Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) is the likely infectious cause of Kaposi’s sarcoma, primary effusion lymphoma, and some cases of multicentric Castleman’s disease. Its latent nuclear antigen (LANA) is expressed in the nuclei of latently infected cells and may play a role in the persistence of episomal viral DNA in dividing cells. Here we report that LANA interacts with RING3, a nuclear protein and member of the Drosophila fsh (female sterile homeotic) family of proteins, some of which have previously been implicated in controlling gene expression. Binding of RING3 to LANA involves the ET domain, characteristic of fsh-related proteins, suggesting that this highly conserved region is involved in protein-protein interactions. The interaction between RING3 and LANA results in phosphorylation of serine and threonine residues located between amino acids 951 and 1107 in the carboxy-terminal region of LANA. However, RING3 is not itself a kinase but appears to recruit an as yet unidentified serine/threonine protein kinase into the complex which it forms with LANA.
Kaposi’s sarcoma-associated virus (KSHV) or human herpesvirus 8 (HHV-8) (6) is a type 2 gammaherpesvirus found in all forms of Kaposi’s sarcoma (KS), in primary effusion lymphoma (PEL/BCBL) (5), and in some cases of multicentric Castleman’s disease (30). KSHV is present in the endothelial and spindle (tumor) cells of KS lesions (4, 31), where it persists in a latent form with limited viral gene expression. Among the latent viral genes expressed in KS spindle cells and PEL cells is the latent nuclear antigen (LANA) encoded by the open reading frame 73 (orf73) (18, 19, 25). LANA is a large nuclear protein (>200 kDa) of 1,162 amino acids (in the case of the prototypic BC-1/HBL-6 sequence) with three distinct domains: a proline rich N-terminal domain, a long acidic internal repeat region which includes an extended leucine zipper motif, and a carboxy-terminal end containing a putative nuclear localization signal (27). In the nuclei of latently KSHV/HHV-8 infected PEL and KS spindle cells, LANA is located in subnuclear bodies of unresolved identity (12, 17, 25).
LANA may be required for the episomal replication of viral genomes (2) by tethering viral DNA to mitotic chromosomes, and some of its sequence features could suggest that LANA may also act as a viral transactivator (27). Several other gammaherpesviruses have positional homologs of the orf73 gene, although their overall sequence homology is low and an internal repeat region is not always present (1, 9, 20, 28, 37).
Here we report that LANA binds to RING3. Originally identified as an open reading frame located within the major histocompatibility complex class II region (3), the RING3 protein belongs to the Drosophila female sterile homeotic (fsh) family of proteins, which are related by the presence of two bromodomains, thought to be involved in protein-protein interactions, and an ET domain (extra terminal) of unknown function (16, 23). Three further human homologs of RING3, orfX, brdt, and HUNKI, have been identified (16, 23) [GenBank accession no. Y12059], and homologs of fsh have also been found in chickens, mice, rats, toads, Caenorhabditis elegans, and Saccharomyces cerevisiae (21, 26, 35, 36, 38). Whether RING3 is a nuclear mitogen-activated kinase with autophosphorylating properties (8, 24) is controversial (26). Drosophila (fsh) and yeast (BDF-1) members of this family have been implicated in controlling gene expression or modulating chromatin structure (7, 10, 22, 33, 34).
MATERIALS AND METHODS
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed with the Matchmaker II system plasmids (Clontech), the reporter strain PJ69-4A (14), and the Y187 strain for β-galactosidase plate assays. A 636-bp PCR fragment encoding amino acids 951 to 1162 of LANA was generated from the BCP-1 cell line by PCR with primers BD1 (5′-GACAGAATTCGATTACCCTGTTGTTAGCACA-3′) and BD3 (5′-GTGTGGATCCTTATGTCATTTCCTGTGGAGAGTC-3′) (restriction sites are underlined) and cloned into the polylinker of the GAL4 binding domain vector, pAS2-1 (Clontech), yielding plasmid pAS2-73c. A trp+ transformant in PJ69-4A, expressing the LANA/GAL4 BD fusion protein, was transformed with a human leukocyte cDNA library in the yeast vector pGAD10 (Clontech). Library plasmids were isolated (39) and cotransformed with pAS73C into Y187 (Clontech) to confirm the interaction in a β-galactosidase colony lift assay with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate. A liquid assay to measure β-galactosidase activity was performed with o-nitrophenyl-β-d-galactopyranoside (ONPG) (Clontech) as a substrate from three independent transformants, each performed in duplicate.
Recombinant baculoviruses.
Spodoptera frugiperda (Sf9) cells were maintained in Grace’s insect medium (Gibco-BRL) containing 10% fetal calf serum. The complete RING3 cDNA (2,262 bp), generated by reverse transcription-PCR with HEK 293 cell RNA and the primers R3.1 (5′-CTCAGATCTGGCTTCGGTGCCTGCTTTG-3′) and R3.2 (5′-CTCGGATCCTTAGCCTGAGTCTGAATCACT-3′), and the EcoRI-BamHI insert from pAS-73C (encodes the C-terminal 212 amino acids) were cloned into the baculovirus transfer vector pVLH6 (a gift from Richard Marais, Institute of Cancer Research), yielding constructs pVLH6-R3 and pVLH6-73C, respectively. A baculovirus expressing full-length LANA (bp 127296 to 123808) (27) was generated by using PCR and BCP-1 DNA and primers Gs10 (5′-GAGAGATCTATTTCCCGAGGATGGCGCCC-3′) and 73p3a (5-CTCGAATTCTTATGTCATTTCCTGTGGAGA-3′) and the transfer vector pVL1392 (PharMingen). To avoid PCR-induced errors within the internal repeat region, this region and the carboxyl-terminal end of the orf73 gene was replaced with the corresponding sequence subcloned from a cosmid (GenBank accession no. AF148805) by using BamHI sites within (bp 11214) and downstream of (bp 88370) the orf73 gene. Recombinant baculoviruses were produced by using the BaculoGold transfection system (Pharmingen) and amplified by infection of Sf9 cells. For routine production of recombinant proteins, 3 × 106 cells were infected with 1 to 10 infectious units per cell of each baculovirus and harvested at 48 h postinfection.
GST fusion proteins and pull-down assays.
Glutathione S-transferase (GST) fusion proteins (29) containing different regions of LANA and RING3 (see Fig. 2 and 3) were produced by using pGEX-1 (Pharmacia) and appropriate PCR primers (details available from the authors on request). Sf9 cells infected with the appropriate recombinant baculovirus or uninfected cells were harvested, washed in phosphate-buffered saline, lysed in 150 μl of lysis buffer (50 mM Tris [pH 7.8], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 [NP-40], 50 μM leupeptin, 1.0 μM pepstatin A, 200 μM benzamidine, 1.0 mM phenylmethylsulfonyl fluoride [PMSF]), incubated on ice for 10 min, and centrifuged (10 min at 13,000 × g and 4°C). Cleared lysates were diluted in 1 ml of binding buffer (lysis buffer with 0.1% NP-40), and equal volumes of lysates were mixed with equal amounts of GST-, GST-LANAC-, or GST-RING3-coated glutathione beads. Samples were incubated for 1 h at room temperature and washed three times in 1 ml of binding buffer before being resuspended in loading buffer and analyzed by Western blot analysis.
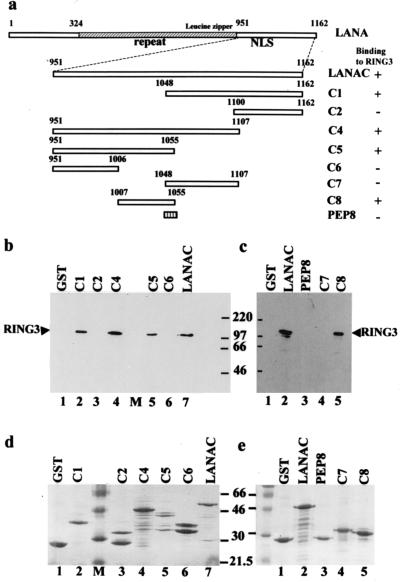
FIG. 2.
Deletion mapping of the LANA domains which interact with RING3. (a) Schematic diagram of the LANA fragments expressed as fusion proteins. The ability to bind to RING3 is summarized on the right. The positions of the internal repeat region (repeat), the nuclear localization signal (NLS), and the leucine zipper motif within LANA are indicated. LANA is numbered 1 to 1162 as in a previous study (25, 27). (b and c) Lysates prepared from Sf9 cells infected with the RING3 recombinant baculovirus were incubated with different GST-LANA deletion proteins, and binding was detected by using the affinity-purified antibody to RING3. (d and e) The GST-LANA fusion proteins used in the binding on a Coomassie blue-stained SDS-polyacrylamide gel.
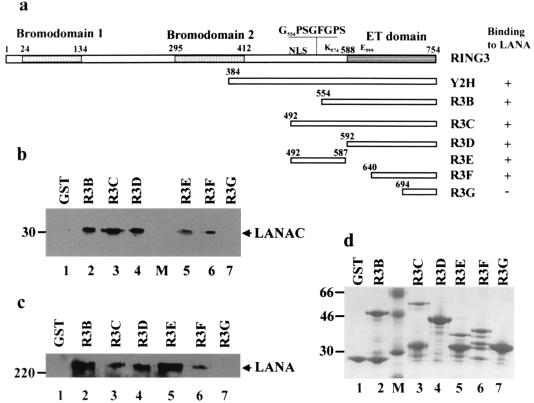
FIG. 3.
Deletion mapping of RING3 domains which interact with LANA. (a) Schematic diagram of the RING3 regions used in the construction of the GST-RING3 deletion proteins. The RING3 protein is numbered 1 to 754 as in a previous study (3). The positions of the two extended bromodomains (15), the ET domain (21), the nuclear localization signal (NLS), the postulated ATP binding motif (G554PSGFGPSG561), the catalytic lysine (K574), and the catalytic glutamic acid (E599) (8) are indicated. The ability of the GST RING3 fusion proteins to bind to LANA is summarized on the right. (b and c) Binding of LANA to the RING3 deletion proteins. Lysates of Sf9 cells expressing the recombinant c-terminal LANA fragment (LANAC) (b) or complete LANA (c) were incubated with the GST-RING3 deletion proteins immobilized on glutathione-Sepharose beads, and binding was detected on Western blots with serum from a patient with KS. The sizes of the protein markers are indicated in kilodaltons on the left. (d) The GST-RING3 fusion proteins used in the binding assays on a Coomassie blue-stained SDS-polyacrylamide gel.
Site-directed Mutagenesis.
A 1,315-bp SphI-EcoRI fragment was subcloned from pVLH6-R3 into pUC18 and mutagenized by using the Quick Change site-directed mutagenesis kit (Stratagene) and the following primers: for K574A, 5′-CCCAAAAAGGCCA CAGCGACAG CCCCACCTGCC-3′ and 5′-GGCAGGTGGGGCTGTCGCTGTGGCCTTTTTGGG-3′ (mutations are underlined); and for E599A, 5′-CCCATGAGTTACGATGCGAAGCGGCAGCTGAGC-3′ and 5′-GCTCAGCTGCC GCTTCGCATCGTAACTCATGGG-3′. Mutations were confirmed by sequencing. A mutated 1,081-bp SacI-BamHI fragment was substituted for the wild-type sequence in pVLH6-R3. To construct an in-frame deletion within the ATP binding domain, two PCRs were performed with pVLH6-R3 as a template and the following primers: reaction 1, R3.1 (5′-CTCAGATCTGGCTTCGGTGCCTGCTTTG-3′) and R3ATPRev (5′-GAGCTTGGTGCCACTTCCTC CTAAAGCAGCACTGCCACC-3′), and reaction 2, R3ATPFor (5′-GGTGGCAGTGCTGCTTTAGGAGGAAGTGGCACCAAGCTC-3′) and R3.2 (5′-CTCGGATCCTTAGC CTGAGTCTGAATCACT-3′). A 50-ng portion of the 600-bp product from reaction 1 and 50 ng of the 1,700-bp product from reaction 2 were mixed and used in a second round of PCR with primers R3.1 and R3.2. The resulting 2,238-bp BglII-BamHI fragment was cloned into pVLH6. The presence of the in-frame deletion was confirmed by sequence analysis.
Immunoprecipitation of RING3 LANA complexes.
BCP-1 cells (a KSHV/HHV-8-infected body cavity-based B-cell lymphoma cell line) were lysed (2 × 107 cells per ml) in phosphate-buffered saline containing 0.5% NP-40, 0.25% sodium deoxycholate, 0.05% sodium dodecyl sulfate (SDS), 50 μM leupeptin, 1.0 μM pepstatin A, 200 μM benzamidine, and 1.0 mM PMSF, incubated on ice for 10 min, and centrifuged at 13,000 × g for 5 min. Lysates were mixed with either rabbit anti-RING3 polyclonal antiserum or prebleed rabbit serum and incubated on ice for 1 h. The protein-antibody complexes were recovered with protein G beads (incubation at 4°C for 3 h), washed five times with 1 ml of PBS–0.1% NP-40, and resuspended in loading buffer. Samples were analyzed on Western blots.
Protein kinase assays.
Uninfected Sf9 cells and Sf9 cells infected with the recombinant RING3 baculovirus were washed twice in PBS, lysed in 150 μl of lysis buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA [pH 7.8], 25 mM β-glycerophosphate, 1 mM sodium vandate, 50 μM leupeptin, 1.0 μM pepstatin A, 200 μM benzamidine, 1.0 mM PMSF), incubated on ice for 10 min, and centrifuged (10 min at 13,000 × g and 4°C). Preimmune beads were prepared by incubating 100 μl of prewashed protein G beads with 10 μl of prebleed serum in 1 ml of binding buffer (lysis buffer with 0.2% NP-40 substituted) overnight and washing three times with binding buffer. A 15-μl volume of lysate was diluted in 500 μl of binding buffer and precleared by incubation with 20 μl of preimmune beads for 1 h. The beads were removed from the lysate by centrifugation (3,000 × g for 1 min twice). The cleared lysates were incubated with rabbit anti-RING3 antiserum or rabbit prebleed serum (45 min at 4°C) and then with protein G beads (2 h at 4°C). The beads were washed five times in 1 ml of wash buffer (lysis buffer with 0.4% NP-40 and 0.05% SDS substituted) and once in 1 ml of kinase buffer (50 mM HEPES [pH 7.4], 10 mM MgCl2, 10 mM β-glycerophosphate, 1 mM sodium vandate). The beads were resuspended in 20 μl of kinase buffer, and 10 μl was added to a kinase reaction mixture containing the above kinase buffer, 2 mM dithiothreitol, 10 μM ATP, 4 μCi of [γ-32P]ATP and 500 ng of GST or GST-LANAC fusion protein as substrates. The reaction mixtures were incubated at 30°C for 30 min before being quenched with SDS loading buffer. The reaction products were resolved on SDS–10% polyacrylamide gels, and the gels were dried and exposed to autoradiography film. Phosphoaminoacid analysis was performed on the GST-LANAC-excised gel slice essentially as previously described (40).
RESULTS
Identification of cellular proteins which interact with LANA.
To identify cellular proteins which interact with LANA, a yeast two-hybrid screen was performed with a carboxy-terminal fragment of LANA (amino acids 951 to 1162) fused to the DNA binding domain of GAL4 as bait. This region of LANA, located carboxy-terminally to the internal repeat region, contains a putative nuclear localization signal and shows some homology to the C-terminal end of orf73 positional homologs in other rhadinoviruses, suggesting a degree of functional conservation. We screened a human leukocyte cDNA library that expressed proteins fused to the GAL4 activation domain. From 3 × 106 transformants, 3 clones which activated reporter gene expression in the presence of the LANA fusion protein (but not in the presence of the unrelated fusion proteins, murine p5372–390 and human lamin C) were isolated.
Sequence analysis showed that one of the clones encoded amino acids 384 to 754 of the RING3 protein. RING3 was originally identified as an open reading frame located within the major histocompatibility complex class II region (3), although it does not have any obvious function within the immune response. The RING3 protein belongs to the Drosophila fsh subclass of proteins, related by the presence of two bromodomains and an ET domain of unknown function (21). The bromodomain is evolutionarily conserved, and although the function remains to be elucidated, it has been suggested that it may mediate protein-protein interactions (13, 15).
The interaction was further confirmed by cotransformation of the Y187 yeast strain with the above RING3 and LANA yeast two-hybrid plasmids and testing for activation of the β-galactosidase reporter gene. A colony lift assay showed that the colonies turned blue in the presence of both plasmids but not when single plasmids were present (data not shown). We attempted to quantify the strength of this interaction in a liquid β-galactosidase assay. The LANA-RING3 interaction resulted in 9.35% of the β-galactosidase activation obtained with a strong positive control (simian virus 40 [SV40]-p53 interaction), compared with a negative control (pAS2-1/pACT-2) value of 0.57% of the SV40-p53 interaction.
LANA binds to RING3.
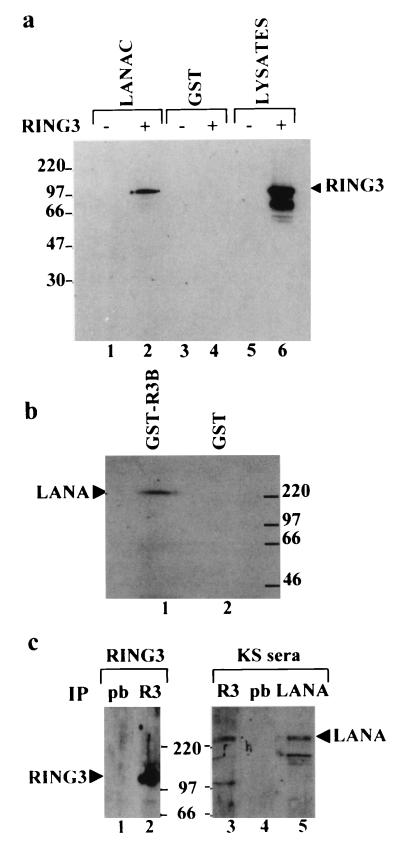
To confirm the interaction biochemically, a GST binding assay was performed. As shown in Fig. 1a, the full-length 105-kDa RING3 protein, expressed in Sf9 insect cells, binds to a recombinant GST-LANA fusion protein (GST-LANAC) containing amino acids 951 to 1162 (lane 2), but not to GST alone (lane 4). A smaller product of about 70 kDa, which reacted with an affinity-purified antibody to RING3 and could be a degradation product or an alternatively processed form of RING3, did not bind to GST-LANAC (Fig. 1a). The complete LANA protein from BCP-1 cells also binds to a recombinant GST RING3 protein (GST-R3B), containing amino acids 554 to 754 of RING3, including the ET domain (amino acids 588 to 754) (Fig. 1b, lane 1), but not to GST alone (lane 2). To investigate whether the RING3-LANA complexes can be detected in vivo, coimmunoprecipitation from BCP-1 cells was performed. Lysates prepared from BCP-1 cells were incubated with either a rabbit RING3 antibody or prebleed rabbit serum. Figure 1c shows that immunoprecipitation with the RING3 antiserum results in the detection of both RING3 (lane 2) and LANA (lane 3). This data indicates that an interaction between RING3 and LANA occurs in the KSHV/HHV-8-infected BCP-1 cell line in vivo.
FIG. 1.
Interaction of RING3 and LANA. (a) RING3 binds to the C-terminal 212 amino acids of LANA. Lysates of uninfected Sf9 cells (lanes 1 and 3) or Sf9 cells infected with the recombinant RING3 baculovirus (lanes 2 and 4) were incubated with either GST-LANAC (lanes 1 and 2) or GST alone (lanes 3 and 4) immobilized on glutathione beads. Binding of RING3 was detected by Western blot analysis with an affinity-purified rabbit antibody raised against the R3B fusion protein (Fig. 3). (b) The complete LANA protein from the BCP-1 cell line binds to RING3. Cellular lysates of BCP-1 cells were incubated with a GST RING3 fusion protein (amino acids 554 to 754) (lane 1) or GST alone (lane 2). Binding of LANA was detected on Western blots with serum from a KS patient. (c) Detection of LANA-RING3 complexes in BCP-1 cells by coimmunoprecipitation. Lysates of BCP-1 cells were incubated with either the RING3 antibody (lanes 2 and 3) or prebleed (pb) rabbit serum (lanes 1 and 4). Immunoprecipitation (IP) with a rabbit antibody raised against the C-terminal LANA region was used as a positive control for the presence of LANA (lane 5). The protein-antibody complexes were isolated by using protein G beads and analyzed by Western blotting with either a rat anti-RING3 peptide antibody (a gift from K. Thorpe, Cambridge University, Cambridge, United Kingdom) (lanes 1 and 2) or human serum from a KS patient (lanes 3 to 5). The positions of the RING3 and LANA proteins are indicated. The sizes of the protein markers are shown in kilodaltons.
Taken together, the results from the yeast two-hybrid screening, GST binding assays and coimmunoprecipitation strongly support the conclusion that LANA and the RING3 protein interact specifically both in vitro and in vivo.
Mapping of binding domains in LANA and RING3.
To define the domains within LANA which bind to RING3, we investigated the ability of a series of GST fusion proteins containing deletions within the LANA carboxy terminus (illustrated in Fig. 2a) to interact with the full-length RING3 protein expressed in insect cells. We detected binding of RING3 to deletions C1, C4, and C5 (Fig. 2b, lanes 2, 4, and 5, respectively) but not to GST (lane 1) or deletions C2 and C6 (lanes 3 and 6, respectively). RING3 binds to both C1 and C5 (amino acids 1048 to 1162 and 951 to 1055 respectively), which span the complete C-terminal region of LANA. The binding domain within C5 was further mapped to C8 (amino acids 1007 to 1055 [Fig. 2c, lane 5]), but the binding domain within C1 could not be further mapped by using C7 (amino acids 1048 to 1107 [lane 4). C1 and C5 overlap by 8 amino acids (RDPKCQWK); however, a GST fusion containing this peptide did not bind RING3 (lane 3), suggesting that the motif (RDPKCQWK) does not represent a minimal region for binding.
Taken together, these results suggest that C1 and C8, corresponding to amino acids 1048 to 1162 and 1007 to 1055, respectively, may represent two nonoverlapping binding domains within LANA which contribute to the interaction with RING3.
A similar approach was used to map the LANA binding site in the C-terminal region of RING3 (Fig. 3). A series of fragments of RING3 were expressed as GST fusion proteins (illustrated in Fig. 3a), and their ability to bind to both the full-length LANA and its C-terminal fragment (amino acids 951 to 1162) expressed in insect cells was determined. LANA (Fig. 3c) and its C-terminal 212 amino acids (Fig. 3b) bound to R3D (amino acids 592 to 754), corresponding to nearly the complete ET domain (amino acids 588 to 754), and a truncated ET domain (R3F, amino acids 640 to 754), but not the last 60 amino acids of the ET domain (R3G). A region upstream of the ET domain (amino acids 492 to 587; R3E) also contributed to this interaction.
Involvement of RING3 in phosphorylation of LANA.
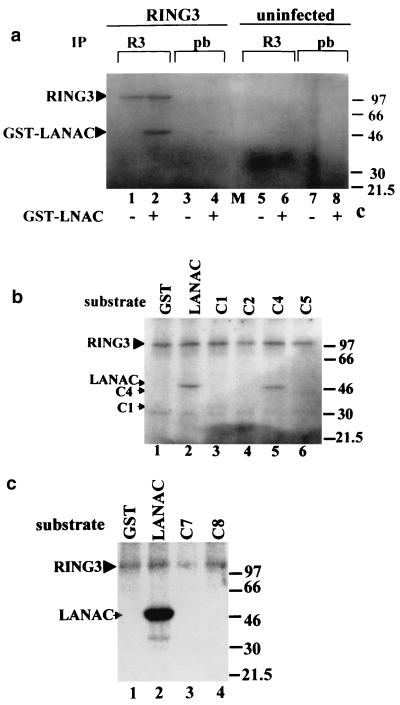
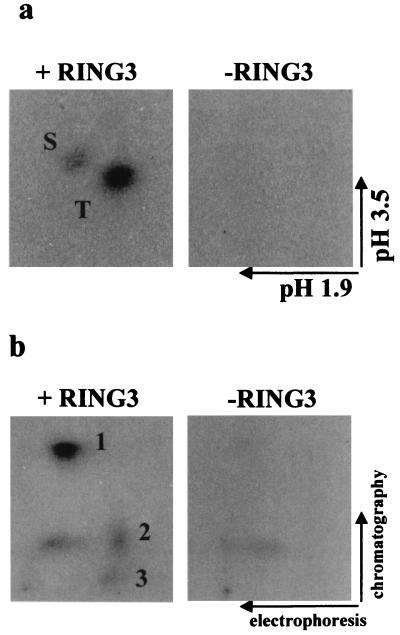
Two recent reports (8, 24) had suggested that RING3 might be a mitogen-activated nuclear serine/threonine kinase. We therefore investigated whether RING3 could phosphorylate LANA and thus perhaps be involved in the regulation of LANA function. Recombinant RING3 protein immunoprecipitated from insect cells was included in an in vitro kinase assay with GST or GST-LANAC as substrates. A 105-kDa phosphorylated protein, corresponding to the immunoprecipitated RING3 protein, was detected on autoradiographs (Fig. 4a, lanes 1 and 2), and a 47-kDa protein, corresponding to GST-LANAC, was phosphorylated strongly in the presence of the immunoprecipitated RING3 protein (lane 2). Weak background phosphorylation of GST-LANAC was observed in the absence of recombinant RING3 (lane 6), but this phosphorylation was greatly increased in the presence of immunoprecipitated RING3. To further confirm that additional sites of GST-LANAC became phosphorylated in the presence of RING3, two-dimensional phosphoamino acid and tryptic phosphoamino acid maps were generated. The phosphoamino acid map (Fig. 5a) indicates that there is increased phosphorylation of the C-terminal LANA fragment on serine and threonine residues in the presence of RING3, and the tryptic peptide map (Fig. 5b) confirms that LANA becomes phosphorylated on specific sites in the presence of RING3. The serine and threonine residues targeted by this RING3-mediated phosphorylation were further investigated by using the GST-LANA fusion proteins (Fig. 2a) as substrates. Figure 4b shows that C4 (amino acids 951 to 1107) is efficiently phosphorylated and C1 (amino acids 1048-1162) becomes weakly phosphorylated; however, C5 (amino acids 951 to 1055) is not phosphorylated. This suggests that the phosphorylation sites of the C-terminal LANA fragment are located between amino acids 951 and 1107 and possibly between amino acids 1055 and 1107. Figure 4c shows that neither C7 (amino acids 1048 to 1107) nor C8 (amino acids 1007 to 1055) becomes phosphorylated. However, since C7 does not bind to RING3 (Fig. 2c) and since binding may be required for phosphorylation, the possibility that the phosphorylation sites do lie between 1055 and 1107 cannot be ruled out.
FIG. 4.
The C-terminal domain of LANA becomes phosphorylated in the presence of RING3. (a) Cellular lysates of SF9 cells infected with the RING3 baculovirus (lanes 1 to 4) or uninfected SF9 cells (lanes 5 to 8) were immunoprecipitated (IP) with either a rabbit antibody to RING3 (lanes 1, 2, 5, and 6) or prebleed (pb) rabbit serum (lanes 3, 4, 7, and 8). The immunoprecipitated proteins were added to an in vitro kinase assay mixture containing either 500 ng of GST-LANAC (lanes 2, 4, 6, and 8) or 500 ng of GST alone (lanes 1, 3, 5, and 7) as substrates and [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography. The positions of the phosphorylated RING3 and GST-LANAC proteins are indicated, and the sizes of the protein markers are indicated in kilodaltons on the right. (b) A 500-ng portion of each of the GST LANA deletion proteins C1 (lane 3), C2 (lane 4), C4 (lane 5), and C5 (lane 6) (Fig. 2) was used as a substrate in an in vitro kinase assay in the presence of the recombinant RING3 protein. GST (lane 1) or GST-LANAC (lane 2) were used as negative and positive controls, respectively. (c) A 500-ng portion of each of the GST-LANA deletion proteins C7 (lane 3) and C8 (lane 4) were used as substrates in an in vitro kinase assay.
FIG. 5.
Phosphorylation of LANA in the presence of RING3 occurs on specific sites. GST-LANAC was phosphorylated by an in vitro kinase assay in the presence or absence of immunoprecipitated recombinant RING3, as in Fig. 4, and eluted from a gel slice (38). (a) Phosphoamino acid analysis by two-dimensional electrophoresis on cellulose-acetate sheets. The positions of the phosphoserine (S) and phosphothreonine (T) and the direction of the first (pH 1.9) and second (pH 3.5) electrophoretic dimensions are indicated. (b) Tryptic phosphopeptide map. Tryptic peptides were separated on thin-layer chromatography plates. The directions of the first (electrophoresis) and second (chromatography) dimensions are indicated. The numbers 1 to 3 denote the specific peptides observed only in the presence of immunoprecipitated RING3.
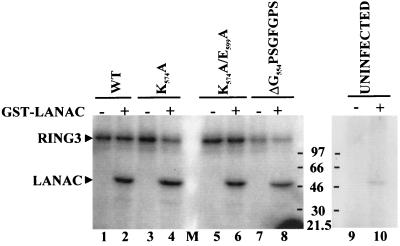
To investigate whether, as suggested previously (8, 24), the phosphorylated 105-kDa RING3 protein observed in our experiments may be due to an autophosphorylating activity of RING3, which in turn could have phosphorylated LANA, we mutagenized RING3 at predicted functionally important sites (8). A postulated catalytic lysine (K574) residue in the predicted protein kinase subdomain II was reported to be essential for RING3-mediated phosphorylation of three common substrates, MBP, kemptide, and MLCK (8). Similarly, deletion of a large section of the carboxy terminus, including the putative catalytic glutamine residue (E599) in the putative subdomain III, also eliminated the renaturable autophosphorylation activity of RING3. In addition, an ATP binding motif, G554PSGFGPSG561, had been predicted in the putative subdomain I (8) (Fig. 3a). Recombinant RING3-expressing baculoviruses containing either a single mutation (K574A), a double mutation (K574A/E599A), or an in-frame deletion within the putative ATP binding motif (G554PSGFGPS561) were constructed. Phosphorylation of the three mutant RING3 proteins (Fig. 6, lanes 2, 4, and 6) and the 47-kDa GST-LANAC fusion protein was still observed. These results strongly imply that binding of RING3 to LANA mediates phosphorylation of the latter on specific sites but that RING3 is not itself a nuclear kinase with autophosphorylation properties. Our findings suggest that an insect protein kinase interacting with RING3 is coimmunoprecipitated with RING3 and is responsible for the phosphorylation of RING3 and LANA.
FIG. 6.
Mutational analysis of the putative protein kinase subdomains of RING3. Three mutant, insect cell-expressed, immunoprecipitated RING3 proteins were added to an in vitro kinase assay mixture in the presence of either GST (lanes 1, 3, 5, and 7) or GST-LANAC (lanes 2, 4, 6, and 8), as in Fig. 4. Wild-type RING3 protein (WT) (lanes 1 and 2), mutation of the postulated catalytic lysine residue to alanine (K574A) (lanes 3 and 4), mutation of the two postulated catalytic residues K574 and E599 to alanine (K574/E599) (lanes 5 and 6), deletion of 8 amino acids within the postulated ATP binding motif (ΔG554PSGFGPS561) (lanes 7 and 8), and uninfected Sf9 lysates (lanes 9 and 10) are shown.
DISCUSSION
In this report we demonstrate that the KSHV LANA protein, encoded by orf73 and a major component of LANA of KSHV-infected cell lines, interacts with the nuclear RING3 protein. This interaction involves two domains in LANA, amino acids 1007 to 1055 and 1049 to 1162. LANA binds to RING3 within its ET domain and to an additional upstream region. The ET domain is characteristic of members of the fsh subclass of proteins (21). Our findings suggest that the ET domain may play a role in protein-protein interactions and that binding of LANA might substitute for the binding of a physiological ligand. To our knowledge, this is the first function attributed to the evolutionarily conserved ET domain. Among the four human fsh homologs, homology is particularly high in the region between amino acids 588 and 671 (72% amino acid identity). It is therefore possible that LANA also binds to the other human fsh homologues, and this is currently under investigation.
We also show that the interaction between RING3 and LANA results in the phosphorylation of LANA but that RING3 is unlikely to be a nuclear kinase, as postulated previously (8, 24). The first of these studies (8) identified RING3 as a 90-kDa autophosphorylated mitogen-inducible nuclear kinase in HeLa cell nuclear extracts. In the second study (24) an 85-kDa renaturable autophosphorylation activity was detected in mouse tissue extracts immunoprecipitated with a RING3 antibody, and the study concluded that RING3 and p85 are identical. However, a more recent study (26) did not find RING3 to be autophosphorylated but reported a 90-kDa band with autophosphorylating properties which was distinct from RING3. In the same study, RING3 had an apparent molecular mass of 110 kDa, similar to our observations.
The presence of several sequence motifs in RING3 with homology to the catalytic domain of protein kinases has suggested that RING3 might be a protein kinase (8). Mutation of the catalytic lysine (K574) in subdomain II had been reported to eliminate the ability of RING3 to phosphorylate the substrates MBP, kemptide and MLCK (8). The same authors reported that deletion of the C-terminal ET domain, including a putative catalytic glutamic acid (E599) residue in the predicted subdomain III, eliminated autophosphorylation activity. In our hands, point mutation of these two residues had no effect on the phosphorylation of RING3 or LANA. In addition, an in-frame deletion of part of a potential ATP binding motif (GXGXXG) did not affect the ability of immunoprecipitated RING3 to become phosphorylated or to mediate LANA phosphorylation. These results support the findings of Rhee et al. (26) that RING3 itself may not be a kinase. The fact that RING3 and the various mutants still become efficiently phosphorylated in our kinase assay suggests that a protein kinase present in insect cells associated with RING3 is coimmunoprecipited with the RING3 protein and consequently phosphorylates both LANA and RING3. As mentioned above, an 85-kDa phosphorylated protein was observed when mouse tissue extracts immunoprecipitated with RING3 antisera were used in an assay to detect autophosphorylation activity (24). It is possible that this phosphorylated protein is the mouse homolog of the insect kinase which forms a complex with RING3. In binding to LANA, the RING3 protein may act as a scaffold between the kinase and the viral protein, allowing the LANA protein to become phosphorylated by the kinase and thus modulating its activity. The identity of this kinase is currently under investigation.
Many of the known bromodomain-containing proteins are involved in controlling signal-dependent but not basal transcription (15, 33, 34). The Drosophila, human (hBrg1), and mouse (mBrg1) brahma proteins form part of the SWI/SNF class of transcription factors which exist within large multiprotein complexes and play a role in chromatin remodelling (10). Very little is known about the precise function of the fsh subclass of proteins, although it has been speculated that they may also play a role in transcriptional regulation (15, 26). Maternal expression of the Drosophila gene female sterile homeotic (fsh) is required for normal embryonic development (11, 13). Genetic studies in Drosophila provide evidence that the fsh protein behaves as a transacting activator of Trithorax (22), which may act by modulating the chromatin structure to alter transcriptional control (reviewed in reference 33). Another member of the fsh subclass of proteins is the product of the Saccharomyces cerevisiae BDF1 gene, which may be required for the transcription of many genes through modulating chromatin structure and for progression through the meiotic cell cycle (7, 22).
Recent evidence suggests that LANA is required for KSHV episome persistence, most probably by binding to an oriP-like region in KSHV DNA and tethering it to mitotic chromosomes (2). It is conceivable that this interaction could involve binding to RING3. Similar to LANA, the Epstein-Barr virus EBNA-1 protein is required for the propagation of viral episomes but also acts as a transcriptional activator (32). Whether LANA has similar properties that might involve an interaction with RING3 is unknown.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council of Great Britain and the North West Cancer Research Fund.
We thank John Trowsdale for helpful discussions, K. Thorpe for the antibody to a RING3 peptide, and Frederic Aurade for comments about the manuscript.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K H. Efficient persistance of extrachromosomal KSHV DNA mediated by latency associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Beck S, Hanson I, Kelly A, Pappin D J, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. doi: 10.3109/10425179209020804. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpes-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Chua P, Roeder G S. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol. 1995;15:3685–3696. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis G V, Green M R. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–71. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 9.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature (London) 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 11.Gans M, Forquignon F, Masson M. The role of dosage of the region 7D1-7D5-6 of the X chromosome in the production of homeotic transformations in Drosophila melanogaster. Genetics. 1980;96:887–902. doi: 10.1093/genetics/96.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion of antibodies to Kaposi’s sarcoma associated herpes virus related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 13.Haynes S R, Mozer B A, Bhatia-Dey N, Dawid I B. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev Biol. 1989;134:246–257. doi: 10.1016/0012-1606(89)90094-8. [DOI] [PubMed] [Google Scholar]
- 14.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeanmougin F, Wurtz J M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;5:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 16.Jones M H, Numata M, Shimane M. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics. 1997;45:529–534. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- 17.Kedes D, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroprevalence of human herpesvirus 8 (HHV8): the distribution of infection in Kaposi’s sarcoma risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 18.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen (LANA) in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 20.Lomonte P M, Bublot, van Santen V, Keil G M, Pastoret P-P, Thiry E. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gamma herpesvirus gene blocks. J Gen Virol. 1995;76:1835–1841. doi: 10.1099/0022-1317-76-7-1835. [DOI] [PubMed] [Google Scholar]
- 21.Lygerou Z, Conesa C, Lesage P, Swanson R N, Ruet A, Carlson M, Sentenac A, Seraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mozer B A, Dawid I B. Cloning and molecular characterization of the trithorax locus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86:3738. doi: 10.1073/pnas.86.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomura N, Nagase T, Miyajima N, Sazuka T, Tanaka A, Sato S, Seki N, Kawarabayasi Y, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041-KIAA0080) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1994;1:223–229. doi: 10.1093/dnares/1.5.223. [DOI] [PubMed] [Google Scholar]
- 24.Ostrowski J, Florio S K, Denis G V, Suzuki H, Bomsztyk K. Stimulation of p85/RING3 kinase in multiple organs after systemic administration of mitogens into mice. Oncogene. 1998;16:1223–1227. doi: 10.1038/sj.onc.1201624. [DOI] [PubMed] [Google Scholar]
- 25.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee K, Brunori M, Besset V, Trousdale R, Wolgemuth D J. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J Cell Sci. 1998;111:3541–3550. doi: 10.1242/jcs.111.23.3541. [DOI] [PubMed] [Google Scholar]
- 27.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Moore P S. Nucleotide sequence of Kaposi’s sarcoma associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14868. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macque rhadinovirus with similarity to the Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 30.Soulier J, Grollet L, Olkenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 31.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamkun J W. The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- 34.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SW12. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi Y, Matsuzaka Y, Fujimoto H, Miyado K, Kohda A, Okumura K, Kimura M, Inoko H. Nucleotide sequence of the ring3 gene in the class II region of the mouse MHC and its abundant expression in testicular germ cells. Genomics. 1998;51:114–123. doi: 10.1006/geno.1998.5262. [DOI] [PubMed] [Google Scholar]
- 36.Thorpe K L, Abdulla S, Kaufman J, Trowsdale J, Beck S. Phylogeny and structure of the RING3 gene. Immunogenetics. 1996;44:391–396. doi: 10.1007/BF02602785. [DOI] [PubMed] [Google Scholar]
- 37.Virgin H W, Latreille P, Wamlsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Dibenedetto A J, Pittman R N. Genes induced in programmed cell death of neuronal PC12 cells and developing sympathetic neurons in vivo. Dev Biol. 1997;188:322–336. doi: 10.1006/dbio.1997.8655. [DOI] [PubMed] [Google Scholar]
- 39.Ward A C. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]