Abstract
In the NL4-3 strain of human immunodeficiency virus type 1 (HIV-1), regulatory elements responsible for the relative efficiencies of alternative splicing at the tat, rev, and the env/nef 3′ splice sites (A3 through A5) are contained within the region of tat exon 2 and its flanking sequences. Two elements affecting splicing of tat, rev, and env/nef mRNAs have been localized to this region. First, an exon splicing silencer (ESS2) in NL4-3, located approximately 70 nucleotides downstream from the 3′ splice site used to generate tat mRNA, acts specifically to inhibit splicing at this splice site. Second, the A4b 3′ splice site, which is the most downstream of the three rev 3′ splice sites, also serves as an element inhibiting splicing at the env/nef 3′ splice site A5. These elements are conserved in some but not all HIV-1 strains, and the effects of these sequence changes on splicing have been investigated in cell transfection and in vitro splicing assays. SF2, another clade B virus and member of the major (group M) viruses, has several sequence changes within ESS2 and uses a different rev 3′ splice site. However, splicing is inhibited by the two elements similarly to NL4-3. As with the NL4-3 strain, the SF2 A4b AG dinucleotide overlaps an A5 branchpoint, and thus the inhibitory effect may result from competition of the same site for two different splicing factors. The sequence changes in ANT70C, a member of the highly divergent outlier (group O) viruses, are more extensive, and ESS2 activity in tat exon 2 is not present. Group O viruses also lack the rev 3′ splice site A4b, which is conserved in all group M viruses. Mutagenesis of the most downstream rev 3′ splice site of ANT70C does not increase splicing at A5, and all of the branchpoints are upstream of the two rev 3′ splice sites. Thus, splicing regulatory elements in tat exon 2 which are characteristic of most group M HIV-1 strains are not present in group O HIV-1 strains.
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus that transcribes its RNA from an integrated proviral genome and uses the host splicing machinery to produce its mRNAs. To generate the more than 30 different singly and multiply spliced mRNAs, the HIV-1 9.2-kb primary transcript undergoes splicing by a complex pathway. The regulatory proteins Tat, Rev, and Nef are encoded by multiply spliced ∼2-kb mRNAs, whereas the Env, Vif, Vpr, and Vpu proteins are encoded by singly spliced mRNAs. In addition, approximately half of the HIV-1 RNA remains unspliced and is used as message for gag and pol gene products. The unspliced RNA is also packaged into progeny virions. The HIV-1 Rev protein, which binds to the Rev-responsive element located in the env gene, facilitates the stabilization and nuclear transport of unspliced and partially spliced mRNAs. However, prior to the action of Rev, the efficiency of viral RNA splicing at the different splice sites is regulated by a number of cis elements within the viral genome.
Several splicing elements have been localized within the first tat coding exon of the NL4-3 HIV-1 strain (tat exon 2). One of these elements, has been termed exon splicing silencer 2 (ESS2), is located 60 to 70 nucleotides (nt) downstream from the 3′ splice site (3′ splice site A3) used to generate Tat mRNA (Fig. 1A) (3, 4). It maps to the 10-nt core sequence CUAGACUAGA and acts specifically to inhibit splicing at this 3′ splice site (18). Mutations within ESS2 result in a selective increase in tat splicing when tested in an in vitro splicing system or in cell culture after transfection of infectious proviral DNA or virus infection (3, 7, 18). Inspection of HIV-1 sequences in the database (9a) indicates that the region of ESS2 in many strains within the M (major) group of HIV-1 strains contain only one copy of the CUAGA sequence, which is repeated in the NL4-3 ESS2. The corresponding sequences of the O (outlier) group of HIV-1 strains are more divergent and do not contain even a single copy of the CUAGA sequence, which suggests that there may be significant differences in the regulation of tat splicing in different HIV strains.
FIG. 1.
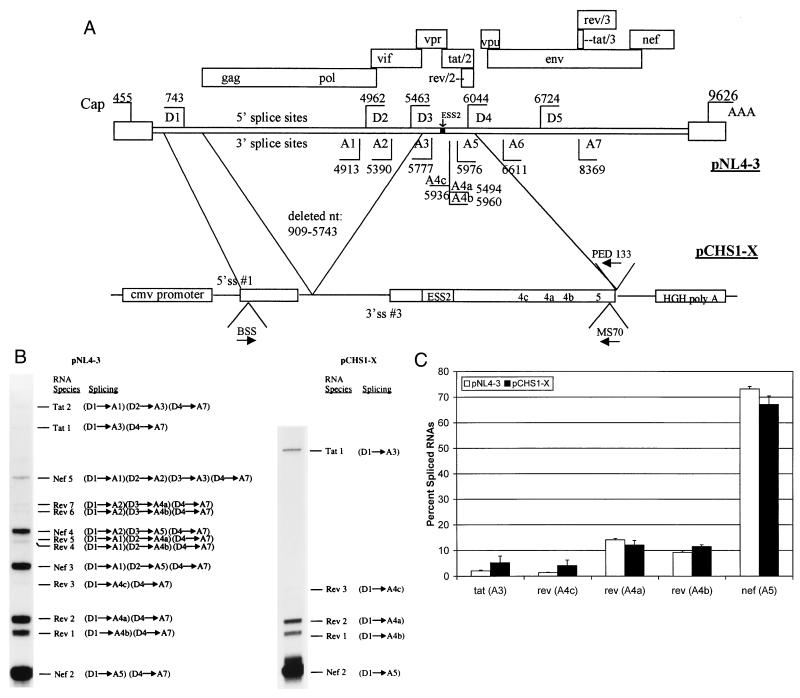
RT-PCR analysis of multiply spliced HIV-mRNAs in cells transfected with wild-type and minigene NL4-3 constructs. (A) Structure of the NL4-3 HIV-1 genome. Locations of the known 5′ (D) and 3′ (A) splice sites (ss) and the ESS2 are shown. Boxes indicate open reading frames. Locations of the RNA initiation (Cap) and poly(A) site (AAA) are shown. Oligonucleotide primers used are indicated with arrows designating position and orientation. The construct pCHS1-X contains the indicated regions of pNL4-3. Location of the cytomegalovirus (cmv) promoter and human growth hormone poly(A) (HGH poly A) site are shown. (B) Representative polyacrylamide gel of products of RT-PCR using RNA from pNL4-3 and pCHS1-X. RT-PCR products of multiply spliced HIV-1 are designated according to the nomenclature of Purcell and Martin (15). “Splicing” indicates the 5′ and 3′ splice sites used to generate each RNA species. (C) Comparison of amounts of spliced products in genomic and minigene constructs based on multiple RT-PCR analyses. Shown are the percentages of products spliced at the following 3′ splice sites: tat (A3), rev (A4c), rev (A4a), rev (A4b), and env/nef (A5).
A second splicing element in tat exon 2 is the most 3′-terminal of the three AG dinucleotides used to generate rev mRNAs (3′ splice site A4b [Fig. 1A]). We have shown that the A4b AG serves as both a splice site for rev mRNA and a branchpoint sequence (BPS) for splicing of single-spliced and double-spliced mRNAs encoding the Env and Nef proteins, respectively (22). Mutagenesis of the A4b AG results in significantly increased splicing at the env/nef 3′ splice site (A5) and a concomitant replication defect (16, 22). We have hypothesized that the increase in splicing at A5 results from relief of competition for AG binding factors. The consequence of this is facilitated binding of a factor(s) to the branchpoint. The A4b 3′ splice site is conserved in all group M HIV-1 strains but not in any group O strains. However, in a fraction of group M and all group O strains, some of the other rev AG dinucleotides are not present and different AG dinucleotides are created that are potential new rev splice sites.
These sequence differences among the HIV-1 strains suggest that regulation of splicing may be different in these strains. To test for this, we have compared alternative splice site usage of NL4-3 in the region of the first tat coding exon to two other virus strains: (i) a diverged group M, clade B virus (SF2 strain) with changes in the ESS2 sequence as well as in the region of the rev 3′ splice sites and (ii) a highly divergent group O virus strain (ANT70C). Our results indicate that regulatory elements in the region of tat exon 2 are characteristic of group M but not group O viruses.
MATERIALS AND METHODS
Plasmids.
Infectious HIV-1 plasmid pNL4-3 (GenBank accession no. M19921) was constructed by Adachi et al. (1) and was obtained from the NIH AIDS Research and Reference Reagent Program. Plasmid pHS1-X was used as template for RNA splicing substrates and has been previously described (3). pHSI-ESS4 is a derivative of pHS1-X containing four mutated bases in ESS2. This plasmid has previously been described in a report by Si et al., where it was named pESS4748+5152 (18). pHS1-SF2 was constructed by replacement of the EcoRI-HindIII fragment of pHS1-X with the corresponding fragment derived from an infectious HIV-1 SF2/ARV-2 plasmid (p9B-18) obtained from Jay Levy, University of California San Francisco (GenBank accession no. K02007). pHS1-A70 was generated by ligation of a 372-nt XcmI-ScaI fragment derived from A7054-17S (a subclone of group O strain ANT70C obtained from Eric Saman, Innogenetics, Ghent, Belgium) into pHS1-X cleaved with XcmI and ScaI. The GenBank accession number of HIV-1 ANT70C is L20587 (23). Mutant plasmid pHS1-SF4b with an AG-to-AC mutation in the SF2 A4b 3′ splice site was created by replacing the region between the EcoRI and KpnI restriction enzyme sites of pHS1-SF2 with a mutated PCR product. The mutant PCR product was synthesized by using a modified megaprimer technique (2). The mutagenic primers were SF4BS (5′AAAAGGCTTACGCATCTCCTA3′) and SF4BA (5′TAGGAGATGCGTAAGCCTTTT3′) (the changed nucleotides are underlined). Mutant plasmid pHS1-A4e, with a mutation in the ANT70C A4e 3′ splice site, was constructed similarly by replacing the region between the EcoRI and KpnI restriction enzyme sites of pHS1-A70 with a mutated PCR product. The mutagenic primers were A4ES (5′TCGTAAGAAACGGTTTGGGAA3′) and A4EA (TTCCCAAACCGTTTCTTACGA3′). The XbaI-ScaI 554-bp fragment of pHS1-X was ligated to the XbaI-SmaI-cleaved pCMV-5 (provided by Mark Stinski, University of Iowa) to create the pCHS1-X construct used for in vivo transfection assays. The XbaI-HindIII fragment of pHS1-ESS4 was ligated into the XbaI and HindIII sites of pCHS1-X to generate pCHS1-ESS4. To generate construct pCHS1-A70, the Eco0109I/BamHI fragment of pHS1-A70 was ligated to the XbaI-BamHI sites of pCHS1-X via an Eco0109I-BamHI adapter. The XbaI/HindIII fragment of pHS1-SF2 was ligated to the XbaI and HindIII sites of pCHS1-X to generate pCHS1-SF2.
Cell transfections.
HeLa cells obtained from American Type Culture Collection were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. HeLa cells in 60-mm-diameter plastic petri dishes were cotransfected by the modified calcium phosphate coprecipitation technique with 12 μg of plasmid DNA and 12 μg of pCMV-110 β-galactosidase reporter plasmid. Plasmid pCMV-110 was obtained from T. Hope (Salk Institute, La Jolla, Calif.).
RNA isolation, reverse transcription, and PCR.
Total cellular RNA was isolated from transfected HeLa cells 48 h posttransfection by extraction with Tri-Reagent (Molecular Research Center, Inc.) according to procedures supplied by the manufacturer. Three micrograms of RNA was reverse transcribed for 1 h in a 30-μl total volume containing 20 mM deoxynucleoside triphosphates, 20 U of Rnasin (Promega, Madison, Wis.), 100 pmol random hexamer (Pharmacia, Piscataway, N.J.), 6 μg of bovine serum albumin, and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL, Grand Island, N.Y.). Semiquantitative PCR analysis of 5 μl of pNL4-3 cDNA was completed with forward oligonucleotide primer BSS (5′GGCTTGCTGAAGCGCGCACGGCAAGAGG; nt 700 to 727) and reverse primer SJ4.7A, which spans splice sites D5 and A7 (5′TTGGGAGGTGGGTTGCTTTGATAGAG; nt 8381 to 8369 and nt 6044 to 6032). These two primers have been previously used to carry out reverse transcription-PCR (RT-PCR) analysis of HIV-1 multiply spliced mRNAs (13). PCR analysis of pCHS1-X, -ESS4, and -SF2 cDNAs was performed in the same manner except that the reverse primer PED133 (5′TCGAGTACGACGTACTGCTTTGATAGAGA; nt 6031 to 6002) was used. The reverse primer MS70 (5′TGCTTTGGTACAGGATCTTTATGATC; nt 6044 to 6018) was used for PCR analysis of pCHS1-A70 with BSS forward primer. PCR amplification was performed essentially as described by Purcell and Martin (15), with modifications. Thirty cycles of PCR (94°C for 30 s, 60°C for 1 min, and 72°C for 2 min) were completed in a total reaction volume of 100 μl containing 75 mM MgCl2, 10 mM deoxynucleoside triphosphates, 25 pmol of each primer, and 0.1 U of Perkin-Elmer Ampli-Taq. Prior to PCR, the reaction mixture was denatured for 5 min at 94°C. After confirmation of amplified spliced product by polyacrylamide gel electrophoresis (PAGE), amplification products (100 ng) were radiolabeled by performing a single round of PCR with the addition of 10 μCi of [32P]dCTP, and the products were analyzed by electrophoresis on a 6% polyacrylamide–7 M urea gel. Bands were visualized by autoradiography and quantitated with a Packard Instant Imager.
RNA substrate synthesis.
In vitro runoff RNA transcripts labeled with [32P]UTP (Amersham Pharmacia Biotech) were carried out as previously described (3). To prepare templates, pHS1-X, pHS1-ESS, pHS1-SF2 were linearized with XhoI and pHS1-A70 was linearized with Eco0109I.
In vitro splicing.
Splicing reactions were carried out as previously described (3). In brief, approximately 8 fmol of RNA was incubated for 2 h at 30°C with 60% (vol/vol) nuclear extract in Dignam’s buffer D (8)–20 mM creatine phosphate–3 mM MgCl2–0.8 mM ATP–2.6% (wt/vol) polyvinyl alcohol. To increase the yield of branched lariat intermediate product for primer extension analysis, approximately 100 fmol of RNA was spliced for 1 h.
Primer extension analysis.
Primer extension analysis of branched lariat-exon intermediates (BLEs) to locate branchpoints was carried out essentially as described previously (17, 22). PED133 was used as the oligonucleotide primer for NL4-3 and SF2, and MS-70 was used as the oligonucleotide primer for ANT70C. Primer extension was carried out with avian myeloblastosis virus reverse transcriptase (Promega). Dideoxynucleotide sequencing of DNA corresponding to the splice site region was performed with the same oligonucleotide primers used for the primer extension.
RESULTS
Regulatory elements determining tat, rev, and env/nef splicing efficiencies are localized to the region of the first tat coding exon.
Previous studies have indicated that the splicing pattern of pNL4-3 in HeLa cells is qualitatively and quantitatively very similar to the pattern seen in T-cell lines (15). We transfected HeLa cells with wild-type pNL4-3 and after 48 h carried out semiquantitative RT-PCR using primers designed to amplify sequences from the multiply spliced HIV-1 mRNAs as described in Materials and Methods (Fig. 1B). The resulting pattern of PCR products was similar to that reported by Purcell and Martin (15) and shows that the level of tat mRNA is lower than that of rev and nef mRNAs.
To define the sequences necessary for regulation of tat, rev, and env/nef splicing, we transfected the minigene construct pCHS1-X shown in Fig. 1A into HeLa cells and analyzed the RNA by RT-PCR. This construct contains tat exon 2 and the splice sites flanking it fused to the region containing the major HIV-1 5′ splice site D1. The minigene was placed downstream of the cytomegalovirus promoter and upstream of the human growth hormone poly(A) processing site. The results are shown in Fig. 1B. As expected, bands corresponding to spliced products which include either one or both of two upstream noncoding exons (Nef 3 to 5; Rev 4 to 7) were not present in cells transfected with pCHS1-X since these noncoding exons are not present in the minigene plasmid. The relative amounts of products spliced at the five alternative 3′ splice sites (A3, A4a, A4b, A4c, and A5) were compared to the levels of products spliced at these splice sites with the genomic plasmid (Fig. 1C). The results in either case were similar. Relatively little RNA was spliced at the tat 3′ splice site A3. Approximately equal amounts of RNA were spliced at the rev 3′ splice sites 4a and 4b, whereas there was relatively little splicing at rev 3′ splice site A4c. A high level of splicing also occurred at the env/nef 3′ splice site A5. These results implied that only the HIV-1 sequences contained in pCHS1-X are required for the appropriate regulation of alternative splicing at the tat, rev, and env/nef mRNA 3′ splice sites.
Spliced products in HIV-1 strains with changes in splicing regulatory elements.
We next tested the effect of substituting the region containing the tat, rev, and env/nef 3′ splice sites with the equivalent sequences of several other divergent HIV-1 strains with changes in tat exon 2. We compared the 3′ splice site region since previous studies have indicated that the major HIV-1 5′ splice site D1 is an efficient 5′ splice site (14). Furthermore, this sequence, which corresponds to a 5′ splice site consensus sequence, is strongly conserved in all HIV-1 strains in the database (11). We compared the NL4-3 strain to two other HIV-1 strains. One strain, SF2, is, like NL4-3, a member of the B clade of viruses and of HIV-1 group M. It contains two changes within the ESS2 core region relative to NL4-3 (Fig. 2A). As shown in Fig. 2A, it also has multiple sequence changes in the region containing the rev and env/nef 3′ splice sites. The effects of these latter changes are to inactivate the rev 3′ splice site A4c and to create a new AG dinucleotide six bases upstream of the rev 3′ splice site A4a. The other strain, ANT70C, is a member of HIV-1 group O. These viruses are highly divergent compared to the group M viruses. As shown in Fig. 2A, the ANT70C HIV strain has seven changes within the 10-nt ESS2 core region compared to NL4-3. It also has multiple sequence changes in the region containing the rev and env 3′ splice sites. The effects of these latter changes are to inactivate the normal NL4-3 rev splice sites A4a, A4b, and A4c and to create two new AG dinucleotides; one AG is 1 nt upstream of A4a, and the other is 6 nt upstream of the A4a splice site.
FIG. 2.
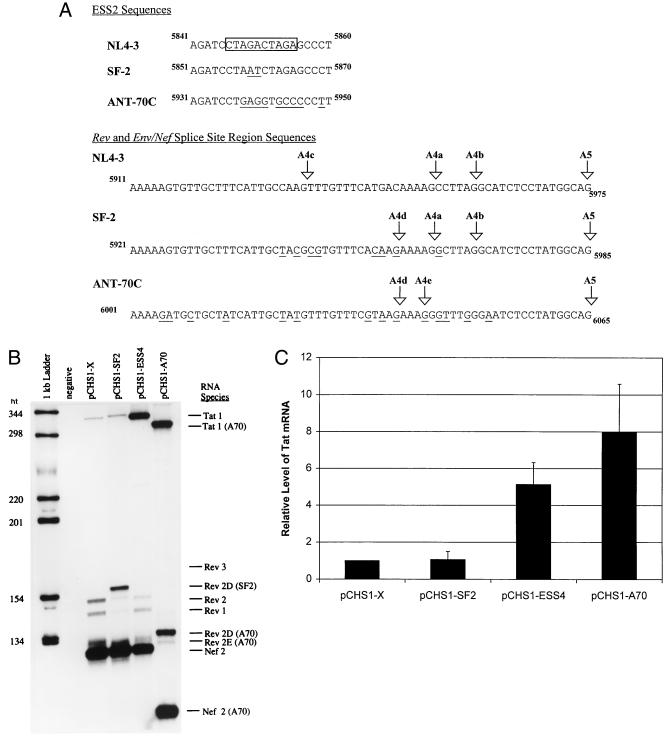
Comparison of tat, rev, and env/nef 3′ splice site usages in different HIV-1 strains by RT-PCR analysis. (A) Comparison of ESS2 sequences and the rev and env/nef splice site regions of HIV-1 strains NL4-3, SF2, and ANT70C. The core element within the ESS of pNL4-3 is boxed. The rev and env/nef splice sites are indicated by arrows, and the nucleotide numbers corresponding to GenBank sequences are shown. The sequence differences between SF-2 or ANT70C compared to the NL4-3 sequences are underlined. (B) RT-PCR analysis of spliced RNAs from mock-transfected HeLa cells or from HeLa cells transfected with pCHS1-X, pCHS1-SF2, pCHS1-ESS4, or pCHS1-A70. Rev 3 in the pCHS1-X lane is faint but is more detectable on longer exposures. The RT-PCR products from pCHS1-A70 migrate faster than those from NL4-3 and SF2 because the reverse primer (MS70) used for amplification of pCHS1-A70 products is located further upstream than the primer used for NL4-3 and SF2. (C) Relative amounts of spliced tat product. Multiple gels were quantitated, and the amounts of product are expressed relative to that of NL4-3 construct (pCHS1-X).
There was an approximately eightfold increase in relative amount of RNA spliced at the ANT70C tat 3′ splice site A3 (pCHS1-A70) compared to the NL4-3 construct (pCHS1-X) (Fig. 2B and C). The level of RNA spliced at the ANT70C tat 3′ splice site was comparable to that for an NL4-3 ESS mutant (pCHS1-ESS4) in which ESS2 was disrupted. In contrast, the level of tat splicing with the SF2 construct (pCHS1-SF2) was only slightly higher than that with pCHS1-X. These results suggested that a single copy of the CUAGA sequence in SF2 is sufficient for ESS activity.
Changes also occurred in the use of the Rev splice sites as predicted by the sequence changes shown above. As expected for both SF2 and ANT70C, the RT-PCR product (Rev 3), corresponding to splicing at 3′ splice site A4c, was not detectable because this AG dinucleotide was mutated. In the case of pCHS1-A70 and pCHS1-SF2, a major RT-PCR product (referred to in Fig. 2B as Rev 2D) results from splicing at the new AG (A4d) which is present 6 nt upstream from the NL4-3 A4a 3′ splice site (Fig. 2A). The Rev 2D PCR products were sequenced to confirm the location of the new rev 3′ splice site. In the case of SF2, products corresponding to mRNAs spliced at the A4a, A4b, and A5 splice sites (Rev 1, Rev 2, and Nef 2) were also present, and these products were identical to those of NL4-3. In the case of ANT70C, products corresponding to splicing at the A4b splice site were not present, as expected, because of the AG-to-GG change. Minor amounts of product (Rev 2E) were spliced at the AG which is 1 nt upstream of the A4a AG in NL4-3 (A4e). Thus, there appears to be considerable plasticity in the locations of the splice sites used to produce rev mRNAs in different HIV-1 strains. On the other hand, the location of the env/nef 3′ splice site A5 is conserved with all strains in the HIV-1 database.
Differences in level of products spliced at the tat 3′ splice site A3 correlate with differences in efficiencies of splicing.
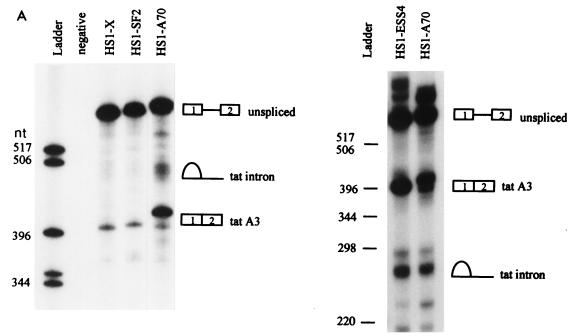
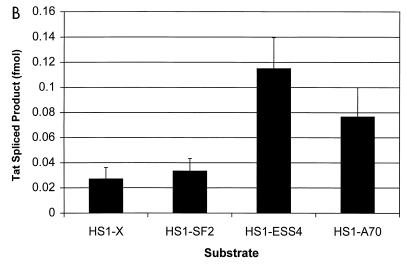
To determine whether the differences detected in the levels of tat mRNA in cells paralleled differences in efficiencies of splicing, we performed in vitro splicing assays using HeLa cell nuclear extracts. The substrates used for these experiments included HIV-1 sequences present in the minigene constructs used in the assays described above and are identical to those we have used in previous experiments to define HIV-1 splicing regulatory elements (3, 4, 18, 19). The experiment shown in Fig. 3A indicated that the amount of spliced product as well as the intron lariat product corresponding to splicing at the tat 3′ splice site (A3) was significantly greater with the ANT70C substrate (HS1-A70) than with the NL4-3 substrate (HS1-X). The radioactivity in the tat spliced product band from a number of experiments was determined. These results (Fig. 3B) indicated that splicing of the ANT70C substrate was elevated approximately fourfold and was comparable to that of an NL4-3 substrate in which ESS2 was inactivated by mutagenesis (HS1X-ESS4). Figure 3 shows that splicing of the SF2 substrate (HS1-SF2) at the tat 3′ splice site A3 was only slightly greater than that of the NL4-3 control. The results agree reasonably well with those in Fig. 2 and suggest that relative levels of spliced tat mRNAs in transfected cells with the different constructs reflect different splicing efficiencies at 3′ splice site A3.
FIG. 3.
Comparison of tat 3′ splice site usage in different HIV-1 strains by in vitro splicing assays. (A) Denaturing PAGE of [32P]UTP-labeled HIV-1 substrates spliced in vitro. The positions of precursors, spliced products, and tat lariat products are indicated. On the left is a 6% gel comparing HS1-X, HS1-SF2, and HS1-A70; on the right is a 4% gel comparing HS1-ESS4 and HS1-A70. HS1-A70 spliced product migrates more slowly because the restriction site used to produce the linearized DNA template for transcription of RNA substrates is 7 nt further downstream than for HS1-X and HS1-SF2, resulting in longer RNA products. tat lariats migrate more slowly than the linear RNA species on 6% compared to 4% gels. (B) Quantitation of spliced tat RNA from the in vitro-spliced substrates shown in panel A. Multiple gels were quantitated, and the amounts of product spliced at the tat 3′ splice site (A3) were calculated based on uridine content of the RNA species.
The A4b rev 3′ splice site plays a regulatory role in determining efficiency of splicing at the A5 env/nef 3′ splice site of the SF2 HIV-1 strain.
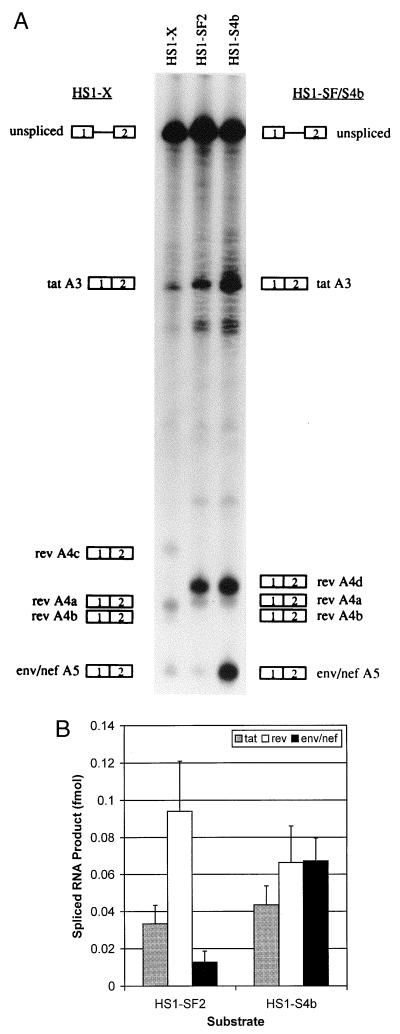
We and others have previously shown the importance of the A4b 3′ splice site as a regulatory element repressing splicing at A5 (16, 22). We first determined if changes in the region of the SF2 rev and env/nef 3′ splice sites were reflected in differences in this regulatory mechanism. We showed in Fig. 2 that these changes resulted in use of a different splice site compared to NL4-3. This was confirmed by the in vitro splicing assays shown in Fig. 4A (HS1-SF2), which demonstrate that there is a major spliced product corresponding to splicing at the new rev A4d 3′ splice site. Only small amounts of spliced products corresponding to products spliced at the A4a and A4b 3′ splice sites in NL4-3 substrate (pHS1-X) were present. As expected for HS1-SF2, there was no detectable splicing at the A4c 3′ splice site.
FIG. 4.
Mutagenesis of the rev A4b 3′ AG dinucleotide enhances splicing of the env/nef 3′ splice site in HS1-SF2. (A) Denaturing PAGE (6% gel) of [32P]UTP-labeled HS1-SF2 and rev A4b splice site mutant (HS1-SF4b) substrates spliced in vitro. Also shown is in vitro splicing of NL4-3 substrate HS1-X. HS1-SF4b is an AG-to-AC mutant at the A4b 3′ splice site. The positions of precursors and products spliced at the indicated splice sites are shown. (B) Quantitation of spliced tat, rev, and env/nef spliced products from the in vitro-spliced substrates shown in panel A. Multiple gels were quantitated, and the amounts of products spliced at the tat, rev, and env/nef 3′ splice sites were calculated based on of uridine content of the different RNA species.
To test for the repressive effect of the SF2 A4b AG dinucleotide on splicing at the env/nef 3′ splice site A5, we mutated it to AC. This change as well as a change to GG or CG results in a significant increase in splicing at A5 in NL4-3 (22). The results in Fig. 4A show that this is indeed the case, and Fig. 4B indicates that an approximate sixfold increase in splicing at A5 occurred with the A4b-mutated SF2 substrate HS1-SF4b. These results imply that the A4b 3′ splice site plays a similar role as a suppressor of splicing at 3′ splice site A5 in the SF2 strain as it does in the NL4-3 strain.
The A4b AG dinucleotide of the SF2 HIV-1 strain is a branchpoint for splicing at A5.
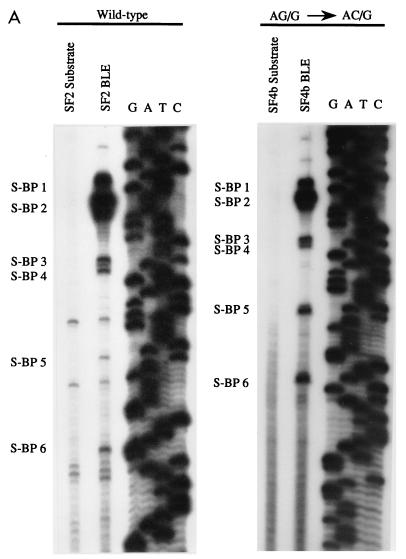
We have previously shown that with the NL4-3 HIV-1 strain, one of the branchpoints for splicing at the env/nef 3′ splice site A5 overlaps the A4b AG dinucleotide (22). To determine if this was the case for the SF2 HIV-1 strain, we performed primer extension analyses of BLEs to map the locations of the branchpoints. As a control for specificity, RNA precursor isolated from the same gel was analyzed in parallel. The experiment for the wild-type SF2 (HS1-SF2) is shown in Fig. 5A. The positions of the branchpoints corresponding to specific bands in the BLE lanes are marked in Fig. 5A, and their locations on the sequence are shown in Fig. 5B. We have previously mapped eight different branchpoints in this region for the NL4-3 strain, and their locations are also shown in Fig. 5B. In a previous study, we also correlated the use of BPSs 5 through 8 with splicing at the env/nef 3′ splice site A5 (22). As shown in Fig. 5A, the most 3′-terminal SF2 branchpoint (S-BPS 6) coincided with the A4b AG dinucleotide and therefore with BPS 8 in the NL4-3 strain. No SF2 branchpoint corresponded to NL4-3 BPS 6 presumably because the NL4-3 sequence UCAUGAC (where the position of the branchpoint is underlined) is changed to UCACAAG; this sequence deviates from the branchpoint consensus sequence YNYURAC at two additional positions. With the NL4-3 strain, we showed there was a minor amount of branching within the A4a AG dinucleotide (BPS 7), but we could not detect this branchpoint with the SF2 strain. Branching did, however, occur at S-BPS 5, which coincides with NL4-3 BPS 5.
FIG. 5.
Enhanced splicing at the env/nef A5 3′ splice site of HS1-SF2 correlates with increased use of downstream branchpoints. (A) Branchpoint location on BLEs of wild-type (HS1-SF2) and AG/G-to-AC/G mutant (HS1-SF4b) was carried out by primer extension analysis as described previously (22). Specific stops corresponding to branchpoints are shown. “Substrate” lanes indicate the same primer extension analysis carried out with unspliced precursor RNAs isolated from splicing gels. A dideoxy sequencing gel of the region was run simultaneously, and the results are shown. (B) Sequence alignment of NL4-3 and SF2 rev/env-nef splice site region. Locations of NL4-3 branchpoints sites have been described elsewhere (22). Branchpoints are indicated in bold underline type, and boxes represent BPSs. Splice sites are labeled by arrows.
We then localized branchpoints in BLEs isolated from the SF2 A4b AG-to-AC mutant (HS1-SF4b [Fig. 5A]). Because of the increased splicing at the A5 3′ splice site with this mutant, we expected an increase in primer extension products arising from branchpoints used for A5 splicing. Consistent with this, there was a relative increase compared to the wild-type SF2 in use of S-BPS 5 and S-BPS 6 versus the more upstream branchpoints. This finding suggests that the upstream branchpoints (S-BP1 through S-BP4), as found for NL4-3, are used for splicing at the rev splice sites A4d and A4a.
All detectable branchpoints in the ANT70C strain are upstream of the rev 3′ splice sites.
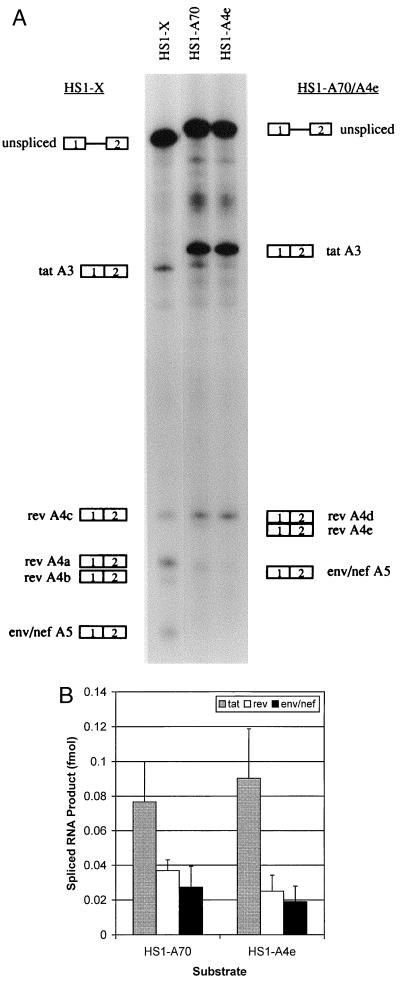
In vitro splicing assays were also used to study rev and env/nef splicing of the ANT70C substrate HS1-A70 (Fig. 6A). A major band corresponding to splicing at the A4d 3′ splice site was present, and little or no splicing occurred at the A4e 3′ splice site. There was also no splicing, as expected, at the position of the A4b splice site since the AG was changed to GG. Because the ANT70C strain as well as other group O HIV-1 strains do not have the A4b 3′ splice site AG dinucleotide, we tested whether the most downstream rev 3′ splice site (A4e) might play the same role in regulation of splicing at env/nef 3′ splice site A5 as does A4b in the group M strains. However, mutagenesis of A4e AG to AC (HS1-A4e) did not result in a significant change in splicing at A5 (Fig. 6). These results indicate that the A4e AG dinucleotide of ANT70C does not substitute for the A4b 3′ splice site as a negative regulatory element.
FIG. 6.
Mutagenesis of rev 3′ splice site A4e of HS1-A70 does not increase splicing at the env/nef 3′ splice site A5. (A) Denaturing PAGE (6% gel) of [32P]UTP-labeled HS1-A70 and rev A4e splice site mutant (HS1-A4e) substrates spliced in vitro. Also shown is in vitro splicing of NL4-3 substrate HS1-X. HS1-A4e is an AG-to-AC mutant at the A4e 3′ splice site. The positions of precursors and products spliced at the indicated splice sites are shown. (B) Quantitation of spliced tat, rev, and env/nef spliced products from the in vitro-spliced substrates shown in panel A. Multiple gels were quantitated, and the amounts of products spliced at the tat, rev, and env/nef 3′ splice sites were calculated based on of uridine content of the different RNA species.
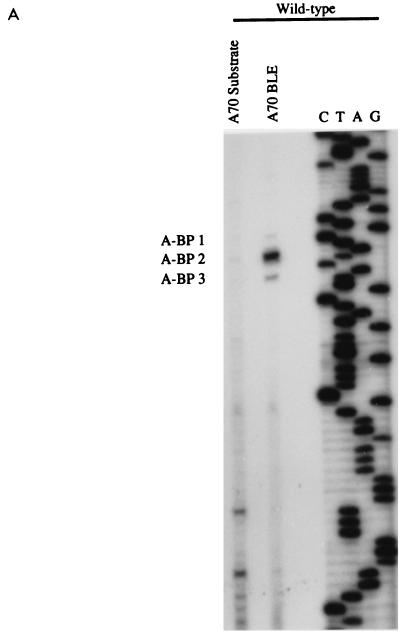
We then localized the branchpoints in BLEs isolated from the ANT70C splicing reactions. The results (Fig. 7A) indicated that there were one major and two minor branchpoints. All of these branchpoints mapped upstream of the two rev 3′ splice sites, and these coincided with the locations of NL4-3 BPS 1 and BPS 2 (Fig. 7B). We did not detect branchpoints within either of the AG dinucleotides of rev 3′ splice sites A4d and A4e.
FIG. 7.
All detectable branchpoints in HS1-A70 are upstream of the rev 3′ splice sites. (A) Branchpoint analysis of BLEs from splicing reactions using the HS1-A70 substrate. (B) ANT70C sequence, location of the 3′ splice sites, and locations of the branchpoints (underlined).
DISCUSSION
Regulation of alternative splicing of HIV-1 RNA is required for efficient virus replication. It has been shown that in the NL4-3 strain, a deletion mutation that includes ESS2 results in oversplicing to tat mRNA and a more defective phenotype than a tat point mutation that terminates translation of Tat at amino acid residue 9 (7). It would therefore be expected that this element would be conserved in different viral strains. The ESS2 core element in the NL4-3 strain maps to the sequence CUAGACUAGA, containing two repeats of the CUAGA sequence (18). Of the sequenced strains in the HIV-1 genome database, 27 of 95 group M strains contain this sequence. Many (54 of 95) group M HIV-1 strains, however, contain only one of the two repeats at this position. We have shown above that the single copy of CUAGA in the SF2 strain is sufficient to elicit ESS2 activity although there were small increases in tat splicing efficiency in SF2 relative to NL4-3 substrates. An additional five strains contain a single copy of the sequence UUAGA, which we have previously shown acts as an ESS element in tat exon 3 (19). Thus, most group M HIV-1 strains contain an ESS in tat exon 2. The group O ANT70C strain, on the other hand, appears not to have an ESS at the location of ESS2 since the level of tat splicing both in vivo and in vitro was comparable to that of an NL4-3 ESS mutant. This is also presumably true for the 14 other sequenced group O strains since their sequences are homologous to that of ANT70C and very different from the sequence of the group M strain ESS2 (5).
The Tat coding sequences of group O HIV-1 strains differ from those of group M strains at a number of positions in addition to those in the region of ESS2. These differences may be reflected in reduced Tat activity compared to the group M strains. Thus, a larger amount of Tat mRNA in the case of group O strains may be necessary in order to produce sufficient Tat protein. On the other hand, it is possible that sequences in the group O viral genomes distant from the tat exon and its flanking sequences can compensate for the lack of ESS2. Studies of tat splicing in the context of the viral genome in lymphoid and nonlymphoid cell types will be necessary to test these possibilities.
Mutation of the rev 4b 3′ splice site also results in an oversplicing at the env/nef 3′ splice site (22) and in a defective viral phenotype (16). We have hypothesized that the regulation occurs because different splicing factors leading to splicing at the rev or env/nef 3′ splice sites compete for binding to the same site on the viral RNA, the rev A4b AG dinucleotide. Mutagenesis of the rev A4b AG dinucleotide relieves the repression and results in an increase in env/nef splicing (22). Splicing at the SF2 3′ splice site A5 is subject to a similar repression, and its A4b AG dinucleotide also contains a branchpoint used for splicing at A5. The repression appears not to be related to the efficiency with which A4b is used as a 3′ splice site since approximately half of NL4-3 rev mRNA but much less SF2 rev mRNA is spliced at this site (Fig. 2B). Consistent with its importance as a splicing regulatory element, the sequence surrounding the A4b splice site is highly conserved within all group M HIV-1 strains.
In contrast to the group M HIV-1 strains, ANT70C as well as all other sequenced group O strains do not contain the A4b 3′ splice site. We considered the possibility that the most 3′ distal ANT70C rev 3′ splice site (A4e) plays a repressive role analogous to that of the A4b splice site in the group M strains, but no significant difference in level of A5 splicing with an A4e AG-to-AC mutant was detected compared to the wild-type ANT70C. Furthermore, in contrast to NL4-3 and SF2, all detectable branchpoints in ANT70C mapped upstream of the rev 3′ splice sites. Thus, the regulation of rev and env/nef 3′ splicing in ANT70C and presumably other group O strains does not appear to involve overlapping cis splicing elements as it does with the group M strains. Based on the splicing of NL4-3 substrates, we previously proposed that several splicesome complexes form in the region of the rev and env/nef 3′ splice sites. These correspond to different sets of branchpoints leading to splicing at the different rev and env/nef 3′ splice sites (22). In contrast, in the case of the ANT70C substrates, we showed above that there is one major branchpoint upstream of the three rev and env/nef 3′ splice sites, suggesting that a single spliceosome complex may predominate. The efficiency of splicing at the three ANT70C 3′ splice sites would then be determined by scanning downstream from the branchpoint and competition for the use of the three AGs as previously proposed by Smith et al. for splicing of mammalian cell mRNA precursors (21).
The group M HIV-1 strains are responsible in large part for the worldwide AIDS pandemic, whereas the group O strains have mainly been found in patients in Cameroon and Gabon (10, 12). Group O strains differ significantly in sequence from group M strains, suggesting that the group M and group O HIV-1 strains arose from two independent zoonotic transmissions of HIV-1 from the chimpanzee subspecies Pan troglodytes troglodytes (9). One major difference that distinguishes the two groups is that in contrast to all group M viruses, group O viruses are not sensitive to the cyclophilin A inhibitor cyclosporin A and thus do not require cellular cyclophilin A for replication (6). Our studies have indicated another striking phenotypic difference in that group O viruses lack two cis-acting splicing regulatory elements within tat exon 2 that appear to be present in most group M viruses. A prototype member of a third HIV-1 group (group N) has recently been isolated from a patient in Cameroon which may represent another independent transmission to humans (20). Interestingly, the ESS2 core sequence of this virus has only a single C-to-U change compared to NL4-3 (CUAGAUUAGA), and it preserves one unchanged copy of the CUAGA motif. Furthermore, this virus contains the conserved A4b 3′ splice site. Based on our results, we postulate that alternative tat, rev, and env/nef mRNA splicing of Group N viruses is regulated by the two tat exon 2 elements similarly to group M viruses and not to group O viruses.
ACKNOWLEDGMENTS
We thank D’Andra Luna for assistance in constructing clones used for some of the experiments. We also thank Stanley Perlman and Lung-Ji Chang for critical reviews of the manuscript.
Cells were obtained from the Cell Culture Center, which is sponsored by the National Center for Research Resources of the NIH. This research was supported by PHS grant AI36073 from the National Institute of Allergy and Infectious Disease.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques. 1993;14:366–369. [PubMed] [Google Scholar]
- 3.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of the human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat/rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibollet-Ruche F, Loussert-Ajaka I, Simon F, Mboup S, Ngole E M, Saman E, Delaporte E, Peeters M. Genetic characterization of accessory genes from human immunodeficiency virus type 1 group O strains. AIDS Res Hum Retroviruses. 1998;14:951–961. doi: 10.1089/aid.1998.14.951. [DOI] [PubMed] [Google Scholar]
- 6.Braaten D, Franke E K, Luban J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZGAB but not group O HIV-1 or other primate immunodeficiency viruses. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang L-J, Zhang C. Infection and replication of Tat− human immunodeficiency viruses: genetic analyses of LTR and tat mutations in primary and long-term human lymphoid cells. Virology. 1995;211:157–169. doi: 10.1006/viro.1995.1388. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao F, Bailes E, Robertson D L, Chen Y, Rodenberg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 9a.HIV Sequence Database. December 1998, revision date. [Online.] http://hiv-web.lanl.gov/ Los Alamos National Laboratory, Los Alamos, N.Mex. [April 1999, last date accessed.]
- 10.Janssens W, Nkengasong J N, Heyndrickx L, Fransen K, Ndumbe P M, Delaporte E, Peeters M, Perret J-L, Ndoumou A, Atende C, Piot P, van der Groen G. Further evidence of the presence of genetically aberrant HIV-1 strains in Cameroon and Gabon. AIDS. 1994;8:1012–1013. doi: 10.1097/00002030-199407000-00022. [DOI] [PubMed] [Google Scholar]
- 11.Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuiken C. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 12.Mauclere P, Lousser-Ajaka I, Damond F, Fagot P, Souquieres S, Lobe M M, Keou F-X M, Barre-Sinoussi F, Saragosti S, Brun-Vezinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Neumann M, Harrison J, Saltarelli M, Hadziyannis E, Erfle V, Felber B K, Pavlakis G N. Splicing variability in HIV type 1 revealed by quantitative RNA polymerase chain reaction. AIDS Res Hum Retroviruses. 1994;10:1531–1542. doi: 10.1089/aid.1994.10.1531. [DOI] [PubMed] [Google Scholar]
- 14.O’Reilly M M, McNally M T, Beemon K L. Two strong 5′ splice sites and competing suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 15.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riggs N L, Little S J, Richman D D, Guatelli J C. Biological importance and cooperativity of HIV-1 regulatory gene splice acceptors. Virology. 1994;202:264–271. doi: 10.1006/viro.1994.1342. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1989. [Google Scholar]
- 18.Si Z-H, Amendt B A, Stoltzfus C M. Splicing efficiency of human immunodeficiency virus type 1 Tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si Z-H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon F, Mauclere P, Roques P, Lousser-Ajaka I, Muller-Turwin M C, Saragosti S, Georges-Courbot M C, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 21.Smith C W J, Chu T T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson A K, Stoltzfus C M. Overlapping cis sites used for splicing of HIV-1 env/nef and rev mRNAs. J Biol Chem. 1998;273:34551–34557. doi: 10.1074/jbc.273.51.34551. [DOI] [PubMed] [Google Scholar]
- 23.vandenHaesevelde M, Decourt J-L, De Leys R J, Vanderborght B, van der Groen G, van Heuverswijn H, Saman E. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol. 1994;68:1586–1596. doi: 10.1128/jvi.68.3.1586-1596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]