Abstract
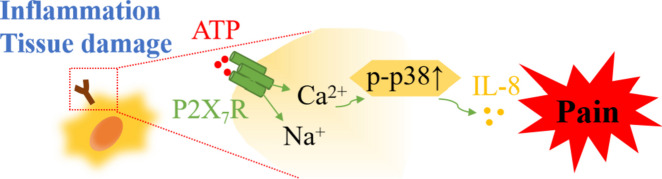
Inflammatory pain, sustained by a complex network of inflammatory mediators, is a severe and persistent illness affecting many of the general population. We explore possible anti-inflammatory pathways of Polyphyllin VI (PPVI) based on our prior study, which showed that PPVI reduces inflammation in mice to reduce pain. Network pharmacology and RNA-Seq identified the contribution of the MAPK signaling pathway to inflammatory pain. In the LPS/ATP-induced RAW264.7 cell model, pretreatment with PPVI for 1 h inhibited the release of IL-6 and IL-8, down-regulated expression of the P2X7 receptor(P2X7R), and decreased phosphorylation of p38 and ERK1/2 components of the MAPK pathway. Moreover, PPVI decreased expression of IL-6 and IL-8 was observed in the serum of the inflammatory pain mice model and reduced phosphorylation of p38 and ERK1/2 in the dorsal root ganglia while the reductions of expression of IL-6 and phosphorylation of ERK1/2 were not observed after the pre-treatment with A740003 (an antagonist of the P2X7R). These results suggest that PPVI may inhibit the release of IL-8 by regulating P2X7R to reduce the phosphorylation of p38. However, the modulation of PPVI on the release of IL-6 and phosphorylation of ERK1/2 may mediated by other P2X7R-independent signals.
Graphical Abstract
Keywords: Polyphyllin VI, Inflammatory pain, Network pharmacology, RNA sequencing, P2X7 receptor
Introduction
International Classification of Diseases (ICD-11) recognizes chronic pain as a separate nosological entity [1]. More than 30% of the world's population currently suffers from chronic pain [2], which has a crucial impact on patients' cognition, mood, emotions, and behavior [3]. Pathogenetic mechanisms indicate chronic pain can be divided into nociplastic, inflammatory, and neuropathic pain [4]. Numerous adverse effects of non-steroidal anti-inflammatory drugs, which are the first-line clinical treatment for inflammatory pain, call for novel analgesics [5–7].
Inflammatory pain is sustained by long-lasting tissue damage and inflammation thus outlasting the resolution of the primary lesion. Inflammatory mediators, in particular, mediate sensitization of both the peripheral and central nervous systems manifested in aberrant neuronal excitability [8, 9]. For example, increased expression of IL-8 in the Complete Freund’s adjuvant (CFA)-induced inflammatory pain mice model enhanced synaptic transmission and caused thermal hyperalgesia; however, thermal hyperalgesia disappeared after local administration of a non-competitive allosteric blocker of IL-8 receptor [10]. The P2X7R is abundantly expressed in macrophages, microglia, and astrocytes [11], all of which are closely related to hyperalgesia [12]. P2X7R keeps low activity under physiological conditions due to the low ATP concentration. However, the concentration of ATP is increased in response to the nociceptive stimulus, which activates the P2X7R [13–15]. In particular, activation of P2X7R mobilizes several intracellular signaling pathways that evoke the release of inflammatory mediators [16, 17]. Genetic deletion of P2X7R decreases the release of IL-1β and reduces pain hypersensitization in a CFA pain animal model [18]. Likewise, pharmacological inhibition of the P2X7R produces antinociceptive effects in various chronic pain models [18–20]. However, increasing studies indicate that the activation of the P2X7R stimulates the release of numerous inflammatory mediators such as IL-1β, IL-6, IL-8, and IL-18 in the pathological state of inflammation [21]. Therefore, the P2X7R is a legitimate and promising target for treating inflammatory pain.
Polyphyllin VI (PPVI), a steroidal saponin, is one of the active ingredients of Paris Polyphylla. Clinical usage of PPVI is mainly focused on diverse types of malignancies such as non-small-cell lung cancer and hepatocellular carcinoma [22, 23]. We found that PPVI exerts an analgesic effect in CFA-induced pain mice. The analgesic function of PPVI is arguably linked to the inhibition of P2X3 purinoceptors alleviating inflammation [24]. In this study, we further clarify the analgesic and anti-inflammatory mechanism of PPIV.
Materials and methods
Data preparation for network pharmacology analysis
The Traditional Chinese Medicine Systems Pharmacology(TCMSP) database (http://lsp.nwu.edu.cn/index.php) was utilized to retrieve the targets of PPVI. Corresponding target names of the target proteins were obtained by using the UniProt protein database. We searched the GeneGards database(https://www.genecards.org) and OMIM databases (http://www.omim.org) for reported disease-related targets by entering the keywords "inflammation" and "pain". The organism was limited to Homo sapiens. After removing the duplicated targets, inflammation and pain-related targets were obtained.
PPI network diagram construction
The intersection targets of the PPVI target, inflammation-related target, and pain-related target were obtained by importing the targets to JVENN: interactive Venn diagram viewer (http://www.bioinformatics.com.cn/static/others/jvenn/example.html) and creating the Venn diagram. Intersection targets were extracted and submitted to the STRING database (https://string-db.org) to build a protein–protein interaction (PPI) network, and the screening condition of organisms was set as "Homo sapiens". The PPI network was imported into Cytoscape 3.8.2 for visualization.
KEGG enrichment analysis
We utilized the KEGG rest API (https://www.kegg.jp/kegg/rest/keggapi.html) to retrieve the most recent gene annotations from the KEGG Pathway as a backdrop for gene set functional enrichment analysis. To obtain the results of gene set enrichment in the background set, the enrichment analysis was performed using the R software package cluster profile (version 3.14.3). p values of 0.05 and an FDR of 0.25 were deemed statistically significant when the minimum gene set was 5 and the largest gene set was 5000.
The docking of molecules
The structure of PPVI was obtained and downloaded from PubChem and converted the files to PDBQT format using Auto Dock Tools. The RCSB Protein Data Bank database was used to download the structure of target proteins. The water and ligands were removed from the target protein by importing its structure to AutoDock Tools and hydrogenation, dehydration, and other pretreatments were conducted to obtain the PDBQT format file. Vina was adopted to dock the PPVI with the target protein to analyze its binding activity. Finally, the results of docking were analyzed with PyMol software.
CFA model and administration
Animal
Adult C57BL/6 J mice(6–9 weeks, 20–25 g) were obtained from Guangdong Yaokang Biotechnology Co., Ltd. All mice were randomly located in Jinan University Laboratory Animal Center at a temperature and light-controlled environment(standard temperature of 24 °C ± 1 °C under a 12 h light–dark cycle) with unrestricted access to food and water. The experiments were conducted mainly with adult female mice. Our research was approved by the Jinan University Animal Ethics Committee.
Mice were divided into six groups randomly (N = 10): Control group, CFA model group, CFA model + Diclofenac sodium(7.5 mg/kg, i.p.) group, and CFA model + different dosages of PolyphyllinVI (1.5, 3, and 6 mg/kg, i.p.) groups. The details of the CFA-induced inflammatory pain mice model and administration are presented in our previous article [24]. In brief, unilateral intraplantar injection at the left hind paw of mice with 30 µL of CFA, the Control group was injected with an equal volume of saline. Behavioral tests including the Vonfery test and hot-plate test were used to evaluate the success of model construction. Five days after the CFA model was established drugs were administered once per day for a total of 7 days.
RNA sequencing (RNA-Seq) analyses
The dorsal root ganglia (DRG) from the (L3-L5) lumbar region of the spinal cord of the mice in the control, CFA, and PPVI (6 mg/kg) groups were separated after the last treatment. DRGs with 4 biological replicate samples for each group were sent to BGI Co., LTD (Shenzhen, China), and the RNA-Seq was performed on the BGIseq500 platform. Principal component analysis (PCA) was performed by the R package stats (version 3.6.0). Limma (linear models for microarray data [25]), a differential expression screening method based on generalized linear models, was processed by the R package limma (version 3.40.6) using the RNA-Seq data which was normalized from FPKM value to log2. In this process, genes with |log2FoldChange|> 1.3, p < 0.05 were considered as differentially expressed genes(DEGs) and showed in the volcano plot. The hierarchical clustering of the top 50 DEGs was visualized in a heatmap using the R package. GO annotation and KEGG pathway enrichment analysis were used to cluster the biological process (BP), cellular component (CC), molecular function (MF), and signaling pathway regulatory networks for DEGs.
Cell culture and cell viability assay
RAW264.7 cells (Procell CL-0190, Wuhan Procell Life Technology Co., Ltd.) were grown in DMEM culture medium (containing 10% FBS, 1% penicillin, and 1% streptomycin) at 37 °C in a 5% CO2 incubator. RAW264.7 cells were plated into 6 well plates and incubated for 24 h. The following experimental groups were set: Control group, LPS/ATP group(LPS 1 μg/ml, ATP 3 μM), A740003 group(10 μM, a P2X7R blocker), and PPVI groups(1.5 μM, 3 μM, 6 μM). RAW264.7 cells were pretreated with 10 μM A740003 or 1.5 μM, 3 μM, 6 μM PPVI for 1 h before LPS/ATP administration. The cells were first incubated with LPS for 24 h and then with ATP for 0.5 h.
The cytotoxicity of PPVI on RAW264.7 cells was assessed by 3-(4,5-di-2-yl)-2,5-tetrazolium bromide (MTT) assay. The cells were seeded in 96-well plates cultured for 24 h and treated with different concentrations of PPVI (0.75, 1.5, 3, 6, and 12 μM) (PPVI, Shanghai Yuanye, concentration ≥ 99%). 24 h later, 10 µL MTT was added to each well and incubated for 4 h. Removed the supernatant and added 150 µL DMSO into the supernatants and the absorbance was measured at 490 nm.
Measurement of inflammatory mediator levels
Determination of the level of IL-6 and IL-8 in CFA mice serum and RAW264.7 cell supernatants using Enzyme-Linked Immunosorbent Assay (ELISA) kits according to kit manufacturer's instructions.
Western blotting
The procedure of protein extraction was described in detail [24]. 20 µg per sample of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and deposited onto a polyvinylidene fluoride (PVDF) membrane after the concentration of proteins was determined. The membranes were blocked with 5% milk for 1 h before incubation with the primary antibody at 4 °C overnight, which is as follows, Anti-p-38(1:2000), Anti-p-p-38(1:2000), Anti-ERK1/2(1:2000), Anti-p-ERK1/2(1:2000), Anti- P2X7R (Abcam, 1:2000). After response with the primary antibody, the blotted PVDF membrane was treated with horseradish peroxidase (HRP) conjugated secondary antibody (Proteintech, 1:2000). The ECL chemiluminescence(Meilunbio) western blotting detection technique was performed with a gel imaging system and the band intensity was analyzed via ImageJ.
Statistical analysis
Protein bands were analyzed in grayscale using ImageJ, and test results were expressed as mean ± standard deviation, one-way ANOVA (0ne-wayANOVA) using GraphPad Prism, and Duncan's method was used for multiple comparisons, with p < 0.05 being considered as a significant difference, and p < 0.01 and p < 0.001 being considered as highly significant differences.
Results
Intersection targets of Polyphyllin VI, inflammation, and pain
A total of 301 potential interaction targets of PPVI were extracted from the TCMSP database. We entered "inflammation" and "pain" into the GeneGards and OMIM databases, removed the duplicated targets, and collected a total of 10,951 targets related to inflammation and 12,251 targets related to pain. The overlap of PPVI, inflammation, and pain targets as revealed is shown in Fig. 1. 102 common targets were thus identified for the construction of the PPI network.
Fig. 1.
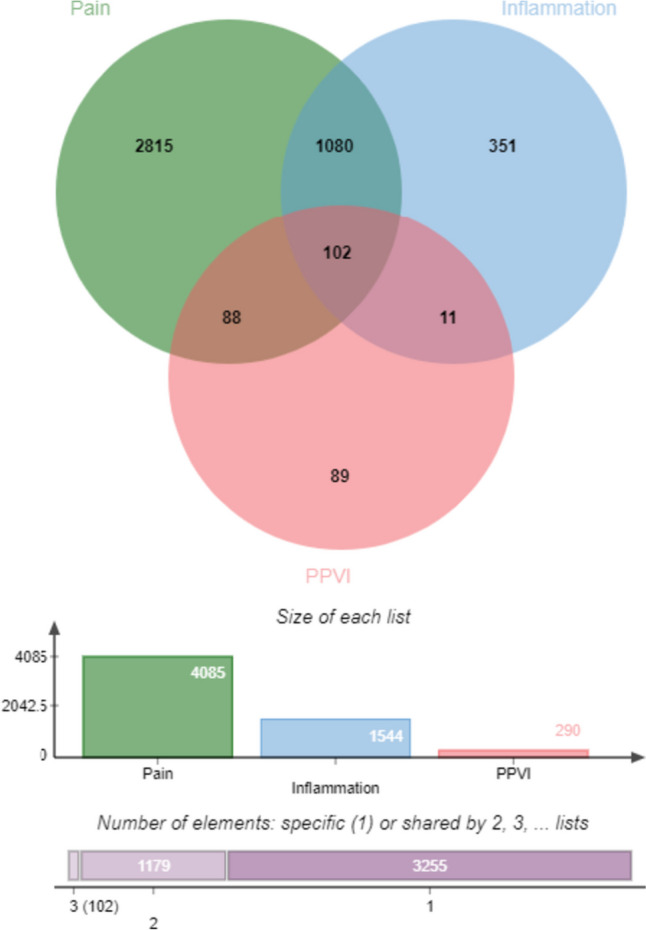
Venn diagram of targets of PPVI, pain, and inflammation
Construction and analysis of PPVI-inflammation-pain targets network
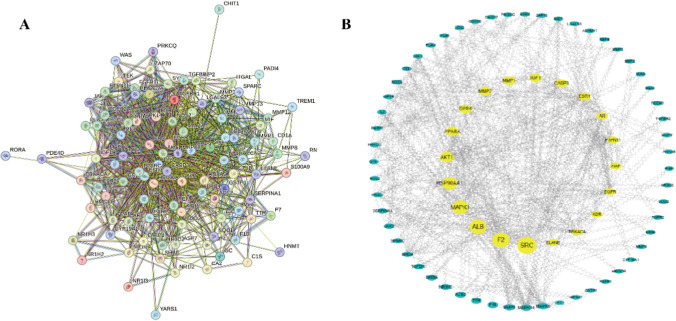
The intersection targets were submitted into the STRING database to construct the PPI network, as shown in Fig. 2A, in which the orb represents the specific target, the connecting line between the orbs represents the correlation between the two targets, and different types of correlation are distinguished by different colors; the higher the correlation, the thicker the edges are. The PPI network was visualized by Cytoscape software; the analysis of the number of edges corresponding to different targets is shown in Fig. 2B, in which the larger the shape and the darker the color shows that the corresponding target has a higher degree of association with other targets in the protein interaction network. The four targets, SRC(Proto-oncogene tyrosine-protein kinase Src), F2(coagulation factor II), ALB(Albumin), and MAPK1(Mitogen-activated protein kinase 1) are located in the core of the network, indicating that they play a key regulatory role in the protein interactions network.
Fig. 2.
The construction and analysis of the intersection targets of PPVI, inflammation, and pain. A Protein–protein interaction (PPI) network of intersection targets of PPVI, pain, and inflammation. B Intersection target network diagram generated using Cytoscape
GO and KEGG enrichment analysis of potential anti-inflammatory and analgesic targets of PPVI
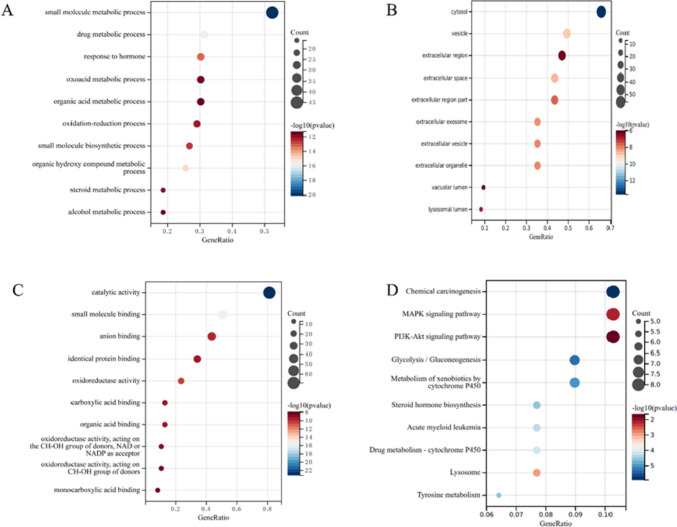
GO enrichment analysis was conducted to reveal the role of PPVI on inflammation and pain. The resulting BP (Fig. 3A) is mainly enriched in small molecule metabolism, drug metabolism, hormone response, and oxidative metabolism. The CC analysis (Fig. 3B), shows enrichment in the cytosol, vesicles, extracellular region, and extracellular space. Finally, MF is shown in Fig. 3C, which mainly involves catalytic activity, small molecule binding, anion binding, and same-protein binding, etc. KEGG-enriched pathways are shown in Fig. 3D and include, chemical carcinogenesis, MAPK signaling pathway, P13K-Akt signaling pathway, glycolysis/glycolysis, cytochrome P450 metabolism of xenobiotics and other biological processes occupied the top.
Fig. 3.
The GO and KEGG enrichment analysis of the common targets of PPVI, inflammation, and pain. A–C GO enrichment analysis of the biological process, molecular function, and all components; X-axis for gene number and p-value for color. D KEGG enrichment analysis of the Common targets of PPVI, inflammation, and pain. Y-axis for pathway, X-axis for gene number, and p-value for color
Molecular docking of PPVI with related target proteins
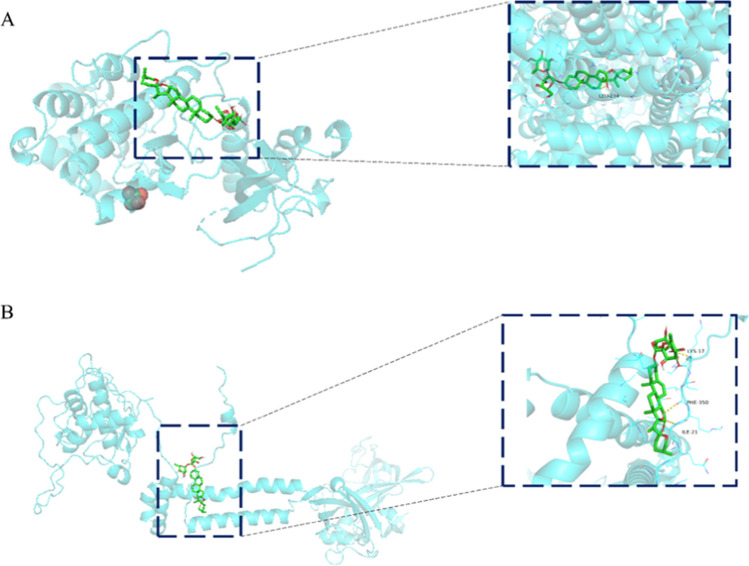
Molecular docking was applied to verify the binding of PPVI to seven key target proteins (Table 1). Results show that PPVI is structurally compatible with the protein, and mostly bound to amino acid residues such as tryptophan, histidine, and leucine in proteins through hydrogen bonding and π-π bonding interactions. ALB was the highest-scoring docking protein, followed by the P2X7R, with binding energies of -9.4 kcal/mol and -9.3 kcal/mol, respectively, indicating excellent binding ability (Fig. 4A, B). Thus PPVI can bind to the active sites of these targets.
Table 1.
Affinities and amino acid sites of ligand–protein detected by molecular docking
| Protein | Ligand | Binding energy (kcal/mol) |
Amino acid binding site |
|---|---|---|---|
| Polyphyllin VI | P2X3 | −7.8 | LYS-151 |
| P2X4 | −7.7 | ALA-297, TRP-164, ARG-309, ARG-295 | |
| P2X7 | −9.3 | LYS-17, PHE-350, ILE-21 | |
| SRC | −7.7 | GLN-228, VAL-177, VAL-233, SER-234 | |
| MAPK1 | −8.0 | THR-206, HIS-299, ASN-47 | |
| ALB | −9.4 | LEV-234 | |
| F2 | −8.4 | GLN-209, ILE-103, MET-201 |
Fig. 4.
Molecular docking models of ALB and P2X7R. A is PPVI and ALB docking mode and the interaction plane diagram. B the PPVI and P2X7R docking model and the interaction plane diagram
Analysis of the DEGs in the DRG during the CFA-induced inflammatory pain mice model
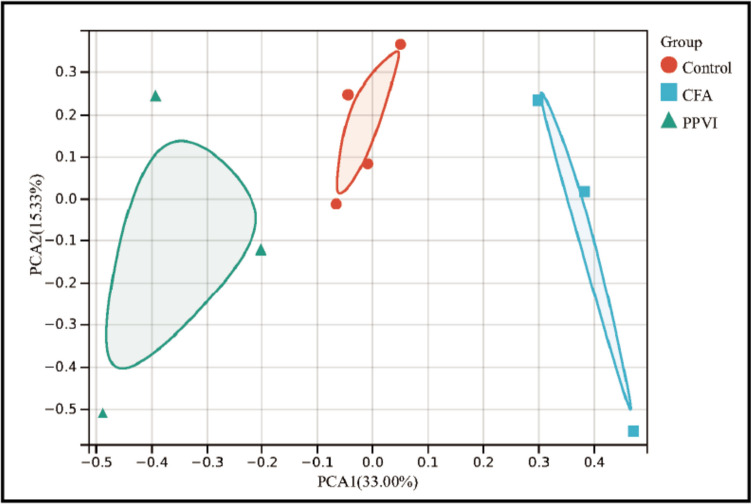
First, we performed PCA on the RNA-Seq data, and the results after excluding abnormal samples are shown in Fig. 5. PCA analysis revealed distinct partitioning of control and PPVI groups with CFA groups.
Fig. 5.
PCA of control、CFA、PPVI group
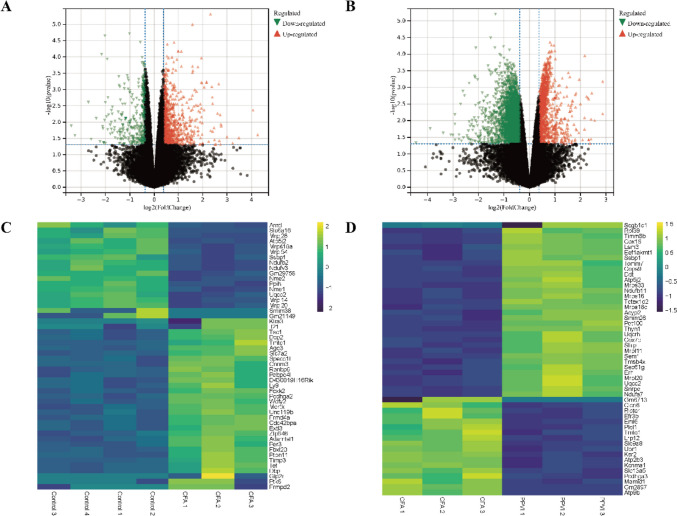
DEGs were identified by the threshold of |log2Foldchange|> 1.3 and p < 0.05 by the limma package in R, as shown in Table 2 and Fig. 6. Compared to the control group, 1332 DEGs were found in the CFA group, the expression levels of 962 genes were up-regulated and 370 genes were down-regulated. Compared with the CFA group, 4291 DEGs were found after PPVI treatment, of which 1164 were up-regulated and 3127 were down-regulated. Volcano curves (Fig. 6A, B) and heatmaps (Fig. 6C, D) were constructed by R to visualize the DEGs.
Table 2.
The number of DEGs in each group
| Group | Total | Up-regulated | Down-regulated | p-value |
|---|---|---|---|---|
| CFA vs. control | 1332 | 962 | 370 | < 0.05 |
| PPVI vs. CFA | 4291 | 1164 | 3127 | < 0.05 |
Fig. 6.
Volcano map and heatmap of DEGs. A Volcano map of our RNA-seq. A total of 962 upregulated and 370 downregulated DEGs were identified between the CFA and the control group. B Volcano map of our RNA-seq. A total of 1164 upregulated and 3127 downregulated DEGs were identified between the PPVI and the CFA group. C A Heatmap of top DEGs was identified between the CFA and the control group. D A Heatmap of top DEGs was identified between the PPVI and the CFA group. Volcano maps exhibit significant DEGs, in volcano maps, red dots mean up-regulated genes, green dots mean down-regulated genes and black dots mean non-significant genes. The dots in the area above the horizontal dotted line have a p-value < 0.05. The dots outside the two vertical dotted lines have a —log2FC— > 1.3. Based on the gene expression matrix, clustering analysis was shown in the heatmap, yellow mean up-regulated genes, purple mean down-regulated genes. (—log2 FC— > 1 and p-value < 0.05)
Enrichment analysis of DEGs
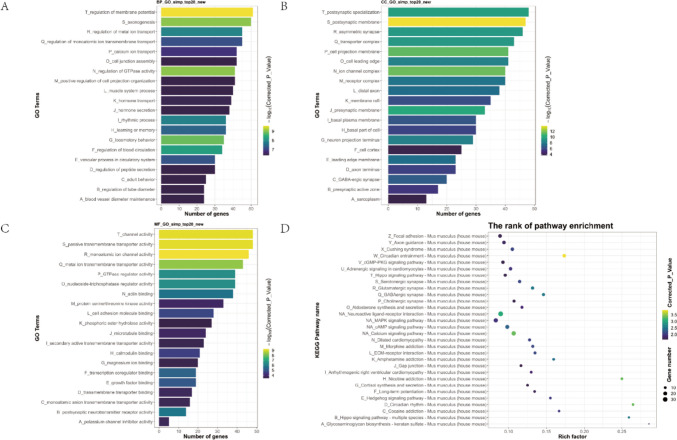
Then, GO and KEGG enrichment analyses were conducted for genes up-regulated by CFA versus control and genes down-regulated by PPVI versus CFA to determine the BP, MF, CC, and signaling pathways associated with the DEGs, as shown in Fig. 7. BP including “regulation of membrane potential”, “axonogenesis”, “regulation of metal ion transport”, “regulation of monoatomic ion transmembrane transport”, and “calcium ion transport” were enriched. The results include “postsynaptic specialization”, “postsynaptic membrane”, “asymmetric synapse”, “transporter complex”, and “cell projection membrane”, which is CC concentrated in. The top 5 of MF are “channel activity”, “passive transmembrane transporter activity”, “monoatomic ion channel activity”, “metal ion transmembrane transporter activity”, and “GTPase regulator activity”. KEGG analysis showed that the signaling pathways enriched mainly included the “cGMP-PKG signaling pathway”, “Hippo signaling pathway”, “MAPK signaling pathway”, “cAMP signaling pathway”, and “Calcium signaling pathway”.
Fig. 7.
GO and KEGG pathway enrichment analysis of DEGs between the CFA and the control group. A, B, C (Top 20) The Bar graph shows that DEGs of the CFA and the control group are enriched in several BP, CC, and MF. D (Top 30) Bubble graph shows that the signaling pathways are enriched by the DEGs of the CFA and the control group. In the bar graph, we sorted the top 20 of BP, CC, and MF by p-value and visualized them. In the bubble graph, the color represents the p-value, and the size of the bubbles represents the gene number
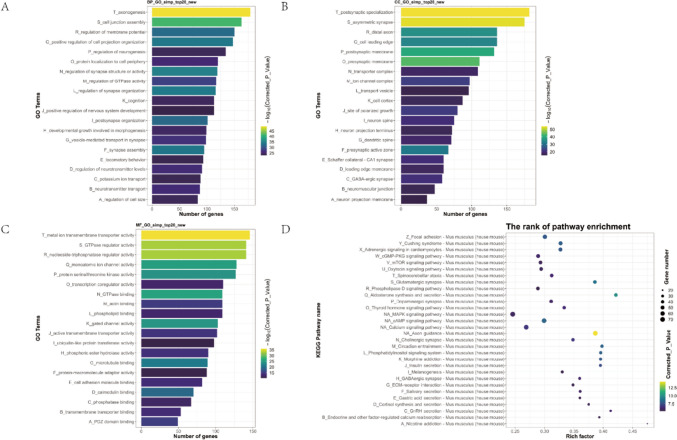
The BP (Fig. 8A) of DEGs was mainly in “axonogenesis”、 “cell junction assembly”, “regulation of membrane potential”, “positive regulation of cell projection organization”, and “regulation of neurogenesis”. The top 5 of CC (Fig. 8B) are “postsynaptic specialization”, “asymmetric synapse”, “distal axon”, “cell leading edge”, and “postsynaptic membrane”. The results of MF (Fig. 8C) include “metal ion transmembrane transporter activity”, “GTPase regulator activity”, “nucleoside-triphosphatase regulator activity”, “monoatomic ion channel activity”, “protein serine/threonine kinase activity”. The results of the KEGG analysis (Fig. 8D) including the “cGMP-PKG signaling pathway”, “mTOR signaling pathway”, “Oxytocin signaling pathway”, “MAPK signaling pathway”, “cAMP signaling pathway” were enriched.
Fig. 8.
GO Annotation and KEGG pathway enrichment analysis of DEGs between the PPVI and the CFA group. A, B, C (Top 20) The Bar graph shows that DEGs of the PPVI and the CFA group are enriched in several BP, CC, and MF. D (Top 30) Bubble graph shows that the signaling pathways are enriched by the DEGs of the PPVI and the CFA group. In the bar graph, we sorted the top 20 of BP, CC, and MF by p-value and visualized them. In the bubble graph, the color represents the p-value, and the size of the bubbles represents the gene number
Analysis revealed substantial similarity in the enriched signaling pathways, whereas the up-regulated pathway in CFA was significantly down-regulated after the treatment of PPVI. Based on the results of RNA-Seq and network pharmacology, MAPK was found to be a common signaling pathway, which may be the key target for PPVI to relieve inflammatory pain.
The effect of PPVI on the viability of RAW264.7 cells
The safe administration concentration range of PPVI to RAW264.7 cells was assessed by using MTT. The results showed that PPVI did not affect RAW264.7 cell viability at concentrations lower than 6 μM, but at 12 μM, cell viability was significantly reduced, as shown in Fig. 9. Therefore, the safe dose range of PPVI for RAW264.7 cells was considered to be 0–6 μM, and the low, medium, and high concentrations of PPVI for further experiments were determined at 1.5 μM, 3 μM, and 6 μM.
Fig. 9.
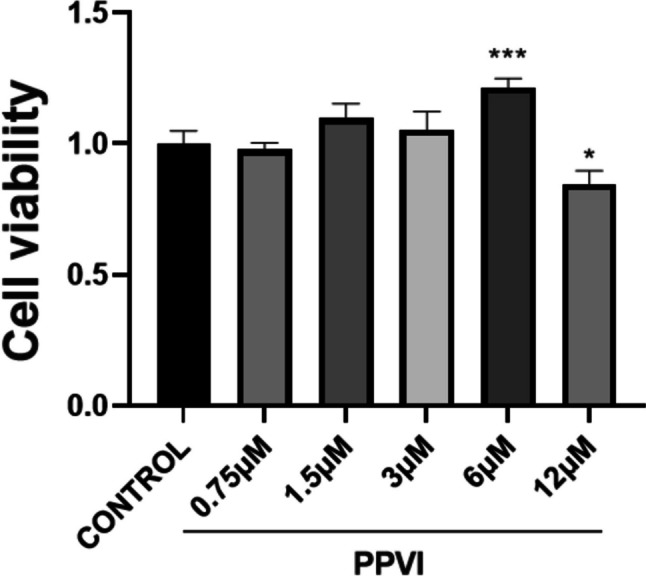
The effect of PPVI on the viability of RAW264.7 cells. Cytotoxic effect of various concentrations of PPVI (0.75, 1.5, 3, 6, and 12 mM) on RAW264.7 cells after 24 h examined by using the MTT assay. Values represent average + S.U. Significant differences among different groups are indicated as *p < 0.05, ***p < 0.001 vs. the control group
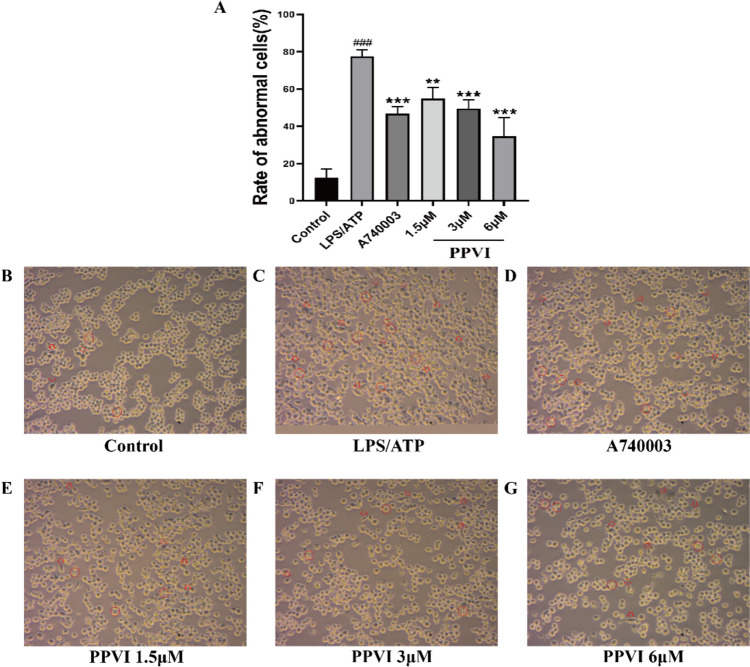
The effect of PPVI on LPS/ATP-induced RAW264.7 cell in cell morphology
Then, we used the LPS/ATP-induced in vitro model of RAW264.7. As shown in Fig. 10B, the RAW264.7 cells were small and round in the Control group. By contrast, after incubation with the LPS/ATP for 24 h cells showed (Fig. 10C) enlarged cell bodies and mostly extended lamellipodia from the entire periphery of the cell. In addition, LPS/ATTP-induced RAW264.7 cells also showed vacuolation and cell membrane rupture. Figure 10A, D–G indicated that compared with the LPS/ATP-treated cells, the RAW264.7 cells pre-treated with A740003 and PPVI (1.5 μM, 3 μM, 6 μM) displayed significant improvement of cell morphology(smaller and rounded) and disappearance of vacuolation. The results showed that PPVI inhibited the morphological changes and vacuolation of RAW264.7 cells induced by LPS/ATP and exhibited inflammatory protective effects at 1.5, 3, and 6 μM.
Fig. 10.
The effect of PPVI pre-treated for 1 h on the cell morphology of the LPS/ATP-induced RAW264.7 cell (n = 3). Red arrows indicate changes in cell morphology, and red circles indicate vacuolation and cell membrane rupture. Each group was visualized by an ordinary light microscope (× 400 magnification). Values represent average ± S.D. Significant differences among different groups are indicated as ###p < 0.001 vs. Control; **p < 0.01, ***p < 0.001 vs. LPS/ATP group
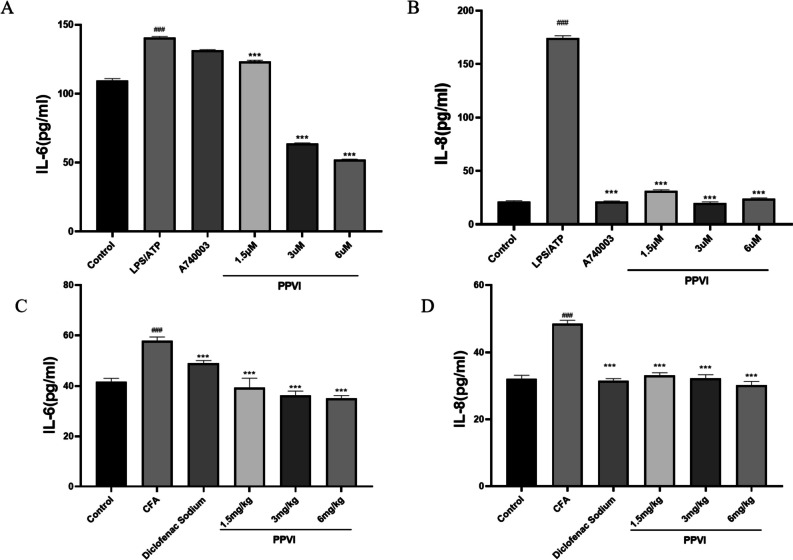
Effect of PPVI on the levels of inflammatory mediators
ELISA kits were used to detect the inflammatory mediators to verify the validity of the inflammation model. The content of IL-6 and IL-8 was significantly elevated in LPS/ATP-treated cells. Compared with the LPS/ATP group, different doses of PPVI significantly inhibited the release of IL-6 and IL-8 after pre-protection. However, the pre-treatment with A740003 significantly reduced the elevation of IL-8 induced by LPS/ATP but not IL-6(Fig. 11A–B), this result may be attributed to PPVI may also modulate targets other than P2X7R;
Fig. 11.
The effects of PPVI on the expression of IL-6 and IL-8 of LPS/ATP-induced RAW264.7 cells and serum of CFA-induced inflammatory pain mice. (A-B) The effects of PPVI on the expression of IL-6 and IL-8 of LPS/ATP-induced RAW264.7 cell (n = 3). (C-D) The effects of PPVI on the expression of IL-6 and IL-8 in serum of CFA-induced inflammatory pain mice (n = 3). Values represent average ± S.D. Significant differences among different groups are indicated as ###p < 0.001 vs. Control; *p < 0.05, **p < 0.01vs. ***p < 0.001 vs. LPS/ATP group or CFA group
In addition, we examined the effect of PPVI on the serum levels of inflammatory mediators induced by CFA in mice. The serum levels of IL-6 and IL-8 inflammatory mediators were significantly increased in the CFA group, whereas PPVI significantly inhibited the release of IL-6 and IL-8 after 7 days of continuous administration (Fig. 11C–D).
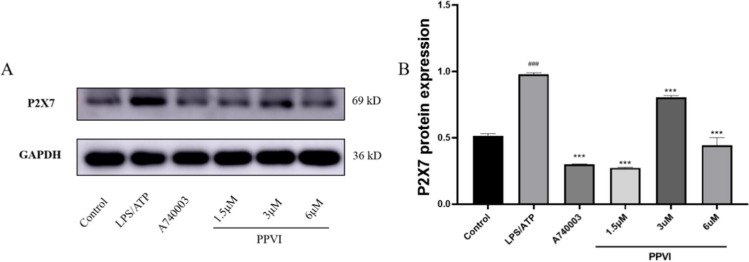
Effect of PPVI on LPS/ATP-induced P2X7R expression in RAW264.7 cells
The results of molecular docking showed that PPVI had extremely strong binding energy with the P2X7R. Thus we used western blotting to analyse the effect of PPVI on the expression of P2X7R in LPS/ATP-induced RAW264.7 cells. The results are shown in Fig. 12: the expression of the P2X7R increased after the treatment of LPS/ATP, while both A740003 and PPVI significantly inhibited this up-regulation.
Fig. 12.
Effects of PPVI on the P2X7R protein expression of LPS/ATP-induced RAW264.7 cell (n = 3). Values represent average ± S.D. Significant differences among different groups are indicated as ##p < 0.01, ###p < 0.001 vs. Control; *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS/ATP group
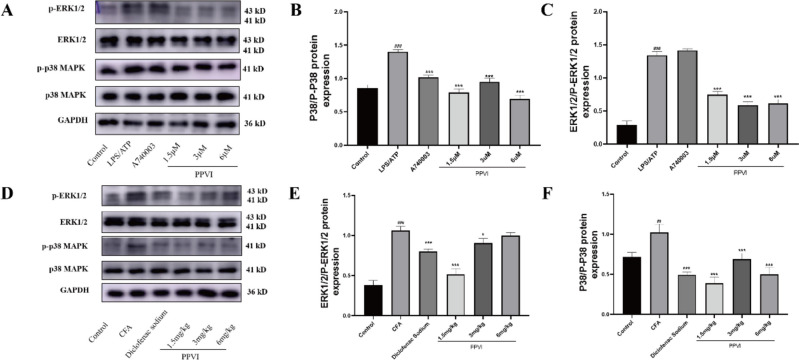
Effect of PPVI on the expression of MAPK signaling cascade
Based on the results of KEGG enrichment analysis in network pharmacology and RNA-Seq, PPVI most likely exerts anti-inflammatory and analgesic effects by modulating the MAPK signaling pathway. Therefore, we examined the effects of PPVI on the MAPK signaling pathway in both in vitro and in vivo models. The levels of p-p38 and p-ERK1/2 increased in LPS/ATP-induced RAW264.7 cells (Fig. 13A–C) and DRGs of mice of the CFA-induced pain model (Fig. 13D–F), whereas PPVI administration significantly reduced the phosphorylation of p38 and ERK1/2. Interestingly, in cell experiments, A740003 significantly reduced the phosphorylation level of P38, but similar results were not observed for p-ERK1/2. These results suggest that P2X7R may modulate the release of inflammatory cytokines through enhanced phosphorylation of p38 rather than ERK1/2.
Fig. 13.
Effects of PPVI on the phosphorylation level of p38 and ERK1/2 of LPS/ATP-induced RAW264.7 cells and CFA-induced inflammatory pain mice. A–C The effects of PPVI on phosphorylation level of p38 and ERK1/2 in LPS/ATP induced RAW264.7 cell (n = 3). D–F The effects of PPVI on phosphorylation level of p38 and ERK1/2 in serum of CFA-induced inflammatory pain mice (n = 3). Values represent average ± S.D. Significant differences among different groups are indicated as ##p < 0.01, ###p < 0.001 vs. Control; *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS/ATP or CFA group
Discussion
Neutrophils, macrophages, and glial cells have a critical role in the development and maintenance of chronic pain, according to research from recent decades [26–28]. These immune cells and neuroglia are affected by nociceptor sensitization caused by nociceptive stimulation and inflammatory mediators, leading to neuroinflammation [29]. The release of cytokines and chemokines by reactive glial cells and system immune cells is necessary for the development of neuroinflammation, and cytokines interact with nociceptors to exacerbate neuropathic pain [30, 31]. In this study, network pharmacology was utilized to combine putative intervention targets of pain and inflammation in order to forecast the targets and mechanisms of the PPVI analgesic and anti-inflammatory activities. SRC, F2, ALB, and MAPK1 were found to be the core targets of the protein interaction network, while KEGG analysis was mainly concentrated on highlighted inflammatory signaling pathways such as MAPK and P13K-Akt. This conclusion is consistent with the animal experiments showing that PPVI alleviates pain sensitization in the CFA-induced pain mice model by reducing the expression of inflammatory mediators [24]. The MAPK signaling pathway was also present in the subsequent KEGG enrichment analysis of DEGs of RNA-Seq.
Due to repeated nociceptive stimulation and infiltration of inflammatory mediators, receptors and ion channels in nociceptive neurons are continuously activated [29, 32]. The activation of these receptors like the P2X7R releases various pro-inflammatory mediators that exacerbate inflammation. Expression of the P2X7R is up-regulated in various pain models such as CFA [33], SNI [34], and CCI [35]. Activation of the P2X7R facilitates the release of various pro-inflammatory mediators and leads to the recruitment of inflammasome [36–38], which are essential for pain sensitization. However, the specific regulatory mechanism of P2X7R on the release of inflammatory mediators remains unclear. Studies have shown that the activation of NF-κB induced by myeloid differentiation primary-response protein 88 (MyD88) was enhanced by P2X7R co-expression after the treatment with LPS/ATP [39]. NF-κB is the important transcriptional factor involved in the production of pro-inflammatory cytokines (IL-1β, IL-18, IL-6, IL-8, and TNF-α). At present, there are numerous of evidence that P2X7R mediates the release of IL-1β and IL-18 through the NLRP3-Caspase-1 signaling pathway [40–42]. On the contrary, the regulation of IL-6 and IL-8 by P2X7R needs to be further clarified. It is widely believed that IL-6 and IL-8 play an important role in inflammatory pain. IL-6 in the red nucleus sustains neuropathic pain by up-regulating the expression of tumor necrosis factor-α (TNF-α) and Interleukin-1β (IL-1β). Intracerebral injection of exogenous IL-6 to naive rats lowered the paw withdrawal threshold(PWT) of the contralateral hind paw and boosted the local levels of TNF-α and IL-1β [43]. In addition, IL-8 modulated thermal hyperalgesia by enhancing prefrontal synaptic transmission in persistent inflammatory pain mice [10].
In this study, we utilized the RAW264.7 cell model induced by LPS/ATP and incubated with a P2X7R inhibitor(A740003) to investigate the mechanism of the release of IL-6 and IL-8, as well as the regulatory effects of PPVI on P2X7R. In recent years, the LPS/ATP-induced RAW264.7 cell model was used to explore the release of IL-1β regulated by NLRP3 inflammasome [44, 45]. In addition, P2X7R was able to activate NF-κB through a MyD88-dependent pathway after the treatment with LPS/ATP [39]. LPS is usually used to induce inflammation model in vivo/vitro, and ATP activates the purinergic receptor P2X7R. The combination of the LPS/ATP may provide new insight into the release of inflammatory mediators caused by the activation of P2X7R.
With the development of bioinformatics and high-throughput omics technology, the characteristics of multi-target, multi-pathway, and multi-link of traditional Chinese medicine are no longer a major problem, and the elucidation of potential targets and mechanisms of action of natural compounds has become relatively simple. At present, PolyphyllinI [46] and PolyphyllinVII [47] have been found from Paris Polyphylla to have anti-inflammatory effects. In our previous studies, PPVI was identified to reduce the expression of inflammatory mediators(TNF-α, IL-1β, IL-6) and the aggregation of inflammatory cells in DRG and spinal cord of CFA-induced inflammatory pain mice [24]. However, the analgesic and anti-inflammatory mechanism of PPIV needs to be further clarified. In this study, we combined network pharmacology, RNA-Seq, and molecular docking to predict and explore PPVI in various aspects. Based on the essential role of the P2X7R in inflammation and pain, the molecular docking with PPVI was analyzed. Subsequent cell experiments also found that PPVI inhibits the release of IL-8 of RAW264.7 induced by LPS/ATP based on the regulation of the phosphorylation of p38 instead of ERK1/2 by P2X7R. However, the modulation of PPVI on the release of IL-6 may be mediated by other P2X7R-independent signals. Though PPVI inhibited the expression of P2X7R in LPS/ATP-induced RAW264.7 cells in vitro, the analgesic effects of PPVI by inhibiting P2X7R needed to be further evaluated by using the DRG and spinal cord of CFA mice.
Conclusion
In summary, by using network pharmacology, molecular docking and RNA-Seq technologies, we identified the possible target genes and related signaling pathways of PPVI action on inflammatory pain which the PPVI showed strong binding energy to P2X7R and KEGG results suggested that PPVI relieves pain by modulating the MAPK signaling pathway. In addition, PPVI down-regulates expression of IL-8 in LPS/ATP-induced RAW264.7 cells and CFA mice, by decreasing the phosphorylation of p-p38 and p-ERK1/2through inhibiting the activation of P2X7R, instead, the down-regulation of IL-6 is in a P2X7R-independent pathway. Our work provides insight into the potential new targets and mechanisms of PPVI action for the treatment of inflammatory pain.
Abbreviations
- BP
Biological process
- CFA
Complete Freund's adjuvant
- CC
Cellular component
- DRG
Dorsal root ganglia
- DEGs
Differentially expressed genes
- ELISA
Enzyme-Linked Immunosorbent Assay
- GO
Gene ontology
- ICD-11
International Classification of Diseases
- KEGG
Kyoto Encyclopedia of Genes and Genome
- MTT
3-(4,5-Di-2-yl)-2,5-tetrazolium bromide
- MF
Molecular function
- MyD88
Myeloid differentiation primary-response protein 88
- P2X7 receptor
P2X7R
- PPVI
Polyphyllin VI
- PCA
Principal component analysis
- PPI
Protein-protein interaction
- PWT
Paw withdrawal threshold
- PVDF
Polyvinylidene fluoride
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCMSP
Traditional Chinese Medicine Systems Pharmacology
- TNF-α
Tumor necrosis factor-α
Zhenglang Zhang
is performing experimental work to get a master’s degree in pharmacology at Jinan University. His research work is focused on the excavation of potential targets and mechanisms of chronic pain including inflammatory pain and neuropathic pain. In addition, He also discover natural compounds from traditional Chinese medicine that exert analgesic effects based on pain-related purinergic receptors such as the P2X3, P2X4, and P2X7 receptors. 
Author’s contribution
Work Zhenglang Zhang and Tingting Wang: Conceptualization, Investigation, data curation, Formal analysis, writing-original draft preparation, Writing-Review and Editing. Zhenhui Luo: Conceptualization, Investigation, Writing-Reviewing and Editing. Muhammad Haris Zaib and Mengqin Yi: Methodology. Hekun Zeng: Investigation. Peiyang Li: Investigation. Dan Tang, Hong Nie and Alexei Verkhratsky: Funding acquisition, Conceptualization, Writing-Reviewing, and Editing. All the authors agreed for the final version to be published. Zhenglang Zhang and Tingting Wang contributed equally to this.
Funding
This research was supported by the National Natural Science Foundation of China. (No. 8181101216).
Data availability
Data can be made available upon reasonable request.
Declarations
Ethical approval
The animal study was reviewed and approved by the Jinan University Animal Ethics Committee. (Approval No. IACUC-20210510-02).
Competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflicts of interest
Zhenglang Zhang declares that he has no conflict of interest.
Tingting Wang declares that she has no conflict of interest.
Zhenhui Luo declares that he has no conflict of interest.
Muhammad Haris Zaib declares that he has no conflict of interest.
Mengqin Yi declares that she has no conflict of interest.
Hekun Zeng declares that he has no conflict of interest.
Peiyang Li declares that he has no conflict of interest.
Dan Tang declares that he has no conflict of interest.
Alexei Verkhratsky declares that he has no conflict of interest.
Hong Nie declares that she has no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alexei Verkhratsky, Email: alexej.verkhratsky@manchester.ac.uk.
Hong Nie, Email: hongnie1970@163.com, Email: tnieh@jnu.edu.cn.
References
- 1.Seidel MF et al (2022) Neurogenic inflammation as a novel treatment target for chronic pain syndromes. Exp Neurol 356:114108. 10.1016/j.expneurol.2022.114108 10.1016/j.expneurol.2022.114108 [DOI] [PubMed] [Google Scholar]
- 2.Cohen SP, Vase L, Hooten WM (2021) Chronic pain: an update on burden, best practices, and new advances. Lancet 397(10289):2082–2097. 10.1016/S0140-6736(21)00393-7 10.1016/S0140-6736(21)00393-7 [DOI] [PubMed] [Google Scholar]
- 3.Meints SM, Edwards RR (2018) Evaluating psychosocial contributions to chronic pain outcomes. Prog Neuropsychopharmacol Biol Psychiatry 87(Pt B):168–182. 10.1016/j.pnpbp.2018.01.017 10.1016/j.pnpbp.2018.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afridi B et al (2021) Pain perception and management: where do we stand? Curr Mol Pharmacol 14(5):678–688. 10.2174/1874467213666200611142438 10.2174/1874467213666200611142438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindu S, Mazumder S, Bandyopadhyay U (2020) Non-steroidal anti-inflammatory drugs (NSAIDs) and organ damage: a current perspective. Biochem Pharmacol 180:114147. 10.1016/j.bcp.2020.114147 10.1016/j.bcp.2020.114147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro H et al (2022) Non-steroidal anti-inflammatory drugs (NSAIDs), pain and aging: Adjusting prescription to patient features. Biomed Pharmacother 150:112958. 10.1016/j.biopha.2022.112958 10.1016/j.biopha.2022.112958 [DOI] [PubMed] [Google Scholar]
- 7.Enthoven WT et al (2016) Non-steroidal anti-inflammatory drugs for chronic low back pain. Cochrane Database Syst Rev 2(2):CD012087. 10.1002/14651858.CD012087 10.1002/14651858.CD012087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes FIF, Cunha FQ, Cunha TM (2020) Peripheral nitric oxide signaling directly blocks inflammatory pain. Biochem Pharmacol 176:113862. 10.1016/j.bcp.2020.113862 10.1016/j.bcp.2020.113862 [DOI] [PubMed] [Google Scholar]
- 9.Li C et al (2021) Common and discrete mechanisms underlying chronic pain and itch: peripheral and central sensitization. Pflugers Arch 473(10):1603–1615. 10.1007/s00424-021-02599-y 10.1007/s00424-021-02599-y [DOI] [PubMed] [Google Scholar]
- 10.Cui GB et al (2012) Elevated interleukin-8 enhances prefrontal synaptic transmission in mice with persistent inflammatory pain. Mol Pain 8:11. 10.1186/1744-8069-8-11 10.1186/1744-8069-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z et al (2021) From purines to purinergic signalling: molecular functions and human diseases. Signal Transduct Target Ther 6(1):162. 10.1038/s41392-021-00553-z 10.1038/s41392-021-00553-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu SQ et al (2022) P2X7 receptor in inflammation and pain. Brain Res Bull 187:199–209. 10.1016/j.brainresbull.2022.07.006 10.1016/j.brainresbull.2022.07.006 [DOI] [PubMed] [Google Scholar]
- 13.Illes P, Khan TM, Rubini P (2017) Neuronal P2X7 receptors revisited: do they really exist? J Neurosci 37(30):7049–7062. 10.1523/JNEUROSCI.3103-16.2017 10.1523/JNEUROSCI.3103-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera A, Vanzulli I, Butt AM (2016) A central role for ATP signalling in glial interactions in the CNS. Curr Drug Targets 17(16):1829–1833. 10.2174/1389450117666160711154529 10.2174/1389450117666160711154529 [DOI] [PubMed] [Google Scholar]
- 15.Faria RX, Freitas HR, Reis RAM (2017) P2X7 receptor large pore signaling in avian Muller glial cells. J Bioenerg Biomembr 49(3):215–229. 10.1007/s10863-017-9717-9 10.1007/s10863-017-9717-9 [DOI] [PubMed] [Google Scholar]
- 16.Calik I et al (2020) P2X7 receptor as an independent prognostic indicator in gastric cancer. Bosn J Basic Med Sci 20(2):188–196. 10.17305/bjbms.2020.4620 10.17305/bjbms.2020.4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Virgilio F et al (2017) The P2X7 receptor in infection and inflammation. Immunity 47(1):15–31. 10.1016/j.immuni.2017.06.020 10.1016/j.immuni.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 18.Chessell IP et al (2005) Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain 114(3):386–396. 10.1016/j.pain.2005.01.002 10.1016/j.pain.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y et al (2021) P2X7 receptor antagonist BBG inhibits endoplasmic reticulum stress and pyroptosis to alleviate postherpetic neuralgia. Mol Cell Biochem 476(9):3461–3468. 10.1007/s11010-021-04169-3 10.1007/s11010-021-04169-3 [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K et al (2011) Induction of the P2X7 receptor in spinal microglia in a neuropathic pain model. Neurosci Lett 504(1):57–61. 10.1016/j.neulet.2011.08.058 10.1016/j.neulet.2011.08.058 [DOI] [PubMed] [Google Scholar]
- 21.Alves LA et al (2013) Physiological roles and potential therapeutic applications of the P2X7 receptor in inflammation and pain. Molecules 18(9):10953–10972. 10.3390/molecules180910953 10.3390/molecules180910953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng JF et al (2020) Polyphyllin VI induces Caspase-1-Mediated pyroptosis via the induction of ROS/NF-kappaB/NLRP3/GSDMD signal axis in non-small cell lung cancer. Cancers (Basel) 12(1):193. 10.3390/cancers12010193 10.3390/cancers12010193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Q et al (2023) High‑throughput screening identification of a small‑molecule compound that induces ferroptosis and attenuates the invasion and migration of hepatocellular carcinoma cells by targeting the STAT3/GPX4 axis. Int J Oncol 62(3). 10.3892/ijo.2023.5490 [DOI] [PMC free article] [PubMed]
- 24.Luo Z et al (2023) Polyphyllin VI screened from Chonglou by cell membrane immobilized chromatography relieves inflammatory pain by inhibiting inflammation and normalizing the expression of P2X(3) purinoceptor. Front Pharmacol 14:1117762. 10.3389/fphar.2023.1117762 10.3389/fphar.2023.1117762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie ME et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. 10.1093/nar/gkv007 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji RR, Chamessian A, Zhang YQ (2016) Pain regulation by non-neuronal cells and inflammation. Science 354(6312):572–577. 10.1126/science.aaf8924 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang JX et al (2022) Potential neuroimmune interaction in chronic pain: a review on immune cells in peripheral and central sensitization. Front Pain Res (Lausanne) 3:946846. 10.3389/fpain.2022.946846 10.3389/fpain.2022.946846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnelly CR et al (2020) Central nervous system targets: glial cell mechanisms in chronic pain. Neurotherapeutics 17(3):846–860. 10.1007/s13311-020-00905-7 10.1007/s13311-020-00905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang XX et al (2023) Inflammation in pathogenesis of chronic pain: Foe and friend. Mol Pain 19:17448069231178176. 10.1177/17448069231178176 10.1177/17448069231178176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer C, Leinders M, Uceyler N (2018) Inflammation in the pathophysiology of neuropathic pain. Pain 159(3):595–602. 10.1097/j.pain.0000000000001122 10.1097/j.pain.0000000000001122 [DOI] [PubMed] [Google Scholar]
- 31.Kong YF et al (2021) CXCL10/CXCR3 signaling in the DRG exacerbates neuropathic pain in mice. Neurosci Bull 37(3):339–352. 10.1007/s12264-020-00608-1 10.1007/s12264-020-00608-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold MS, Gebhart GF (2010) Nociceptor sensitization in pain pathogenesis. Nat Med 16(11):1248–1257. 10.1038/nm.2235 10.1038/nm.2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neves AF et al (2020) Peripheral inflammatory hyperalgesia depends on P2X7 receptors in satellite glial cells. Front Physiol 11:473. 10.3389/fphys.2020.00473 10.3389/fphys.2020.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu X et al (2020) Inhibition of P2X7R in the amygdala ameliorates symptoms of neuropathic pain after spared nerve injury in rats. Brain Behav Immun 88:507–514. 10.1016/j.bbi.2020.04.030 10.1016/j.bbi.2020.04.030 [DOI] [PubMed] [Google Scholar]
- 35.Yang R et al (2021) Gallic acid alleviates neuropathic pain behaviors in rats by inhibiting P2X7 receptor-mediated NF-kappaB/STAT3 signaling pathway. Front Pharmacol 12:680139. 10.3389/fphar.2021.680139 10.3389/fphar.2021.680139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson KV, Deng M, Ting JP (2019) The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19(8):477–489. 10.1038/s41577-019-0165-0 10.1038/s41577-019-0165-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz FM et al (2020) Reactive oxygen species play a role in P2X7 receptor-mediated IL-6 production in spinal astrocytes. Purinergic Signal 16(1):97–107. 10.1007/s11302-020-09691-5 10.1007/s11302-020-09691-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying YL et al (2014) Over-expression of P2X7 receptors in spinal glial cells contributes to the development of chronic postsurgical pain induced by skin/muscle incision and retraction (SMIR) in rats. Exp Neurol 261:836–843. 10.1016/j.expneurol.2014.09.007 10.1016/j.expneurol.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Xiao Y, Li Z (2011) P2X7 receptor positively regulates MyD88-dependent NF-kappaB activation. Cytokine 55(2):229–236. 10.1016/j.cyto.2011.05.003 10.1016/j.cyto.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 40.Hayashi K et al (2023) P2X7-NLRP3-Caspase-1 signaling mediates activity-induced muscle pain in male but not female mice. Pain. 10.1097/j.pain.0000000000002887 10.1097/j.pain.0000000000002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahlenberg JM et al (2005) Potentiation of caspase-1 activation by the P2X7 receptor is dependent on TLR signals and requires NF-kappaB-driven protein synthesis. J Immunol 175(11):7611–7622. 10.4049/jimmunol.175.11.7611 10.4049/jimmunol.175.11.7611 [DOI] [PubMed] [Google Scholar]
- 42.Sun S et al (2023) Ligand-gated ion channel P2X7 regulates NLRP3/Caspase-1-mediated inflammatory pain caused by pulpitis in the trigeminal ganglion and medullary dorsal horn. Brain Res Bull 192:1–10. 10.1016/j.brainresbull.2022.10.020 10.1016/j.brainresbull.2022.10.020 [DOI] [PubMed] [Google Scholar]
- 43.Yang QQ et al (2020) Red nucleus IL-6 mediates the maintenance of neuropathic pain by inducing the productions of TNF-alpha and IL-1beta through the JAK2/STAT3 and ERK signaling pathways. Neuropathology 40(4):347–357. 10.1111/neup.12653 10.1111/neup.12653 [DOI] [PubMed] [Google Scholar]
- 44.Li W et al (2022) A novel drug combination of mangiferin and cinnamic acid alleviates rheumatoid arthritis by inhibiting TLR4/NFkappaB/NLRP3 activation-induced pyroptosis. Front Immunol 13:912933. 10.3389/fimmu.2022.912933 10.3389/fimmu.2022.912933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hseu YC et al (2022) Coenzyme Q(0) inhibits NLRP3 inflammasome activation through mitophagy induction in LPS/ATP-stimulated macrophages. Oxid Med Cell Longev 2022:4266214. 10.1155/2022/4266214 10.1155/2022/4266214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S et al (2021) Polyphyllin I inhibits propionibacterium acnes-induced IL-8 secretion in HaCaT cells by downregulating the CD36/NOX1/ROS/NLRP3/IL-1beta pathway. Evid Based Complement Alternat Med 2021:1821220. 10.1155/2021/1821220 10.1155/2021/1821220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C et al (2019) In vitro and in vivo anti-inflammatory effects of Polyphyllin VII through Downregulating MAPK and NF-kappaB pathways. Molecules 24(5):875. 10.3390/molecules24050875 10.3390/molecules24050875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be made available upon reasonable request.