Abstract
Although viral gene expression occurs in the peripheral nervous system during acute infection, bovine herpesvirus 1 (BHV-1) gene expression is extinguished, many neurons survive, and latency ensues. The only abundant viral transcript expressed during latency is the latency-related (LR) RNA, which is alternatively spliced in trigeminal ganglia during acute infection (L. Devireddy and C. Jones, J. Virol. 72:7294–7301, 1998). A subset of neurons express a protein encoded by the LR gene and the LR protein (LRP) is associated with cyclin-dependent kinase 2 (Cdk2)/cyclin complexes during productive infection (Y. Jiang, A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones, J. Virol. 72:8133–8142, 1998). LR gene products inhibit cell cycle progression, perhaps as a result of LRP interacting with Cdk2/cyclin complexes. During acute infection, expression of cyclin A occurs in trigeminal ganglionic neurons (L. M. Schang, A. Hossain, and C. Jones, J. Virol. 70:3807–3814, 1996). Inappropriate expression of G1- and S-phase cyclins can initiate programmed cell death (PCD), apoptosis, in neurons, suggesting that LR gene products inhibit PCD. To test this hypothesis, we modified an assay to measure PCD frequency in transiently transfected cells. C6-ceramide, fumonisin B1 (FB1), or etoposide was used to initiate PCD following transfection of cells with plasmids expressing LR gene products and the β-galactosidase gene. Transfected cells that survived were quantified by counting β-galactosidase-positive cells. Plasmids that expressed LR gene products promoted survival of monkey kidney (CV-1), human lung (IMR-90), or mouse neuroblastoma (neuro-2A) cells after induction of PCD. Plasmids with termination codons at the beginning of LR open reading frames or deletion of sequences that mediate splicing of LR RNA did not promote cell survival following PCD induction. We hypothesize that LR gene products play a role in promoting survival of postmitotic neurons during acute infection or reactivation.
Bovine herpesvirus 1 (BHV-1) is a significant bovine pathogen, which causes respiratory disease, abortion, genital disease, or occasionally encephalitis (reviewed in reference 29). Like other members of the Alphaherpesvirinae subfamily, BHV-1 establishes latent infection in sensory ganglionic neurons (reviewed in reference 29). Viral DNA persists in these neurons for the lifetime of infected cattle but can periodically reactivate and spread. In contrast to the 70 to 80 viral genes expressed during productive infection, latency-related (LR) RNA is the only abundant viral transcript detected in latently infected neurons. A small fraction of LR RNA is polyadenylated and alternatively spliced in trigeminal ganglia (TG), suggesting that this RNA is translated into an LR protein (LRP) (8, 21, 27). LR gene products inhibit S-phase entry, and LRP is associated with cyclin-dependent kinase 2 (Cdk2)/cyclin complexes (27, 46). Cdk2/cyclin complexes regulate the transition from G1 to S to G2 (reviewed in reference 19) and are required for DNA replication (30). Cdk2 activity is stimulated after herpes simplex virus type 2 (HSV-2) infection (22) and is important for HSV-1 infection (47, 48). It is reasonable to hypothesize that members of the Alphaherpesvirinae subfamily utilize Cdk2 and perhaps other Cdks to stimulate viral DNA replication and transcription. Although the functional significance of the interactions between LRP and Cdk2/cyclin is not known, these interactions are likely to be important.
Herpesviruses can induce programmed cell death (PCD), or apoptosis, when cultured cells are infected (reviewed in references 18 and 51). BHV-1 also induce PCD after infection of cultured cells (9, 14–17) or calves (53). Neuronal PCD occurs during neurodegenerative disorders, trauma, or imbalances of growth factors and cytokines (reviewed in references 10, 25, and 54). Expression of cell cycle regulatory proteins is frequently observed in neurons undergoing PCD, suggesting that altered Cdk activity initiates PCD (reviewed in reference 49). The HSV-1 US3 kinase plays a role in preventing PCD (1, 33), suggesting that regulation of PCD is crucial for pathogenesis. Inhibiting neuronal damage and/or PCD may also be important because the primary site of latency for BHV-1 is sensory neurons.
This study demonstrated that LR gene products inhibit or delay PCD following transient transfection of CV-1 cells, low-passage human fibroblasts, or mouse neuroblastoma cells. Although there was a correlation between LR protein expression and cell survival, we cannot exclude the possibility that LR RNA by itself is important. We hypothesize that LR gene products promote neuronal survival by inhibiting PCD.
MATERIALS AND METHODS
Cells.
Cells were plated at a density of 5 × 105/100-mm-diameter plastic dish in Earle’s modified Eagle’s medium supplemented with 5% fetal bovine serum (FBS). CV-1 cells were split at a 1:5 ratio every 4 or 5 days. Human primary lung fibroblasts (IMR-90 cells) were obtained from the American Type Culture Collection (ATCC; Rockville, Md. and split in a 1:3 ratio every 5 days. IMR-90 cells were split five to seven times and then discarded. Mouse neuroblastoma (neuro-2A; ATCC CCL131) cells were grown in Earle’s minimal essential medium supplemented with 5% FBS. All media contained penicillin (10 U/ml) and streptomycin (100 μg/ml).
β-Gal cotransfection and analysis of cell death.
To quantitatively measure cell death, we modified a previously described assay (23, 24, 31, 35) that entails cotransfecting a β-galactosidase (β-Gal) expression plasmid (pCMV-β-gal) and a gene of interest by calcium phosphate precipitation (5, 13). If a gene induces PCD or is toxic to cells, the number of β-Gal-positive cells decrease. Conversely, a gene that inhibits PCD maintains the number of β-Gal-positive cells after inducing PCD. Cells were plated at a density of 2 × 105/well in six-well plastic plates (35 mm/well) 12 to 16 h prior to transfection. After transfection, a glycerol shock (20% glycerol–phosphate-buffered saline [PBS]) was performed for 4 min, followed by two PBS washes. Fresh medium containing 5% FCS and 5 mM sodium butyrate was added to the cells to facilitate transfection efficiency. Cells were then treated (37°C for 48 h) with 25 μM fumonisin B1 (FB1) (5, 52) or 15 μM C6-ceramide (2, 40) to initiate PCD.
Neuro-2A cells were transfected as described above except that the glycerol shock was not performed and cultures were treated with 2.5 mM sodium butyrate. Cultures were subsequently treated with 15 μM etoposide (catalogue no. E1383; Sigma) to induce PCD. Etoposide is an anticancer agent that inhibits topoisomerase II. Cells treated with etoposide have higher levels of DNA damage (double- and single-stranded DNA), especially cells that are in late S and G2 (reviewed in references 13a and 39). At 48 h after etoposide treatment, β-Gal-positive cells were observed microscopically. The number of stained blue cells was counted by identifying the same area of each plate. At least five fields per plate were counted (>500 cells), and the average number of cells per field was calculated.
Plasmids.
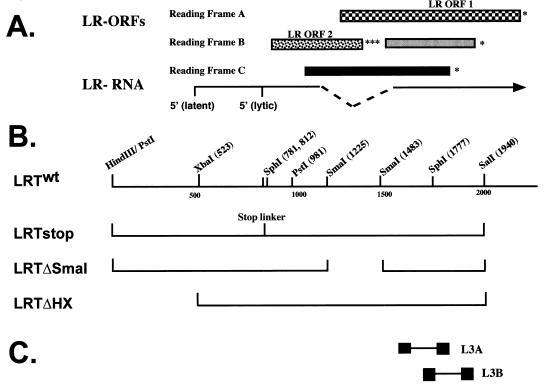
The various constructs (see Fig. 3) were generated by standard recombinant techniques and as previously described (21, 46). To construct LRTΔSmaI, plasmid LRTwt was digested with SmaI, and the large fragment was purified and religated. LRTstop contains a stop codon linker in the SphI sites located at positions 781 and 812 of the LR gene. This was accomplished by insertion of the PstI fragment containing the first 981 nucleotides (nt) of the LR gene into the pBlueBacHis vector and digestion with SphI; large fragment was purified, and an SphI linker containing stop codons in all three open reading frames (ORFs) (5′-CAGAATTCTAGTTAGTTAGCATG-3′) was ligated into the amino terminus of LR ORF2. This linker also contains an EcoRI site to facilitate screening.
FIG. 3.
Schematic of LR gene and mutants. (A) Locations of the LR gene and ORFs. The 5′ ends of the LR RNA were mapped by RACE (rapid amplification of cDNA ends) PCR or primer extension (3, 21). The vertical lines denote the 5′ termini of the transcripts. In latently infected cattle, the 5′ terminus of LR RNA has additional leader sequences. Splicing of LR RNA (dashed lines) occurs within the region of the transcript that would eliminate the three stop codons of LR ORF2 (8, 21) and may yield transcripts that encode LRP isoforms. The two major ORFs are marked LR ORF 1 and LR ORF 2 (32). The other regions that have the potential to encode a protein (░⃞ and ■) are in reading frames B and C respectively, but do not have a methionine residue at their amino termini. Asterisks indicate where in-frame stop codons are located. (B) Partial restriction map of the 2-kb LR gene. Construction of the mutants is described in Materials and Methods. (C) Locations of the LR primers used to detect LR RNA (21). Sequences of these primers are given in Materials and Methods.
pCMVCpIAP (hereafter referred to as CpIAP), a plasmid that contains the baculovirus antiapoptotic gene iap (6), was obtained from Lois Miller (University of Georgia, Athens). The adenovirus E1A gene was obtained from E. White (Rutgers University, Piscataway, N.J.). pCMV-β-gal was purchased from Clontech (Palo Alto, Calif.). Plasmid E2.6 contains the BHV-1 ICP0 gene (bICP0) and was obtained from M. Schwyzer (Zurich, Switzerland). Two rounds of cesium chloride centrifugation were used to purify plasmids after bacteria were lysed with alkali and sodium dodecyl sulfate (SDS).
Preparation of RNA and RT-PCR.
RNA from transfected CV-1 cells was prepared as described previously (8, 21). RNA was quantified spectrophotometrically (optical density at 260 nm) and stored at −80°C in 3 volumes of ethanol. Reverse transcriptase PCR (RT-PCR) and LRT primers were described previously (21). The L3A upstream sense primer spans nt 1672 to 1693 (5′-CGCTCCCCTTCGTCCCTCCTCA-3′). The L3A downstream antisense primer is complementary to nt 1835 to 1815 (5′-GACGAGACCCCCGATTGCCG-3′). The L3B upstream sense primer spans nt 1755 to 1775 (5′-TTCTCTGGGCTCGGGGCTGC-3′). The downstream antisense primer is complementary to 1924 to 1947 (5′-AGAGGTCGACAAACACCCGCGGT-3′). The nucleotide numbering system for the LR gene was previously described (32). Actin primers to screen RT reactions were derived from rat sequences. The Actin+ primer is (5′-GTGGGGCGCCCCAGGCACCA-3′). The Actin− primer is (5′-CTCCTTAATGTCACGCACGATTTC-3′). RNA samples were treated with 2 U of DNase 1 (RNase free) and 10 U of RNasin (Promega, Madison, Wis.) per μg of total RNA for 30 min at 20°C. RT-PCR was performed essentially as described previously (21, 46). Amplified products were detected by 2% agarose gel electrophoresis.
Western blot analysis.
Preparation of extracts and Western blot analysis were performed as described previously (4, 21, 22, 46). The P2 antibody is directed against an 18-amino-acid peptide near the N terminus of LR ORF2 and specifically recognizes a 40 kDa protein in infected or transiently transfected cells (21, 27).
Statistical analysis.
Statistical analysis was performed with the SSPS program, student version. Values shown in Fig. 2 and 6 are normalized to those for the vector-alone transfected controls (defined as 0). Data are average mean differences from vector-alone control (n = 7). P values represent the probability that the result occurred by chance, using 95% confidence; P < 0.05 is statistically significant. Error bars represent the standard error of the mean differences.
FIG. 2.
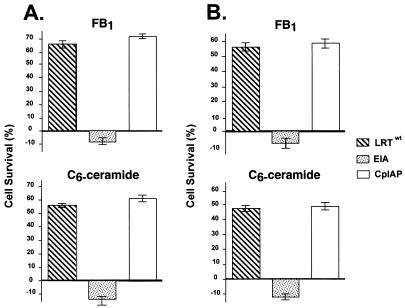
Regulation of PCD by LRT. CV-1 cells (A) or IMR-90 cells (B) were transfected with pCMV-β-gal (2 μg) and the designated plasmid (4 μg). After transfection, cells were treated with 25 μM FB1 or 15 μM C6-ceramide for 48 h, and β-Gal-positive cells were identified. The number of blue cells in five fields was counted. The blue cells that survived FB1 or C6-ceramide treatment were divided by the blue cells in control cultures to yield percent cell survival; the data presented are normalized to that for empty vector control (pCDNA 3.1). For each transfection, the percent cell survival for pCDNA3.1 was subtracted from values for the other plasmid transfections because previous studies have demonstrated that not all cells undergo apoptosis when they are treated with FB1 or C6-ceramide (5, 52). The resulting values were averaged, thus providing the mean difference for cell survival (n = 7). Error bars represent the standard error of the mean difference. (A) P values for CV-1 cells treated with the indicated plasmids and treated with C6-ceramide were as follows: LRTwt, 0.0001; CpIAP, 0.0001; and EIA, 0.101. P values for transfected CV-1 cells treated with FB1 were as follows: LRTwt, 0.0001; CpIAP, 0.0001; and EIA, 0.150. (B) P values for transfected IMR-90 cells treated with C6-ceramide were as follows: LRTwt, 0.0001; CpIAP, 0.0001; and EIA, 0.072. P values for transfected IMR-90 cells treated with FB1 were as follows: LRTwt, 0.0001; CpIAP, 0.0001; and EIA, 0.080. The difference in values between LRTwt and CpIAP in panels A and B had a P value of 0.619 (average).
FIG. 6.
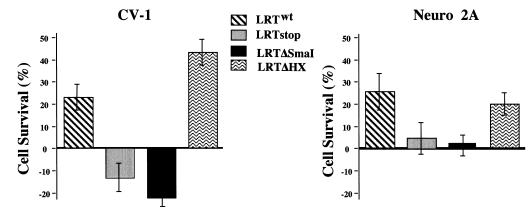
Survival of CV-1 and neuro-2A cells after transfection with LR gene mutants. CV-1 and neuro-2A cells were transfected with pCMV-β-gal (2 μg) and the designated plasmid (4 μg). After transfection, cells were treated with 15 μM C6-ceramide (CV-1) or etoposide (neuro-2A) for 48 h, fixed, and stained for β-Gal expression. The number of blue cells was counted in exactly the same areas of each well. The number of blue cells that survived treatment was divided by the number of blue cells in each respective nontreated control culture to yield percent cell survival. The data shown are the mean differences as determined by normalizing to pCDNA3.1 alone control as described for Fig. 2. P values for CV-1 cells transfected with the indicated plasmids and treated with C6-ceramide were as follows: LRTwt, 0.0020; LRTstop, 0.0680; LRTΔSmaI, 0.0010; and LRTΔHX, 0.00015. P values for transfected neuro-2A cells treated with etoposide were as follows: LRTwt, 0.0110; LRTstop, 0.524; LRTΔSmaI, 0.801; and LRTΔHX, 0.003.
RESULTS
Analysis of PCD in cells transfected with the LR gene.
Previous studies concluded LR gene products inhibit G1-to-S transition (46) and LRP is associated with Cdk2/cyclin complexes (27). Several independent studies have concluded that cell cycle factors, in addition to regulating cell cycle progression, play a role during PCD (11, 38, 41–43, 49). To test whether the LR gene influences cell survival, we modified a β-Gal cotransfection assay (24, 27, 31, 35) to measure the effects of various genes on PCD. Two sphingoid bases, FB1 or C6-ceramide, can induce PCD in mammalian cells (2, 5, 40, 52). Cells treated with FB1 or C6-ceramide exhibit the hallmarks of PCD: DNA laddering, formation of apoptotic bodies, and condensation of chromatin. Both agents kill approximately 80% of treated cells (5, 52). C6-ceramide is one of the central regulators of the sphingomyelin signal transduction pathway, a ubiquitous signaling system that links specific cell surface receptors and environmental stresses to the nucleus. The sphingomyelin pathway is crucial during PCD initiated by tumor necrosis factor alpha, FAS, and ionizing radiation (2, 26, 40). FB1 inhibits ceramide synthase, the enzyme that synthesizes complex sphingolipids (including ceramide). We have characterized the differences between the mechanism of PCD initiated by these two sphingoid bases (5, 52) and thus are useful reagents for analyzing PCD. Monkey kidney (CV-1) and primary human lung (IMR-90) cells were used for these studies because these cell types are nontumorigenic and susceptible to PCD.
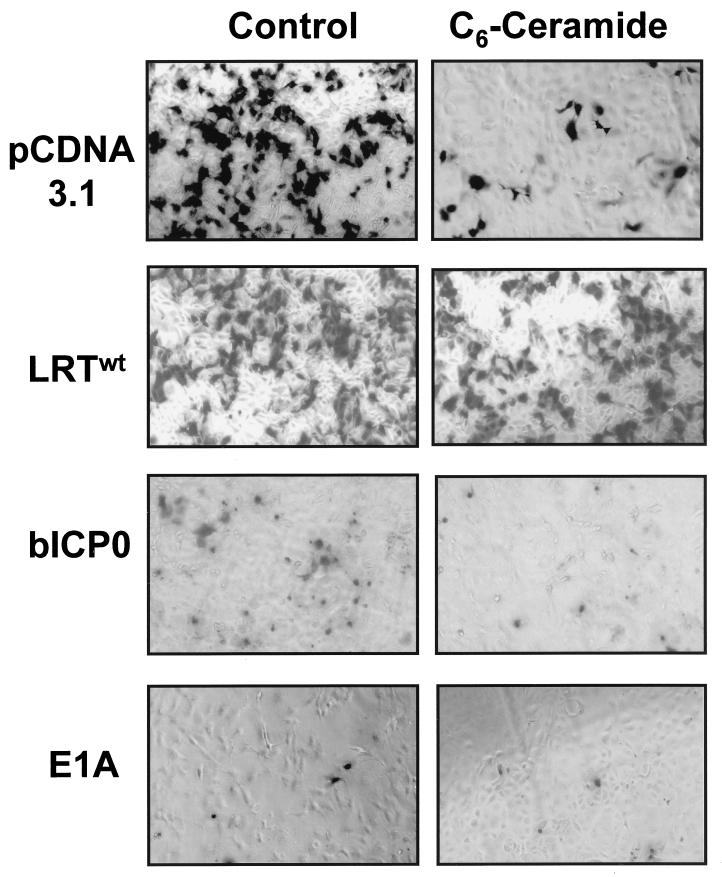
A plasmid that expresses LR gene products (LRTwt) enhanced cell survival after treatment with C6-ceramide or FB1, as judged by an increase in the number of surviving β-Gal-positive CV-1 cells (Fig. 1 and 2A). As expected, the frequency of β-Gal-positive cells was reduced dramatically when CV-1 cells were cotransfected with pCDNA/3.1 and pCMV-β-Gal followed by treatment with C6-ceramide or FB1 (5) (Fig. 1). A plasmid that expresses bICP0 was used as a control because it contains sequences that overlap the LR gene. Overexpression of bICP0 in CV-1 cells reduced the number of β-Gal-positive CV-1 cells (Fig. 1) independent of treatment with C6-ceramide or FB1, suggesting that it was toxic. After transfection with bICP0, the β-Gal-positive cells were smaller and rounded compared to those transfected with LRTwt or pCDNA/3.1 (Fig. 1). Further studies will be necessary to determine whether bICP0 induces PCD or was merely toxic. The adenovirus E1A gene, which induces PCD (7, 45), reduced the number of β-Gal-positive cells independent of treatment with FB1 or C6-ceramide (Fig. 2A). As expected, CpIAP enhanced the survival of cells after treatment with C6-ceramide or FB1 (Fig. 2A).
FIG. 1.
Induction of PCD by FB1 or C6-ceramide. CV-1 cells were cotransfected with plasmid pCMV-β-gal (2 μg) and the designated plasmid (4 μg). After transfection, cells were treated with 20 μM C6-ceramide for 48 h, fixed, and stained with Bluo-Gal 5-bromo-3-indolyl-β-d-galactopyranoside for 24 h. As a control, some cultures were treated with PBS. Morphology of typical blue cells is shown.
To ensure that these findings were not a peculiarity of CV-1 cells, this study was repeated with IMR-90 cells. After transfection with LRTwt or CpIAP, a higher frequency of cells survived C6-ceramide or FB1 treatment relative to cultures transfected with the blank expression vector (Fig. 2B). LRTwt and CpIAP were statistically different from the vector alone. However, the difference between LRTwt and CpIAP was not statistically significant. E1A reduced the number of blue cells independent of treatment with C6-ceramide or FB1. In summary, these studies indicated that LR gene products promoted survival of CV-1 and IMR-90 cells after treatment with C6-ceramide or FB1.
Identification of LR gene sequences that inhibit PCD.
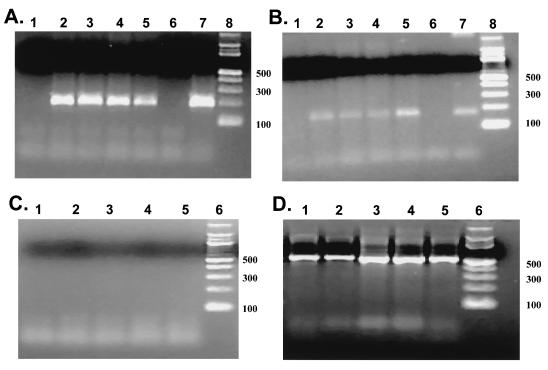
Three LR gene mutants were constructed to further characterize the sequences that were necessary to inhibit PCD (Fig. 3). LRTstop contains three in-frame stop codons at the amino terminus of LR ORF2 and thus should prevent translation of any ORF encoded by a LR RNA. Insertion of stop codons at this position was previously shown to prevent expression of a 40-kDa protein that was recognized by a LR ORF2-specific antibody P2 (21). Plasmid LRTΔSmaI has a 258-bp SmaI deletion that spans the intron/exon borders of LR RNA (8, 21). In LRTΔHX, 523 bp of the LR promoter was deleted. To test whether these constructs synthesized LR RNA, CV-1 cells were transfected, RNA was prepared 48 h after transfection, and RT-PCR was performed with LR-specific primers L3A and L3B (Fig. 3C). As expected, LRTwt synthesized RNA that was amplified with primers L3B (Fig. 4A, lane 5) and L3A (Fig. 4B, lane 5). L3A (Fig. 4B) and L3B (Fig. 4A) primers also amplified a similar-sized cDNA fragment, using RNA prepared from CV-1 cells transfected with LRTΔHX (lane 2), LRTstop (lane 3), or LRTΔSmaI (lane 4). These bands were amplified cDNA because no bands were observed when RT was omitted from the reaction (Fig. 4C). As expected, β-actin RNA was amplified in every sample (Fig. 4D). Since primers L3A and L3B are downstream of the SmaI restriction site (Fig. 3C), the SmaI deletion was not expected to interfere with amplification of the 3′ terminus of LR RNA.
FIG. 4.
Detection of LR RNA in CV-1 cells transfected with the LR gene deletion plasmids. RT-PCR was performed with RNA prepared from CV-1 cells transfected with pCDNA3.1 (lane 1), LRTΔHX (lane 2), LRTstop (lane 3), LRTΔSmaI (lane 4), and LRTwt (lane 5). In panels A and B, lane 6 was the no-template reaction, lane 7 was a reaction containing LATwt plasmid DNA, and lane 8 was a 100-bp molecular weight marker. Numbers on the right are sizes of the markers in base pairs. RT-PCR was performed as described in Materials and Methods. Amplified products were electrophoresed on 2% agarose gels. In panels A and B, RT-PCR was performed with primers L3B (1.5 mM MgCl2) and L3A (1.0 mM MgCl2), respectively. Primers L3B will amplify a 197-bp product, and primer L3A will amplify a 187-bp product (21). (C) The no-RT reaction using primers L3B. A similar result was obtained with primer L3A (50). (D) RT-PCR reactions amplified with β-actin primers. The actin primers span an intron, which allows identification of genomic DNA and cDNA in the same sample. Genomic DNA yields a 1,091-bp amplicon, whereas cDNA yields a 540-bp amplicon.
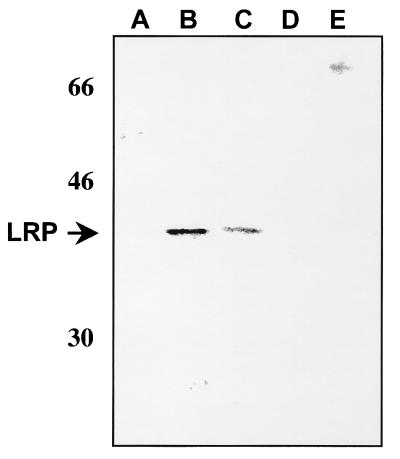
Transient transfection of COS-7 (21, 46), U2-OS (27), and 293 (Fig. 5) cells with the LR gene leads to expression of a 40-kDa protein that is recognized by the P2 antibody. We believe that the difficulty in detecting LRP in other transiently transfected cells (CV-1, neuro-2A, and primary human fibroblasts) is due to lower transfection efficiency. However, it cannot be ruled out that factors in these cell lines inhibit stable expression of LRP or the protein is not expressed at detectable levels (50). 293 cells transfected with LRTwt and LRTΔHX expressed a 40-kDa protein that was recognized by the P2 antibody (Fig. 5, lanes B and C, respectively). The same antibody did not recognize a 40-kDa protein in cells transfected with LRTstop (lane D) or LRTΔSmaI (lane E) or in mock-transfected cells (lane A). Since COS-7, U2-OS, and 293 cells are highly transformed and readily form tumors in immunodeficient mice, they are more resistant to apoptotic agents such as FB1 and C6-ceramide. Consequently, these cell lines are not good models for studies of apoptosis. In summary, all four plasmids containing the LR gene constructs synthesized RNA in CV-1 cells that was amplified by primers L3A and L3B. Only LRTwt and LRTΔHX expressed a 40-kDa protein in transiently transfected 293 cells.
FIG. 5.
Detection of a 40-kDa protein in cells transfected with LR gene constructs. Human (293) cells were transfected with the designated plasmids, whole-cell extract was prepared 48 h after transfection, and 50 μg of protein was electrophoresed on an SDS–12% polyacrylamide gel. Extracts were prepared from mock-transfected cells (A) and cells transfected with LRTwt (B), LRTΔHX (C), LRTstop (D), and LRTΔSmaI (E). Locations of molecular weight markers are shown at the left in kilodaltons. The arrow indicates the position of LRP.
The β-Gal assay was then used to determine the effectiveness of the different LR gene deletion constructs with respect to protection against apoptotic agents (Fig. 6). Two different cell lines, CV-1 and neuro-2A, were used for these studies. CV-1 cells were treated with C6-ceramide whereas neuro-2A cells required etoposide for induction of efficient PCD. LRTwt and LRTΔHX enhanced cell survival compared to pCDNA3.1 in CV-1 and neuro-2A cells. In contrast, LRTstop and LRTΔSmaI did not protect cells better then the vector alone in neuro-2A cells. In CV-1 cells, LRTstop was not significantly different from the vector control. The difference between LRTΔSmaI and the vector control was significantly different in CV-1 cells, suggesting that truncated LR gene products promoted PCD or cooperated with C6-ceramide to enhance cell death. Etoposide and the transcription factor E2F cooperate to induce PCD (39), indicating that these interactions can occur. There was also an increase in CV-1 cell survival after transfection with LRTΔHX compared to LRTwt. This was not observed in neuro-2A cells treated with etoposide and may reflect differences in cell types or the mechanism by which etoposide kills cells. In summary, this study demonstrated that LRTstop and LRTΔSmaI did not protect CV-1 and neuro-2A cells from PCD but LRTwt and LRTΔHX did.
DISCUSSION
This study provided evidence that LR gene products inhibited PCD induced by C6-ceramide, FB1, or etoposide. Three different cell types, including a cell line of neuronal origin, were used. LRTstop and LRTΔSmaI were unable to prevent PCD in any cell type tested and did not express a 40-kDa protein in 293 cells that was recognized by the P2 antibody. Although it is tempting to speculate that expression of LRP is required for preventing PCD, it is clear that LRTΔSmaI and LRTstop would express transcripts that are different from those expressed by LRTwt and LRTΔHX. Consequently, it cannot be ruled out that subtle quantitative or qualitative differences in the transcripts encoded by LRTΔSmaI and LRTstop play a role in cell survival. Further studies are necessary to prove that continual expression of LRP is necessary for inhibiting PCD.
The ability of LR gene products to inhibit cell cycle progression (46) and LRP to bind Cdk2/cyclins (27) may play a role in preventing PCD. A link between cell cycle regulatory proteins and PCD has been established. For example, cyclin A-dependent kinase activity is stimulated during PCD (37), and PCD is suppressed by dominant negative mutants of Cdk2 or Cdc2 (38). Second, cell cycle inhibitors promote survival of postmitotic neurons (41–43), and the Cdk inhibitor p21 can protect cells from PCD (12, 36). Third, proteolytic enzymes that are activated during PCD (caspases) induce cdk/cyclin activity in the early stages of PCD (34, 55). Genes that regulate PCD, Bcl-2 and BAX, modulate cdk2 activation during thymocyte apoptosis (11). Finally, FB1 treatment of CV-1 cells induces a transient increase in Cdk2 and Cdk4 activity (5). At this time, the factors that direct cell cycle regulators to initiate PCD but not cell cycle progression have not been identified.
BHV-1 induces PCD in lymphocytes (14–17), bovine kidney cells (9), and acutely infected cattle (53), suggesting that cells in the peripheral nervous system undergo PCD. During pathological states, neurons undergo PCD (10, 49, 54), indicating they are usually resistant to PCD. LR RNA and LRP may promote neuronal survival during establishment and maintenance of latency (outlined in Fig. 7). The LR gene inhibits the transactivation potential of bICP0 in transient transfection assays presumably because LR RNA interferes with bICP0 expression (3) and thus may interfere with productive infection. The interaction between LRP and Cdk2 (28) may also repress productive infection because roscovitine, a chemical that inhibits Cdk2, Cdc2, and Cdk5 activity (47, 48), inhibits HSV transcription and DNA replication. Cdk2 activity is also required for initiation of cellular DNA replication (30). Based on these observations, we hypothesize that LR RNA and LRP act in concert to inhibit productive viral gene expression and neuronal PCD during establishment and maintenance of latency.
FIG. 7.
Hypothetical model summarizing the effects that LR gene products have on neuronal survival. For details, see Discussion.
During reactivation, LR gene products may promote neuronal survival in the face of productive viral gene expression and DNA replication, thus maximizing virion production. Only 20% of neurons latently infected with BHV-1 actually reactivate following dexamethasone injection (44), suggesting that 80% of latently infected neurons resume latency. Thus, LR gene products may enhance neuronal survival in the event of incomplete reactivation. During reactivation, TG neurons may be more vulnerable to PCD because dexamethasone inhibits LR promoter activity (28), represses LR RNA expression in TG but initiates viral gene expression (44), and induces PCD (20). Considering LR RNA is alternatively spliced in neurons (8), it is possible that reactivation-specific factors facilitate reactivation. This hypothesis can be tested directly in cattle when a LR-negative mutant is constructed.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
This research was supported by grants from the USDA (9702394 and 9802064) and the Center for Biotechnology.
We thank Martin Schwyzer (Zurich, Switzerland) for E2.6, Eileen White (University of New Jersey Medical Center) for the E1A plasmid, L. Miller (University of Georgia) for the CpIAP plasmid, and Gerald Kutish and Gary Stevens for help with statistical analysis.
REFERENCES
- 1.Asano S, Honda T, Nishiyama T. US3 protein kinase of herpes simplex virus type 2 plays a role in protecting corneal epithelial cells from apoptosis in infected mice. J Gen Virol. 1999;80:51–56. doi: 10.1099/0022-1317-80-1-51. [DOI] [PubMed] [Google Scholar]
- 2.Bose R, Varheij M, Haimovitz A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates dauorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82:405–414. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- 3.Bratanich A C, Hanson N, Jones C. The latency related gene of bovine herpesvirus 1 inhibits the activity of immediate early transcription unit 1. Virology. 1992;191:988–991. doi: 10.1016/0042-6822(92)90278-w. [DOI] [PubMed] [Google Scholar]
- 4.Ciacci-Zanella J R, Merrill A H, Jr, Wang E, Jones C. Characterization of cell cycle arrest by fumonisin B1in CV-1 cells. Food Chem Toxicol. 1998;10:1–13. doi: 10.1016/s0278-6915(98)00034-9. [DOI] [PubMed] [Google Scholar]
- 5.Ciacci-Zanella, J. R., and C. Jones. Tumor necrosis factor but not p53 plays an important role in fumonisin B1 induced apoptosis. Food Chem. Toxicol. in press.
- 6.Clem R J, Miller L K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 8.Devireddy L R, Jones C. Alternative splicing of the latency-related transcript of bovine herpesvirus type 1 yields RNAs containing unique open reading frames. J Virol. 1998;72:7294–7301. doi: 10.1128/jvi.72.9.7294-7301.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devireddy L R, Jones C. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J Virol. 1999;73:3778–3788. doi: 10.1128/jvi.73.5.3778-3788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Mello S R. Molecular regulation of neuronal apoptosis. Curr Top Dev Biol. 1998;39:187–213. doi: 10.1016/s0070-2153(08)60456-1. [DOI] [PubMed] [Google Scholar]
- 11.Gil-Gomez G, Berns A, Brady H J M. A link between cell cycle and cell death: Bax and Bcl-2 modulate cdk2 activation during thymocyte apoptosis. EMBO J. 1998;17:7209–7218. doi: 10.1093/emboj/17.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorospe M, Wang X, Guyton K Z, Holbrook N J. Protective role of p21Waf1/Cip1against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol Cell Biol. 1996;16:6654–6660. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham T L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 13a.Hainsworth J D, Greco F A. Etoposide: twenty years later. Ann Oncol. 1995;6:323–341. doi: 10.1093/oxfordjournals.annonc.a059180. [DOI] [PubMed] [Google Scholar]
- 14.Hanon E, Vanderplasschen A, Lyaku J, Keil G, Denis M, Pastoret P P. Inactivated bovine herpesvirus 1 induces apoptotic cell death of mitogen-stimulated bovine peripheral blood mononuclear cells. J Virol. 1995;70:4116–4120. doi: 10.1128/jvi.70.6.4116-4120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanon E, Vanderplasschen A, Lyaku J, Keil G, Denis M, Pastoret P-P. Inactivated bovine herpesvirus 1 induces apoptotic cell death of mitogen-stimulated bovine peripheral blood mononuclear cells. J Virol. 1996;70:4116–4120. doi: 10.1128/jvi.70.6.4116-4120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanon E, Hoornaert S, Dequiedt F, Vanderplasschen A, Lyaku J, Willems L, Pastoret P-P. Bovine herpesvirus 1-induced apoptosis occurs at the G0/G1 phase of the cell cycle. Virology. 1997;232:351–358. doi: 10.1006/viro.1997.8562. [DOI] [PubMed] [Google Scholar]
- 17.Hanon E, Meyer G, Vanderplasschen A, Dessy-Doize C, Thiry E, Pastoret P-P. Attachment but not penetration of bovine herpesvirus 1 is necessary to induce apoptosis in target cells. J Virol. 1998;72:7638–7641. doi: 10.1128/jvi.72.9.7638-7641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardwicke J M. Viral interference with apoptosis. Cell Dev Biol. 1998;9:339–349. doi: 10.1006/scdb.1998.0243. [DOI] [PubMed] [Google Scholar]
- 19.Heichman K A, Roberts J M. Rules to replicate by. Cell. 1994;79:557–562. doi: 10.1016/0092-8674(94)90541-x. [DOI] [PubMed] [Google Scholar]
- 20.Heimberg A, Auphan N, Caelles C, Karin M. Glucocorticoid-induced apoptosis of human leukemic cells is caused by the repressive function of the glucocorticoid receptor. EMBO J. 1995;14:452–460. doi: 10.1002/j.1460-2075.1995.tb07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hossain A, Schang L, Jones C. Identification of gene products encoded by the latency-related gene of bovine herpesvirus type 1. J Virol. 1995;69:5345–5352. doi: 10.1128/jvi.69.9.5345-5352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hossain A, Holt T, Ciacci-Zanella J, Jones C. Analysis of cyclin dependent kinase activity after herpes simplex virus type 2 infection. J Gen Virol. 1997;78:3341–3348. doi: 10.1099/0022-1317-78-12-3341. [DOI] [PubMed] [Google Scholar]
- 23.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 24.Hsu H, Hong-Bing S, Ming-Gui P, Goeddel D V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 25.Hu S, Peterson P K, Chao C C. Cytokine-mediated neuronal apoptosis. Neurochem Int. 1997;30:427–431. doi: 10.1016/s0197-0186(96)00078-2. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis W D, Kolesnick R N, Fornari F A, Traylor R S, Gerwitz D A, Grant S. Induction of apoptotic DNA damage and cell death by activation of the sphinomyelin pathway. Proc Natl Acad Sci USA. 1994;91:73–77. doi: 10.1073/pnas.91.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Hossain A, Winkler M T, Holt T, Jones C. Interaction between cyclin-dependent kinases and the bovine herpesvirus 1 latency-related protein. J Virol. 1998;72:8133–8142. doi: 10.1128/jvi.72.10.8133-8142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones C, Delhon G, Bratanich A, Rock D. Analysis of the transcriptional promoter which regulates the latency-related transcript of bovine herpesvirus 1. J Virol. 1990;64:1164–1170. doi: 10.1128/jvi.64.3.1164-1170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones C. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv Virus Res. 1998;51:47–99. doi: 10.1016/s0065-3527(08)60784-8. [DOI] [PubMed] [Google Scholar]
- 30.Krude T, Jackman M, Pines J, Laskey R A. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Kinoshita M, Noda M, Copeland N G, Jenkins N A. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1β-converting enzyme. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 32.Kutish G, Mainprize T, Rock D L. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J Virol. 1990;64:5730–5737. doi: 10.1128/jvi.64.12.5730-5737.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leopardi R, Vansant C, Roizman B. The herpes simplex virus 1 protein kinase U(S)3 is required for protection from apoptosis induced by the virus. Proc Natl Acad Sci USA. 1997;94:7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levkau B, Koyama H, Raines E W, Clurman B E, Herrem B, Orth K, Roberts J M, Ross R. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of cdk2: role of a caspase cascade. Mol Cell. 1998;4:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z-G, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Yamagishi N, Yagi T, Takeba H. Mutated p21WAF/1/CIP1/SDI1 lacking CDK-inhibitory activity fails to prevent apoptosis in human colorectal carcinoma cells. Oncogene. 1998;16:705–712. doi: 10.1038/sj.onc.1201585. [DOI] [PubMed] [Google Scholar]
- 37.Meikrantz W, Geisselbrecht S, Tam S W, Schlegel R. Activation of cyclin A-dependent protein kinases during apoptosis. Proc Natl Acad Sci USA. 1994;91:3754–3758. doi: 10.1073/pnas.91.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meikrantz W, Schlegel R. Suppression of apoptosis by dominant negative mutants of cyclin-dependent protein kinases. J Biol Chem. 1996;271:10205–10209. doi: 10.1074/jbc.271.17.10205. [DOI] [PubMed] [Google Scholar]
- 39.Nip J, Strom D K, Fee B E, Zambetti G, Cleveland J L, Hiebert S W. E2F-1 cooperates with topoisomerase II inhibition and DNA damage to selectively augment p53-independent apoptosis. Mol Cell Biol. 1997;17:1049–1056. doi: 10.1128/mcb.17.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obeid L M, Linardic C M, Karolak L A, Hannun Y A. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 41.Park D S, Farinellis S E, Greene L A. Inhibitors of cyclin-dependent kinases promote survival of post-mitotic neuronally differentiated PC12 cells and sympathetic neurons. J Biol Chem. 1996;14:8161–8169. doi: 10.1074/jbc.271.14.8161. [DOI] [PubMed] [Google Scholar]
- 42.Park D S, Morris E J, Greene L A, Geller H M. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neural apoptosis. J Neurosci. 1997;17:1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park D S, Levine B, Ferrari G, Greene L A. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-derived sympathetic neurons. J Neurosci. 1997;17:8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rock D, Lokensgard J, Lewis T, Kutish G. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J Virol. 1992;66:2484–2490. doi: 10.1128/jvi.66.4.2484-2490.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabbatini P, Lin J, Levine A J, White E. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 46.Schang L M, Hossain A, Jones C. The latency-related gene of bovine herpesvirus type 1 encodes a factor which inhibits cell cycle progression. J Virol. 1996;70:3807–3814. doi: 10.1128/jvi.70.6.3807-3814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schang L M, Phillips J, Schaffer P A. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J Virol. 1998;72:5626–5637. doi: 10.1128/jvi.72.7.5626-5637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schang L M, Rosenberg A, Schaffer P A. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J Virol. 1999;73:2161–2172. doi: 10.1128/jvi.73.3.2161-2172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirvan A, Ziv I, Zilkha-Falb R, Machlin T, Barzilai A, Melamed E. Expression of cell cycle-related genes during neuronal apoptosis: is there a distinct pattern? Neurochem Res. 1998;23:767–777. doi: 10.1023/a:1022415611545. [DOI] [PubMed] [Google Scholar]
- 50.Stone, M., and C. Jones. Unpublished data.
- 51.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, Jones C, Zanella J, Holt T, Gilchrist D, Dickman M. Fumonisins and Alternaria alternata lycopersici toxins: sphinganine analog mycotoxins induce apoptosis in monkey kidney cells. Proc Natl Acad Sci USA. 1996;93:3461–3465. doi: 10.1073/pnas.93.8.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winkler M T C, Doster A, Jones C. Bovine herpesvirus 1 can infect CD4+T lymphocytes and induce programmed cell death during acute infection of cattle. J Virol. 1999;73:8657–8668. doi: 10.1128/jvi.73.10.8657-8668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan J, Yanker B A. Caspase activity sows the seeds of neuronal death. Nat Cell Biol. 1999;1:44–45. doi: 10.1038/10037. [DOI] [PubMed] [Google Scholar]
- 55.Zhou B B, Li H, Yuan J, Kirschner M W. Caspase-dependent activation of cyclin-dependent kinases during Fas-induced apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 1998;95:6785–6790. doi: 10.1073/pnas.95.12.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]