Abstract
The herpesvirus saimiri open reading frame (ORF) 50 produces two transcripts. The first is spliced, contains a single intron, and is detected at early times during the productive cycle, whereas the second is expressed later and is produced from a promoter within the second exon. Analysis of their gene products has shown that they function as sequence specific transactivators. In this report, we demonstrate that the carboxy terminus of ORF 50b contains an activation domain which is essential for transactivation. This domain contains positionally conserved hydrophobic residues found in a number of activation domains, including the herpes simplex virus VP16 and the Epstein-Barr virus R proteins. Mutational analysis of this domain demonstrates that these conserved hydrophobic residues are essential for ORF 50 transactivation capability. Furthermore, this domain is required for the interaction between the ORF 50 proteins and the basal transcription factor TATA-binding protein.
Herpesvirus saimiri (HVS) is a gammaherpesvirus of the Rhadinovirus genus that persistently infects squirrel monkeys (Saimiri sciureus) without causing an overt manifestation of disease. However, infection of a number of New World primate species results in fulminant polyclonal T-cell lymphomas and lymphoproliferative diseases (11). In addition, HVS is capable of transforming simian and human lymphocytes to continuous growth in vitro (4). The genome of HVS (strain A11) consists of a unique internal low-G+C DNA segment (L-DNA) of approximately 110 kbp, flanked by a variable number of 1,444-bp high-G+C tandem repetitions (H-DNA) (2). Sequence analysis indicates it shares significant homology with the herpesviruses Epstein-Barr virus (EBV), bovine herpesvirus 4, Kaposi’s sarcoma-associated herpesvirus (or human herpesvirus 8), and murine gammaherpesvirus 68 (1, 2, 5, 13, 14, 39, 40, 46, 51). The genomes of these viruses are generally colinear, with large blocks of conserved genes interspersed by relative small regions of sequence unique to each virus (2, 5, 39, 46, 51).
Gene expression in HVS is modulated by the two major transcriptional regulating genes encoded by open reading frame (ORF) 50 and ORF 57 (41, 42, 52, 54, 55). The ORF 57 gene product encodes a multifunctional protein capable of both transactivation and repression of viral gene expression. Transactivation of late viral genes occurs at a posttranscriptional level, whereas repression of gene expression appears to correlate with the presence of introns (54, 55). The ORF 50 or R gene produces two transcripts. The first is spliced, contains a single intron, and is detected at early times during the productive cycle, whereas the second is expressed later and is produced from a promoter within the second exon. The spliced transcript is fivefold more potent in activating the delayed-early ORF 6 promoter. However, the function of the nonspliced transcript is unclear (41, 52). Further analysis of the ORF 50 gene products have demonstrated that they activate transcription directly following interactions with promoters containing a specific sequence motif. Deletion and gel retardation analysis have identified the consensus ORF 50 recognition sequence, CCN9GG, required for ORF 50 binding (53). These response elements have significant homology to the EBV R response element consensus sequence, GNCCN9GGNG. It has been shown by guanine methylation studies that the CCN9GG motif is essential for EBV R binding and suggests that R binds to adjacent major grooves of the DNA (16–18).
In addition to the DNA motif, to which sequence specific transactivators bind, at least two functional components are essential. These are inherent to the protein itself and include structural domains which (i) direct the protein to its target, the DNA-binding domain, and (ii) facilitate the initiation of RNA transcription, the activation domain, by recruiting cellular proteins at the promoter (38, 43). The latter is facilitated by the interaction of the viral activation domain(s) with basal transcription factors of the host, which has been demonstrated for several viral transactivators, including adenovirus E1A (26, 36), herpes simplex virus VP16 (23, 48), the EBV Z (28) and R (34) proteins, human T-cell leukemia virus (HTLV-1) Tax1 protein (6), and human immunodeficiency virus type 1 (HIV-1) Tat (25).
Analysis of the ORF 50 homologue, the EBV R protein, demonstrated that the deletion of amino acid sequences near the carboxy terminus ablated its transactivation activity without affecting its ability to bind DNA (21, 33). This finding suggested that the domain(s) of EBV R, which contact cellular transcription factors to facilitate viral transcription, are contained in the carboxy terminus of the protein. To specifically identify the activation domain(s), different segments of the EBV R were linked to the DNA-binding domain of the yeast transactivator GAL4 (20). These GAL4-R fusion proteins were then assayed for the ability to activate the adenovirus E1B promoter with an upstream GAL4 DNA-binding site, linked to the chloramphenicol acetyltransferase (CAT) reporter gene. The carboxy terminus of the EBV R gene was demonstrated to be a potent activator of transcription. Characterization of the activation domain of EBV R revealed three overlapping copies of a motif containing positionally conserved hydrophobic amino acids present in a number of other transactivators, including ORF 50, GAL4 (24), HSV-1 VP16 (10), and adenovirus E1A (26). Furthermore, Hardwicke et al. (20) showed that the carboxy terminus of the HVS ORF 50 protein was capable of activating transcription in the assay described. However, it was not determined whether these sequences were essential to activate transcription. In this report, we demonstrate that the carboxy terminus is essential for ORF 50 transactivation and for the interaction between the ORF 50 proteins and the general cellular transcription factor TATA-binding protein (TBP).
MATERIALS AND METHODS
Plasmid constructs.
The 3′ deletion series which contained the putative ORF 50b transcription start site and 71,188 bp of the published sequence (2) but removed larger portions of the carboxy terminus was constructed by PCR amplification from viral DNA (strain A11) by using the forward primer 5′-CAG AAT TCG ATG CAG CGC CTT GTA TAT ACT and a series of reverse primers consisting of Δ1 (5′-CAG AAT TCC TAG CCA AGG TCT TCA ATA TCT AC), Δ2 (5′-CAG AAT TCC TAT GAA CAT AAA ACT GGA GGT GC), Δ3 (5′-CAG AAT TCC TAG TCA TCT GTT TCT GCT TCG T), Δ4 (5′-CAG AAT TCC TAC ACA GAT GAT GAA GTA CAT GG), and Δ5 (5′-CAG AAT TCC TAT GGT ACT GTA GGT AAC ATT TCA G); these oligonucleotides incorporated EcoRI restriction sites for the convenient cloning of the PCR products. Each fragment was inserted into the eukaryotic expression vector pBKCMV (Stratagene) to derive pBK50Δ1-5.
A range of mutants containing site-directed changes within the hydrophobic residues of the carboxy terminus of ORF 50b were generated by a PCR-based method which incorporated the alteration of one or more of the conserved residues to glycine (underlined) in the 3′ primer: M1 (5′-CGC GAA TTC TTC ATC ATT TAA AAA ATC TTG TAA AGA CAT AGG AAA TGA GCC GCC), M2 (5′-CGC GAA TTC TTC ATC ATT TAA AAA ATC TTG GCC AGA CAT AGG AAA TGA TAA GCC), M3 (5′-CGC GAA TTC TTC ATC ATT TAA GCC ATC TTG TAA AGA CAT AGG AAA TGA TAA GCC), M4 (5′-CGC GAA TTC TTC ATC ATT GCC AAA ATC TTG TAA AGA CAT AGG AAA TGA TAA GCC), M5 (5′-CGC GAA TTC TTC ATC ATT GCC AAA ATC TTG GCC AGA CAT AGG AAA TGA TAA GCC), M6 (5′-CGC GAA TCC TTC ATC ATT GCC GCC ATC TTG TAA AGA CAT AGG AAA TGA TAA GCC), M7 (5′-CGC GAA TTC TTC ATC ATT GCC GCC ATC TTG GCC AGA CAT AGG AAA TGA TAA GCC), and M8 (5′-CGC GAA TTC TTC ATC ATT GCC GCC ATC TTG GCC AGA CAT AGG AAA TGA GCC GCC). In addition, a site-directed mutant containing alterations in residues flanking the conserved hydrophobic residue domain (M9 [5′-CGC GAA TTC TTC ATC ATT TAA AAA ATC TTG TAA AGA CAT AGG AAA TGA TAA GCC AAG GTC TTC AAT TAC TAC GCC GCC]) was used as a control. These oligonucleotides incorporated EcoRI restriction sites for the convenient cloning of the PCR products. Each fragment was inserted into the transfer vector pBKCMV to derive pBK50M1-9.
In producing the construct pGST50, a 332-bp fragment containing bp 71845 to 72177 of the published sequence was generated by PCR using the primers 5′-CCG GAA TCC GCC AGC CCT AGA AAG CTT and 5′-CCG GAA TTC GGT TGA ATG TTC GAT GAG; these oligonucleotides incorporated EcoRI restriction sites to facilitate subcloning into the expression vector pGEX-2T, yielding pGST50. In addition, pGST50C contains the last 50 amino acids of the ORF 50 carboxy terminus fused with glutathione S-transferase (GST). The carboxy-terminal ORF 50 fragment was generated by PCR amplification from viral DNA (strain A11), using the forward primer 5′-CGC GGA TCC ACA GAT GAC AAT ATA TTA GCT and reverse primer 5′-CCG GAA TTC TAG TTA GAC ATT ACA C; these oligonucleotides incorporated BamHI and EcoRI restriction sites, respectively, to facilitate subcloning into the expression vector pGEX-2T to derive pGST50C.
In vitro transcription-translation.
Protein expression from individual clones of the pBK50 deletion series was analyzed by in vitro transcription-translation using the TNT system (Promega) according to the manufacturer’s instructions. Transcription was initiated from the bacteriophage T3 promoter situated upstream of the cloned ORF 50b fragments in pBKCMV. The synthesized products were then separated on a 12% polyacrylamide gel and detected by autoradiography.
Polyclonal antibody generation.
Polyclonal antisera was raised against a portion of recombinant ORF 50 protein. The ORF 50 fragment was expressed as a GST fusion protein in Escherichia coli DH5α and purified from crude lysates by affinity chromatography with glutathione-Sepharose 4B as specified by the manufacturer (Pharmacia Biotech). The purified recombinant protein was used to generate a polyclonal antibody in New Zealand White rabbits by using standard protocols.
Viruses, cell culture, and transfections.
HVS (strain A11) was propagated in owl monkey kidney (OMK) cells which were maintained in Dulbecco modified Eagle medium (Life Technologies) supplemented with 10% fetal calf serum (FCS). Plasmids used in the transfections were prepared by using Qiagen plasmid kits according to the manufacturer’s directions. OMK cells were seeded at 5 × 105 cells per 35-mm-diameter petri dish 24 h prior to transfection. Transfections were performed with DOTAP (Boehringer Mannheim) as described by the manufacturer, using 2 μg of the appropriate DNAs.
Immunofluorescence analysis.
Cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS), washed in PBS, and permeabilized in 0.5% Triton X-100 for 5 min. The cells were rinsed in PBS and blocked by preincubation with 1% (wt/vol) nonfat milk powder for 1 h at 37°C. A 1:20 dilution of anti-ORF 50 antibody was layered over the cells and incubated for 1 h at 37°C. Fluorescence-conjugated anti-rabbit immunoglobulin (1:50 dilution; Dako) was added for 1 h at 37°C. After each incubation step, cells were washed extensively with PBS. The immune fluorescence slides were observed in a Zeiss Axiovert 135TV inverted microscope with a Neofluar 40× oil immersion lens.
CAT assay.
Cell extracts were prepared 48 h after transfection and incubated with [14C]chloramphenicol in the presence of acetyl coenzyme A as described previously (15). The percentage acetylation of chloramphenicol was quantified by scintillation counting (Packard) of appropriate regions of the thin-layer chromatography plate.
Immunoprecipitation analysis.
OMK cells were seeded at 106 cells per 35-mm-diameter petri dish and washed in labelling medium (minimum essential medium minus methionine and cysteine plus 2% FCS). Controls remained untransfected, were transfected with 2 μg of the appropriate DNAs, or were infected with HVS at a multiplicity of infection (MOI) of 1. The cells were incubated with 2 ml of the labelling medium containing 200 μCi of Pro-mix 35S in vitro cell labelling mix plus 10% FCS (Amersham) for 24 h. Cells were harvested and lysed with lysis buffer (0.3 M NaCl, 1% Triton X-100, 50 mM HEPES buffer [pH 8.0]) containing protease inhibitors (leupeptin and phenylmethylsulfonyl fluoride [PMSF]). For each immunoprecipitation, 20 μl of the anti-ORF 50 polyclonal antibody was incubated with protein A-Sepharose beads (Pharmacia Biotech) for 16 h at 4°C. The beads were then pelleted and washed four times in PBS. Each cell lysate was then added to the beads and incubated for 16 h at 4°C. The beads were then pelleted, washed four times in lysis buffer, and resuspended in Laemmli buffer; precipitated polypeptides were resolved on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and analyzed by autoradiography.
Gel retardation assay.
Gel retardation assays were performed as previously described (53). Briefly, two oligonucleotides encoding the ORF 50 response elements, 5′-TTA AAA ATT TCC TGT CAA TGT GGT TTG CTT GG and 5′-CCA AGC AAA CCA CAT TGA CAG GAA ATT TTT AA, were annealed and labelled by using T4 polynucleotide kinase in the presence of [γ-32P]dATP. The radiolabelled oligonucleotides were incubated for 20 min with nuclear extracts of untransfected OMK cells or cells transfected with the appropriate DNAs prepared by the method of Andrews and Faller (3). The binding reactions were performed in 20 μl of binding buffer (100 mM KCl, 20 mM HEPES [pH 7.3], 1% glycerol, 0.2 mM EDTA, 5 mM MgCl2, 4 mM dithiothreitol, 0.5 mM PMSF) with 1 μg of poly(dI-dC) as an unspecific competitor. The protein-nucleic acid complexes were separated on a 5% polyacrylamide gel, run in 1% Tris-borate-EDTA (TBE) buffer, and detected by autoradiography.
Immunoblot analysis.
Immunoblot analysis was performed with the immunoprecipitation samples described above. Precipitated polypeptides were resolved on an SDS–12% polyacrylamide gel and then soaked for 10 min in transfer buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol, 0.1% SDS). The proteins were transferred to nitrocellulose membranes by electroblotting for 3 h at 250 mA. After transfer, the membranes were soaked in PBS and blocked by preincubation with 2% (wt/vol) nonfat milk powder for 2 h at 37°C. Membranes were incubated with a 1/1,000 dilution of the anti-TFIID monoclonal antibody (Promega), washed with PBS, and incubated for 1 h at 37°C with a 1/1,000 dilution of anti-mouse immunoglobulin conjugated with horseradish peroxidase (Dako) in blocking buffer. After five washes with PBS, the nitrocellulose membranes were developed by using enhanced chemiluminescence (Pierce) according to the manufacturer’s directions.
GST pulldown assay.
The ORF 50 carboxy terminus was expressed as a GST fusion protein in E. coli DH5α. A fresh overnight culture of transformed E. coli was diluted 1 in 20 with Luria-Bertani medium containing ampicillin (100 μg/ml). After growth at 37°C for 2 h, the culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside and grown at 37°C for a further 4 h. The cells were harvested by centrifugation and resuspended in 0.1× lysis buffer (100 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.5 mM PMSF). Cells were sonicated and stored on ice for 30 min, and cellular debris was pelleted. The recombinant protein was purified from crude lysates by incubation with glutathione-Sepharose 4B affinity beads as specified by the manufacturer (Pharmacia Biotech). The beads containing the GST-ORF 50 carboxy-terminus fusion were then incubated with OMK cell lysates previously lysed with lysis buffer (0.3 M NaCl, 1% Triton X-100, 50 mM HEPES buffer [pH 8.0]) containing protease inhibitors (leupeptin and PMSF) for 16 h at 4°C. The beads were then pelleted, washed four times in lysis buffer, and resuspended in Laemmli buffer; precipitated polypeptides were resolved on an SDS–12% polyacrylamide gel. The proteins were then transferred to nitrocellulose membranes by electroblotting and probed for TFIID as described above.
RESULTS
Deletion analysis of the carboxy terminus of ORF 50.
To analyze the sequences which are essential for transactivation by the ORF 50 protein, a 3′ deletion series of the ORF 50b coding region was produced. The carboxy-terminal deletion series contained the putative ORF 50b transcription start site at 71,188 bp of the published sequence (2) but removed larger portions of the carboxy terminus. Each fragment was inserted into the transfer vector pBKCMV (Stratagene) to derive pBK50Δ1-5 (Fig. 1). All constructs were confirmed by DNA sequencing (data not shown). In addition, the complete coding region of ORF 50b contained as a HincII cassette was excised from pAWHincII (9) and inserted into pBKCMV to derive the control plasmid pBK50b.
FIG. 1.
Schematic representation of the carboxy-terminus deletion series of the ORF 50b protein. A series of 3′ mutants were constructed by PCR amplification and ligated into the eukaryotic expression vector pBKCMV to derive pBK50Δ1-5.
To investigate protein expression from each clone of the pBK50 deletion series, in vitro transcription-translation was performed with the TNT system (Promega). Transcription was initiated from the bacteriophage T3 promoter situated upstream of the cloned ORF 50b fragments in pBKCMV. Results showed that pBK50b generated a product of approximately 41 kDa and that each ORF 50b deletion construct supported the production of a major protein species. Moreover, the apparent molecular weights of these proteins were consistent with consecutive deletions of the ORF 50b gene product, ranging from approximately 38.5 kDa following expression from pBK50Δ1 to 29 kDa for pBK50Δ5 (Fig. 2), although the proteins from pBK50b and pBK50Δ1 were produced at low levels.
FIG. 2.
In vitro transcription-translation analysis of ORF 50 deletion series. Protein expression from pBK50b (lane 1), pBK50Δ1 (lane 2), pBK50Δ2 (lane 3), pBK50Δ3 (lane 4), pBK50Δ4 (lane 5), and pBK50Δ5 (lane 6) was analyzed by in vitro transcription-translation. Transcription was initiated from the bacteriophage T3 promoter situated upstream of the cloned ORF50b fragments in pBKCMV. The synthesized products (indicated by arrows) were then separated on a 12% polyacrylamide gel and detected by autoradiography. Sizes are indicated in kilodaltons.
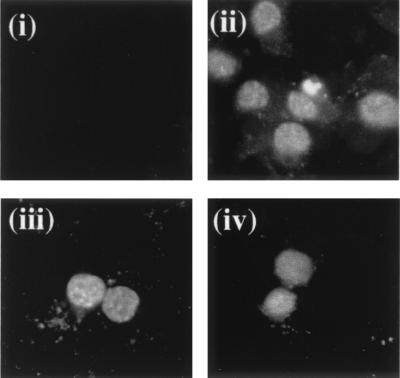
Furthermore, to determine the expression levels and subcellular localization of ORF 50b and the deletion series, a polyclonal antiserum was raised against a portion of recombinant ORF 50 protein. Unfortunately, the ORF 50 polyclonal antiserum did not react on a Western blot of HVS-infected or ORF 50-transfected cells (data not shown). Therefore, to determine if each deletion produced a protein product, immunofluorescence analysis of HVS-infected and transient transfected cells was performed. Immunofluorescence analysis of HVS-infected cells resulted in a strong fluorescence of the nuclei of infected cells. Similar results were observed with pBK50b- and pBK50Δ1-transfected cells; no reaction was observed with untransfected cells (Fig. 3). Similar nuclear immunofluorescence staining was observed with pBK50Δ2-5-transfected cells (data not shown). This finding suggested that each deletion construct produced a protein which localized to the cell nucleus.
FIG. 3.
Expression levels and subcellular localization of ORF 50b and 50Δ1 proteins. A polyclonal antiserum was raised against a portion of recombinant ORF 50 protein and used in immunofluorescence analysis of cells mock transfected (i), HVS infected (MOI of 1) (ii), and transiently transfected with pBK50b (iii) and pBK50Δ1 (iv).
The carboxy terminus is essential for ORF 50 transactivation of the ORF 6 promoter.
To identify the sequences essential for ORF 50 transactivation activity, 1 μg of each deletion plasmid was cotransfected with 1 μg of pAWCAT2. This plasmid contains the CAT coding region under the control of the ORF 50-responsive ORF 6 promoter (52). Plasmid pBK50b was also used in the assay as a positive control. Cells were harvested after 48 h and assayed for CAT activity by standard methods (15) (Fig. 4). Reduced CAT activity was observed when the deletion constructs were used to transactivate pAWCAT2. However, pBK50b was shown to transactivate the ORF 6 promoter to levels similar to those found previously (52). This suggested that the sequences contained within bp 72350 to 72402 of the published sequence, which were deleted in pBK50Δ1, are essential for transactivation by the ORF 50b gene product.
FIG. 4.
Analysis of the ORF 50b carboxy-terminus deletion series. OMK cell monolayers were transfected with 1 μg of pAWCAT2 or cotransfected 1 μg of each deletion plasmid, using DOTAP transfection reagent (Boehringer Mannheim) according to the manufacturer’s instructions. Cells were harvested at 48 h posttransfection, and cell extracts were assayed for CAT activity. Percentages of acetylation were calculated by the scintillation counting of the appropriate regions of the chromatography plate and are shown in graphical format; the variations between three replicated assays are indicated.
The carboxy-terminal 50Δ1 mutant produces a stable protein which binds to the ORF 50 response elements.
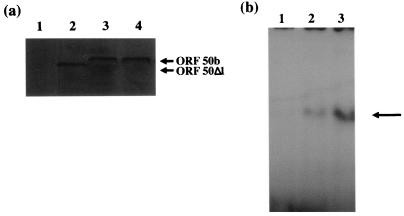
To determine whether the lack of transactivation of the ORF 6 promoter was due to the removal of the ORF 50b carboxy terminus, or whether the removal of these sequences affected the protein stability or DNA-binding ability of the ORF 50b protein, immunoprecipitation and gel retardation assays were performed with pBK50Δ1. First, to determine whether the expression vector pBK50Δ1 produced a stable protein compared to the wild-type 50b protein in transfected and infected cells, immunoprecipitation analysis was performed. OMK cells remained untransfected, were transfected with 2 μg of pBK50b or pBK50Δ1, or were infected with HVS at an MOI of 1. The cells were then incubated in the presence of 35S in vitro cell labelling mix for 24 h and harvested, and cell lysates were utilized in immunoprecipitation analysis using the anti-ORF 50 polyclonal antibody. The results demonstrate that a protein of the correct size is produced and precipitated with the anti-ORF 50 antibody from cell lysates transfected with pBK50Δ1 (Fig. 5a). In addition, the precipitated protein was produced in quantities similar to those for the wild-type protein, suggesting that deletion of the last 14 amino acids does not affect ORF 50 protein stability.
FIG. 5.
The carboxy-terminal ORF 50Δ1 construct produces a stable protein which binds to the ORF 50 response elements. (a) OMK cells were seeded at 106 cells per 35-mm-diameter petri dish and washed in labelling medium. Controls remained untransfected (lane 1) or were transfected with 2 μg of pBK50Δ1 (lane 2), pBK50 (lane 3) or infected with HVS (lane 4). The cells were incubated in labelling medium, harvested, and then lysed after 24 h. For each immunoprecipitation, 20 μl of the anti-ORF 50 polyclonal antibody was incubated with protein A-Sepharose beads for 16 h at 4°C. Immunoprecipitations were then performed with each cell lysate, using the anti-ORF 50 antibody. Beads were then pelleted, washed, and resuspended in Laemmli buffer; precipitated polypeptides were resolved on an SDS–12% polyacrylamide gel and analyzed by autoradiography. (b) Gel retardation assays were performed as previously described (53). Briefly, the ORF 50 response elements contained in a set of oligonucleotides were annealed and radiolabelled. These were incubated with nuclear extracts of untransfected OMK cells (lane 1) or cells transfected pBK50b (lane 2) and pBK50Δ1 (lane 3). The protein-nucleic acid complexes (indicated by arrow) were separated on a 5% polyacrylamide gel, run in 1% TBE buffer, and detected by autoradiography.
We have previously determined, using deletion analysis and gel retardation assays, the ORF 50 response elements contained within the ORF 6 promoter to which ORF 50b binds (54). To determine if the 50Δ1 protein could bind to these sequences, gel retardation assays were performed. Radiolabelled probes encoding the ORF 50 response elements were incubated with DNA-binding protein extracts from untransfected cells and cells transfected with pBK50b or pBK50Δ1. The protein-nucleic acid complexes were then separated on a polyacrylamide gel (Fig. 5b). Results show the formation of a retarded complex using the extracts of cells transfected with pBK50b and pBK50Δ1. Unlabelled oligonucleotide was shown to compete with this reaction (data not shown), further indicating that the protein produced from pBK50Δ1 has the ability to specifically bind to the ORF 50 response elements. It must be noted that the 50Δ1 protein seems to bind the oligonucleotides at a greater affinity; this observation is currently under investigation. However, these two experiments demonstrate that pBK50Δ1 produced a stable protein which could bind to the ORF 50 response elements. This result further suggests that the sequences contained within the carboxy-terminal 14 amino acids are essential for transactivation by the ORF 50b gene product.
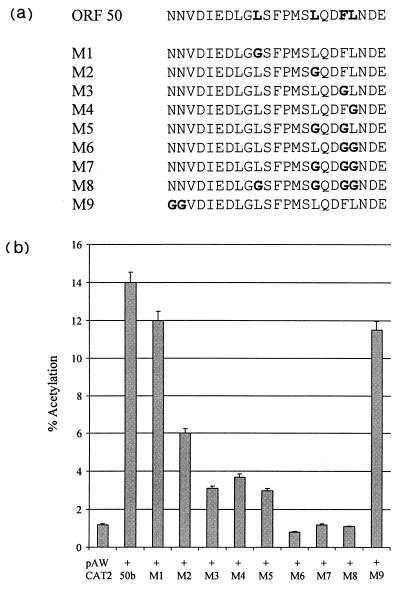
Mutational analysis of the ORF 50b transactivation domain.
The deletion of the carboxy-terminal 14 amino acids abrogates the transactivating capability of ORF 50b. As previously described, the carboxy terminus of ORF 50 contains a motif of positionally conserved hydrophobic amino acids homologous with the EBV R protein. To determine the importance of the conserved hydrophobic amino acids for ORF 50b transactivation activity, we constructed a range of site-directed mutations (Fig. 6a) by a PCR-based method which incorporated the alteration of one or more of the conserved residues to glycine. In addition, residues flanking the hydrophobic residue domain were altered to glycine to serve as an appropriate control. Each fragment was inserted into the transfer vector pBKCMV to derive pBK50M1 to pBK50M9. All constructs were confirmed by DNA sequencing (data not shown). To identify the residues within the ORF 50b transactivation domain which are essential for ORF 50 transactivation, 1 μg of each construct was cotransfected with 1 μg of pAWCAT2. Cells were harvested after 48 h and assayed for CAT activity (Fig. 6b). Results show that the mutations of asparagine residues at bp 72334 to 72337 of the published sequence, which flank the conserved hydrophobic residues, had little, if any, effect on ORF 50 transactivation capability. However, mutation of the conserved leucine residue at bp 72361 of the published sequence reduced CAT activity by approximately 10%. Moreover, single mutations at the conserved leucines at bp 72379 and 72391 and phenylalanine at bp 72388 of the published sequence proved highly detrimental to ORF 50b transactivation, reducing CAT activity by 57, 75, and 72%, respectively. Furthermore, multiple substitutions of these residues completely abrogated biological activity, showing that these conserved hydrophobic residues are essential for ORF 50 transactivation activity.
FIG. 6.
Mutational analysis of the ORF 50b transactivation domain. (a) The carboxy-terminal 14 amino acids contain a motif of positionally conserved hydrophobic amino acids homologous with the EBV R protein (boldface). A range of site directed mutations were constructed such that one or multiple conserved hydrophobic residues were replaced with a glycine residue (boldface). (b) OMK cell monolayers were transfected with 1 μg of pAWCAT2 or cotransfected with 1 μg of each 50b mutation plasmid, using DOTAP transfection reagent. Cells were harvested at 48 h posttransfection, and cell extracts were assayed for CAT activity. Percentages of acetylation were calculated by scintillation counting of the appropriate regions of the chromatography plate and are shown in graphical format; the variations between three replicated assays are indicated.
The carboxy-terminal mutation 50M7 produces a stable protein which binds to the ORF 50 response elements.
To determine whether these mutations within the hydrophobic residues, specifically M7, affect protein stability or the ability of ORF 50 to bind to the response elements’ immunofluorescence, immunoprecipitations and gel retardation assays were performed as described above. To determine if pBK50M7 produced a protein product which localized to the nucleus, immunofluorescence analysis of pBK50M7-transfected cells was performed. A strong nuclear fluorescence was observed in cells transfected with pBK50M7, as previously described (Fig. 7a). Similar results were observed with the remaining mutant constructs in transfected cells (data not shown). To determine whether the expression vector pBK50M7, which abrogated transactivation capability of the ORF 50b protein, produced a stable protein compared to wild-type 50b, immunoprecipitations were performed. Results demonstrate that a stable protein product is produced, at levels similar to those for the wild-type protein, from pBK50M7 (Fig. 7b). Furthermore, to determine if the 50M7 protein could bind to these sequences, gel retardation assays were performed. Results show the formation of a retarded complex, using extracts of cells transfected with pBK50b and pBK50M7 (Fig. 7c). Unlabeled oligonucleotide was shown to compete with this reaction (data not shown), further indicating that the protein produced from pBK50M7 has the ability to specifically bind to the ORF 50 response elements. These three experiments demonstrate that the mutations within the hydrophobic residues, specifically the mutations contained within 50M7, of the ORF 50 transactivation domain are responsible for the lack of transactivation by the ORF 50b gene product.
FIG. 7.
The ORF 50 carboxy-terminal mutation, 50M7, produces a stable protein which binds to the ORF 50 response elements. (a) Subcellular localization of ORF 50M7 protein, determined by immunofluorescence analysis of cells mock transfected (i) or transfected with pBK50M7 (ii). (b) OMK cells were seeded at 106 cells per 35-mm-diameter petri dish and washed in labelling medium. Controls remained untransfected (lane 1) or transfected with 2 μg of pBK50b (lane 2) or pBK50M7 (lane 3). The cells were incubated in labelling medium, harvested, and then lysed after 24 h. For each immunoprecipitation, 20 μl of the anti-ORF 50 polyclonal antibody was incubated with protein A-Sepharose beads for 16 h at 4°C. Immunoprecipitations were then performed with each cell lysate, using the anti-ORF 50 antibody. These samples were then pelleted, resuspended in Laemmli buffer, resolved on an SDS–12% polyacrylamide gel, and analyzed by autoradiography. (c) Gel retardation assays were performed as previously described (53). Briefly, the ORF 50 response elements contained in a set of oligonucleotides were annealed, radiolabelled and then incubated with nuclear extracts of untransfected OMK cells (lane 1) or cells transfected pBK50b (lane 2) and pBK50M7 (lane 3). The protein-nucleic acid complexes (indicated by the arrow) were separated on a 5% polyacrylamide gel, run in 1% TBE buffer, and detected by autoradiography.
The hydrophobic residues contained within the ORF 50 transactivation domain, specifically 50M7, are essential for the interaction with TBP.
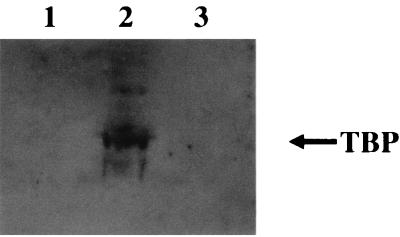
Hydrophobic residues have been proposed to be important in protein-protein interactions between activating proteins and the cellular transcription machinery. To determine if the conserved hydrophobic residues within the carboxy-terminal activation domain were essential for interactions with cellular transcription factors, immunoblot analysis was performed with pBK50M7-transfected cells. Radiolabelled cell lysates from either untransfected cells or cells transfected with pBK50b or pBK50M7 were incubated with protein A-Sepharose beads, previously bound to the anti-ORF 50 polyclonal antibody. Immunofluorescence on duplicate samples was performed to ensure transfection efficiency (data not shown). Polypeptides precipitated from transfected cellular extracts by the anti-ORF 50 antibody were then resolved by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane, and immunoblot detection was performed with an anti-TFIID monoclonal antibody which specifically recognizes TBP. Immunoprecipitations using the pBK50b-transfected cell lysate specifically recognized the TBP. However, the pBK50M7 (containing mutations within the acidic transactivation domain) cell lysate did not react with the antibody (Fig. 8). This finding suggests that the hydrophobic residues contained within the carboxy-terminal acidic transactivation domain, specifically the mutations containing within M7, are essential for the interaction of ORF 50b with the cellular transcription factor TBP.
FIG. 8.
The hydrophobic residues contained within the transactivation domain are required for the interaction with TBP. Immunoblot analysis was performed with the immunoprecipitation samples, untransfected control cells (lane 1) and cells transfected with 2 μg of pBK50b (lane 2) or pBK50M7 (lane 3). Polypeptides were resolved on an SDS–12% polyacrylamide gel and then transferred to nitrocellulose membranes. After transfer, the membranes were blocked, incubated with a 1/1,000 dilution of the anti-TFIID monoclonal antibody, washed, and incubated for 1 h at 37°C with a 1/1,000 dilution of anti-mouse immunoglobulin conjugated with horseradish peroxidase in blocking buffer. After five washes with PBS, the nitrocellulose membranes were developed, using enhanced chemiluminescence.
The carboxy-terminal domain of ORF 50 is sufficient for the interaction with TBP.
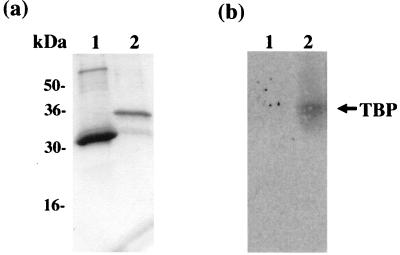
To determine whether the carboxy-terminal domain is sufficient for the interaction between the ORF 50b protein and TBP, GST pulldown analysis was performed. The ORF 50 carboxy terminus was expressed as a GST fusion protein, or the control GST alone, in E. coli and purified from crude lysates by incubation with glutathione-Sepharose 4B affinity beads (Fig. 9a). These protein bound beads were then incubated with an OMK cell lysate. The beads were then pelleted and washed, and the cellular proteins precipitated by GST or the GST-ORF 50 fusion protein were resolved on an SDS–12% polyacrylamide gel. The proteins were then transferred to nitrocellulose membranes by electroblotting and probed for TBP as previously described. Results demonstrate that GST alone did not precipitate TBP but TBP did interact with the GST-ORF 50 carboxy terminus fusion protein (Fig. 9b). This finding suggests that the ORF 50 carboxy terminus is responsible for the interaction with the cellular transcription factor TBP.
FIG. 9.
The ORF 50 transactivation domain is sufficient for the interaction with TBP. (a) The control GST alone (lane 1) and the GST-ORF 50 carboxy terminus fusion protein (lane 2) were expressed in E. coli DH5α and purified from crude lysates by incubation with glutathione-Sepharose 4B affinity beads. (b) The beads containing GST alone (lane 1) or the GST-ORF 50 carboxy terminus fusion protein (lane 2) were then incubated with OMK cell lysates. The beads were then pelleted, washed, resuspended in Laemmli buffer, and resolved on an SDS–12% polyacrylamide gel. The proteins were then transferred to nitrocellulose membranes by electroblotting and probed for TFIID.
DISCUSSION
Transcriptional activators have the ability to stimulate in vitro the assembly of transcription preinitiation complexes (9, 31) as well as transcriptional elongation by RNA polymerase II (57). This activation directly or indirectly is dependent on the interaction between the general transcriptional machinery and the transcriptional activators. Transcriptional activators have at least two distinct domains, the DNA-binding domain and the activation domain (43). The activation domain enhances the transcription of target genes. Three major classes of activation domains have been identified according to their amino acid composition: acidic negatively charged, proline rich, and glutamine rich (38).
In this report, we demonstrate that the HVS ORF 50 protein contains an acidic transactivation domain within the carboxy terminus, which is essential for the transactivating capability of the protein. Analysis of the HVS ORF 50 activation domain indicates that it contains positionally conserved hydrophobic residues with activation domains found in a variety of viral, yeast, and mammalian transcriptional activators, including HSV-1 VP16, EBV R, GAL4, GCN4, Sp1, and CTF (10, 20). Site-directed mutagenesis indicates that these conserved hydrophobic residues are required for full transactivation activity of the ORF 50b protein, further suggesting that these hydrophobic residues are essential components of the ORF 50 activation domain. Moreover, extensive mutational analysis of the activation domains present in a number of other proteins, such as VP16 (10), p53 (30), Sp1 (12), c-Fos (37), E1a (29), and GAL4 (27), have shown that these conserved bulky hydrophobic residues are also essential for transcriptional activation. Hydrophobic residues may be involved in the direct protein-protein interactions between the activation domain and the cellular transcription machinery and/or in maintaining the structure of the activation domain. Circular dichroism spectroscopy has demonstrated, however, that the spectra of the wild-type p53 activation domain and a mutated activation domain had no significant difference in structure (7). This finding suggests that the hydrophobic residues play a role in the interaction of the activation domain and cellular transcription factors.
It is interesting that the activation domain of the ORF 50 homologue, EBV R protein, is comprised of two domains, a potent acidic activation domain and a proline-rich domain. It has been shown that the acidic region, which contains three overlapping copies of a motif containing the conserved hydrophobic residues, plays a central role in transcriptional activation (20). However, although the proline-rich domain has alone no activity, it increases the R protein-activating potential in a cell-specific manner (34). This finding suggests that this proline-rich domain may be required for stabilizing the interaction of the EBV R transactivation domain and target molecules. However, although analysis of the HVS ORF 50 carboxy terminus shows that a number of proline residues are present, we believe that this region does not constitute a proline-rich domain. Mutational analysis of these proline residues will help in addressing their role, if any, in ORF 50’s transactivation capability.
In addition, we have preliminary data to suggest that the ORF 50 transactivation domain is required for the interaction of ORF 50 with TBP. A key role in transcription initiation by RNA polymerase II is the binding of a multisubunit complex, TFIID, to the TATA element close to the transcription start site. The major component of the TFIID complex is TBP, which is also required for transcription of RNA polymerase I and III promoters. In addition, a number of TBP-associated factors are assembled into the TFIID (reviewed in references 45 and 50) and interact with more distant transcription factors, binding to enhancer elements and to RNA polymerase II and its accessory proteins, allowing transcription initiation. Many activation domains have been shown to interact with TBP. These include the acidic activation domains of VP16 (23, 48), p53 (8, 32, 35, 47, 49), c-Fos and c-Jun (44), c-Myc (22), v-Rel and c-Rel (56), E2F-1 (19), the EBV Z (28) and R (34) proteins, HTLV-1 Tax1 protein (6), and HIV-1 Tat (25). The direct interaction between the ORF 50 transactivation domain and a component of the cellular transcription machinery may have several functions. It may be involved in the stabilization of the interaction of TFIID with promoter DNA, as shown for EBV Zta (28). Second, recruitment of TFIIB into the initiation complex may be enhanced in the presence of ORF 50, as demonstrated in assays using the VP16 transactivation domain, or in the recruitment of other general transcription factors into the initiation complex. Alternatively, it may play a role in enhancing the rate of transcription elongation. The data reported herein suggests a preliminary finding of the interaction between ORF 50 and TBP. The specific role of the ORF 50-TBP interaction requires further investigation.
In summary, we have demonstrated that the carboxy terminus of ORF 50 is essential for transactivation. It contains a motif of positionally conserved hydrophobic amino acids found in a number of activation domains. Mutational analysis has shown that these conserved hydrophobic domains are essential for transcriptional activation and for the interaction between the ORF 50 proteins and the cellular transcription factor TBP.
ACKNOWLEDGMENTS
This work was supported in part from grants from the Medical Research Council (MRC), Yorkshire Cancer Research, and the West Riding Medical Research Trust. A.W. is a recipient of an MRC fellowship.
REFERENCES
- 1.Albrecht J C, Fleckenstein B. Structural organization of the conserved gene block of herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology. 1990;174:533–542. doi: 10.1016/0042-6822(90)90107-3. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beisinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bublot M, Manet E, Lequarre A S, Albrecht J C, Nicholas J, Fleckenstein B, Pastoret P P, Thiry E. Genetic relationships between bovine herpesvirus 4 and the gamma-herpesviruses Epstein-Barr and herpesvirus saimiri. Virology. 1992;190:654–665. doi: 10.1016/0042-6822(92)90903-3. [DOI] [PubMed] [Google Scholar]
- 6.Caron C, Rousset R, Beraud C, Moncollin V, Egly J-M, Jalinot P. Functional and biochemical interaction of the HTLV-1 tax1 transactivator with TBP. EMBO J. 1993;12:4269–4278. doi: 10.1002/j.1460-2075.1993.tb06111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J, Kim D-H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;42:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Farmer G, Zhu H, Prywes R, Prives C. Cooperative binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- 9.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature (London) 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 10.Cress W D, Triezenberg S J. Critical structure elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 11.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 253–332. [Google Scholar]
- 12.Gill G E, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gompels U A, Craxton M A, Honess R W. Conservation of gene organization in the lymphotrophic herpesvirus saimiri. J Virol. 1988;62:757–767. doi: 10.1128/jvi.62.3.757-767.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gompels U A, Craxton M A, Honess R W. Conservation of glycoprotein H (gH) in herpesvirus: nucleotide sequence of the gH gene from herpesvirus saimiri. J Gen Virol. 1988;69:2819–2829. doi: 10.1099/0022-1317-69-11-2819. [DOI] [PubMed] [Google Scholar]
- 15.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruffat H, Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994;22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence specific DNA binding protein. Nucleic Acids Res. 1990;18:6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardwicke J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activator domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardwicke J M, Liberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hateboer G, Timmers H T M, Rustgi A K, Billaud M, van’t Veer L J, Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci USA. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature (London) 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 24.Johnston S A, Zavortink M J, Debouck C, Hopper J E. Functional domains of the yeast regulatory protein GAL4. Proc Natl Acad Sci USA. 1986;83:6553–6557. doi: 10.1073/pnas.83.17.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C-M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature (London) 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus EIA transactivation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 27.Leuther K K, Salmeron J M, Johnston S A. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a β sheet. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman P M, Berk A J. The Zta transactivator protein stabilises TFIID association with promoter DNA by direct protein-protein interactions. Genes Dev. 1991;5:2441–2554. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 29.Lillie J W, Green M R. Transcription activation by the adenovirus E1A protein. Nature (London) 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 30.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Miller C W, Koeffler P H, Berk A J. The p53 activation domain binds the TATA-box binding polypeptide in holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manet E, Rigolet A, Gruffat H, Giot J F, Sergeant A. Domains of the Epstein-Barr virus transcription factor R required for dimerisation, DNA binding and activation. Nucleic Acids Res. 1991;19:2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manet E, Allera C, Gruffat H, Mikaelian I, Rigolet A, Sergeant A. The acidic activation domain of the EBV transcription factor interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 1993;3:49–59. [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D W, Munoz R M, Subler M A, Deb S. p53 binds to the TATA-binding protein-TATA complexes. J Biol Chem. 1993;268:13062–13067. [PubMed] [Google Scholar]
- 36.Mazzarelli J M, Mengus G, Davidson I, Ricciardi R P. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAFII135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz R, Bannister A J, Sutherland J A, Hagemeier C, O’Rourke E C, Cook A, Bravo R, Kouzarides T. c-Fos-induced activation of a TATA-box-containing promoter involves direct contact with TATA-binding protein. Mol Cell Biol. 1994;14:6021–6029. doi: 10.1128/mcb.14.9.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 39.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in Kaposi’s sarcoma associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas J, Cameron K R, Coleman H, Newman C, Honess R W. Analysis of nucleotide sequence of the rightmost 43 kbp of herpesvirus saimiri (HVS) L-DNA: general conservation of genetic organization between HVS and Epstein-Barr virus. Virology. 1992;188:296–310. doi: 10.1016/0042-6822(92)90759-i. [DOI] [PubMed] [Google Scholar]
- 41.Nicholas J, Coles L S, Newman C, Honess R W. Regulation of the herpesvirus saimiri (HVS) delayed-early 110-kilodalton promoter by HVS immediate-early gene products and a homolog of the Epstein-Barr virus R trans activator. J Virol. 1991;65:2457–2466. doi: 10.1128/jvi.65.5.2457-2466.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholas J, Gompels U A, Craxton M A, Honess R W. Conservation of sequence and function between the product of the 52-kilodalton immediate-early gene of the herpesvirus saimiri and the BMLF1-encoded transcriptional effector (EB2) of the Epstein-Barr virus. J Virol. 1988;62:3250–3257. doi: 10.1128/jvi.62.9.3250-3257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ptashne M. How eukaryotic transcriptional activators work. Nature (London) 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 44.Ransone L J, Kerr L D, Schmitt M J, Wamsley P, Verma I M. The bZIP domains of Fos and Jun mediate a physical association with the TATA box-binding protein. Gene Expr. 1993;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- 45.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 46.Russo J J, Bohenzhy R A, Chein M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seto E, Usheva A, Zambetti P, Momand J, Horikoshi N, Weinmann R, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stringer K F, Ingles C J, Greenbalt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 49.Truant R, Xiao H, Ingles C J, Greenblat J. Direct interaction between the transcriptional activation domain of human p53 and the TATA-box binding protein. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 50.Verrijer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 51.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehouse A, Carr I M, Griffiths J C, Meredith D M. The herpesvirus saimiri ORF 50 gene, encoding a major transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J Virol. 1997;71:2550–2554. doi: 10.1128/jvi.71.3.2550-2554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitehouse A, Stevenson A J, Cooper M, Meredith D M. Identification of a cis-acting element within the herpesvirus saimiri ORF6 promoter that is responsive to the HVS.R transactivator. J Gen Virol. 1997;78:1411–1415. doi: 10.1099/0022-1317-78-6-1411. [DOI] [PubMed] [Google Scholar]
- 54.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitehouse A, Cooper M, Hall K T, Meredith D M. The open reading frame (ORF) 50a gene product regulates ORF 57 gene expression in herpesvirus saimiri. J Virol. 1998;72:1967–1973. doi: 10.1128/jvi.72.3.1967-1973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gelinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733–6741. doi: 10.1128/mcb.13.11.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]