Abstract
Clostridium botulinum is an important foodborne pathogen capable of forming heat resistant endospores and producing deadly botulinum neurotoxins (BoNTs). In 2006, C. botulinum was responsible for an international outbreak of botulism attributed to the consumption of commercially pasteurized carrot juice. The purpose of this study was to isolate and characterize strains of C. botulinum from the adulterated product. Carrot juice bottles retrieved from the manufacturing facility were analyzed for the presence of BoNT and BoNT-producing isolates using DIG–ELISA. Toxigenic isolates from the carrot juice were analyzed using pulsed-field gel electrophoresis (PFGE) and DNA microarray analysis to determine their genetic relatedness to the original outbreak strains CDC51348 and CDC51303. PFGE revealed that isolates CJ4-1 and CJ10-1 shared an identical pulsotype with strain CDC51303, whereas isolate CJ5-1 displayed a unique restriction banding pattern. DNA microarray analysis identified several phage related genes unique to strain CJ5-1, and Southern hybridization analysis of XhoI digested and nondigested DNA showed their chromosomal location, while a homolog to pCLI_A009 of plasmid pCLI of C. botulinum serotype Langeland F, was located on a small plasmid. The acquisition or loss of bacteriophages and other mobile genetic elements among C. botulinum strains has epidemiological and evolutionary implications.
Keywords: Clostridium botulinum DNA microarray, PFGE (pulsed-field gel electrophoresis), Botulinum neurotoxin (BoNT), Plasmids, Bacteriophage
1. Introduction
Clostridium botulinum is a genetically diverse species that produces seven distinct serotypes of the neuroparalytic protein toxin called, botulinum neurotoxin (BoNTs/A–G). Recently, an eighth serotype (type H) of botulinum neurotoxin was discovered (Barash and Arnon, 2014; Dover et al., 2014). BoNTs are the causative agents of the foodborne intoxication known as botulism, which results from the oral ingestion of preformed toxin in an adulterated food product. C. botulinum is categorized into four groups (I–IV), where strains of groups I (proteolytic) and II (nonproteolytic) produce BoNT serotypes A, B, E and F which are associated with causing human botulism (Collins and East, 1998). BoNTs are also produced by rare strains of Clostridium baratii (BoNT/F) (Hall et al., 1985) and Clostridium butyricum (BoNT/E) (McCroskey et al., 1986). Additionally, bivalent C. botulinum strains carry and express more than one toxin serotype. Typically, one toxin serotype is produced in higher quantities than the other and is denoted with a capital letter (e.g. Ab, Af, Ba, Bf) (Franciosa et al., 2004; Gimenez and Ciccarelli, 1978; Hatheway et al., 1981; Hatheway and McCroskey, 1987). Certain strains that have been identified and classified as A(B), express BoNT/A and carry a silent or, unexpressed bont/B gene (Hutson et al., 1996).
BoNTs are naturally produced as toxin complexes in which the neurotoxin is associated with the nontoxic nonhemagglutinin (NTNH), hemagglutinins (HA) or other uncharacterized proteins (ORFX), and RNA. The genes encoding the neurotoxin, the regulatory component and associated proteins of the progenitor toxin complex are arranged in two primary types of toxin gene clusters, ha+/orfX− and ha −/orfX+. The ha+/orfX− gene cluster is found in serotype A1, B, C, D and G strains (Peck, 2009). The ha−/orfX+ toxin gene clusters are found in C. botulinum serotype A1, A2, A3, A4, E, and F strains (Peck, 2009). In these strains hemagglutinin genes are absent and the cluster instead contains three open reading frames orfX1, orfX2 and orfX3, which encode for uncharacterized proteins. The location of these toxin gene clusters varies among strains, and although previously believed to be chromosomally located in serotype A, B, E and F strains, toxin gene clusters have been discovered to reside on plasmids in some serotype A, B, and E strains, and in all serotype G strains (Zhou et al., 1995; Marshall et al., 2007; Franciosa et al., 2009; Smith et al., 2007; Zhang et al., 2013). Some of these plasmids have been found to be mobile via conjugation (Marshall et al., 2010). In serotype C and D strains the toxin gene clusters are carried by bacteriophages (Iida et al., 1974; Sakaguchi et al., 2005). Whole genome sequencing has revealed bacteriophages as well as small cryptic plasmids, such as pBot3502 in serotype A strain ATCC 3502 (AM412317, Sebaihia et al., 2007) and pCLI (CP000729) in serotype F strain Langeland F (CP000728). These potentially mobile genetic elements, specific to C. botulinum strains, are not well characterized and likely contribute to the phylogenetic and evolutionary diversity of this foodborne pathogen.
Cases of botulism caused by commercially prepared foods are fairly uncommon. Sevenier et al. (2012) examined 316 samples of raw carrots and green beans collected from canned food manufacturers in France for the incidence and prevalence of anaerobic mesophiles, thermophiles and specifically spores of C. botulinum. It was concluded from that study that raw carrots had a higher incidence of these organisms than raw green beans, which is likely due to carrots being grown directly in the soil (Sevenier et al., 2012). Sheth et al. (2008) reported a botulism outbreak in the US and Canada that was due to the consumption of commercially pasteurized carrot juice. During the outbreak investigation the FDA retrieved 20 unopened bottles of implicated carrot juice from the manufacturer, which were incubated at 35 °C for 5 days and tested for the presence of BoNT/A, B, E and F using ELISA and the mouse bioassay (Sheth et al., 2008). BoNT/A was only detected in bottles numerically labeled 1 (<10 to <20 MIPLD50/ml), 4 (>2000 MIPLD50/ml), 5 (>2000 MIPLD50/ml), 6 (~2000 MIPLD50/ml) and 10 (~200 MIPLD50/ml) (Sheth et al., 2008). Although, no botulinum neurotoxigenic strains were described as being isolated from any of these bottles, two C. botulinum strains were isolated during the outbreak; CDC51303 (also called CDC-CR1; Reddy et al., 2013) was isolated from the carrot juice belonging to the patient identified in Georgia, and CDC51348 (also called CDC-CR2; Reddy et al., 2013) was isolated from carrot juice that belonged to the Florida patient (Raphael et al., 2008). Genetic analysis of these serotype A1 outbreak strains revealed that CDC51348 contained an unexpressed bont/B gene. The bont/A1 of CDC51303 was reported to contain unique nucleotide polymorphisms in relation to other bont/A1 sequences, and resided within a ha−/orfX+ cluster (Raphael et al., 2008). The purpose of this study was to isolate botulinum neurotoxigenic strains from the carrot juice bottles retrieved from the manufacturer and compare these isolates with previously isolated C. botulinum strains CDC51303 and CDC51348 using PFGE.
2. Materials and methods
2.1. Isolation of botulinum neurotoxin-producing strains from carrot juice
In 2006, the FDA received twenty 1-L bottles of commercially pasteurized carrot juice from implicated lots retrieved from the manufacturer (Sheth et al., 2008). During that study, ten of the carrot juice bottles were incubated at 37 °C for 5 days, tested for botulinum neurotoxin using the ELISA and mouse bioassays and then stored at 4 °C. The remaining ten bottles were not incubated at 37 °C for 5 days nor tested for toxin production using ELISA and mouse bioassays, but instead were stored at 4 °C (Sheth et al., 2008). All twenty bottles of carrot juice were used in the current study to isolate botulinum neurotoxin-producing species of clostridia. The lot codes on the bottles were as follows: bottles 1 and 2, #451500, bottles 3 and 4, #451800, bottles 5 and 6, #450000, bottles 7 and 11–20, #450100, bottle 8, #451000, and bottles 9 and 10, #452100.
Enrichment of botulinum neurotoxin-producing clostridia from the implicated carrot juice was performed following the procedures described in the Bacteriological Analytical Manual (Solomon and Lilly, 2001). Briefly, 20 ml of carrot juice was removed from each of the bottles and added to individual bottles containing 100 ml of TPGY broth (50 g/L trypticase peptone, 5 g/L Bacto peptone, 4 g/L dextrose, 20 g/L yeast extract, 1 g/L sodium thioglycollate) and 10 g of cooked meat medium (CMM). The bottles were incubated anaerobically for at least 5 days at 37 °C and examined for turbidity and digestion of the cooked meat particles.
Isolated colonies were obtained by mixing 2.5 ml of the carrot juice enrichment culture with an equal volume of filter sterilized ethanol. The mixture was incubated at room temperature for one hour, and streaked for isolation onto egg yolk agar (EYA) (75 g/L McClung Toabe Agar, 80 ml/L egg yolk enrichment) and EYA supplemented with d-cycloserine (250 μg/ml), sulfamethoxazole (76 μg/ml) and trimethoprim (4 μg/ml) (EYA + CST). The plates were incubated anaerobically at 37 °C for 2 days. Individual colonies were selected and streaked for isolation onto duplicate EYA plates and incubated anaerobically and aerobically at 37 °C for at least 3 days. Isolated colonies showing strict anaerobic growth were selected and inoculated into 10 ml of TPGY and incubated at 37 °C prior to ELISA.
2.2. Digoxigenin – Enzyme Linked Immunosorbant Assay (DIG–ELISA)
DIG–ELISA was performed to test for the presence of botulinum neurotoxin types A, B, E and F in the carrot juice samples, the carrot juice enrichment cultures and strict anaerobic colonies isolated from the carrot juice enrichment cultures. The DIG–ELISA assay was performed as described in Ferreira et al. (2003) and Sharma et al. (2006) with some modifications. Samples were diluted 1:10 using gel phosphate buffer and centrifuged in a Sorvall Evolution RC ultracentrifuge (Thermo Fisher Scientific, Waltham, MA) for 20 min at 20,000 × g at 4 °C. The supernatants were removed and tested using DIG–ELISA. Purified BoNT/A, /B, /E, and /F were purchased from Metabiologics (Madison, WI) and used as positive controls.
Microtiter plates (ImmunoChemistry Technologies, Bloomington, MN) were coated with affinity-purified capture antibody from the FDA Southeast Regional Laboratory (Atlanta, GA) (100 μl/well) in bicarbonate buffer (Sigma, 0.1 M Na2CO3, pH 9.6) containing 2 μg/ml BoNT/A, /B, or /E or 1 μg/ml BoNT/F. The microtiter plates were sealed and stored overnight at 4 °C. The plates were washed 5 times with phosphate buffered saline Tween-20 (PBST) and blocked with 5% skim milk buffer (50 g/L skim milk buffer, 10 mM phosphate buffer saline, pH 7.4) (300 μl/well) for 60 min at 35 °C. The blocking reagent was discarded, and the plate was tapped over absorbent paper to remove the remaining residual skim milk buffer. Serial dilutions of the purified BoNT positive control samples were prepared in skim milk buffer. The test samples, the skim milk buffer (negative control), and toxin standards (positive controls) were added (100 μl) to the designated wells of the microtiter plates. The plates were incubated for 2 h at 37 °C and washed five times with PBST. The dilutions of the DIG-labeled affinity-purified antibodies prepared in skim milk buffer were optimized according to each antibody lot and dispensed (100 μl/well) onto the plate, which was subsequently incubated at 20–25 °C in the dark on a Delfia Plate Shaker Model 1296–004 (PerkinElmer, Waltham, MA) on low speed for 120 min. Horseradish peroxidase-conjugated antidigoxigenin antibody (antidigoxigenin–peroxidase [polyclonal], fab fragment) was diluted (1:5000) in skim milk buffer and added (100 μl/well) to each well. The plates were incubated for an additional 60 min. The plates were washed 5 times with PBST, the substrate solution containing 3,3’,5,5’-tetramethylbenzidine (TMB) was added to the individual wells (100 μl/well), and the plates were incubated for 15 min at 20–25 °C in the dark on a Delfia shaker on low speed. The reaction was quenched with 1 N sulfuric acid (100 μl/well), and the color was monitored by measuring the absorbance at 450 nm using a Bio-Tek ELx808 Ultra Microplate Reader (Winooski, VT).
2.3. Genomic DNA extraction
C. botulinum isolates were inoculated (1/100) into 10 ml of TPGY broth and incubated anaerobically overnight at 37 °C. From the overnight cultures 1.5 ml was centrifuged at 10,000 × g for 5 min to pellet the bacteria. The cell pellet was resuspended in 300 μl of TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) buffer containing 30 mg/ml of lysozyme and incubated at 37 °C for 30 min. Genomic DNA was extracted using the MasterPure DNA purification kit (Epicentre Biotechnologies, Madison, WI) following the manufacturer’s instructions. To the lysed cell suspension 300 μl of 2× tissue and cell lysis buffer containing proteinase K (167 mg/ml) was added and the suspension was incubated at 65 °C for 15 min; vortexing every 5 min. The suspension was cooled to 37 °C, then 1.5 μl of RNase A (100 mg/ml) (Qiagen, Valencia, CA) was added and incubated for 30 min. The cells were incubated on ice for 3–5 min prior to adding 350 μl of MPC protein precipitation buffer and vortexing vigorously for 10 s. The cell debris was pelleted by centrifugation at 10,000 × g for 10 min at 4 °C, and the supernatant was transferred to a clean microcentrifuge tube containing 500 μl of isopropanol. The microcentrifuge tube was inverted several times to mix and the DNA was pelleted by centrifugation for 10 min at 4 °C. The isopropanol was decanted and the DNA pellet was washed twice with 70% ethanol and resuspended in TE buffer. The concentration and quality of each DNA sample were evaluated using a NanoDrop2000 (Thermo Scientific, Wilmington, DE) and the DNA was stored at 4 °C prior to further analysis.
2.4. DNA microarrays
A previously described 225-probe Group I C. botulinum subtyping microarray (Raphael, 2012) was used to evaluate gene content among genomic DNA isolated from test samples. Briefly, genomic DNA was labeled with Cy-5 random primers and allowed to hybridize for 16 h at 42 °C. Probes with a log ratio (signal/background) ≥1 were considered positive. The microarray data obtained were deposited into the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession number GSE51155.
The results of the DNA microarray data were validated by polymerase chain reaction (PCR) using the same primers employed for the design of the Southern blot hybridization probes (Table 2). PCR reactions were prepared using the GeneAmp High Fidelity PCR system (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. The gDNA from isolates CJ4-1 and CJ5-1 were used as templates to evaluate the presence or absence of the genes listed in Table 3. The PCR reactions were cycled utilizing a Mastercycler pro S (Eppendorf, Hauppauge, NY), with cycling conditions optimized for each primer pair. The PCR products were electrophoresed on a 1% TAE (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) gel and stained with 0.5 μg/ml of ethidium bromide and imaged using a GelDocXRS+ camera (BioRad, Hercules, CA).
Table 2.
Primers used for PCR DIG labeling of probes used in Southern hybridization analyses.
| Gene probe | Primer | Sequence (5’–3’) | Amplicon size (bp) |
|---|---|---|---|
| 2952 (CLB_2952) | Forward | ATGGTACAGTAAATTTCAGTGC | 451 |
| Reverse | GGAAATCCTGTTTGCATAGG | ||
| B28 (CBO_1744) | Forward | GAATATCGGCTTATGACTTTGG | 565 |
| Reverse | CAACCATCTTTGTCTGATCC | ||
| BF9 (CBO_2394) | Forward | TATATGAGGATGAAGGATACTCAGG | 556 |
| Reverse | GACATGGATGCAAATTCTCC | ||
| Okra14 (CLD_2430) | Forward | TCGAGAGGGATTCTATTGCTTTCA | 305 |
| Reverse | GCATGGGACATGGTGAGGAC | ||
| Okra17 (CLD_2442) | Forward | ATTATCATACTTTCTACCACGC | 571 |
| Reverse | ACAGTTTACTATTGCTGGGG | ||
| PL7 (pCLI_A009) | Forward | ATGAATAACGTAGAGCCTATTAGAG | 470 |
| Reverse | ATTGCAGGATTAAAGCCAGC |
Table 3.
Gene content differences between Clostridium botulinum isolates CJ4-1 and CJ5-1 identified by DNA microarray analysis.
| Probe | Strain | Probe properties | |||
|---|---|---|---|---|---|
| CJ4-1 | CJ5-1 | Gene locus | Genome | Annotated function | |
| BF9 | .082306b | 2.174096a | CBO_2394 | ATCC 3502 (AM412317) |
Putative resolvase |
| B28 | .077197 | 2.328237 | CBO_1744 | ATCC 3502 (AM412317) |
Putative phage tail fiber protein |
| CLB_2952 | −0.01081 | 2.457756 | CLB_2952 | ATCC 19397 (CP000726) |
Glycosyl hydrolase |
| Okra14 | .402789 | 2.597363 | CLD_2430 | Okra (CP000939) |
Phage related protein |
| Okra17 | .422569 | 2.014994 | CLD_2442 | Okra (CP000939) |
Hypothetical protein |
| PL7 | .111761 | 2.660995 | pCLI_A009 | Langeland plasmid pCLI (CP000729) |
Phage integrase |
Log (signal/background) ≥1.0; probe sequence is present and/or highly conserved.
Log (signal/background) <0.5; probe sequence is absent or highly divergent.
2.5. Pulsed-field gel electrophoresis (PFGE)
Botulinum neurotoxigenic isolates obtained from carrot juice bottles labeled 4, 5 and 10 and C. botulinum strains CDC51303 (CDC-CR1) and CDC51348 (CDC-CR2) were cultured in TPGY broth to an OD600nm of 0.6. PFGE plug samples were prepared using 1.5 ml culture aliquots as described in Reddy et al. (2013). Restriction digestion of the DNA samples embedded in agarose plugs was performed using restriction endonucleases XhoI (at 37 °C) and SmaI (at 25 °C) as previously described (Reddy et al., 2013). The sample plugs were rinsed with 0.5× Tris–borate–EDTA (TBE) (45 mM Tris–borate, 1 mM EDTA, pH 8.0), loaded onto a 1% agarose gel, and electrophoresed in 0.5× TBE containing thiourea (4.5 mg/ml) using a clamped homogenous electric field system (CHEF Mapper; Bio-Rad, Hercules, CA) with the following parameters: initial switch time of 0.5 s, final switch time of 40 s, at 6 V/cm, at 14 °C for 22 h. The gels were stained for 20 min with ethidium bromide (1 μg/ml), destained in ultrapure water for 1 h, and visualized using a GelDocXRS+ camera. The restriction banding patterns generated using XhoI and SmaI were analyzed using the BioNumerics Software, version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium) according to the standardized PulseNet protocol (Ribot et al., 2006; Tenover et al., 1995). Similarity between the restriction banding patterns was determined using the Dice coefficient correlation. The dendrograms were constructed using the average of the composite data sets generated by SmaI and XhoI and the unweighted-pair group (UPGMA) method using average UPGMA linkages clustering tool. The optimization value and position tolerance was 1.5%.
2.6. Southern blot hybridization
Hybridization probes: B28 (CBO_1744), BF9 (CBO_2394), Okra14 (CLD_2430), Okra17 (CLD_2442), PL7 (pCLI_A009) and CLD_2952 were PCR labeled with digoxigenin (DIG) using the primers listed in Table 2 and the PCR DIG Probe Synthesis Kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions on a Mastercycler pro S (Eppendorf, Hauppauge, NY). The DNA was transferred from the PFGE gels to a positively charged nylon membrane (Hybond-N+, Roche, Indianapolis, IN) by overnight downward capillary action in 0.4 N NaOH, 1.5 M NaCl for 18 h. The membranes were neutralized in 0.5 M Tris–HCl, pH 7.0 for 15 min, rinsed briefly in 2× SSC, and baked at 80 °C for 2 h. The membranes were stored at 4 °C prior to hybridization. The membranes were prehybridized in DIG EasyHyb solution for 1 h at 42 °C and hybridized with 2.5 ng of the DIG-labeled probe in DIG EasyHyb solution for 18 h at 42 °C. The membranes were washed once with 1× SSC, 0.1% SDS for 5 min at room temperature and washed again for 15 min at 42 °C with gentle shaking.
Probe-target hybrids were detected using a chemiluminescence based technology according to the manufacturer’s instructions (Roche, Indianapolis, IN). The membranes were incubated in blocking solution for up to 3 h at room temperature with gentle rocking. Anti-Digoxigenin-AP, Fab fragment antibody (75 mU/ml) in blocking buffer solution was added to the membranes which were then incubated for 30 min at room temperature with gentle rocking. The membranes were washed twice, equilibrated in detection buffer for 3 min, and incubated with the chemiluminescent substrate CDP-Star for 5 min at room temperature. Visualization of the membranes was achieved using a ChemiDocXRS+ camera and Image Lab software version 4.1 (BioRad, Hercules, CA).
3. Results and discussion
3.1. Detection of BoNT and BoNT-producing isolates in carrot juice samples
During the 2006 botulism outbreak investigation, 20 bottles of carrot juice were retrieved from the manufacturer by the US FDA. Botulinum neurotoxin was previously detected in bottles numerically labeled 1, 4, 5, 6, and 10 (Sheth et al., 2008), and the bottles were stored at 4 °C. In the present study the carrot juice was retested for the presence of botulinum neurotoxin serotypes A, B, E and F using DIG–ELISA. As expected, only BoNT/A was detected in carrot juice bottles 1, 4, 5, 6 and 10. Bottles labeled 11–20 all tested negative for BoNT/A, B, E or F.
Prior to our study, no botulinum neurotoxin-producing clostridia had been isolated from any of the 20 bottles of carrot juice retrieved from the manufacturer. However, two C. botulinum strains had been isolated from carrot juice during the outbreak; one isolate (CDC51303; CDC-CR1) was obtained from the carrot juice implicated in the three cases in Georgia, and the other isolate (CDC51348; CDC-CR2) was found in the carrot juice from the Florida case (Raphael et al., 2008; CDC, 2006). In order to isolate botulinum neurotoxigenic strains from the carrot juice, 20 ml of carrot juice from each bottle was used to inoculate individual bottles of TPGY broth containing cooked meat particles. The enrichment cultures inoculated with carrot juice from bottles 4, 5, 6 and 10 were positive for BoNT/A by ELISA (Table 1). The TPGY enrichment inoculated with carrot juice from bottle 1 never became turbid and tested negative for BoNT/A, B, E and F, and no botulinum neurotoxigenic isolates were recovered. This suggests that any organism previously present in the carrot juice is no longer viable for culturing or that only BoNT/A was present when the carrot juice was bottled for that particular bottle. Repeated attempts to obtain a turbid and toxigenic culture from carrot juice extracted from bottle 1 were unsuccessful. The carrot juice and the enrichment culture from bottle number 6 tested positive for BoNT/A, and isolates were recovered from the carrot juice enrichment, however, none of the isolates produced BoNT/A (Table 1). Sheth et al. (2008) reported that the carrot juice in bottle 6 contained high quantities of BoNT/A. It is possible that upon transfer of 20 ml of the carrot juice aliquot to the TPGY enrichment broth, enough toxin was still present to be detected by DIG–ELISA. Similar to bottle 1, any botulinum neurotoxigenic organisms previously present in the carrot juice of bottle 6 are either no longer viable or were outcompeted by other microbes, or the carrot juice was bottled with preformed toxin. PFGE was performed on toxigenic isolates obtained from bottles 4, 5 and 10 that produced BoNT/A to assess their genetic diversity and compare their pulsotypes with the DNA banding patterns of the outbreak strains.
Table 1.
Detection of BoNT in carrot juice and carrot juice enrichment and isolation of botulinum neurotoxigenic isolates from carrot juice.
| Bottle | Lot code | BoNT serotype detected by ELISA | |||
|---|---|---|---|---|---|
| Carrot juice | Carrot juice enrichmenta | Isolates obtained | Toxigenic isolates | ||
| 1 | 451500 | BoNT/A | –b | No | – |
| 2 | 451500 | – | – | Yes | No |
| 3 | 451800 | – | – | Yes | No |
| 4 | 451800 | BoNT/A | BoNT/A | Yes | Yes |
| 5 | 450000 | BoNT/A | BoNT/A | Yes | Yes |
| 6 | 450000 | BoNT/A | BoNT/A | Yes | No |
| 7 | 450100 | – | – | Yes | No |
| 8 | 451000 | – | – | Yes | No |
| 9 | 452100 | – | – | Yes | No |
| 10 | 452100 | BoNT/A | BoNT/A | Yes | Yes |
| 11–20 | 450100 | – | – | No | – |
TPGY, CMM and carrot juice incubated anaerobically for 5 days at 37 °C.
No botulinum neurotoxin or toxigenic isolates detected.
3.2. Pulsed-field gel electrophoresis (PFGE) analysis of isolates extracted from carrot juice
PFGE is a molecular subtyping tool that has been used extensively to study the genetic diversity of C. botulinum (Leclair et al., 2006; Nevas et al., 2005; Johnson et al., 2005). Several restriction enzymes have been tested and reported to produce a sufficient number of DNA fragments to allow reliable genomic analysis (Leclair et al., 2006; Nevas et al., 2005). Of those reported the enzymes SmaI and XhoI are most commonly utilized. PFGE was performed on XhoI and SmaI digested DNA samples of neurotoxigenic isolates obtained from carrot juice bottles labeled 4, 5 and 10, as well as the outbreak C. botulinum strains CDC51303 and CDC51348 for comparison (Fig. 1). The pulsotypes of isolates from carrot juice bottles 4 (CJ4-1, CJ4-3) and 10 (CJ10-1, CJ10-2) were identical to that of CDC51303 (CDC-CR1) when either SmaI or XhoI were used as the restriction endonuclease (RE). The restriction banding patterns of the isolates from bottle 5 (CJ5-1, CJ5-2) were similar to that of CDC51303 (CDC-CR1) when SmaI was used as the RE, but additional bands migrating between ~80 and 100 kb were observed. The unique restriction banding patterns exhibited by CJ5-1 and CJ5-2 were more pronounced when XhoI was used as the RE (Fig. 1) and unique DNA bands migrating to a position in the gel of ~336 kb and 130 kb were visualized. As a result isolates CJ5-1 and CJ4-1 were selected for DNA microarray analysis to identify differences in gene content between these isolates. Surprisingly, none of the isolates shared a similar pulsotype with the C. botulinum outbreak strain, CDC51348 (CDC-CR2). A single pathogenic strain is usually the cause of a particular foodborne outbreak. However, the 2006 botulism outbreak involving carrot juice is interesting because multiple serotype A strains likely contaminated the commercially prepared carrot juice as the data presented in this study suggests. Two additional botulism cases were reported in Toronto, Canada during the 2006 outbreak; however it is unknown whether PFGE was performed on any toxigenic isolates that may have been cultured from the carrot juice associated with those cases.
Fig. 1.
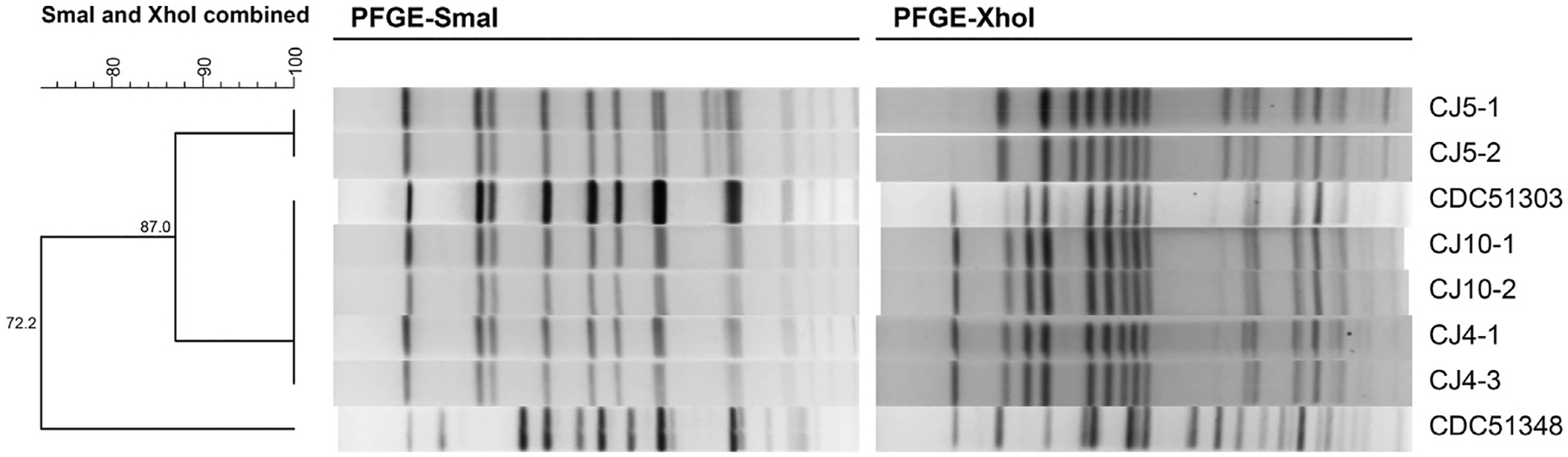
Cluster analysis of the combined SmaI and XhoI PFGE pulsotypes of botulinum neurotoxigenic isolates obtained from commercially prepared carrot juice, CDC51303, and CDC51348.
3.3. DNA microarray analysis
A custom focused oligonucleotide array which featured 225 probes (Raphael et al., 2010) was used to detect gene content differences between CJ4-1 and CJ5-1. This microarray contained probes which targeted proteolytic strain variable regions, markers for Group I C. botulinum, C. botulinum plasmid specific genes designed from the nucleotides sequences of plasmids pBot3502, pCLK, pCLI, and pCLD, the botulinum neurotoxin genes and the neurotoxin gene cluster components (Raphael et al., 2010). Six probes hybridized with the genomic DNA of CJ5-1, as indicated by a log (signal/background) ratio greater than 1.0 (Table 3). The log ratios for all six probes were below 0.5 for strain CJ4-1, suggesting that the probe sequence is absent or highly divergent for that particular strain.
PCR was performed to validate the DNA microarray results. PCR amplification of CBO_2394, CLB_2952, CLD_2430, CLD_2442, and pCLI_A009 resulted in products of the expected size when genomic DNA of CJ5-1 was used as the template, but no amplification product was observed when genomic DNA of CJ4-1 was used as the template (data not shown). PCR amplification of a region of CBO_1744 resulted in the expected 565 bp product when the gDNA of CJ5-1 and CJ4-1 was used as the template, suggesting that both strains contain a gene similar to CBO_1744, but the sequence may be highly divergent in CJ4-1 because only a faint band was visible (data not presented).
3.4. Southern hybridization analysis of C. botulinum isolates CJ4-1 and CJ5-1
To determine the genomic location of each of the six genes identified by DNA microarray analysis Southern hybridization analysis was performed on XhoI digested and nondigested DNA samples of CJ4-1 and CJ5-1 using DIG-labeled DNA probes specific for each of the six genes. PFGE of XhoI digested DNA of CJ5-1 revealed a DNA fragment of ~336 kb present in CJ5-1, but absent in CJ4-1 (Fig. 2A). Both the Okra14 (CLD_2430) and Okra17 (CLD_2442) probes hybridized with this ~336 kb DNA band in the XhoI digested DNA of CJ5-1 (Fig. 2B and C). A BLAST search was conducted for both CLD_2430 and CLD_2442 and these genes appear to be unique to C. botulinum serotype B strain, OkraB, and are located within an ~40 kb phage region of the chromosome (nucleotides 2309384–2349516). The Okra14 and Okra17 probes produced hybridization signals at the well position and at the level of sheared chromosomal DNA known as the ‘compression zone’ (Fig. 2B and C), confirming the chromosomal location of genes homologous to CLD_2430 (Okra14) and CLD_2442 (Okra17) in CJ5-1. Whether CJ5-1 contains the entire 40 kb region of phage genes found in strain OkraB is unknown at this time.
Fig. 2.
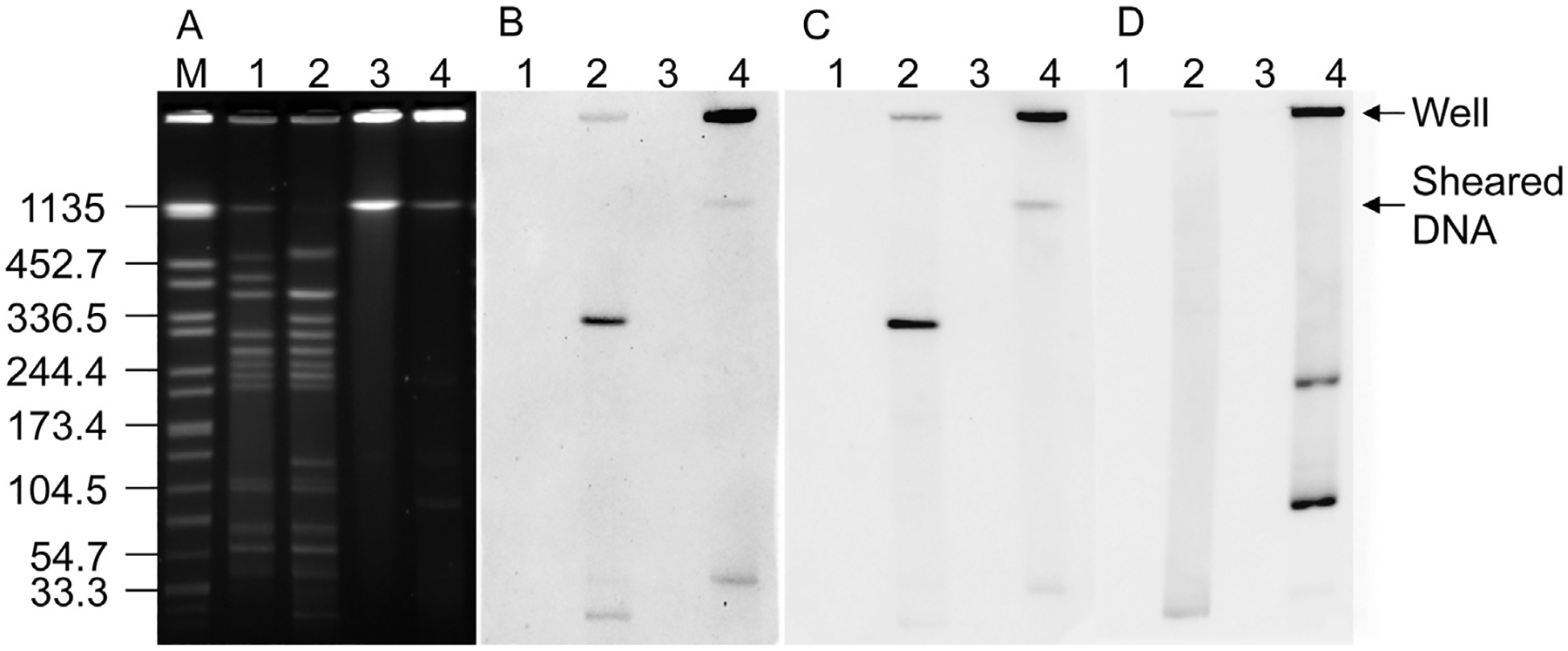
(A) PFGE of XhoI (lanes 1–2) and nondigested (lanes 3–4) DNA of Clostridium botulinum carrot juice isolates CJ4-1 (lanes 1 and 3) and CJ5-1 (lanes 2 and 4) and Southern hybridization analysis using (B) Okra17 (C) Okra14 and (D) PL7 gene specific probes.
Whole genome sequencing of C. botulinum serotype F strain, Langeland F (CP000728), revealed a 17.5 kb plasmid, pCLI (CP000729). The PL7 gene probe contains a region of the putative phage integrase gene pCLI_A009 which resides on pCLI. Faint bands are observed in the nondigested DNA sample of CJ5-1, which appear to be extrachromosomal DNA elements, and are absent in CJ4-1 (Fig. 2A). Hybridization signals were detected with the PL7 probe and the nondigested DNA of CJ5-1 at the well position and with three DNA bands of ~215 kb, 90 kb and 30 kb in size. No hybridization signals were detected with the PL7 probe and the DNA in the compression zone of nondigested DNA of CJ5-1, suggesting that these DNA bands may correspond to the relaxed, supercoiled and linear forms of the plasmid. The extent of supercoiling of plasmid DNA has been found to dramatically affect plasmid migration in PFGE (Hightower et al., 1987; Simske and Scherer, 1989). Large plasmids (>200 kb) migrate into the gel matrix during PFGE, but display a complex banding pattern of at least three DNA bands (Cole and Canard, 1997). Plasmid species migrate into the gel during PFGE in the following order from fastest to slowest; linear > supercoiled > relaxed > nicked open circular (Hightower et al., 1987). In the XhoI digested DNA of CJ5-1 hybridization signals were only detected with a DNA band of ~25 kb, which is larger than plasmid pCLI (~17.5 kb), but is likely the linear form of the plasmid because of its fast migration. The linear forms of plasmids can arise spontaneously during the extraction of the DNA from the agarose embedded bacteria during preparation of the PFGE sample plugs and have been shown to be pulse-time dependent and correspond with a linear sized marker (Cole and Canard, 1997; Hightower et al., 1987). A restriction enzyme analysis was conducted on the nucleotide sequence of pCLI using VectorNTI version 11 (Life Technologies, Carlsbad, CA), and XhoI was not found to cleave pCLI, however it is possible that the plasmid in CJ5-1 has a XhoI restriction site allowing its linearization during the restriction digestion reaction. Further studies aimed at nucleotide sequencing of the plasmid found in CJ5-1, are necessary to determine its level of similarity to pCLI.
Two prophage regions (CBO_1679–CBO_1755 and CBO_2312–CBO_2394) have been identified in C. botulinum serotype A1 strain ATCC 3502 (Sebaihia et al., 2007; Lindström et al., 2009). The BF9 probe contains a region of the putative resolvase gene (CBO_2394) of strain ATCC 3502. BLAST analysis indicated that homologues of CBO_2394 are common in proteolytic Group I C. botulinum serotype A, B and F strains. Hybridization of the BF9 probe with the XhoI digested DNA of CJ5-1 produced a signal with a ~453 kb DNA fragment (Fig. 3B). Hybridization signals were also detected at the well position and the compression zone of nondigested DNA of CJ5-1 indicating the chromosomal location of a gene homologous to CBO_2394. No hybridization of the BF9 probe was detected with the DNA of CJ4-1.
Fig. 3.
(A) PFGE of XhoI (lanes 1–2) and nondigested (lanes 3–4) DNA of Clostridium botulinum carrot juice isolates CJ4-1 (lanes 1 and 3) and CJ5-1 (lanes 2 and 4) and Southern hybridization analysis using (B) BF9 (C) CLB_2952 and (D) B28 gene specific probes.
CLB_2952 encodes for a glycosyl hydrolase and is located within a prophage region (CLB_2938 (hypothetical protein) – CLB_2998 (phage integrase)) in C. botulinum serotype A1 strain ATCC 19397. The probe for the glycosyl hydrolase gene, CLB_2952, hybridized with the ~380 kb chromosomal DNA band in the XhoI digested DNA of strain CJ5-1, but not with CJ4-1 (Fig. 3C). This suggests that strain CJ5-1 may have a similar prophage region within its genome. Whole genome sequencing of CJ5-1 would need to be completed to examine the phage related gene content of this strain more closely.
The B28 probe was amplified from CBO_1744 which is within the prophage region CBO_1679–CBO_1755 of strain ATCC 3502. Although the DNA microarrays suggested that the B28 probe (CBO_1744) was absent in CJ4-1, a region of CBO_1744 was amplified by PCR when the genomic DNA of CJ4-1 was used as the template (data not shown). Southern hybridization analysis of the XhoI digested DNA of CJ4-1 and CJ5-1 produced hybridization signals with a ~55 kb DNA fragment (Fig. 3D). However, in CJ5-1 hybridization of the B28 probe was also detected with two additional chromosomal DNA fragments (~453 kb, and ~380 kb), suggesting there are three copies of the gene in CJ5-1. CBO_1744 is a putative phage tail fiber protein and a BLASTn search indicated that CBO_2331 shared 70% nucleotide identity with CBO_1744. CBO_2331 is a putative phage tail fiber protein in prophage region CBO_2312–CBO_2391 of strain ATCC 3502. This suggests that strain CJ5-1 may contain both prophage regions that are present in strain ATCC 3502 as well as a third prophage region that may share similarity with the phage region of strain ATCC 19397.
4. Conclusions
A new serotype A strain of C. botulinum was isolated from commercially pasteurized carrot juice that was implicated in a 2006 botulism outbreak and was analyzed using pulsed-field gel electrophoresis. Several phage genes and a plasmid were identified in C. botulinum isolate CJ5-1 using DNA microarray analysis and their genomic location was revealed using Southern hybridization analysis. Based on the data presented in this study it appears that CJ5-1 potentially contains four distinct phage regions previously identified in C. botulinum strains ATCC 3502, ATCC 19397 and OkraB. Interestingly, the plasmid identified in CJ5-1 contains at least one gene that appears to be homologous to pCLI_A009 of plasmid, pCLI of strain Langeland F. The use of higher resolution molecular subtyping tools such as whole genome sequencing as a replacement to PFGE is needed to analyze strains CJ5-1 and CJ4-1 in order to investigate the degree of genetic rearrangement of CJ5-1 and to understand how bacteriophages and other mobile genetic elements play a role in the evolutionary and phylogenetic diversity of botulinum neurotoxin-producing clostridia. Throughout this study we have also identified the need for improved methods for the detection and extraction of botulinum neurotoxin-producing clostridia from a food product. Current methods involve culturing the organism which can take days and the growth and survival of C. botulinum is often outcompeted by other indigenous microflora in the food. Therefore, to improve the accurate and timely detection of a botulism outbreak improved methods which would reduce the enrichment or culturing time of C. botulinum from foods implicated in a botulism outbreak is warranted in order to perform epidemiological subtyping methods and respond to a botulism outbreak more rapidly.
Footnotes
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessary express the views of the United States Food and Drug Administration or the Centers for Disease Control and Prevention.
References
- Barash JR, Arnon SS, 2014. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J. Infect. Dis 209 (2), 183–191. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC), October 13, 2006. Botulism associated with commercial carrot juice – Georgia and Florida, September 2006. MMWR Morb. Mortal. Wkly. Rep 55, 1098–1099. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5540a5.htm. [PubMed] [Google Scholar]
- Cole ST, Canard B, 1997. Structure, organization and evolution of the genome of Clostridium perfringens. In: Rood JI, McClane BA, Songer JG, Titball RW (Eds.), The Clostridia: Molecular Biology and Pathogenesis. Academic Press, Inc., San Diego, pp. 49–64. [Google Scholar]
- Collins MD, East AK, 1998. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol 84, 5–17. [DOI] [PubMed] [Google Scholar]
- Dover N, Barash JR, Hill KK, Xie G, Arnon SS, 2014. Molecular characterization of a novel botulinum neurotoxin type H gene. J. Infect. Dis 209 (2), 192–202. [DOI] [PubMed] [Google Scholar]
- Ferreira JL, Maslanka S, Johnson E, Goodnough M, 2003. Detection of botulinal neurotoxins A, B, E, and F by amplified enzyme-linked immunosorbent assay: collaborative study. J. AOAC Int 86 (2). [PubMed] [Google Scholar]
- Franciosa G, Floridi F, Maugliani A, Aureli P, 2004. Differentiation of the gene clusters encoding botulinum neurotoxin type A complexes in Clostridium botulinum type A, Ab, and A(B) strains. Appl. Environ. Microbiol 70, 7192–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciosa G, Maugliani A, Scalfaro C, Aureli P, 2009. Evidence that plasmid-borne botulinum neurotoxin type B genes are widespread among Clostridium botulinum serotype B strains. PLoS One 4 (3), e4829. 10.1371/journal.pone.0004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez DF, Ciccarelli AS, 1978. New strains of Clostridium botulinum subtype Af. Zbl. Bakt. Hyg 240, 215–220. [PubMed] [Google Scholar]
- Hall JD, McCroskey LM, Pincomb BJ, Hatheway CL, 1985. Isolation of an organism resembling Clostridium barati which produces type F botulinal toxin from an infant with botulism. J. Clin. Microbiol 21, 654–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatheway CL, McCroskey LM, 1987. Examination of feces and serum for diagnosis of infant botulism in 336 patients. J. Clin. Microbiol 25, 2334–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatheway CL, McCroskey LM, Lombard GL, Dowell VR Jr., 1981. Atypical toxin variant of Clostridium botulinum type B associated with infant botulism. J. Clin. Microbiol 14, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower RC, Metge DW, Santi DV, 1987. Plasmid migration using orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res 15, 8387–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson RA, Zhou Y, Collins MD, Johnson EA, Hatheway CL, Sugiyama H, 1996. Genetic characterization of Clostridium botulinum type A containing silent type B neurotoxin gene sequences. J. Biol. Chem 271, 10786–10792. [DOI] [PubMed] [Google Scholar]
- Iida H, Oguma K, Inoue K, 1974. Phage-conversion of toxigenicity in Clostridium botulinum types C and D. Jpn. J. Med. Sci. Biol 27, 101–103. [PubMed] [Google Scholar]
- Johnson EA, Tepp WH, Bradshaw M, Gilbert RJ, Cook PE, McIntosh EDG, 2005. Characterization of Clostridium botulinum strains associated with an infant botulism case in the United Kingdom. J. Clin. Microbiol 43, 2602–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclair D, Pagotto F, Farber JM, Cadieux B, Austin JW, 2006. Comparison of DNA fingerprinting methods for use in investigations of type E botulism out-breaks in the Canadian arctic. J. Clin. Microbiol 44, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M, Hinderink K, Somervuo P, Kiviniemi K, Nevas M, Chen Y, Auvinen P, Carter AT, Mason DR, Peck MW, Korkeala H, 2009. Comparative genomic hybridization analysis of two predominant Nordic group I (proteolytic) Clostridium botulinum type B clusters. Appl. Environ. Microbiol 75, 2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KM, Bradshaw M, Pellett S, Johnson EA, 2007. Plasmid encoded neurotoxin genes in Clostridium botulinum serotype A subtypes. Biochem. Biophys. Res. Commun 361, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KM, Bradshaw M, Johnson EA, 2010. Conjugative botulinum neurotoxin encoding plasmids in Clostridium botulinum. PLoS One 5 (6), e11087. 10.1371/journal.pone.0011087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCroskey LM, Hatheway CL, Fenicia L, Pasolini B, Aureli P, 1986. Characterization of an organism that produces type E botulinal toxin but which resembles Clostridium butyricum from the feces of an infant with type E botulism. J. Clin. Microbiol 23, 201–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevas M, Lindstrom M, Hautamaki K, Puoskari S, Korkeala H, 2005. Prevalence and diversity of Clostridium botulinum types A, B, E and F in honey produced in the Nordic countries. Int. J. Food Microbiol 105, 145–151. [DOI] [PubMed] [Google Scholar]
- Peck MW, 2009. Biology and genomic analysis of Clostridium botulinum. In: Poole RK (Ed.), Advances in Microbial Physiology, vol. 55. Elsevier, The Netherlands, pp. 183–265. [DOI] [PubMed] [Google Scholar]
- Raphael BH, 2012. Exploring genomic diversity in Clostridium botulinum using DNA microarrays. Botulinum J 2, 99–108. [Google Scholar]
- Raphael BH, Luquez C, McCroskey LM, Joseph LA, Jacobson MJ, Johnson EA, Maslanka S, Andreadis J, 2008. Genetic homogeneity of Clostridium botulinum type A1 strains with unique toxin gene clusters. Appl. Environ. Microbiol 74, 4390–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael BH, Joseph LA, McCroskey LM, Luquez C, Maslanka S, 2010. Detection and differentiation of Clostridium botulinum type A strains using a focused DNA microarray. Mol. Cell. Probes 24, 146–153. [DOI] [PubMed] [Google Scholar]
- Reddy NR, Marshall KM, Morrissey TM, Loeza V, Patazca E, Skinner GE, Krishnamurthy K, Larkin JW, 2013. Combined high pressure and thermal processing on inactivation of type A and proteolytic type B spores of Clostridium botulinum. J. Food Prot 8, 1384–1392. [DOI] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ, 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis 3, 59–67. [DOI] [PubMed] [Google Scholar]
- Sakaguchi Y, Hayashi, Kurokawa T, Nakayama K, Oshima K, 2005. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc. Nat. Acad. Sci. U. S. A 102, 17472–17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M, Peck MW, Minton NP, Thompson NR, Holden MTG, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MHJ, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J, 2007. Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17, 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenier V, Delannoy S, Andre S, Fach P, Remize F, 2012. Prevalence of Clostridium botulinum and thermophilic heat resistant spores in raw carrots and green beans used in French canning industry. Int. J. Food Microbiol 155 (3), 263–268. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Ferreria JL, Eblen BS, Whiting RC, 2006. Detection of type A, B, E, and F Clostridium botulinum neurotoxins in foods by using an amplified enzyme-linked immunosorbent assay with digoxigenin-labeled antibodies. Appl. Environ. Microbiol 72, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth AN, Wiersma P, Atrubin D, Dubey V, Zink D, Skinner G, Doerr F, Juliao P, Gonzales G, Burnett C, Drenzek C, Shuler C, Ausin J, Ellis A, Maslanka S, Sobel J, 2008. International outbreak of severe botulism with prolonged toxemia caused by commercial carrot juice. Clin. Infect. Dis 47, 1245–1251. [DOI] [PubMed] [Google Scholar]
- Simske JS, Scherer S, 1989. Pulsed-field gel electrophoresis of circular DNA. Nucleic Acids Res 17, 4359–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Hill KK, Foley BT, Detter JC, Munk AC, Bruce DC, Doggett NA, Smith LA, Marks JD, Xie G, Brettin TS, 2007. Analysis of the neurotoxin complex genes in Clostridium botulinum A1–A4 and B1 strains: BoNT/A3, /Ba4 and /B1 clusters are located within plasmids. PLoS One 2 (12), e1271. 10.1371/journal.pone.0001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon HM, Lilly T Jr., 2001. Clostridium botulinum, chapter 17. In: Jackson GJ, Merker RI, Bandler R (Eds.), Bacteriological Analytical Manual, eighth ed. U. S. Food and Drug Administration, Center for Food Safety and Applied Nutrition, College Park, M.D. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/usm070879.htm (accessed 11.07.13.). [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B, 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Sugiyama H, Nakano H, Johson EA, 1995. The genes for Clostridium botulinum type G toxin complex are on a plasmid. Infect. Immun 63, 2087–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hintsa H, Chen Y, Korkeala H, Lindström M, 2013. Plasmid-borne type E neurotoxin gene clusters in Clostridium botulinum strains. Appl. Environ. Microbiol 79, 3856–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]


