Abstract
Viral phenotype, tropism, coreceptor usage, and envelope gene diversity were examined in blood isolates collected from 27 individuals at different stages of human immunodeficiency virus type 1 (HIV-1) disease and tissue derived isolates from 10 individuals with AIDS. The majority (89%) of blood and all tissue HIV-1 isolates from all stages of infection were non-syncytium inducing and macrophage (M) tropic. Tropism and productive infection by HIV isolates in both monocytes and monocyte-derived macrophages (MDM) increased in advanced disease (HIV tropism for monocytes, 1 of 6 from categories I and II versus 11 of 21 [P = 0.05] from category IV and II [CD4 < 250]; and high-level replication in MDM, 1 of 6 from categories I and II versus 16 of 21 from categories IV and II [P = 0.015]). There was a high level of replication of blood and tissue isolates in T lymphocytes without restriction at any stage. Overall, the level of replication in MDM was 5- to 10-fold greater than in monocytes, with restriction in the latter occurring mainly at entry and later stages of replication. Only three blood isolates were identified as syncytium inducing, and all had a dualtropic phenotype. There was a significant increase of HIV envelope gene diversity, as shown by a heteroduplex mobility assay, in advanced disease; this may partly underlie the increase of HIV replication in MDM. Unlike blood isolates (even those from patients with advanced disease), tissue isolates displayed greater similarities (90%) in productive infection between MDM and monocytes. The majority (87%) of all isolates, including those from patients with advanced disease, used CCR5, and only 5 of 37 isolates showed expanded coreceptor usage. These results indicate that in the late stage of disease with increasing viral load and diversity, CCR5 utilization and M-tropism persist in blood and tissue and the replicative ability in macrophages increases. This suggests that these characteristics are advantageous to HIV and are important to disease progression.
Macrophages act as major reservoirs for human immunodeficiency virus type 1 (HIV-1) in tissues of the body (32, 47). HIV-infected macrophages may be found in the brain, lungs, lymph nodes, skin, bone marrow, and blood of seropositive individuals (10, 31, 32, 41, 63). They are the main source of productive infection in the brain, and HIV-infected macrophages in lymph nodes persist for weeks or months after commencement of highly active antiretroviral therapy. Monocytes/macrophages are believed to serve as vehicles for dissemination of HIV between different tissues of the body (43, 55). Macrophages may play a key role in regulating the intensity and progression of disease in HIV infection, and their secretory products have been implicated in the pathogenesis of AIDS dementia complex (43, 48).
Until recently the biological properties of HIV strains were classified as macrophage (M)-tropic non-syncytium-inducing (NSI) or T-cell-line (T)-tropic syncytium-inducing (SI) strains. M-tropic NSI (CCR5-using) HIV-1 strains are the predominant viral population immediately after seroconversion and during asymptomatic HIV infection, and they play a crucial role in the initiation of new infections through sexual and vertical transmission. M-tropic NSI HIV strains have no syncytia formation in MT2 lymphoblastoid cells and can infect macrophages and primary T lymphocytes but not T-cell lines (17, 63, 81). T-cell-line-tropic (primary and laboratory-adapted) HIV strains are characterized by higher viral replication in peripheral blood mononuclear cells (PBMCs), preferential infection of primary T cells and T-cell lines, with syncytium formation in MT2 cells and an inability to infect macrophages (31, 64, 67). In 40% of HIV-infected individuals with AIDS, the emergence of T-tropic HIV-1 variants in the advanced stage of disease coincides with the decline in the number of CD4+ T cells, which is predictive of disease progression (40, 63, 71).
Studies of chemokine receptor utilization by HIV has resolved many of the mysteries involved in differential tropism and biological properties displayed by many HIV-1 isolates (5). The chemokine receptor CCR5 is the major coreceptor used by M-tropic NSI isolates, while CXCR4 is used primarily by T-tropic SI isolates (3, 5, 22–24, 28). Numerous other chemokine receptors, including CCR3 (2, 4, 13, 23, 34, 58, 59), CCR2b (1, 23), STRL33 (Bonzo) (21, 44, 46), GPR15 (Bob) (21, 25), and GPR1 (25), which may be essential for some isolates have also been identified. Dualtropic viruses have also been identified which may utilize CCR5 and/or CXCR4 for entry (16, 23, 68). Dualtropic viruses have been proposed as the transitional phenotype during the evolution of viral populations from NSI to SI variants (16, 23, 57, 66). An evolving increase in viral genome diversity is believed to underlie the majority of changes in the biological characteristics of HIV. Despite several hypotheses and more recent data, the relationship of viral genomic diversity to viral load, CD4 count, and biological changes in the viral quasispecies in blood during disease progression remains unclear (7, 19, 52, 65, 76).
In this study, we examined the tropism, coreceptor usage, and viral phenotype of primary HIV-1 isolates collected from the blood of individuals at different stages of HIV infection and disease (27) and from tissue at autopsy (10). Although essentially a cross-sectional study, these findings, using many primary isolates, contribute to an understanding of the parallel evolutionary biological changes of HIV quasispecies which may be associated with disease progression. Viral envelope diversity in relation to disease stage was also examined by a heteroduplex mobility assay (HMA) and sequencing to determine the mechanisms underlying biological changes in tropism and productivity of infection in various cell types.
MATERIALS AND METHODS
Description of patients and viruses.
A total of 27 blood-derived and 10 tissue-derived HIV-1 isolates were obtained from HIV-1-infected patients in Sydney, Australia (for the disease stage and CD4+ T-cell counts, see Table 1). Blood-derived isolates were from 2 acute seroconverters, 6 patients with asymptomatic infections (4 with blood CD4 counts of >500 and 2 with counts of <250), and 19 individuals with AIDS. Tissue isolates were obtained at autopsy from the brain (NT1, NT2, NT4 to NT6, NT8, and NT9), cerebrospinal fluid (NT7), lungs (NT10) and spleen (NT3) from different individuals. Brains and other tissues were washed with sterile phosphate-buffered saline until all the blood had been removed and then were either mashed (brain) or cut into small pieces (other tissues) and ground with pestle and mortar. Sample suspension was added to 107 donor PBMCs in 2 ml of growth medium incubated initially for 30 min, the volume was topped up to 10 ml into cell culture flasks, and cultures were maintained for 14 days and then examined by a reverse transcription (RT) assay for virus isolation.
TABLE 1.
Phenotypic characteristics of blood and tissue HIV-1 primary isolates from patients at different stages of infection and their level of replication in monocytes, macrophages, and T lymphocytes as measured by EC p24 antigen levels
| Isolate | Stage of diseasea (category) | CD4 countsb | Level of replication
inc:
|
MT-2 cell assay | CPE in PBMC | ||
|---|---|---|---|---|---|---|---|
| Monocytes | Macrophages | Lymphocytes | |||||
| NB1 | IV | <50 | ++ | +++ | +++ | NSI | − |
| NB2 | IV | <50 | ++ | +++ | +++ | NSI | − |
| NB3 | IV | <50 | ++ | +++ | +++ | SI | +++ |
| NB4 | IV | <50 | ++ | +++ | +++ | NSI | − |
| NB5 | IV | <100 | − | +++ | +++ | SI | ++ |
| NB6 | IV | <50 | − | +++ | +++ | NSI | + |
| NB7 | IV | <50 | + | +++ | +++ | NSI | − |
| NB8 | IV | <50 | ++ | +++ | ++ | NSI | − |
| NB9 | IV | <100 | − | ++ | +++ | NSI | + |
| NB10 | IV | <50 | ++ | ++ | ++ | NSI | + |
| NB11 | IV | <50 | + | ++ | +++ | NSI | − |
| NB12 | IV | 200 | − | ++ | +++ | NSI | + |
| NB13 | IV | <50 | − | + | +++ | NSI | + |
| NB14 | IV | <50 | − | + | ++ | NSI | − |
| NB15 | IV | <100 | − | + | ++ | NSI | + |
| NB16 | IV | <50 | − | − | ++ | NSI | + |
| NB17 | IV | 200 | − | + | +++ | NSI | − |
| NB18 | IV | <50 | ++ | +++ | +++ | NSI | − |
| NB19 | IV | <100 | + | ++ | ++ | NSI | − |
| NB20 | II | 200 | − | +++ | +++ | NSI | − |
| NB21 | II | 250 | − | +++ | +++ | SI | +++ |
| NB22 | II | 550 | + | ++ | +++ | NSI | − |
| NB23 | II | 600 | − | + | +++ | NSI | + |
| NB24 | II | 650 | − | − | ++ | NSI | + |
| NB25 | II | 500 | − | − | +++ | NSI | + |
| NB26 | I | >750 | − | + | +++ | NSI | − |
| NB27 | I | >750 | + | + | +++ | NSI | − |
| HIV-BaL | NA | NA | ++ | +++ | +++ | NSI | − |
| HIV-JRFL | NA | NA | + | +++ | +++ | NSI | − |
| NT1 | IV | <50 | ++ | + | +++ | NSI | − |
| NT2 | IV | <50 | +++ | +++ | +++ | NSI | + |
| NT3 | IV | <50 | ++ | ++ | +++ | NSI | − |
| NT4 | IV | <50 | − | − | + | NSI | − |
| NT5 | IV | <50 | +++ | +++ | +++ | NSI | ++ |
| NT6 | IV | <50 | +++ | +++ | +++ | NSI | ++ |
| NT7 | IV | <50 | − | ++ | ++ | NSI | + |
| NT8 | IV | <50 | + | +++ | +++ | NSI | − |
| NT9 | IV | <50 | ++ | +++ | +++ | NSI | − |
| NT10 | IV | <50 | + | ++ | +++ | NSI | − |
According to CDC classification (category IV, AIDS; category II, asymptomatic; category I, acute seroconversion).
Absolute count of CD4 lymphocytes per microliter of blood at the time of virus isolation.
Level of replication: −, no productive infection; +, low (<1 ng/ml); ++, intermediate (1 to 49 ng/ml), +++, high (>50 ng/ml).
Peripheral blood samples were collected in heparin and processed within 2 to 3 h of phlebotomy. Isolates used in this study were from a single passage of cocultivation with phytohemagglutinin (PHA)-stimulated PBMC to produce virus stocks. These isolates were tested for their ability to infect 16-h-old monocytes and 5-day-old monocyte-derived macrophages (MDM) from the same donor and their ability to infect autologous T cells. Virus stocks were subjected to titer determination by end-point dilution in 96-well tissue culture plates with PHA-stimulated PBMC. The 50% tissue culture infective dose was determined by measuring the level of the extracellular (EC) HIV p24 antigen in culture supernatants by using a commercial ELISA kit (Abbott Laboratories) and the Spearman-Korber formula. Virus inocula which were used in DNA experiments were first filtered through a 0.22-μm-pore-size filter (Millex-GS; Millipore, Bedford, Mass.) and then treated with DNase (20 μg/ml) for 1 h at room temperature in the presence of 10 mM MgCl2 to decontaminate the inoculum of HIV-1 DNA (50). Approximately equal numbers of monocytes/MDM and T cells (106) from two donors were exposed to the same virus inoculum (multiplicity of infection, 0.025/cell as measured by determining the 50% tissue culture infective dose in PBMC) of clinical isolates, HIV-BaL, and HIV-JRFL. HIV-BaL (31) and HIV-JRFL (41), obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, were used as control M-tropic strains. Statistical analysis was performed by Fisher’s exact test.
Monocyte isolation.
Blood-derived monocytes were isolated from 400 ml of whole blood from healthy HIV seronegative donors as previously described (38). Briefly, PBMC were obtained by differential centrifugation on Ficoll-Hypaque (Pharmacia-AMRAD, Sydney, Australia). Monocytes were separated from PBMC by countercurrent elutriation (Beckman J-6M/E centrifuge fitted with a JE 5.0 elutriation rotor) and OKT3/complement lysis as an extra step to purify monocytes from contamination with T cells. The isolated monocyte populations were routinely >96% positive for nonspecific esterase. Cells were cultured in the absence of growth factors in 1 ml of 10/10 medium, containing RPMI 1640 supplemented with antibiotics, 10% heat-inactivated fetal bovine serum, and 10% heat-inactivated pooled AB+ human serum at a density of 106 cells per well in a 48-well tissue culture plate (Nunc, Sydney, Australia). The cultures were replenished with fresh 10/10 medium every 3 or 4 days. Monocytes were allowed to adhere for 5 days to differentiate into a cell population rich with macrophages, as judged by cell morphology and maturation cell surface markers (CD14 and CD68). Primary lymphocytes were isolated from whole blood, stimulated with PHA (5 μg/ml) for 48 h, and then replenished with fresh medium supplemented with 10% human interleukin-2 (Roche/Boehringer Mannheim, Sydney, Australia). To minimize differences in replication due to donor cell effects, all isolates were assayed on the same day by using monocytes, MDM, and lymphocytes from the same donor PBMCs. Monocytes and MDM were infected with the same panel of HIV-1 clinical isolates and HIV-BaL after 16 h and 5 days of adherence, respectively. Culture supernatants were assayed for EC p24 antigen, and cells were lysed for DNA determination on days 0, 7, and 14 postinfection. The levels of the EC p24 antigen in culture supernatants were determined by a commercial ELISA as specified by the manufacturer (Abbott Laboratories, Sydney, Australia). Antigen amounts (in nanograms per milliliter) were calculated, and values below 25 pg/ml was designated nonproductive; other values were designated low (<1 ng/ml), intermediate (1 to 49 ng/ml), or high (>50 ng/ml).
Determination of HIV-1 coreceptor usage by using chemokine receptor-transfected human osteosarcoma (HOS.CD4) cells.
HOS cells transfected with the chemokine receptors CCR1, CCR2, CCR3, CCR4, CCR5, CXCR4, GPR15/Bob, STRL33/Bonzo, and the parental HOS.CD4 (generously provided by D. Littman, Howard Hughes Institute, The New York University Medical Center, New York, N.Y.) were used. These cells were cultured at 5 × 104 cells/well in a 24-well tissue plate (Nunc) with Dulbecco’s modified Eagle’s medium (Sigma Chemical, St. Louis, Mo.) enriched with glucose and supplemented with 10% fetal bovine serum. Cultures were treated with a mixture of 1 μg of puromycin dihydrochloride (Calbiochem, Melbourne, Australia) per ml, 50 μg of hygromycin B (Roche/Boeringher Mannheim) per ml, and 500 μg of G418 sulfate (Calbiochem, Melbourne, Australia) per ml to maintain stable expression of chemokine receptors and CD4. These cells were infected with HIV-1 clinical isolates from different stages of infection and with HIV-BaL (as an M-tropic control) and NL-43 (as a T-tropic control) to determine their coreceptor usage. Infection was examined by DNA PCR and EC p24 antigen measurement.
DNA extraction and PCR amplification.
Monocyte and MDM DNA lysates were prepared as previously described (51) by incubation at 60°C for 2 h and at 95°C for 15 min in a DNA lysis buffer containing proteinase K and stored at −20°C until used for PCR. The HIV-1 DNA content in monocytes and MDM was amplified by PCR with 2.5 U of Taq polymerase, 0.2 mM each deoxyribonucleotide, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 0.001% gelatin. Primers M667 (78) and gag1 (51) were used to amplify a 320-bp region encompassing from the R region within the 5′ long terminal repeat to the gag region, representing full-length synthesis of HIV cDNA. Each test sample was subjected to 30 cycles of amplification (1 min at 95°C, 2 min at 60°C, and 3 min at 72°C) in a Perkin-Elmer Cetus thermal cycler with a final extension at 72°C for 7 min. Concurrent reactions were also performed with primers PCO3 and PCO4 to amplify 110-bp DNA fragments of the human β-globin gene (60) to ensure that equivalent amounts of DNA were used in each sample reaction. PCR products were electrophoresed on a 2% agarose gel, visualized by ethidium bromide staining under a UV transilluminator, and then photographed.
Southern liquid hybridization.
The specificity of PCR products was confirmed by using oligonucleotide probes from within the amplified regions, using liquid Southern hybridization (39, 51). This method allows direct in situ detection of PCR products on an agarose gel with a 32P-labelled oligonucleotide. Briefly, 15 μl of PCR products was used in liquid hybridisation with addition of 100 cpm of [γ-32P]ATP-labelled oligonucleotide probe per reaction and incubated at 95°C for 5 min and then at 55°C for 10 min in a thermal cycler (Perkin-Elmer Cetus). This mixture was loaded on a gel, electrophoresed at 60 V for 1.5 h, stained with ethidium bromide, and visualized under a UV transilluminator. Agarose gels were semidried with a gel drier (Bio-Rad, Sydney, Australia) and exposed overnight against X-ray films (Cronex; DuPont, Sydney, Australia) at −80°C.
By using the same DNA lysate extraction, DNA from 8E5 cells, containing one integrated copy of HIV-1 DNA per cell (29), was used to construct a standard curve for quantification of DNA from monocytes, MDM, and T lymphocytes. Tenfold dilutions (0, 10, 102, 103, and 104 cells/reaction) of 8E5 cells were prepared, with uninfected PBMCs making up a total of 105 cells/reaction. HIV DNA levels were classified as 0 (undetectable), + (low copy number, 10 to 102), ++ (intermediate copy number, 102 to 103), and +++ (high copy number, >103) as normalized to the HIV DNA standard curve. For HIV DNA PCR, parallel cell cultures were treated with zidovidine (20 mM) for 30 min at 37°C to control for de novo HIV DNA synthesis. Treatment with zidovidine reduced HIV-BaL DNA levels from high (+++) to undetectable (0).
V3 and V1-V2 amplification and sequencing.
The V3 and V1-V2 regions of HIV DNA from blood and tissue isolates cultured in PBMCs were amplified by nested PCR. The first round of amplification with V3 external primers (NV3-1 and NV3-2) was carried out for 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. Aliquots (5 μl) of the amplified products were included in the second round of amplification with V3 internal primers (NV3-A and NV3-B) for 25 cycles under the same conditions as for the first round. Both rounds of PCR were preceded by a denaturation step at 94°C for 5 min and ended by extension at 72°C for 7 min. The V3 external primer pair was NV3-1 (5′-CAACTGCTGTTAAATGGCAGTCT-3′; positions 6985 to 7008 using HIV-1 pnl43) and NV3-2 (5′-ACTGTGCATTACAATTTCTGGGTC-3′; positions 7316 to 7339). The V3 internal primer pair was NV3-A (5′-GCAGTCTAGCAGAAGAAG-3′; positions 7002 to 7019) and NV3-B (5′-TGGGTCCCCTCCTGAGGA-3′; positions 7304 to 7321). The same amplification conditions were also used to amplify the V1-V2 region with an external primer pair, NVV3 (5′-GCCTGTGTACCCACAGACCCCAA-3′; positions 6437 to 6459) and NVVC (5′-CTGGCCTAATTCCATGTGTACATTG-3′, positions 6950 to 6974), and an internal primer pair, NVV4 (5′-CAGTTTATGGGATCAAAGCCT-3′; positions 6547 to 6567) and NVVD (5′-GCACAATAATGTATGGGAATTGG-3′; positions 6848 to 6870). Samples (10 μl) of DNA from the second round of PCR were electrophoresed on 1.5% agarose gel, detecting 350- and 254-bp DNA fragments of the V3 and V1-V2 regions, respectively. PCR products were precipitated with polyethylene glycol solution (containing 26.7% polyethylene glycol 8000, 0.6 M sodium acetate [pH 5.2], and 6.5 M MgCl2), washed twice with 95% ethanol, air dried, and reconstituted with an appropriate amount of sterile water. The DNA concentration was spectrophotometrically quantitated, and 100 to 300 ng was used for sequencing with 10 pmol of either sense or antisense primer. The sequence was obtained by sequencing of purified PCR products (population sequence) with a Dye-Deoxy terminator and automated DNA sequencer (373A; ABI DNA sequencer). Sequence alignment was performed with Clustal W from the Australian National Genomic Information Service [ANGIS] facility, University of Sydney.
HMA.
DNA was extracted from PBMCs infected with blood and tissue primary isolates by using the QIAamp Blood Kit (Qiagen, Melbourne, Australia). Nested PCR was used to amplify the V3-V5 region of gp120 from these isolates. First-round amplification was performed with 200 nM primers NV3A and V5B (5′-ATAGTGCTTCCTGCTGCTCCCAAGAACC-3′; positions 7776 to 7803), and different amounts of input DNA (500 to 300 ng) were used for each PCR. PCR was performed as follows: 4 min at 94°C (1 cycle); 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C (3 cycles); 30 s at 94°C, 45 s at 62°C, and 1 min at 72°C (32 cycles); and 10 min at 72°C (1 cycle). The nested round was performed with internal primers ES7 and ES8 (20), using 2 μl of the first-round product as template and the same PCR amplification conditions. Second-round products were adjusted to 0.1 M NaCl–10 mM Tris-HCl (pH 7.8)–2 mM EDTA and denatured at 94°C for 2 min. Samples were then immediately placed in wet ice to promote heteroduplex formation. A 10-μl volume of each sample was loaded on a 5% polyacrylamide gel (30:0.8 acrylamide/bisacrylamide) in Tris-borate-EDTA (TBE) and resolved with a V16 vertical gel apparatus (Gibco/BRL, Gaithersburg, Md.) with 1.5-mm-thick spacers at 250 V for 2.5 h. The gel was stained with ethidium bromide and visualised with using Fluor-S (Bio-Rad).
RESULTS
M-tropism and NSI phenotype are common characteristics of primary blood isolates.
Primary HIV-1 isolates exhibited different levels of replication in MDM, monocytes, and T lymphocytes. In general, the level of replication was classified into four categories, i.e., nonproductive (below detection), low, intermediate, and high, as measured by using p24 antigen from culture supernatants at the peak of the infection. Of 27 isolates, 24 (89%) showed productive infection in MDM according to results with the EC p24 antigen (Table 1). The majority (17 of 27; 63%) replicated to high or intermediate levels (Table 1; Fig. 1A and B). Only three isolates (NB16, NB24, and NB25) showed undetectable levels of p24 antigen in MDM culture supernatants (Table 1). When HIV DNA and EC p24 antigen concentrations were compared, one of these isolates (NB25) showed inhibition prior to complete RT (probably at viral entry) and one (NB16) was nonproductive due to restriction after RT. In general, there was good correlation between HIV DNA and p24 antigen levels. However, the high HIV-DNA levels and low p24 antigen concentrations obtained with NB17 and NB19 suggest some restriction to viral replication at a stage after RT.
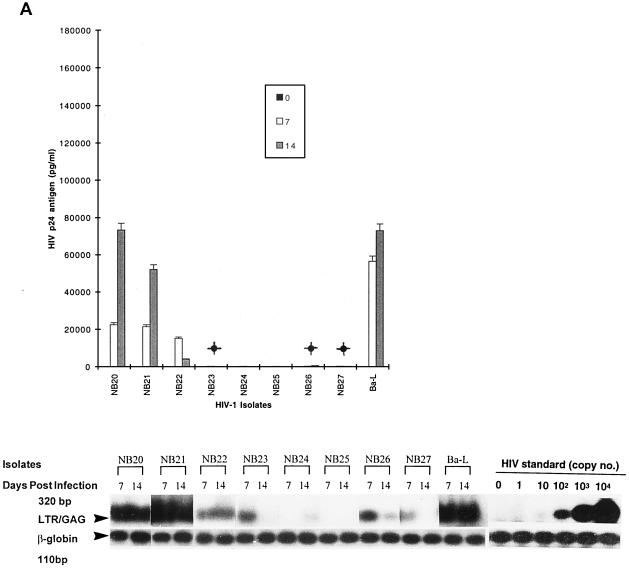
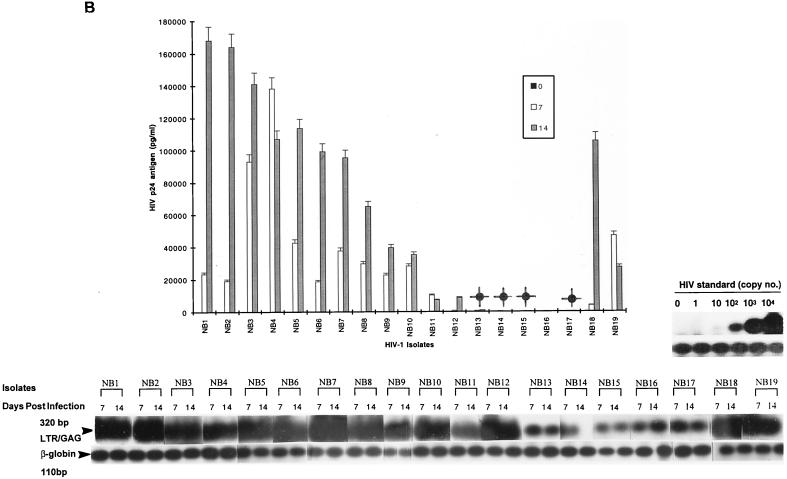
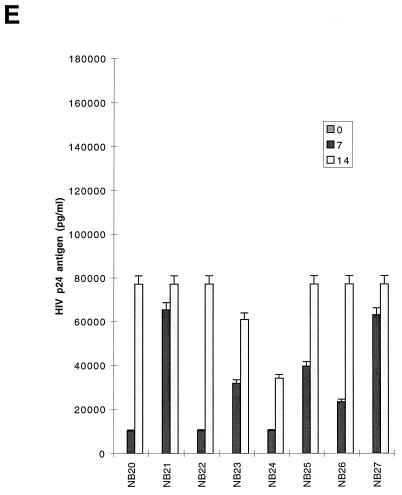
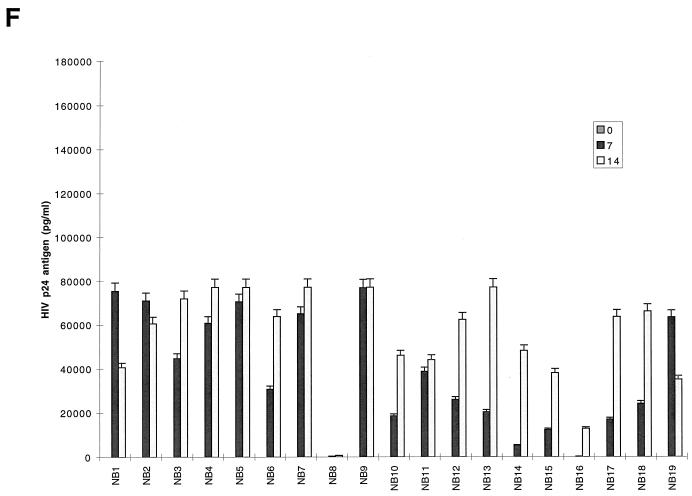
FIG. 1.
Replication kinetics of infection in 5-day-old MDM (A and B) and 16-h-old monocytes (C and D) and T lymphocytes (E and F) by blood-derived HIV-1 isolates from patients at early (categories I and II) (A, C and E) and late (category IV) (B, D, and F) stages of disease. (Top) HIV-1 replication as assessed by p24 antigen ELISA (Coulter Electronics) in culture supernatants on days 0, 7, and 14 (error bars represent standard deviation; P < 0.05). (Bottom) DNA PCR products for full-length HIV DNA on days 7 and 14 of infection. |, productive infection with detectable low levels of p24 antigen on day 14 of infection.
Fewer isolates were able to infect monocytes productively, as shown by both DNA PCR and p24 antigen levels, and also showed 5- to 10-fold-lower levels than MDM, by p24 antigen. For monocytes, comparison of HIV DNA and p24 antigen levels showed nonproductive infection for a majority of blood isolates (15 of 27; 56%) (Table 1). The remaining 12 isolates with productive infection in monocytes had low or intermediate levels of replication (Fig. 1C and D; Table 1). Full-length viral DNA was detectable in 21 isolates on days 7 and 14 after infection (Fig. 1C and D, bottom). The remaining six isolates (NB9, NB12, NB16, NB21, NB24, and NB25) showed a complete block prior to RT (probably at entry), where no DNA was detected. With several isolates (NB23, NB26, and NB27), a decline or loss of HIV DNA from days 7 to 14 was observed in monocytes, suggesting that infection did not spread.
In T lymphocytes, productive infection with replication levels ranging from high to intermediate was observed for all blood isolates without any restriction, as shown by both p24 antigen and HIV DNA levels (Table 1; Fig. 1E and F [DNA results not shown]). All isolates from individuals in stage IV disease which replicated to high levels in MDM replicated at two- to threefold-lower levels in lymphocytes (Fig. 1B and F). Only three isolates (NB3, NB5, and NB21) (11%) were found to induce syncytia in MT2 cells. Two were from patients with clinical stage IV disease, and one (NB21) was from a patient with advanced stage II disease with a CD4+-T-cell count of 250 cells/μl. The ability of these viruses to produce productive infections in macrophages indicates that they are dually tropic. The majority of NSI variants were able to induce cytopathic effects (CPE) in PBMCs (Table 1).
Monocyte/macrophage tropism and levels of replication increase in the advanced stages of HIV infection.
The capacity of primary isolates to infect MDMs, monocytes, and T lymphocytes was analyzed according to the clinical stage of the source patient (Fig. 1; Table 1). Of 11 high-replication isolates in MDM, 9 (82%) were from patients with category IV disease (AIDS) (Table 1). The two (NB20 and NB21) of six patients with category II infection who had isolates which replicated to high levels in MDM had relatively advanced HIV infection with low CD4+-T-cell counts of 200 and 250 (i.e., close to the Centers for Disease Control and Prevention [CDC] definition of AIDS) (Table 1). Similarly, five of six isolates able to replicate to intermediate levels were from patients with AIDS. A similar pattern was seen in monocytes, where all the intermediate replicating isolates were from patients with category IV disease (Table 1). In contrast, most isolates (five of six) obtained from patients with category II infection replicated to undetectable levels in monocytes. Isolates obtained from two patients during the seroconversion stage (category I) replicated to low levels both in MDM and monocytes, while one had no detectable level of infection in monocytes. No correlation was observed between disease stage and level of replication in T lymphocytes. Therefore, most isolates that replicated at high or intermediate levels in both monocytes and MDM with similar input multiplicities were from individuals with advanced infection, including the two who had low CD4 counts (monocytes, 0 of 6 from categories I and II versus 7 of 21 from categories IV and II (CD4 < 250), P = 0.057; MDM, 1 of 6 from categories I and II versus 16 of 21 from categories IV and II (CD4 < 250), P = 0.015). Taken together, these data suggest that the level of replication of HIV-1 primary isolates in cultured MDM increases with advancing HIV disease.
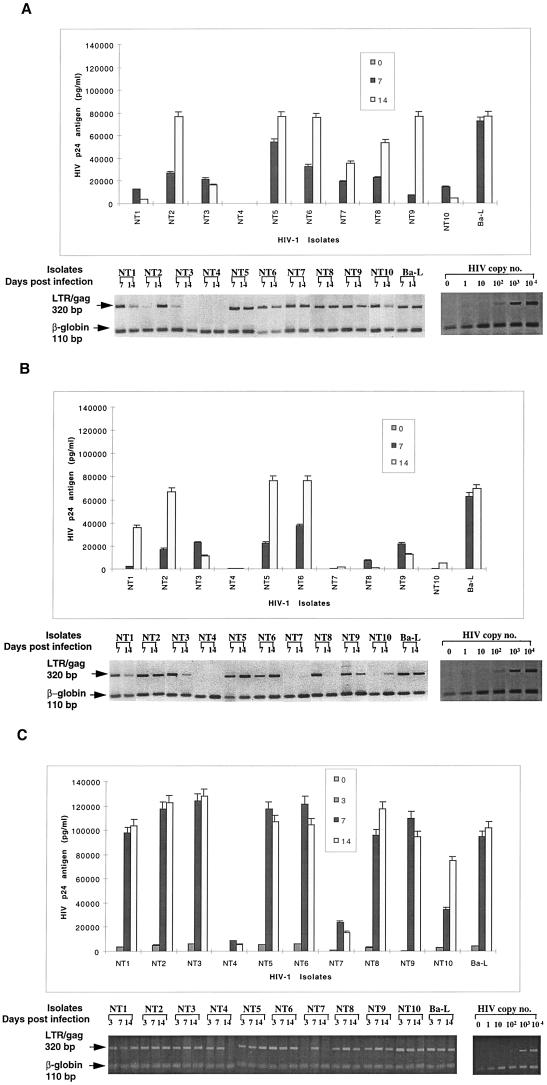
Tissue isolates display similar replication levels in monocytes and MDM.
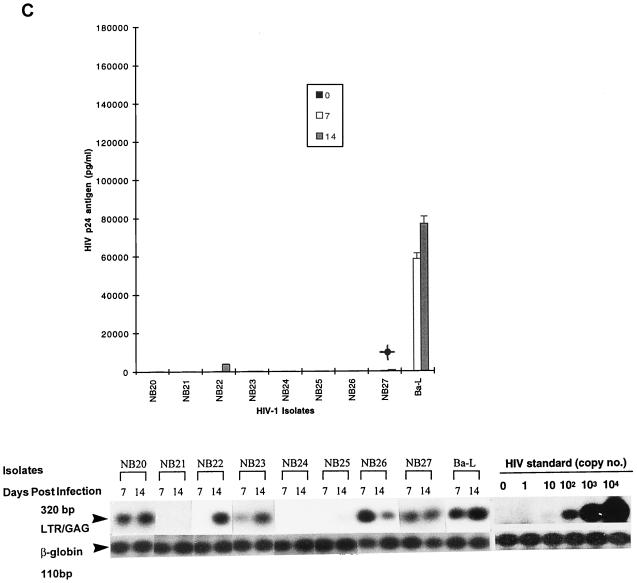
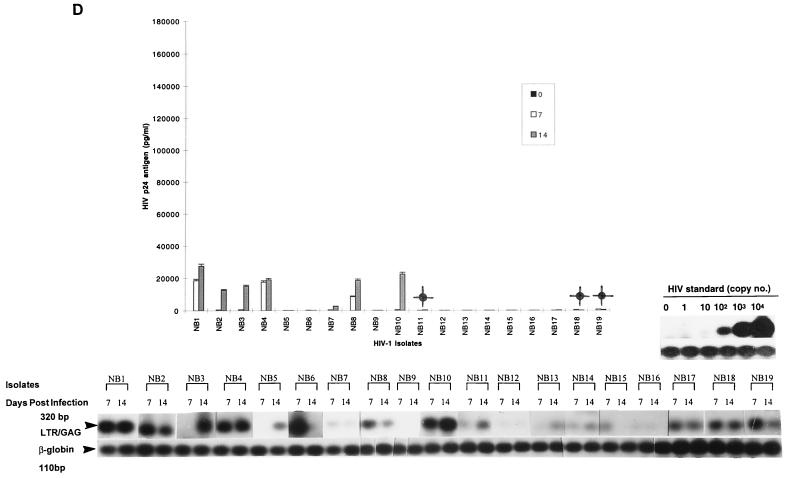
Tissue isolates were examined by similar approaches as described above for blood-derived isolates (i.e., by comparison of HIV DNA and EC p24 antigen levels). The same semiquantitative levels for replication were also assigned to categorize the different isolates. Unlike blood isolates, most tissue isolates replicated productively in both MDMs and monocytes (9 of 10 and 8 of 10, respectively), as measured by p24 antigen levels and HIV DNA (Fig. 2A and B; Table 1). In fact, there were no major differences in the level of replication between MDMs and monocytes (Table 1). Highly to intermediately replicating isolates in MDM generally showed corresponding virus productivity in monocytes without restriction at any stage (i.e., HIV p24 antigen and DNA levels were similar). No HIV DNA was detected for one isolate (NT4) in both monocytes and MDM, indicating that restriction occurred at entry. Brain-derived isolates (NT1, NT2, NT4 to NT6, NT8, and NT9) were among the most highly replicating tissue isolates in both MDMs and monocytes, with the exception of one isolate (NT4), which exhibited undetectable levels of replication in both MDM and monocytes. Conversely, brain isolate NT1 consistently replicated at an intermediate level in monocytes but at lower levels in MDMs. Tissue isolates showed similar patterns of replication in T lymphocytes to blood isolates, with the majority of isolates (9 of 10) also showing high to intermediate levels of replication (Table 1; Fig. 2C), indicating no restriction of replication. The similarities in the levels of replication of tissue isolates in MDMs and monocytes, in contrast to blood isolates, even those from category IV, suggest that infectivity of tissue isolates was not restricted by the state of monocyte maturation.
FIG. 2.
Replication kinetics of tissue-derived HIV-1 isolates obtained at autopsy from different tissue compartments of patients with AIDS, after infection of 5-day-old MDM (A), monocytes (B), and T lymphocytes (C). (Top) HIV-1 replication as assessed by p24 antigen ELISA (Coulter Electronics) in culture supernatants on days 0, 7, and 14 after infection (error bars represent standard deviation; P < 0.05). (Bottom) DNA PCR products for full-length HIV DNA on days 7 and 14 of infection.
Persistent CCR5 usage of blood and tissue isolates.
The blood- and tissue-derived isolates were examined to determine trends in coreceptor usage which may be associated with disease stage. The majority of blood isolates from asymptomatic patients (7 of 8; 88%) or those with AIDS (16 of 19; 84%) used only CCR5 for entry (Table 2). Four isolates were found to have an apparently expanded coreceptor usage, including those other than CCR5, when tested in chemokine receptor-transfected HOS.CD4 cells. Three of these used CXCR4 but not CCR5. These three isolates appeared to use more than one coreceptor; two (NB3 and NB5) used both CCR3 and CXCR4, and one (NB21) used CCR3, Bob, and Bonzo as well as CXCR4. However, infection of HOS.CD4.CCR3/Bonzo/Bob cells could be blocked by pre- and coincubation with stromal-derived factor SDF-1α (data not shown), indicating that low levels of endogenous CXCR4 were being used for entry and that CCR3, Bonzo, and Bob were not being used. This isolate was from one of the two asymptomatic individuals with low CD4 T-cell counts. This indicates that switch in coreceptor usage can also occur at early stages of disease. Only one isolate (NB7) used CCR3 and CCR5. All tissue isolates uniformly used CCR5 as a major coreceptor, with the exception of one brain isolate (NT1), which also used CCR3 (Table 2). There was no CXCR4 usage by tissue isolates.
TABLE 2.
Coreceptor usage of blood and tissue HIV-1 primary isolates in HOS.CD4 cellsa
| Isolate | Usage of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2 | CCR3 | CCR4 | CCR5 | CXCR4 | Bob | Bonzo | CNT | |
| NB1 | − | − | − | − | + | − | − | − | − |
| NB2 | − | − | − | − | + | − | − | − | − |
| NB3 | − | − | + | − | − | + | − | − | − |
| NB4 | − | − | − | − | + | − | − | − | − |
| NB5 | − | − | + | − | − | + | − | − | − |
| NB6 | − | − | − | − | + | − | − | − | − |
| NB7 | − | − | + | − | + | − | − | − | − |
| NB8 | − | − | − | − | + | − | − | − | − |
| NB9 | − | − | − | − | + | − | − | − | − |
| NB10 | − | − | − | − | + | − | − | − | − |
| NB11 | − | − | − | − | + | − | − | − | − |
| NB12 | − | − | − | − | + | − | − | − | − |
| NB13 | − | − | − | − | + | − | − | − | − |
| NB14 | − | − | − | − | + | − | − | − | − |
| NB15 | − | − | − | − | + | − | − | − | − |
| NB16 | − | − | − | − | + | − | − | − | − |
| NB17 | − | − | − | − | + | − | − | − | − |
| NB18 | − | − | − | − | + | − | − | − | − |
| NB19 | − | − | − | − | + | − | − | − | − |
| NB20 | − | − | − | − | + | − | − | − | − |
| NB21 | − | − | + | − | − | + | + | + | − |
| NB22 | − | − | − | − | + | − | − | − | − |
| NB23 | − | − | − | − | + | − | − | − | − |
| NB24 | − | − | − | − | + | − | − | − | − |
| NB25 | − | − | − | − | + | − | − | − | − |
| NB26 | − | − | − | − | + | − | − | − | − |
| NB27 | − | − | − | − | + | − | − | − | − |
| NT1 | − | − | + | − | + | − | − | − | − |
| NT2 | − | − | − | − | + | − | − | − | − |
| NT3 | − | − | − | − | + | − | − | − | − |
| NT4 | − | − | − | − | + | − | − | − | − |
| NT5 | − | − | − | − | + | − | − | − | − |
| NT6 | − | − | − | − | + | − | − | − | − |
| NT7 | − | − | − | − | + | − | − | − | − |
| NT8 | − | − | − | − | + | − | − | − | − |
| NT9 | − | − | − | − | + | − | − | − | − |
| NT10 | − | − | − | − | + | − | − | − | − |
−, undetectable HIV DNA or p24 antigen concentrations of <50 pg/ml; +, detectable HIV DNA or p24 antigen concentrations of >50 pg/ml.
Sequence analysis of the V3 region demonstrates a macrophage-tropic genotype.
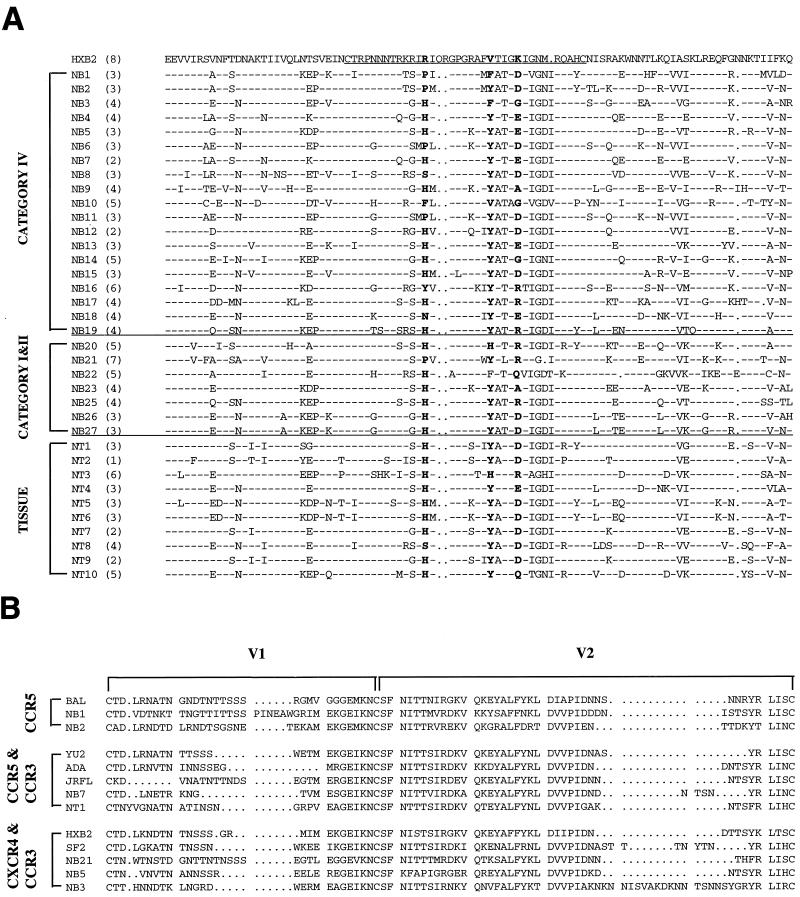
Substitutions of amino acid at positions 11, 13, and 25 of the V3 loop have been implicated in viral tropism (11, 67, 75). The blood isolates showed (changes in) either serine or glycine at position 11, histidine or proline at position 13, or aspartic acid, glutamic acid, or glycine at position 25 (Fig. 3A). Approximately half of the high-replication isolates had a proline at position 13, whereas a serine appeared in some of the low-replication isolates. The crown of the V3 loop occurred as five motifs: GPGK (NB5, NB6, NB9, NB11, and NB23), GPGQ (NB12), GLGR (NB15), and APGR (NB22), but none were specifically linked with other phenotypic characteristics. The net charge of V3 loop residues was usually low, ranging from 2 to 4, although two isolates (NB16 and NB21) showed higher positive charges of 6 and 7, respectively, due to arginine substitution at position 25. Only NB21, however, was shown to induce syncytia in MT2 cells, while NB16 did not do so. The other two SI isolates (NB3 and NB5) both had low net charges (4 and 3, respectively). Sequencing of the V3 region of tissue isolates revealed that the majority of isolates contained serine residues at position 11. All isolates except for NT8 possessed histidine residues at position 13. Aspartic acid was the predominant residue at position 25. The NT3 isolate, derived from spleen tissue, contained an arginine residue at position 25 which contributed to its overall high net charge of 6. However, it did not exhibit an SI phenotype in MT2 cells. Six of seven brain and CSF isolates displayed changes in the crown (GPGS and GPGK) of the V3 loop (Fig. 3A). Phylogenetic analysis of blood and tissue V3 sequences revealed no clear segregation according to the stage of disease or the level of replication, although tissue isolates tended to cluster with blood isolates from advanced stages of infection (data not shown). Note that as population rather than clonal sequencing was performed, it is possible that changes in minor variants for some isolates were missed.
FIG. 3.
(A) Deduced amino acid sequence analysis of the V3 loop and flanking sequences derived from PBMC infected with blood and tissue primary isolates. Sequences are compared with HIV HXB2. The net positive charge of each sequence is shown in parentheses next to the name of the isolate. NB24 was not sequenced. Dots indicate amino acid deletion, dashes indicate an amino acid that is identical to HIV-HXB2 sequence. Horizontal lines separate blood from tissue isolates. (B) Amino acid sequence alignment of the V1-V2 region of isolates utilizing coreceptors other than CCR5 (NB3, NB5, NB7, NB21, and NT1) and those displaying the SI phenotype (dual-tropic) compared with primary and laboratory-adapted (HIV-Bal) NSI isolates using CCR5 alone, laboratory-adapted (HXB2 and SF2) SI isolates using only CXCR4, or laboratory-adapted using both CCR5 and CCR3 (ADA, YU2, and JRFL). Laboratory-adapted amino acid sequences were obtained from the Los Alamos HIV sequence database.
Sequence analysis of V1-V2 does not reveal any distinct patterns correlating with expanded coreceptor usage.
Since the V1-V2 region may also interact with CXCR4 and CCR3, the V1-V2 region of isolates utilizing coreceptors other than CCR5 (NB3, NB5, NB7, NB21, and NT1) and those displaying the SI phenotype (dually tropic) were sequenced and compared with primary and laboratory adapted (Bal) NSI isolates using only CCR5, laboratory adapted (HXB2 and SF2) SI isolates using only CXCR4, or laboratory adapted isolates using both CCR5 and CCR3 (ADA, YU2, and JRFL) (58) to detect distinct signatures of amino acids which may be involved in altered coreceptor usage or phenotype. Various deletions, insertions, substitutions, and duplications were present in both V1 and V2 of all isolates. The length of V1 ranged from 25 to 36 amino acids, while that of V2 ranged from 39 to 42 amino acids, with the exception of NB3, which displayed the greatest length of 57 amino acids due to an insertion of an asparigine-, serine-, and lysine-rich string of 16 amino acids (KN, KNN, SNN). However, no distinct pattern of amino acids, which might be linked to specific coreceptor usage or phenotype, was observed. There were more glycosylation sites observed in the V1 region (ranging from two to five) due to the higher frequency of asparigine, serine, and threonine. The number of sites in the V2 region ranged from one to three, with the exception of NB3, which contained five potential glycosylation sites due to the 16-amino-acid insertion. Analysis of the charge characteristics of both V1 and V2 also did not reveal any distinct patterns that may be associated with altered coreceptor usage or phenotype.
Viral diversity is increased in isolates from patients with advanced disease.
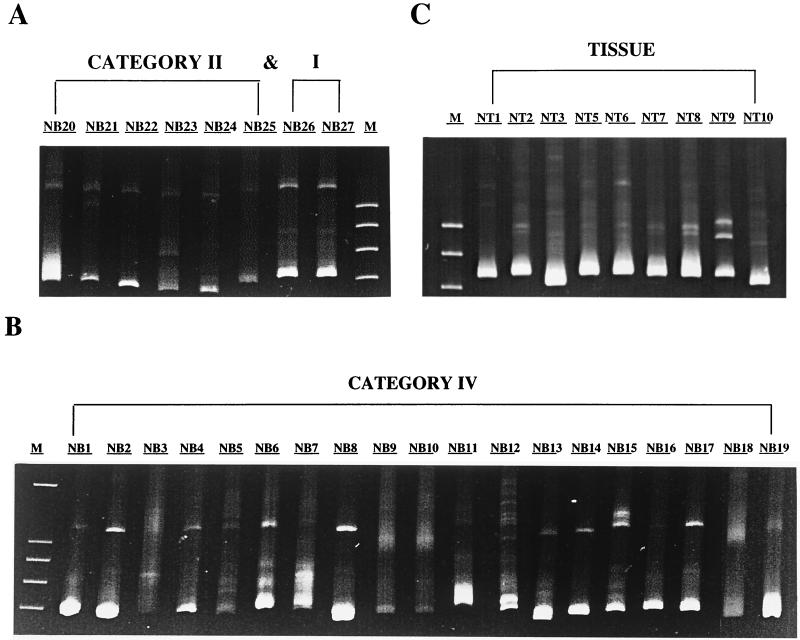
Quasispecies diversity was assessed for each isolate by HMA to determine patterns of viral diversity associated with different stages of HIV-1 infection. The degree of diversity of envelope gene diversity was semiquantified according to the following scale: +, predominantly homoduplex visible; ++, homoduplex and heteroduplex bands of approximately equal intensity; and +++, widely dispersed heteroduplex bands. The envelope gene diversity of blood isolates from patients with category IV disease was greater overall than that of isolates category I/II from patients (mean, 2.5+ and 1.5+, respectively (Fig. 4A and B). The diversity of all tissue isolates was uniformly high (mean, 3.0+), greater than that of category IV blood isolates (Fig. 4C). Overall, there was a trend (P = 0.09, Spearman’s rank correlation) between the degree of envelope gene diversity measured by HMA and productive replication in MDMs. There was less of a trend toward such a correlation in monocytes (P = 0.46), but neither were significant.
FIG. 4.
Hetroduplex mobility analysis of proviral variants from PBMCs of 27 blood-derived isolates from categories I and II (A) and category IV (B) of HIV infection and from tissue (C). A region spanning V3 to V5 was analyzed for tissue and blood isolates. Tissue isolate NT4 was not amplified. The fastest migrating bands represent homoduplexes. Slowly migrating heteroduplexes represent coamplified sequence variants. Each quasispecies on the gel represents 20 to 50 proviruses as determined by end-point dilution.
DISCUSSION
The HIV-1 quasispecies constantly evolve in response to their dynamic relationship with host cells and the immune system. Attempts to determine viral factors which influence the course of infection have led to investigations into the various genetic and biological characteristics of HIV, such as quasispecies diversity, phenotype, coreceptor usage, cellular tropism, and replicative ability. The complex relationships between these factors and disease stage have often been addressed separately. In this study, these factors have been compared in a cross-sectional study of blood isolates of HIV collected at various stages of disease, and in tissue isolates collected from brain, cerebrospinal fluid, lungs, and spleen at autopsy.
Most previous studies which have investigated viral factors such as viral phenotype and cellular tropism have focused on the coincident switch in phenotype and tropism from NSI/M-tropic to SI/T-cell-line tropic (usually associated with a switch from CCR5 to CXCR4 utilization) that is accompanied by a decline in the number of CD4+ T lymphocytes and disease progression in almost half of normal progressors (8, 17, 26, 37, 40, 62, 71). These properties have been measured by measuring HIV tropism and cytopathicity for lymphoblastoid T-cell lines (MT2 cells) and macrophages in vitro and, most recently, by determining changes in coreceptor usage. However, this study also shows a marked and significant increase in replication as well as tropism for MDM and monocytes by HIV-1 strains isolated from blood at the advanced stages compared with the early stages of HIV infection. A much higher proportion of isolates from patients with advanced disease than from those with early stages of infection had intermediate to high levels of both EC p24 antigen concentrations and HIV cDNA after infection of MDM and monocytes (for monocytes, 0 of 6 from categories I and II versus 7 of 21 from advanced disease [P = 0.057], and for MDM, 1 of 6 from categories I and II versus 16 of 21 from advanced disease [P = 0.015]). This also extends previous observations that MDM generally show a greater susceptibility to and support higher levels of replication of primary strains of HIV than do monocytes (50).
In MDMs, there was good correlation between HIV DNA and EC p24, indicating that both tropism and productivity of infection were controlled prior to RT, probably at the stage of viral entry or uncoating. In monocytes, where such correlation was not as high as in MDM, restriction to replication occurred both before and after RT. These results confirmed our previous report with blood isolates, which showed that this restriction to viral entry (or uncoating) may be due predominantly to viral factors with some strains or to an interaction between viral and host factors with others (49).
Viral entry is the primary determinant of tropism for macrophages, T lymphocytes, and T-cell lines and also the major bottleneck for productivity of infection in MDM (49). Therefore, the assessment of coreceptor usage of primary isolates from the different stages of infection and disease is essential to understanding the relationship between viral tropism, productivity, and disease progression. Examination of the coreceptor usage of blood- and tissue-derived isolates as assessed by using HOS.CD4 cells revealed that the majority of blood isolates used CCR5 in these cells (85%), even in the advanced stages (84%). The proportion of blood isolates using chemokine receptors other than CCR5 was slightly greater in advanced stages of disease (3 of 19) than in early stages (1 of 8). However, this one isolate with diverse chemokine receptor usage in the asymptomatic group came from one of the patients with low CD4+-T-cell counts (CD4+ = 250). Interestingly, all three SI and CXCR4-using isolates (NB3, NB5, and NB21) were dually tropic, since they exhibited productive infections in MDM. The apparently expanded coreceptor usage of CXCR4-using isolates (CCR3, Bonzo, and Bob) was not substantiated by ligand-blocking studies and presumably represents the entry of these isolates via variable endogenous levels of CXCR4 on HOS.CD4.CCR3/Bonzo/Bob cells. CXCR4 has been recently reported to mediate the entry of some SI primary but not laboratory-adapted CXCR4 strains into MDM (69, 73). Only the three CXCR4-using viruses demonstrated an SI phenotype in MT2 cells. They also produced the greatest CPE in PBMCs. However, approximately one-third of all strains, whether isolated from late or early stages of disease and which used CCR5, produced some degree of CPE in PBMCs. The V3 sequences of the NSI isolates generally showed low charges due to amino acid substitutions reflecting the M-tropic genotype. Furthermore, analysis of the V1-V2 amino acid sequences did not reveal any distinct amino acid patterns associated with expanded coreceptor usage or altered phenotype including length, location of potential glycosylation sites, and number of positively charged residues. There was no clear phylogenetic segregation (data not shown) between SI and NSI isolates or between those displaying expanded coreceptor utilization according to sites of glycosylation or charge. This suggests that characteristics of isolates defined by V1-V2 probably relates more to the combined conformational structure of V1, V2, and V3 rather than to its isolated amino acid sequence.
The development of blood-derived HIV-1 strains able to use coreceptors other than CCR5 has also been observed by various groups and has been shown to be associated with disease progression (18, 61). These groups have shown that broadened coreceptor usage coincides with increasing resistance to blocking by beta-chemokines in vitro and hypothesize that the level of beta-chemokines in vivo is a major selective pressure which promotes promiscuous coreceptor usage (and then only for CXCR4). Furthermore, only a few changes in the HIV envelope are required for a switch in viral phenotype (30, 35) and coreceptor usage (6, 12, 14, 74, 77). Therefore, the selective pressure of beta-chemokine levels in vivo in addition to the inherent viral mutation rate of HIV should facilitate this switch earlier (15, 42) to produce the expanded coreceptor usage and altered phenotype. The limited number of isolates which showed expanded coreceptor usage, in this study and others (79, 80), during the asymptomatic and terminal stages of infection suggests that there are opposing selective forces and that this phenomenon may not be essential for disease progression. Our results also show that expanded coreceptor usage is not necessary for an increase in productivity from infected MDMs or enhanced tropism for monocytes.
The emergence of CXCR4 and SI phenotype viruses only in patients with advanced disease suggests that strong selective pressures against these variants must first be overcome before they can emerge. Ida et al. (36) have shown that SI T-cell-line-tropic variants, in comparison to NSI M-tropic variants, occur transiently during the asymptomatic stage of infection and are characterized by greater nonsynonymous substitution per synonymous substitution in the V3 region and greater branch lengths in phylogenetic tree analysis. This suggests that strong immune pressures on these CXCR4 variants are present during the asymptomatic stages of infection and are effective at controlling the replication of these variants until the immune system has been compromised by infection with M-tropic NSI viruses (9, 54).
An evolving increase in HIV genomic diversity, especially in the envelope gene, underlies all of the changes in the biologic characteristics of HIV, including altered cytopathic ability, cell tropism, coreceptor usage, and immune system evasion during disease progression (5, 12, 14, 35, 74, 77). The drivers of this genomic diversity and the consequent biologic changes must be the interactions between HIV-1 and the host genetic factors, especially those controlling the relevant proteins interacting during viral replication within the predominant infected cell types in blood and tissues (19, 45, 49).
In this study, genomic diversity of HIV isolates was examined according to disease stage by HMA of the envelope gene, which is the HIV gene showing greatest variability. Isolates from later stages (category IV) of infection showed greater heterogeneity than did isolates from early stages (categories I and II) of infection. These results are consistent with the longitudinal studies (19) which showed an overall increase in HIV genomic diversity with progression and decline in CD4+-T-cell counts. However, that rapid decline in CD4+ T cells was often, but not always, associated with lower quasispecies complexity.
In this study there was an overall correlation of both envelope gene diversity and productivity of infection of MDM each individually with advanced disease. However, the trend toward a correlation between diversity and productivity of infection was not significant, suggesting that other factors, such as viral load and cytokine concentrations, are also important determinants of MDM infection. Nevertheless, this enhanced envelope sequence and structural diversity in advanced disease probably allow the selection of strains which are able to enter more readily via enhanced affinity for CCR5 and subsequent (obscure) steps of entry/uncoating, especially since the predominant restriction to both tropism and productivity of infection in MDM occurred before RT. In monocytes restriction after RT was observed, and this is likely to be influenced by other factors such as spreading and envelope gene sequence. Strains using multiple coreceptors, including CXCR4 strains, showed high envelope gene diversity, but such high diversity in other strains did not lead to multiple coreceptor usage.
All the tissue isolates obtained at autopsy were found to be NSI M tropic. Tissue isolates were not the phenotypic equivalent of blood isolates from patients with advanced disease. They displayed far greater similarity in their tropism for monocytes and MDM and the subsequent level of productive infection than did blood isolates from patients with advanced disease, where most isolates (>80%) replicated to high or intermediate levels in both cell types. All used CCR5 to enter HOS.CD4 cells, and none were shown to use CXCR4, although one isolate from spleen (NT3) had a high V3 positive charge. Only one tissue isolate (NT1) showed expanded coreceptor usage (CCR5 and CCR3), but it was blocked by anti-CCR5 antibodies in MDM (data not shown). These results extend our previously reported results (27, 49) and those of others (56), using laboratory-adapted strains originally isolated from tissues (such as HIV-1 BaL), which also demonstrated greater similarity in tropism and productivity of infection of monocytes and MDM compared with blood isolates. Differences in the differentiation of tissue macrophages in a different microenvironment, especially different chemokine and cytokine levels, may affect the different patterns of replication of blood and tissue isolates in monocytes and MDM. These tissue and laboratory-adapted isolates probably have greater affinity for and can utilize lower levels of CCR5 to facilitate entry into low-CCR5-expressing monocytes (50). Alternatively, these isolates may use both CCR5 and another unidentified coreceptor present in both monocytes and MDMs. Two recent reports also addressed tropism or chemokine receptors utilization by tissue isolates. van’t Wout et al. (72) identified both SI and NSI isolates in various tissues (lymph nodes, spleen, bone marrow, kidney, liver, testes, lung, and brain) by using biological clones, but they did not examine coreceptor utilization. Chan et al. (9) examined coreceptor utilization by tissue isolates by using V3 substitutions into an isogenic background. Only CCR5 utilization was observed, suggesting that the coreceptor switch from CCR5 to CXCR4 does not usually occur in tissue-specific isolates. Our results extend and resolve the apparent conflict between these two reports. In some tissues where T lymphocytes are dominant and trafficking of blood T cells is common, it is not surprising to find M-tropic, T-tropic, and dual-tropic strains. In other tissues, such as the brain, where macrophages are the predominant productively infected cells, CCR5-using isolates would be expected. The CCR5- and CCR3-utilizing isolate NT1 was from brain, but entry was inhibited only by anti-CCR5 inhibitors. There has been considerable debate about the relative importance of these two coreceptors in HIV entry into brain microglial cells (33, 34).
All tissue isolates showed uniformly high envelope gene diversity, greater than that of category IV blood isolates. This diversity may explain the high monocyte tropism by allowing selection of strains able to enter monocytes via CCR5 and other restricting steps in the replicative cycle.
The cross-sectional design of this study allowed the examination of a greater number of primary blood isolates from the early and late stages of infection and also of tissue isolates. Its validity can be assessed by the consistency in results with NSI/SI phenotype, envelope genetic diversity by HMA, and chemokine receptor utilization between these and the recently published longitudinal studies (18, 61). However, in addition, our results show that NSI M-tropic viruses with persistent CCR5 coreceptor usage predominate in late as well as early stages of infection in both blood and tissue compartments. Furthermore, isolates from advanced rather than early disease stages show a marked enhancement of the levels of HIV replication in blood macrophages and monocytes. Interestingly, tissue isolates differed from category IV blood isolates in their greater capacity to infect blood monocytes at an early stage of maturation, suggesting increased gp120 affinity for CCR5 or other mechanisms of viral entry.
The enhancement in HIV replication in these cell types probably occurs via selection from a larger pool of envelope variants in advanced disease and in turn may contribute to progression by increasing the local viral load or transmission to T lymphocytes (53, 70). However, the limited number of blood isolates which showed a switch in phenotype or coreceptor usage in this study suggests that these events are not essential for the enhanced replication or disease progression. Indeed, the maintenance of M-tropism, NSI phenotype, and CCR5 usage is probably advantageous. The strong selective forces against phenotypic switch or broader coreceptor usage need to be better defined. Finally, the increase in the replicative ability of M-tropic strains in monocytes and macrophages at the later stages of infection suggests that they may play a substantial role in general or local disease progression, especially in the brain.
ACKNOWLEDGMENTS
We thank Eric Delwart and Belinda Herring for technical advice with HMA. We also thank Dan Littman, Howard Hughes Medical Institute, New York Medical Center, for generous donation of HOS cells.
This work was supported by ANCARD through the Australian National Center for HIV Virology Research (ANCHVR).
REFERENCES
- 1.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. CC chemokine receptors 5-mediated signaling and HIV-1 coreceptor activity share common structural determinants. Critical residues in the third extracellular loop support HIV-1 fusion. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Berger E A, Murphy P M, Pease J E. Determinants of HIV-1 coreceptor function on CC chemokine receptor 3. Importance of both extracellular and transmembrane/cytoplasmic regions. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1a, MIP-1b receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger E A, Doms R W, Fenyo E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonhoeffer S, Holmes E C, Nowak M A. Cause of HIV diversity. Nature. 1995;376:125. doi: 10.1038/376125a0. [DOI] [PubMed] [Google Scholar]
- 8.Bozzette A A, McCutchan J A, Spector S A, Wright B, Richman D D. A cross-sectional comparison of persons with syncytium- and non-syncytium-inducing human immunodeficiency virus. J Infect Dis. 1993;168:1374–1379. doi: 10.1093/infdis/168.6.1374. [DOI] [PubMed] [Google Scholar]
- 9.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Weiss C, Seto D, Levy J A. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc Natl Acad Sci USA. 1989;86:8575–8579. doi: 10.1073/pnas.86.21.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rowllins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Neuman W, Gerard N, Gerard G, Sodroski J. The beta chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:621–628. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 15.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 16.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delwart E L, Busch M P, Kalish M L, Mosley J W, Mullins J I. Rapid molecular epidemiology of human immunodeficiency virus transmission. AIDS. 1995;11:1081–1092. doi: 10.1089/aid.1995.11.1081. [DOI] [PubMed] [Google Scholar]
- 21.Deng H, Unutmaz D, Kewal-Armani V N, Littman D R. Expression, cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 22.Deng H, Liu R, Ellmeier W, Choc S, Unutmaz D, Burkhart M, DiMarizio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Litman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 23.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual tropic isolate that uses fusin and the beta chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 24.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 25.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan 7-transmembrane segment receptors which are expressed on CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauci A S, Pantaleo G, Stanley S, Weissman D. Immunopathogenic mechanisms of HIV infection. Ann Intern Med. 1996;124:654–663. doi: 10.7326/0003-4819-124-7-199604010-00006. [DOI] [PubMed] [Google Scholar]
- 27.Fear W R, Kesson A M, Naif H M, Lynch G W, Cunningham A L. Differential tropism and chemokine receptor expression of human immunodeficiency virus type 1 in neonatal monocytes, monocyte-derived macrophages, and placental macrophages. J Virol. 1998;72:1334–1344. doi: 10.1128/jvi.72.2.1334-1344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 29.Folks T M, Powel D, Lightfoote M, Koenig S, Fauci A S, Hogan S, Venatesan S, Martin M A. Biological and biochemical characterization of a cloned Leu-3-cell surviving infection with the acquired immunodeficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. Virus isolation and identification of HTLV-III/LAV producing cells in brain tissue from a patient with AIDS. Science. 1986;233:215–219. [Google Scholar]
- 32.Gendelman H E, Orenstein J M, Baca L M, Weiser B, Burger H, Kalter D C, Meltzer M S. The macrophage in the persistence and pathogenesis of HIV-1 infection. AIDS. 1990;3:475–495. doi: 10.1097/00002030-198908000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Ghorpade A, Xia M Q, Hyman B T, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman H E, Mackay C R. Role of the beta-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72:3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Neuman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are coreceptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 35.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–73. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 36.Ida S, Gatanaga H, Shioda T, Nagai Y, Kobayasi K, Shimada K, Kimura S, Iwamato A, Oka S. HIV type 1 V3 variation dynamics in vivo: long term persistence of non-syncytium inducing genotypes and transient presence of syncytium inducing genotypes during the course of progressive AIDS. AIDS. 1997;13:1597–1609. doi: 10.1089/aid.1997.13.1597. [DOI] [PubMed] [Google Scholar]
- 37.Kaneshima H, Su L, Bonyahadi M L, Connor R I, Ho D D. Rapid-high, syncytium-inducing isolates of human immunodeficiency virus type 1 induce cytopathicity in the human thymus of SCID-hu mouse. J Virol. 1991;68:8188–8192. doi: 10.1128/jvi.68.12.8188-8192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazazi F, Mathijs J-M, Foley P, Cunningham A L. Variations in CD4 expression by human monocytes and macrophages and their relationship to infection with the human immunodeficiency virus. J Gen Virol. 1989;70:2661. doi: 10.1099/0022-1317-70-10-2661. [DOI] [PubMed] [Google Scholar]
- 39.Kellog D E, Sninsky J J, Kwok S. Quantitation of HIV-1 proviral DNA relative to cellular DNA by the polymerase chain reaction. Anal Biochem. 1990;189:202–208. doi: 10.1016/0003-2697(90)90108-l. [DOI] [PubMed] [Google Scholar]
- 40.Koot M, Keet I P, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 41.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 42.Kuiken C, De Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S-L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loetscher M, Amara A, Oberlin E, Brass N, Legler D F, Loetscher P, Seisdedos M, Moser B. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Curr Biol. 1997;7:652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- 47.Massari F E, Poli G, Schnittman S M, Psallidopolous M C, Davey V, Fauci A S. In vitro T lymphocyte origin of macrophage tropic strains of HIV. Role of monocytes during in vitro isolation and in vivo infection. J Immunol. 1990;144:4628–4632. [PubMed] [Google Scholar]
- 48.Meltzer M S, Skillman D R, Gomatos P J, Kalter D C, Gendelman H E. Role of mononuclear phagocytes in the pathogenesis of human immunodeficiency virus infection. Annu Rev Immunol. 1990;8:169–194. doi: 10.1146/annurev.iy.08.040190.001125. [DOI] [PubMed] [Google Scholar]
- 49.Naif H M, Li S, Alali M, Chang J, Mayne C, Sullivan J, Cunningham A L. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocyte-derived macrophages using twins. J Virol. 1999;73:4866–4881. doi: 10.1128/jvi.73.6.4866-4881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naif H M, Li S, Alali M, Sloane A, Wu L, Kelly M, Lynch G, Lloyd A, Cunningham A L. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naif H M, Li S, Ho-Shon M, Mathijs J-M, Cunningham A L. The state of maturation of monocytes into macrophages determines the effects of IL-4 and IL-13 on HIV replication. J Immunol. 1997;158:501–511. [PubMed] [Google Scholar]
- 52.Nowak M A, May R M, Anderson R M. The evolutionary dynamics of HIV-1 quasispecies and development of immunodeficiency disease. AIDS. 1990;4:1095–1103. doi: 10.1097/00002030-199011000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Orenstein J M, Fox C, Wahl S M. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1860. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 54.Pantaleo G, Cohen O J, Schacker T, Vaccarezza M, Graziosi C, Rizzardi G P, Kahn J, Fox C H, Schnittman S M, Schwartz D H, Corey L, Fauci A S. Evolutionary pattern of HIV-1 replication and distribution in lymph nodes following primary infection: implications for antiviral therapy. Nat Med. 1998;4:341–345. doi: 10.1038/nm0398-341. [DOI] [PubMed] [Google Scholar]
- 55.Phillips D M, Tan X, Perotti M, Zacharopoulos V R. Mechanism of monocyte-macrophage-mediated transmission of HIV. AIDS Res Hum Retroviruses. 1998;14:S67–S70. [PubMed] [Google Scholar]
- 56.Platt E J, Wehrly K, Kuhnman S E, Chesbro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J, Guo H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by M-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the CCR5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubbert A, Combadiere C, Ostrowski M, Arthos J, Dybul M, Machado E, Cohn M A, Hoxie J A, Murphy P M, Fauci A S, Weissman D. Dendritic cells express multiple chemokine receptors used as coreceptors for HIV entry. J Immunol. 1998;160:3933–3941. [PubMed] [Google Scholar]
- 60.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullins K B, Erlich H A. Primer-directed enzyme amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 61.Scarlatti G, Tresold E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 62.Schellekens P T, Tersmette M, Roos M T, Keet R P, de Wolf F, Coutinho R A, Miedema F. Biphasic rate of CD4+ cell decline during progression to AIDS correlates with HIV-1 phenotype. AIDS. 1992;6:665–669. doi: 10.1097/00002030-199207000-00008. [DOI] [PubMed] [Google Scholar]
- 63.Schuitemaker H, Koot M, Kootstra K N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange T M A, Eeftink Schattenkerk T K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz S, Felber B K, Fenyo E M, Pavlakis G N. Rapidly and slowly replicating human immunodeficiency virus type 1 isolates can be distinguished according to target cell tropism and monocyte cell lines. Proc Natl Acad Sci USA. 1989;86:7200–7203. doi: 10.1073/pnas.86.18.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shankarappa R, Gupta P, Learn G H, Jr, Rodrigo A G, Rinaldo C R, Jr, Gorry M C, Mullins J I, Nara P L, Ehrlich G D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 66.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing macrophage/T cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons G, Reeves J D, McKnight A, Dejucq N, Hibitts S, Power C A, Aarons E, Schols D, De Clercq E, Proudfoot A E, Clapham P R. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevenson M. Molecular mechanisms for the regulation of HIV replication, persistence and latency. AIDS. 1997;11:S25–S33. [PubMed] [Google Scholar]
- 71.Tersmette M, Gruters R, de Wolf F, de Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential isolates. J Virol. 1989;63:2118–2155. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van’t Wout A B, Ran L J, Kuiken C L, Kootstra N A, Pals S T, Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verani A, Pesenti E, Polo S, Tresoldi E, Scarlatti G, Lusso P, Siccardi A G, Vercelli D. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998;161:2084–2088. [PubMed] [Google Scholar]
- 74.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of HIV-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 75.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstsman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adoptive evolution of human immunodificiency virus type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 77.Xiao-Fang Y, Wang Z, Vlahov D, Markham R B, Farzadegan H, Margolick J B. Infection with dual-tropic human immunodeficiency virus type 1 variants associated with rapid total T cell decline and disease progression in injection drug users. J Infect Dis. 1998;178:388–396. doi: 10.1086/515646. [DOI] [PubMed] [Google Scholar]
- 78.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 79.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kuntsman K J, Brown R C, Phair J P, Neumann A U, Ho D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, Kewalramani V N, Moore J P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]