451–9. 10.1016/j.yexcr.2014.09.008 [DOI] [PubMed] [Google Scholar]
- 18.Pang LY, Hurst EA, Argyle DJ. Cyclooxygenase-2: a role in Cancer Stem Cell Survival and Repopulation of Cancer cells during therapy. Stem Cells Int. 2016;2016:1–11. 10.1155/2016/2048731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunsu VO, Facey COB, Fields JZ, Boman BM. Retinoids as Chemo-Preventive and Molecular-targeted anti-cancer therapies. Int J Mol Sci 2021, 22. [DOI] [PMC free article] [PubMed]
- 20.Liu K, Zhao T, Wang J, Chen Y, Zhang R, Lan X, Que J. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 2019;28:21–8. 10.1016/j.canlet.2019.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbarzadeh M, Maroufi NF, Tazehkand AP, Akbarzadeh M, Bastani S, Safdari R, Farzane A, Fattahi A, Nejabati HR, Nouri M, Samadi N. Current approaches in identification and isolation of cancer stem cells. J Cell Physiol 2019. [DOI] [PubMed]
- 22.Ishiguro T, Ohata H, Sato A, Yamawaki K, Enomoto T, Okamoto K. Tumor-derived spheroids: relevance to cancer stem cells and clinical applications. Cancer Sci. 2017;108:283–9. 10.1111/cas.13155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nio K, Yamashita T, Kaneko S. The evolving concept of liver cancer stem cells. Mol Cancer. 2017;16:4. 10.1186/s12943-016-0572-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masciale V, Grisendi G, Banchelli F, D’Amico R, Maiorana A, Sighinolfi P, Stefani A, Morandi U, Dominici M, Aramini B. Isolation and identification of cancer stem-like cells in adenocarcinoma and squamouscell carcinoma of the lung: a pilot study. Front Oncol. 2019;18:1394. 10.3389/fonc.2019.01394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu SS, Cirillo N. The molecular markers of cancer stem cells in head and neck tumors. J Cell Physiol. 2020;235:65–73. 10.1002/jcp.28963 [DOI] [PubMed] [Google Scholar]
- 26.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. 10.1084/jem.183.4.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris MA, Yang H, Low BE, Mukherjee J, Guha A, Bronson RT, Shultz LD, Israel MA, Yun K. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68:10051–9. 10.1158/0008-5472.CAN-08-0786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M, Bai H, Jethava Y, Wu Y, Zhu Y, Yang Y, Xia J, Cao H, Franqui-Machin R, Nadiminti K, et al. Identification and characterization of tumor-initiating cells in multiple myeloma. J Natl Cancer Inst. 2020;112:507–15. 10.1093/jnci/djz159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo M, Lian J, Liu Y, Dong B, He Q, Zhao Q, Zhang H, Qi Y, Zhang Y, Huang L. Loss of miR-637 promotes cancer cell stemness via WASH/IL-8 pathway and serves as a novel prognostic marker in esophageal squamous cell carcinoma. Biomark Res. 2022;10:77. 10.1186/s40364-022-00424-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietra G, Manzini C, Vitale M, Balsamo M, Ognio E, Boitano M, Queirolo P, Moretta L, Mingari MC. Natural killer cells kill human melanoma cells with characteristics of cancer stem cells. Int Immunol. 2009;21:793–801. 10.1093/intimm/dxp047 [DOI] [PubMed] [Google Scholar]
- 31.Ferreira-Teixeira M, Paiva-Oliveira D, Parada B, Alves V, Sousa V, Chijioke O, Munz C, Reis F, Rodrigues-Santos P, Gomes C. Natural killer cell-based adoptive immunotherapy eradicates and drives differentiation of chemoresistant bladder cancer stem-like cells. BMC Med. 2016;14:163. 10.1186/s12916-016-0715-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weng J, Han X, Liu K, Yang J, Wei S, Zhang Y, Zeng F, Li Y, Shen L, Gao Y. CD44 3’-untranslated region functions as a competing endogenous RNA to enhance NK sensitivity of liver cancer stem cell by regulating ULBP2 expression. Int J Biol Sci. 2019;15:1664–75. 10.7150/ijbs.35216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Wang Q, Wang Z, Jiang J, Yu SC, Ping YF, Yang J, Xu SL, Ye XZ, Xu C, et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014;74:5746–57. 10.1158/0008-5472.CAN-13-2563 [DOI] [PubMed] [Google Scholar]
- 34.She M, Niu X, Chen X, Li J, Zhou M, He Y, Le Y, Guo K. Resistance of leukemic stem-like cells in AML cell line KG1a to natural killer cell-mediated cytotoxicity. Cancer Lett. 2012;318:173–9. 10.1016/j.canlet.2011.12.017 [DOI] [PubMed] [Google Scholar]
- 35.Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol. 2018;50:102–11. 10.1016/j.coi.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan S, Guo W, Xu Z, He Y, Liang C, Mo Y, Wang Y, Xiong F, Guo C, Li Y, et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18:29. 10.1186/s12943-019-0956-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tallerico R, Todaro M, Di Franco S, Maccalli C, Garofalo C, Sottile R, Palmieri C, Tirinato L, Pangigadde PN, La Rocca R, et al. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J Immunol. 2013;190:2381–90. 10.4049/jimmunol.1201542 [DOI] [PubMed] [Google Scholar]
- 38.Koh J, Lee S-b, Park H, Lee HJ, Cho NH, Kim J. Susceptibility of CD24(+) ovarian cancer cells to anti-cancer drugs and natural killer cells. Biochem Biophys Res Commun. 2012;427:373–8. 10.1016/j.bbrc.2012.09.067 [DOI] [PubMed] [Google Scholar]
- 39.Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–21. 10.1056/NEJMoa1300874 [DOI] [PubMed] [Google Scholar]
- 40.Wu MJ, Kim MR, Chen YS, Yang JY, Chang CJ. Retinoic acid directs breast cancer cell state changes through regulation of TET2-PKCζ pathway. Oncogene. 2017;36:3193–206. 10.1038/onc.2016.467 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 41.Zhang Y, Guan DX, Shi J, Gao H, Li JJ, Zhao JS, Qiu L, Liu J, Li N, Guo WX, et al. All-trans retinoic acid potentiates the chemotherapeutic effect of cisplatin by inducing differentiation of tumor initiating cells in liver cancer. J Hepatol. 2013;59:1255–63. 10.1016/j.jhep.2013.07.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.