Abstract
Introduction:
Biomarkers have been essential to understanding Alzheimer’s disease (AD) pathogenesis, pathophysiology, progression, and treatment effects. However, each biomarker measure is a representation of the biological target, the assay used to measure it, and the variance of the assay. Thus, biomarker measures are difficult to compare without standardization, and the units and magnitude of effect relative to the disease are difficult to appreciate, even for experts. To facilitate quantitative comparisons of AD biomarkers in the context of biologic and treatment effects, we propose a biomarker standardization approach between normal ranges and maximum abnormal AD ranges, which we refer to as CentiMarker, similar to the Centiloid approach used in PET.
Methods:
We developed a standardization scale that creates percentile values ranging from 0 for a normal population to 100 for the most abnormal measures across disease stages. We applied this scale to CSF and plasma biomarkers in autosomal dominant AD, assessing the distribution by estimated years from symptom onset, between biomarkers, and across cohorts. We then validated this approach in a large national sporadic AD cohort.
Results:
We found the CentiMarker scale provided an easily interpretable metric of disease abnormality. The biologic changes, range, and distribution of several AD fluid biomarkers including amyloid-β, phospho-tau and other biomarkers, were comparable across disease stages in both early onset autosomal dominant and sporadic late onset AD.
Discussion:
The CentiMarker scale offers a robust and versatile framework for the standardized biological comparison of AD biomarkers. Its broader adoption could facilitate biomarker reporting, allowing for more informed cross-study comparisons and contributing to accelerated therapeutic development.
1. Introduction
The quantification and comparison of Alzheimer’s disease (AD) biomarkers is crucial for understanding disease progression and the effectiveness of interventions. However, the current measures of fluid biomarker concentration, such as mass or moles per volume, are unique to each analyte, assay, and study.1–4 This lack of standardization makes it challenging to compare results across different studies, laboratories, and biomolecules. Therefore, standardization efforts are crucial to allow for quantitative comparison of the AD biomarkers in the context of both biologic and treatment effects.
One kind of standardization is an absolute reference standard for the molar amount of analyte per volume unit. Current practice and requirements by Health Authorities advise or require standardization of assays based on the metrology approach with traceability back to the SI-unit through certified reference materials (CRM) and reference measurement procedures for diagnostic biomarkers for clinical use, as has been done for cerebrospinal fluid (CSF) amyloid-beta 42 (Aβ42) by the International Federation of Clinical Chemistry (IFCC) Work Group.5 The final Aβ42 CRMs have also been used for re-calibrating commercial immunoassays for CSF Aβ42, including those that are FDA approved.6 However, this approach is very laborious and time-consuming.
Other standardization methods, such as z-score normalization, have been employed to facilitate comparisons of biomarkers within or across studies.7 Z-score normalization typically involves utilizing a “control” group, such as a baseline or a reference group, like young healthy controls, to convert raw values or log-transformed values into z-scores. In this case, 1 unit change in z-score represents 1 SD change in the original scale. In our study, we aim to introduce an alternative, the concept of CentiMarkers, which is designed to transform fluid biomarker values onto a scale from 0 to 100. CentiMarkers aim to provide a common metric that allows for quantitative comparisons of AD biomarkers between normal and near maximum abnormal ranges. This would be helpful for comparing the same analyte and assay across different studies and cohorts. Additionally, this would be helpful in comparing fluid biomarker values across different assays measuring the same analytes. For instance, the measurement of amyloid-β 42/40 using different antibodies, immunoassays, or mass spectrometry methods can be standardized, enabling meaningful comparisons both within and across studies.
Recent evidence from interventional trials suggests that some AD biomarkers that reach normal levels, such as achieving amyloid PET negativity, have predictive value for clinical benefit.2,3,8 Additionally, other biomarkers such as p-tau217 may be associated with cognitive impairment and response to treatment.9 However, comparing different disease cohorts, various forms of the disease, and diverse populations is often impractical due to the lack of a standardized biologic metric for fluid biomarker measurements. Having a common metric for these diverse analytes enables the interpretation of their biologic effects in relation to disease progression or response to interventions. Tracking disease progression or modification often involves monitoring multiple biomarkers, such as amyloid, tau, and neurodegeneration markers. The different AD biomarkers change sequentially over a 30-year disease span (20 years before symptom onset through 10 years after symptom onset) and may not be monotonic.10–13 Having a common metric for these diverse analytes enables the interpretation of their biologic effects in relation to disease progression or response to interventions.
The objectives of this study are the following: (i) to propose a mechanism for calculating CentiMarkers and showcase its applicability in different study cohorts; and (ii) to illustrate the usefulness of CentiMarkers values in facilitating the comparison of treatment effects across various fluid biomarkers for dominantly inherited AD (DIAD). This publication details the scope of use, methodology, contrasts approach for other purposes, addresses potential limitations, and explores alternative approaches. Future work will focus on how to generalize and apply the CentiMarker approach for other studies and purposes.
2. Methods
2.1. Study Oversight
The DIAN-TU study was conducted following the principles outlined in the Declaration of Helsinki and adhered to the guidelines set by the International Council for Harmonization and Good Clinical Practice. Ethical approval from the respective ethics committees at each participating site was obtained. Prior to participating in the study, all individuals provided written informed consent.
2.2. Study Participants
DIAN-TU-001 is a randomized, placebo-controlled, multi-arm trial of gantenerumab or solanezumab in participants with DIAD across asymptomatic and symptomatic disease stages (NCT01760005). Mutation carriers were assigned 3:1 to either drug or placebo and received treatment for 4–7 years. The information about these participants as well as the dosing schedule has been published in previous studies.1,4,8 Briefly, the trial included 193 participants, consisting of 144 mutation carriers (MCs) and 49 non-mutation carriers (NCs). The participants in the trial were either cognitively normal (Clinical Dementia Rating® [CDR = 0]) or had early-stage disease (CDR 0.5 or 1, indicating very mild or mild dementia) at the time of enrollment. The DIAN-TU trial presents a unique opportunity to utilize a subject set of young to middle-aged healthy controls. These controls consist of family members of mutation carriers without mutations (referred to as non-carriers or NCs). These NCs are young, healthy individuals without AD-related disease pathologies.
ADNI is a longitudinal study conducted in the United States focusing on sporadic Alzheimer’s disease and individuals at risk who undergo neuroimaging and biomarker evaluations. The study data utilized for our analysis ranged from 2008 to 2022. We specifically selected a subset of ADNI participants with derived estimated years from symptom onset (EYO), as reported in a previous study.14 Two fluid biomarkers, which are common to both the DIAN-TU-001 and ADNI studies, were converted into CentiMarkers to facilitate illustration and comparison.
2.3. Statistical Methods
The parameters for calculating CentiMarker values in each of the two cohorts (DIAN-TU and ADNI) were established using their respective data. Subsequently, in the re-analysis of the DIAN-TU-001 trial, we interpreted the treatment effect of gantenerumab based on the CentiMarker unit, which provided a meaningful measure for assessing the efficacy across different fluid biomarkers.
2.3.1. CentiMarker Calculation
In order to convert the raw biomarker values to CentiMarker values, we utilized a similar methodology as the Centiloid conversion approach.15 For CentiMarker calculations, more abnormal biomarker raw values indicate worse disease stages as defined by the direction of disease vs. normal groups. In summary, the process involves the following steps:
Identification of the CentiMarker-0 data set and determination of the CM-0 CentiMarker anchor value based on the CentiMarker-0 cohort.
Identification of the CentiMarker-100 data set and determination of the CM-100 CentiMarker anchor value based on the CentiMarker-100 cohort.
Calculation of the CentiMarkers.
Identification of the CentiMarker-0 data set
To establish the CentiMarker-0 anchor , all the data from the asymptomatic mutation non-carriers enrolled in the DIAN-TU-001 trial is utilized. The first step involves calculating the interquartile range (IQR) of the data. Then, any outliers that fall outside the range of or are excluded from the dataset.15 Finally, the mean of the remaining data points, was established as the CentiMarker-0 anchor for subsequent CentiMarker calculations.
Identification of the CentiMarker-100 data set
To determine the CentiMarker-100 anchor , data from all mutation carriers enrolled in the DIAN-TU trial who did not receive active treatments were used. This includes both the baseline data from participants who were assigned to the active treatment as well as the baseline and post-baseline data from those who were assigned to placebo.
The same approach applied in the identification of the CentiMarker-0 data set, is again employed to identify and remove outliers from this dataset. The CentiMarker-100 anchor, denoted as , is established as the 95th percentile of the most abnormal value across the spectrum of disease stages in the CentiMarker-100 dataset. With this approach, when the higher values indicate more severe disease stage, the 95th percentile of the most abnormal value corresponds to the 95th percentile of the highest values; when the lower values indicate more severe disease stage, the 95th percentile of the most abnormal value corresponds to the 5th percentile of the lowest values. The bootstrapping method was used to calculate the standard deviation of the 95th percentile of the most abnormal value. When a biomarker demonstrates a monotonic disease progression trajectory, higher CentiMarkers consistently indicate a more severe disease stage, irrespective of the direction of its raw values. However, for biomarkers that do not exhibit a monotonic decline (e.g., decreasing after reaching a certain disease stage), higher values of the CentiMarker do not necessarily correspond to a more severe disease stage.
Calculation of CentiMarkers
The CentiMarker is calculated as:
| (1) |
where y is the raw biomarker value, is the CentiMarker-0 anchor, is the CentiMarker-100 anchor
2.3.2. Benefit of Using the Same CentiMarker-0 Anchor Cohort
Due to the two-anchor approach involved in CentiMarker conversion, namely the 0 and 100 value, maintaining consistency in the CentiMarker-0 subject set is crucial. This not only facilitates the comparability of CentiMarker calculated using different CentiMarker-100 anchors, but also makes them exchangeable. By keeping the CentiMarker-0 cohort constant, we can utilize the same CentiMarker-0 anchor when calculating CentiMarkers values regardless how the is determined. Assuming the CentiMarkers is calculated by two different CentiMarker-100 anchors as following:
where and are the two different CentiMarker-100 anchors. Rearranging these two formulas, we have:
| (2) |
Therefore, by using the same CentiMarker-0 cohort, the method to determine the is inherently exchangeable. In this study, we selected the 95th percentile of the most abnormal value as the CentiMarker-100. CentiMarkers obtained through alternative methods, such as using the 90th percentile of the most abnormal value or the mean of all mutation carriers, can be converted into each other using formula (2).
3. Results
Figure 1 illustrates the disease progression trajectories utilizing the CentiMarker concept. In this approach, each fluid biomarker is rescaled to a range of 0 to 100, enabling a straightforward, intuitive, and direct comparison across different biomarkers.
Figure 1:
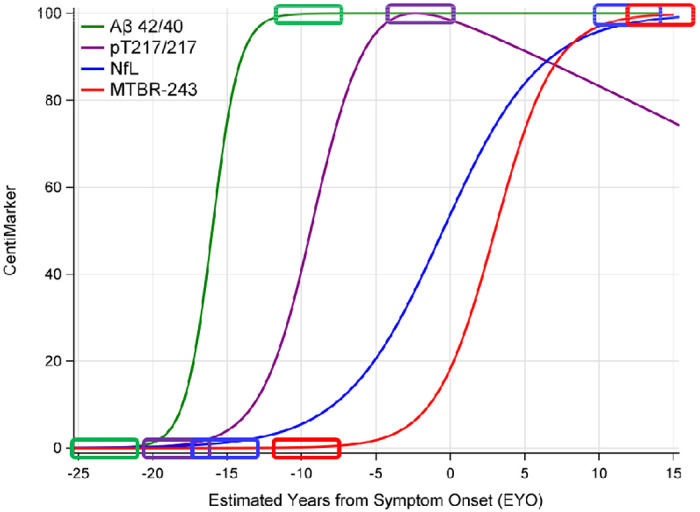
Illustration of CentiMarkers Approach. The rectangles indicate the CentiMarker 0 and CentiMarker 100 for each biomarker
Additionally, Table 1 provides the baseline demographics of all participants included in this study for each respective cohort.
Table 1:
Baseline demographics for each cohort.
| DIAN-TU 001 mutation carriers | ADNI | |||||
|---|---|---|---|---|---|---|
| Active gantenerumab N=52 | Active solanezumab N=50 | Active placebo N=40 | Controls N=69 | MCI group N=335 | AD group N=160 | |
| Age (years) | 46.0 ± 10.8 | 42.5 ± 9.5 | 44.2 ± 9.6 | 74.9 ±5.4 | 73.3 ± 7.2 | 73.9 ± 8.0 |
| Female (n(%)) | 21 (40) | 29 (58) | 22 (55) | 37 (54) | 130 (39) | 67 (42) |
| Education (years) | 14.8 ± 3.1 | 14.9 ± 2.9 | 15.5 ± 3.1 | 16.2 ± 2.6 | 15.9 ± 2.8 | 15.7 ± 2.7 |
| CDR 0 (n (%)) | 31 (60) | 30 (60) | 22 (55) | 69 (100) | 1 (0.3) | NA |
| CDR 0.5 (n (%)) | 15 (29) | 13 (26) | 15 (38) | NA | 334 (100) | 68 (43) |
| CDR 1 (n (%)) | 6 (12) | 7 (14) | 3 (8) | NA | NA | 91 (57) |
| CDR >= 2 (n (%)) | NA | NA | NA | NA | NA | 1 (0.6) |
| CDR-SB (n (%)) | 1.33 ± 2.08 | 1.37 ± 2.01 | 1.43 ± 1.87 | 0.07 ± 0.17 | 1.62 ± 0.89 | 4.52 ± 1.64 |
| MMSE | 27.10 ± 3.45 | 26.72 ± 4.11 | 26.68 ± 3.97 | 29.09 ± 1.04 | 27.58 ± 1.76 | 23.14 ± 2.09 |
3.1. Using the 95 Percentile of the Most Abnormal Value as the CentiMarker-100 Anchor
The two anchor values, CentiMarker 0 and CentiMarker 100, which were established using the DIAN-TU-001 data for each biomarker, are provided in Table 2. These values were then utilized in formula (1) to calculate the CentiMarkers for each biomarker accordingly.
Table 2:
CentiMarker-0 and CentiMarker-100 values for each biomarker within the DIAN-TU-001 population
| Biomarker | CentiMarker 0 values (Mean ± SD (n)) of NMCs | CentiMarker 100 values (95 Percentile of the Most Abnormal Score ± SD (n)) of MCs |
|---|---|---|
| NTK CSF Tau (pg/mL) | 135.20 ± 30.87 (109) | 399.90 ± 7.68 (197) |
| NTK CSF p-Tau181 (pg/mL) | 11.95 ± 2.45 (93) | 52.93 ± 1.41 (194) |
| NTK CSF Neurofilament Light Chain Protein (pg/mL) | 67.69 ± 20.33 (113) | 242.60 ± 10.27 (203) |
| NTK CSF Neurogranin (pg/mL) | 842.82 ± 223.63 (119) | 2066.00 ± 68.77 (205) |
| NTK CSF YKL-40 Protein (ng/mL) | 105.35 ± 33.71 (117) | 247.20 ± 8.71 (205) |
| NTK CSF Glial Fibrillary Acidic Protein (ng/mL) | 5.14 ± 1.88 (116) | 13.75 ± 0.80 (209) |
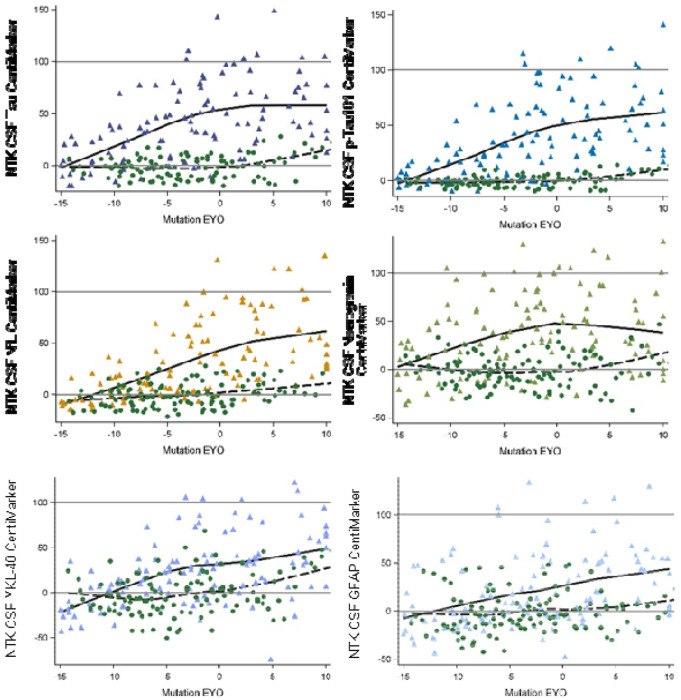
Figure 2 illustrates the CentiMarkers for some of the fluid biomarkers published within the DIAN-TU-001 trial.16 As anticipated, most CentiMarkers are observed to fall within the range of 0 to 100 and demonstrate an increasing trend as the disease progresses. Table 3 presents the means (SD) of CentiMarkers for both the CentiMarker-0 and CentiMarker-100 datasets. As anticipated, the means for the CM-0 dataset are all zero. In contrast, the means for the CentiMarker-100 dataset change as the disease advances from asymptomatic (CDR 0) to symptomatic (CDR>0). Note that not all measures go up, and some measures are not monotonic, as they increase and then decrease as the disease advances (e.g., CSF tau).11,17 By utilizing CentiMarkers in this way, the maximum and minimum abnormalities of the biomarker measures can be expressed on a common and simple scale to enable easy interpretation.11,18
Figure 2:
Comparison of different CentiMarkers in the same study by stage of disease comparing normal controls (green circles) to mutation carriers (color triangles). The mutation carriers included only the baseline data from the treatment groups and all the data from the placebo group in the DIAN-TU-001 trial. The x-axis represents the Estimated Years to Symptom Onset (EYO) based on mutation information, with zero indicating the expected onset of symptoms, covering a range of 25 years to represent disease progression.
Table 3:
Mean ± SD (N) of CentiMarkers for the CentiMarker-0 dataset and CentiMarker-100 dataset by CDR global for the DIAN-TU-001 cohort
| Biomarker | CM-0 (NMCs), mean±SD (n) | CM-100 (MCs), mean±SD (N) | |||
|---|---|---|---|---|---|
| CDR = 0 | CDR = 0 | CDR = 0.5 | CDR = 1 | CDR >= 2 | |
| NTK CSF Tau (pg/mL) | 0.00 ± 11.66 (109) | 25.31 ± 28.50 (119) | 56.72 ± 34.53 (55) | 68.39 ± 32.60 (18) | 42.37 ± 49.43 (5) |
| NTK CSF p-Tau181 (pg/mL) | 0.00 ± 5.98 (93) | 22.95 ± 26.73 (116) | 52.84 ± 30.48 (54) | 68.82 ± 33.99 (19) | 28.50 ± 35.27 (5) |
| NTK CSF neurofilament light chain (Log) | 0.00 ± 23.18 (118) | 14.55 ± 27.81 (123) | 56.64 ± 24.32 (63) | 82.65 ± 28.26 (21) | 76.60 ± 31.46 (5) |
| NTK CSF Neurogranin (pg/mL) | 0.00 ± 18.28 (119) | 24.77 ± 35.08 (123) | 45.23 ± 36.61 (59) | 52.11 ± 32.45 (18) | 11.74 ± 50.52 (5) |
| NTK CSF YKL-40 Protein (ng/mL) | 0.00 ± 23.77 (117) | 13.04 ± 31.27 (122) | 36.87 ± 37.88 (60) | 60.50 ± 38.72 (18) | 50.86 ± 45.53 (5) |
| NTK CSF Glial Fibrillary Acidic Protein (ng/mL) | 0.00 ± 21.81 (116) | 9.50 ± 32.89 (123) | 35.04 ± 39.92 (61) | 37.15 ± 28.95 (20) | 38.23 ± 58.91 (5) |
3.2. Treatment Effect in CentiMarkers
Figure 3 illustrates the change from baseline to year 4 for both the gantenerumab and the placebo groups as measured in both CentiMarker units and in raw values (as previously published16). CentiMarkers facilitate the interpretation and comparison of the treatment effect compared to the raw values. The change from baseline to year 4 in both the gantenerumab and the placebo groups are comparable in CentiMarkers across all the biomarkers, which contrasts with those using raw values.16 Supplemental Figure 1 illustrates the comparison, including GFAP. It is worth noting that GFAP exhibits larger variability in the gantenerumab group, making it less comparable to other biomarkers. However, the change observed in the placebo group remains consistent across all biomarkers, including GFAP.
Figure 3:
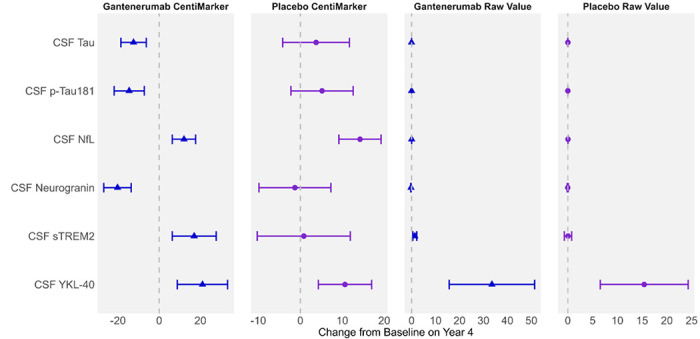
Utilizing CentiMarkers facilitates the interpretation and comparison across biomarkers by converting them to a similar scale, while using raw values is more difficult to compare as units are different and not scaled to disease ranges. Estimated mean change from baseline in CentiMarkers with 95% confidence intervals for the gantenerumab and placebo groups using MMRM analyses in the DIAN-TU-001 trial. These results demonstrate the magnitude of disease normalization compared to normal (CM 0) vs. fully abnormal (CM 100) states. Raw values are in the unit of ng/mL
Supplemental Figure 2 illustrates the change from baseline over a 4-year treatment period. The CentiMarkers clearly depict the distance of participants from the normal level at baseline and the extent to which the treatment has brought them closer to the normal level after 4 years. For instance, when considering CSF tau, the administration of gantenerumab resulted in a decrease of 12 units in CentiMarker, reducing it from 46 to 34. However, it is important to note that even at the end of the treatment, participants still remained 34 units above the normal CentiMarker value.
Figure 4 illustrates the comparison of disease progression for each biomarker across a time span of 15 years preceding symptom onset to 10 years following symptom onset. This comparison included three groups: mutation non-carriers (i.e. cognitively normal individuals), mutation carriers treated with gantenerumab, and mutation carriers not receiving gantenerumab treatment. CentiMarkers allow us to quickly identify biomarkers that exhibit a similar pattern of progression and evolve at a similar rate.
Figure 4:
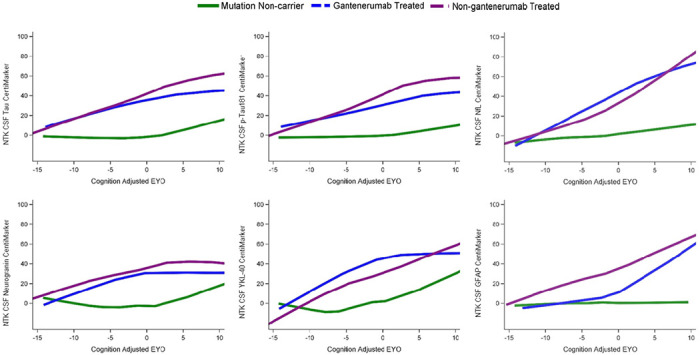
Illustration of treatment effects over EYO relative to mutation non-carriers (green line). Participants treated with gantenerumab (blue line) showed slower progression than those non-gantenerumab treated (purple line) across multiple biomarkers. The relative differences of treatment effect indicate that although there were improvements in multiple biomarkers, the measures did not reach normal CentiMarker levels for some measures.
3.3. CentiMarkers in ADNI
In the ADNI study, the calculation of CentiMarker 0 (normal cohort) incorporated datasets from the clinical normal cohort. This cohort was defined as individuals with a CDR score of 0, indicating no significant cognitive impairment. Additionally, participants in this cohort had a diagnosis status of cognitive normal and fell within the age range of 50 to 89 years old. The CentiMarker 100 cohort in the study consisted of individuals with cognitive impairment, specifically those with a CDR score greater than 0. This cohort included individuals diagnosed with mild/moderate cognitive impairment or AD and had an age range of 50 to 89 years old. The CentiMarker 0 and CentiMarker 100 for CSF total tau are 240.03 ± (80.76) and 527.50 (8.44); and 21.57 ± (7.65) and 54.28 (1.03) for CSF p-tau181. Figure 6 illustrates the CentiMarkers of the two shared fluid biomarkers collected in both the DIAN-TU and ADNI studies. Similar to the findings in the DIAN-TU studies, the CentiMarkers in the ADNI study mostly fall below 100 and tend to cluster within the range of 0 to 100.
4. Discussion
AD researchers have developed biomarkers of AD pathogenesis, pathophysiology, progression, and treatment effects that have become amongst the most advanced in both breadth and accuracy of common diseases. However, because each biomarker measure is a representation of the biological target, the assay used to measure it, and the variance of the assay, these biomarker measures are difficult to compare between biomarkers, across studies, and even within a biomarker type (e.g., amyloid-β or phospho-tau concentrations). To facilitate quantitative biological comparison of AD biomarkers in the context of biologic and treatment effects, in this paper we propose an approach, which we refer to as CentiMarker, to re-scale biomarker levels for standardization between normal ranges and maximum abnormal AD ranges similar to the Centiloid approach used for amyloid PET. Other standardization efforts are focused on reference standards to make raw values of the same biological measure comparable across labs (e.g., IFCC standardization), or harmonizing one measure across assay run and labs per Good Laboratory Practices (GLP). These standardization projects typically take years to complete, and some of the biomarkers may be hard to standardize due to molecular heterogeneity of the analyte to be standardized, which may result in non-commutable reference materials.19 In contrast, the goal of the CentiMarker standardization is to have a biologically relevant scale that relates to the disease stage from normal to abnormal to facilitate biologic interpretation. Thus, these multiple standardization approaches are useful in different contexts: comparing the same measure across labs, harmonizing across runs and labs, and comparing the magnitude of biological effects across biomarker measures in an easy-to-understand scale.
AD is a complex neurodegenerative disease characterized by the accumulation of amyloid-beta plaques and tau tangles in the brain with associated inflammation and neuronal injury. These pathological changes are often accompanied by measurable brain atrophy and cognitive decline. However, the relationship between the magnitude of change in biomarkers and the clinical symptom of AD is not linear, making it challenging to interpret the disease progression and the effects of potential interventions.
Rationale for establishing CentiMarkers:
We have developed a conceptual framework to facilitate the interpretation of biomarker changes in AD studies. This approach, called CentiMarkers, aims to mathematically transform biomarker measurements onto a standardized scale, with 0 representing normal and 100 representing nearly (95%) maximally abnormal levels, conceptionally similar but not identical to Centiloid scales used in PET imaging.15 This transformation addresses several challenges in AD biomarker research.
Firstly, it provides a more intuitive measure of the magnitude of biomarker change. This is essential for both observational studies and clinical trials, facilitating comparisons between drugs and their effects at different stages of the disease and the comparison of stage of disease across cohorts. Secondly, this approach partly addresses the lack of comparability across biomarker assays. While no standardization can provide cross-assay equivalence, it enables a reasonable approximation for understanding overall trends in biomarker dynamics. Importantly, this concept was developed through extensive discussion and collaboration with experts across research groups in biomarkers, imaging, clinical studies, and interventional trials. This manuscript introduces the CentiMarker method, offering a conceptual framework and practical application for its use in observational studies and interventional trials. The method is demonstrated using large AD cohorts from the ADNI and DIAN-TU-001 studies, providing real-world examples. We anticipate that this approach will enhance the communication of AD biomarker results, making them more readily understood by the broader research community. Wide adoption of this approach could lead to improved comparability between studies and potentially aiding in a more unified understanding of AD pathophysiology as reflected by biomarkers.
Defining normal and abnormal ranges for normalization:
The CentiMarker 0 is the mean value in a normal cohort, defined by each study. The CentiMarker approach uses the 95th percentile most abnormal value (i.e., the value defining the most abnormal 5% of the affected population) to define the CentiMarker 100, which is different from amyloid PET Centiloid, which uses the mean of individuals with early symptomatic AD to define Centiloid 100 (50% percentile). Unlike amyloid PET, which has a monotonic change until very late in the course of AD, fluid biomarkers may reach their maximum earlier in the course of AD, and importantly reach maximums at different stages of disease (see Figure 1). Therefore, CentiMarker 100 is defined by the 95th percentile because of the more complex trajectories of these biomarkers. By establishing CentiMarker 100 as the upper abnormal value for biomarkers across the entire range of disease stages and time, a maximum abnormal upper range is set, irrespective of when and how the biomarker changes or whether it exhibits non-monotonic behavior. In cases where only a small cohort is available for deriving the CentiMarker 100 value or concerns exist regarding non-representative outliers, an alternative approach can be implemented. Rather than relying on the maximum upper range, the CentiMarker 100 value can be determined using either the 90th percentile abnormal value within the available cohort or by referencing the CentiMarker 100 value reported in other studies utilizing the same assay.
To derived the CentiMarker scale,15 we recommend having a minimum of 30 data points for the CM 0 group in order to obtain a relatively accurate estimation of the mean. Through extensive bootstrapping estimation, our analysis suggests that having 30 to 50 data points for the CM 100 groups can yield a relatively stable value for the 95th most abnormal value. Notably, 50% to 70% of the bootstrapped 95th most abnormal values fall within 10% of the overall group’s 95% most abnormal value, varying depending on the biomarkers’ variability.
Another issue is that as the disease advances, some research participants become too impaired to continue in research studies. Some biomarkers (e.g., amyloid PET) may plateau, but other biomarkers (e.g., brain atrophy) likely continue to progress even after participants are too impaired to participate in studies, thus precluding defining the full disease course for CM 100. Future work would need to address this for studies that extend beyond the defining CM 0 and 100 groups.
Use of CentiMarkers:
CentiMarkers offer a solution for ensuring comparability across different biomarker measures and units, as well as accounting for disease effects. By transforming biomarker measurements onto a common scale ranging from normal to maximally abnormal, CentiMarkers provide a standardized framework for accurate comparison and assessment. This allows for a more intuitive understanding of the magnitude of biomarker changes and their relationship with the clinical symptoms of AD. For instance, a CentiMarker value of 50 would indicate that a biomarker is halfway between the normal and maximum abnormal ranges, providing a clear and straightforward interpretation of the biomarker status. Moreover, CentiMarkers can be used to compare the effects of different interventions on AD biomarkers. This is particularly important in the context of clinical trials, where multiple biomarkers are often assessed simultaneously. For instance, let’s consider a scenario where a CentiMarker originally measured 50 and after treatment, it decreased to 0. In this case, the biological measure abnormality has been effectively corrected. On the other hand, if the CentiMarker decreased from 100 to 50, despite a similar magnitude of change, the measure has not been biologically corrected. By providing a common metric for all biomarkers, CentiMarkers allow us to differentiate between treatments that fully normalize the biological measure and those that only produce partial improvement, and also facilitate direct comparisons of the effects of different biological effects.
Future Research and Application:
The application of CentiMarkers is not limited to AD research. The concept can be generalized to other neurodegenerative diseases, such as Parkinson’s disease and Huntington’s disease, and even to other fields of medicine, such as cardiovascular disease, cancer, and infectious diseases. This broad applicability makes CentiMarkers a potentially powerful tool for effective communication of biomedical research results.
Challenges and Limitations:
However, the implementation of CentiMarkers also has its challenges. It requires a well-characterized disease cohort and a normal control to define CentiMarkers. Each assay must be tested on these cohorts, which can be resource intensive. Furthermore, different ways to choose maximum and minimum abnormality may lead to different scales, limiting comparisons. Other harmonization techniques, like the use of z-scores, have been employed to facilitate direct comparisons across fluid biomarkers. However, it is important to note that the z-score approach does not incorporate disease normality into its scale. Additionally, a one-unit change in z-score corresponds to a one standard deviation (SD) change in the original raw values. Therefore, the comparability of z-scores relies on measurement variability, which may be influenced by factors such as assay precision and biological variation. Other efforts utilize certified reference standards to make individual assays or analytes comparable across studies, but this only works within an analyte, not across analytes, and again does not provide relative disease information.
Future research directions include testing the approach in more studies, cohorts, and individual biomarkers to validate and refine the method.7 Additionally, collaboration among various research groups will be crucial in establishing bridging cohorts similar to the Centiloid’s approach.20 This collaborative effort aims to facilitate the conversion of CentiMarker measurements across various cohorts and assays, thereby enhancing comparability across different fluid biomarkers. An important future goal and challenge will be to ensure researcher understanding, uptake, and utilization of CentiMarkers in their studies. The coordination and harmonization of specific approaches are necessary to enable the comparability of findings. With these combined efforts, CentiMarkers have the potential to revolutionize our understanding of AD and other diseases, expediting the development of effective treatments.
Supplementary Material
Figure 5:
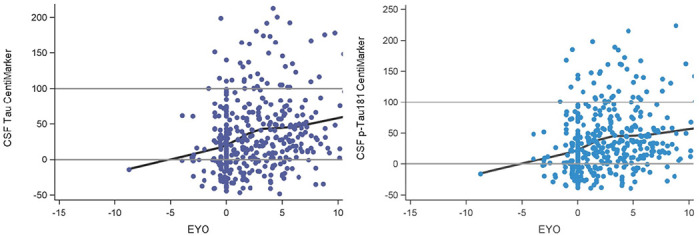
Illustration of CentiMarkers in ADNI. EYO is calculated by using the individual’s own age of symptom onset as EYO of 0. These distributions replicate increasing CSF tau and p-tau181 by stage of disease.
Acknowledgements
We gratefully acknowledge the outstanding commitment of the participants, family members, and caregivers whose participation was critical to the success of the DIAN-TU trial. We thank the DIAN-TU study team for their exceptional dedication and accomplishments, which ensured the success of the trial. We thank the DIAN-EXR and DIAN-OBS study teams for their support on recruitment and commitment to family members. We appreciate the robust intellectual collaboration between the DIAN-TU investigators, participants and family members, F. Hoffmann-La Roche, Ltd./Genentech, and Eli Lilly and Company, the DIAN-TU Pharma Consortium (https://dian.wustl.edu/our-research/the-pharma-consortium/), the NIH, and regulatory representatives who were critical in making this study possible. We thank the Alzheimer’s Association, GHR Foundation, an anonymous organization, other industry partners (Avid Radiopharmaceuticals, a wholly-owned subsidiary of Eli Lilly and Company, Signant, and Cogstate), and regulatory representatives for their support. We also acknowledge Dr. Laurie Ryan from the National Institute on Aging for her key contributions in leadership and scientific guidance on this project.
Funding Sources
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Numbers U01AG042791, U01AG042791-S1 (FNIH and Accelerating Medicines Partnership), R01AG046179, R01AG053267-S1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also supported by the Alzheimer’s Association, Eli Lilly and Company, F. Hoffman-LaRoche Ltd., Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly and Company, GHR Foundation, an anonymous organization. Cogstate, and Signant offered in-kind support. The DIAN-OBS was supported by the National Institute on Aging of the National Institutes on Aging (DIAN, U19AG032438), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), partial support by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). The methodology development of this paper is also supported by The Foundation for Barnes-Jewish Hospital, WU Institute of Clinical and Translational Sciences, and the NIH/National Center for Advancing Translational Sciences (NCATS), grant UL1 TR002345.
Disclosures/Conflicts
Guoqiao Wang, PhD, is the biostatistics core co-leader for the DIAN-TU. He discloses serving on the DSMB for Eli Lilly and Company, Amydis Corporate, and Abata Therapeutics, as well as working as a statistical consultant for Alector, Inc. and Pharmapace, Inc. He also serves as DSMB member for another five studies funded by NIH.
Randall J. Bateman, M.D., is the Director of the DIAN-TU and Principal Investigator of the DIAN-TU-001. He co-founded C2N Diagnostics. Washington University and R.J.B. have equity ownership interest in C2N Diagnostics and receive royalty income based on technology (stable isotope labeling kinetics, blood plasma assay and methods of diagnosing Alzheimer’s disease with phosphorylation changes) that is licensed by Washington University to C2N Diagnostics. R.J.B. receives income from C2N Diagnostics for serving on the scientific advisory board. R.J.B. has received research funding from Avid Radiopharmaceuticals, Janssen, Roche/Genentech, Eli Lilly, Eisai, Biogen, AbbVie, Bristol Myers Squibb and Novartis. He receives research support from the National Institute on Aging of the National Institutes of Health, DIAN-TU Trial Pharmaceutical Partners (Eli Lilly and Company, F. Hoffman-La Roche, Ltd., and Avid Radiopharmaceuticals), Alzheimer’s Association, GHR Foundation, Anonymous Organization, DIAN-TU Pharma Consortium (Active: Biogen, Eisai, Eli Lilly and Company, Janssen, F. Hoffmann-La Roche, Ltd./Genentech. Previous: AbbVie, Amgen, AstraZeneca, Forum, Mithridion, Novartis, Pfizer, Sanofi, United Neuroscience). He has been an invited speaker for Novartis and serves on the Advisory Board for F. Hoffman La Roche, Ltd.
Eric McDade, D.O., is the Associate Director of the DIAN-TU. He reports serving on a Data Safety Committee for Eli Lilly and Company and Alector; scientific consultant for Eisai and Eli Lilly and Company; institutional grant support from Eli Lilly and Company, F. Hoffmann-La Roche, Ltd. and Janssen.
J.L.D. is an inventor on patents or patent applications of Eli Lilly and Company relating to the assays, methods, reagents and / or compositions of matter for P-tau assays and Aβ targeting therapeutics. J.L.D. has served as a consultant or on advisory boards for Eisai, Abbvie, Genotix Biotechnologies Inc, Gates Ventures, Karuna Therapeutics, AlzPath Inc., Cognito Therapeutics, Inc., Prevail Therapeutics, and received research support from ADx Neurosciences, Fujirebio, AlzPath Inc., Roche Diagnostics and Eli Lilly and Company in the past two years. J.L.D. has received speaker fees from Eli Lilly and Company. J.L.D. is a founder and advisor for Monument Biosciences. J.L.D. has stock or stock options in Eli Lilly and Company, Genotix Biotechnologies, AlzPath Inc. and Monument Biosciences. SES has served as a consultant or an advisory board or received speaker fees from Eisai, Eli Lilly and Novo Nordisk.
KB has served as a consultant and at advisory boards for Abbvie, AC Immune, ALZPath, AriBio, BioArctic, Biogen, Eisai, Lilly, Moleac Pte. Ltd, Neurimmune, Novartis, Ono Pharma, Prothena, Roche Diagnostics, and Siemens Healthineers; has served at data monitoring committees for Julius Clinical and Novartis; has given lectures, produced educational materials and participated in educational programs for AC Immune, Biogen, Celdara Medical, Eisai and Roche Diagnostics; and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program, outside the work presented in this paper.
HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, Alzinova, ALZPath, Amylyx, Annexon, Apellis, Artery Therapeutics, AZTherapies, Cognito
Therapeutics, CogRx, Denali, Eisai, LabCorp, Merry Life, Nervgen, Novo Nordisk, Optoceutics, Passage Bio, Pinteon Therapeutics, Prothena, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Alzecure, Biogen, Cellectricon, Fujirebio, Lilly, Novo Nordisk, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work).
Carlos Cruchaga (CC) has received research support from GSK and EISAI. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of Circular Genomics and owns stocks in these companies. CC of the advisory board of ADmit.
Collaborators
DIAN-TU Study Team
The DIAN-TU acknowledges the many individuals who have contributed to the DIAN-TU Trials including funding partners, leadership team, core leaders, project arm leaders, study sites and institutional study partners listed in the following pages.
DIAN-TU Leadership Team
Randall Bateman, MD, Director and Principal Investigator
Eric McDade, DO, Associate Director
David Clifford, MD, Associate Director and Medical Director
Stephen Salloway, MD, Project Arm Leader, Gantenerumab
Martin Farlow, MD, Project Arm Leader, Solanezumab
Lon Schneider, MD, Project Arm Leader
DIAN-TU Core Leaders
Randall Bateman, MD, Administrative Core Leader, Washington University School of Medicine
Anne Fagan, PhD, Biomarker Core Leader, Washington University School of Medicine
Chengjie Xiong, PhD, Biostatistics Core Leader, Washington University School of Medicine
Guoqiao Wang, PhD, Biostatistics Co-Core Leader, Washington University School of Medicine
Jason Hassenstab, PhD, Cognition Core Leader, Washington University School of Medicine
Alison Goate, DPhil, Genetics Core Leader, Mt. Sinai School of Medicine
Carlos Cruchaga, PhD, Genetics Core Leader, Washington University School of Medicine
Tammie Benzinger, MD, Imaging Core Leader, Washington University School of Medicine
Rick Perrin, MD, Neuropathology Core Leader, Washington University School of Medicine
Jorge Llibre-Guerra, MD, MSc, Post-Doctoral Associate
Cliff Jack, MD, MRI, Mayo Clinic
Robert Koeppe, MD, PET Imaging, University of Michigan
Nigel Cairns, MD (retired), Neuropathology Core Leader, Washington University School of Medicine
Peter Snyder, PhD, (former Cognition Core Leader), Brown University
DIAN Expanded Registry (DIAN-EXR)
Eric McDade - Director
Randall Bateman – Associate Director
Jorge Llibre-Guerra – Post-Doctoral Associate
Ellen Ziegemeier - Senior Clinical Research Coordinator
Jennifer Petranek - Clinical Coordinator I
Sarah Adams - Clinical Research Coordinator II
Susan Brandon - Clinical Research Coordinator IIDIAN-TU Staff
DIAN-TU Faculty and Staff
Amanda Fulbright, Grant Specialist, Administration Core
Ron Hawley, IT Audiovisual and Interactive web designer, Administration Core
Jacki Mallmann, Senior Grant Specialist, Administration Core
Karen McCann, Financial Accounting Assistant, Administration Core
Julie Murphy, Accounting/Purchasing Assistant, Administration Core
Anna Santacruz, Administrative Director, Administration Core
Jeanette Schillizzi, Research Administrator, Administration Core
Wendy Simpson, Financial Accounting Assistant, Administration Core
Shannon Sweeney, Finance Analyst, Administration Core
Kelley Coalier, Lead Scientist, Biomarker Core
Fatima Amtashar, QC Technician, Biomarker Core
Sushila Sathyan, Archivist, Biomarker Core
Jennifer Stauber, Research Specialist, Biomarker Core
Susan Mills, Director of Clinical Operations
Nicole Kelley, Associate Director of Clinical Operations
Stephanie Belyew, Clinical Trial Manager, Site, Vendor and GCP Management, Clinical Operations
Angela Fuqua, Clinical Trial Manager, Clinical Scale, Drug Supply and Vendor Management, Clinical Operations
Inbal Meshulam, Clinical Trials Manager, Clinical Operations
Annette Stiebel, Clinical Trial Manager, Regulatory, Clinical Operations
Jeanine Portell, Clinical Trial Manager, MRI Sites, Clinical Operations
Bettina Bell, Clinical Trial Manager, Brain Donation, Clinical Operations
Caryll Bentley, Contract Project Manager, Clinical Operations
Sharon Cirello, Senior Contract Data Manager, Clinical Operations
Nithyanjali Devarapalli, Contract Specialist Clinical Data Manager, Clinical Operations
Arthur Gipson, Contract Specialist Clinical Data Manager, Clinical Operations
JaNeen Wisner, Contract Project Coordinator, Clinical Operations
Tayona Mayhew, Data Management Specialist, Clinical Operations
Zenobia Bridgewater, Clinical Research Coordinator, Clinical Operations
Dana Burgdorf, Research Nurse Coordinator II, Clinical Operations
Molly Fitzgerald, Clinical Research Study Assistant I, Clinical Operations
Erica Fowler, Clinical Research Coordinator I, Clinical Operations
Dottie Heller, Research Nurse Coordinator II, Clinical Operations
Miranda Jany, Clinical Research Coordinator I, Clinical Operations
Latoya Jones, Clinical Research Coordinator II, Clinical Operations
Michelle Jorke, Clinical Research Coordinator II, Clinical Operations
Paulette MacDougall, Research Nurse Coordinator II, Clinical Operations
Eugene Rubin, QC, Clinical Operations
Jessi Smith, Senior Clinical Research Coordinator, Clinical Operations
Mary Wolfsberger, Clinical Research Coordinator, Clinical Operations
Andy Aschenbrenner, Assistant Professor, Cognition Core
Jennifer Smith, Professional Rater III, Cognition Core
Marisol Tahan, Clinical Research Coordinator, Cognition Core
Theresa Butler, Research Lab Manager, Imaging Core
Lisa Cash, Senior Clinical Research Coordinator, Imaging Core
Jon Christensen, Staff Scientist, Imaging Core
Aylin Dince, Research Assistant, Imaging Core
Tony Durbin, Senior Clinical Research Coordinator, Imaging Core
Shaney Flores, Research Assistant, Imaging Core
Karl Friedrichsen, Research Assistant, Imaging Core
Brian Gordon, Co-Investigator, Imaging Core
Russ Hornbeck, Project Manager, Imaging Core
Nelly Joseph-Mathurin, Post-doc, Imaging Core
Sarah Keefe, Research Assistant, Imaging Core
Lakisha Lloyd, Research Coordinator, Imaging Core
Laura Marple, Research Technician II, Imaging Core
Austin McCullough, Graduate Research Assistant, Imaging Core
Stephanie Schultz, Pre-Doctoral Trainee, Imaging Core
Sally Schwarz, Co-Investigator, Imaging Core
Yi Su, Co-Investigator, Imaging Core
Andrei Vlassenko, Co-Investigator, Imaging Core
Qing Wang, Post-doc, Imaging Core
Jinbin Xu, Co-Investigator, Imaging Core
Erin Franklin, Research Coordinator, Neuropathology Core
Eli Lilly and Company and Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly and Company
Eli Lilly and Company Avid Radiopharmaceuticals
John Sims, MD, Eli Lilly and Company. Michael Devous
Karen Holdridge, MPH, Eli Lilly and Company. Erica Elephant
Cheryl Brown, BS, Eli Lilly and Company. Laura Harper
Roy Yaari, MD, Eli Lilly and Company Marybeth Howlett
Isabella Velona, Clinical Trial Project Manager Mark Mintun
Scott Andersen, Biostatistician Michael Pontecorvo
Michele Mancini, GPS
Brian Willis, PK/PD Project Leader
Jillian Venci Fuhs, Global Regulatory Affairs
Julie Bush, Product Delivery
Shamrock Garrett, Sr. CTMA
Traci Peddie, Data Sciences & Solutions
Natalie Vantwoud, Data Management
Barbara Lightfoot-Owens, Medical Writer
John Brad-Holmes, Central Lab
Former Team Members
Phyllis Ferrell Barkman, Russ Barton, Lauren Brunke, Robert Dean, Deanilee Deckard, Ann Catherine
Downing, Ganapathy Goppalrathnam, David Henley, Janice Hitchcock, Sonia Nijampatnam,Tracie
Peddie, Melissa Pugh, Tami Jo Rayle, Shiloh Scott, James Senetar, Gopalan Sethuraman, Eric
Siemers, Brian Steuerwald, Connie Tong, Jim Vandergriff
F. Hoffman-LaRoche Team
Monika Baudler, LifeCycle Leader
Rachelle Doody, Global Head of Neurodegneration
Paul Delmar, Principal Statistical Scientist
Carsten Hofmann, Clinical Pharmacologist
Michaela Jahn, Global Biometrics Team Leader
Geoff Kerchner, Global Development Leader
Gregory Klein, Biomarker Experimental Medicine Leader
Smiljana Ristic, Associate Group Medical Director
Alison Searle, Operations Program Leader
Marco Sonderegger, Technical Development Leader
Roz Sutton, EU Regulatory Partner
Janette Turner, US Regulatory Partner
Jaku Wojtowicz, Safety Science Director
Susan Yule, Global Regulatory Leader
Former Team Members
Elizabeth Ashford, Operations Program Leader; Bogdon Balas, Safety Science Leader; Estelle Vester-Blokland, LifeCycle Leader; Stephanie Capo-Chichi, EU Regulatory Partner; David Agnew, Global Study Manager; Ernest Dorflinger, Translational Medicine Leader; Efe Egharevba, Global Study Manager; Christelle Laroche, Technical Development Leader; Isabelle Bauer Dauphin, Technical Development Leader; Rob Lasser, Global Development Leader; Ferenc Martenyi, Global Development Leader; Glenn Morrison, Global Development Leader; Tania Nikolcheva, Biomarker Experimental Medicine Leader; Michael Rabbia, Statistical Scientist; Juha Savola, Project Leader; Janice Smith, Clinical Science Leader; Dietmar Volz, Statistical Scientist
DIAN-TU DSMB Members
Gary Cutter, PhD, DSMB Chairperson, University of Alabama at Birmingham
Steve Greenberg, MD, PhD, Massachusetts General Hospital, Boston
Scott Kim, MD, PhD, National Institutes of Health, Bethesda
David Knopman, MD, Mayo Clinic, Rochester
Willis Maddrey, MD (retired), UT Southwestern, Dallas
Kristine Yaffe, MD, University of California, San Francisco
Karl Kieburtz, MD, PhD, (DSMB Chairperson, retired) University of Rochester, NY
Allan Levey, MD, PhD (retired), Emory University, Atlanta
DIAN-TU Therapy Evaluation Committee
Randall Bateman, MD, Chair, Washington University School of Medicine, St. Louis
Eric McDade, DO, Co-Chair, Washington University School of Medicine, St. Louis
Paul Aisen, MD, Alzheimer’s Therapeutic Research Institute, USC
Jasmeer Chhatwal, MD, PhD, MMSc, Massachusetts General Hospital, Harvard Medical School
David Clifford, MD, Washington University School of Medicine, St. Louis
David Cribbs, MD, UC Irvine, CA
Nick Fox, MD, FRCP, FMedSci, Dementia Research Centre
Serge Gauthier, Serge Gauthier, CM, MD, FRCPC, Director, AD & Related Disorders Unit
McGill Centre for Studies in Aging
David Holtzman, MD, Washington University School of Medicine
Matthias Jucker, PhD, Hertie Institute for Clinical Brain Research, DZNE, Germany
Jeff Kelly, MD, Scripps University, California
Virginia Lee, PhD, University of Pennsylvania, Perelman School of Medicine
Simon Mead, FRCP, PhD, Institute of Prion Diseases, London
Cath Mummery, PhD, FRCP, Dementia Research Centre, London
Erik Musiek, MD, PhD, Washington University School of Medicine
Erik Roberson, MD, PhD, University of Alabama
Mathias Staufenbiel, PhD, Hertie Institute for Clinical Brain Research, DZNE, Tubingen, Germany
Robert Vassar, PhD, Northwestern University, IL
Former TEC Members
Bart DeStrooper, PhD; William Klunk, MD, PhD; Cynthia Lemere, MD; John C. Morris, MD
DIAN Clinical Trials Committee (CTC) Members
Randall Bateman, Washington University in St. Louis School of Medicine
John Morris, Washington University in St. Louis School of Medicine
Chengjie Xiong, Washington University in St. Louis School of Medicine
Denise Heinrichs, DIAN Family Representative
John Ringman University of California, Los Angeles
Laurie Ryan, Division of Science, National Institute on Aging
Neil Buckholtz, National Institute on Aging
Reisa Sperling, Director, Center for Alzheimer Research and Treatment
Stephen Salloway, Butler Hospital
Paul Aisen, University of California, San Diego
Anna Santacruz, Washington University in St. Louis School of Medicine
Gabrielle Strobel, Alzheimer Research Forum
Bill Klunk, University of Pittsburgh
William Thies, Alzheimer’s Association
Anne Fagan, Washington University in St. Louis School of Medicine
Mark Mintun, Washington University in St. Louis School of Medicine
Natalie Ryan, University College London
Virginia Buckles, Washington University in St. Louis School of Medicine
David Hawver, Food and Drug Administration
Martin Farlow, Indiana University
Maritza Ciliberto, DIAN Family Representative
Ralph Martins, Edith Cowan University
Jennifer Williamson, Columbia University
Study Sites
Australia: Neuroscience Research Australia - W Brooks, MJ Fulham, J Bechara, D Foxe; Australian Alzheimer’s Research Foundation – R Clarnette, N Reynders, P Mather; University of Melbourne – C Masters, C Rowe, B Clinch, D Baxendale
Canada: McGill University – S Gauthier, P Rosa-Neto, C Mayhew, L Robb; University of British Columbia – R Hsiung, D Worsley, M Assaly, E Nicklin; Sunnybrook Research Institute – M Masellis, K Sharp, S Hetherington
France: Hopital Charles Nicolle – D Wallon, D Hannequin, A Morin, A Zarea, E Gerardin, P Bohn, M Chastan, P Vera, M Colnot, N Donnadieu, M Quillard-Muraine, C Bergot, S Jourdain; Groupe Hospitalier Pitie-Salpetriere – B Dubois, M Habert, N Younsi; Hopital Pierre Wertheimer – M Formaglio, D Lebars, N El Kfif, A Jullien; Hopital Purpan – J Pariente, P Payoux, C Thalamas, A Driff, E Pomies, P Gauteul; Hôpital Roger Salengro – F Pasquier, A Rollin-Sillaire, F Semah, L Breuilh, M Laforce; Orsay Imaging – M Bottlaender
Spain: Hospital Clinic i Provincial de Barcelona – R Sanchez-Valle, M Balasa, A Lladó, B Bosch, N Bargalló, I Banzo, A Perisinotti
United Kingdom: University College London Hospital – C Mummery, I Kayani, J Douglas, M Grilo
United States: Washington University School of Medicine – BJ Snider, T Benzinger, W Sigurdson, T Donahue, P Kelly; Emory University – J Lah, C Meltzer, G Schwartz, P Vaughn, L Piendel; University of Pittsburgh – S Berman, J Mountz, L Macedonia, S Ikonomovic, S Goldberg, E Weamer, J Ruskiewiecz, S Hegedus, L Tarr, T Potter, G Valetti; University of Alabama at Birmingham – E Roberson, D Geldmacher, M Love, A Watkins, L Ashley; Indiana University – J Brosch, A Kohn, N McClaskey, J Buck, J Fletcher; Butler Hospital – G Surti, R Noto, C Bodge, W Menard; University of Puerto Rico – I Jimenez Valazquez, J Diaz, K Aleman; Yale University – C van Dyck, M Chen, N Diepenbrock, A Mecca, S Good; University of California San Diego – D Galasko, C Hoh, D Szpak, S Peackock; University of Washington – S Jayadev, D Lewis, Y Tutterow
DIAN-TU Collaborators and Advisers
John Morris, MD, Senior Advisor, Washington University School of Medicine, St. Louis, MO
David Holtzman, MD, Senior Advisor, Washington University School of Medicine, St. Louis, MO
Laura Swisher, MS, Deputy Director, Washington University School of Medicine, St. Louis, MO
Alisha Daniels, MD, MHA, Executive Director, DIAN, Washington University School of Medicine, St. Louis, MO
Janice Hitchcock, PhD, Hitchcock Regulatory Consulting Inc.
Thomas Bird, MD, University of Washington, Seattle
Dennis Dickinson, MD, Mayo Clinic, Jacksonville, FL
M. Marsel Mesulam, MD, Cognitive Neurology and Alzheimer’s Disease Center, Northwestern University, Chicago, IL
Cornelia Kamp, MBA, University of Rochester, New York
Ron Thomas, PhD, ADCS, University of California, San Diego
Paul Aisen, MD, ADCS, University of Southern California, Los Angeles
DIAN Observational Study Site Investigators
James Noble, MD, Columbia University, New York
Martin Farlow, MD, Indiana University, Indianapolis, IN
Jasmeer Chhatwal, MD, PhD, Brigham and Women’s Hospital-Massachusetts GH, Charlestown, MA
Stephen Salloway, MD, Butler Hospital, Warren Alpert School of Medicine, Brown University
Sarah Berman, MD, PhD, University of Pittsburgh, PA
Gregg Day, MD, Mayo Clinic Jacksonville, FL
Hiroyuki Shimada, MD, PhD, Osaka City University, Japan
Takeshi Ikeuchi, MD, PhD, Brain Research Institute, Nigata, Japan
Kazushi Suzuki, MD, PhD, The University of Tokyo, Japan
Peter Schofield, PhD, DSc, Neuroscience Research Australia, Sydney
Ralph Martins, BSc, PhD, Edith Cowan University, Nedlands, Western Australia
Nick Fox, MD, FRCP, FMedSci, Dementia Research Centre, University College London, United Kingdom
Johannes Levin, MD, PhD, German Center for Neurodegenerative Diseases (DZNE), Munich, Germany
Mathias Jucker, PhD, German Center for Neurodegenerative Diseases (DZNE), Tubingen, Germany
Raquel Sanchez Valle, MD, Hospital Clinic i Provincial de Barcelona, Spain
Patricio Chrem, MD, Fundación para la Lucha contra las Enfermedades Neurológicas de la Infancia (FLENI), Buenos Aires, Argentina
DIAN EXR Referring Clinicians, Researchers and Partner Sites
Neelum T. Aggarwal, MD, Rush University Medical Center, Chicago IL
Tom Ala, Center for Alzheimer’s Disease and Related Disorders, Southern Illinois University School of Medicine
Thomas Bird, University of Washington, Seattle
Sandra E. Black, Sunnybrook Health Sciences Centre, University of Toronto, Canada
William J. Burke, MD, Banner Alzheimer’s Institute
Cynthia M. Carlsson, MD, MS, University of Wisconsin School of Medicine and Public Health
Andrew Frank M.D. B.Sc.H. F.R.C.P.(C), Bruyere Continuing Care, Ottawa, Ontario, Canada
James E. Galvin, MD, MPH, Charles E. Schmidt College of Medicine, Florida Atlantic University
Alvin C Holm, MD, Bethesda Hospital, St. Paul, MN
John S.K. Kauwe, Brigham Young University
David Knopman MD, Mayo Clinic, Rochester MN
Sarah Kremen, MD, University of California, Los Angeles
Alan J. Lerner, University Hospitals Cleveland Medical Center
Barry S. Oken, MD, PhD, Oregon Health & Science University
Hamid R. Okhravi, Eastern Virginia Medical School
Ronald C. Petersen, Mayo Clinic, Rochester, MN
Aimee L. Pierce, MD, University of California Irvine
Marsha J. Polk, MED, University of Texas Health Science Center at San Antonio
John M. Ringman, MD, MS, University Southern California
Peter St. George Hyslop, MD, FRS, FRSC, FRCPC, University of Toronto
Sanjeev N. Vaishnavi, MD, PhD, University of Pennsylvania
Sandra Weintraub, Northwestern University Feinberg School of Medicine, IL
Footnotes
All the other authors reported no conflicts of interest.
Consent Statement
The DIAN-TU study was conducted in accordance with the Declaration of Helsinki and the International Council for Harmonization and Good Clinical Practice guidelines and had ethics committee approval at each participating site. All participants provided written informed consent.
5 References
- 1.Salloway S, Farlow M, McDade E, et al. A trial of gantenerumab or solanezumab in dominantly inherited Alzheimer’s disease. Nature Medicine. 2021:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Dyck CH, Swanson CJ, Aisen P, et al. Lecanemab in early Alzheimer’s disease. New England Journal of Medicine. 2023;388(1):9–21. [DOI] [PubMed] [Google Scholar]
- 3.Sims JR, Zimmer JA, Evans CD, et al. Donanemab in early symptomatic Alzheimer disease: the TRAILBLAZER-ALZ 2 randomized clinical trial. JAMA. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G, Li Y, Xiong C, et al. Evaluation of dose-dependent treatment effects after mid-trial dose escalation in biomarker, clinical, and cognitive outcomes for gantenerumab or solanezumab in dominantly inherited Alzheimer’s disease. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring. 2022;14(1):e12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhlmann J, Andreasson U, Pannee J, et al. CSF Aβ1–42–an excellent but complicated Alzheimer’s biomarker–a route to standardisation. Clinica Chimica Acta. 2017;467:27–33. [DOI] [PubMed] [Google Scholar]
- 6.Keshavan A, O’Shea F, Chapman MD, et al. CSF biomarkers for dementia. Practical Neurology. 2022;22(4):285–294. [DOI] [PubMed] [Google Scholar]
- 7.Timsina J, Ali M, Do A, et al. Harmonization of CSF and imaging biomarkers in Alzheimer’s disease: Need and practical applications for genetics studies and preclinical classification. Neurobiology of Disease. 2024;190:106373. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Li Y, Xiong C, et al. Examining amyloid reduction as a surrogate endpoint through latent class analysis using clinical trial data for dominantly inherited Alzheimer’s disease. Alzheimer’s & Dementia. 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pontecorvo MJ, Lu M, Burnham SC, et al. Association of donanemab treatment with exploratory plasma biomarkers in early symptomatic Alzheimer disease: a secondary analysis of the TRAILBLAZER-ALZ randomized clinical trial. JAMA neurology. 2022;79(12):1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. New England Journal of Medicine. 2012;367(9):795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDade E, Wang G, Gordon BA, et al. Longitudinal cognitive and biomarker changes in dominantly inherited Alzheimer disease. Neurology. 2018;91(14):e1295–e1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindler SE, Li Y, Todd KW, et al. Emerging cerebrospinal fluid biomarkers in autosomal dominant Alzheimer’s disease. Alzheimer’s & Dementia. 2019;15(5):655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Coble D, McDade EM, et al. Staging biomarkers in preclinical autosomal dominant Alzheimer’s disease by estimated years to symptom onset. Alzheimer’s & Dementia. 2019;15(4):506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckles VD, Xiong C, Bateman RJ, et al. Different rates of cognitive decline in autosomal dominant and late-onset Alzheimer disease. Alzheimer’s & Dementia. 2022;18(10):1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.1–15 e1–4. In. Klunk W, Koeppe R, Price J, Benzinger T, Devous M, Jagust W. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagemann O, Liu H, Wang G, et al. Downstream Biomarker Effects of Gantenerumab or Solanezumab in Dominantly Inherited Alzheimer Disease: The DIAN-TU-001 Randomized Clinical Trial. JAMA neurology. 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barthélemy NR, Li Y, Joseph-Mathurin N, et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nature medicine. 2020;26(3):398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Yen D, Hendrix RD, et al. Timing of Biomarker Changes in Sporadic Alzheimer’s Disease in Estimated Years from Symptom Onset. Annals of Neurology. 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giangrande C, Delatour V, Andreasson U, Blennow K, Gobom J, Zetterberg H. Harmonization and standardization of biofluid-based biomarker measurements for AT (N) classification in Alzheimer’s disease. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring. 2023;15(3):e12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong C, Schindler SE, Henson RL, et al. Correlational analyses of biomarkers that are harmonized through a bridging study due to measurement errors. Statistical methods in medical research. 2024;33(2):185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.