Figure 1.
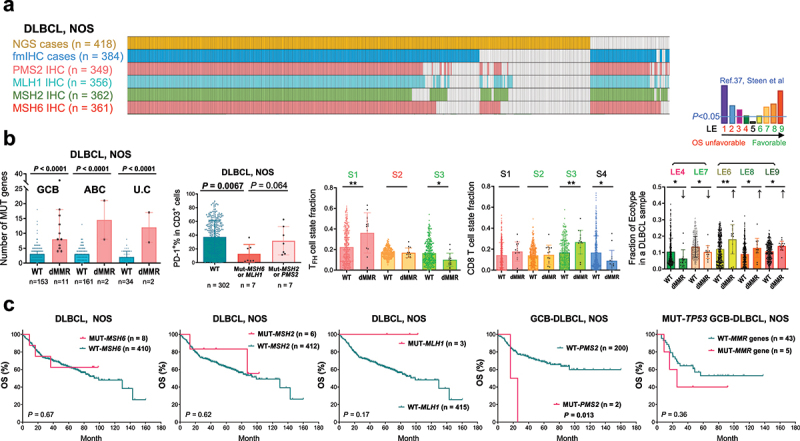
The DLBCL-NOS study cohort and analyses for mutations in four essential DNA mismatch repair (MMR) genes.
(a) Illustration of DLBCL-NOS case numbers successfully analyzed by next-generation sequencing (NGS), immunohistochemistry (IHC), and fluorescent multiplex IHC (fmIHC). Each column represents one patient. (b) Scatter plots showing the value for each patient (each dot) and mean values of patient groups (bars). MMR gene mutation (defective MMR, dMMR) was associated with increased non-silently mutated (MUT) gene numbers in GCB, ABC, and unclassifiable (U.C) subtypes of DLBCL classified by GEP. PD-1+ expression in T cells, which were quantified by fmIHC, was significantly lower in DLBCL cases with MSH6 or MLH1 mutation. Ecotyper analysis results showed that dMMR was associated with higher fractions of prognosis-favorable LE6, LE8, and LE9 ecotypes, TFH cell state S1 and CD8 T cell state S3, and lower fractions of prognosis-unfavorable LE4 ecotype, favorable LE7 ecotype, favorable TFH cell state S3, and CD8 T cell state S4. Greenish LE6-9, S1, S2 and S3 indicate favorable prognostic associations, and reddish LE4 and S2 indicate unfavorable prognostic associations according to the Ecotyper study in DLBCL by Steen et al. P values are by two-tailed unpaired t test. Significance: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (c) MSH6, MSH2, and MLH1 mutations had no significant prognostic impact in patients with DLBCL-NOS. PMS2 mutations were associated with significant poorer OS in the GCB subtype (and the overall DLBCL-NOS cohort; figure not shown). In the subset of patients with GCB-DLBCL and TP53 gene mutation, combined dMMR patients showed a non-significant trend of adverse prognostic effect.
