Abstract
Mus dunni endogenous virus (MDEV) is an apparently intact retrovirus that normally lies transcriptionally silent in cultured M. dunni cells, but the provirus can be activated by treatment of the cells with hydrocortisone or 5-iodo-2′-deoxyuridine. Sequence analysis of a molecular clone of the replicating virus revealed a simple retrovirus with a chimeric VL30/GALV-like structure. Interestingly, in the region of the long terminal repeat (LTR) that typically contains the retroviral transcription enhancers, we found over six 80-bp repeats with only a single mismatch, indicating that acquisition of the repeats was a recent event. Here we provide evidence for the following model of MDEV activation and replication. The MDEV provirus in M. dunni cells has a chimeric structure similar to that of the molecular clone but has only 1.15 copies of the 80-bp repeat sequence found in the molecular clone. Activating chemicals directly stimulate transcription from the LTR, allowing a low level of virus replication. Copying errors made during reverse transcription allow multimerization of the 80-bp enhancer region, resulting in viruses with higher transcriptional rates and improved fitness, but increased enhancer copy number is likely balanced by the natural instability of retroviral repeats and constraints imposed by virion packaging limits. The resultant population of replicating MDEV is widely heterogeneous, having from 2.15 to 13.15 enhancer repeats in the LTR. These results reveal a novel mechanism for regulation of transcription and replication of an endogenous retrovirus, in terms of both activation of the virus by the steroid hydrocortisone and the large number and variation in enhancer repeats observed.
A defining feature of retroviruses is their ability to integrate into the host cell genome, resulting in high-fidelity inheritance of the integrated provirus in the progeny of the cell. Diverse animal species have accumulated many such events in their germ cells. Most of these endogenous proviruses are defective, while some are intact but transcriptionally inactive. These intact proviruses can activate spontaneously or can be activated by treatment of animals or cultured cells with a variety of agents, including halogenated pyrimidines, ionizing radiation, chemical carcinogens, protein synthesis inhibitors, and chemicals that induce DNA demethylation (reviewed in reference 9). Because of the pleiotropic effects of these agents, the mechanisms underlying provirus activation have been difficult to determine.
Mus dunni endogenous virus (MDEV) is one such virus that is present in the germ line of a wild mouse species found in Asia. MDEV normally lies transcriptionally inactive in cultured M. dunni cells but can be activated by treating the cells with 5-iodo-2′-deoxyuridine (IdU) or hydrocortisone 21-succinate (HC) (13). Once activated, MDEV can replicate in M. dunni tail fibroblasts (dunni cells) and can infect cells from many mammalian and at least one avian species (8). Interference analysis demonstrates that MDEV uses a novel receptor among murine retroviruses (14).
We obtained molecular clones of the replicating form of MDEV to further study its envelope and receptor usage (8). Sequence analysis revealed several interesting features in addition to a distinct envelope (21). MDEV has a hybrid structure, with the majority of the coding regions derived from a virus similar to gibbon ape leukemia virus (GALV) and long terminal repeats (LTRs) derived from virus-like 30S (VL30) elements, replication-defective retroelements that are similar in structure and replication cycle to retroviruses. The U3 region of the MDEV LTR, which contains the retroviral promoter and enhancers, is unusual in two respects. First, the sequence of the MDEV U3 defines a novel, fifth VL30 family. Second, the MDEV U3 region contains more than six 80-bp repeats, which is likely the highest U3 repeat number observed in a retrovirus. Except for a single nucleotide in the sixth repeat, all 6.15 repeats are identical. This means that only a single mutation had occurred in the 500-bp region since the repeats were generated. Because the error rate of retroviral reverse transcriptase is high (10−4 mutations per nucleotide per round of replication), we hypothesized that the repeats in the molecular clone were of recent origin and were generated during or after the activation of the MDEV.
Here we provide evidence that the native MDEV provirus has only 1.15 repeats (two 12-bp minirepeats separated by 68 bp of intervening sequence), that HC and IdU directly activate transcription of the provirus, that LTR expansion is due to enhancer multimerization and is a common event occurring during propagation of the virus, and that the LTR expansion provides MDEV a replicative advantage. This model does not require a mechanism for epigenetic suppression of virus expression, as found for other endogenous retroviruses, and efficient virus activation by HC indicates that DNA damage resulting from treatment of cells with other virus activators, such as halogenated pyrimidines or radiation, is not required for MDEV activation.
MATERIALS AND METHODS
Plasmids and viruses.
Plasmids pMDEV9 and pMDEV have been described previously (8, 21). pMDEV9 is the original circularly permuted clone of the replicating form of MDEV and contains a frameshift mutation in the env gene, while pMDEV contains a replication-competent copy of MDEV and was made from pMDEV9 by depermutation and correction of the frameshift mutation. LAPSN is a retroviral vector that encodes a heat-stable human placental alkaline phosphatase (AP) and neomycin phosphotransferase (15). The pSEAP plasmid contains a heat-stable human placental AP cDNA with a synthetic stop codon engineered to prevent translation of the 3′ transmembrane tail such that the translated product is a secreted AP (SEAP) (4). The SEAP cDNA is followed by a human growth hormone polyadenylation sequence (1), and the plasmid does not include a promoter. pM1SEAP and pM6SEAP are derived from pSEAP and have MDEV LTRs with 1.15 or 6.15 repeats, respectively, inserted in the proper orientation upstream of the SEAP cDNA. pMoSEAP contains a Moloney murine leukemia virus (MoMLV) LTR promoter upstream of the SEAP cDNA. The MoMLV LTR was truncated on the 5′ end up to the NheI site and was truncated on the 3′ end to the BanI site in the R region, which leaves the enhancers and promoter intact. Plasmid pEQ176 contains a cytomegalovirus immediate-early promoter driving a cDNA encoding bacterial β-galactosidase (β-Gal).
Cell culture.
G355 feline embryonic glioma cells (10) were grown in McCoy’s medium with 15% fetal bovine serum. D17 dog cells (ATCC CCL 183) and dunni cells (12) were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Two M. dunni tail fibroblast cell strains derived from the same mouse are available, and they can be distinguished by whether medium exposed to the cells becomes viscous (dunni-v cells) or nonviscous (dunni-nv cells) (14). Unless specifically indicated, dunni-v cells were used to produce MDEV and for the other experiments described here. Activation of MDEV was performed as previously described (13).
SEAP assay.
Culture medium was incubated at 68°C for 30 min to inactivate background heat-labile AP activity, and the medium was clarified by centrifugation at 13,000 × g for 5 min at 4°C. SEAP activity was measured by mixing 100 μl of test medium, 100 μl of 2× SEAP buffer (2 M diethanolamine, 1 mM MgCl2, 10 mM l-homoarginine), and 5 μl of 4-methylumbelliferyl phosphate (MUP) solution (11.4 mg of MUP [Sigma] per ml in dimethyl sulfoxide) in wells of a 96-well plate. The plate was incubated at 37°C, 10-μl samples were periodically removed and mixed with 100 μl of stop solution (100 mM glycine [pH 10]), and fluorescence due to production of 4-methylumbelliferone was measured with a microfluorometer. Alternatively, fluorescence was measured directly in the wells of the plate at several time points. Product production was calibrated with a 4-methylumbelliferone (Sigma) standard.
β-Gal assay.
Transfected cells were rinsed with phosphate-buffered saline, rinsed with PE buffer (10 mM NaPO4 [pH 7.5], 1 mM EDTA), and lysed by adding 1 ml of PE buffer containing 1% NP-40 (Sigma). Cells were harvested by scraping with a rubber policeman, and the cell lysates were centrifuged at 13,000 × g for 10 min to remove particulate material. For measurement of β-Gal activity (5), 100 μl of test lysate was mixed with 100 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, 1 mg of bovine serum albumin per ml) and 1 μl of 4-methylumbelliferyl-β-d-galactoside (MUG) solution (15 mg of MUG [Sigma] per ml in dimethyl sulfoxide) in wells of a 96-well plate. The plate was incubated at 37°C, and fluorescence due to production of 4-methylumbelliferone was measured with a microfluorometer.
RESULTS
Unactivated dunni cells contain elements that have the same recombinant structure as MDEV.
We used PCR to determine whether there exists in the M. dunni genome an element that has the VL30/GALV-like chimeric structure of the molecular clones. The 5′ PCR primer corresponded to the beginning of the envelope within sequence similar to that of GALV and the 3′ primer corresponded to VL30-like sequence in the LTR downstream of the repeat region (Fig. 1). Unambiguous amplification products were obtained with DNA from two samples of dunni cells (dunni-v and dunni-nv cells [14]) and from G355 cat cells containing the LAPSN vector and infected with MDEV (G355/LAPSN+MDEV cells), while no specific amplification products were obtained with DNA from G355 cells containing the LAPSN vector but not infected with MDEV (G355/LAPSN cells) (Fig. 1). No differences were observed between the dunni-v or dunni-nv samples, consistent with the cells having been derived from the same mouse (14). The dunni cells used in this experiment had not been activated with HC or IdU and were not expressing MDEV. Thus, the dunni PCR products were most likely amplified from the native provirus(es), demonstrating that at least one element with a chimeric VL30/GALV-like structure exists in the M. dunni genome.
FIG. 1.
Elements with a chimeric VL30/GALV-like structure exist in the M. dunni genome. Samples of DNA from the indicated cell lines and from a pMDEV9 plasmid preparation were subjected to PCR using the primers shown, which flank the MDEV 3′ recombination breakpoint. The PCR products were subjected to Southern analysis using the indicated MfeI-BsaBI env fragment of pMDEV9 as a probe.
Close inspection of the ethidium-stained gel showed two closely spaced dominant bands present in the dunni cell lanes, consistent with previous Southern blotting evidence demonstrating additional elements related to MDEV in the M. dunni genome (8). Inspection of the ethidium-stained gel also revealed multiple, evenly spaced bands in the G355/LAPSN+MDEV lanes but only a single band in the pMDEV9 lane. Additionally, the products derived from the dunni DNA were about 400 bp smaller than those derived from the infected cells or pMDEV9 (Fig. 1). Both the multiple bands in the G355/LAPSN+MDEV lanes and the size discrepancies can be explained by differences in U3 repeat numbers, described in detail below.
To molecularly clone a 3′ breakpoint region (the junction between GALV- and VL30-like sequences) from the provirus present in dunni cells, PCR products amplified from dunni DNA were digested with SalI (located in env) and XbaI (located in the repeat region in the LTR) and separated on an agarose gel. The band corresponding to that expected to contain the breakpoint region based on the MDEV sequence (400 bp) was isolated and ligated into the appropriate restriction sites of pBSIIKS+. Sequence analysis demonstrated that three of five of these clones had sequences 98 to 99% identical to that of the molecular clone pMDEV9. These clones likely represent the native MDEV provirus; the few differences may be due to errors by Pwo polymerase during PCR amplification or due to errors by reverse transcriptase during passage of MDEV prior to molecular cloning. Two of five clones had more divergent sequences but also contained the breakpoint region and likely represent related elements.
An element consistent with an MDEV provirus containing 1.15 of the U3 repeats, but not 6.15 repeats, exists in M. dunni cells.
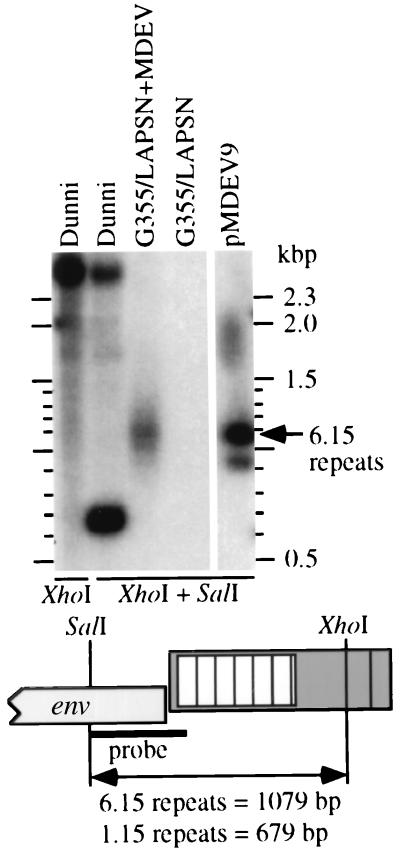
We hypothesized that the native MDEV provirus contains 1.15 repeats rather than 6.25 repeats as present in the molecular clone. A prediction of this hypothesis is that the native proviral SalI-XhoI env-LTR fragment is 679 bp, rather than 1,079 bp as for the molecular clone. To examine this possibility, we performed Southern blot analyses using DNA from dunni, G355/LAPSN+MDEV, and G355/LAPSN cells (Fig. 2). Supporting our hypothesis, a strongly hybridizing fragment in dunni DNA digested with SalI-XhoI has a size consistent with 679 bp, and there is no fragment consistent with 1,079 bp. Fragments of higher molecular weight also hybridized with the probe, consistent with previous evidence that there are additional elements in the M. dunni genome that are related to MDEV. The hybridization signal seen in the G355/LAPSN+MDEV lane is smeared but is centered around the 1,079-bp fragment observed with the plasmid. Note that there are two bands in the plasmid lane (and an artifactual spot at ≈1.8 kb) because pMDEV9 has two adjacent LTRs; the relevant SalI-XhoI env-LTR fragment is 1,079 bp, and an irrelevant XhoI-XhoI fragment is 928 bp. The smearing observed in the G355/LAPSN+MDEV lane prompted additional experiments to determine whether it represented a range of fragment sizes.
FIG. 2.
DNA sequences consistent with the presence of an MDEV provirus containing 1.15 LTR repeats in the M. dunni genome. Genomic DNA (10 μg) or pMDEV9 plasmid DNA (100 pg) was digested with XhoI or XhoI plus SalI and subjected to Southern analysis using the indicated SalI-XbaI fragment of pMDEV9 as a probe. Ethidium bromide staining of the gel prior to blotting indicated approximately equal loading of genomic DNA samples (data not shown). The two panels are from the same gel and blot, with irrelevant plasmid-containing central lanes removed.
MDEV populations have variable numbers of repeats.
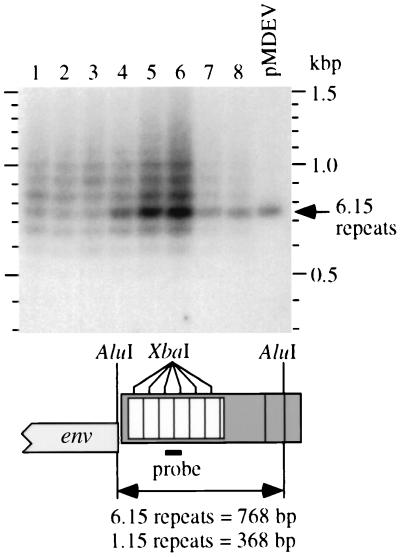
For greater resolution in examining the MDEV U3 repeats, we subjected AluI-digested cellular DNA to Southern analyses using a U3 repeat probe (Fig. 3). Analysis of DNA from G355/LAPSN cells infected under various conditions with MDEV revealed multiple bands consistent with the presence of proviruses with variable numbers of 80-bp repeats (Fig. 3, lanes 1 to 6). In contrast, analysis of plasmid pMDEV revealed a single band consistent with the size of 768 bp predicted by sequence analysis (Fig. 3, rightmost lane).
FIG. 3.
MDEV populations have variable numbers of repeats. Genomic DNA samples (10 μg of each) from MDEV-infected (lanes 1 to 6) or MDEV-transfected (lanes 7 and 8) G355/LAPSN cells or pMDEV plasmid DNA (10 pg) (rightmost lane) were digested with AluI and subjected to Southern analysis using an 80-bp XbaI fragment cut from the pMDEV9 repeat region as a probe. The ethidium bromide stain of the gel prior to blotting indicated approximately equal loading of genomic DNA samples (not shown). See Results for descriptions of the samples analyzed.
For lanes 1 to 4 of Fig. 3, G355/LAPSN cells were infected with 100, 10, 1, or 0.1 μl of MDEV-containing medium to see whether infection with limiting amounts of virus would reveal a stronger band at a particular size indicative of a founder effect. Cellular DNA was harvested after passaging the cells for 2 weeks to allow virus spread. All of these cultures had become positive for LAPSN vector production at this time (≈5 × 104 AP+ focus-forming units/ml), indicating a productive MDEV infection, while cells initially infected with 0.01 μl of MDEV did not produce the LAPSN vector; thus, 0.1 μl of the MDEV stock represents a limiting dilution of the virus. The Southern analysis reveals similar patterns for all of these samples: a repeat range of 4.15 to 11.15, and the most abundant species having 7.15 (lanes 1,2), 8.15 (lane 3), or 6.15 (lane 4) repeats (Fig. 3). Longer autoradiographic exposures revealed the range of repeats to be from 2.15 to at least 13.15 (not shown). There did seem to be a slight founder effect in cells infected with a limiting dilution of virus (0.1 μl [lane 4]) that resulted in a predominance of MDEV virus with 6.15 repeats, which was also detected in two additional serial passages at limiting dilution (Fig. 3, lanes 5 and 6). Each of these cultures presumably received 1 to 10 infectious units of MDEV, yet each culture displays a full complement of repeat numbers, indicating that the repeat number can easily change during virus replication.
We also analyzed DNA from two separate cultures of G355/LAPSN cells that had been transfected with pMDEV and were passaged for 24 days to allow for virus spread (Fig. 3, lanes 7 and 8). Although the source of MDEV in this case had 6.15 repeats, the repeat array was again observed, demonstrating that variants with different repeat numbers can be generated from a virus with a defined number during a short period of virus replication. The predominant band in the DNA from the transfected cells corresponded to the 6.15 repeats of the transfected plasmid, but variants with 4.15 to 10.15 repeats were also present.
Figure 3 and additional similar analyses provided evidence that MDEV populations can have variable numbers of repeats. This prompted us to reexamine the molecular clones that we had previously isolated from a library of extrachromosomal DNA from infected G355 cells. Six molecular clones were obtained during the cloning of MDEV (8), and the prototype clone pMDEV9 was originally sequenced (accession no. AF053745) and found to have 6.15-bp repeats (21). Sequence analysis of the other clones demonstrated that we had isolated clones containing 3.15 and 4.15 repeats as well (data not shown). No U3 alterations were observed other than the number of repeats, providing additional evidence that the ladders observed are due only to variation of the repeat number.
Despite different passage histories, all of the MDEV-infected cells analyzed in Fig. 3 were infected with virus that originated from the same MDEV producer cell line, G355/LAPSN+MDEV clone 16. This virus was also used originally to isolate MDEV molecular clones (8). Additional samples of activated MDEV were required to determine whether the expanded LTR with variable numbers of repeats is a general feature of activated MDEV or was specific to this activant. Independent cultures of dunni cells transduced with the LAPSN vector (dunni/LAPSN cells) were exposed to HC or IdU as described previously (13) to activate the endogenous MDEV. Activation of MDEV was ascertained by measuring production of the LAPSN vector. Southern analysis of viruses from these cultures, performed as described for Fig. 3, revealed an 80-bp ladder in DNA from independently cultured cells exposed to HC or IdU (data not shown), indicating that enhancer multimerization is a general feature of MDEV activated from dunni cells. However, we did observe IdU activation of a virus that gave a single band migrating between the 5.15- and 6.15-repeat MDEV bands (data not shown), showing that other events are possible.
Expanded MDEV LTRs exhibit greater promoter activity in G355 and dunni cells.
We hypothesized that expanded MDEV LTRs are selected due to higher promoter strength. To test this, a series of constructs were made with different promoters driving the expression of a SEAP reporter cDNA. Each plasmid was introduced with the β-Gal expression plasmid pEQ176 into G355 cells by CaPO4-mediated transfection and into dunni cells by lipofection. Two days after plasmid introduction, both the medium containing the SEAP and the cells containing β-Gal were harvested separately. Fluorometric assays were performed on the medium samples by using the substrate MUP, which fluoresces after cleavage of the phosphate group by SEAP, and were performed on the cell lysates with the substrate MUG, which fluoresces after cleavage of the galactoside group by β-gal.
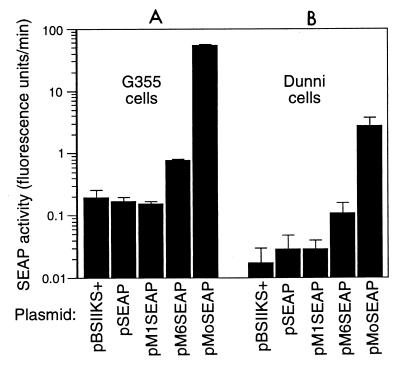
No significant differences in SEAP activity were detected in G355 cells containing plasmid pBSIIKS+, which contains no SEAP cDNA; pSEAP, which contains a SEAP cDNA but no promoter; or pM1SEAP, which contains an MDEV LTR with 1.15 repeats as a promoter to drive expression of the SEAP cDNA (Fig. 4A). The same was true for dunni cells containing these plasmids (Fig. 4B). However, introduction of plasmid pM6SEAP, which contains an MDEV LTR with 6.15 repeats driving expression of the SEAP cDNA, led to a three- to fivefold increase in SEAP activity from G355 and dunni cells compared to that measured for pM1SEAP (Fig. 4). Note that the true increase may be larger since the pM1SEAP activity was not detectable above background levels in both cell types. The activity of the MoMLV LTR was 30- to 60-fold greater than that of the MDEV LTR with 6.15 repeats in G355 and dunni cells (Fig. 4). We found equivalent levels of β-Gal activity in all cellular lysates, indicating that the differences in SEAP activity were not due to differences in transfection efficiency.
FIG. 4.
The expanded MDEV LTR has greater promoter strength than the unexpanded LTR in G355 and dunni cells. Cells were seeded at 2 × 105 per 3.5-cm-diameter well of six-well plates on day 1. On day 2, the medium was replaced and 4 μg of test plasmid mixed with 1 μg of the β-Gal expression plasmid pEQ176 was introduced into the G355 cells by CaPO4 precipitation or into the dunni cells by using Lipofectamine (Gibco). The medium was replaced on day 3, and medium and cells were harvested separately on day 4 for SEAP and β-Gal assays. β-Gal activities were similar for all assays, indicating similar gene transfer efficiencies. Results shown are means from one experiment, with error bars indicating standard deviations. This experiment was repeated several times, and also with different culture media and substrate concentrations, with consistent results.
To rule out the possibility that the undetectable activity of plasmid pM1SEAP was due to defects in the plasmid, we sequenced the promoter region of pM1SEAP to demonstrate that no errors had occurred during plasmid construction. Two additional clones of pM1SEAP were tested to rule out mutation of the SEAP cDNA during growth in bacteria, and again neither produced SEAP activity detectable above background when tested in G355 cells. Together these results show a strong increase in promoter strength resulting from multimerization of the 80-bp repeat region in the MDEV LTR. Even so, the activity of the 6.15-repeat MDEV LTR promoter is still well below that of the well characterized, highly active MoMLV LTR promoter.
The MDEV LTR is responsive to HC and IdU in dunni cells.
MDEV production from dunni cells can be activated by treatment of the cells with HC or IdU (13). Therefore we tested for the responsiveness of the 1.15- and 6.15-repeat MDEV LTR promoters to these drugs to see if these agents would have a direct stimulatory effect on MDEV transcription. To perform this experiment, the SEAP constructs were stably introduced into dunni cells by cotransfection with a plasmid that expressed neomycin phosphotransferase. G418-resistant clones were selected and pooled for each construct, and two pools of transfectants were separately generated for each construct. We found that SEAP production by a given construct could vary between the two sets of transfectants, but Southern blot analysis demonstrated that the inconsistencies could be mostly explained by the different copy numbers of the SEAP constructs introduced into the cells. Thus, for the following assays we have corrected the SEAP activity by dividing the SEAP activity in the culture medium by the plasmid copy number per cell and the cell number to obtain the SEAP activity per gene copy.
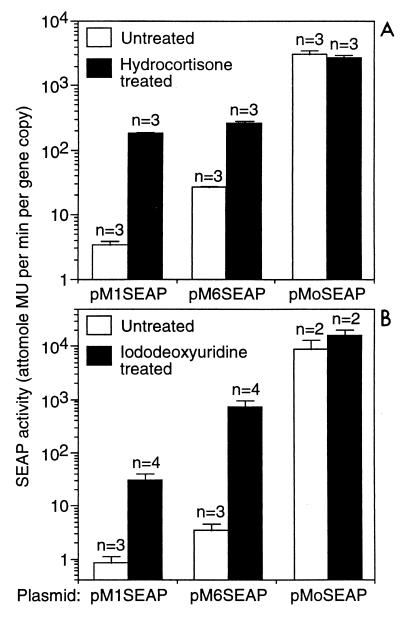
HC treatment increased markedly (10- to 50-fold) SEAP production from the cells containing the pM1SEAP and pM6SEAP constructs, while no increase was observed for the cells containing the MoMLV promoter plasmid, showing that the response was specific to the MDEV LTR constructs (Fig. 5A). HC slowed cell growth such that there were approximately 2.5-fold more cells in the untreated dishes than in the treated dishes at the time of medium harvest, which was taken into account in the normalization to gene copy number.
FIG. 5.
The MDEV LTR is responsive to HC and IdU in dunni cells. Each SEAP construct was mixed at a 20:1 ratio with the selectable plasmid pSV2neo and introduced into dunni cells by electroporation. Drug-resistant colonies were selected by using 0.7 mg of active G418 (Gibco) per ml, and at least 16 clones for each construct were combined to make a polyclonal population. For assay, cells were plated at 2 × 105 per 3.5-cm-diameter well of six-well dishes on day 1. On days 2 and 3, cells were fed with medium with or without 10−4 M HC (A) or medium with or without a saturating concentration of IdU (B). The saturated solution of IdU was made by incubating medium with an excess of IdU crystals at 37°C and filtering the medium to remove undissolved IdU. Solutions containing IdU and cells treated with IdU were kept in the dark, or under low light conditions when necessary for manipulation. On day 4, the medium samples were harvested and assayed for SEAP activity. Gene copy number was determined by Southern analysis using a SEAP cDNA probe followed by quantitation of band intensities by phosphorimaging. Band intensities were compared to that obtained from cells having a single integrated copy of a retrovirus vector containing the SEAP cDNA to determine copy number. SEAP activity is expressed as the amount of enzyme product (4-methylumbelliferone [MU]) produced per cell divided by the SEAP gene copy number per cell, or enzyme product per gene copy. Each experiment was performed in triplicate, and the data represent means ± standard deviations of from two to four experiments for each condition.
IdU also stimulated SEAP production from the pM1SEAP and pM6SEAP constructs markedly (40- to >200-fold), while there was no significant increase in production from the pMoSEAP construct, showing that the response was specific to the MDEV LTR constructs (Fig. 5B). As with HC, treatment with IdU slowed cell growth such that there were about three- to fourfold more cells in the untreated dishes than in the treated dishes. Together these results show that both HC and IdU have direct effects on transcription from the MDEV LTRs with either 1.15 or 6.15 repeats.
DISCUSSION
Based on our results, we propose the following model of MDEV activation and replication: (i) the MDEV provirus native to the M. dunni genome has a chimeric VL30/GALV-like structure but contains only 1.15 80-bp repeats (two 12-bp minirepeats separated by 68 bp of intervening sequence) in the U3 region, rather than the 6.15 repeats contained by our molecular clone of the replicating virus; (ii) the MDEV provirus normally lies transcriptionally quiescent in cultured dunni cells, but treatment with HC or IdU induces a low level of replication by activating transcription of the LTR; (iii) the initial replication offers the opportunity for enhancer multimerization during reverse transcription; and (iv) the repeat number of a replicating population is dynamic, with selection for higher rates of virus transcription due to increased enhancer multimerization likely being balanced by repeat deletions and viral genomic RNA packaging constraints.
The first hypothesis of the model, that the native provirus has a chimeric VL30/GALV-like structure like the replicating virus but has only 1.15 U3 repeats, is supported by Southern analysis, PCR analysis, restriction analysis of bulk PCR products, and sequence analysis of clones generated by PCR (Fig. 1 and 2 and data not shown). The data provide no evidence of an endogenous provirus containing 6.15 repeats but cannot rule out the possibility of the existence of an element with multiple enhancer repeats flanked by sequences different from MDEV such that the restriction enzyme and PCR analysis did not detect it.
The second hypothesis of the model is that the MDEV LTR is responsive to HC or IdU, agents that activate MDEV. To address this possibility, we have created dunni cells that have been stably transfected with constructs containing the MDEV LTRs promoting a cDNA encoding SEAP. Fluorometric assays demonstrated that the MDEV LTR with 1.15 repeats showed 40- to 50-fold-greater activity in the presence of concentrations of HC or IdU that activate MDEV (Fig. 5). Thus, the initial steps in MDEV activation are likely due to the stimulatory effects of these drugs on the promoter activity of the endogenous MDEV provirus. Of particular interest is the fact that HC and IdU have similar abilities to activate MDEV production from dunni cells, indicating that the ability of IdU to incorporate into DNA and act as a mutagen is not critical component of MDEV activation, since HC does not share this property.
The third hypothesis of the model is that expanded LTRs containing multimerized enhancers can be generated from the endogenous MDEV provirus. We have not shown this directly but have shown that the number of enhancer repeats can rapidly diverge in replicating MDEV virus populations, from 2.15 to 13.15 repeats, and have observed the appearance of multiple repeats in virus produced by treatment of dunni cells with HC or IdU in several independent experiments (data not shown). Sequencing of additional molecular clones of MDEV revealed that only the repeat number varied among the clones, thus confirming that the laddering observed in Fig. 3 is likely due to variation in repeat number and not to other changes in the virus. The simplest model to explain enhancer multimerization involves homologous misannealing during reverse transcription leading to increased or decreased enhancer number. The MDEV provirus already contains two 12-bp repeats flanking the enhancer region that could serve to initiate enhancer multimerization. Any number of repeats could be generated by such a mechanism, and indeed retroviruses have been observed to generate tandem repeats from sequences surrounded by small repeats (19).
It is still possible that repeated enhancer elements similar to those of MDEV exist in the dunni cell genome and play a role in the generation of the multiple enhancer repeats in the replicating populations of MDEV. However, MDEV populations with widely variable repeat numbers were generated in G355 cat cells (Fig. 3), which have no MDEV-related endogenous elements as determined by Southern analysis using a probe for the entire MDEV provirus (8), a probe specific for the enhancer repeats (data not shown), or by PCR analysis, indicating that participation of endogenous elements in the generation of enhancer multimers is not required.
The fourth hypothesis is that the expanded LTRs offer a selective advantage through promoter strength. That the expanded LTRs are selected is supported by several observations. First, repeats in retroviruses are unstable (18) and would not be expected to be maintained in the absence of selection. Second, the MDEV populations with multiple U3 repeats are likely derived from a provirus that has 1.15 repeats, and we have observed no evidence of an actively replicating MDEV containing only 1.15 repeats. Because the U3 region is that which contains the promoter and enhancers, the selective advantage of LTR expansion is likely due to promoter strength. Indeed, we could not detect any promoter activity from the MDEV LTR with 1.15 repeats in the absence of HC or IdU but could detect at least three- to fivefold greater activity from the expanded LTR (Fig. 4). Increase in promoter strength due to enhancer multimerization is likely offset by viral RNA packaging constraints and the natural instability of repeats in retroviruses, creating a distribution centered around an optimal number of repeats.
Others have observed that increased leukemogenicity of mink cell focus-forming (MCF) retroviruses is associated with and appears to be dependent on duplication of the retroviral enhancer (2, 11, 20). LTRs with up to three copies of the enhancer have been found in tumors. These results can be explained by a model similar to that proposed for the enhancer multimerization in MDEV, that more enhancers leads to higher viral transcription and replication, and in the case of MCF viruses, increased leukemogenicity. However, the enhancer multimerization during MDEV replication is much more extensive and likely reflects the fact that the 1.15-repeat MDEV LTR is a very poor promoter, while MCF virus LTRs having a single enhancer allow relatively efficient MCF virus replication.
Restriction enzyme analysis of the replicating form of the endogenous BALB/c MLV WN1802N has also revealed LTRs with variable size and shown that the size variations occurred in a region of the LTR that corresponds with the position of the enhancers in MoMLV (7). The restriction pattern suggests that duplication of an enhancer is involved, but the altered LTRs were not sequenced to verify this hypothesis. Most of the clones with small LTRs were found to be infectious, while only one of the five viruses with larger LTRs was infectious. Similarly, variations in enhancer number have been observed in different isolates of MoMLV, but again, only a single enhancer repeat is necessary for virus infectivity (3). In contrast to these examples, we are unable to detect promoter activity from an MDEV LTR with one enhancer repeat, and the majority of replicating MDEV populations have many repeats, indicating the one-repeat MDEV is poorly infectious.
Synergistic cooperation among three U3 repeats of other VL30 LTRs has been observed, both at the level of binding factors in nuclear extracts (17) and at a functional level (6). Although these repeats are different in sequence from the MDEV repeats, the MDEV repeats may work in a cooperative manner as well. We have inspected the MDEV repeats for the presence of consensus enhancer elements and have found only a potential retinoic acid receptor binding site (21).
The promoter assays demonstrate that even the expanded MDEV LTR has 65-fold-lower activity than the MoMLV promoter in G355 cells. This is consistent with relative titers of the parental viruses, since the titer of MDEV is generally about 100-fold lower than that of MoMLV. Since VL30 LTRs show tissue-specific activity (16), it is possible that the MDEV LTR does not require a high degree of expansion to be effective in some cell types.
In conclusion, analysis of MDEV has revealed an interesting example of virus adaptation to the requirements of an endogenous lifestyle, where viruses often remain inactive passengers in the host, and the requirements of an exogenous lifestyle, involving high rates of virus transcription and virus replication. These needs are met by transcriptional regulation both at the level of promoter activation and at the level of enhancer multimerization. In particular, initial activation of the endogenous MDEV provirus by the steroid hydrocortisone and the role played by the large variation in enhancer repeats in virus replication are novel findings. Further studies of mice will be useful to determine the roles of these regulatory mechanisms in the control of MDEV replication in the host organism.
ACKNOWLEDGMENTS
We thank Adam Geballe for the use of the fluorescence plate reader and for the β-Gal expression plasmid pEQ176, and we thank the Biocomputing Shared Resource for assistance with sequence analysis.
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL36444 and HL54881) and the National Institute of Diabetes and Digestive and Kidney Diseases (DK47754).
REFERENCES
- 1.Abraham K M, Levin S D, Marth J D, Forbush K A, Perlmutter R M. Thymic tumorigenesis induced by overexpression of p56lck. Proc Natl Acad Sci USA. 1991;88:3977–3981. doi: 10.1073/pnas.88.9.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belli B, Patel A, Fan H. Recombinant mink cell focus-inducing virus and long terminal repeat alterations accompany the increased leukemogenicity of the Mo+PyF101 variant of Moloney murine leukemia virus after intraperitoneal inoculation. J Virol. 1995;69:1037–1043. doi: 10.1128/jvi.69.2.1037-1043.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns A J, Lai M H, Bosselman R A, McKennett M A, Bacheler L T, Fan H, Maandag E C, van der Putten H, Verma I M. Molecular cloning of unintegrated and a portion of integrated Moloney murine leukemia viral DNA in bacteriophage lambda. J Virol. 1980;36:254–263. doi: 10.1128/jvi.36.1.254-263.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 5.Biegalke B J, Geballe A P. Translational inhibition by cytomegalovirus transcript leaders. Virology. 1990;177:657–667. doi: 10.1016/0042-6822(90)90531-u. [DOI] [PubMed] [Google Scholar]
- 6.Bohm S M, Bakke, Nilsson M, Zanger U M, Spyrou G, Lund J. Cooperating nonconsensus cAMP-responsive elements are mediators of adrenocorticotropin-induced VL30 transcription in steroidogenic adrenal cells. J Biol Chem. 1993;268:3952–3963. [PubMed] [Google Scholar]
- 7.Boone L R, Myer F E, Yang D M, Kiggans J O, Koh C, Tennant R W, Yang W K. Analysis of recombinant DNA clones of the endogenous BALB/c murine leukemia virus WN1802N: variation in long terminal repeat length. J Virol. 1983;45:484–488. doi: 10.1128/jvi.45.1.484-488.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonham L, Wolgamot G, Miller A D. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin J. Endogenous viruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 1122–1132. [Google Scholar]
- 10.Dunn K J, Yuan C C, Blair D G. A phenotypic host range alteration determines RD114 virus restriction in feline embryonic cells. J Virol. 1993;67:4704–4711. doi: 10.1128/jvi.67.8.4704-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland C A, Thomas C Y, Chattopadhyay S K, Koehne C, O’Donnell P V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989;63:1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Bonham L, Alfano J, Kiem H-P, Reynolds T, Wolgamot G. A novel murine retrovirus identified during testing for helper virus in human gene transfer trials. J Virol. 1996;70:1804–1809. doi: 10.1128/jvi.70.3.1804-1809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller A D, Wolgamot G. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson M, Bohm S. Inducible and cell type-specific expression of VL30 U3 subgroups correlate with their enhancer design. J Virol. 1994;68:276–288. doi: 10.1128/jvi.68.1.276-288.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pribnow D S, Chen L-Y, Zhang Y, Magun B E. Complex interactions with direct repeats of a mitogen-responsive VL30 enhancer. Biochim Biophys Acta. 1996;1307:55–65. doi: 10.1016/0167-4781(95)00218-9. [DOI] [PubMed] [Google Scholar]
- 18.Rhode B W, Emerman M, Temin H. Instability of large direct repeats in retrovirus vectors. J Virol. 1987;61:925–927. doi: 10.1128/jvi.61.3.925-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimotohno K, Temin H M. Spontaneous variation and synthesis in the U3 region of the long terminal repeat of an avian retrovirus. J Virol. 1982;41:163–171. doi: 10.1128/jvi.41.1.163-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoye J P, Moroni C, Coffin J M. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolgamot G, Bonham L, Miller A D. Sequence analysis of Mus dunni endogenous virus (MDEV) reveals a hybrid VL30/GALV-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]