Abstract
Lead affects photosynthesis and growth and has serious toxic effects on plants. Here, the differential expressed proteins (DEPs) in D. huoshanense were investigated under different applications of lead acetate solutions. Using label-free quantitative proteomics methods, more than 12,000 peptides and 2,449 proteins were identified. GO and KEGG functional annotations show that these differential proteins mainly participate in carbohydrate metabolism, energy metabolism, amino acid metabolism, translation, protein folding, sorting, and degradation, as well as oxidation and reduction processes. A total of 636 DEPs were identified, and lead could induce the expression of most proteins. KEGG enrichment analysis suggested that proteins involved in processes such as homologous recombination, vitamin B6 metabolism, flavonoid biosynthesis, cellular component organisation or biogenesis, and biological regulation were significantly enriched. Nearly 40 proteins are involved in DNA replication and repair, RNA synthesis, transport, and splicing. The effect of lead stress on D. huoshanense may be achieved through photosynthesis, oxidative phosphorylation, and the production of excess antioxidant substances. The expression of 9 photosynthesis-related proteins and 12 oxidative phosphorylation-related proteins was up-regulated after lead stress. Furthermore, a total of 3 SOD, 12 POD, 3 CAT, and 7 ascorbate-related metabolic enzymes were identified. Under lead stress, almost all key enzymes involved in the synthesis of antioxidant substances are up-regulated, which may facilitate the scavenging of oxygen-free radical scavenging. The expression levels of some key enzymes involved in sugar and glycoside synthesis, the phenylpropanoid synthesis pathway, and the terpene synthesis pathway also increased. More than 30 proteins involved in heavy metal transport were also identified. Expression profiling revealed a significant rise in the expression of the ABC-type multidrug resistance transporter, copper chaperone, and P-type ATPase with exposure to lead stress. Our findings lay the basis for research on the response and resistance of D. huoshanense to heavy metal stress.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05476-9.
Keywords: Heavy metal, Stress tolerance, Free radical burst, Antioxidant, Anabolism
Introduction
Lead (Pb), a heavy metal, does not easily decompose and significantly exhibits toxicity to organisms in nature, resulting in long-lasting toxic effects. Lead has serious effects on plant growth. The root system is the first to sense heavy metals in the soil and is highly susceptible to their effects of heavy metals Heavy metal ions first enter the plant through the absorption of plant rhizomes. When the heavy metals in the soil reach a certain concentration, they will inhibit the elongation of plant root tip cells and accelerate the lignification of the cells, resulting in poor root growth, short plants, and yellow leaves [1]. It will wither and affect the growth and development of the whole plant. Roots not only absorb water and nutrients from the environment but also secrete organic compounds such as sugars and organic acids into the growth medium. These organic compounds can chelate, complex, and precipitate heavy metals in the environment, thereby affecting the availability of heavy metals in the soil [2]. Heavy metals primarily exist in ion form or bind to cellulose, lignin, etc. Lead has very little mobility in plants. A small amount of lead can be transferred upward or downward; however, it rarely enters the inside of fruits or the starch of roots. Heavy metal stress affects the germination potential, germination rate, main root length, epicotyl length, plant height, and plant biomass of seeds to varying degrees [3]. Although heavy metals cause certain damage to plant growth, over the long-term evolution or due to plant factors, plants have developed specific tolerance mechanisms to heavy metal stress environments. The response of plants to heavy metal stress varies depending on their type, variety, and growth period. The sustained tolerance to heavy metals is not only related to the variety but also to the type and concentration of heavy metals, and it shows a certain dose effect.
Plants gradually develop a series of physiological defense mechanisms to deal with the accumulation and positioning of heavy metals. They have formed a dynamic regulatory system that controls the absorption, accumulation, distribution, and detoxification. In recent years, research on screening plants for tolerance to heavy metals has made some progress, but the criteria used to screen each variety are different [4]. Low concentrations of Pb2 + promote the germination of wheat seeds, while high concentrations inhibit the germination of seeds. The germination rate is significantly affected, and the higher the concentration, the more obvious the inhibitory effect. When the Pb2 + concentration is 50 mg/L, the germination rate and germination potential of the durum wheat variety “Zhongyin 1320” are 12.9% and 4.4% higher than the control, respectively; when the Pb2 + concentration reaches 200 mg/L, the germination rate and germination potential drop by 0.5% and 2.2%, respectively [5]. Hydroponic experiments revealed that wheat’s chlorophyll content in the early growth stage can be promoted by low concentrations of Cd and Pb, while high concentrations inhibit chlorophyll synthesis. Additionally, different heavy metals exhibit varying degrees of inhibition. Under lead stress above 500 mg/L, the leaves of wheat seedlings may even develop chlorosis [6].
Cell wall is the first barrier for toxic and harmful substances to enter the cells, and it also plays a pivotal role in their resistance to heavy metals. Lead is mainly distributed in the cell wall and the soluble part of the cell. The pectin component in the cell wall provides a large number of exchange sites for heavy metal binding [7]. When the binding of heavy metal ions on the cell wall reaches saturation, most of the heavy metals entered into the cell will be transported into the vacuole. They complex with various substances, such as inorganic salts, sugars, amino acids, and organic acids contained in the vacuoles, to achieve compartmentalization and thereby reduce damage to other cell structures [8]. Among the various subcellular components in Hemerocallis fulva leaves, the vacuole and cell-soluble parts contained the most Pb, followed by the cell wall and its residue parts, and the cell membrane and organelle parts were the least distributed. Cell wall and vacuole compartmentalization are important tolerance mechanisms for plants to cope with heavy metal stress. The localization at the subcellular level is necessary to clarify its role in plant resistance to Cd and Pb toxicity. Low-concentration Pb treatment of liquidambar seeds can stimulate the synthesis of ester compounds in membrane lipids and cell wall pectin, while high-concentration Pb and Cd treatment both promote the synthesis of organic matter such as cellulose, hemicellulose, polysaccharides, and proteins, thus increasing plant’s stress tolerance [9].
The accumulation of heavy metals in leaves cause the disintegration of chloroplasts and pigments, reduce photosynthetic efficiency, and thus affect the nutritional metabolism of plants [10]. Leaves often turn yellow when stressed by heavy metals. Plants absorb heavy metal ions, which replace iron, magnesium, and other ions in the leaves, changing the configuration of key enzymes and inhibiting enzyme activity, thereby hindering chlorophyll synthesis. As a non-essential element for plants, lead has a certain promoting effect at a low level. However, a certain accumulation of lead content outside the plant inhibits photosynthesis. The combined stresses of Pb2 + and Cd2 + lower the activity of the RuBP enzyme, make carboxylation less efficient, and drastically reduce the content of chlorophyll a, chlorophyll b, and chlorophyll a/b ratio. The fluorescence parameter Fv/Fm decreases with the increase in Cd and Pb concentrations [11]. Additionally, the net photosynthetic rate Pn, Fv/Fm, ΦPSII, and qN of rapeseed increased under low concentration Pb stress. Low-concentration stress activates the energy dissipation pathway, disperses excess light energy, and protects photosynthetic machinery from damage. Cd and Pb can also affect the absorption and transportation of mineral nutrients by roots. Some studies have demonstrated that radish treatment with cadmium inhibits the absorption and accumulation of Cu, Mn, Zn, K, and Ca in leaves. Pb significantly inhibited the absorption of Zn, K, Ca, and Mn. Pb and Cd composite treatment significantly inhibited the absorption of K, Mn, Zn, and Fe. It is indicated that high concentrations of Pb significantly inhibit the absorption of Cu, Ca, Mg, Zn, and Fe in soybean plants while increasing the accumulation of Mn [12].
Once heavy metals enter the plant, they will cause membrane lipid peroxidation. This releases large amounts of superoxide anions (O2−), hydrogen peroxide (H2O2), hydroxyl radicals (.OH−), singlet oxygen (1O2 ) and other reactive oxygen-free radicals, systematically causing chaos in the redox reactions [13]. Mmembrane lipid peroxidation is a significant cause of membrane damage. Plant antioxidants mainly include enzymatic and non-enzymatic antioxidants.Among them, enzymatic antioxidants mainly include SOD, APx (ascorbate peroxidase), GPx (glutathione peroxidase), CAT, and GR (glutathione reductases); non-enzymatic antioxidants include glutathione, NP-SH (non-protein thiols), cysteine, ascorbic acid, and proline. This antioxidant systems including enzymes such as POD, CAT, and SOD, is produced in significant quantities to scavenge free radicals generated during the process of membrane lipid peroxidation. Intracellular heavy metals significantly affect the activities of SOD and POD [14]. The activities of SOD, CAT, and POD in plants were enhanced when exposed to low concentrations of lead ions. These enzymes played a role in eliminating reactive oxygen-free radicals. The treatment of lead nitrate solution will cause substantial alterations in the antioxidant enzyme activity within plant cells. Pb also affects the nucleic acid metabolism, respiration, nitrogen metabolism, and carbohydrate metabolism by affecting the activity of some important enzymes in the plant metabolism process [15].
Due to environmental pollution and the abuse of organic fertilizers, pesticides, etc., the content of harmful heavy metal elements in Chinese medicines has increased significantly. The excessive levels of Cd, Pb, and other heavy metals in Dendrobium cultivars from different production areas are associated with the local soil and planting environment [16]. At present, the underlying clues of absorption, enrichment, transport, and detoxification responses to heavy metal pollution are still unclear. In this study, variations at the proteome level were examined in D. huoshanense grown in the Ta-pieh Mountains in response to lead stress. GO and KEGG enrichment analyses showed that lead acetate treatment first increased the expression of genes responsible for RNA, DNA, and protein synthesis and transport. Lead acetate treatment also induced genes involved in photosynthesis and oxidative phosphorylation. The expression of antioxidant-related proteins in vitamin B6 metabolism, thiamine metabolism, ascorbate and aldarate metabolism, and glutathione metabolism increased. Lead stress stimulates the biosynthesis of some key enzymes realted to sucrose and glycosides. It also elevated the enzyme levels of some secondary metabolites such as flavonoids, phenylpropanoids, terpenes, and ubiquinone. Through the above-mentioned regulatory mechanism, D. huoshanense has completed the resistance mechanism of lead stress.
Materials and methods
Lead treatment and plant sampling
The D. huoshanense tissue culture seedlings used for the experiment all from the Plant Cell Engineering Center of West Anhui University (Luan, China). After twenty-five days of propagation on the medium, the seeds undergo a color change from pale yellow to green. After a period of 30 days, the seedlings were moved to the germination medium for further growth. A 40-day period was established for subcultures. The chosen materials were grown for three successive generations. The samples in the control group were added with the same amount of water daily, and the samples in the Pb group were added with a solution containing 200 mg/L Pb(NO3)2 and sampling on day 7 and 15. We harvested foliage from the perennial plants in order to acquire three biological duplicates.
Extraction of total proteins
We extracted the total protein using a previous method [17]. We added the SDT protein lysate to the protein solution and kept it in an ice bath for 5 min. The reaction was then centrifuged at 95 °C for 8 min. We add DTT reagent to the supernatant, and add pre-cooled acetone to the reaction solution. After two hours, the precipitate was obtained by centrifugation at 12,000 g for 15 min. The precipitate was rinsed multiple times with acetone and freeze-dried to obtain a lyophilized product. For protein trypsinolysis and extraction, the sample was first added to a DB protein lysis solution (8 M urea, 100 mM TEAB, pH = 8.5) to make up 100 µL. Trypsin and 100 mM TEAB buffer were added to the lysis solution, mixed, and digested at 37 °C for 4 h. The solution was then supplemented with trypsin and CaCl2 solutions, which were digested overnight. Formic acid was added to the digestion solution to adjust the pH to less than 3. The digestion solution was centrifuged at 12,000 g for 5 min at room temperature. The supernatant was slowly passed through a C18 desalting column and then washed three times with a cleaning solution (0.1% formic acid, 3% acetonitrile). The C18 desalting column was added with an eluent (0.1% formic acid, 70% acetonitrile), and the filtrate was collected and freeze-dried to obtain a protein sample.
LC-MS/MS conditions
Liquid chromatography mass spectrometry analysis used a UHPLC-Q Exactive tandem mass spectrometry system and a Nanospray ESI ion source [8]. Xcalibur 2.2 software was used for mass spectrometry analysis. We used a data-dependent acquisition mode to acquire mass spectrometry data, setting the mass detection range at m/z 350–1500. For secondary fragment acquisition, high-energy collision fragmentation selects the top forty most ionized quasi-molecular ions. We set the dynamic exclusion range to 20 s.
Raw data processing and functional enrichment analysis
To identify and decipher proteins, all spectra obtained were searched using the search software Proteome Discoverer 2.2, which was aligned with a public database. The search parameters were established in the following manner: The precursor ion had a mass tolerance of 10 parts per million (ppm), while the fragment ion had a mass tolerance of 0.02 Daltons (Da). The fixed modification involved the alkylation of cysteine, whereas the variable modification consisted of the oxidation of methionine. The N-terminus was acetylated, and a maximum of two missed cleavage sites were permitted. To enhance the accuracy of the analysis results, the PD2.2 software applied further filtering to the search results. Peptide spectrum matches (PSMs) with a credibility over 99% were considered credible PSMs, while proteins that contained at least one unique peptide were deemed credible proteins. Only peptides and proteins that were deemed trustworthy were kept, and a verification process was conducted to eliminate peptides and proteins with a false discovery rate (FDR) exceeding 1%. The protein quantification results were subjected to statistical analysis using the T-test. Proteins exhibiting significant quantitative differences between the experimental group and the control group (p < 0.05, FC ≤ 0.25) were classified as differentially expressed proteins. The interproscan program was employed for functional annotation of GO and IPR databases. The discovered proteins went through functional protein pathway and family studies utilizing the COG and KEGG databases.
Results
Data quality and protein identification
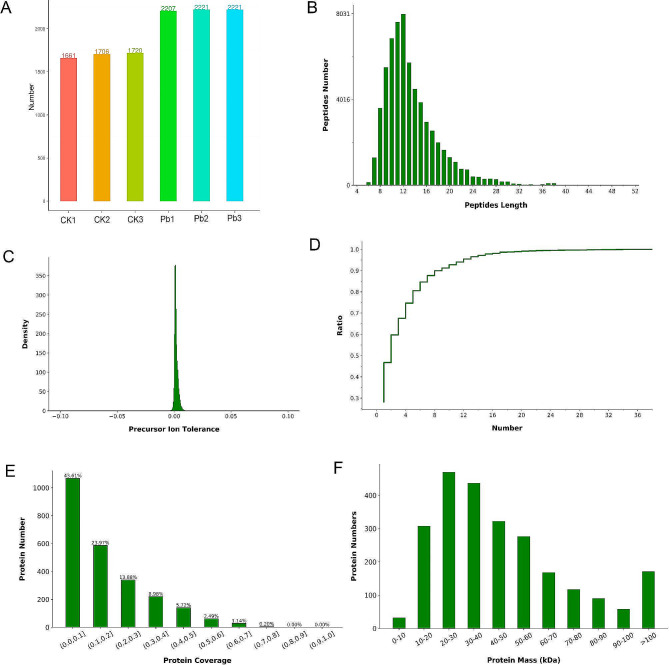
We obtained a total of 63,194 matched spectra, 12,402 identified peptides, and 2,449 identified proteins using the label-free quantitative method. Among them, the number of proteins identified in the control group was 1,696, and the average number of proteins identified in the lead acetate-treated group was 2,216 (Fig. 1A). Further quality assessment of our proteomics data was performed, including principal component analysis, precursor ion mass tolerance distribution, protein sequence coverage distribution, peptide length, and protein molecular weight distribution. The results showed that the peptide length was mainly distributed between 10 and 16 amino acid residues (Fig. 1B). The mass error distribution diagram of precursor ions shows the accuracy of mass spectrometry data. Most proteins have smaller precursor ion mass errors and higher mass spectrometry accuracy (Fig. 1C). The distribution of the number of unique peptides shows that the proportion of proteins containing unique peptides in the total protein gradually both increases. When the number of unique peptides equals 16, the ratio tends to 1 (Fig. 1D). Protein coverage shows that for proteins whose full length accounts for less than 10%, the number of peptides containing this protein is larger, accounting for 43.61% of the total protein. Peptide coverage of less than 30% was observed in 81.46% of the identified proteins, indicating the need for improvement in the depth of protein identification (Fig. 1E). The protein molecular weight distribution showed that the identified proteins mainly ranged in molecular weight from 10 to 60 kDa (Fig. 1F).
Fig. 1.
Data quality assessment and protein identification. (A) Number of proteins identified in the CK group and Pb(NO3)2 group. (B) Peptide length distribution and number. (C) Precursor ion mass tolerance distribution. (D) Number distribution of unique peptides in identified proteins. (E) Distribution and number of protein coverages. (F) Protein molecular weight distribution and quantity
Annotation of identified proteins
The functional annotation of the discovered proteins was performed using databases such as GO, KEGG, and COG. The Interproscan software utilized the six databases of Pfam, PRINTS, ProDom, SMART, ProSite, and PANTHER to accurately identify the gene families and structures of all discovered proteins. Sum of 1,170 proteins have been annotated by the four databases, including GO, KEGG, COG, and IPR (Fig. 2A; Table S1). The COG annotation reveals the greatest abundance of homologous genes associated with protein turnover, chaperones, translation, ribosomal structure and biogenesis, posttranslational modification, glucose transport, and metabolism (Fig. 2B). The GO functional annotation results indicate that the discovered proteins primarily participate in biological activities such as oxidation and reduction, as well as metabolic processes. They are primarily found in cellular structures, such as ribosomes, and within cells. GO functions primarily encompass biological activities such as protein binding, ATP binding, nucleic acid binding, oxidoreductase activity, and nucleic acid binding (Fig. 2C). The KEGG pathway annotation results indicate that the main biochemical metabolic pathways include carbohydrate metabolism, energy metabolism, protein folding, sorting, and degradation activities (Fig. 2D). The IPR annotation indicated that all the proteins that discovered possessed an abundance of RNA recognition domains, protein kinase domains, ATPase domains, and several other domains (Fig. 2E). The subcellular localization study revealed that 20.8% of the protein was localized in the cytoplasm, 19.27% in the chloroplasts, 13.97% in the mitochondria, and 12.64% in the nucleus (Fig. 2F).
Fig. 2.
Protein functional annotation based on database search. (A) Venn diagram of proteins annotated by different databases. (B) COG functional classification. (C) GO functional annotation. (D) KEGG pathway annotation. (E) IPR protein domain annotation. (F) Protein subcellular localization analysis
Protein quantification and differential protein analysis
A total of 636 DEGs were identified, including 518 significantly different upregulated proteins (fold change > 4, p < 0.05) and 118 down-regulated proteins with significant differences (fold-change < 0.25, p < 0.05). The volcano plot indicated the distribution of the two groups of differential proteins (Fig. 3A). Differential protein clustering analysis shows the up-regulation and down-regulation of different proteins between different samples. Heavy metal lead caused the up-regulation of most differential proteins (Fig. 3B). GO enrichment analysis results show that the proteins are mainly involved in cellular component organization or biogenesis, biological regulation, regulation of cellular processes, oxidoreductase activity, ligase activity, carbon-nitrogen lyase activity, and protein disulfide oxidoreductase activity (Fig. 3C). KEGG enrichment analysis showed significant enrichment in homologous recombination, vitamin B6 metabolism, flavonoid biosynthesis, and other processes (Fig. 3D). In addition, we screened out nearly 40 proteins involved in DNA replication and repair, RNA synthesis, transport, and splicing from these differential proteins (Table 1). Replication factors A1 and A2 are crucial for homologous recombination and DNA replication, or mismatch repair. These proteins showed up-regulated expression under lead stress. A total of 15 small and large subunit ribosomal proteins are involved in protein processing in ribosomes, and most of them are up-regulated after lead acetate treatment. Translation initiation factor 3, translation initiation factor 1 A, translation initiation factor 4E, polyadenylate-binding protein, etc. showed increased expression levels during the RNA transport process. A total of ten splicing body proteins were identified. Except for U5 small nuclear ribonucleoprotein, splicing factor 3B, and FUS-interacting serine-arginine-rich protein 1, the expression of other proteins was up-regulated after lead treatment. We also found that the effect of D. huoshanense on heavy metal lead stress was mainly achieved by enhancing photosynthesis, oxidative phosphorylation, and the synthesis of antioxidant substances. A total of nine genes involved in photosynthesis were identified. The expression levels of cytochrome b6-f complex, F-type h+-transporting ATPase, photosystem II oxygen-evolving enhancer, photosystem II Psb27 protein, F-type H+-transporting ATPase, ferredoxin, etc. were up-regulated under lead stress. Twelve genes involved in the oxidative phosphorylation pathway were identified, and V-type ATPase, F-type ATPase, NADH dehydrogenase 1, ubiquinol-cytochrome c reductase, succinate dehydrogenase, etc. showed up-regulated expression (Table 2).
Fig. 3.
Difference analysis and functional enrichment analysis of proteins under lead stress. (A) Differential protein volcano plot. (B) Hierarchical clustering analysis of differential proteins. (C) GO enrichment analysis of differential proteins. (D) KEGG enrichment analysis of differential proteins
Table 1.
DEGs involved in DNA replication and repair, RNA synthesis, transport and splicing
| KEGG term | Protein ID | log2fc | p-Value | Regulated | KEGG description |
|---|---|---|---|---|---|
| Homologous recombination | Dhu000026760 | Infinity | 0.00 | Up | Replication factor A1 |
| Dhu000026446 | Infinity | 0.00 | Up | Replication factor A2 | |
| Dhu000017350 | -Infinity | 0.00 | Down | Recombination factor Rad54 | |
|
DNA replication/ Mismatch repair |
Dhu000002634 | 2.0956 | 0.00038 | Up | Proliferating cell nuclear antigen |
| RNA transport | Dhu000007551 | 2.3068 | 0.00657 | Up | Elongation factor 1-alpha |
| Dhu000025977 | -3.3673 | 1.40e-05 | Down | Importin subunit beta-1 | |
| Dhu000006080 | -2.4857 | 0.00178 | Down | Translation initiation factor 3 subunit E | |
| Dhu000009649 | 3.5484 | 0.00939 | Up | Translation initiation factor 3 subunit A | |
| Dhu000013536 | Infinity | 0.00 | Up | Translation initiation factor 3 subunit A | |
| Dhu000022628 | Infinity | 0.00 | Up | Translation initiation factor 1 A | |
| Dhu000028824 | Infinity | 0.00 | Up | Translation initiation factor eIF-2B subunit gamma | |
| Dhu000010912 | 2.0183 | 0.00782 | Up | Translation initiation factor 4E | |
| Dhu000002565 | 2.3284 | 0.00034 | Up | Polyadenylate-binding protein | |
| Dhu000014780 | Infinity | 0.00 | Up | THO complex subunit 2 | |
| Dhu000017521 | Infinity | 0.00 | Up | THO complex subunit 4 | |
| Dhu000012452 | 2.2565 | 0.00048 | Up | Pinin | |
| Ribosome | Dhu000009495 | 2.6292 | 2.31e-05 | Up | Small subunit ribosomal protein S10 |
| Dhu000021321 | -5.4675 | 2.76e-06 | Down | Large subunit ribosomal protein L4 | |
| Dhu000011193 | 3.2359 | 7.82e-06 | Up | Large subunit ribosomal protein L35e | |
| Dhu000005346 | 3.2041 | 0.00152 | Up | Large subunit ribosomal protein L24 | |
| Dhu000016133 | 2.0349 | 0.00203 | Up | Large subunit ribosomal protein L26e | |
| Dhu000005028 | Infinity | 0.00 | Up | Small subunit ribosomal protein S2e | |
| Dhu000013939 | 2.9020 | 1.25e-05 | Up | Small subunit ribosomal protein S14e | |
| Dhu000019946 | 3.1997 | 0.00121 | Up | Large subunit ribosomal protein L7 | |
| Dhu000024895 | 2.6700 | 0.00109 | Up | Large subunit ribosomal protein L10 | |
| Dhu000023682 | 3.6982 | 2.91e-06 | Up | Large subunit ribosomal protein L11 | |
| Dhu000023585 | 2.1081 | 0.00034 | Up | Small subunit ribosomal protein S13e | |
| Dhu000003738 | 2.3842 | 0.00011 | Up | Large subunit ribosomal protein L21 | |
| Dhu000025515 | -4.0546 | 0.00045 | Down | Large subunit ribosomal protein L15e | |
| Dhu000017134 | 2.6055 | 0.00066 | Up | Large subunit ribosomal protein L31e | |
| Dhu000001456 | 4.2940 | 0.00309 | Up | Small subunit ribosomal protein S19e | |
| Spliceosome | Dhu000019670 | Infinity | 0.00 | Up | U6 snRNA-associated Sm-like protein LSm3 |
| Dhu000016515 | -2.7027 | 4.96e-05 | Down | U5 small nuclear ribonucleoprotein component | |
| Dhu000006224 | Infinity | 0.00 | Up | U4/U6 small nuclear ribonucleoprotein PRP3 | |
| Dhu000006583 | Infinity | 0.00 | Up | U4/U6 small nuclear ribonucleoprotein PRP4 | |
| Dhu000003217 | -2.4813 | 5.96e-07 | Down | Splicing factor 3B subunit 3 | |
| Dhu000027568 | Infinity | 0.00 | Up | RNA-binding protein 25 | |
| Dhu000017002 | Infinity | 0.00 | Up | ATP-dependent RNA helicase DDX5/DBP2 | |
| Dhu000014916 | Infinity | 0.00 | Up | Heterogeneous nuclear ribonucleoprotein G | |
| Dhu000026442 | -4.708 | 0.00136 | Down | FUS-interacting serine-arginine-rich protein 1 | |
| Dhu000001640 | Infinity | 0.00 | Up | Intron-binding protein aquarius | |
| RNA degradation | Dhu000004787 | Infinity | 0.00 | Up | CCR4-NOT transcription complex subunit 3 |
| Dhu000002565 | 2.3284 | 0.00034 | Up | Polyadenylate-binding protein | |
| Dhu000019670 | Infinity | 0.00 | Up | U6 snRNA-associated Sm-like protein LSm3 | |
| Dhu000004415 | Infinity | 0.00 | Up | ATP-dependent DNA helicase RecQ | |
| Dhu000002005 | -2.9928 | 0.00227 | Down | 6-Phosphofructokinase 1 | |
| Dhu000003802 | -2.0687 | 0.04845 | Down | Molecular chaperone DnaK |
Table 2.
DEGs involved in photosynthesis and oxidative phosphorylation under pb stress
| KEGG term | Protein ID | log2fc | p-Value | Regulated | KEGG description |
|---|---|---|---|---|---|
| Photosynthesis | Dhu000018784 | 3.5639 | 2.76e-08 | Up | Cytochrome b6-f complex iron-sulfur subunit |
| Dhu000002238 | 4.9756 | 0.00673 | Up | Cytochrome b6-f complex iron-sulfur subunit | |
| Dhu000022570 | 2.9183 | 0.00666 | Up | F-type h+-transporting atpase subunit b | |
| Dhu000027584 | 2.4682 | 0.00197 | Up | Photosystem II oxygen-evolving enhancer protein 3 | |
| Dhu000018717 | 3.1809 | 0.00178 | Up | F-type h+-transporting atpase subunit δ | |
| Dhu000002825 | 2.8333 | 0.00075 | Up | Photosystem II Psb27 protein | |
| Dhu000016663 | Infinity | 0.00 | Up | Plastocyanin | |
| Dhu000028844 | 4.6143 | 0.00128 | Up | Ferredoxin | |
| Dhu000002675 | 2.5765 | 1.56e-05 | Up | F-type H+-transporting ATPase subunit beta | |
| Oxidative phosphorylation | Dhu000008527 | Infinity | 0.00 | Up | V-type h+-transporting atpase 16kda proteolipid subunit |
| Dhu000010659 | Infinity | 0.00 | Up | Nadh dehydrogenase (ubiquinone) 1 beta subcomplex subunit 7 | |
| Dhu000028439 | Infinity | 0.00 | Up | Nadh dehydrogenase (ubiquinone) fe-s protein 6 | |
| Dhu000014507 | 4.7665 | 4.80E-05 | Up | Nadh dehydrogenase (ubiquinone) 1 α subcomplex subunit 5 | |
| Dhu000000294 | 3.1013 | 0.00165 | Up | Nadh dehydrogenase (ubiquinone) 1 β subcomplex subunit 9 | |
| Dhu000006207 | 2.5308 | 0.00019 | Up | Ubiquinol-cytochrome c reductase cytochrome c1 subunit | |
| Dhu000000527 | 2.8147 | 0.00127 | Up | Succinate dehydrogenase (ubiquinone) Iron-sulfur subunit | |
| Dhu000017283 | 6.1260 | 4.67E-07 | Up | V-type h+-transporting atpase subunit g |
Peroxidases family involved in scavenging oxygen-free radicals and the differential expression of key enzymes involved in antioxidant were identified. Superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) are the main antioxidases (Table 3). We identified a total of 3 SODs, 6 PODs, 5 L-ascorbate peroxidase (APx),1 glutathione peroxidase (GPx), 1 polyphenol oxidase (PPO) and 3 CATs. All SOD genes had significantly up-regulated expression. There are 2 PODs, 2 APx, 1 GPx gene, and 1 CAT whose expression is up-regulated under lead stress. In addition to activating the expression of POD, lead stress also increased the synthesis of antioxidants such as vitamin B6, vitamin C, thiamine, glutathione. The DEGs showed that lead stress can raise the level of pyridoxal 5’-phosphate synthase, an important enzyme in the biosynthesis pathway for vitamin B6, and directly raise the amount of pyridoxal 5-phosphate. Lead stress downregulated pyridoxine 4-dehydrogenase and phosphoserine aminotransferase, as well as decreased pyridoxine 4-dehydrogenase’s level. Ascorbic acid is one of the strongest natural antioxidants, and the heavy metal lead can stimulate the accumulation of root apoplastic ascorbyl radicals. A total of 7 ascorbate-related metabolic enzymes were identified, among which dehydroascorbic reductase, L-galactose dehydrogenase, and a few APx genes were up-regulated, and 1 APx gene was down-regulated. A total of 11 glutathione-related synthases were identified in this study, including glutathione S-transferase (GST), GPx, glutamate-cysteine ligase (GCL), and glutathione reductase (GR). Among them, four GSTs and one GPx were induced by membrane lipid peroxidation caused by lead stress, and their expression levels increased, while the expression level of GCL was down-regulated.
Table 3.
Key enzyme related to antioxidant biosynthesis involved in ROS scavenging
| KEGG term | Protein ID | log2fc | p-Value | Regulated | KEGG description |
|---|---|---|---|---|---|
| Antioxidases | Dhu000028022 | 1.8872 | 1.27E-05 | Nonsignificant | Superoxide dismutase |
| Dhu000015323 | 2.0411 | 0.00641 | up | Superoxide dismutase | |
| Dhu000020666 | Infinity | 0.00 | up | Copper chaperone for superoxide dismutase | |
| Dhu000016068 | Infinity | 0.00 | up | Superoxide dismutase | |
| Dhu000021108 | -0.1656 | 0.02582 | Nonsignificant | Catalase | |
| Dhu000010726 | 0.5575 | 0.00012 | Nonsignificant | Catalase | |
| Dhu000014602 | 3.6757 | 0.00185 | up | Catalase | |
| Dhu000003920 | -0.2587 | 0.01211 | Nonsignificant | Peroxidase | |
| Dhu000011608 | -0.4039 | 0.06789 | Nonsignificant | Peroxidase | |
| Dhu000027806 | Infinity | 0.00 | up | Peroxidase | |
| Dhu000005526 | Infinity | 0.00 | up | Peroxidase | |
| Dhu000012112 | 0.3981 | 0.25471 | Nonsignificant | Peroxidase | |
| Dhu000018249 | -2.0542 | 0.09989 | Nonsignificant | Peroxidase | |
| Dhu000016586 | -1.1297 | 0.05514 | Nonsignificant | Polyphenol oxidase | |
| Vitamin B6 metabolism | Dhu000007824 | 3.1005 | 0.00271 | Up | Pyridoxal 5’-phosphate synthase |
| Dhu000002640 | Infinity | 0.00 | Up | Pyridoxal 5’-phosphate synthase | |
| Dhu000028434 | -6.5826 | 0.00924 | Down | Pyridoxine 4-dehydrogenase | |
| Dhu000010367 | Infinity | 0.00 | Up | Phosphoserine aminotransferase | |
| Thiamine metabolism | Dhu000026717 | 2.4968 | 0.00069 | Up | Cysteine desulfurase |
| Ascorbate and aldarate metabolism | Dhu000017762 | 3.2938 | 1.81E-06 | Up | Dehydroascorbic reductase |
| Dhu000012810 | Infinity | 0.00 | Up | L-galactose dehydrogenase | |
| Dhu000018823 | 3.4734 | 2.38E-07 | Up | L-ascorbate peroxidase | |
| Dhu000018044 | 2.0075 | 3.48E-06 | Up | L-ascorbate peroxidase | |
| Dhu000028960 | -5.1318 | 0.01809 | Down | L-ascorbate peroxidase | |
| Dhu000028634 | 1.1110 | 0.00023 | Nonsignificant | L-ascorbate peroxidase | |
| Dhu000019246 | 1.8758 | 9.05E-06 | Nonsignificant | L-ascorbate peroxidase | |
| Glutathione metabolism | Dhu000023012 | 2.1224 | 0.00558 | Up | Glutathione S-transferase |
| Dhu000021472 | -2.0814 | 0.00148 | Down | Glutathione S-transferase | |
| Dhu000010171 | 2.7129 | 0.00073 | Up | Glutathione peroxidase | |
| Dhu000020272 | 3.5236 | 6.86E-05 | Up | Glutathione S-transferase | |
| Dhu000028205 | -2.6148 | 0.00139 | Down | Glutamate–cysteine ligase | |
| Dhu000001976 | 0.7666 | 0.00036 | Nonsignificant | Glutathione reductase | |
| Dhu000025514 | 0.3992 | 0.01215 | Nonsignificant | Glutathione reductase |
D. huoshanense is rich in polysaccharides, alkaloids, flavonoids, and other active substances. In this study, we identified several differential proteins in sucrose metabolism and glycoside synthesis. Under lead stress, the levels of chitinase, UDP-apiose/xylose synthase, fructokinase, GDP-mannose 4,6-dehydratase, mannose−6-phosphate isomerase, and raffinose synthase all went up, while the levels of hexokinase went down. For N-glycan biosynthesis, dolichyl-phosphate beta-glucosyltransferase, the oligosaccharyltransferase complex, and arabinosyltransferase showed down-regulated expression (Table 4). In addition to primary metabolites, many key enzymes for the synthesis of secondary metabolites are also responsive to lead stress. Chalcone synthase (CHS), caffeoyl-CoA O-methyltransferase (CCoAOMT), chalcone isomerase (CHI), cinnamyl-alcohol dehydrogenase (CAD), ferulate-5-hydroxylase (F5H), and caffeoyl shikimate involved in the biosynthesis of phenylpropanoid, and flavonoid esterase is up-regulated under lead stress. For terpenoid biosynthesis, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MCS) and hydroxymethylglutaryl-CoA reductase (HMGR) were up-regulated under lead stress, while geranylgeranyl pyrophosphate synthase (GGPPS) and mevalonate kinase (MVK) decreased (Table 5).
Table 4.
DEGs involved in sugar and glycoside biosynthesis
| KEGG term | Protein ID | log2fc | p-Value | Regulated | KEGG description |
|---|---|---|---|---|---|
| Amino sugar and nucleotide sugar metabolism | Dhu000006991 | Infinity | 0.00 | Up | Chitinase |
| Dhu000006916 | 2.9940 | 8.47E-05 | Up | UDP-apiose/xylose synthase | |
| Dhu000007460 | 2.3095 | 0.00060 | Up | UDP-glucose 4,6-dehydratase | |
| Dhu000024131 | 3.1023 | 0.01471 | Up | Fructokinase | |
| Dhu000001044 | -2.0457 | 1.24E-05 | Down | Hexokinase | |
| Dhu000006988 | Infinity | 0.00 | Up | Chitinase | |
| Dhu000010306 | Infinity | 0.00 | Up | GDPmannose 4,6-dehydratase | |
| Dhu000017859 | Infinity | 0.00 | Up | Mannose-6-phosphate isomerase | |
| Dhu000025211 | Infinity | 0.00 | Up | Phosphoacetylglucosamine mutase | |
| Galactose metabolism | Dhu000001044 | -2.0457 | 1.24E-05 | Down | Hexokinase |
| Dhu000023690 | Infinity | 0.00 | Up | Raffinose synthase | |
| N-Glycan biosynthesis | Dhu000002200 | Infinity | 0.00 | Up | Dolichyl-phosphate beta-glucosyltransferase |
| Dhu000008194 | Infinity | 0.00 | Up | Oligosaccharyltransferase complex subunit gamma | |
| Dhu000013620 | Infinity | 0.00 | Up | Arabinosyltransferase |
Table 5.
DEGs involved in secondary metabolite biosynthesis
| KEGG term | Protein ID | log2fc | p-Value | Regulated | KEGG description |
|---|---|---|---|---|---|
| Flavonoid biosynthesis | Dhu000026929 | Infinity | 0.00 | Up | Chalcone synthase |
| Dhu000023635 | Infinity | 0.00 | Up | Caffeoyl-CoA O-methyltransferase | |
| Dhu000001795 | -2.0838 | 0.00306 | Down | Chalcone isomerase | |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | Dhu000023635 | Infinity | 0.00 | Up | Caffeoyl-CoA O-methyltransferase |
| Terpenoid backbone biosynthesis | Dhu000023600 | Infinity | 0.00 | Up | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| Dhu000019364 | -3.0844 | 0.00156 | Down | Geranylgeranyl pyrophosphate synthase | |
| Dhu000015319 | -8.4440 | 0.00071 | Down | Mevalonate kinase | |
| Dhu000004523 | Infinity | 0.00 | Up | Hydroxymethylglutaryl-CoA reductase | |
| Phenylpropanoid biosynthesis | Dhu000027474 | -2.7092 | 5.30E-07 | Down | Cinnamyl-alcohol dehydrogenase |
| Dhu000019210 | Infinity | 0.00 | Up | Cinnamyl-alcohol dehydrogenase | |
| Dhu000014285 | Infinity | 0.00 | Up | Ferulate-5-hydroxylase | |
| Dhu000006901 | Infinity | 0.00 | Up | Caffeoylshikimate esterase | |
| Dhu000023635 | Infinity | 0.00 | Up | Caffeoyl-CoA O-methyltransferase | |
| Ubiquinone biosynthesis | Dhu000013985 | 2.5468 | 6.33E-05 | Up | NAD(P)H dehydrogenase |
| Dhu000020416 | 2.4765 | 9.34E-05 | Up | NAD(P)H dehydrogenase |
Expression profiles of proteins involved in lead detoxification and transport
Thirty proteins involved in heavy metal transport, including ABC transporter, P-type ATPase, NRAMP, HMA, and antioxidantase, were selected from these DEGs to further determine their potential involvement in the extracellular and intracellular transport and enrichment of lead ions. Expression profile analysis results showed that Dhu000027032 (an ABC-type multidrug resistance transporter) was almost not expressed before lead stress, and the expression level surged rapidly after treatment (Fig. 4A). Similar results are shown for Dhu000009772 (an ABC-type uncharacterized transporter) and Dhu000001566 (a forkhead- associated domain protein). HMA is widely involved in the migration and accumulation of heavy metals in plants. We identified a total of three HMA proteins, but they showed differential expression patterns before and after lead stress (Fig. 4B). The expression level of Dhu000025719 (copper chaperone, CopZ) increased significantly after exposure to lead stress. Interestingly, Dhu000018148 (NRAMP) showed downregulated expression after lead treatment (Fig. 4C). Expression profile analysis revealed that Dhu000014277 (Ca2+-transporting ATPase) and Dhu000014567 (manganese-transporting P-type ATPase) were sensitive to lead stress, among the 5 identified P-type ATPases (Fig. 4D). The expression of antioxidant enzymes was mentioned earlier. After lead stress, the expression profile analysis further determined significant effects on Dhu000016068 (superoxide dismutase), Dhu000005526 (peroxidase), Dhu000020666 (copper chaperone for superoxide dismutase), Dhu000027806 (peroxidase), and Dhu000014602 (catalase). up-regulated expression (Fig. 4E).
Fig. 4.
Expression profiling of proteins involved in lead ion detoxification and transport. (A) ATP-binding cassette (ABC) transporter. (B) heavy metal-associated (HMA) protein. (C) natural resistance-associated macrophage protein (NRAMP). (D) P-type ATPase. (E) antioxidantase. The abundance of all proteins was normalized by log10 calculation
Analysis of differential protein interaction
Protein interaction network was constructed using significantly DEGs and analyzed the regulation between these proteins (Table S2). The results showed that there were 278 pairs DEGs with score values more than 600 (Fig. 5). Dhu000002840 (RNA polymerases I and III subunit RPAC1) interacts strongly with other proteins and shows opposite expression to Dhu000016515 (translation elongation factor EF-G). Dhu000008563 (nucleoside-diphosphate kinase) interacts most strongly with Dhu000008599 (CTP synthase). Dhu000015455 (3-hydroxyacyl-CoA dehydrogenase) interacts with Dhu000018551 (carnithine racemase) but exhibits opposite expression to Dhu000015562 (3-hydroxyacyl-CoA dehydrogenase).
Fig. 5.
Protein-protein interaction analysis. Node size represents the number of interacting proteins. The color of the node indicates the expression level of the protein
Discussion
Pb pollution in soil changes the chemical composition of the soil, destroys the physical structure of the soil, and directly affects the quality and safety of agricultural products [18]. Pb have toxic effects on plant morphogenesis, physiology, biochemistry, and cell genetics at different levels. The impact of Pb on plant morphology mostly manifests in seed germination, root elongation, and seedling development.It affects plant physiology by impairing photosynthesis and respiration, as well as reducing and blocking organic matter transportation and absorption [19]. In addition, heavy metal pollution can also produce some other negative effects, such as inhibition of cell division and genotoxicity [20]. In our study, the toxic effects of lead on Dendrobium seedling are mainly manifested by destroying the permeability of cell membranes and affecting the balance of mineral element absorption, transportation, penetration, and regulation (Figs. 1 and 2). Pb also cause mineral element metabolism disorders and inhibit the metabolism of nucleic acids, carbohydrates, proteins, etc. [21]. The impact of heavy metal lead ions on plant cell membranes is that lead ions can change the physical structure of the cell wall and affect the cell membrane. After the lead ions interact with the cell membrane, they destroy the structure of the cell membrane and increase the permeability of the cell membrane [22]. As lead ions enter the cells through the cell wall, they can change the ultrastructure of the chloroplasts in the plant cells, thereby affecting the synthesis of chlorophyll [23].
In order to cope with a series of adverse effects of the heavy metal Pb on plants, plants have developed tolerance mechanisms at different levels, including morphology, cytology, physiology, etc. Typically, plants adopt an “avoidance strategy” toward Pb [24]. Through the activation of the antioxidant enzyme system and the adsorption, separation, and excretion of lead, they can tolerate Pb stress (Fig. 3). These tolerance mechanisms collectively reduce or even prevent Pb uptake at the source. During the transport process, Pb is bound or retained in the cell wall and vacuoles. The enhanced efflux reduces the accumulation of Pb concentrations. Physical barriers, such as thick cuticles, are the first line of defense for plants against heavy metal stress [25]. Exposure to heavy metal toxicity slows root growth, reducing contact with heavy metal pollution and minimizing harm caused by heavy metals. Furthermore, plants respond to stress by changing their growth direction and redirecting their growth to minimize stress exposure [26]. Here, 30 proteins involved in heavy metal transport, including ABC transporter, P-type ATPase, NRAMP, HMA, and antioxidantase were identified and used for expression pattern analysis (Fig. 4).
To prevent heavy metal ions from causing damage to plants, plants have built-in antioxidant defense systems and antioxidants. The antioxidant defense system is based on enzymes that regulate cellular redox status and reduce metabolites—GSH, PCs, and metallothioneins—that are involved in the detoxification of heavy metal ions in plant cells [27]. Lead-induced toxicity can inhibit the activity of the above enzymes or induce their synthesis. Higher Pb concentrations decrease enzyme activity due to enzyme inhibition caused by reactive oxygen species, reduced enzyme synthesis, or changes in its subunit assembly. When bamboo and cypress seedlings were stressed by different concentrations of lead nitrate solutions, the activities of SOD, CAT, and POD in their leaves all went up and then down as the treatment concentration went up [28]. Lead ions enhance lipid peroxidation of cell membranes in bamboo and cypress leaves, resulting in the production of a large amount of reactive oxygen species and the destruction of cell structures. The roots retained most of the lead ions absorbed under the stress of a lead nitrate solution, resulting in less transportation to the stems and leaves [29].
The accumulation and distribution of heavy metals in subcellular cells is one of the important factors that determine the tolerance and detoxification ability of plants to heavy metals [30]. Induced adsorption of lead on the cell wall is a common cellular mechanism to prevent the toxic effects of lead. Polysaccharides from cell walls contain a large number of negatively charged groups that can react with metal cations to immobilize harmful metal ions. Extracellular lead isolation mainly refers to cell wall isolation [31]. Some functional groups (-COOH, -OH, -SH) in the cell wall provide the ability to bind to divalent metal cations. In addition, plant root exudates such as malic acid, oxalic acid, and other organic anions can provide extracellular isolation of heavy metal ions. Heavy metals in cells are complexed with different polymers (pectin, lignin, cellulose, hemicellulose, etc.) or organic anions. The combined Pb is sequestered in vacuoles, endoplasmic reticulum vesicles, and plasma vesicles. This blocks lead transport across the membrane. Some transporters can excrete metal ions into the extracellular space and play an important role in metal detoxification [32]. ATPases, COPT-type transporters, ZIP transporters, and cation antiporters control metal transport in plasmodesmata, enhancing their efflux between cells when heavy metal transport into the endoderm is blocked (Fig. 4).
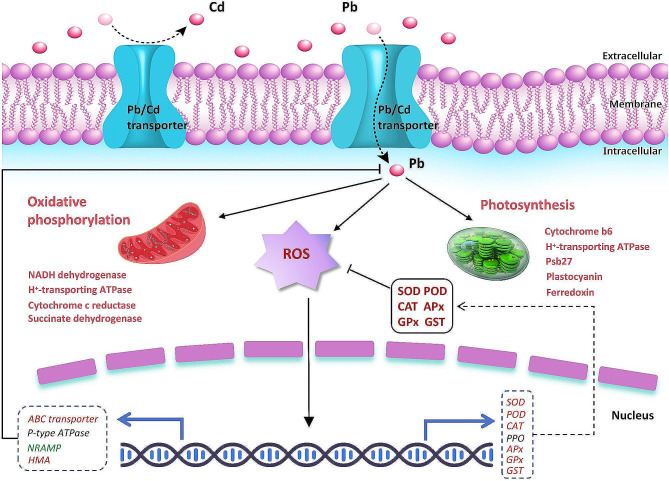
Phytochelatins (PCs) are non-protein thiols known to have metal-binding capabilities and can form sulfhydryl bonds with various metals. To reduce induced cell damage, toxic metals trigger this defense mechanism [33]. First, metal ions activate PC synthase. After chelating with metals, the synthesized PCs are transported to the vacuoles, where they form complexes with sulfides or organic acids, hence stabilizing the metal ion complexes. Some organic anions (such as tartrate, aconitate, oxalate, malate, malonate, or citrate) form strong bonds by chelating with the metal of the carboxylate group as an electron donor. Plants reduce the concentration of heavy metals in the body by reducing the intake of heavy metals [34]. Plants can store heavy metals in trichomes on the leaf epidermis, thereby avoiding direct damage to mesophyll cells by heavy metals. Plants can detoxify heavy metals that enter their cells through compartmentalization, precipitation, and chelation [35]. Heavy metals such as Cr, Cd, Hg, and Ni generate many reactive oxygen radicals. These free radicals cause membrane lipid peroxidation of major biological macromolecules. Various antioxidant defense systems in plants can scavenge free radicals and protect cells from oxidative stress. Plants can also get rid of reactive oxygen species by making metallothioneins, phytochelins, amino acids, and organic acids. This lowers the damage that reactive oxygen species do to cells [36, 37]. Using label-free quantitative proteomics methods, we engaged in uncovering the underlying mechanisms by which D. huoshanense responds to and alleviates lead stress (Fig. 6). Excessive lead enters cells via the Pb/Cd transporter, causing membrane lipid peroxidation and the explosion of reactive oxygen species. Pb uptake simultaneously activates photosynthetic system-related proteins and oxidative phosphorylation. Reactive oxygen species signals are transmitted into cells, causing the transcription levels of antioxidant enzymes and related heavy metal (especially Pb) transporter genes to further aggravate the expression of these enzymes, thus inhibiting the generation of ROS and promoting the efflux and transfer of lead.
Fig. 6.
Putative regulatory mechanism of D. huoshanense against lead stress. Red genes represent significantly up-regulated expression, green represents significantly down-regulated expression, and black represents no significant expression
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- APx
ascorbate peroxidase
- GPx
glutathione peroxidase
- CAT
catalase
- GR
glutathione reductase
- HCD
high-energy collision dissociation
- SOD
superoxide dismutase
- POD
peroxidase
- APx
L-ascorbate peroxidase
- GPx
glutathione peroxidase
- PPO
polyphenol oxidase
- FDR
false discovery rate
- FC
fold change
- DEP
differential expressed protein
Author contributions
CS and JD conceived and designed the experiments. CS and YYZ wrote the draft manuscript. JD performed the experiments and prepared the materials. CS, JD and YSR analyzed the data. CS and MAM revised the manuscript. CS and YYZ acquired the funding. All the authors approved the final manuscript.
Funding
This work was supported by the Open Fund of Anhui Engineering Research Center for Eco-agriculture of Traditional Chinese Medicine (WXZR202318), Demonstration Experiment Training Center of Anhui Provincial Department of Education (2022sysx033), National Health Commission Scientific Research Fund (SBGJ202301010) and Startup fund for high-level talents of West Anhui University (WGKQ2022025).
Data availability
The data sets are publicly available in ProteomeXchange at https://proteomecentral.proteomexchange.org/cgi/GetDataset? ID=PXD047050 and iProX at https://www.iprox.cn/page/project.html? id=IPX0007578000.
Declarations
Ethics approval and consent to participate
The authors declared that permission to collect D. huoshanense material has been obtained and that it complies with institutional, national, and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Park W, Feng Y, Ahn S-J. Alteration of leaf shape, improved metal tolerance, and productivity of seed by overexpression of CsHMA3 in Camelina sativa. Biotechnol Biofuels. 2014;7:96. 10.1186/1754-6834-7-96. 10.1186/1754-6834-7-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu R, Jiang Q, Xv C, Li L, Bu S, Shi G. Comparative proteomics analysis of peanut roots reveals differential mechanisms of cadmium detoxification and translocation between two cultivars differing in cadmium accumulation. BMC Plant Biol. 2019;19:137. 10.1186/s12870-019-1739-5. 10.1186/s12870-019-1739-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasnaoui S, El, Fahr M, Zouine M, Smouni A. De Novo Transcriptome Assembly, Gene Annotations, and characterization of functional profiling reveal key genes for lead alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L). Curr Issues Mol Biol. 2022;44:4658–75. 10.3390/cimb44100318. 10.3390/cimb44100318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal-Alvarado DA, Estrella-Maldonado H, Sáenz-Carbonell L, Ramírez-Prado JH, Zapata-Pérez O, Santamaría JM. Genes coding for transporters showed a rapid and sharp increase in their expression in response to lead, in the aquatic fern (Salvinia minima Baker). Ecotoxicol Environ Saf. 2018;147:1056–64. 10.1016/j.ecoenv.2017.09.046. 10.1016/j.ecoenv.2017.09.046 [DOI] [PubMed] [Google Scholar]
- 5.Bali S, Jamwal VL, Kaur P, Kohli SK, Ohri P, Gandhi SG, et al. Role of P-type ATPase metal transporters and plant immunity induced by jasmonic acid against lead (pb) toxicity in tomato. Ecotoxicol Environ Saf. 2019;174:283–94. 10.1016/j.ecoenv.2019.02.084. 10.1016/j.ecoenv.2019.02.084 [DOI] [PubMed] [Google Scholar]
- 6.Tefera W, Liu T, Lu L, Ge J, Webb SM, Seifu W, et al. Micro-XRF mapping and quantitative assessment of cd in rice (Oryza sativa L.) roots. Ecotoxicol Environ Saf. 2020;193:110245. 10.1016/j.ecoenv.2020.110245. 10.1016/j.ecoenv.2020.110245 [DOI] [PubMed] [Google Scholar]
- 7.Song C, Cao Y, Dai J, Li G, Manzoor MA, Chen C, et al. The multifaceted roles of MYC2 in plants: toward transcriptional reprogramming and stress tolerance by Jasmonate Signaling. Front Plant Sci. 2022;13:1–14. 10.3389/fpls.2022.868874. 10.3389/fpls.2022.868874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song C, Zhang Y, Chen R, Zhu F, Wei P, Pan H, et al. Label-free quantitative proteomics unravel the impacts of salt stress on Dendrobium huoshanense. Front Plant Sci. 2022;13:1–12. 10.3389/fpls.2022.874579. 10.3389/fpls.2022.874579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, Zhang W, Sabir IA, Jiao C, Li G, Wang Y, et al. The spatiotemporal profile of Dendrobium huoshanense and functional identification of bHLH genes under exogenous MeJA using comparative transcriptomics and genomics. Front Plant Sci. 2023;14:1169386. 10.3389/fpls.2023.1169386. 10.3389/fpls.2023.1169386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M, Xu L, Wang Y, Dong J, Zhang X, Wang K, et al. Melatonin-induced DNA demethylation of metal transporters and antioxidant genes alleviates lead stress in radish plants. Hortic Res. 2021;8:124. 10.1038/s41438-021-00561-8. 10.1038/s41438-021-00561-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Methela NJ, Islam MS, Lee D-S, Yun B-W, Mun B-G. S-Nitrosoglutathione (GSNO)-Mediated lead detoxification in soybean through the regulation of ROS and Metal-related transcripts. Int J Mol Sci. 2023;24. 10.3390/ijms24129901. [DOI] [PMC free article] [PubMed]
- 12.Tian J, Wang L, Hui S, Yang D, He Y, Yuan M. Cadmium accumulation regulated by a rice heavy-metal importer is harmful for host plant and leaf bacteria. J Adv Res. 2023;45:43–57. 10.1016/j.jare.2022.05.010. 10.1016/j.jare.2022.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y-Y, Choi H, Segami S, Cho H-T, Martinoia E, Maeshima M, et al. AtHMA1 contributes to the detoxification of excess zn(II) in Arabidopsis. Plant J. 2009;58:737–53. 10.1111/j.1365-313X.2009.03818.x. 10.1111/j.1365-313X.2009.03818.x [DOI] [PubMed] [Google Scholar]
- 14.Han B, Jing Y, Dai J, Zheng T, Gu F, Zhao Q, et al. A Chromosome-Level Genome Assembly of Dendrobium Huoshanense using long reads and Hi-C Data. Genome Biol Evol. 2020;12:2486–90. 10.1093/gbe/evaa215. 10.1093/gbe/evaa215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanter U, Hauser A, Michalke B, Dräxl S, Schäffner AR. Caesium and strontium accumulation in shoots of Arabidopsis thaliana: genetic and physiological aspects. J Exp Bot. 2010;61:3995–4009. 10.1093/jxb/erq213. 10.1093/jxb/erq213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song C, Zhang Y, Zhang W, Manzoor MA, Deng H, Han B. The potential roles of acid invertase family in Dendrobium huoshanense: identification, evolution, and expression analyses under abiotic stress. Int J Biol Macromol. 2023;253:127599. 10.1016/j.ijbiomac.2023.127599. 10.1016/j.ijbiomac.2023.127599 [DOI] [PubMed] [Google Scholar]
- 17.Song C, Jiao C, Jin Q, Chen C, Cai Y, Lin Y. Metabolomics analysis of nitrogen-containing metabolites between two Dendrobium plants. Physiol Mol Biol Plants Int J Funct Plant Biol. 2020;26:1425–35. 10.1007/s12298-020-00822-1. 10.1007/s12298-020-00822-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan T, Yang L, Wu X, Ni J, Jiang H, Zhang Q, et al. The PSE1 gene modulates lead tolerance in Arabidopsis. J Exp Bot. 2016;67:4685–95. 10.1093/jxb/erw251. 10.1093/jxb/erw251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma B, Shukla P. Lead bioaccumulation mediated by Bacillus cereus BPS-9 from an industrial waste contaminated site encoding heavy metal resistant genes and their transporters. J Hazard Mater. 2021;401:123285. 10.1016/j.jhazmat.2020.123285. 10.1016/j.jhazmat.2020.123285 [DOI] [PubMed] [Google Scholar]
- 20.Kim D-Y, Bovet L, Maeshima M, Martinoia E, Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–18. 10.1111/j.1365-313X.2007.03044.x. 10.1111/j.1365-313X.2007.03044.x [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, Ozaki H, Matsui M, Sonoike K. A T-DNA insertion mutant of AtHMA1 gene encoding a Cu transporting ATPase in Arabidopsis thaliana has a defect in the water-water cycle of photosynthesis. J Photochem Photobiol B. 2009;94:205–13. 10.1016/j.jphotobiol.2008.12.002. 10.1016/j.jphotobiol.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Jam M, Alemzadeh A, Tale AM, Esmaeili-Tazangi S. Heavy metal regulation of plasma membrane H+-ATPase gene expression in halophyte Aeluropus littoralis. Mol Biol Res Commun. 2014;3:129–39. [PMC free article] [PubMed] [Google Scholar]
- 23.Niu L, Li H, Song Z, Dong B, Cao H, Liu T, et al. The functional analysis of ABCG transporters in the adaptation of pigeon pea (Cajanus cajan) to abiotic stresses. PeerJ. 2021;9:e10688. 10.7717/peerj.10688. 10.7717/peerj.10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Yang F, Liu J-L, Wu H-T, Yang H, Shi Y, et al. Heavy metal transporters: functional mechanisms, regulation, and application in phytoremediation. Sci Total Environ. 2022;809:151099. 10.1016/j.scitotenv.2021.151099. 10.1016/j.scitotenv.2021.151099 [DOI] [PubMed] [Google Scholar]
- 25.Zhu F-Y, Li L, Lam PY, Chen M-X, Chye M-L, Lo C. Sorghum extracellular leucine-rich repeat protein SbLRR2 mediates lead tolerance in transgenic Arabidopsis. Plant Cell Physiol. 2013;54:1549–59. 10.1093/pcp/pct101. 10.1093/pcp/pct101 [DOI] [PubMed] [Google Scholar]
- 26.Lee M, Lee K, Lee J, Noh EW, Lee Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005;138:827–36. 10.1104/pp.104.058107. 10.1104/pp.104.058107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D-Y, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006;140:922–32. 10.1104/pp.105.074146. 10.1104/pp.105.074146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Do THT, Martinoia E, Lee Y, Hwang J-U. 2021 update on ATP-binding cassette (ABC) transporters: how they meet the needs of plants. Plant Physiol. 2021;187:1876–92. 10.1093/plphys/kiab193. 10.1093/plphys/kiab193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi R, Bashir K, Ishimaru Y, Nishizawa NK, Nakanishi H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal Behav. 2012;7:1605–7. 10.4161/psb.22454. 10.4161/psb.22454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012;35:1948–57. 10.1111/j.1365-3040.2012.02527.x. 10.1111/j.1365-3040.2012.02527.x [DOI] [PubMed] [Google Scholar]
- 31.Migocka M, Papierniak A, Maciaszczyk-Dziubinska E, Posyniak E, Kosieradzka A. Molecular and biochemical properties of two P1B2-ATPases, CsHMA3 and CsHMA4, from cucumber. Plant Cell Environ. 2015;38:1127–41. 10.1111/pce.12447. 10.1111/pce.12447 [DOI] [PubMed] [Google Scholar]
- 32.Das S, Sen M, Saha C, Chakraborty D, Das A, Banerjee M, et al. Isolation and expression analysis of partial sequences of heavy metal transporters from Brassica juncea by coupling high throughput cloning with a molecular fingerprinting technique. Planta. 2011;234:139–56. 10.1007/s00425-011-1376-1. 10.1007/s00425-011-1376-1 [DOI] [PubMed] [Google Scholar]
- 33.Ilyas MZ, Sa KJ, Ali MW, Lee JK. Toxic effects of lead on plants: integrating multi-omics with bioinformatics to develop Pb-tolerant crops. Planta. 2023;259:18. 10.1007/s00425-023-04296-9. 10.1007/s00425-023-04296-9 [DOI] [PubMed] [Google Scholar]
- 34.Xu B, Wang Y, Zhang S, Guo Q, Jin Y, Chen J, et al. Transcriptomic and physiological analyses of Medicago sativa L. roots in response to lead stress. PLoS ONE. 2017;12:e0175307. 10.1371/journal.pone.0175307. 10.1371/journal.pone.0175307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Liu J, Dong D, Jia X, McCouch SR, Kochian LV. (2014). Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. U. S. A 111, 6503–6508. 10.1073/pnas.1318975111 [DOI] [PMC free article] [PubMed]
- 36.Catty P, Boutigny S, Miras R, Joyard J, Rolland N, Seigneurin-Berny D. Biochemical characterization of AtHMA6/PAA1, a chloroplast envelope Cu(I)-ATPase. J Biol Chem. 2011;286:36188–97. 10.1074/jbc.M111.241034. 10.1074/jbc.M111.241034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno I, Norambuena L, Maturana D, Toro M, Vergara C, Orellana A, et al. AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J Biol Chem. 2008;283:9633–41. 10.1074/jbc.M800736200. 10.1074/jbc.M800736200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets are publicly available in ProteomeXchange at https://proteomecentral.proteomexchange.org/cgi/GetDataset? ID=PXD047050 and iProX at https://www.iprox.cn/page/project.html? id=IPX0007578000.