Abstract
The orchestrated liberation of viral proteins by 3Cpro-mediated proteolysis is pivotal for gene expression by picornaviruses. Proteolytic processing is regulated either by the amino acid sequence at the cleavage site of the substrate or by cofactors covalently or noncovalently linked to the viral proteinase. To determine the role of the amino acid sequence at cleavage sites 3A/3B and 3B/3C that are essential for the liberation of 3Cpro from its precursors and to assess the function of the stable processing intermediates 3AB and 3ABC, we studied the effect of cleavage site mutations on hepatitis A virus (HAV) polyprotein processing, particle formation, and replication. Using the recombinant vaccinia virus system, we showed that the normally retarded cleavage at the 3A/3B junction can be improved by altering the amino acid sequence at the scissile bond such that it matches the preferred HAV 3C cleavage sites. In contrast to the processing products of the wild-type polyprotein, 3ABC was no longer detectable in the mutant. VP0 and VP3 were generated less efficiently, implying that processing of the structural protein precursor P1-2A depends on the presence of stable 3ABC and/or 3AB. In addition, cleavage of 2BC was impaired in 3AB/3ABC-deficient mutants. Formation of HAV particles was not affected in mutants with blocked 3A/3B and/or 3B/3C cleavage sites. However, 3ABC-deficient mutants produced small numbers of HAV particles, which could be augmented by coexpressing 3AB or 3ABC. The hydrophobic domain of 3A that has been proposed to mediate membrane anchorage of the replication complex was crucial for restoration of defective particle formation. In vitro transcripts of the various cleavage site mutants were unable to initiate an infectious cycle, and no progeny viruses were obtained even after blind passages. Taken together, the data suggest that accumulation of uncleaved HAV 3AB and/or 3ABC is pivotal for both viral replication and efficient particle formation.
Picornavirus gene expression requires the orchestrated liberation of viral proteins by proteolysis from a single polyprotein which comprises three major domains, P1, P2, and P3 (for nomenclature of the hepatitis A virus [HAV] polyprotein, see Fig. 1A). For optimal use of the low coding capacity of the RNA genome, it seems that not only mature but also intermediate products of proteolytic processing play important roles during the viral life cycle. In the past few years, it also became evident that picornavirus proteins can function in alternate processing forms or as aggregates with viral or host proteins (5, 22, 27, 36, 40). The virus-encoded proteinase 3Cpro is responsible for the regulated processing of the picornavirus polyprotein at most cleavage sites whose primary amino acid sequences are highly conserved. The best studied substrate is the polyprotein of poliovirus (PV), where Gln-Gly pairs are found at all 3C-cleaved scissile bonds. In addition to the primary amino acid sequence at the cleavage site, extrinsic factors, such as exposure and environment, might determine the cleavability of the substrate (5, 19, 22). It was clearly shown that the functional proteinase for cleavage at some sites—in particular within the P1 domain—is proteinase 3CD, a stable precursor form of proteinase 3C (43). In fact, it seems that the liberation of PV proteins is controlled by both the form of the proteinase and the primary structure of the substrate.
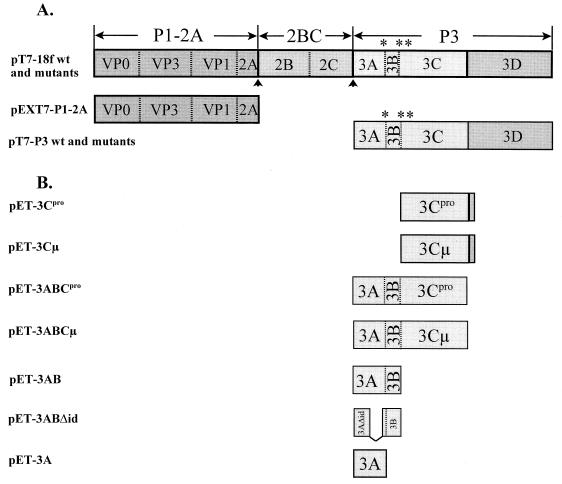
FIG. 1.
Schematic presentation of wild-type (wt) and mutated HAV cDNA constructs encoding the complete polyprotein (pT7-18f), P1-2A (pEXT7-P1-2A), or P3 (pT7-P3) (A) and of HAV cDNA clones used in trans-complementation experiments (B). ∗ and ∗∗ indicate the 3A/3B and 3B/3C cleavage sites mutated in the HAV genome, respectively. The primary cleavage sites are marked by arrowheads. μ denotes the proteolytically inactive form of the proteinase due to the Cys-to-Ala mutation at the active site of the enzyme.
In HAV, liberation of viral proteins differs in several aspects from that in other picornaviruses, and this might be the reason why this virus is distinct in its structure and growth pattern (1, 15–17, 21, 28–32, 34). 3C, as the proteolytic entity of the HAV P3 domain, first cleaves at two sites within the polyprotein, resulting in the liberation of the HAV primary cleavage products P1-2A, 2BC, and P3 (Fig. 1A), which are further processed in secondary cleavages. Detailed studies of the processing of HAV P3 revealed that only the 3C/3D junction is efficiently cleaved whereas the retarded cleavage at sites 3A/3B and/or 3B/3C gives rise to the stable intermediates 3BC and 3ABC (29). In contrast to the enteroviruses and rhinoviruses, the amino acid sequence at the HAV 3C-cleaved junctions varies widely. Based on studies with synthetic peptides, a hierarchy of preferred 3C cleavage sites in the HAV polyprotein was proposed and could be further substantiated when the topology of the substrate binding pocket of HAV 3C was resolved by determination of the crystal structure (4, 14). The collective results of these studies suggest that a consensus sequence at the scissile bond might be a prerequisite for optimal cleavage by HAV 3C (Table 1). The amino acid sequence at the 3A/3B junction differs significantly from those at other sites, providing an explanation for the stability of 3A-containing intermediate processing products. These observations suggest that cleavability of the HAV 3A/3B site might be determined predominantly by the primary sequence and, furthermore, that the retarded cleavage might be essential for the viral life cycle.
TABLE 1.
Amino acid sequence at the wild-type and mutated cleavage sites of the HAV polyprotein
| Cleavage site or mutant | Amino acid at cleavage sitea:
|
Cleavage efficiency at indicated site(s)b | |||
|---|---|---|---|---|---|
| P4 | P3 | P2 | P1/P1′ | ||
| Cleavage sites within HAV polyprotein | |||||
| VP0/VP3 | L | S | T | Q/M | |
| VP3/VP1 | V | T | T | Q/V | |
| VP1/2A | L | S | T | E/S | |
| 2A/2B | L | F | S | Q/A | |
| 2B/2C | L | R | T | Q/S | |
| 2C/3A | L | W | S | Q/G | |
| 3A/3B | I | P | A | E/G | |
| 3B/3C | V | E | S | Q/S | |
| 3C/3D | L | E | S | Q/R | |
| Consensus sequence | L(V) | x | T(S) | Q(E)/x | |
| 3A/3B cleavage site and mutants | |||||
| wt | I | P | A | E/G | ± |
| 1 | I | P | A | v/G | − |
| 2 | I | P | A | q/G | ++ |
| 3 | I | t | A | E/G | ++ |
| 4 | I | t | A | q/G | +++ |
| 3B/3C cleavage site and mutant | |||||
| wt | V | E | S | Q/S | ± |
| 5 | V | E | S | Q/l | − |
| Double mutantc | |||||
| 6 | − | ||||
As an intermediate product of PV and HAV P3 processing, protein 3AB was found to exhibit diverse properties and multiple functions (3, 7, 26, 35, 40, 41). A prominent structural feature of 3AB is a stretch of hydrophobic amino acids that mediates its interaction with membranes and viral proteins. Interaction of PV 3AB with 3CD is particularly pronounced, since it stimulates the autoproteolytic activity of 3CD and cleavage of 2BC (19, 22, 27, 41). Unlike the PV protein (40), HAV 3AB interacts specifically with viral RNA when studied in vitro (2, 18). Furthermore, a central role of 3A and/or its precursor polypeptides in HAV replication is underlined by the observation that adaptation to rapid growth in cell culture is often accompanied by mutations in 3A, in addition to mutations in the P2 domain (11, 23, 44).
The role of particular cleavage site sequences in polyprotein processing and replication was studied for a number of RNA viruses, such as Sindbis virus, yellow fever virus, hepatitis C virus (HCV), coxsackievirus, and PV, by using specifically mutated cleavage sites (6, 8, 20, 24, 37, 38). From these studies, it was concluded that blocking the liberation of nonstructural proteins always resulted in a loss of RNA replication but usually did not prevent cleavage at other sites in the polyprotein. Although differential cleavage by a single proteolytic entity (e.g., PV 3Cpro) has been described in detail, no attempts were made to enhance the processing of unfavorable sites in a polyprotein in order to elucidate the role of cleavage retardation. To test the assumption that the retarded cleavage of 3A-containing precursor polypeptides of HAV is due to the noncanonical amino acid sequence at the 3A/3B junction and to elucidate the role of 3A-containing polypeptides in HAV polyprotein processing and replication, we assayed for the cleavability of several mutants with preferred cleavage site sequences at the 3A/3B junction. In addition, we determined the processing products of mutants resistant to cleavage at the 3A/3B and/or 3B/3C sites. Our results show that processing of the structural proteins and subsequent particle assembly was severely compromised when accumulation of 3ABC was impaired. In contrast to wild-type RNA, none of the mutated in vitro transcripts were replication competent. Defective particle formation could be restored in vivo by coexpression of 3AB or 3ABC, suggesting that these intermediate products play an essential role in processing, particle formation, and virus replication.
MATERIALS AND METHODS
Plasmid constructions.
The parent plasmid for our studies, p5′P2P3-3′, is a chimeric infectious cDNA containing the P1 region of the attenuated HAV strain HM175 in the background of the cytopathic HAV strain 18f (44). To place the HAV cDNA under the control of the T7 promoter, pT7-18f was prepared by inserting the HAV-encoding HindIII-SphI fragment of p5′P2P3-3′ into pGEM2 (Promega) digested with the same enzymes. For mutagenesis, the EcoRI-SphI fragment (nucleotide [nt] 4960 to the end of the genome) of pT7-18f was inserted into the multiple-cloning site of pBS-KS(+) (Stratagene). The two-primer site-directed mutagenesis system (QuikChange; Stratagene) was used to create substitutions at the proteolytic cleavage sites within the P3 region. After mutagenesis, the BsgI-BbsI fragment (nt 5087 to 5764) was reinserted into the wild-type P3 region and the sequence was verified by the dideoxynucleotide method. To prepare P3 plasmids mutated at two cleavage sites, the cDNA containing a single nucleotide change was mutated at the second site by the same procedure. To construct mutated full-length HAV genomes, the P3 region of pT7-18f (EcoRI-SphI fragment) was replaced by the mutated P3 fragments. Plasmid pT7-P3, encoding only the HAV P3 region under control of the T7 promoter, was prepared by excising the P1-P2 domain from pT7-18f with HindIII and EcoRI. After blunt ending with the Klenow enzyme and religation, wild-type and mutated pT7-P3 were generated. pEXT7-P1-2A (E/S, 273/274), encoding the P1-2A region of the attenuated HAV strain HM175, was described previously (28). Constructs pET-3C, pET-3Cμ (17) (formerly pET-3CD* and pET-3CD*μ), pET-3ABC, pET-3ABCμ (18), pET-3AB, pET-3AB PV, pET-3ABΔid, pET-3A (3, 7, 26), and pGEM1-lacZ (13) were described previously.
Description of mutants.
For the construction of mutants, we compared the primary amino acid sequences with the observed processing efficiency at the various cleavage sites in the polyprotein. Positions P1 and/or P3 at the 3A/3B and P1′ at the 3B/3C cleavage site were exchanged such that cleavage was abrogated (Table 1, mutants 1, 5, and 6) or enhanced (mutants 2, 3, and 4). To block cleavage, the glutamate at the P1 position of the wild-type amino acid sequence (IPAE/G) at the 3A/3B site was replaced by a valine, resulting in mutant 1 (IPAv/G; mutated residues are shown in lowercase letters). To enhance the cleavability of the 3A/3B junction, the sequence was modified by replacing the P1 glutamate by a glutamine and the P3 proline by a threonine, resulting in mutants 2 (IPAq/G) and 3 (ItAE/G) and in mutant 4, carrying both altered residues (ItAq/G). To abrogate cleavage at the 3B/3C site, the P1′ serine was replaced by a leucine, resulting in mutant 5 (VESQ/l). Cleavage at both the 3A/3B and 3B/3C sites was blocked by combining mutations 1 (IPAv/G) and 5 (VESQ/l), resulting in mutant 6. The sequences of the primers used to create the indicated mutations are listed in Table 2.
TABLE 2.
Primers used to create cleavage site mutants
| Site of mutationa | Amino acid sequence of mutated siteb | Nucleotide sequence of primer pairsc |
|---|---|---|
| 3A/3B (IPAE/G) | IPAv/G | Sense, 5′ gaa cca atc cca gct gTa ggg gta tat cat ggt gta act aag cc 3′ |
| Antisense, 5′ gg ctt agt tac acc atg ata tac ccc tAc agc tgg gat tgg ttc 3′ | ||
| IPAq/G | Sense, 5′ gag gaa gaa cca atc cca gct Caa ggg gta tat cat ggt gt 3′ | |
| Antisense, 5′ ac acc atg ata tac ccc ttG agc tgg gat tgg ttc ttc ctc 3′ | ||
| ItAE/G | Sense, 5′ gag gaa gaa cca atc Aca gct gaa ggg gta tat cat ggt gt 3′ | |
| Antisense, 5′ ac acc atg ata tac ccc ttc agc tgT gat tgg ttc ttc ctc 3′ | ||
| ItAq/G | Sense, 5′ gag gaa gaa cca atc Aca gct Caa ggg gta tat cat ggt gt 3′ | |
| Antisense, 5′ ac acc atg ata tac ccc ttG agc tgT gat tgg ttc ttc ctc 3′ | ||
| 3B/3C (VESQ/S) | (VESQ/l | Sensed, 5′ gat gca gat cca gta gaa tct cag tTa act ttg gaa ata gca gga c 3′ |
| Antisensed, 5′ g tcc tgc tat ttc caa agt tAa ctg aga ttc tac tgg atc tgc atc 3′ |
The wild-type amino acid sequence is shown in parentheses.
Mutated amino acids are shown in bold lowercase letters.
Mutated nucleotides are shown in bold capital letters.
New HpaI restriction site is shown in italics.
Transinfection of COS-7 cells.
COS-7 cells were transfected for 3 h at 37°C with 1 μg of DNA and 9 μl of LipofectAmine (Gibco-BRL) as previously described (9, 29, 30). Subsequently, transfected cells were infected with the recombinant vaccinia virus vTF7-3 (multiplicity of infection, 1) and incubated for 24 h at 37°C. After being washed with phosphate-buffered saline (PBS), the cells were scraped into 0.25 ml of PBS containing 0.05% Tween 20 (PBS-Tween) and lysed by three freeze-thaw cycles. An aliquot of the total extract was used for immunoblotting or, after clarification by centrifugation, in the particle-specific enzyme-linked immunosorbent assay (ELISA) (see below). The total amount of transfected DNA did not exceed 1 μg in transinfection experiments with two or three cDNAs, except when otherwise stated.
β-Galactosidase activity.
pGEM1-lacZ in combination with other cDNAs was transfected in duplicate into vTF7-3-infected COS-7 cells as described above. After 20 to 24 h, the cells were washed and disrupted in 0.25 ml of lysis buffer (25 mM Tris [pH 7.8], 1 mM dithiothreitol, 1 mM EDTA, 15% glycerol, 1% Triton X-100) and centrifuged for 1 min (13,000 × g). The clarified cell lysate (40 μl) was assayed in duplicate for β-galactosidase activity by using para-nitrophenyl-β-d-galactopyranoside (PNPG) as a substrate (13).
Immunological detection of viral proteins and particles.
Recombinantly expressed proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes (Schleicher & Schuell), and detected with monospecific antisera directed against HAV proteins 3A, 3B, 3C, 2C-3A, 2B, VP0, VP3, and VP1, as described elsewhere (13, 29, 30). For the detection of subviral particles, the particle-specific ELISA described previously was used (29, 30). Briefly, plates coated with the monoclonal anti-HAV antibody K2-4F2 (25) and blocked with 1% bovine serum albumin were incubated for 2 h at 37°C with the appropriately diluted clarified extracts of transinfected cells. After the plates were washed with PBS-Tween, the same antibody conjugated to horseradish peroxidase was added and incubated for 2 h at 37°C. For the color reaction, 3,3′,5,5′-tetramethylbenzidine (Sigma) was added to the washed plates and after incubation for 30 min at 37°C, the reaction was read at 450 nm. The linear relationship of the ELISA signal to the concentration of subviral particles was confirmed by a serial dilution of a well-documented HAV preparation (data not shown).
RNA transcription and transfection.
RNA was transcribed from SphI-linearized DNA with the MEGAscript kit (Ambion) as recommended by the manufacturer. After the transcription mixture was treated with DNase I (for 15 min at 37°C), the RNA was precipitated with LiCl, washed with 70% ethanol, and quantitated by spectrophotometry. The quality of runoff transcripts was estimated on an agarose gel under denaturing conditions. By electroporation (960 μF, 100 Ω, 150 V; Bio-Rad Gene Pulser), 10 μg of RNA was transfected into 106 BS-C-1 cells suspended in 100 μl of PBS. After incubation at 20°C for 30 min with occasional shaking, the cells were suspended in 2 ml of Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and seeded into six-well plates. The cells were further incubated at 37°C for 2 to 3 weeks before they were assayed for virus replication. Viral antigen was determined by the particle-specific ELISA and expressed in relative units (29, 30). To assay for viral infectivity, the end-point titer was determined with the lysates of RNA-transfected cells (passage 0; 50% tissue culture infective dose [TCID50] per microgram of RNA) and cells derived from the second blind passage (TCID50 per milliliter).
RT-PCR.
HAV RNA present in a total RNA preparation (RNeasy; Qiagen) of RNA-transfected BS-C-1 cells was subjected to reverse transcription (RT) at elevated temperature (70°C) for 15 min with recombinant Thermus thermophilus DNA polymerase (GeneAmp thermostable rTth reverse transcriptase RNA PCR kit; Perkin-Elmer) and the antisense primer 5′-gta aac tcc act ttc ata att ctc tta ctt tca att ttc tta tc-3′ (nt 5951 to 5908). After addition of the sense primer 5′-gat gca gat cca gta gaa tct cag tta act ttg gaa ata gca gga c-3′ (nt 5248 to 5293), the cDNA product was PCR amplified in the same tube in 35 cycles (94°C for 1 min 20 s, 60°C for 2 min, and 72°C for 3 min).
RESULTS
Processing within P3 mutated at the 3A/3B and/or 3B/3C cleavage site.
In an initial attempt to test for the role of the primary structure at the 3A/3B cleavage site, we mutated the amino acid sequence such that the cleavage efficiency would be either reduced or enhanced (see Materials and Methods) (Table 1). In addition, site 3B/3C was mutated to make it resistant to cleavage. These site-specific mutations were engineered into cDNA constructs bearing either the complete HAV genome (pT7-18f) or its P3 domain (pT7-P3), which were placed under the control of the T7 promoter (Fig. 1A).
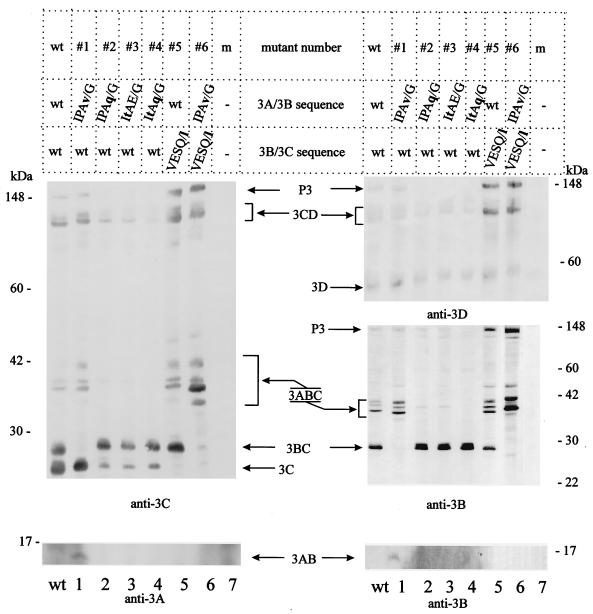
To directly show that the site-specific mutations had the expected effects on P3 cleavage, equal amounts of wild-type and mutated T7-promoted P3 constructs were expressed in the vaccinia virus system with vTF7-3 as the helper virus (9). P3 cleavage products were analyzed by immunoblotting with anti-3A, anti-3B, anti-3C, and anti-3D antibodies (Fig. 2). 3C-containing products of wild-type P3 and polypeptides P3, 3CD, 3ABC, 3BC, and 3C were detected (Fig. 2, anti-3C panel; confirmed in part in the anti-3D and the upper anti-3B panels, lanes wt). As previously reported (29), 3ABC appeared in three forms (Fig. 2, anti-3C and upper anti-3B panel) and 3AB was found in small amounts under some experimental conditions. Here, 3AB liberated from the wild-type polyprotein was undetectable by anti-3A or anti-3B (Fig. 2, lanes wt). Replacing the E/G dipeptide sequence at the 3A/3B junction by v/G (mutant 1) completely prevented cleavage at this site, which was obvious from the prominent 3AB band that was detected by anti-3A and anti-3B (Fig. 2, anti-3A and lower anti-3B panels, lanes 1). The generation of 3C, 3CD, and 3ABC was only slightly affected, but the complete lack of 3BC confirmed that the 3A/3B site was uncleavable in mutant 1 (Fig. 2, anti-3C and upper anti-3B panels, lanes 1). Blocking the 3B/3C cleavage site by replacing serine at position P1′ with leucine (Q/l in mutant 5) resulted in the loss of 3C (Fig. 2, anti-3C panel, lane 5) without having major effects on the formation of other P3 cleavage products. Minute amounts of 3BC and 3C were found when both sites (3A/3B and 3B/3C) were mutated, and 3ABC was the predominant anti-3C reactive product of P3 (mutant 6, Fig. 2, anti-3C and anti-3B panels, lanes 6). Among the products of mutants 5 and 6, the larger polypeptides (P3 and 3CD) were detected in larger amounts, which might be due to their altered conformation rendering them less susceptible to 3C-mediated cleavage at sites remote from the mutated site.
FIG. 2.
Proteolytic processing pattern of HAV P3 mutated at the 3A/3B and/or 3B/3C sites. The HAV P3 domain containing the wild-type (wt) or mutated sequences (#1 to #6) (see Table 1) was transiently expressed in COS-7 cells. Equal amounts of each cell extract were analyzed by immunoblotting with anti-3A, anti-3B, anti-3C, and anti-3D antibody. The anti-3B immunoblot analysis was performed with 12 and 15% polyacrylamide gels to ensure optimal separation of small (3AB) and large (3BC and 3ABC) polypeptides. Molecular mass markers are shown on the margin, and HAV P3 cleavage products are indicated. m, mock-transfected extracts.
In cells expressing the mutants that contain amino acid sequences at the 3A/3B site which should allow for enhanced cleavage (mutants 2, 3, and 4), very little or no P3 and 3ABC accumulated (Fig. 2, anti-3C, anti-3D, and anti-3B panels, lanes 2 to 4). For unknown reasons, 3CD derived from these mutated P3 polypeptides was found in slightly smaller amounts than was 3CD derived from wild type P3 (Fig. 2, the anti-3C and anti-3D panels, lanes 2 to 4). Formation of 3C was reduced in these mutants, possibly due to the accumulation of 3BC, which is autoproteolytically less active, as reported previously (29). Taken together, the data suggest that cleavability of the 3A/3B and 3B/3C junctions can be effectively abrogated by replacing the P1 glutamate by a valine (mutant 1) or the P1′ serine by a leucine (mutant 5), respectively. In contrast, cleavage at the 3A/3B site could be drastically enhanced by changing the P3 proline to a threonine and the P1 glutamate to the favoured glutamine (mutant 4; Table 1). The complete lack of 3ABC and P3 after expression of mutant 4 indicates that the retarded cleavage at the 3A/3B site is due to the unfavorable wild-type amino acid sequence at this site. Mutant 4 now provides the tool to directly assess the role of 3ABC as stable processing intermediate, whereas the other mutants that accumulate different proportions of proteins 3ABC, 3BC, and 3C can be used to estimate their impact on polyprotein processing and particle formation.
Polyprotein processing.
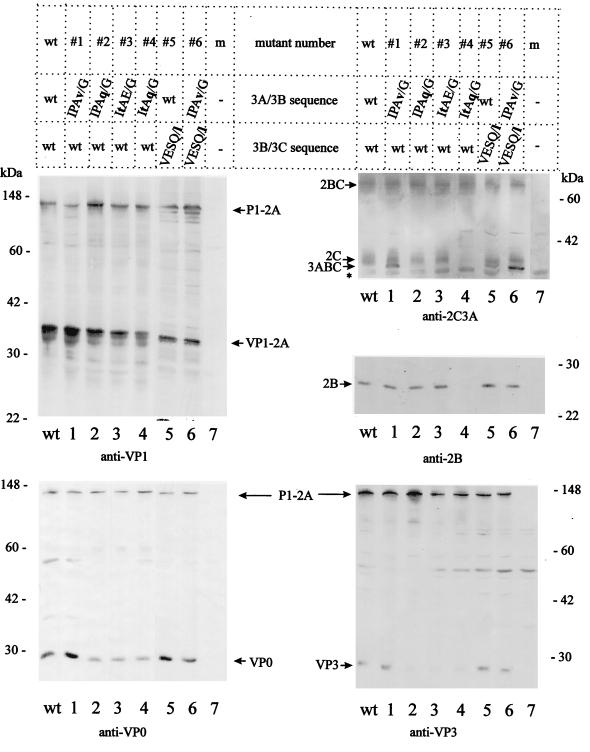
To study the proteolytic activity of the various forms of the proteinase at distal sites, processing of the complete HAV polyprotein carrying the various mutations was assessed after transient expression of the HAV genome in mammalian cells. For this, COS-7 cells were transfected with cDNAs encoding the complete wild-type or mutated HAV genome and infected with the recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase (9). The efficiency of processing was deduced from the amounts of end products which were transferred to membranes and detected by immunoblotting. The amounts of precursor polypeptides were mostly unaffected, possibly because only a relatively small portion of the precursors (e.g., P1-2A and 2BC) was cleaved. To detect intermediate products arising early in the processing cascade (the primary cleavage products P1-2A, 2BC, and P3), we performed kinetic analyses over the 24-h expression period. In these experiments, the extent of cleavage at the primary sites of the polyprotein (2A/2B and 2C/3A) was not different among the mutants tested (data not shown). The extracts of a 24-h expression were analyzed (Fig. 3). In immunoblots with anti-VP1, anti-VP0, anti-VP3, anti-2B, and anti-2C3A, polypeptides P1-2A and 2BC were detected in similar amounts in all extracts, demonstrating again that the primary cleavages at the 2A/2B and the 2C/3A sites were essentially unaffected by the P3 mutations. However, some of the P3 mutants had a pronounced effect on the liberation of products of secondary cleavages, which were detected in reduced amounts. Mutants 1, 5, and 6 accumulating 3ABC but with small amounts of 3BC and/or 3C due to blocked 3A/3B and/or 3B/3C sites, showed essentially the same pattern of the structural proteins VP1-2A, VP0, and VP3 as the wild type (Fig. 3, anti-VP1, anti-VP0, and anti-VP3 panels, lanes 1, 5, and 6). However, structural proteins, in particular VP0 and VP3, were generated in smaller amounts from the mutants containing preferred 3A/3B cleavage sites (mutants 2, 3, and 4 in Fig. 3, lanes 2 to 4) than from the wild-type polyprotein (lanes wt). In contrast to other expression strategies (29, 30), VP1 was liberated in very small amounts when the complete polyprotein was expressed. Cleavage of 2BC was not affected by most P3 mutants, which was evident in the anti-2C3A and anti-2B blots, where both 2C and 2B were found in similar amounts as derived from the wild-type polyprotein (Fig. 3, lanes 1 to 3 and lanes 5 to 7). Mutant 4 (lane 4) is an exception in that neither 2C nor 2B was produced. Although the anti-2C3A serum recognizes both 3ABC and 2C, which often comigrate, it is obvious in lane 4 of the anti-2C3A blot that 2C was not liberated from mutant 4, which does not accumulate 3ABC. Taken together, the data suggest that the intermediate product 3ABC is essential for the efficient liberation of both the structural and nonstructural proteins, since mutants that accumulate small amounts of or no 3ABC produce less VP0, VP3, 2B, and 2C.
FIG. 3.
Processing products derived from domains P1-2A and 2BC of the HAV polyprotein mutated at the 3A/3B and/or 3B/3C site. The complete HAV genomes of wild-type (wt) and mutated sequence (mutants 1 to 6) was expressed in COS-7 cells. Equal amounts of cells extract were analyzed by immunoblotting with anti-VP1, anti-VP0, anti-VP3, anti-2C3A, and anti-2B antibodies. The asterisk indicates an unidentified host protein immunodetected by anti-2C3A. Molecular mass markers and cleavage products of P1-2A and 2BC are indicated. m, mock-transfected extracts.
Effect of P3 cleavage site mutants on HAV particle formation.
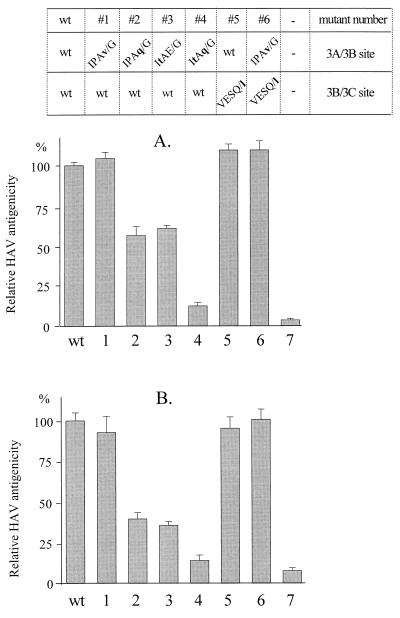
Assembly of HAV particles is tightly linked to processing of the P1-2A precursor and seems to be dependent on the presence of proteins flanking 3Cpro. As shown previously, various subviral particles are formed in the recombinant system used (29). To test whether the cleavage site mutations had any effect on capsid assembly, HAV particle formation was quantitatively assessed by an ELISA. In this assay, a neutralizing monoclonal antibody that detects only virions (160S), procapsids (70S), and processed pentamers (14S), but not uncleaved pentameric structures and protomers (5S) was used (1, 25, 29, 30). The complete HAV genome containing either wild-type or mutated 3A/3B and/or 3B/3C cleavage sites (pT7-18f in Fig. 1A) was expressed, and the cell extracts were tested by the particle-specific ELISA. Blocking cleavage at sites 3A/3B (mutant 1) and/or 3B/3C (mutants 5 and 6) had no effect on HAV particle formation compared with the wild type (Fig. 4A, columns 1, 5, 6, and wt). In extracts of mutants that contained low levels of 3ABC (mutants 2 and 3), particle formation was 50 to 60% of the wild-type level (columns 2 and 3), whereas in extracts of mutant 4, where 3ABC was completely absent (Fig. 2, anti-3C and anti-3B panels, lanes 4), only small amounts (approximately 10%) of subviral particle were detectable (Fig. 4A, column 4). We had shown by immunoblotting (Fig. 3, anti-VP0 and anti-VP3 panels, lanes 2 to 4) that liberation of the structural proteins VP0 and VP3 was reduced in mutants 2, 3, and 4, suggesting that particle formation and liberation of structural proteins are directly correlated. However, the defect of mutant 4 in particle assembly was much more pronounced (reduction to 10%) (Fig. 4) than was expected from the reduced amounts of structural proteins (about 50% of the wild-type level) (Fig. 3). This suggests that 3ABC not only functions as a proteinase on P1-2A but also might be involved in particle assembly (see below). Neither 3C nor 3BC seems to be essential for capsid formation, since expression of mutant 6 yielded similar levels to those obtained with the wild-type polyprotein. Particle formation was also tested after coexpression of P1-2A with mutated P3 encoded on separate plasmids (pEXT7-P1-2A and pT7-P3, respectively [Fig. 1A]). Under these conditions, capsid assembly (Fig. 4B) and liberation of structural proteins (data not shown) was affected similarly to that described above for the expression of genome length cDNAs, indicating that the effects of P3 proteins on the precursor of the structural proteins P1-2A can be exerted both in cis and in trans. Similar data (not shown) were obtained when P1-P2 was coexpressed with the various mutants of P3.
FIG. 4.
HAV particle formation after recombinant expression of HAV genomes mutated at the 3A/3B and/or 3B/3C cleavage sites. Either the complete genome (A) or P1-2A with P3 (B) was expressed in COS-7 cells. The extracts in panel A are identical to those in Fig. 3. Particle formation was determined by the particle-specific ELISA and is expressed as percentage of the antigenicity produced by the wild-type genome (column wt). In column 7 of panel A, the mock extract is shown. In column 7 of panel B, the relative antigenicity of P1-2A-expressing cells is shown and is used as control to demonstrate the specificity of the ELISA for processed and particulate material.
Rescue of defective particle formation by 3AB and 3ABC.
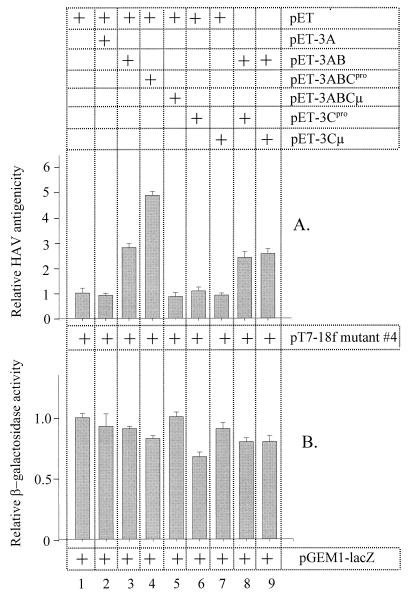
Analysis of processing within the P3 and P1-2A domains of the HAV polyprotein had shown that improving the cleavage efficiency at the 3A/3B site and thus preventing the accumulation of 3A-containing intermediates, in particular 3ABC, strongly impaired HAV particle formation (mutant 4 in Fig. 3 and 4). To examine whether defective capsid assembly of mutant 4 could be rescued by supplementing 3AB or 3ABC in trans and to show that 3AB or 3ABC are directly involved in viral assembly, we coexpressed the cDNA encoding the complete HAV polyprotein with mutation 4 together with cDNAs coding for either 3A, 3AB, proteolytically active and inactive (μ) 3ABC, or 3C (Fig. 1B). To quantitate the effect and to ensure equal cDNA concentrations in transfection, cotransfections were done in triplicate and with the empty vector as a control (Fig. 5A). In a parallel experiment, the reporter protein β-galactosidase was coexpressed with these cDNAs to control for a possible nonspecific effect on overall protein synthesis (Fig. 5B). As shown by the particle-specific ELISA and the β-galactosidase activity, 3AB specifically enhanced the particle formation of the defective mutant (mutant 4) whereas 3A had no effect (Fig. 5A, columns 3 and 2). The effect of proteolytically active 3ABCpro was even more pronounced than that of 3AB. The subviral particles in extracts of cells expressing mutant 4 and 3ABCpro reached concentrations (Fig. 5A, column 4) which were comparable to those of the wild-type HAV (see Fig. 6, columns 1 and 5). Proteolytically inactive 3ABCμ had no effect, implying that either the proteinase activity and/or the liberated 3AB is involved in the rescuing effect of 3ABCpro (Fig. 5A, column 5). The involvement of 3AB was confirmed by coexpression of mutant 4 with both 3AB and 3Cμ. In this case, HAV particle formation was rescued to an extent similar to that observed when 3AB was expressed alone (columns 9 and 3, respectively). However, no synergistic effect was achieved after coexpression of the mutated polyprotein 4 with 3AB and proteolytically active 3Cpro (column 8), despite the ability of both proteins to interact (2). Based on the observation that cells cotransfected with 3Cpro, but not those cotransfected with 3Cμ, expressed the reporter gene at somewhat reduced levels (30%) (Fig. 5B, columns 6 and 7), we assume that a possible synergistic effect of 3Cpro on the partial rescue by 3AB was compensated by its cytotoxic effect. To determine proteolytic liberation of the structural proteins, all coexpression extracts were examined by immunoblot analyses. These experiments demonstrated that coexpression of the polyprotein carrying mutation 4 with 3AB or 3ABC resulted in significantly improved processing of P1-2A (see Fig. 7, anti-VP0 panel, and data not shown). These data suggest that the precursor polypeptides 3AB and, in particular, 3ABC are required for efficient processing of the capsid precursor and for viral assembly.
FIG. 5.
Effect of P3 proteins on defective recombinant-particle formation from pT7-18f mutant 4 (A) and on protein synthesis (B). pT7-18f mutant 4 (A) and pGEM1-lacZ (B) were coexpressed in COS-7 cells with cDNAs indicated on the top right. To ensure equal protein levels of the trans-complementing polypeptides, equal amounts of cDNA encoding the same promoter and initiation region of protein synthesis were used. Particle formation was determined in the cell extracts by the particle-specific ELISA (A) or protein synthesis was assessed by the β-galactosidase activity (B), both presented in arbitrary units. μ denotes the proteolytically inactive form of the proteinase.
FIG. 6.
Specificity of HAV 3AB and 3ABC as the cofactor in HAV particle formation. pT7-18f or pT7-18f mutant 4 in the presence of cDNAs as marked on the top right was expressed in COS-7 cells. The effect on particle formation was determined in the cell extracts by the particle-specific ELISA and is expressed as percentage of the antigenicity produced in cells expressing the wild-type (wt) genome (pT7-18f, set at 100%).
FIG. 7.
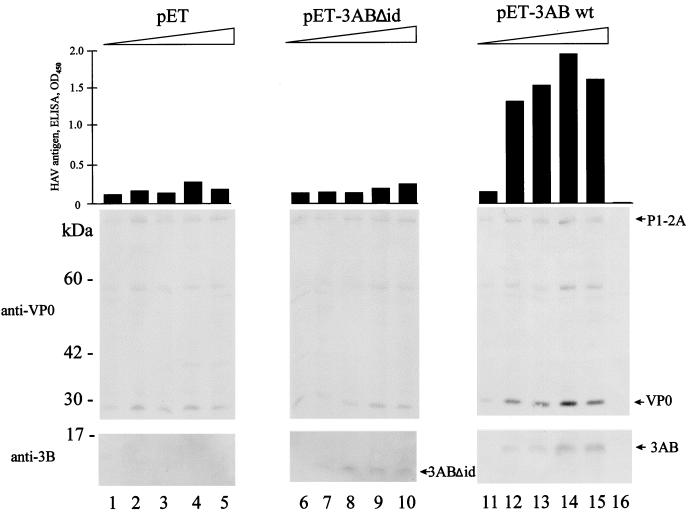
Dose-dependent stimulation of P1-2A processing and particle formation by 3AB. pT7-18f mutant 4 (0.3 μg) was coexpressed in the presence of increasing amounts (0, 0.2, 0.4, 0.6, and 0.8 μg) of cDNAs pET (lanes 1 to 5), pET-3ABΔid (lanes 6 to 10), and pET-3AB wt (lanes 11 to 15). Cell extracts were analyzed by the particle-specific ELISA (top) and by immunoblotting with anti-VP0 (middle) and anti-3B (bottom). In lane 16, the extract of mock-infected cells is shown. Molecular mass markers and HAV polypeptides are indicated. OD450, optical density at 450 nm.
For PV and HAV, biochemical evidence demonstrating that polypeptide 3AB interacts with membranes, viral RNA, and other viral proteins has been provided (2, 3, 7, 19, 26, 27, 35, 40, 41). The domain responsible for membrane binding and homodimerization was mapped to the hydrophobic region located in the C-terminal half of HAV 3A (amino acid residues 40 to 60) (7). To directly assess whether this region might also be essential for the function of 3AB as cofactor in capsid assembly, a 3AB deletion mutant was analyzed for its ability to rescue defective HAV capsid assembly. The HAV polyprotein carrying mutation 4 was coexpressed with 3AB of PV and with 3ABC, 3AB, and 3ABΔid of HAV (3ABΔid is deleted in the hydrophobic domain of 3A) (Fig. 1B). The number of HAV particles formed by recombinant expression was determined by the particle-specific ELISA (Fig. 6). Coexpression of 3ABC restored the defective particle formation of mutant 4 (Fig. 6, column 2) to almost wild-type levels (column 1). Among the 3AB constructs tested, only wild-type HAV 3AB rescued defective particle formation to 50% of the wild-type level (columns 3) whereas neither PV 3AB nor HAV 3ABΔid was effective (columns 4 and 6).
The specificity of the HAV 3AB was further documented in an experiment which showed that the effect of 3AB on P1-2A processing and defective particle formation was dose dependent. Mutant 4 was coexpressed with increasing amounts of cDNA of vector pET, wild type pET-3AB, or pET-3ABΔid. The amount of cDNA transfected directly correlated with the amounts of expressed 3AB (Fig. 7, anti-3B panels, lanes 12 to 15) and 3ABΔid (lanes 7 to 10). In a dose-dependent manner, liberation of VP0 (anti-VP0 panel) and particle assembly (upper panel) was augmented when 3AB was coexpressed (Fig. 7, lanes 12 to 15). This effect was not observed when the cDNAs of the vector (lanes 2 to 5) or 3ABΔid (lanes 7 to 10) were coexpressed in equal amounts, suggesting that HAV 3AB is a specific cofactor for both P1-2A processing and particle formation. The reduction in liberation of VP0 and particle formation in lane 15 might be due to the large amounts of transfected DNA. Membranes seem to be involved in both processes, since the membrane binding domain of 3AB was required for efficient rescue.
Replication of mutated transcripts.
To test whether the accumulation of HAV 3AB and 3ABC was a prerequisite for virus replication, genome-length RNA transcripts were prepared from the wild-type and all mutant plasmids and transfected into BS-C-1 cells. As expected, the wild-type transcript was infectious, as was evident from the cytopathic effect (CPE) exerted by this viral strain on BS-C-1 cells, by high levels of HAV antigenicity determined by ELISA, and by the presence of plus-strand RNA detected by RT-PCR (Table 3). Irrespective of whether 3AB and 3ABC were stable or unstable processing products, none of the mutated RNA transcripts was able to replicate, since no viable virus could be recovered 10 to 14 days after transfection. Furthermore, two blind passages, each extending for 21 days, did not force the appearance of HAV antigen or RNA, demonstrating the inability of the mutated genomes to replicate and to produce revertants. Since both types of mutants with either preferred or blocked cleavage sites were replication defective, we conclude that the finely tuned balance of 3ABC and its cleavage products is absolutely required for HAV replication.
TABLE 3.
Replication of mutated HAV RNAs
| Mutant | Cleavage site | Amino acid sequence at sitea | HAV antigen (ELISA, relative units)b | Infectivityc
|
RT-PCR (passage 0d) | CPE | |
|---|---|---|---|---|---|---|---|
| Passage 0 (TCID50/μg) | Passage 2 (TCID50/ml) | ||||||
| wt | 3A/3B | IPAE/G | |||||
| 3B/3C | VESQ/G | 100 | 106.8 | 107.5 | + | + | |
| 1 | 3A/3B | IPAv/G | <2 | <101 | <101 | − | − |
| 2 | IPAq/G | <2 | <101 | <101 | − | − | |
| 3 | ItAE/G | <2 | <101 | <101 | − | − | |
| 4 | ItAq/G | <2 | <101 | <101 | − | − | |
| 5 | 3B/3C | VESQ/l | <2 | <101 | <101 | − | − |
| 6 | 3A/3B | IPAv/G | |||||
| 3B/3C | VESQ/l | <2 | <101 | <101 | − | − | |
Mutated amino acids are shown in bold lowercase letters.
Particle-specific ELISA units (see Materials and Methods) detected in a cell lysate prepared 2 weeks after RNA transfection into BS-C-1 cells; the result of wild-type RNA transfection was set to 100.
TCID50/μg, 50% infectivity per microgram of transfected RNA; TCID50/ml, 50% infectious titer in the cell lysate of passage 2.
Presence (+) or absence (−) of the 0.7-kb amplicon which was produced from the total RNA fraction prepared 2 weeks after transfection of wild-type or mutated RNA transcripts into BS-C-1 cells.
DISCUSSION
3ABC is a predominant and distinct product in the P3 processing pathway of HAV; however, its function in the viral life cycle is still enigmatic (15, 16, 29, 34). Here, a genetic approach was chosen to determine the importance of precursor proteins 3ABC and 3AB in HAV polyprotein processing, particle formation, and replication. The data presented in this study show that both blocking and enhancing the normally retarded cleavage at the 3A/3B junction completely abrogated the infectivity of the mutated RNA. No revertant progeny virus could be isolated, even after two extended blind passages. The ability to generate revertant progeny virus depends on various aspects of the natural history of the virus, in particular on several of the central processes of viral replication whose individual role in reversion has not been elucidated. The availability of replication components through protein processing, the efficiency of RNA replication, and the accuracy of the replication machinery all seem to be essential parameters to permit reversion.
In the experiments described here, we have mutated HAV cleavage sites 3A/3B and 3B/3C and thus affected the accessibility of some of the components of the viral replication machinery derived from the P3 region. For encephalomyocarditis virus (EMCV), similar mutants which all were quasiinfectious and resulted in revertant progeny viruses were studied previously (12). Compared with the reported EMCV transcript, the wild-type HAV RNA transcript used for transfection was of similar specific infectivity (106.8/μg; Table 3), however, the period required to detect viral replication was much longer for HAV (>7 days) than for EMCV (24 h). Since, unlike EMCV, no revertants were produced from any of the mutated HAV genomes (even after extended incubation periods), we conclude that the high efficiency of the replication machinery is an essential factor for reversion to occur. As suggested for the origin of PV pseudorevertants and the heterogeneity of a virus population as a quasi-species (10), our data support the notion that genetic plasticity of the picornavirus genomes is largely dependent on rapid replication.
Proteolytic cleavage at the various sites in the picornavirus polyprotein occurs with different kinetics and efficiencies and, among other factors, is determined by the primary amino acid sequences at the scissile bonds. This fact was also inferred for HAV 3C in studies with synthetic peptides and some cleavage site mutants (14, 21, 34). We now extended these investigations and analyzed the role of the amino acid sequence at the 3A/3B scissile bond, which is unique among all HAV 3C cleavage sites (4). Replacing the amino acid residues at positions P3 and/or P1 of the 3A/3B junction by residues found at preferred sites dramatically enhanced its cleavage and led to a loss of the otherwise stable intermediate 3ABC (Fig. 2). Our data thus directly indicate that the regulated liberation of individual polypeptides from the polyprotein is governed by the amino acid sequence at the scissile bonds.
Cleavage by a proteolytic activity contained in a polyprotein can be regulated by polypeptides that flank the proteinase region and that can either be covalently or noncovalently linked to the proteinase (24). From the experimental data described here, it can be concluded that the specificity for the various HAV cleavage sites is indeed affected by the polypeptides attached to 3Cpro. HAV 2BC was poorly cleaved by HAV 3C but clearly preferred by 3ABC as substrate, a conclusion also inferred by other experiments (13). Accumulation of 3ABC also seemed to be essential for efficient production of VP0 and VP3 (Fig. 2 and 3). In a similar fashion, it was shown previously for PV that the two prominent proteolytic forms, 3Cpro and 3CDpro, differ in their substrate specificities (5, 22, 43). Whereas P1, 2BC, and 3AB were found to be poor substrates for PV 3Cpro in vitro, they were cleaved efficiently by 3CDpro.
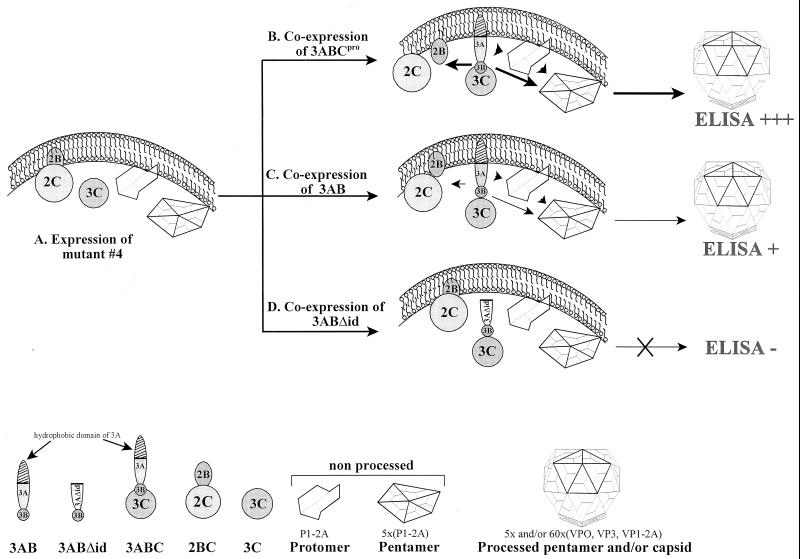
HAV particle formation by all mutants was investigated and clearly shown to be reduced in mutant 4, which is completely defective in accumulating 3ABC. The negative effect on capsid formation was substantially more pronounced than expected from the reduced liberation of structural proteins (Fig. 3 and 4), implying that 3ABC or 3AB not only are involved in P1-2A proteolysis but also might participate in the assembly of HAV particles. The putative role of 3AB or 3ABC in particle formation is shown in Fig. 8, which presents a working model for the rescue of defective HAV polyprotein processing and particle formation as determined by the particle-specific ELISA (for details, see the legend to Fig. 8). It has been proposed that the initial step in HAV assembly is the aggregation of pentamers through the interaction of five molecules of uncleaved P1-2A (1, 30). Although both 3C and 3ABC cleave P1-2A with apparently similar efficiency (29), it is possible that 3ABC is the polypeptide form of 3C with specificity for P1-2A assembled into pentamers and hence that 3ABC drives the assembly process from pentamers to procapsids (Fig. 8B, thick arrows). Based on its proposed ability to bind to membranes, it is also conceivable that 3ABC might function as the membrane-bound form of the proteinase, which is required for colocalizing the proteinase with its substrate P1-2A and in tethering the assembly complex to membranes (7, 13, 29). This concept is strengthened by our observation that coexpression only of wild-type 3AB (Fig. 8C) but not of 3ABΔid (carrying a deletion of the hydrophobic, membrane-anchoring domain [Fig. 8D]) is able to rescue defective particle formation (Fig. 6 and 7). Possibly, HAV 3ABC plays a regulatory role in coordinating the sequential steps of capsid assembly. We hypothesize that aggregation of P1-2A prior to its cleavage may depend on the presence of 3AB-containing proteins and that 3AB or 3ABC might be essential in coordinating both early steps of capsid formation, namely aggregation of five molecules of P1-2A and their subsequent cleavage.
FIG. 8.
Working model for the rescue of defective HAV polyprotein processing and particle formation of mutant 4 by 3AB and 3ABC. (A) Expression of mutant 4 that is unable to accumulate 3AB and 3ABC results in uncleaved 2BC and mostly unprocessed P1-2A and thus yields very low levels of HAV particles, as determined by the particle-specific ELISA. (B) Coexpression of 3ABCpro tethers the assembly complex to membranes, resulting in coordinated pentamer cleavage and assembly (thick arrows), leading to high levels of antigenically reactive particles (ELISA +++). 2BC is efficiently cleaved (thick arrow) (reference 13 and data not shown). (C) Coexpression of 3AB helps to bring 3C derived from the polyprotein of mutant 4 close to the membrane-associated assembly complex, resulting in somewhat enhanced particle formation (thin arrows, ELISA +). As shown recently by coimmunoprecipitation and by Far Western and affinity chromatography, HAV 3C can interact with cognate 3AB (2). (D) Coexpression of 3ABΔid, which is deleted in the hydrophobic domain of 3A and unable to bind to membranes (7), does not bring 3C close to the membrane-associated assembly complex. Therefore, particle formation is not enhanced over that of mutant 4 (ELISA −). Note that the particle-specific ELISA detects processed pentamers and capsids (1, 25, 29, 30), as shown at the bottom.
The role of virus-encoded cofactors in controlled processing has been described for several viral proteinases contained in a polyprotein, in particular for HCV proteinase NS3 (33, 39, 42). HCV peptide NS4A forms a noncovalent complex with NS3 and is an essential cofactor for processing of the HCV nonstructural proteins (42). For HCV NS4A, which is highly hydrophobic, it was clearly shown that it stabilizes the conformation of NS3 (33). The functional and biochemical similarities of HCV NS4A and HAV 3AB are striking, in particular their hydrophobicity and membrane association. Further experiments will be performed to directly assess the biochemical and functional interaction of HAV 3AB and 3C.
ACKNOWLEDGMENTS
The recombinant vaccinia virus vTF7-3 was kindly provided by B. Moss. We thank S. Lemon for providing p5′-P2P3-3′, M. Jecht for providing pT7-18f and pGEM1-lacZ, and G. Morace for providing pET-3A, pET-3AB, pET-3ABΔid, and pET-3AB PV. We are grateful to D. Reinhardt for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 367, project B7).
REFERENCES
- 1.Anderson D A, Ross B. Morphogenesis of hepatitis A virus: isolation and characterisation of subviral particles. J Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beneduce, F., and G. Morace. Personal communication.
- 3.Beneduce F, Ciervo A, Morace G. Site-directed mutagenesis of hepatitis A virus protein 3A: effects on membrane interaction. Biochim Biophys Acta. 1997;1326:157–165. doi: 10.1016/s0005-2736(97)00023-0. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann E M, Mosimann S C, Chernaia M M, Malcolm B A, James M N G. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair W S, Li X, Semler B L. A cellular cofactor facilitates efficient 3CD cleavage of the poliovirus P1 precursor. J Virol. 1993;67:2336–2343. doi: 10.1128/jvi.67.4.2336-2343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers T J, Nestrorowicz A, Rice C. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J Virol. 1995;69:1600–1605. doi: 10.1128/jvi.69.3.1600-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciervo A, Beneduce F, Morace G. Polypeptide 3AB of hepatitis A virus is a transmembrane protein. Biochem Biophys Res Commun. 1998;249:266–274. doi: 10.1006/bbrc.1998.9121. [DOI] [PubMed] [Google Scholar]
- 8.Cohen L, Kean K M, Girard M, van der Werf S. Effects of P2 cleavage site mutations on poliovirus polyprotein processing. Virology. 1996;224:34–42. doi: 10.1006/viro.1996.0504. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic expression system based on a recombinant vaccinia virus that synthesize bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gmyl A P, Pilipenko E V, Maslova S V, Belov G A, Agol V I. Functional and genetic plasticity of the poliovirus genome quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J Virol. 1993;67:6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graff J, Kasang C, Normann A, Pfisterer-Hunt M, Feinstone S M, Flehmig B. Mutational events in consecutive passages of hepatitis A virus strain GBM during cell culture adaptation. Virology. 1994;204:60–68. doi: 10.1006/viro.1994.1510. [DOI] [PubMed] [Google Scholar]
- 12.Hall D J, Palmenberg A C. Cleavage site mutations in the encephaliomyocarditis virus P3 region lethally abrogate the normal processing cascade. J Virol. 1996;70:5954–5961. doi: 10.1128/jvi.70.9.5954-5961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jecht M, Probst C, Gauss-Müller V. Membrane permeability induced by hepatitis A virus (HAV) proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology. 1998;252:218–227. doi: 10.1006/viro.1998.9451. [DOI] [PubMed] [Google Scholar]
- 14.Jewell D A, Swietnicki W, Dunn B M, Malcolm B A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 15.Jia X Y, Ehrenfeld E, Summers D F. Proteolytic activity of hepatitis A virus 3C protein. J Virol. 1991;65:2595–2600. doi: 10.1128/jvi.65.5.2595-2600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jürgensen D, Kusov Y Y, Fäcke M, Kräusslich H G, Gauss-Müller V. Cell free translation and proteolytic processing of the hepatitis A virus polyprotein. J Gen Virol. 1993;74:677–683. doi: 10.1099/0022-1317-74-4-677. [DOI] [PubMed] [Google Scholar]
- 17.Kusov Y Y, Sommergruber W, Schreiber M, Gauss-Müller V. Intermolecular cleavage of hepatitis A virus (HAV) precursor protein P1-P2 by recombinant HAV proteinase 3C. J Virol. 1992;66:6794–6796. doi: 10.1128/jvi.66.11.6794-6796.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusov Y Y, Morace G, Probst C, Gauss-Müller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5′ and 3′ termini of the HAV RNA. Virus Res. 1997;51:151–157. doi: 10.1016/s0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 19.Lama J, Paul A V, Harris K S, Wimmer E. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- 20.Lemm J A, Rice C. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol. 1993;67:1916–1926. doi: 10.1128/jvi.67.4.1916-1926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin A, Escriou N, Chao S F, Girard M, Lemon S M, Wychowski C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 22.Molla A, Harris K S, Paul A V, Shin S H, Mugavero J A, Wimmer E. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB*. J Biol Chem. 1994;269:27015–27020. [PubMed] [Google Scholar]
- 23.Morace G, Pisani G, Beneduce F, Divizia M, Pana A. Mutations in the 3A genomic region of two cytopathic strains of hepatitis A virus isolated in Italy. Virus Res. 1993;28:187–194. doi: 10.1016/0168-1702(93)90135-a. [DOI] [PubMed] [Google Scholar]
- 24.Paul A V, Mugavero J, Molla A, Wimmer E. Internal ribosomal entry site scanning of the poliovirus polyprotein: implications for proteolytic processing. Virology. 1998;250:241–253. doi: 10.1006/viro.1998.9376. [DOI] [PubMed] [Google Scholar]
- 25.Ping L-H, Lemon S M. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J Virol. 1992;66:2208–2216. doi: 10.1128/jvi.66.4.2208-2216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pisani G, Beneduce F, Gauss-Müller V, Morace G. Recombinant expression of hepatitis A virus protein 3A: interaction with membranes. Biochem Biophys Res Commun. 1995;211:627–638. doi: 10.1006/bbrc.1995.1859. [DOI] [PubMed] [Google Scholar]
- 27.Plotch S J, Palant O. Poliovirus 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J Virol. 1995;69:7169–7179. doi: 10.1128/jvi.69.11.7169-7179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Probst C, Jecht M, Gauss-Müller V. Proteinase 3C-mediated processing of VP1-2A of two hepatitis A virus strains: in vivo evidence for cleavage at amino acid position 273/274 of VP1. J Virol. 1997;71:3288–3292. doi: 10.1128/jvi.71.4.3288-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Probst C, Jecht M, Gauss-Müller V. Processing of proteinase precursors and their effect on hepatitis A virus particle formation. J Virol. 1998;72:8013–8020. doi: 10.1128/jvi.72.10.8013-8020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probst C, Jecht M, Gauss-Müller V. Intrinsic signals for the assembly of hepatitis A virus particles—role of structural proteins VP4 and 2A. J Biol Chem. 1999;274:4527–4531. doi: 10.1074/jbc.274.8.4527. [DOI] [PubMed] [Google Scholar]
- 31.Schultheiß T, Kusov Y Y, Gauss-Müller V. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology. 1994;198:275–281. doi: 10.1006/viro.1994.1030. [DOI] [PubMed] [Google Scholar]
- 32.Schultheiß T, Sommergruber T W, Kusov Y Y, Gauss-Müller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanji Y, Hijikata M, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesar M, Pak I, Jia X Y, Richards O C, Summers D F, Ehrenfeld E. Expression of hepatitis A virus precursor protein P3 in vivo and in vitro: polyprotein processing of the 3CD cleavage site. Virology. 1994;198:524–533. doi: 10.1006/viro.1994.1063. [DOI] [PubMed] [Google Scholar]
- 35.Towner J S, Ho T V, Semler B L. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 36.Towner J S, Mazanet M M, Semler B L. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol. 1998;72:7191–7200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbani A, Bianchi E, Narjes F, Tramontana A, De Francesco R, Steinkühler C, Pessi A. Substrate specificity of the hepatitis C virus serine protease NS3. J Biol Chem. 1997;272:9204–9209. doi: 10.1074/jbc.272.14.9204. [DOI] [PubMed] [Google Scholar]
- 38.van Kuppefeld F J M, van den Hurk P J J, Zoll J, Galama J M D, Melcher W J G. Mutagenesis of the coxsackie B3 virus 2B/2C cleavage site: determinants of processing efficiency and effects on viral replications. J Virol. 1996;70:7632–7640. doi: 10.1128/jvi.70.11.7632-7640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang W, Cuconati A, Paul A P, Cao W, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol. 1998;72:6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y, Li Y, Munshi S, Sardana V, Cole J L, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo L C, Chen Z. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Chao S F, Ping L H, Grace K, Clarke B, Lemon S M. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effects. Virology. 1995;212:686–697. doi: 10.1006/viro.1995.1526. [DOI] [PubMed] [Google Scholar]