Abstract
Herpes simplex virus (HSV) codes for several envelope glycoproteins, including glycoprotein G-2 (gG-2) of HSV type 2 (HSV-2), which are dispensable for replication in cell culture. However, clinical isolates which are deficient in such proteins occur rarely. We describe here five clinical HSV-2 isolates which were found to be unreactive to a panel of anti-gG-2 monoclonal antibodies and therefore considered phenotypically gG-2 negative. These isolates were further examined for expression of the secreted amino-terminal and cell-associated carboxy-terminal portions of gG-2 by immunoblotting and radioimmunoprecipitation. The gG-2 gene was completely inactivated in four isolates, with no expression of the two protein products. For one isolate a normally produced secreted portion and a truncated carboxy-terminal portion of gG-2 were detected in virus-infected cell medium. Sequencing of the complete gG-2 gene identified a single insertion or deletion of guanine or cytosine nucleotides in all five strains, resulting in a premature termination codon. The frameshift mutations were localized within runs of five or more guanine or cytosine nucleotides and were dispersed throughout the gene. For the isolate for which a partially inactivated gG-2 gene was detected, the frameshift mutation was localized upstream of but adjacent to the nucleotides coding for the transmembranous region. Thus, this study demonstrates the existence of clinical HSV-2 isolates which do not express an envelope glycoprotein and identifies the underlying molecular mechanism to be a single frameshift mutation.
Glycoprotein G-2 (gG-2) of herpes simplex virus type 2 (HSV-2) is a viral envelope protein with a feature unique among HSV proteins in the form of cleavage of the gG-2 precursor during processing to a secreted amino-terminal portion (50, 51) and to a cell- and virion-associated, heavily O-glycosylated carboxy-terminal portion that constitutes the mature gG-2 (3, 11, 32, 38, 43, 51). Mature gG-2 has been identified as a major target for the human antibody response (1) and, in contrast to other HSV membrane proteins, this protein has been shown to be an ideal antigen for type-discriminating serodiagnosis (2, 19, 20, 25, 52) since only type-specific epitopes have been described (28). Conservation of the gene coding for the mature portion of gG-2 among clinical HSV-2 isolates is a factor of essential importance for the reliability of serological determination of gG-2-specific antibodies from patient sera and also for the correct typing of HSV-2 isolates by anti-gG-2 monoclonal antibodies (MAbs). However, eventual differences in the phenotypic expression of this portion of gG-2 have seldom been described among clinical isolates.
In search of HSV variants, we studied a large number of clinical isolates by investigating antibody reactivity for the purpose of determining the difference in frequency of expression of a type-specific glycoprotein C-1 (gC-1) and of a gG-2 MAb epitope (28, 39). Altogether, 13 MAb escape mutants were found among the 2,400 HSV-2 isolates tested, whereas none were detected in an equal number of HSV-1 isolates (29). This indicated that the variability of the gG-2 epitope was significantly higher than that of the gC-1 epitope. Of these 13 HSV-2 isolates, 5 were in addition unreactive with two other type-specific anti-gG-2 MAbs, which had been previously mapped to different epitopes (28), and were therefore defined as phenotypically gG-2 negative. The function of the two gG-2 protein products is not known. However, as described for other HSV genes coding for envelope proteins such as glycoprotein C (gC) (21, 58), gE (30), or gI (23), the gG-2 gene is dispensable for virus propagation in cell cultures; i.e., viable gG-2-deficient mutant viruses have been constructed in vitro (16). In contrast, isolation of clinical HSV mutants lacking a dispensable gene product occurs rarely (13, 18, 40, 41), indicating that the viral proteins contribute significant functions in the natural infection of the host. We identified here five nonimmunocompromised patients with recurrent HSV-2 infections caused by isolates which harbored a completely or partially inactivated gG-2 gene. Sequencing of the gG-2 gene identified single frameshift mutations as the molecular mechanism underlying the lack of expression of the two gG-2 protein products in four of the isolates. The same mechanism also accounted for a normally produced secreted portion and a truncated mature gG-2 in the fifth isolate.
MATERIALS AND METHODS
Cells and virus strains.
African green monkey kidney (GMK-AH1) and human epidermoid carcinoma-2 (HEp-2; ATCC CCL 23) cells were cultured in Eagle minimal essential medium supplemented with 2% calf serum and antibiotics. Rabbit kidney cells (RK13; ATCC CCL 37) were cultured in Eagle minimal essential medium supplemented with 10% fetal calf serum and antibiotics. A local wild-type HSV-2 strain, B4327UR (S. Jeansson, Göteborg, Sweden), was used as a control virus. The original specimens of vesicle fluid from five HSV-2-positive patients were passaged once in GMK-AH1 cells for production of viral stocks and kept at −70°C. All experiments, including the gene sequencing, were performed with these viral stocks.
MAbs.
Three type-specific anti-gG-2 MAbs reactive to the carboxy-terminal portion of gG-2, used as reagents in this study, were produced according to standard hybridoma techniques. The MAb epitopes have previously been localized to the following amino acids: 552PPPPEHR558 for MAb O1.B9.E5, 557HRGGPEE563 for MAb O1.C5.B2, and 579ATGLAFRTP587 for MAb O3.G11.H7 (28).
EIA on HSV-2-infected cells.
In a recent study (29), 13 of 2,400 clinical HSV-2 strains isolated from patients with clinical lesions were unreactive with the anti-gG-2 MAb O1.C5.B2 when infected GMK-AH1 cells were tested by an enzyme immunoassay (EIA). In the current study these mutant strains were tested for reactivity by the same method by using the anti-gG-2 MAbs O1.B9.E5 and O3.G11.H7. Briefly, confluent monolayers of GMK-AH1 cells were infected with the respective isolate and when complete cytopathic effect was seen the cells were fixed in 0.25% glutaraldehyde for 30 min, the MAbs were added separately, and the culture was incubated for 1 h at room temperature. Alkaline phosphatase-conjugated F(ab′)2 goat anti-mouse immunoglobulin G (IgG) at a 1:2,000 dilution (Jackson ImmunoResearch Laboratories, Inc.) was used as conjugate, with p-nitrophenyl dissolved in carbonate buffer (pH 9.8) as a substrate.
DNA sequencing of the gG-2 gene.
Viral DNA was prepared from stock viruses by using the QIAmp Blood Kit (Qiagen) method prior to PCR amplification of the complete gG-2 gene. Since the gene has an overall G+C content of 71.3% (33), several sets of primers were tested to optimize the amplification and sequencing signals. Nine overlapping oligonucleotide pairs were used as primers (Table 1), and amplified products were separated on a 1% agarose gel prior to extraction of the amplicon bands with the QIAquick Gel Extraction Kit (Qiagen). PCR cycle sequencing was carried out by using fluorescent labeled stop nucleotides with the dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems), and unidirectional extension was performed with sense or antisense primers in separate reaction mixtures. After precipitation with ethanol, the labeled samples were analyzed on an automated sequencer (ABI Prism 310 Genetic Analyzer; Perkin-Elmer). The sequences were compared with the HindIII l fragment containing the gG-2 gene (US4) for the HSV-2 reference strain HG52 (33).
TABLE 1.
The primer sequences used for amplification and sequencing of the complete gG-2 gene
| Primer type and position (nt)a | Sequence |
|---|---|
| Sense primers | |
| −57–(−39) | 5′ GCACAAAAAGACGCGGCCC |
| 222–240 | 5′ CGTCGTCCGTCACGAGCCC |
| 498–517 | 5′ GCTGGTGCCGATCTGGGACC |
| 847–866 | 5′ TTTATTCGGATGGCACGACC |
| 1052–1071 | 5′ CCTCCGATTCGCCTACGTCC |
| 1372–1392 | 5′ CCCACGTCTACCCACGCGACC |
| 1530–1548 | 5′ CGCCAACGTTTCGGTCGCC |
| 1723–1740 | 5′ GACGACGACAGCGCCACC |
| 1761–1780 | 5′ GAACCCCAACAAACCACCCC |
| Antisense primers | |
| 306–287 | 5′ TGCGCCAAATCCGCGTACC |
| 600–582 | 5′ CTCCCCGCCCACCTCTACC |
| 926–907 | 5′ GGACCGTCATCTAGGGCCCC |
| 1196–1177 | 5′ GTTGCGGCTTGTGTGGCCAT |
| 1430–1413 | 5′ GGAGGGGTTGTTTGGGGCC |
| 1583–1567 | 5′ GCGGTGCCCCGGGTTCC |
| 1780–1761 | 5′ GGGGTGGTTTGTTGGGGTTC |
| 1907–1894 | 5′ TGTTGGGGTGTGGGGCCC |
| 2140–2119 | 5′ TCCCGTCCTTCATCGTTTCTC |
The nt 2842 to 4938 within the HSV-2 HindIII l fragment, encompassing the gG-2 gene (US4), for the reference HSV-2 strain HG52 (33) are numbered here as 1 to 2097.
Production of hyperimmune sera.
Rabbit hyperimmune serum was produced by using a synthetic peptide (247RFRERCLPPQTPAA260) representing part of the secreted portion of gG-2 (51). The peptide was synthesized by using Fmoc (9-fluorenylmethoxy carbonyl) chemistry, purified by high-pressure liquid chromatography (99% purity), and covalently coupled to bovine serum albumin fraction V (Sigma Chemical Co.) at an approximately 20:1 (peptide-bovine serum albumin) molar ratio by using N-succinimidyl 3-(2-pyridyldithio)propionate (Pharmacia Biotech) according to conditions given by the manufacturer. The rabbit was immunized with 200 μg of the peptide emulsified in 0.5 ml of Freund complete adjuvant for priming (first injection) and incomplete adjuvant for booster doses (second and third injections) at 3-week intervals. Rabbit hyperimmune serum directed to the mature gG-2 was prepared by immunization of a rabbit, as described earlier, with 500 μg of Helix pomatia lectin-purified gG-2 antigen (37) produced from RK13 cells infected with the B4327UR strain.
Detection of the carboxy-terminal portion of gG-2 by immunoblotting.
Cell lysate antigens from strain B4327UR and respective clinical mutant strains were prepared by infecting HEp-2 cells. When complete cytopathic effect was seen, the cells were harvested and lysed in Tris-buffered saline and 1% Nonidet P-40, followed by ultrasonication. The samples were mixed with sample buffer containing 2% sodium dodecyl sulfate (SDS) and 5% mercaptoethanol and then subjected to polyacrylamide gel electrophoresis (PAGE) by using a 10% Tris-glycine gel (Novex) and Tris-glycine-SDS as the running buffer. The proteins were electrotransferred to an Immobilon-P transfer membrane (Millipore Corp.). The gG-2-reactive MAb O1.C5.B2 and a type-common anti-gD MAb C4.D5 (6), at a final concentration of 16 μg/ml, were incubated overnight with strips containing the blotted HSV-2 antigen. Peroxidase-labeled rabbit anti-mouse IgG (Dako) at a 1:100 dilution was used as conjugate with 4-chloro-1-naphthol as the substrate.
Detection of the secreted portion of gG-2 by immunoblotting.
GMK-AH1 cells were infected with strain B4327UR and the respective clinical mutant strains. When complete cytopathic effect was seen, the media were harvested and centrifuged at 2,000 × g for 10 min before ultracentrifugation at 100,000 × g for 1.5 h, followed by centrifugation until dry at 5,000 × g, by using Microsep microconcentrator tubes with a 10-kDa cutoff (Filtron Skandinavia AB). Proteins were resuspended in 200 μl of phosphate-buffered saline, mixed with sample buffer as described above, separated on a 4 to 12% NuPAGE Bis-Tris gradient gel (Novex) with 2-(N-morpholino)ethanesulfonic acid–SDS as the running buffer, followed by immunoblotting to an Immobilon-P transfer membrane. Rabbit hyperimmune serum was added at a 1:20 dilution, and peroxidase-labeled goat anti-rabbit IgG (Dako) at a 1:100 dilution was used as conjugate with 4-chloro-1-naphthol as the substrate.
Amino acid sequencing of the secreted portion of gG-2.
The proposed secreted portion of gG-2 detected by immunoblotting was localized from the same Immobilon-P transfer membrane stained with Coomassie blue solution. This band was used for amino acid sequencing with an automatic sequencer (Applied Biosystems model 470A). Ten cycles were acquired in which the first eight amino acids were unambiguously determined.
Radioimmunoprecipitation.
Confluent GMK-AH1 cells in 50-mm petri dish cultures were infected with the gG-2-negative mutant strains and labeled with 40 μl of d-[6-3H]glucosamine hydrochloride (28 Ci/mmol) (Amersham Life Science) at 4 h postinfection. When complete cytopathic effect was seen, the media were harvested as described above except for the concentration step. The rabbit hyperimmune serum was mixed at a 1:100 dilution with the medium, and the antigen-antibody complexes were precipitated with Staphylococcus aureus (strain Cowan 1) solution as described previously (38). After SDS-PAGE with a 10% Tris-glycine gel as described above, the gel was soaked in amplifier (Amersham Life Science) for 15 min before it was dried overnight, and subsequent autoradiography was performed with Kodak XRP-1-Omat film.
Type-specific serology.
An indirect enzyme-linked immunosorbent assay (ELISA) designed to detect type-specific antibodies against mature gG-2 and gG-1 was performed with sera from patients from whom the gG-2-negative HSV-2 strains had been isolated. H. pomatia lectin-purified gG-2 (100 μg/ml), coated at a 1:6,000 dilution in carbonate buffer (pH 9.6) on Maxisorp microtiter plates (Nalge Nunc International), was used as the antigen for the assessment of anti-gG-2 antibodies, with peroxidase-conjugated goat anti-human IgG (Jackson ImmunoResearch) as the conjugate, at a 1:3,000 dilution, and O-phenylenediamine as the substrate as described previously (28). Similarly, a truncated recombinant-produced gG-1, at a concentration of 180 μg/ml (kindly provided by SmithKline Beecham Biologicals), was coated in phosphate-buffered saline at a 1:400 dilution. Alkaline phosphatase-conjugated goat anti-human IgG (Jackson ImmunoResearch) was used as conjugate at a 1:3,500 dilution with Sigma 104 phosphatase substrate tablets as the substrate. Sera were obtained from patient 2434 3 months after and from the other patients ≥3 years after the gG-2-negative HSV-2 isolates were collected. Endpoint titers were expressed as the reciprocal of the dilution giving an absorbance value greater than the cutoff. The cutoffs were defined as the mean absorbance values of HSV-1- and HSV-2-negative sera, respectively, plus 0.2 optical density (OD) units.
Nucleotide and protein sequence accession numbers.
The gG-2 gene sequences have been assigned GenBank accession no. AF141854, AF141855, AF141858, AF141856, and AF141857 for strains VI-2434, VI-512, VI-453, VI-147, and VI-4444, respectively. The protein sequence data reported here will appear in the SWISS-PROT Protein Data Bank under accession no. P81780.
RESULTS
gG-negative HSV-2 isolates.
Thirteen clinical HSV-2 isolates, which were earlier shown to be unreactive with the anti-gG-2 MAb O1.C5.B2 in EIA (29), were tested for reactivity with the two additional anti-gG-MAbs, O1.B9.E5 and O3.G11.H7. Eight isolates were clearly reactive with these antibodies and were shown to harbor point mutations within the anti-gG-2 MAb O1.C5.B2 epitope (unpublished data). These isolates were therefore excluded from further characterization in the present study. Five isolates showed low reactivity with all three MAbs tested (Table 2) and were considered gG-2 negative. These strains had been isolated from vesicular lesions from five different patients with variable duration of the clinical HSV-2 infection, as well as variable frequency of recurrences (Table 3). None of the patients were immunocompromised, nor were they on any medication.
TABLE 2.
Enzyme immunoassay reactivity of three anti-gG-2 MAbs to GMK-AH1 cells infected with five gG-2-negative clinical HSV-2 isolates
| Isolate | EIA reactivity (OD values) of MAba:
|
||
|---|---|---|---|
| O1.B9.E5 | O1.C5.B2 | O3.G11.H7 | |
| B4327URb | 2.58 | 2.55 | 2.36 |
| GMK-AH1 cellsc | 0.14 | 0.13 | 0.20 |
| VI-2434 | 0.18 | 0.20 | 0.16 |
| VI-512 | 0.31 | 0.31 | 0.21 |
| VI-453 | 0.18 | 0.15 | 0.16 |
| VI-147 | 0.28 | 0.22 | 0.27 |
| VI-4444 | 0.16 | 0.18 | 0.23 |
OD values were calculated as the mean values from duplicate well testing.
A local clinical HSV-2 strain was used as positive control virus.
Reactivity with uninfected cells.
TABLE 3.
Clinical characterization of five patients harboring gG-2-negative HSV-2 isolates
| Patient no., gender (age [yrs]) | Duration of recurrent HSV-2 infection | Site of lesions | Frequency of recurrences |
|---|---|---|---|
| 2434, F (34) | 4 mos | Vulva | 3/4 mos |
| 512, M (39) | 8 yrs | Penis | 4/yr |
| 453, M (26) | 4 yrs | Penis | 1/yr |
| 147, F (35) | 4 yrs | Vulva | 2/yr |
| 4444, F (51) | 3 yrs | Buttock | 1/yr |
Frameshift mutations within the gG-2 gene.
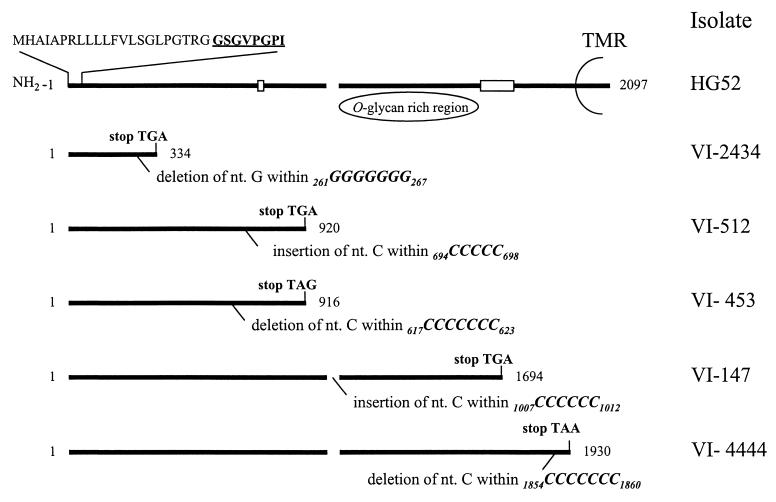
Cyclic gene sequencing of the complete gG-2 gene was performed by using overlapping sense and antisense primers. Except for a region encompassing nucleotides (nt) 518 to 614 and 604 to 906, where only one unidirectional sequence was successfully achieved, identical sequences were obtained for both the sense and the antisense primers. nt 590 to 630, in which the overlapping sequencing signals faded, showed a G+C content of 88%, which may have contributed to the difficulties resulting in termination. This region was therefore sequenced in several runs from at least three different preparations for each isolate, all showing identical sequences. The isolates harbored a single frameshift mutation with an insertion or deletion of the cytosine or guanine nucleotides compared to the previously published gG-2 gene sequence for reference HSV-2 strain HG52 (33). The mutations of the different strains were localized within runs of ≥5 guanine (one isolate) or cytosine (four isolates) nucleotides (Fig. 1). Since unequivocal determination of the precise site of the mutated base was precluded, only the stretch of reiterated nucleotides could be localized. The mutations of the different strains were found at different localities throughout the gene, and the predicted lengths of the respective truncated transcripts were therefore variable (Fig. 1).
FIG. 1.
Schematic representation of the gG-2 gene for HSV-2 reference strain HG52, coding for the secreted gG-2; the proposed cleavage sites (broken line); and the carboxy-terminal high-mannose intermediate which is highly O-glycosylated, generating the mature gG-2 and the transmembranous region (TMR). nt 2842 to 4938 within the HSV-2 HindIII l fragment of strain HG52 coding for gG-2 (33) are numbered 1 to 2097. Localization of frameshift mutations (deletion or insertion) within runs of reiterated nucleotides and the resulting premature termination codons are depicted for five clinical gG-2-negative HSV-2 isolates. Boxed areas show localization of binding of reagents utilized for detection of respective gG-2 protein products. Deduced signal sequence and sequenced amino-terminal amino acids (boldface and underlined) are aligned for the secreted gG-2.
In addition, the isolates showed the following genetic differences compared with strain HG52 (33): strain VI-2434 harbored seven single nucleotide substitutions, as well as a deletion of nucleotides 877GTC879 and 1282GCG1284. Strain VI-512 showed eight single-nucleotide substitutions and a deletion of nt 1282GCG1284. Strain VI-453 displayed seven single-nucleotide substitutions and deletion of nt 877GTC879 and 1282GCG1284. Strain VI-147 harbored 11 single-nucleotide substitutions as well as deletions of nt 1282GCG1284 and 1360ACGACCCCC1368, and, finally, strain VI-4444 contained five single-nucleotide substitutions and a deletion of nt 1282GCG1284. Taken together, the only mutations detected within the gG-2 gene which could explain the lack of reactivity with the three anti-gG-2 MAbs used in EIA were the single −1 or +1 frameshift mutations.
Expression of the gG-2 protein products by clinical HSV-2 isolates.
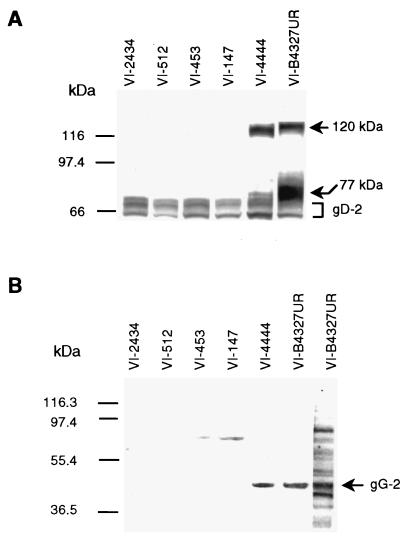
The gG-2 precursor protein was reported to be cotranslationally glycosylated, generating a high-mannose intermediate (3, 50, 55) which was cleaved during processing to a secreted and a carboxy-terminal portion (50, 51). The carboxy-terminal high-mannose intermediate was shown to be further processed by O-glycosylation to give the mature gG-2 (3, 11, 32, 38, 43, 51). Accumulation of the carboxy-terminal high-mannose intermediate was observed in HEp-2 cells (3) and was therefore detectable by immunoblotting (28). Cell lysates of HEp-2 cells infected with strain B4327UR and the gG-2-negative HSV-2 isolates were subjected to SDS-PAGE for detection by immunoblotting. For strain B4327UR, both the carboxy-terminal high-mannose intermediate with an apparent molecular mass of 77 kDa and the fully glycosylated mature gG-2 with a molecular mass of approximately 120 kDa were identified with the anti-gG-2 MAb O1.C5.B2 as described previously (28) (Fig. 2A). These two gG-2 portions were also recognized from isolate VI-4444, of which the mature gG-2 was most prominent, showing a slight reduction of the apparent molecular mass (115 kDa). No reactivity with the anti-gG-2 MAb was identified for any of the other four mutant strains. The anti-gG-2 and anti-gD MAbs were unreactive to mock-infected cell antigen (data not shown).
FIG. 2.
Immunoblot analysis of the gG-2 protein products. (A) HEp-2 cells were infected with a local wild-type HSV-2 strain (B4327UR) and five clinical gG-2-negative HSV-2 isolates. At the point of complete cytopathic effect, cell lysates were subjected to SDS-PAGE and electrotransferred to membranes. The carboxy-terminal high-mannose intermediate (77 kDa) and mature (120 kDa) gG-2 for strain B4327UR, detected by using the type-specific anti-gG-2 MAb O1.C5.B2, are marked with arrows. HSV-2 antigen was visualized by using a type-common anti-gD MAb. (B) Detection of the secreted gG-2 in GMK-AH1 virus-infected cell media by using rabbit hyperimmune serum. The upper band was reactive with rabbit preimmune serum and was considered nonspecific. The lane to the right was from the same membrane, stained with Coomassie blue solution. The arrowed band was subjected to amino acid sequencing. The positions of protein standards are indicated at the left.
To investigate the production of the normally secreted portion of gG-2, virus-infected cell medium was concentrated and subjected to SDS-PAGE. Rabbit hyperimmune serum recognized a band on immunoblotting that had an apparent molecular mass of 40 kDa for the strains B4327UR and VI-4444 (Fig. 2B), indicating that that portion of gG-2 was expressed by the isolate VI-4444. No secreted portion of gG-2 could be detected for the four other mutant strains. The rabbit hyperimmune serum was obtained after immunization with a peptide containing amino acids localized within the carboxy-terminal end of the secreted gG-2 (Fig. 1). Since the frameshift mutations detected for strains VI-2434, VI-512, and VI-453 were found to be localized upstream of the immunoreactive region, we cannot exclude the possibility that shorter fragments of the secreted gG-2 were expressed. It has been proposed that the posttranslational cleavage of gG-2 precursor protein occurs between the amino acids arginine 321 and alanine 322 and between arginine 342 and leucine 343 (10). Since strain VI-147 harbored a frameshift mutation between these cleavages sites, the nucleotides coding for residues reactive with rabbit hyperimmune serum were in frame. Despite this, no secreted gG-2 was detected, suggesting that both cleavage sites may be critical for correct processing and posttranslational cleavage of the precursor gG-2.
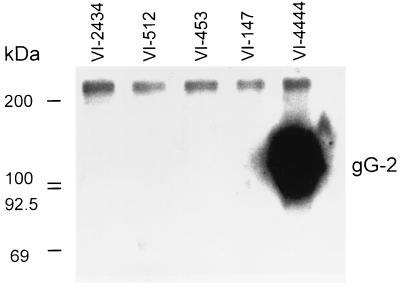
Since the frameshift mutation detected in strain VI-4444 was localized upstream of but adjacent to the nucleotides coding for the transmembranous region (Fig. 1), we investigated whether a truncated form of the mature gG-2 was secreted into the medium. When the polyclonal rabbit anti-gG-2 serum was used for radioimmunoprecipitation of the medium of cells infected with respective mutant isolates, strain VI-4444 expressed a truncated mature gG-2 with an apparent molecular mass of 116 kDa (Fig. 3). In agreement with the localization of the frameshift mutations found for the other gG-2-negative isolates, no truncated and secreted forms of the mature portion of gG-2 were detected in any of these strains.
FIG. 3.
Radioimmunoprecipitation of mature gG-2 in GMK-AH1 virus-infected cell media for five clinical gG-2-negative HSV-2 isolates. Proteins were labeled with d-[6-3H]glucosamine hydrochloride and mixed with rabbit anti-gG-2 polyclonal serum, followed by precipitation with Staphylococcus aureus solution. Antigens were separated by SDS-PAGE with subsequent autoradiography on Kodak XRP-1-Omat film. [14C]methylated proteins were used as molecular mass markers.
Amino acid sequencing of the secreted portion of gG-2.
The band of the secreted portion of gG-2 identified in the immunoblot was clearly recognized by Coomassie blue staining solution for strain B4327UR (Fig. 2B). This band was subjected to amino acid sequencing, and the N-terminal amino acids were as follows: GSGVPGPI. The residues were at positions 23 to 30 after the start codon (Fig. 1) and were identical to the deduced amino acid sequence of the gG-2 gene for strain HG52 (33). This confirmed the identification of the secreted portion of gG-2. Consequently, from the evidence presented here, the first N-terminally localized stretch of 22 residues of gG-2 appeared to constitute the signal sequence, which is in agreement with a previous proposal (33).
Seroreactivity in ELISA.
Since the mature gG-2 antigen usually is used for detection of HSV-2 type-specific antibodies as a marker of infection (2, 19, 20, 25, 52), it was of interest to assess whether sera from patients carrying the characterized viral isolates contained gG-2 antibodies. In addition, the seroreactivity to the HSV-1 type-specific gG-1 antigen was investigated in parallel. The absorbance values were expressed as endpoint titers (Table 4) in an indirect ELISA, and, as shown, three of five patients had detectable serum antibodies against gG-2.
TABLE 4.
Endpoint titers to the type-specific gG-2 and gG-1 antigens in ELISA of sera from patients harboring gG-2-negative clinical HSV-2 isolates
| Patient no. | Endpoint titera to:
|
|
|---|---|---|
| gG-2 antigenb | gG-1 antigenc | |
| 2434 | 400 | 1,600 |
| 512 | 400 | 100 |
| 453 | – | 800 |
| 147 | – | 100 |
| 4444 | 200 | 200 |
–, endpoint titer of <100.
H. pomatia lectin-purified gG-2.
Recombinant-produced gG-1.
DISCUSSION
Although point mutations of HSV genes coding for membrane glycoproteins have been described earlier in clinical HSV isolates (44, 48, 54), the complete inactivation of an HSV gene coding for a virus envelope protein in clinical HSV strains, with subsequent lack of protein expression, has to our knowledge hitherto not been reported. We describe here five gG-2-negative clinical HSV-2 isolates detected among patients with recurrent HSV-2 infection, of which four were shown to have an inactivated gG-2 gene with no expression of the gG-2 protein products. These isolates were found in the search for clinical HSV variants lacking type-specific epitopes of either gG-2 or gC-1, two glycoproteins reported to be dispensable for in vitro infection (9, 16, 17, 21). In contrast, no gC-1-negative strains were recognized among a large number of clinical HSV-1 isolates investigated in the same study (29). These results suggest that HSV-2 strains can reactivate in vivo to induce clinical lesions in immunocompetent patients despite the lack of functional gG-2 proteins, i.e., the gG-2 gene may be classified as nonessential also in vivo at least in some hosts.
An interesting question is whether these patients mostly reactivate wild-type strains expressing the two gG-2 protein products and whether the gG-2-negative strains described here thus could be regarded as single events within each host. During our further studies, two additional isolates were retrieved from patients 512 and 453, after 2 and 4 years, respectively. Sequencing of these two isolates identified frameshift mutations within the gG-2 gene identical to those described here (unpublished observation), suggesting that these gG-2-negative strains could be repeatedly reactivated in vivo. Moreover, the finding of identical frameshift mutations of these additional isolates as described for the original isolates strengthens the evidence that the detected frameshift mutations were not merely an artifact of cell culture.
The molecular basis for lack of expression of the mature gG-2 on virus-infected cells from the five clinical HSV-2 isolates investigated was found to be due to frameshift mutations. In addition, the strains harbored single nucleotide substitutions and deletions of the codons 877GTC879 (two isolates) and 1282GCG1284 (five isolates) compared to the HSV-2 reference strain HG52 (33). These alterations were also found in clinical gG-2-positive HSV-2 isolates (unpublished observation) and were therefore considered as genetic variants present in a Swedish population. The lack of nt 1360ACGACCCCC1368 detected for strain VI-147 was the only alteration which was not found in gG-2-positive isolates. Since the deletion consisted of 9 nt and consequently the reading frame was retained, it seems unlikely that this deletion could have contributed to the inactivation of the gene.
The mechanism of silenced expression or truncation of the coded protein due to frameshift mutations has been described for different microbiological agents such as yeasts (26, 47), bacteria (22, 35), and the bacteriophage T4 (42, 49), as well as for various human viruses. Single neutralization-resistant viral plaques have been selected after serial cell culture passage of a respiratory syncytial virus isolate; these were shown to harbor frameshift mutations within the G gene coding for an envelope glycoprotein (14). Such mutations have also been identified within the early region in polyomavirus after selection of revertants by the use of hydroxyurea (57). A spontaneously originated gC-negative HSV-1 mutant (MP strain) from cell culture (21) was shown to harbor a frameshift mutation within the gC-1 gene (12). Moreover, frameshift mutations responsible for the inactivation of the thymidine kinase gene have been described for clinical HSV-1 and HSV-2 isolates (15, 46) as well as for varicella-zoster virus isolates (7, 53) from immunocompromised patients. One novel observation in this study was that clinical HSV-2 strains from immunocompetent patients could harbor frameshift mutations within the gG-2 gene, coding for an envelope protein, resulting in complete inactivation of the gene. This contrasts with previously described and characterized mutants where prior selection had been exerted via in vitro cell culture conditions or where isolates were obtained from patients with severe immune system dysfunctions.
The detected frameshift mutations were all due to single-base insertion or deletion of cytosine or guanine nucleotides, introducing a premature termination codon within the gG-2 gene. In agreement with other studies, spontaneous frameshift mutations are especially prone to occur at regions of reiterated bases (4, 14, 15, 36, 46, 49, 57), and these homopolymer nucleotide stretches are usually found to be mutational hot spots. The mutations described in the present study were all localized within either oligo(C) or oligo(G) tracts, a result which may be due to the fact that the gG-2 gene has an overall high G+C content (33). The described mutations were preferably detected within runs of cytosine nucleotides (four of five mutants), which also may be due to the nucleotide composition of the gG-2 gene since the gG-2 gene contains a total of 28 reiterations of ≥5 cytosine nucleotides compared to two reiterations of ≥5 guanine repeats. The high number of such repeats may also explain why the frameshift mutations in the different isolates were found to be dispersed throughout the gene.
The biological significance of the described gG-2-negative clinical HSV-2 isolates is currently obscure, and further studies are hampered because of the hitherto unknown function of the two gG-2 protein products. The clinical characterization of the hosts with regard to the site of lesions or the frequency of recurrences did not reveal any obvious discrepancy compared to the described natural history of HSV-2 infection (5, 24). Clinical HSV-1 isolates which harbored a partially inactivated gC-1 gene and expression of a truncated gC-1 protein found in the virus-infected cell medium have been described for a patient with a recurrent eye infection (18). Since this phenotype was suggested to have maintained gC-1-associated functions both in vitro and in vivo with regard to cell penetration ability, as well as with regard to the induction of hemagglutination inhibition antibodies (34), it is possible that the secreted mature gG-2 expressed for strain VI-4444 also retained functional activity.
The detection of antibodies against the mature portion of gG-2 for patient 4444 suggests that the HSV-2 isolate VI-4444, which produced a secreted mature gG-2, had induced a B-cell immune response. Sera from patients 147 and 453 lacked antibodies against gG-2. Since this protein is used as antigen in type-discriminating serodiagnosis, it is therefore notable that a few HSV-2-infected patients can lack antibodies due to an inactivation of the gG-2 gene. Interestingly, sera from patients 2434 and 512 contained antibodies against gG-2, indicating that these patients also harbored gG-2-positive HSV-2 virus capable of inducing an antibody response to the mature gG-2. This observation implied that these patients might have carried multiple HSV-2 strains, as has been described earlier for patients infected with HSV-1 (27, 56) or with HSV-2 (8, 31, 45). However, further studies are needed to clarify the mechanisms behind this observation, and both reinfection with a gG-2-positive HSV-2 strain and the selection of different viral clones from a heterogeneous primary HSV-2 population should be considered as possible explanations.
In conclusion, this study identifies clinical HSV-2 isolates which lack the expression of a viral envelope glycoprotein due to a single frameshift mutation. These strains may prove to be valuable tools for further study of the function of the secreted as well as of the mature portion of the gG-2 protein.
ACKNOWLEDGMENTS
We thank Johan Hoebeke for assistance with the amino acid sequencing and Ann-Sofi Andersson, Carolina Gustafsson, Maria Johansson, and Anette Roth for skillful technical assistance. We also thank Nancy Nenonen for critical reading of the manuscript.
This work was supported by grants from the Medical Society of Göteborg, Swedish Medical Research Council (MFR, grant 11225), the LUA Foundation at Sahlgren’s Hospital, the Central Committee for Animal Research (CFN, Centrala Försöksdjursnämnden), and the Swedish Society for Medical Research.
REFERENCES
- 1.Ashley R L, Militoni J. Use of densitometric analysis for interpreting HSV serologies based on Western blot. J Virol Methods. 1987;18:159–168. doi: 10.1016/0166-0934(87)90121-2. [DOI] [PubMed] [Google Scholar]
- 2.Ashley R L, Militoni J, Lee F, Nahmias A, Corey L. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol. 1988;26:662–667. doi: 10.1128/jcm.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balachandran N, Hutt-Fletcher L M. Synthesis and processing of glycoprotein gG of herpes simplex virus type 2. J Virol. 1985;54:825–832. doi: 10.1128/jvi.54.3.825-832.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 5.Benedetti J K, Zeh J, Selke S, Corey L. Frequency and reactivation of nongenital lesions among patients with genital herpes simplex virus. Am J Med. 1995;98:237–242. doi: 10.1016/S0002-9343(99)80369-6. [DOI] [PubMed] [Google Scholar]
- 6.Bergström T, Sjögren-Jansson E, Jeansson S, Lycke E. Mapping neuroinvasiveness of the herpes simplex virus type 1 encephalitis-inducing strain 2762 by the use of monoclonal antibodies. Mol Cell Probes. 1992;6:41–49. doi: 10.1016/0890-8508(92)90070-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boivin G, Edelman C K, Pedneault L, Talarico C L, Biron K K, Balfour H H., Jr Phenotypic and genotypic characterization of acyclovir-resistant varicella-zoster viruses isolated from persons with AIDS. J Infect Dis. 1994;170:68–75. doi: 10.1093/infdis/170.1.68. [DOI] [PubMed] [Google Scholar]
- 8.Buchman T G, Roizman B, Nahmias A J. Demonstration of exogenous genital reinfection with herpes simplex virus type 2 by restriction endonuclease fingerprinting of viral DNA. J Infect Dis. 1979;140:295–304. doi: 10.1093/infdis/140.3.295. [DOI] [PubMed] [Google Scholar]
- 9.Cassai E, Manservigi R, Corallini A, Terni M. –1976. Plaque dissociation of herpes simplex viruses: biochemical and biological characters of the viral variants. Intervirology. 1975;6:212–223. doi: 10.1159/000149476. [DOI] [PubMed] [Google Scholar]
- 10.Courtney, R. J. Personal communication.
- 11.Dall’Olio F, Malagolini N, Campadelli-Fiume G, Serafini-Cessi F. Glycosylation pattern of herpes simplex virus type 2 glycoprotein G from precursor species to the mature form. Arch Virol. 1987;97:237–249. doi: 10.1007/BF01314424. [DOI] [PubMed] [Google Scholar]
- 12.Draper K G, Costa R H, Lee G T-Y, Spear P G, Wagner E K. Molecular basis of the glycoprotein-C-negative phenotype of herpes simplex virus type 1 macroplaque strain. J Virol. 1984;51:578–585. doi: 10.1128/jvi.51.3.578-585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman H M, Glorioso J C, Cohen G H, Hastings J C, Harris S L, Eisenberg R J. Binding of complement component C3b to glycoprotein gC of herpes simplex virus type 1: mapping of gC-binding sites and demonstration of conserved C3b binding in low-passage clinical isolates. J Virol. 1986;60:470–475. doi: 10.1128/jvi.60.2.470-475.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Barreno B, Portela A, Delgado T, Lopez J A, Melero J A. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO J. 1990;9:4181–4187. doi: 10.1002/j.1460-2075.1990.tb07642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudreau A, Hill E, Balfour H H, Jr, Erice A, Boivin G. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J Infect Dis. 1998;178:297–303. doi: 10.1086/515626. [DOI] [PubMed] [Google Scholar]
- 16.Harland J, Brown M. Generation of a herpes simplex virus type 2 variant devoid of XbaI sites: removal of the 0.91 map coordinate site results in impaired synthesis of glycoprotein G-2. J Gen Virol. 1988;69:113–124. doi: 10.1099/0022-1317-69-1-113. [DOI] [PubMed] [Google Scholar]
- 17.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hidaka Y, Sakuma S, Kumano Y, Minagawa H, Mori R. Characterization of glycoprotein C-negative mutants of herpes simplex virus type 1 isolated from a patient with keratitis. Arch Virol. 1990;113:195–207. doi: 10.1007/BF01316673. [DOI] [PubMed] [Google Scholar]
- 19.Ho D W T, Field P R, Sjögren-Jansson E, Jeansson S, Cunningham A L. Indirect ELISA for the detection of HSV-2 specific IgG and IgM antibodies with glycoprotein G (gG-2) J Virol Methods. 1992;36:249–264. doi: 10.1016/0166-0934(92)90056-j. [DOI] [PubMed] [Google Scholar]
- 20.Ho D W T, Field P R, Irving W L, Packham D R, Cunningham A L. Detection of immunoglobulin M antibodies to glycoprotein G-2 by Western blot (immunoblot) for diagnosis of initial herpes simplex virus type 2 genital infections. J Clin Microbiol. 1993;31:3157–3164. doi: 10.1128/jcm.31.12.3157-3164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoggan M D, Roizman B. The isolation and properties of a variant of herpes simplex virus producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazaki Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafferty W E, Coombs R W, Benedetti J, Critchlow C, Corey L. Recurrences after oral and genital herpes simplex virus infection. Influence of site of infection and viral type. N Engl J Med. 1987;316:1444–1449. doi: 10.1056/NEJM198706043162304. [DOI] [PubMed] [Google Scholar]
- 25.Lee F K, Coleman M, Pereira L, Bailey P D, Tatsuno M, Nahmias A J. Detection of herpes simplex virus type 2-specific antibody with glycoprotein G. J Clin Microbiol. 1985;22:641–644. doi: 10.1128/jcm.22.4.641-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemaire C, Robineau S, Netter P. Molecular and biochemical analysis of Saccharomyces cerevisiae cox1 mutants. Curr Genet. 1998;34:138–145. doi: 10.1007/s002940050378. [DOI] [PubMed] [Google Scholar]
- 27.Lewis M E, Leung W C, Jeffrey V M, Warren K G. Detection of multiple strains of latent herpes simplex virus type 1 within individual human hosts. J Virol. 1984;52:300–305. doi: 10.1128/jvi.52.1.300-305.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liljeqvist J-Å, Trybala E, Svennerholm B, Jeansson S, Sjögren-Jansson E, Bergström T. Localization of type-specific epitopes of herpes simplex virus type 2 glycoprotein G recognized by human and mouse antibodies. J Gen Virol. 1998;79:1215–1224. doi: 10.1099/0022-1317-79-5-1215. [DOI] [PubMed] [Google Scholar]
- 29.Liljeqvist J-Å, Svennerholm B, Bergström T. Typing of clinical herpes simplex virus type 1 and 2 isolates with monoclonal antibodies. J Clin Microbiol. 1999;37:2717–2718. doi: 10.1128/jcm.37.8.2717-2718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longnecker R, Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the α47 gene. J Virol. 1986;58:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maitland N J, Smith I W, Peutherer J F, Robertson D H, Jones K W. Restriction endonuclease analysis of DNA from genital isolates of herpes simplex virus type 2. Infect Immun. 1982;38:834–842. doi: 10.1128/iai.38.3.834-842.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsden H S, Buckmaster A, Palfreyman J W, Hope R G, Minson A C. Characterization of the 92,000-dalton glycoprotein induced by herpes simplex virus type 2. J Virol. 1984;50:547–554. doi: 10.1128/jvi.50.2.547-554.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGeoch D J, Moss H W M, McNab D, Frame M C. DNA sequence and genetic content of the HindIIIl region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68:19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- 34.Minagawa H, Liu Y, Yoshida T, Hidaka Y, Toh Y, Mori R. Pathogenicity of glycoprotein C-deficient herpes simplex virus 1 strain Tn-1 which encodes truncated glycoprotein C. Microbiol Immunol. 1997;41:545–551. doi: 10.1111/j.1348-0421.1997.tb01890.x. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y, Nakano Y, Yamashita Y, Koga T. Identification of a frameshift mutation resulting in premature termination and loss of cell wall anchoring of the PAc antigen of Streptococcus mutans GS-5. Infect Immun. 1997;65:794–797. doi: 10.1128/iai.65.2.794-797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada Y, Streisinger G, Owen J E, Newton J, Tsugita A, Inouye M. Molecular basis of a mutational hot spot in the lysozyme gene of bacteriophage T4. Nature. 1972;236:338–341. doi: 10.1038/236338a0. [DOI] [PubMed] [Google Scholar]
- 37.Olofsson S, Jeansson S, Lycke E. Unusual lectin-binding properties of a herpes simplex virus type 1-specific glycoprotein. J Virol. 1981;38:564–570. doi: 10.1128/jvi.38.2.564-570.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olofsson S, Lundström M, Marsden H, Jeansson S, Vahlne A. Characterization of a herpes simplex virus type 2-specified glycoprotein with affinity for N-acetylgalactosamine-specific lectins and its identification as g92K or gG. J Gen Virol. 1986;67:737–744. doi: 10.1099/0022-1317-67-4-737. [DOI] [PubMed] [Google Scholar]
- 39.Olofsson S, Bolmstedt A, Biller M, Mårdberg K, Leckner J, Malmström B G, Trybala E, Bergström T. The role of a single N-linked glycosylation site for a functional epitope of herpes simplex virus type 1 envelope glycoprotein gC. Glycobiology. 1999;9:73–81. doi: 10.1093/glycob/9.1.73. [DOI] [PubMed] [Google Scholar]
- 40.Pereira L, Cassai E, Honess R W, Roizman B, Terni M, Nahmias A. Variability in the structural polypeptides of herpes simplex virus 1 strains: potential application in molecular epidemiology. Infect Immun. 1976;13:211–220. doi: 10.1128/iai.13.1.211-220.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira L, Dondero D V, Gallo D, Devlin V, Woodie J D. Serological analysis of herpes simplex virus types 1 and 2 with monoclonal antibodies. Infect Immun. 1982;35:363–367. doi: 10.1128/iai.35.1.363-367.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripley L S, Clark A, deBoer J G. Spectrum of spontaneous frameshift mutations. Sequences of bacteriophage T4 rII gene frameshifts. J Mol Biol. 1986;191:601–613. doi: 10.1016/0022-2836(86)90448-1. [DOI] [PubMed] [Google Scholar]
- 43.Roizman B, Norrild B, Chan C, Pereira L. Identification and preliminary mapping with monoclonal antibodies of a herpes simplex virus type 2 glycoprotein lacking a known type 1 counterpart. Virology. 1984;133:242–247. doi: 10.1016/0042-6822(84)90447-1. [DOI] [PubMed] [Google Scholar]
- 44.Rozenberg F, Lebon P. Analysis of herpes simplex virus type 1 glycoprotein D nucleotide sequence in human herpes simplex encephalitis. J Neurovirol. 1996;2:289–295. doi: 10.3109/13550289609146892. [DOI] [PubMed] [Google Scholar]
- 45.Sakaoka H, Aomori T, Gouro T, Kumamoto Y. Demonstration of either endogenous recurrence or exogenous reinfection by restriction endonuclease cleavage analysis of herpes simplex virus from patients with recrudescent genital herpes. J Med Virol. 1995;46:387–396. doi: 10.1002/jmv.1890460416. [DOI] [PubMed] [Google Scholar]
- 46.Sasadeusz J J, Tufaro F, Safrin S, Schubert K, Hubinette M M, Cheung P K, Sacks S L. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schricker R, Magdolen V, Strobel G, Bogengruber E, Breitenbach M, Bandlow W. Strain-dependent occurrence of functional GTP:AMP phosphotransferase (AK3) in Saccharomyces cerevisiae. J Biol Chem. 1995;270:31103–31110. doi: 10.1074/jbc.270.52.31103. [DOI] [PubMed] [Google Scholar]
- 48.Sivadon V, Lebon P, Rozenberg F. Variations of HSV-1 glycoprotein B in human herpes simplex encephalitis. J Neurovirol. 1998;4:106–114. doi: 10.3109/13550289809113488. [DOI] [PubMed] [Google Scholar]
- 49.Streisinger G, Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su H K, Eberle R, Courtney R J. Processing of the herpes simplex virus type 2 glycoprotein gG-2 results in secretion of a 34,000-Mr cleavage product. J Virol. 1987;61:1735–1737. doi: 10.1128/jvi.61.5.1735-1737.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su H K, Fetherston J D, Smith M E, Courtney R J. Orientation of the cleavage site of the herpes simplex virus glycoprotein G-2. J Virol. 1993;67:2954–2959. doi: 10.1128/jvi.67.5.2954-2959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svennerholm B, Olofsson S, Jeansson S, Vahlne A, Lycke E. Herpes simplex virus type-selective enzyme-linked immunosorbent assay with Helix pomatia lectin-purified antigens. J Clin Microbiol. 1984;19:235–239. doi: 10.1128/jcm.19.2.235-239.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talarico C L, Phelps W C, Biron K K. Analysis of the thymidine kinase genes from acyclovir-resistant mutants of varicella-zoster virus isolated from patients with AIDS. J Virol. 1993;67:1024–1033. doi: 10.1128/jvi.67.2.1024-1033.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terhune S S, Coleman K T, Sekulovich R, Burke R L, Spear P G. Limited variability of glycoprotein gene sequences and neutralizing targets in herpes simplex virus type 2 isolates and stability on passage in cell culture. J Infect Dis. 1998;178:8–15. doi: 10.1086/515590. [DOI] [PubMed] [Google Scholar]
- 55.Weldon S K, Su H K, Fetherston J D, Courtney R J. In vitro synthesis and processing of herpes simplex virus type 2 gG-2, using cell-free transcription and translation systems. J Virol. 1990;64:1357–1359. doi: 10.1128/jvi.64.3.1357-1359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitley R, Lakeman A D, Nahmias A, Roizman B. DNA restriction-enzyme analysis of herpes simplex virus isolates obtained from patients with encephalitis. N Engl J Med. 1982;307:1060–1062. doi: 10.1056/NEJM198210213071706. [DOI] [PubMed] [Google Scholar]
- 57.Wilson J B, Hayday A, Courtneidge S, Fried M. A frameshift at a mutational hotspot in the polyoma virus early region generates two new proteins that define T-antigen functional domains. Cell. 1986;44:477–487. doi: 10.1016/0092-8674(86)90469-1. [DOI] [PubMed] [Google Scholar]
- 58.Zezulak K M, Spear P G. Mapping of the structural gene for the herpes simplex virus type 2 counterpart of herpes simplex virus type 1 glycoprotein C and identification of a type 2 mutant which does not express this glycoprotein. J Virol. 1984;49:741–747. doi: 10.1128/jvi.49.3.741-747.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]