Abstract
Mouse mammary tumor virus (MMTV) has frequently been used as an insertional mutagen to identify provirally activated mammary proto-oncogenes. To expedite and facilitate the process of cloning MMTV insertion sites, we have introduced a bacterial supF suppressor tRNA gene into the long terminal repeat (LTR) of MMTV, thus allowing selection of clones containing it in lambda vectors bearing amber mutations. The presence of supF in the LTR should circumvent the screening process for proviral insertion sites, since only those lambda clones with supF-containing proviral-cellular junction fragments should be able to form plaques on a lawn of wild-type Escherichia coli (i.e., lacking supF). The resulting virus (MMTVsupF) induced mammary tumors at the expected rate in infected mice, deleted the appropriate T-cell population by virtue of its superantigen gene, and stably retained the supF gene after passage via the milk to female offspring. To test the selective function of the system, size-selected DNA containing two proviral-cellular junction fragments from an MMTV supF-induced mammary tumor was ligated into λgtWES.λB, packaged, and plated on a supF-deficient bacterial host for selection of supF-containing clones. All plaques tested contained the desired cloned fragments, thus demonstrating the utility of this modified provirus for the rapid cloning of MMTV insertion sites.
Mouse mammary tumor virus (MMTV) is a milk-transmitted, replication-competent retrovirus that causes mammary adenocarcinomas in female mice with an extended latency. It can also cause a low incidence of mammary tumors in males and lymphomas (4, 16, 22). In addition, a variety of premalignant structures in the mammary gland can be induced by MMTV infection, which enables the study of the multistep neoplasic process (23). Moreover, MMTV-induced mammary tumors often begin as hormone-dependent neoplasms in that they grow and regress depending on the pregnancy status of the female, thus allowing the hormonal regulation of tumorigenesis, and the progression to hormone independence, to be studied in this model as well (24).
Like other slowly oncogenic animal retroviruses, MMTV causes tumors via an insertional mutagenesis mechanism. The cellular proto-oncogenes found to be transcriptionally activated or directly mutated by MMTV proviral insertion mutations are often members of either the Wnt or fibroblast growth factor family but may also include Notch4/int3, Cyp19/int5, Int6, and Int41 (5–7, 15, 19, 20, 25–27, 30). The frequency with which genes are targeted in tumors varies with the genetic background of the host (21). Studies of MMTV insertional mutagenesis have been very useful in discovering novel genes involved in mammary tumorigenesis: of the 10 MMTV-activated or -mutated genes reported thus far, 7 were first discovered during such studies. This collection of MMTV-activated genes, however, is small compared to the approximately 50 identified for murine leukemia virus (11).
Insertional mutagenesis studies of MMTV have been hampered in the past by the difficulty in cloning the gag portion of MMTV proviruses into plasmids and lambda vectors (3). This is problematic during the cloning of insertion sites, since the 5′ ends of MMTV proviruses are usually located nearest to transcriptionally activated proto-oncogenes in an enhancer insertion orientation (25, 32). This problem has been solved by the introduction of clonable gag sequences from the endogenous provirus Mtv1 into MMTV (33). However, the cloning of proviral insertion sites from tumor DNAs remains a labor-intensive task. To address this problem, we have now further modified MMTV to include a bacterial supF suppressor tRNA gene in the long terminal repeat (LTR). The presence of supF precludes the lambda plaque screening process and allows selection of plaques containing proviral-cellular junction fragments when lambda vectors that require supF for suppression of mutations in genes necessary for lytic growth are used.
MATERIALS AND METHODS
MMTVsupF provirus construction.
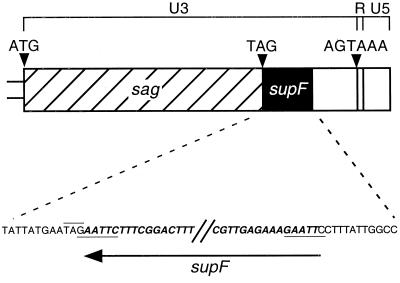
A 0.2-kb EcoRI fragment containing the bacterial SuIII tyrosine suppressor tRNA gene and its bacterial promoter was removed from plasmid pin31suIII (18) (gift of Pat Brown) and cloned into a PCR-generated EcoRI site in the MMTV LTR. The four primers used for introducing the EcoRI site were 5′-TTTAGTCATAGTGCTTA-3′ (primer 1), 5′-GGGAATTCTATTCATAATAACTCA-3′ (primer 2), 5′-TTGAATTCCTTTATTGGCCCA-3′ (primer 3), and 5′-AATAGAACACTCAGAG-3′ (primer 4). Primers 1 and 2 were used to amplify the region 5′ of the EcoRI site, and the primers 3 and 4 were used to amplify the region 3′ of the EcoRI site. Primers 2 and 3 contained the new EcoRI site. The two resulting PCR products and the supF gene fragment were then used in the reconstruction a full-length MMTV provirus containing supF in the 3′ LTR (MMTVsupF [Fig. 1]). No MMTV sequence was lost or duplicated during the cloning process. The MMTV plasmid used in this construction was pUVEH-N, which contains a clonable, full-length hybrid provirus [MMTV(C3H)hyb] consisting of the 5′ half of Mtv1 and the 3′ half of MMTV(C3H) (33). A total of 207 bp was added to MMTV in the process of inserting the supF gene, including the 4 bp added to create the EcoRI site. The supF sequence used here corresponds to bases 113 through 315 of plasmid pπVX (GenBank accession no. X14353), which is the source of supF sequences used to produce pin31suIII (18).
FIG. 1.
Placement of the supF gene in the MMTV LTR. A 203-bp fragment containing a bacterial supF gene was cloned into an EcoRI site (underlined sequence) engineered into the 3′ LTR of MMTV immediately downstream of the sag stop codon (overlined sequence). MMTV sequence is shown in plain font; supF sequence is shown in bold with italics. The transcriptional orientation of the supF gene is opposite that of MMTV. Arrowheads denote the positions of the sag start and stop codons, as well as MMTV’s polyadenylation signal.
Cell culture.
All cells were grown in Dulbecco’s modified Eagle medium with 10% fetal bovine serum (FBS), antibiotics, and 10−7 M dexamethasone except where noted. The MMTVsupF plasmid was introduced with a neomycin resistance plasmid, pMP1-neo, into rat XC cells by calcium phosphate cotransfection, and transfectants were selected with G418 (Geneticin; GIBCO) at 400 μg/ml (net concentration). Thirty colonies were picked and analyzed for MMTV expression by Northern and Western blotting. Two clones with the highest expression were chosen, mixed, and subjected to fluorescence activated cell sorting (FACS) with a FACScan flow cytometer (Becton Dickinson) to further select cells with the highest level of expression. Sorting was performed with a goat antibody to the MMTV envelope surface (SU) protein (previously called gp52) (National Cancer Institute) and fluorescein isothiocyanate-labeled mouse anti-goat antibody (Sigma). The sorted cells were used in this study.
Animal infection.
Female BALB/cJ mice (The Jackson Laboratory) were infected at 3 to 4 weeks of age with MMTV by intraperitoneal (i.p.) injection of approximately 107 rat XC cells producing MMTVsupF or MMTV(C3H)hyb as previously described (33). A negative control group was injected with normal rat XC cells. Offspring of injected mice were infected naturally during nursing via the milk. After infection, mice were bred and observed for tumor formation at weekly intervals or were otherwise analyzed as described in the text.
Milk collection and immunoblotting.
Two to five minutes after the i.p. injection of 1 U of oxytocin (Sigma), approximately 50 μl of milk was pumped out under gentle vacuum from two to six lactating mammary glands per mouse. The milk was then diluted 1:20 in phosphate-buffered saline (PBS) and centrifuged at 3,000 rpm for 10 min at 4°C to skim. The milk serum was filtered through a 0.45-μm-pore-size syringe filter (Costar) and stored at −70°C or analyzed immediately. Dilutions of these milk samples (representing 5, 0.5, 0.05, or 0.005 μl, respectively, of the original milk) were mixed with 200 μl of buffer A (25 mM Tris, 190 mM glycine, 20% methanol, 0.05% sodium dodecyl sulfate) and dot blotted to Hybond-ECL nitrocellulose membranes (Amersham). The blot was blocked with 5% nonfat dry milk in PBS for 1 h at room temperature, incubated with goat anti-MMTV SU envelope antibody (diluted 1:500 in 2% nonfat dry milk in PBS) for 2 h, washed, incubated with peroxidase-labeled rabbit anti-goat immunoglobulin G secondary antibody (diluted 1:1,000) for 1 h, and washed. The signals were detected with ECL Western detection reagent (Amersham) according to the manufacturer’s protocol. Gel electrophoresis and Western blotting of proteins were performed essentially as described elsewhere (31), using the antibodies and conditions noted above except that the secondary antibody was diluted 1:10,000. Per lane, cell extract samples were derived from approximately 5 × 104 cells, and medium samples were derived from 20 μl of medium from 18-h cell cultures.
Lymphocyte isolation and FACS assay.
Mice were sacrificed for T-cell analysis when tumors reached 1.0 to 1.5 cm in diameter in virus-infected mice or at 1 year of age for the uninfected group. One to three iliac lymph nodes (sometimes para-aortic or axillary nodes) were taken and ground gently on a piece of Spectra/Mesh macroporous filter in PBS containing 2.5% FBS to make a single lymphocyte suspension. About 106 cells were then washed twice with same buffer, stained with 1 μg of fluoresceinated rat anti-Vβ14 antibody and phycoerythrin-labeled hamster anti-CD4 antibody (Pharmingen Inc.) in 100 μl of PBS containing 2.5% FBS for 45 min at 4°C, washed twice, and finally analyzed on a FACScan flow cytometer utilizing FACScan software.
RNA and DNA isolation and analysis.
Total cellular RNA was isolated and analyzed by Northern blotting as previously described (33). High-molecular-weight DNAs were isolated from cells and tissues and analyzed by Southern blotting as described elsewhere (33). We used a 1.2-kb BamHI fragment from the envelope region of MMTV to detect the MMTV genomic and envelope RNAs. The MMTV LTR probe was a 1.1-kb PstI-SacI fragment.
λ vector and host bacteria.
λgtWES.λB (14) contains an amber mutation in its S gene, a gene required for lytic growth. The two Escherichia coli strains used in this study as hosts for λgtWES.λB were LE392 and W3110. LE392 is a supF-containing amber-suppressing strain which permits the growth of all recombinant λgtWES.λB phages; W3110 is a nonsuppressing host which allows plaque formation only by phages carrying a supF gene. SacI fragments in the 2 to 3-kb size range from a supF-induced mammary tumor were purified and ligated into SacI-digested and phosphatase-treated λgtWES.λB arms. The ligation products were packaged (Gigapack Gold; Stratagene) and titered on LE392, and supF-containing clones were selected on W3310. Lambda DNA was produced by standard plate lysis methods (31).
RESULTS AND DISCUSSION
MMTV supF construction.
To facilitate the cloning of MMTV insertion sites from tumors of infected mice, we introduced a bacterial supF suppressor tRNA gene into the LTR of the MMTV(C3H)hyb provirus (33). This supF gene carries its own procaryotic promoter and should not be expressed in mammalian cells. Since the supF tRNA suppresses amber stop codons by inserting a tyrosine, selection for DNA fragments containing the supF gene can be accomplished in lambda vectors that contain amber mutations in genes required for phage replication. In an attempt to avoid disrupting important MMTV transcriptional regulatory elements, we cloned supF into a PCR-generated restriction site placed immediately downstream of the MMTV superantigen gene (sag) stop codon at base −234 relative to the U3/R boundary (Fig. 1). This site is not within any known positive or negative regulatory elements or promoters of MMTV (2, 17, 29).
MMTVsupF gene expression in cell culture.
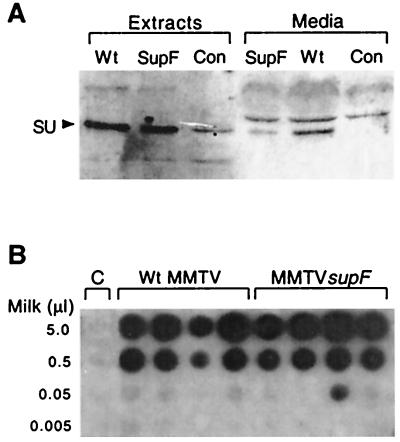
We initially tested the MMTVsupF in cell culture to determine if the new supF sequences present in the LTR would disrupt normal viral gene expression or virion production. Plasmids containing the MMTVsupF provirus and a neomycin resistance gene were cotransfected into rat XC cells and selected in G418, and the resulting clones were isolated. Northern blot analysis of total RNAs from several clones showed that abundant MMTVsupF genomic and spliced envelope RNAs were produced (not shown). MMTV SU protein was also detected in cell extracts and culture media of MMTVsupF cells in levels similar to those of cells producing wild-type MMTV(C3H)hyb virions (Fig. 2A), suggesting that proviral gene expression, RNA stability and processing, and virion production are not negatively affected by the presence of the supF gene.
FIG. 2.
Expression of MMTVsupF in cell culture and in vivo. (A) Western blot analysis (anti-MMTV SU antibody) of cell extracts and media from control XC cells (Con) or XC cells producing MMTV(C3H)hyb (Wt) or MMTVsupF (SupF). A minor background band comigrating with SU was observed in control extracts but not in control medium; control XC cells do not contain MMTV sequences (33). (B) Immunodot blot (anti-MMTV SU antibody) of milk collected from the first lactation of BALB/cJ mice that were either uninfected (Con) or infected with the indicated viruses.
MMTVsupF is superantigen-positive and infectious in vivo.
The sag gene of MMTV encodes a superantigen which stimulates a large subset of T cells when it is expressed on the surface infected B cells or other antigen-presenting cells (1). Activated T cells then stimulate the infected B cells to proliferate, thus producing an expanded reservoir of infected cells (8, 9). The specificity of the T-cell reaction is dictated by the interaction of the Sag protein with specific T-cell receptor β chains (28). The Sag encoded by MMTVs of the C3H strain-the strain used in this study-reacts with T cells bearing the Vβ14 chain of the T-cell receptor (10). During the course of infection, clonal elimination of the activated T cells causes a reduction in the number of cells in the affected subset (13).
We attempted to infect mice with MMTVsupF to determine whether the insertion of supF gene adjacent to sag gene stop codon in MMTVsupF negatively affected superantigen expression. Two cohorts of female BALB/cJ mice were given i.p. injections of either the MMTVsupF producer cells described above or the MMTV(C3H)hyb producer cells; a third cohort (uninfected group) was injected with normal XC cells. Upon tumor development or at 1 year of age, whichever was earliest, analysis of Vβ14 T-cell populations of these mice showed that those injected with MMTVsupF producer cells experienced Vβ14 T-cell deletions to an extent similar to those injected with wild-type MMTV(C3H)hyb producer cells, suggesting that MMTVsupF sag gene expression is normal and sufficient for T-cell activation and deletion (Table 1).
TABLE 1.
Vβ14+ T-cell deletion by MMTVsupFa
| Group | % of T cells (mean ± SD)
|
|
|---|---|---|
| Vβ14+ CD4+ | Vβ14+ CD3+ | |
| Uninfected | 9.76 ± 1.63 | 6.01 ± 0.98 |
| Infected with: | ||
| MMTVsupF | 3.77 ± 1.22 | 2.06 ± 0.65 |
| MMTV(C3H)hyb | 3.76 ± 0.94 | 2.70 ± 0.73 |
Lymph node T cells from four to six BALB/cJ mice per group were analyzed as described in Materials and Methods.
To test whether the life cycle of MMTVsupF was affected by the presence of supF, we bred the injected mice and allowed them to infect their offspring naturally via the milk. We then examined whether virus particles could be detected in the milk of these female offspring by immunodot blot analysis of milk samples using anti-MMTV SU antibody. Offspring in both the MMTVsupF and MMTV(C3H)hyb groups produced similar amounts of SU antigen into their milk, suggesting that the mammary glands were successfully infected by MMTVsupF and were producing virus particles at levels similar to those for glands infected with the wild-type virus (Fig. 2B).
Tumorigenesis by MMTVsupF.
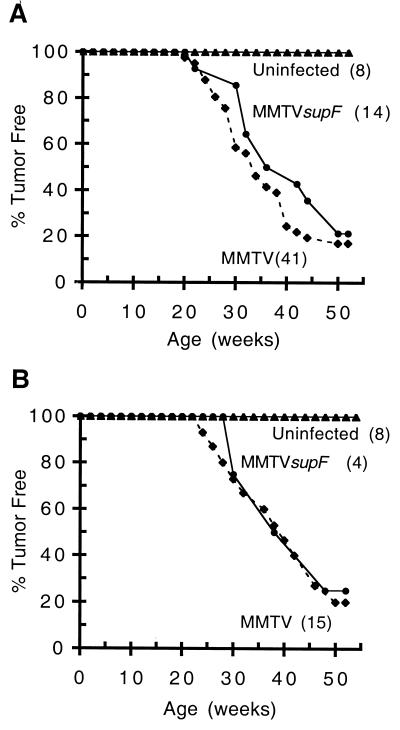
Mammary tumors induced by MMTV occur as a result of somatic insertional mutagenesis, with a median latency of approximately 8 months of age in breeding females (32). We compared the tumor incidence of mice infected with MMTVsupF to the tumor incidence of mice infected with MMTV(C3H)hyb, using both the i.p. injection and natural milk routes of infection. We found that the median age of tumor formation in mice infected with either virus and by either route of infection was approximately 8 months of age as expected (Fig. 3). Together with the results above, these data suggest that MMTVsupF is as infectious and oncogenic as wild-type MMTV(C3H)hyb. The similarity in tumor formation rates also suggests that the presence of supF sequences in the proviral LTRs does not inhibit the activation of neighboring proto-oncogenes by integrated proviruses.
FIG. 3.
Kinetics of tumor formation in MMTVsupF-infected mice. BALB/cJ female mice were infected with either MMTV(C3H)hyb (⧫) or MMTVsupF (●) at 3 weeks of age or left uninfected (▴). They were then allowed to breed freely, and the percentages of mice remaining tumor free at the indicated ages were plotted. (A) Mice infected by i.p. injection of virus-producing XC cells; (B) naturally infected (via milk) offspring of females infected by the i.p. route. The kinetics of tumor formation were similar for MMTV(C3H)hyb and MMTVsupF by either route of infection.
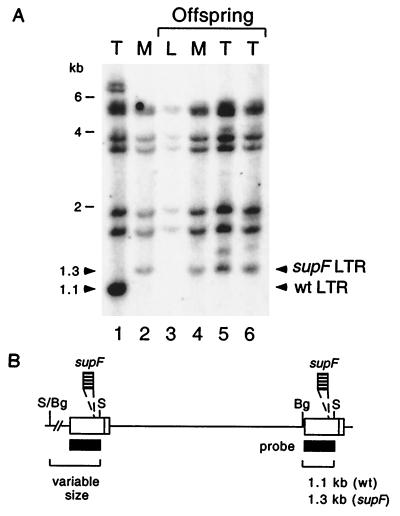
Insertion of additional sequences, such as supF, into viral genomes can result in deletion of the foreign sequences if they present a negative influence on viral replication (18). To determine whether supF was deleted from the MMTVsupF LTR, either in virus-injected animals or during passage from mother to offspring, we analyzed the DNAs of mammary glands and tumors derived from both generations of mice. In Southern blots of these DNAs, capable of detecting internal 3′ LTR restriction fragments of both MMTVsupF and MMTV(C3H)hyb, the larger (by ∼0.2 kb) LTR fragment of MMTVsupF was detected in the infected mother’s mammary gland DNA as well as in her offspring’s mammary gland and tumor DNAs (Fig. 4). The lack of smaller, wild-type LTR fragments in the offspring’s DNAs suggests that deletions did not occur at a significant level during passage via the milk. The minimum number of reverse transcription events required for the infection of the parent mouse and her offspring is three: one for the parent, if the injected virus directly and efficiently infects the mammary gland, and two for the sequential infection of her daughter’s lymphocytes and mammary cells. The number of replication cycles is probably greater, however, due to viral spread among lymphocytes and mammary epithelial cells in both animals. The inability to observe supF deletions in MMTV proviruses in either the parent or her offspring confirms that deletions did not occur at a detectable level and that the supF gene was stable in the MMTV LTR through multiple rounds of infection. Moreover, this result suggests that if deletions do occur, the resulting virus does not have a significant replication advantage over supF-containing viruses.
FIG. 4.
Stability of the supF gene in MMTVsupF during passage in mice. (A) Southern blot analysis (MMTV LTR probe) of tissue DNAs digested with BglII and SacI from the following samples: tumor (T) from mouse infected with MMTV(C3H)hyb by the i.p. route (lane 1); mammary gland (M) from mouse infected with MMTVsupF by the i.p. route (lane 2); liver (L; uninfected control tissue) (lane 3), mammary gland (lane 4), and two mammary tumors (lanes 5 and 6) from one naturally infected female offspring of an MMTVsupF-infected female. The mouse represented in lane 2 is the mother of the mouse represented by lanes 3 to 6. Note the presence of the larger (by 0.2 kb) LTR from MMTVsupF in mammary glands and tumors of the offspring, indicating the stability of the supF gene after passage through mice. wt, wild type. (B) Origins of the probe used (black boxes) and the proviral fragments that hybridize with it (brackets); diagrams of endogenous proviruses are not shown. The fragments of variable size in the tumor samples (e.g., the fragments of ∼7 kb in lane 1 and the fragments of ∼1.5 kb in lanes 5 and 6) are 5′ LTR junction fragments which extend into flanking cellular regions. S, SacI; Bg, BglII.
The lower intensity of the 1.3-kb signals in lanes 5 and 6 of Fig. 4 compared to the 1.1-kb signal in lanes 1 suggested that MMTVsupF-induced tumors may contain fewer proviral insertions than those induced by wild-type MMTV. However, when a larger number of tumors were tested by Southern blotting, we did not detect a significant difference in average insertion number between the two viruses (MMTVsupF average = 3.6 proviruses per tumor in 13 tumors; MMTV(C3H)hyb average = 3.4 proviruses per tumor in 10 tumors).
The presence of a foreign sequence in the MMTV LTR could potentially affect the insertional activation of cellular genes, which for MMTV occurs primarily by an enhancer insertion mechanism. To look for evidence of such an effect, we tested 20 MMTVsupF-induced tumors for transcriptional activation of Wnt1 and Fgf3, the two genes most commonly activated by MMTV insertion mutations. Northern blotting of tumor RNAs showed that Wnt1 was activated in 11 tumors (55%), Fgf3 was activated in 12 tumors (60%), and among these, both genes were activated in seven tumors (35%). These frequencies agree well with those observed previously for MMTV(C3H)hyb, which was found to activate Wnt1, Fgf3, or both at frequencies of 31 to 59%, 72 to 88%, or 28 to 50%, respectively (12). These data indicate that the presence of supF in the MMTV LTR does not significantly affect the insertional activation of these genes.
Facilitated cloning of MMTVsupF integration sites.
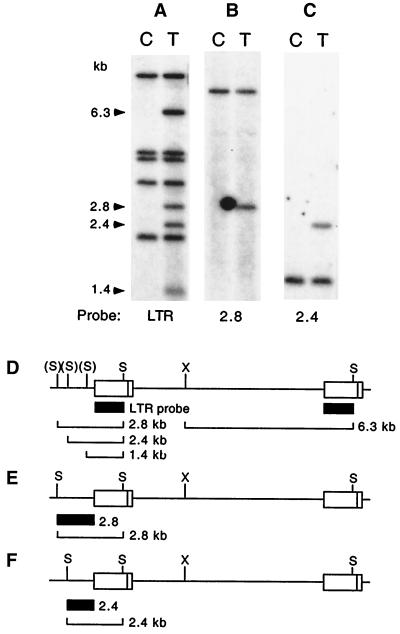
To demonstrate that the supF gene in the LTR of our modified provirus could facilitate the cloning of proviral insertion sites, we attempted to clone two such sites from a single tumor of an MMTVsupF-infected mouse. Southern blot analysis of this tumor using an MMTV LTR probe shows that it harbors three newly integrated proviruses whose SacI-digested proviral-host junction fragments are approximately 2.8, 2.4, and 1.4 kb in size (Fig. 5A). SacI-digested tumor DNA in the 2- to 3-kb size range was isolated from an agarose gel and ligated to SacI arms of λgtWES.λB, and the packaged phage were titered on a nonselective supF+ E. coli host (LE392). Approximately 5 × 105 PFU was then plated on a supF-deficient host (W3110) for the selection of phage containing the supF gene. Fifty plaques resulted, four of which were analyzed by restriction enzyme digestion and Southern blotting. Two of the four were found to contain the 2.4-kb junction fragment, and two contained the 2.8-kb fragment (data not shown). Finally, the cellular portions of these fragments were isolated and used sequentially as probes on the DNA blot used above (from Fig. 5A) after removal of the previous probe. These cellular DNA probes hybridized to the 2.8- and 2.4-kb junction fragments (Fig. 5B and C, respectively) in the tumor samples, as expected, as well as to the normal cellular fragment from the unmutated paired chromosome present in both the tumor and control samples. Thus, the inclusion of the supF gene in MMTV functioned as designed to facilitate the cloning of proviral integration sites.
FIG. 5.
Cloning supF-containing proviral-cellular junction fragments from a mammary tumor. (A to C) DNAs from the liver negative control (lanes C) and a tumor (lanes T) from an MMTVsupF-infected mouse were digested with both SacI and XbaI and analyzed by Southern blotting using an MMTV LTR probe (A), a probe (termed 2.8) from the cellular portion of the cloned 2.8-kb fragment (B), or a probe (termed 2.4) from the cellular portion of the cloned 2.4-kb fragment (C). The same blot was hybridized in each case after removal of any previous probe. The tumor DNA lane in panel A shows 2.8-, 2.4-, and 1.4-kb 5′ junction fragments from newly integrated MMTVsupF proviruses, as well as 6.3-kb XbaI-to-SacI internal fragments from all three proviruses. The bands present in both lanes of panels B and C represent the normal cellular fragments (i.e., those uninterrupted by a proviral insertion) hybridizing to cellular probes 2.8 and 2.4, respectively. The 2.8- and 2.4-kb fragments were known from other experiments (not shown) to be generated by SacI alone; XbaI was added here to provide a better display of all fragments. (D to F) Origins of the probes used (black boxes) and the proviral fragments that hybridize with them (brackets); diagrams of endogenous proviruses are not shown. Diagrams D to F refer to Southern blots A to C, respectively. The parentheses surrounding the SacI sites in diagram D denote that these three sites are each at three different proviral insertion loci (one per locus). S, SacI; X, XbaI.
The modified MMTV provirus described here should significantly speed the labor-intensive process of cloning proviral insertion sites in MMTV-induced neoplasias. It allows for direct selection of MMTVsupF proviral-cellular junction fragments, thus obviating the need to screen for junction fragments using radiolabeled probes. Furthermore, since the supF gene is located in both LTRs of integrated proviruses, either junction fragment may be cloned with equal facility. Many lambda vectors contain amber mutations suitable for use with MMTVsupF, including vectors in the Charon and λgt series, EMBL3a, as well as more modern vectors such as λZAP (Stratagene), which, after selection, can be induced to undergo in vivo excision resulting in an insert-containing plasmid (31, 34). Finally, supF may also be used as a selectable marker in plasmids, thus presenting the possibility of selectively cloning MMTVsupF-containing junction fragments directly from tumor DNAs into plasmids.
ACKNOWLEDGMENTS
We thank Siu-on Jason Chan for technical assistance and figure constructions, members of the laboratory for helpful discussions, and Pat Brown for reagents.
This work was supported by grants to G.M.S. from the Department of Defense Breast Cancer Research Program (DAMD 17-96-1-6039) and the California Breast Cancer Research Program (1RB-0484) and in part by the T. J. Martell Foundation.
REFERENCES
- 1.Acha Orbea H, MacDonald H. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 2.Bramblett D, Hsu C-L, Lozano M, Earnest D, Fabritius C, Dudley J. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brookes S, Placzek M, Moore R, Dixon M, Dickson C, Peters G. Insertion elements and transitions in cloned mouse mammary tumour virus DNA: further delineation of the poison sequences. Nucleic Acids Res. 1986;14:8231–8245. doi: 10.1093/nar/14.21.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudley J, Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984;49:92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia M, Wellinger R, Vessaz A, Diggelmann H. A new site of integration for mouse mammary tumor virus proviral DNA common to BALB/cf(C3H) mammary and kidney adenocarcinomas. EMBO J. 1986;5:127–134. doi: 10.1002/j.1460-2075.1986.tb04186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray D, McGrath C, Jones R, Morris V. A common mouse mammary tumor virus integration site in chemically induced precancerous mammary hyperplasias. Virology. 1986;148:360–368. doi: 10.1016/0042-6822(86)90332-6. [DOI] [PubMed] [Google Scholar]
- 8.Held W, Shakhov A, Izui S, Waanders G, Scarpellino L, MacDonald H, Acha Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Held W, Waanders G, Shakhov A, Scarpellino L, Acha Orbea H, MacDonald H. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 10.Ignatowicz L, Kappler J, Marrack P. The effects of chronic infection with a superantigen-producing virus. J Exp Med. 1992;175:917–923. doi: 10.1084/jem.175.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;16:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 12.Kapoun A M, Shackleford G M. Preferential activation of Fgf8 by proviral insertion in mammary tumors of Wnt1 transgenic mice. Oncogene. 1997;14:2985–2989. doi: 10.1038/sj.onc.1201146. [DOI] [PubMed] [Google Scholar]
- 13.Kappler J, Staerz U, White J, Marrack P. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 14.Leder P, Tiemeier D, Enquist L. EK2 derivatives of bacteriophage lambda useful in the cloning of DNA from higher organisms: the λgtWES system. Science. 1977;196:175–177. doi: 10.1126/science.322278. [DOI] [PubMed] [Google Scholar]
- 15.Lee F S, Lane T F, Kuo A, Shackleford G M, Leder P. Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci USA. 1995;92:2268–2272. doi: 10.1073/pnas.92.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W T, Prakash O, Klein D, Sarkar N H. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987;159:39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S, Dudley J. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobel L I, Patel M, King W, Nguyen-Huu M C, Goff S P. Construction and recovery of viable retroviral genomes carrying a bacterial suppressor transfer RNA gene. Science. 1985;228:329–332. doi: 10.1126/science.2984770. [DOI] [PubMed] [Google Scholar]
- 19.MacArthur C A, Shankar D B, Shackleford G M. Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol. 1995;69:2501–2507. doi: 10.1128/jvi.69.4.2501-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith G H, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchetti A, Robbins J, Campbell G, Buttitta F, Squartini F, Bistocchi M, Callahan R. Host genetic background effect on the frequency of mouse mammary tumor virus-induced rearrangements of the int-1 and int-2 loci in mouse mammary tumors. J Virol. 1991;65:4550–4554. doi: 10.1128/jvi.65.8.4550-4554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalides R, Wagenaar E, Hilkens J, Hilgers J, Groner B, Hynes N E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982;43:819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris D, Cardiff R. Multistep model of mouse mammary tumor development. In: Klastersky J, Staquet M J, editors. Advances in viral oncology. Vol. 7. New York, N.Y: Raven Press; 1987. pp. 123–140. [Google Scholar]
- 24.Nandi S, McGrath C. Mammary neoplasia in mice. Adv Cancer Res. 1973;17:353–414. [Google Scholar]
- 25.Nusse R, Varmus H E. Many tumors induced by mouse mammary tumor virus contain a provirus integrated in the same region of the host chromosome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 26.Peters G, Brookes S, Smith R, Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33:369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- 27.Peters G, Brookes S, Smith R, Placzek M, Dickson C. The mouse homolog of the hst/k-FGF gene is adjacent to int-2 and is activated by proviral insertion in some virally induced mammary tumors. Proc Natl Acad Sci USA. 1989;86:5678–5682. doi: 10.1073/pnas.86.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pullen A, Wade T, Marrack P, Kappler J. Identification of the region of T cell receptor beta chain that interacts with the self-superantigen Mls-1a. Cell. 1993;61:1365–1374. doi: 10.1016/0092-8674(90)90700-o. [DOI] [PubMed] [Google Scholar]
- 29.Reuss F, Coffin J. Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene. J Virol. 1998;72:6073–6082. doi: 10.1128/jvi.72.7.6073-6082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelink H, Wagenaar E, Lopes da Silva S, Nusse R. Wnt-3, a gene activated by proviral insertion in mouse mammary tumors, is homologous to int-1/Wnt-1 and is normally expressed in mouse embryos and adult brain. Proc Natl Acad Sci USA. 1990;87:4519–4523. doi: 10.1073/pnas.87.12.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Shackleford G M, MacArthur C A, Kwan H C, Varmus H E. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 1993;90:740–744. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shackleford G M, Varmus H E. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Short J, Fernandez J, Sorge J, Huse W. λZAP: A bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7588. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]