Abstract
Intrapartum antibiotic prophylaxis (IAP) is commonly used during C-section delivery and in Group B Streptococcus-positive women before vaginal delivery. Here, we primarily aimed to investigate the effect of IAP on the neonatal oral and fecal bacteriomes in the first week of life. In this preliminary study, maternal and neonatal oral swabs and neonatal fecal (meconium and transitional stool) swabs were selected from a pool of samples from healthy mother-neonate pairs participating in the pilot phase of CELSPAC: TNG during their hospital stay. The DNA was extracted and bacteriome profiles were determined by 16S rRNA amplicon sequencing (Illumina). In the final dataset, 33 mother-neonate pairs were exposed to antibiotics during C-section or vaginal delivery (cases; +IAP) and the vaginal delivery without IAP (controls, -IAP) took place in 33 mother-neonate pairs. Differences in alpha diversity (Shannon index, p=0.01) and bacterial composition (PERMANOVA, p<0.05) between the +IAP and -IAP groups were detected only in neonatal oral samples collected ≤48 h after birth. No significant differences between meconium bacteriomes of the +IAP and -IAP groups were observed (p>0.05). However, the IAP was associated with decreased alpha diversity (number of amplicon sequence variants, p<0.001), decreased relative abundances of the genera Bacteroides and Bifidobacterium, and increased relative abundances of genera Enterococcus and Rothia (q<0.01 for all of them) in transitional stool samples. The findings of this study suggest that exposure to IAP may significantly influence the early development of the neonatal oral and gut microbiomes. IAP affected the neonatal oral bacteriome in the first two days after birth as well as the neonatal fecal bacteriome in transitional stool samples. In addition, it highlights the necessity for further investigation into the potential long-term health impacts on children.
Keywords: Microbiome, Infant, Mother, Next-generation sequencing, Antibiotics, 16S rRNA, Diversity
Subject terms: Microbiology, Molecular biology
Introduction
Birth events and factors unique to the labor may influence the composition of the neonate’s microbiome. The mode of delivery and use of antibiotics count among the factors most affecting neonate colonization by microorganisms1–4. In the last decade, the research in gut microbiome acquisition has been consistently reporting that C-section (cesarean section, CS) decreases the relative abundance of Bacteroides and Bifidobacterium spp. and increases the abundance of Enterococcus, Staphylococcus, and/or Clostridioides spp. in the gut microbiome of CS-delivered neonates5–7. A similar pattern was observed in the neonatal oral microbiome in children delivered by CS, which mirrors communities similar to those found on the mother’s skin8. Vaginal mode of delivery (VD) is considered an important primary source of microbial communities for the children´s long-term health because it promotes a more diverse and beneficial microbiota than CS1,9–11. However, microbial profile differs even among neonates after vaginal births. One of the causes for this may lie in intrapartum antibiotics prophylaxis (IAP), the most frequent cause of exposure to antibiotics during the perinatal period and labor12.
The IAP treatment is an important intervention to reduce the risk of maternal and neonatal postnatal infection. It is commonly used in CSs but also important in VDs to prevent the transmission of any pathogens in suspected infection, mostly due to maternal group B Streptococcus (GBS) positivity or, in some cases, due to premature rupture of membranes13. The incidence of CSs globally increases, as well as the IAP use. According to research by the World Health Organization14, the global CS rate increased from approx. 7 % in 1990 to 21 % in 2021. The estimated worldwide prevalence of GBS during pregnancy is 18 %, despite regional variability ranging from 11 to 35%15. Even though the screening procedures for GBS contributed to reducing the incidence of early-onset GBS sepsis in neonates16,17, it also increased the use of IAP18. In 2002, the Center for Disease Control and Prevention (CDC) recommended a universal screening-based strategy in their guidelines, where all pregnant women are screened for GBS between 36 and 38 weeks of pregnancy17,19. In effect, an increase of IAP from 26.8 % in 1998–1999 to 31.7 % in 2003–2004 was reported in the USA20. In contrast, some countries (Denmark, UK, New Zealand) employed GBS-mitigating strategies based on the presence of risk factors for early-onset GBS disease in neonates, while not clearly lowering the antibiotic administration rates19. IAP, however, changes the microbiome acquisition trajectory. It may directly affect microbial colonization by passing antibiotics into the fetal/neonatal bloodstream through placenta or through breast milk. Moreover, it reduces the transmission of susceptible bacterial groups from the mother to the neonate10,21,22.
The gut microbiome has been shown to affect childhood development. Antibiotic-mediated gut dysbiosis in neonates is associated with increased health risks, such as allergy23,24 and obesity25. Despite this knowledge, the effects of IAP on neonatal oral microbiome in the first week of life remains underexplored26 and of the available studies, most focused on the changes of the neonatal fecal bacteriome only two days or later after birth11,12,21,27–30. To this date, only two studies describing the impact of IAP on the bacteriome of meconium samples have been published27,31. The presented preliminary study aimed to investigate the effect of IAP on the bacteriome profiles of the (i) maternal oral mucosa, (ii) neonatal oral cavity, as well as the neonatal (iii) meconium and (iv) transitional stool in the first week after birth.
Methods
Study population
The Central European Longitudinal Studies of Parents and Children: The Next Generation (CELSPAC: TNG) study is designed as a new prospective birth cohort which will follow up on 2,000 children from their prenatal period to adolescence with the aim of assessing exposome factors affecting children’s health32. A pilot phase of CELSPAC: TNG study was initiated to evaluate feasibility of the protocol for collection, processing and storing of biological samples including oralswabs. The mothers (age ≥18 years) were recruited in their 38th week of pregnancy at University Hospital Brno, Czech Republic, in 2015 and 2016. In line with Helsinki declaration, all pregnant women involved in that study were willing to participate and gave informed consent for themselves and prospective neonates. Data related to the pregnancy, maternal IAP, birth, and mother’s and neonate’s health characteristics were retrieved from hospital records.
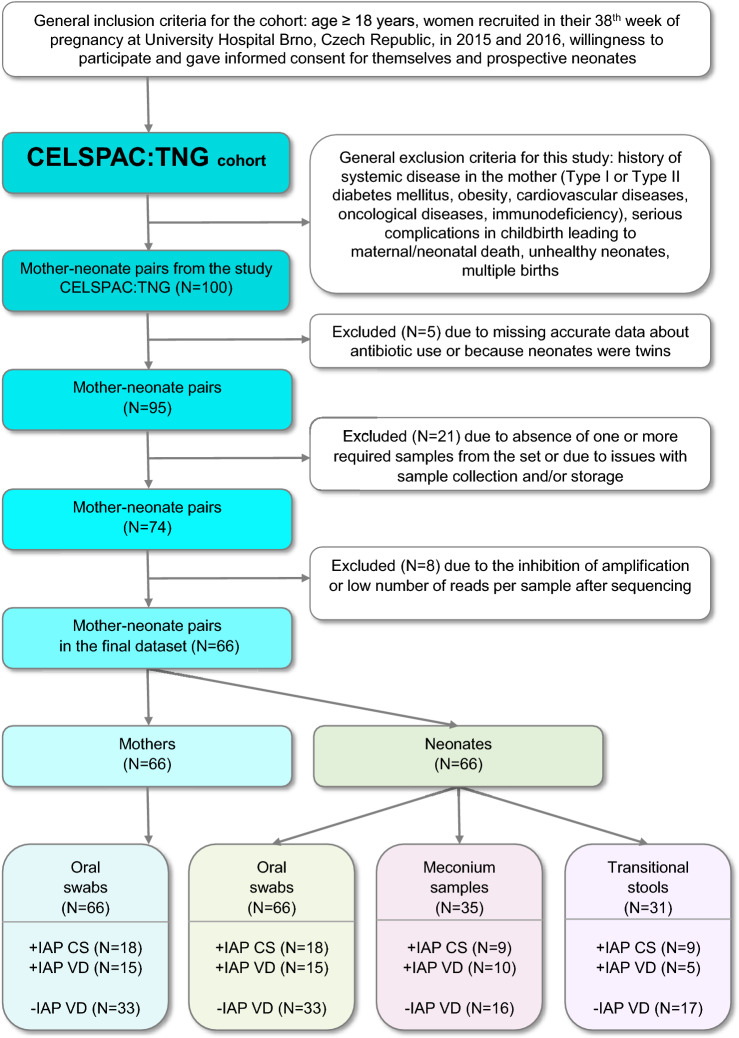
From that cohort, we retrospectively selected 100 mother-neonate pairs. For mothers, exclusion criteria comprised a history of systemic disease (Type I or Type II diabetes mellitus, obesity, cardiovascular diseases, oncological diseases, immunodeficiency; gestational diabetes and asthma were, however, not considered exclusion criteria) and serious complications in childbirth leading to maternal/neonatal death. For neonates, the inclusion criteria were: birth in gestational weeks 38–42 by CS or VD and good health without congenital defects. Multiple births were not included in the study. From thus acquired cohort of 100 mother-neonate pairs, 26 pairs were removed based on additional exclusion criteria, i.e., the absence of crucial data (IAP type and dose, details about sample collection, and sample storing) or the absence of one or more samples from the pair. Lastly, additional eight mother-neonate pairs were removed from the cohort due to the lack of sequencing depth analysis in one or more samples from the mother-neonate pair, yielding a final cohort of 66 pairs, see Fig. 1.
Figure 1.
Inclusion and exclusion criteria flowchart and overview of analyzed samples. CELSPAC: TNG Central European Longitudinal Studies of Parents and Children: The Next Generation Study, N number of participants.
The current preliminary study, focusing on the transmission and development of bacterial community in relation to exposure factors in children in the first week of their life, was designed as a retrospective case-control association study. Mother-neonate pairs were retrospectively classified into 2 groups according to the IAP administration, i.e., cases with IAP (+IAP), and controls without IAP (-IAP).
Sample collection
Healthcare professionals collected all specimens in a sterile and uniform manner within the first 5 days of postpartum hospitalization. Swabs from the oral cavity of each mother (buccal mucosa) and neonate were collected into sterile 1.5 mL tubes by the nylon swab FLOQSwabs (Copan, CA, USA) and stored at −80 ℃ until DNA extraction.
The swabs from the single-use diaper were collected by FLOQSwabs (Copan, CA, USA) and classified as meconium or transitional stool by experienced healthcare professionals based on fecal characteristics (color and consistency). Therefore, hereinafter, we strictly distinguish between the expressions “meconium” and “transitional stool” describing a non-meconium sample.
DNA extraction
All samples were processed using the DNeasy® PowerSoil® kit (Qiagen, Germany), which had been proven suitable for clinical sample extraction before33–35. 750 uL of bead solution and 60 uL of C1 buffer heated to 64 ℃ were added to the broken swabbing head. The FastPrep-24 (MP Biomedicals, USA) was used for sample homogenization and set to 6.5 m/s for 45 s. The next steps were performed according to the manufacturer’s manual. The genomic DNA concentration was determined spectrophotometrically using Synergy Mx (BioTek, USA). The quality of purified genomic DNA was determined after electrophoresis on 1% agarose gel. Extracted DNA was stored at −20 ℃.
16S rRNA amplicon sequencing
Substantial improvement was made to the library preparation process, when the polymerase chain reaction (PCR) reagents´ decontamination step using the 8-methoxypsoralen (8-MOP) was applied, as used earlier for next-generation sequencing (NGS) analysis of low-abundant samples36. Prepared PCR mixtures with 8-MOP (0.16 mM, Sigma-Aldrich, USA) were incubated at 4 ℃ for 1.5 h and exposed to UVA (365 nm) for 7 min (30 J/cm2) in UV-crosslinker. After decontamination, the template and the artificial spike-in standard (SIS) were added to each PCR. The spike-in standard (SIS)37 was used as the internal control and consisted of the synthetic 16S rDNA gene (1525 bp). The synthetic gene was inserted into the pUC57-Amp vector (GenWiz, Germany), transformed into Dh5α Escherichia coli cells, and cloned. The plasmids with artificial sequence were extracted and the exact copy numbers per 1 μL were assessed. Each PCR was spiked with 200 copies of SIS. The sequencing library was prepared according to the Illumina 16S Metagenomic Sequencing Library Preparation Protocol with some deviations regarding the widely used double barcoding strategy, as described before33. The V3-V4 hypervariable region of the bacterial 16S rDNA gene approx. 290 bp long was amplified using the previously published degenerated primers38 with the inner tags for distinguishing the particular samples. The total volume of 31 µL consisted of 15 uL of Q5® High Fidelity Master Mix (New England Biolabs, MA, USA), 1.5 µL of each primer (10 uM), 2 µL of 8-MOP 10x diluted, 1 µL of SIS and 5 µL of extracted DNA and 5 µL of sterile DNA-free water (Qiagen, Germany). The initial denaturation (15 min at 95 ℃) was followed by 30 cycles consisting of denaturation at 94 ℃ for 35 s, primer annealing at 55 ℃ for 35 s, and extension at 72 ℃ for 45 s. The final extension at 72 ℃ lasted 10 min. The PCR negative (sterile DNA-free water used as a template) and in-house positive controls (mixture of stool samples with high load of bacterial DNA) were included in each PCR batch. PCR products were visualized after electrophoresis on 1.5 % agarose gel.
SPRIselect beads (Beckman Coulter Genomics, USA) were used to clean the PCR products according to the manufacturer’s recommendations. Quant-iT (Thermo Fisher Scientific, USA) and microplate reader Synergy Mx (BioTek, USA) were used to fluorochemically assess the concentration of cleaned PCR products to pool those with different inner tags equimolarly. Pools were indexed with Nextera XT indexes (Illumina, USA), quantified fluorochemically, and pooled equimolarly. The quality and concentration of the prepared library was assessed using a 2100 Bioanalyzer Instrument (Agilent Technologies, USA) and qPCR (KAPA Library Quantification Complete Kit, Kapa Biosystems, USA) shortly before sequencing. The library was diluted to a final concentration of 8 pM, and 20% of PhiX DNA (Illumina, USA) was added. Sequencing was performed with the Miseq reagent kit V3 using a MiSeq 2000 instrument according to the manufacturer’s instructions (Illumina, USA).
Bioinformatics processing
Paired reads from 16S rRNA amplicon sequencing were first processed using an in-house pipeline implemented in Python 3. Our in-house script used Trimmomatic to trim bases with a quality score below Q20, to remove adapters and barcodes, while maintaining only paired-end reads with minimum overlap of 20 with the trimming settings set to—forward trimmed to 236 bp and—reverse trimmed to 168 bp. In order to minimize sequencing and PCR-derived errors, forward and reverse reads were denoised using the DADA2 amplicon denoising R package39. Following denoising, the forward and reverse reads were joined using the fastq-join read joining utility. To be joined, reads in pairs had to overlap in at least 20 base pairs with no mismatches allowed. Pairs in which this was not the case were discarded. As the final step, chimeric sequences were removed from the joined reads using the remove Bimera function of the DADA2 R package40. Subsequent taxonomic assignment was conducted by the uclust-consensus method from the QIIME41 microbial analysis framework using the Silva v. 123 reference database42. Samples with number of reads <1000 were removed from the analysis.
Statistical methods
Four different matrices (maternal oral swabs, neonatal oral swabs, neonatal meconium, and neonatal transitional stool) were independently evaluated.
Both maternal and neonatal oral swabs were divided into those collected ≤48 h and those collected >48 h after childbirth. This cut-off was chosen to correspond to the meconium/transitional stool classification as meconium should pass ideally within the first 48 h in healthy full-term neonates43.
Contaminant ASVs were identified using the Decontam R package (v. 1.10.0)44. Neonatal samples were assumed to be low-abundant, hence the function IsNotContaminant with the default setting was used. For the mother’s oral swabs, the function IsContaminant with the default setting was used. Only the prevalence method was applicable. ASVs defined as contaminants were removed from all samples. Additionally, Cyanobacteria, mitochondria, and bacteria unassigned on the phylum level were filtered out. Finally, only genera with a relative abundance ≥0.5 % in at least one sample or with a relative abundance <0.5 % in at least three samples were included in the subsequent analysis.
All analyses were performed on genus, family, order, class, and phylum level. Prior to statistical analysis, data were treated as compositional and transformed using the centered log-ratio (CLR) transformation45 on the raw read abundance matrix. All zeroes in the original dataset were replaced by a constant of 0.65.
Fisher’s exact test (categorical variables) and Mann-Whitney U test (continuous variables) were used for the comparison of demographic and clinical characteristics among the groups of interest as well as for the comparison of alpha diversity indices.
Variability in the bacterial composition of different matrices was first visualized using multivariable approaches, namely Principal Component Analysis (PCA). The permutational multivariate analysis of variance (PERMANOVA; 9999 permutations) was performed to test for differences in the dispersion and centroids of the groups of bacterial communities, based on the Euclidean distance.
Bacterial co-occurrence was displayed by the UpSet plots. The presence of specific genera was considered as more than five reads in at least one sample in a defined material. Mann-Whitney U test was used to compare the differences in abundances of individual taxa between the groups. The comparison was performed only if the genus was present in at least three individuals in at least one of the compared groups. The resulting p-values were adjusted for multiple hypotheses testing using the Benjamini-Hochberg procedure (BH). Results were considered significant at FDR<0.1.
To evaluate the statistical power of this study, we conducted a simulation based on the Shannon index data across all four distinct sample types: maternal oral swabs, neonatal oral swabs, and neonatal meconium and transitional stool samples. A kernel density estimator was developed utilizing the measured Shannon index data, employing a Gaussian kernel with 214 points, a bandwidth of 0.33, and an interval of [0, 6]. Subsequently, 1000 artificial datasets were generated for sample sizes ranging from 5 to 150 (in increments of 5) for both the +IAP and -IAP groups. For each artificial dataset, the Mann-Whitney U test was performed to determine statistical significance. The proportion of tests that yielded significance at the 0.05 level was then calculated to assess the study’s power.
All statistical analyses were performed in R (v. 4.0.5)46 using additional R packages: compositions (v. 2.0-4) for CLR transformation47; vegan (v. 2.6-2) for PERMANOVA48; factoextra (v. 1.0.7.) for PCA49 ggplot2 (v. 3.3.6) for box and whiskers plots and barplots50; FDRestimation (v. 1.0.1) for Benjamini-Hochberg correction51; UpSetR (v. 1.4.0.) for UpSet plots52.
Ethics approval and consent to participate
The CELSPAC: TNG study was approved by the Multicentre and Local Ethical Committee of University Hospital Brno, Czech Republic (No. 20140409-01, date 09/04/2014) and performed according to relevant ethical regulations. Mothers gave informed consent for themselves and prospective neonates.
Results
Participant’s characteristics
Out of 100 pregnant women originally recruited from the CELSPAC: TNG, 66 mother-neonate pairs and their 198 samples met all inclusion and exclusion criteria and were included in this preliminary study (see the flowchart in Fig. 1), i.e., the drop-out was 34%. Of these, N=33 neonates were delivered by CS or vaginally with IAP (cases; 18 +IAP CS and 15 +IAP VD, respectively) and N=33 neonates were delivered vaginally with no IAP (controls, -IAP VD). We have also acquired data on the feeding mode from the hospital and found out that the number of exclusively breastfed, partially breastfed and formula-fed infants did not differ between the IAP groups during the hospital stay, with a clear majority (72%, 48 out of 66 neonates) exclusively breastfed (see Table 1).
Table 1.
Demographic characteristics of the 66 mothers and neonates and characteristics of their samples.
| Characteristics of mothers | +IAP | -IAP | p-value | Statistical test | +IAP CS | +IAP VD | -IAP VD | p-value | Statistical test |
|---|---|---|---|---|---|---|---|---|---|
| N | 33 | 33 | 18 | 15 | 33 | ||||
| Maternal age (in years)—median (min, max) | 32 (19, 38) | 33 (21, 43) | 0.344 | Mann-Whitney | 30 (19, 37) | 32 (28, 38) | 33 (21, 43) | 0.269 | Kruskal-Wallis ANOVA |
| Gravidity (number of pregnancies in life) (%) | |||||||||
| I | 12 (36.4) | 11 (33.3) | 1.000 | Fisher‘s exact | 9 (50.0) | 3 (20.0) | 11 (33.3) | 0.475 | Fisher‘s exact |
| II | 13 (39.4) | 13 (39.4) | 5 (27.8) | 8 (53.3) | 13 (39.4) | ||||
| III and more | 8 (24.2) | 9 (27.3) | 4 (22.2) | 4 (26.7) | 9 (27.3) | ||||
| Parity (number of parturitions) (%) | |||||||||
| I | 16 (48.5) | 13 (39.4) | 0.376 | Fisher‘s exact | 12 (66.7) | 4 (26.7) | 13 (39.4) | 0.092 | Fisher‘s exact |
| II | 15 (45.5) | 14 (42.4) | 6 (33.3) | 9 (60.0) | 14 (42.4) | ||||
| III | 2 (6.1) | 6 (18.2) | 0 (0.0) | 2 (13.3) | 6 (18.2) | ||||
| Induced delivery (%) | 13 (39.4) | 7 (21.2) | 0.180 | Fisher‘s exact | 8 (44.4) | 5 (33.3) | 7 (21.2) | 0.204 | Fisher‘s exact |
| Rupture of the amniotic sac (%) | 5 (15.2) | 2 (6.1) | 0.427 | Fisher‘s exact | 2 (11.1) | 3 (20.0) | 2 (6.1) | 0.384 | Fisher‘s exact |
| Asthma (%) | 2 (6.1) | 3 (9.1) | 1.000 | Fisher‘s exact | 1 (5.6) | 1 (6.7) | 3 (9.1) | 1.000 | Fisher‘s exact |
| Gestational diabetes (%) | 2 (6.1) | 3 (9.1) | 1.000 | Fisher‘s exact | 1 (5.6) | 1 (6.7) | 3 (9.1) | 1.000 | Fisher‘s exact |
| Meconium in amniotic fluid (%) | 4 (12.1) | 3 (9.1) | 1.000 | Fisher‘s exact | 1 (5.6) | 3 (20.0) | 3 (9.1) | 0.441 | Fisher‘s exact |
| Group B Streptococcus positive* (%) | 7 (21.2) | 0 (0.0) | 0.011 | Fisher‘s exact | 2 (11.1) | 5 (33.3) | 0 (0.0) | 0.001 | Fisher‘s exact |
| Oral swab collection (%) | |||||||||
| Within 48 h after birth of her child | 16 (48.5) | 25 (75.8) | 0.041 | Fisher‘s exact | 4 (22.2) | 12 (80.0) | 25 (75.8) | <0.001 | Fisher‘s exact |
| After 48 h after birth of her child | 17 (51.5) | 8 (24.2) | 14 (77.8) | 3 (20.0) | 8 (24.2) | ||||
| Characteristics of neonates | +IAP | -IAP | p-value | Statistical test | +IAP CS | +IAP VD | -IAP VD | p-value | Statistical test |
|---|---|---|---|---|---|---|---|---|---|
| N | 33 | 33 | 18 | 15 | 33 | ||||
| Gestational age (weeks) (%) | |||||||||
| 38–39 | 17 (51.5) | 8 (24.2) | 0.041 | Fisher‘s exact | 9 (50.0) | 8 (53.3) | 8 (24.2) | 0.076 | Fisher‘s exact |
| 40–41 | 16 (48.5) | 25 (75.8) | 9 (50.0) | 7 (46.7) | 25 (75.8) | ||||
| Sex (%) | |||||||||
| Female | 15 (45.5) | 17 (51.5) | 0.806 | Fisher‘s exact | 9 (50.0) | 6 (40.0) | 17 (51.5) | 0.766 | Fisher‘s exact |
| Male | 18 (54.5) | 16 (48.5) | 9 (50.0) | 9 (60.0) | 16 (48.5) | ||||
| Birth weight (g) median (min, max) | 3600 (2350, 4340) | 3390 (2630, 4630) | 0.366 | Mann-Whitney | 3635 (2680, 4340) | 3500 (2350, 4050) | 3390 (2630, 4630) | 0.298 | Kruskal-Wallis ANOVA |
| Apgar score (%) | |||||||||
| Low (<7) | 2 (6.1) | 1 (3.0) | 1.000 | Fisher‘s exact | 2 (11.1) | 0 (0.0) | 1 (3.0) | 0.305 | Fisher‘s exact |
| Normal (>7) | 31 (93.9) | 32 (97.0) | 16 (88.9) | 15 (100.0) | 32 (97.0) | ||||
| Basal excess (Astrup) median (min, max) | −4.9 (−11.9, −0.7) | −5.3 (−13.6, −0.9) | 0.119 | Mann-Whitney | −4.9 (−11.9, −0.7) | −5.0 (−8.1, −1.6) | −5.3 (−13.6, −0.9) | 0.257 | Kruskal-Wallis ANOVA |
| pH median (min, max) | 7.29 (7.07, 7.42) | 7.31 (7.00, 7.47) | 0.714 | Mann-Whitney | 7.29 (7.07, 7.36) | 7.33 (7.19, 7.42) | 7.31 (7.00, 7.47) | 0.377 | Kruskal-Wallis ANOVA |
| Newborn immunization (%) | 4 (12.1) | 1 (3.0) | 0.355 | Fisher‘s exact | 3 (16.7) | 1 (6.7) | 1 (3.0) | 0.172 | Fisher‘s exact |
| Newborn infection (%) | 1 (3.0) | 3 (9.1) | 0.613 | Fisher‘s exact | 1 (5.6) | 0 (0.0) | 3 (9.1) | 0.802 | Fisher‘s exact |
| Newborn conjunctivitis (%) | 3 (9.1) | 6 (18.2) | 0.475 | Fisher‘s exact | 3 (16.7) | 0 (0.0) | 6 (18.2) | 0.227 | Fisher‘s exact |
| Newborn jaundice (%) | 4 (12.1) | 5 (15.2) | 1.000 | Fisher‘s exact | 1 (5.6) | 3 (20.0) | 5 (15.2) | 0.467 | Fisher‘s exact |
| Feeding mode | |||||||||
| Exclusively breastfed | 24 (72.7) | 24 (72.7) | 0.564 | Fisher’s exact | 14 (77.8) | 10 (66.7) | 24 (72.7) | 0.644 | Fisher’s exact |
| Partially breastfed | 2 (6.1) | 1 (3.0) | 1 (7.1) | 1 (6.7) | 1 (3.0) | ||||
| Formula fed | 6 (18.2) | 4 (12.1) | 2 (14.3) | 4 (26.7) | 4 (12.1) | ||||
| Missing | 1 (3.0) | 4 (12.1) | 1 (7.1) | 0 | 4 (12.1) | ||||
| Oral swab collection (%) | |||||||||
| Within 48 h after birth | 16 (48.5) | 27 (81.8) | 0.009 | Fisher‘s exact | 4 (22.2) | 12 (80.0) | 27 (81.8) | <0.001 | Fisher‘s exact |
| After 48 h after birth | 17 (51.5) | 6 (18.2) | 14 (77.8) | 3 (20.0) | 6 (18.2) | ||||
| Fecal samples (%) | |||||||||
| Meconium | 19 (57.6) | 16 (48.5) | 0.622 | Fisher‘s exact | 9 (50.0) | 10 (66.7) | 16 (48.5) | 0.554 | Fisher‘s exact |
| Transitional stool | 14 (42.4) | 17 (51.5) | 9 (50.0) | 5 (33.3) | 17 (51.5) | ||||
Data is presented as counts or median (percentage or minimum and maximum in brackets).
*Group B Streptococcus positive status includes those with positive screening status.
N number of cases, +IAP with intrapartum antibiotic prophylaxis, -IAP without intrapartum antibiotic prophylaxis, CS C-section, VD vaginal delivery, N number of participants.
The average maternal age was 32 ± 4 years. Even though mothers with gestational diabetes and asthma were enrolled in the study, these variables were represented equally among groups. Overall, there were no significant differences (p>0.05) in the mother’s or neonate’s health and birth characteristics between the +IAP and -IAP groups. There were significant differences in the numbers of collected oral swabs, from both mothers and neonates (p<0.05, p<0.01, respectively), grouped according to the sampling period; in mothers and neonates without IAP, oral swabs were collected predominantly ≤48 h after birth (75.8% and 81.8%, respectively) while oral swabs were collected >48 h after birth in 51.5% of both mothers and neonates with IAP. Neonatal birth weight ranged from 2350 to 4630 grams.
Administered antibiotics varied among the delivery modes. In women with CS delivery, cephalosporins (N=14), penicillins (N=3) or lincosamides (N=1, specifically clindamycin) were used. In the VD group, penicillins (N=14) were by far the most common antibiotics; cephalosporin was used only in one case with a premature rupture of membranes.
Bacteriome analysis—general results
In total, 206 samples were analyzed, of which 198 met the quality criteria for further statistical analysis. After quality filtering and chimeras removal, 12,229,051 reads (median=56,823 reads per sample; interquartile range, IQR=25,835) were obtained, of which 4,045,330 reads (median=55,217.50 reads per sample; IQR=23,999.75) originated from the maternal oral swabs (N=66); 3,821,525 reads (median= 55,157 reads per sample; IQR=27,384.25) from the neonatal oral swabs (N=66); 2,255,835 reads (median=59,802 reads per sample; IQR= 28,281) from the meconium (N=35); and 2,106,361 reads (median=63,584 reads per sample; IQR= 34,087) from the transitional stools (N=31). After the decontamination step and filtering out ASV unassigned at the phylum level as well as Cyanobacteria- and mitochondria-assigned ASVs, most phyla (8 out of 12, i.e., 67%) were present at low abundances (median of relative abundance <5%).
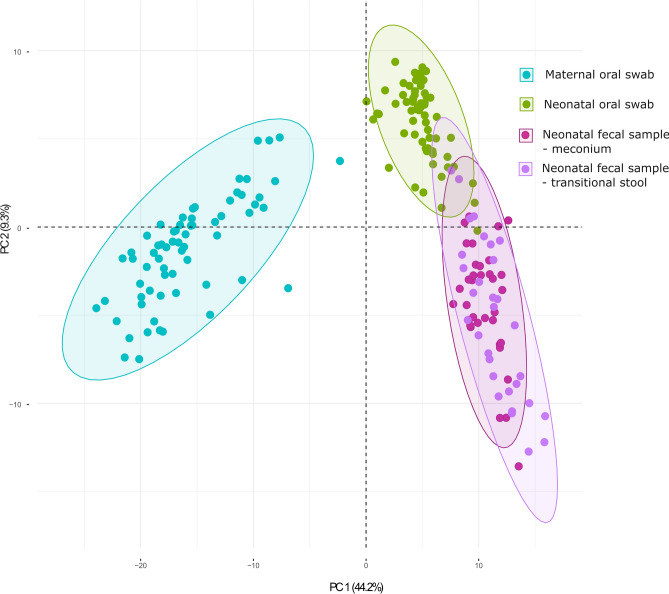
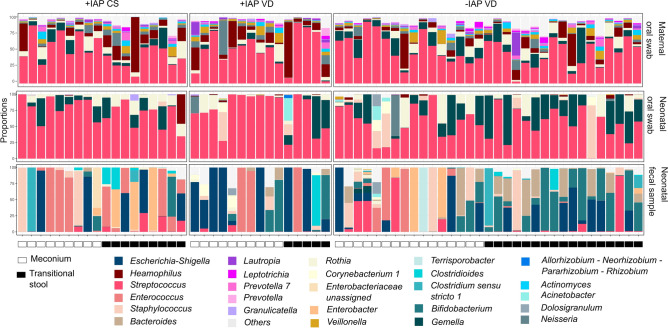
Principal Component Analysis (PCA, Fig. 2) revealed the presence of three distinct bacterial communities representing the maternal oral, neonatal oral, and neonatal fecal bacteriomes. The bacteriome composition of the most abundant bacterial genera in maternal oral swabs, neonatal oral swabs, and fecal samples is shown in Fig. 3. Firmicutes, Proteobacteria, and Actinobacteria were the three most abundant phyla, with median relative abundances of 63.1, 12.5, and 9.2%, respectively. The numbers of genera were as follows: mothers’ oral swabs: 114 genera; neonates’ oral swabs: 28 genera; neonates’ meconium: 35 genera; neonates´ transitional stool: 25 genera.
Figure 2.
Principal component analysis (PCA, genus level) of maternal oral bacteriomes, neonatal oral and fecal bacteriomes.
Figure 3.
Bacteriome composition (genus level) in maternal oral swabs, neonatal oral swabs, and fecal samples. Samples classified according to intrapartum antibiotic exposure +IAP with intrapartum antibiotic prophylaxis, -IAP without intrapartum antibiotic prophylaxis, CS C-section, VD vaginal delivery.
Initial analyses of the samples revealed no statistically significant differences in the number of ASVs or Shannon index between the +IAP CS and +IAP VD groups in any of the neonatal matrices (see Table S1). For this reason, both these groups whom IAP was administered were combined into one for this type of analyses.
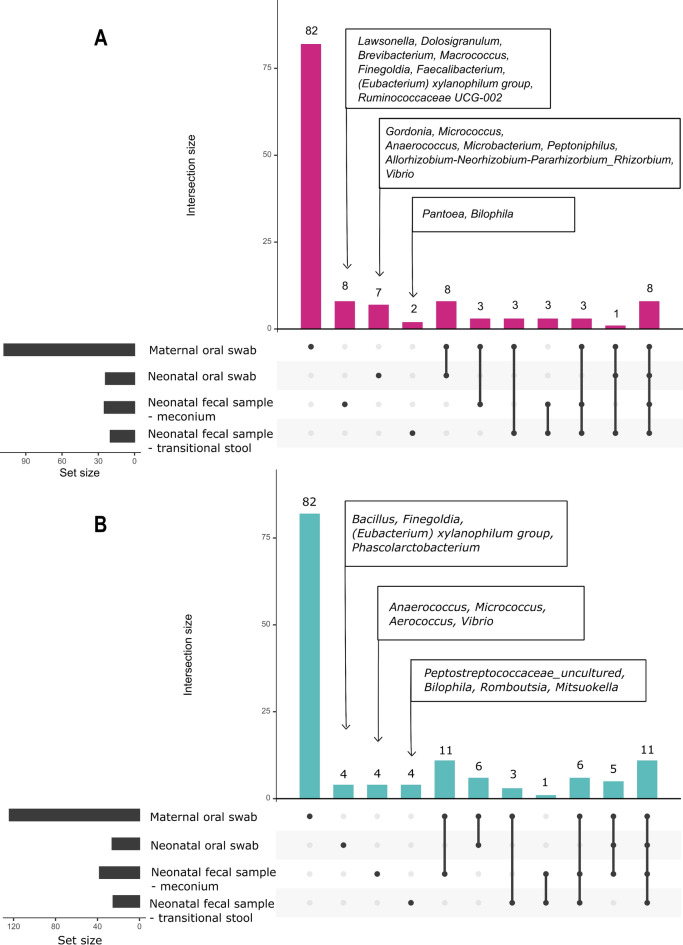
To gain further insights into the bacterial composition overlap among these populations, an UpSet plot (Fig. 4) was constructed. An intersection between maternal and neonatal oral swabs was observed in 17 genera in the +IAP group and 22 genera in the -IAP group. In addition, Haemophilus and Neisseria were also among the most commonly observed genera in both maternal and neonatal oral samples. Furthermore, the overlap between maternal oral swabs and neonatal fecal samples revealed 18 genera present in the +IAP group and 36 genera in the -IAP group. Bifidobacterium and Bacteroides were among the dominant genera in this shared bacterial community.
Figure 4.
Upset plot of bacterial genera in maternal and neonatal samples. In the figure, dots signify the number of unique bacterial genera specific to each sample matrix. Connecting lines between dots indicate the number of genera shared among different sample matrices. (A) with administration of intrapartum antibiotic prophylaxis (+IAP) and (B) without administration of intrapartum antibiotic prophylaxis (-IAP).
In the maternal oral swabs, total of 82 unique bacterial genera were coincidentally observed both in +IAP and -IAP groups. Some examples of genera unique to maternal oral swabs to be named were Prevotella, Corynebacterium, and Fusobacterium. Lastly, there were bacterial genera that were not found in maternal oral swabs but were present in neonatal oral swabs (7 genera in +IAP and 4 genera in -IAP) and neonatal fecal samples (10 genera in +IAP and 8 in -IAP).
Effect of IAP on the maternal oral bacteriome
The comparison of maternal oral bacteriomes after giving birth with or without IAP did not reveal any significant differences in alpha diversity of oral bacterial representatives (p>0.05, Fig. S1). In addition to dominating genera Streptococcus, Rothia, and Gemella (Fig. 3), common oral genera such as Prevotella, Veillonella, Fusobacterium, and Actinomyces were found in maternal oral swabs as well. The PCA showed differences between +IAP and -IAP maternal oral samples; however, the PERMANOVA test was not significant (p=0.056).
Effect of IAP on the neonatal oral bacteriome
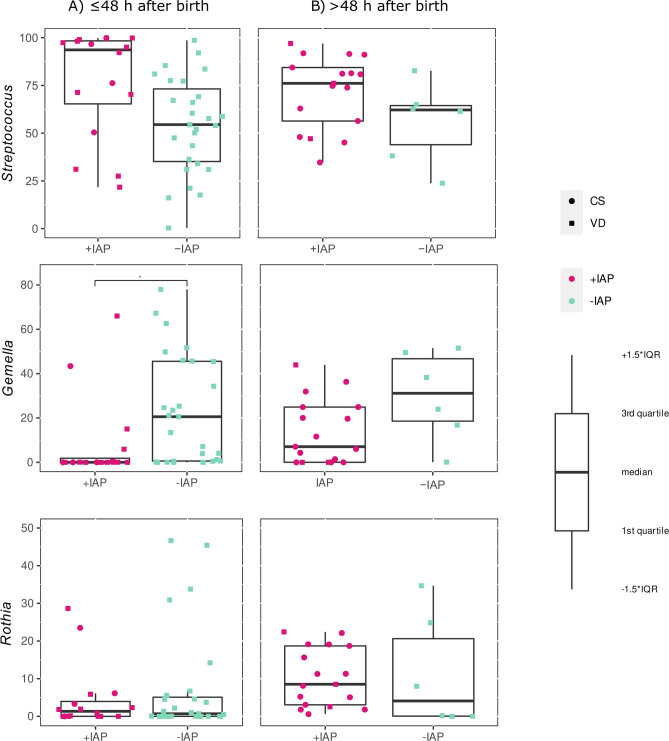
The effect of IAP on the neonatal bacteriome was tested separately for oral samples collected ≤48 h and >48 h after birth. IAP was associated with significantly reduced bacteriome diversity in neonatal oral samples (Shannon index; p=0.01) ≤48 h after birth (Fig. S2). In samples collected >48 h after birth, an insignificant decrease (p>0.05) in both the number of amplicon sequence variants (ASVs) and the Shannon index was observed in oral neonatal samples in the +IAP group compared to the control (-IAP) group. The PERMANOVA test revealed a significant impact of IAP on neonatal oral bacteriomes ≤48 h after birth (p=0.038) but no difference was observed in samples collected >48 h after birth (p>0.05).
Bacterial genera Streptococcus, Gemella, and Rothia dominated in neonatal oral bacteriomes in both groups. In the +IAP group, the relative abundance of the genus Gemella was significantly lower in neonatal oral samples collected ≤48 h after birth (q=0.08) than in the controls (-IAP). This was, however, not true of samples collected >48 h after birth (p>0.05, q>0.1, Fig. 5). Significant differences in relative abundances between cases and controls were also observed on the family level in Neisseriaceae (q=0.09) and Streptococcaceae (q=0.09), data are not shown.
Figure 5.
Relative abundance of specific bacteria in neonatal oral swabs. This figure examines the relative abundance of Streptococcus, Gemella, and Rothia in neonatal oral swabs according to the intrapartum antibiotic prophylaxis: (A) samples collected ≤48 h after birth, (B) samples collected >48 h after birth; +IAP, with intrapartum antibiotic prophylaxis; -IAP without intrapartum antibiotic prophylaxis, IQR interquartile range, CS,C-section, VD vaginal delivery. *q<0.1.
Effect of IAP on the neonatal fecal bacteriomes
When all neonate fecal (both meconium and transitional stool) samples (N=66) were stratified according to the antibiotic exposure during delivery, a statistically significant difference in the number of ASVs (p<0.001) between +IAP and -IAP group was observed.
However, meconium samples alone did not show any significant difference in the number of observed ASVs (p>0.05) or in the Shannon index (p>0.05) between the tested groups. On the other hand, transitional stool samples differed significantly in the number of observed ASVs in +IAP neonates compared to the -IAP neonates (p=0.02, Fig. S3).
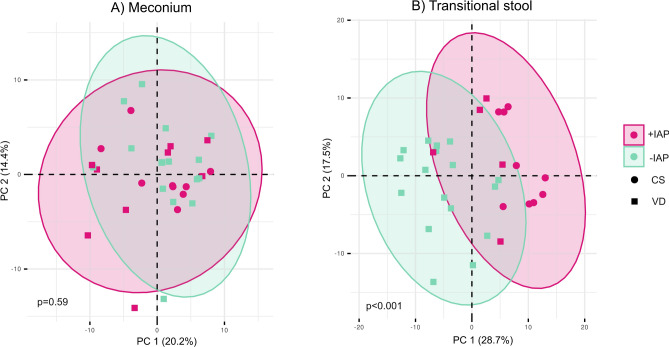
The PERMANOVA test on the bacterial profiles assessing the meconium and transitional stool separately revealed significant differences in neonatal transitional stool bacteriome at the genus level between the two studied groups (p<0.001, Fig. 6), while no significant differences were observed in meconium samples (p=0.59).
Figure 6.
Principal component analysis (PCA) of neonatal meconium and transitional stool samples. Displaying a PCA (genus level) of neonatal (A) meconium and (B) transitional stool samples according to the intrapartum antibiotic prophylaxis; +IAP with intrapartum antibiotic prophylaxis, -IAP without intrapartum antibiotic prophylaxis, CS C-section, VD vaginal delivery.
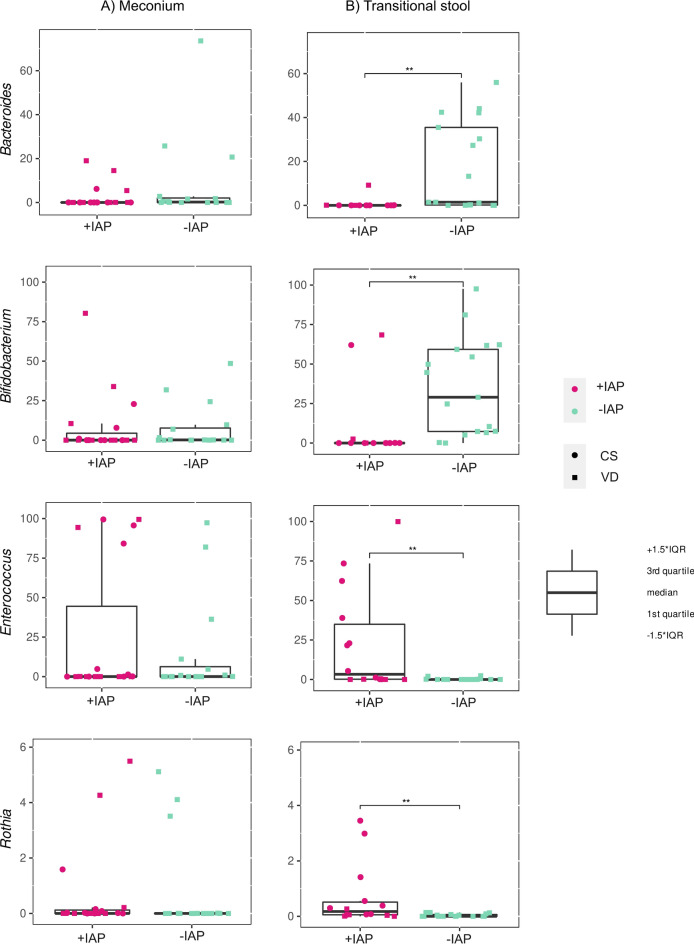
IAP administration affected the abundance of several bacteria in transitional stool samples, but not in meconium samples. The taxonomical analysis identified the five most abundant genera in meconium samples as Staphylococcus, Escherichia-Shigella, Streptococcus, Bifidobacterium, and Enterococcus (Fig. 3). Bifidobacterium, Streptococcus, Escherichia-Shigella, Staphylococcus, and Enterococcus were the most abundant genera in transitional stool samples. IAP was significantly associated with decreased relative abundances of the genera Bifidobacterium (q=0.002) Bacteroides (q=0.003) and Parabacteroides (q=0.07) and significantly higher relative abundances of Rothia (q=0.005), Enterococcus (q=0.01, see Fig. 7), and Clostridioides (q=0.09) in transitional stool samples (Mann-Whitney U test).
Figure 7.
Relative abundance of specific bacteria in stool samples: This figure illustrates the relative abundance of Bacteroides, Bifidobacterium, Enterococcus, and Rothia in neonatal (A) meconium and (B) transitional stool according to the intrapartum antibiotic prophylaxis; +IAP with intrapartum antibiotic prophylaxis, -IAP without intrapartum antibiotic prophylaxis, IQR interquartile range, CS C-section, VD vaginal delivery; **q<0.01.
In addition, we have separately analyzed the representation of individual bacterial genera among all three groups (-IAP VD, +IAP VD and +IAP CS). This analysis revealed that in transitional stool samples (but not meconium samples), the relative abundance of the genera Rothia, Enterococcus, Bifidobacterium, and Bacteroides significantly differed among the groups (Kruskal-Wallis test, q<0.05). This difference was clearly driven by the increased relative abundance of Rothia and Enterococcus in +IAP CS neonates compared to the -IAP VD group (Dunn’s post hoc test, q<0.05) as well as by the higher relative abundances of the genera Bifidobacterium and Bacteroides in the -IAP VD group than in the +IAP CS group (q<0.05). Neither IAP nor mode of delivery affected the relative abundance of Lactobacillus or Clostridioides in the meconium and transitional stool samples.
Discussion
In this article, our primary objective was to investigate the impact of IAP on neonatal gastrointestinal bacteriomes, with a particular focus on the first week of life. We sought to elucidate this relationship by characterizing the maternal and neonatal oral bacteriomes and the bacteriomes of neonatal meconium and transitional stool. This approach allowed us to understand how IAP might shape early neonatal microbiota development and, therefore, improve our understanding of the foundational days of the human gastrointestinal microbiome.
Maternal oral swabs displayed a higher diversity in bacterial composition than neonatal oral swabs, which is consistent with previous research that has suggested the adult oral microbiota is typically more diverse compared to the neonatal one53,54. As expected, no effect of IAP on the maternal oral bacteriome was found in our study. This aligns with the concept that established microbial communities, such as that of the adult oral cavity, are more resilient to disturbances, including antibiotic interventions55,56. However, the antibiotic exposure during delivery affected the diversity of neonatal oral and transitional stool bacteriomes in the first week of life, reflecting the susceptibility of these developing bacterial communities to external interventions.
Impact of IAP on the neonatal oral bacteriome in the first week of life
The oral cavity presents an initial entry point to microbial colonization of gastrointestinal tract. Nevertheless, there is a limited understanding to the effect of IAP on neonatal oral microbiota development in the first week after birth. Our study showed significant changes in bacterial diversity and abundance in the first 48 h after IAP exposure. However, our data suggests that ≤48 h after birth, the impact of IAP on neonatal oral bacteriome is suppressed.
In the first 48 h after birth, IAP exposure affected the abundances of the genus Gemella and families Neisseriaceae and Streptococcaceae in the neonatal oral cavity. This is in agreement with the study by Gomez-Arango et al.26 who investigated the effect of IAP on neonatal oral bacteriome within the first 3 days after birth by CS in 36 neonates. Their results pointed to a decreased relative abundance of Streptococcaceae, Gemellaceae, and Lactobacillales, and an increased abundance of Neiseeriaceae or Prevotellaceae in the oral cavity of neonates exposed to IAP26. The genera Streptococcus, Staphylococcus, Veillonella, and Lactobacillus are recognized as the first oral colonizers55,57,58. Streptococcus is one of the most dominant genera found in the oral microbiome of neonates during their first days of life59–61. This is due to several factors, including the fact that Streptococcus species are naturally present in the maternal vaginal microbiota, which is one of the first microbial communities that the neonates are exposed to during VD10. Additionally, Streptococcus species are well-adapted to the oral cavity environment62,63. The genus Gemella, together with Granulicatella, Haemophilus, and Rothia, count among later colonizers, the abundance of which grows with the increasing age of the infant57,58,62. The exact role of Gemella and Neisseria in the oral cavity of neonates is not yet fully understood, but it is believed that they may play a role in the early colonization of the oral cavity and contribute to the development of a healthy microbial community26,64. Neisseria species are Gram-negative bacteria that are commonly found on the mucosa of the respiratory tract and oral cavity and may play also an important role in the early development of a healthy oral microbial community61,65.
Studies exploring the impact of IAP administered during CS have shown that IAP can have short-term effects on the oral microbiota of neonates8,10,26,66–68. An Irish birth cohort study with 84 neonates highlighted the differences in bacteriomes in the first week of the lives of neonates delivered by CS and VD; later, the fingerprint of CS vanished67. A Swedish birth cohort study with 59 neonates68 did not reveal any association between the delivery mode and neonatal oral bacteriome composition. Unfortunately, they did not investigate differences caused by application of IAP in VD neonates.
It is important to note that the oral microbiota is dynamic and unstable in the first days after birth and can change over time due to various factors. With time, the differences in oral microbiota associated with IAP seem to diminish as other factors come into play in shaping the oral microbiota in infants, such as diet and environmental influence.
Impact of IAP on the neonatal fecal bacteriome in the first week of life
Our study demonstrates that IAP can influence the microbiota of transitional stool but not that of the meconium. Specifically, IAP was associated with decreased relative abundances of the genera Bifidobacterium and Bacteroides in the transitional stool samples in both CS and VD neonates exposed to IAP compared to VD neonates without IAP exposure. Our results correspond to the evidence of the IAP effect on gut bacterial community 3–7 days after birth69. One of the first culture-based bacteriological studies, performed by Jauréguy et al.70 in 2004, examined amoxicillin used in GBS prophylaxis and its effect on neonates 3 days after birth, predominantly in neonates born vaginally. Their results showed that colonization by Bifidobacteria was detected in fewer No-IAP neonates than in IAP neonates. The topic was followed by more than 20 culture-independent bacteriological studies investigating IAP and gut microbiota development since 201571. Only some of the studies evaluated the effect of IAP in the first days after birth in full-term neonates12,21,27,30,31,69,70,72–74. Stearns et al.21, and Dierikx et al.75 presented delayed colonization by Actinobacteria in both CS and VD with IAP neonates on the postpartum Days 3 and 7. Stearns et al.21 observed a decrease in Bifidobacterium and Bacteroides spp. followed by a significant increase in Lachnospiraceae in CS neonates compared to VD neonates unexposed to IAP. Bacteriomes of neonates with IAP did not differ between those born through CS and VD. Furthermore, Dierikx et al.75 found no differences in the bacteriome on the phylum level between vaginally and CS-born neonates on Days 1 and 7. However, on the genus level, they point to a decreased abundance of Bacteroides with a concurrent increase in Enterococcus in CS neonates compared to VD neonates on Day 7.
Studies focusing entirely on VD neonates of mothers positive for GBS screening present similar findings. Mazzola et al.76 found differences within the Enterobacteriaceae family and within the Bifidobacterium and Bacteroides genera in stool samples 7 days after birth. By qPCR quantification of selected bacteria, Aloisio et al.30 and Corvaglia et al.74 also showed a significant average reduction in the counts of Bifidobacterium spp. in neonates whose mothers received IAP. Nogacka et al.12 observed a reduction in the levels of Actinobacteria and Bacteroidetes and an increase of Proteobacteria and Firmicutes in IAP neonates in comparison to those without IAP. However, at 10 days of age, these differences reached statistical significance only for Actinobacteria. Last but not least, Tapianinen et al.27 observed a significant difference between the IAP and control groups in the relative abundance of Bacteroides on the third day, but no significant differences in any species in the first day after birth.
Furthermore, our study also revealed an increased relative abundance of the genus Rothia in CS neonates compared to VD neonates. In addition, the relative abundance of Enterococcus was significantly increased in transitional stool samples in +IAP CS neonates compared to -IAP VD neonates and borderline significantly increased when comparing +IAP CS to the +IAP VD neonates. Whether any of these changes are associated with the general fact that IAP was used, with specific antibiotics administered during the labor, or with the route of exposure to the microorganisms during the delivery needs to be further investigated. Future studies should ideally stratify the number of neonates according to the mode of delivery and IAP.
In line with our expectations, IAP treatment did not affect bacterial abundance or diversity in meconium samples in our study. More frequent meconium/transitional stool sampling in the first hours and days after birth could further help in explaining the early impact of the delivery mode and IAP on neonatal gut microbiota development. Meconium samples from the first two days after birth were investigated in a few other studies27,31,73,75. Similar to our findings, none of the previous studies found significant differences in any of the abovementioned bacterial groups in IAP-exposed neonates. The impact of IAP is, however, commonly reflected in gut microbiota later than two days after the birth.
Strengths and limitations of the study
This is the first study examining the effect of IAP on neonatal bacteriome of the oral cavity, meconium, as well as transitional stool in the first week after birth. The accumulated evidence from previous metagenomic studies has shown inconsistent findings that may be attributed to factors such as varying antibiotic regimens, routes of administration, statistical analytical methods employed, or uncontrolled factors. Even though meconium and transitional stool samples are characterized by low microbial load, the use of an internal standard in our study offsets the analytical challenges. Furthermore, in this study, several negative controls were implemented to detect bacterial contaminants. Moreover, the quality of the data from marker-gene sequencing was further improved by removing contaminant DNA sequences by Decontam45.
In our study, we strictly adhered to rigorous inclusion and exclusion criteria to ensure the reliability and validity of our findings. Out of the 100 pregnant women originally recruited, only 66 mother-neonate pairs and their 198 samples met all the specified criteria, leading to a drop-out rate of 34%. One of the primary challenges we encountered was the logistical difficulty in collecting all required samples, particularly from mothers in the Intensive Care Unit after C-section. Another common cause of exclusion was the absence of crucial data (type of antibiotics provided) or samples. This selection process was necessary to control for various confounding factors and ensure a homogeneous study population of overall healthy mothers with no significant differences (p>0.05) in the mother’s or neonate’s health and birth characteristics between the +IAP and -IAP groups. Even though mothers with asthma and gestational diabetes were included in the study, they were equally distributed in both groups. It might be also objected that the difference in antibiotic treatment between the groups is also a limitation of the study. However, as cephalosporins and penicillins that were used in a vast majority of mothers in both groups have similar modes of action and, in effect, similar antimicrobial profiles, this limitation should not pose a problem for the validity of our results.
Our study describes the period when lactation is initiated and breastfeeding (including the contact with the maternal breast or the additional contact with other materials) can affect the neonates’ microbiota. Nogacka et al. observed different responses of Bacteroides to IAP depending on the presence or absence of breastfeeding12. In our study, only ten neonates evenly distributed among study groups were formula-fed. The low number of formula-fed neonates in individual groups precluded the subanalysis of feeding mode in the relation to neonatal oral and fecal bacteriomes. However, the even distribution of feeding modes ensures that our results are not bias by this factor.
The relatively small sample size in both the -IAP and +IAP groups is another limitation of our analysis. To help future studies similar to the presented one, we have performed a power analysis based on our results to determine the sample size for neonatal oral swabs and transitional stool samples that is necessary for achieving 80% power to detect a statistically significant difference in the Shannon index in neonatal oral swabs (85 individuals in each group) and transitional stool samples (53 individuals in each group).
Conclusions
The current preliminary study describes bacteriomes of maternal and neonatal oral swabs and neonatal meconium and transitional stool to characterize the relationship between IAP and neonatal microbiota development in the first week of their life. No significant effects of IAP on maternal oral bacteriome in the first week after delivery were found. Exposure to IAP influences the oral bacteriome of neonates within the first 48 h after birth. However, the effect of IAP seems to diminish later in the first week of their life. Where meconium and stool are concerned, the differences in bacterial abundances due to IAP exposure are not reflected in meconium samples but transitional stool samples are affected by IAP. These findings highlight how antibiotics influence neonatal early bacterial development and point to the need for more research to understand the impact on children’s health in the long term.
Supplementary Information
Acknowledgements
We would like to thank Dr Jaroslav Janosek for his valuable comments. We acknowledge the CF Genomics CEITEC MU supported by the NCMG research infrastructure (LM2018132 funded by MEYS CR) for their support in obtaining scientific data presented in this paper.
Abbreviations
- CS
Cesarean section
- GBS
Group B Streptococcus
- IAP
Intrapartum antibiotic prophylaxis
- NGS
Next-generation sequencing
- PCR
Polymerase chain reaction
- PCA
Principal component analysis
- VD
Vaginal delivery
- 8-MOP
8-methoxypsoralen
Author contributions
Eliska Pivrncova: Writing—original draft, Investigation, Visualization. Lucie Buresova: Formal analysis, Visualization. Iva Kotaskova: Investigation, Methodology. Petra Videnska: Conceptualization, Supervision. Lenka Andryskova: Resources, Data curation. Pavel Piler: Resources, Data curation. Petr Janku: Resources. Ivo Borek: Resources. Jan Bohm: Data analysis, Critical revision of the manuscript. Jana Klanova: Conceptualization, Funding acquisition. Eva Budinska: Formal analysis, Data curation. Petra Borilova Linhartova: Writing—original draft, Supervision, Conceptualization. All authors revised the final version of the manuscript. Each author made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; or have drafted the work or substantively revised it.
Funding
The authors thank the RECETOX Research Infrastructure (ID LM2023069) and the project CETOCOEN EXCELLENCE (No CZ.02.1.01/0.0/0.0/17_043/0009632) financed by the Ministry of Education, Youth and Sports for supportive background. This work was carried out with the support of the Ministry of Health, the Czech Republic (FNBr, 65269705). Computational resources were supplied by the project “e-Infrastruktura CZ” (e-INFRA CZ LM2018140) supported by the Ministry of Education, Youth and Sports. This work was supported from the European Union’s Horizon 2020 research and innovation program under grant agreement No 857560 and from the Horizon Europe program under grant agreement No 101136566. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the granting authority, European Health and Digital Executive Agency (HADEA). Neither the European Union nor the granting authority can be held responsible for any use that may be made of the information it contains.
Data availability
The data set from 16S rRNA amplicon sequencing can be accessed at the National Center for Biotechnology Information (NCBI) under the accession number PRJNA1036118 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1036118). Other data are available upon request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/14/2024
The original online version of this Article was revised: the original version of this Article was published incorrectly under licence CC BY-NC-ND. The licence has been corrected to CC BY.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68953-z.
References
- 1.Shao, Y. et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature.574, 117–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z. & Dominguez-Bello, M. G. The infant microbiome development: Mom matters. Trends Mol. Med.21, 109–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grech, A. et al. Maternal exposures and the infant gut microbiome: A systematic review with meta-analysis. Gut Microbes13, 1–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe17, 690–703 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rutayisire, E., Huang, K., Liu, Y. & Tao, F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review. BMC Gastroenterol.16, 86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaterian, N., Abdi, F., Ghavidel, N. & Alidost, F. Role of cesarean section in the development of neonatal gut microbiota: A systematic review. Open Med. Wars Pol.16, 624–39 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pivrncova, E., Kotaskova, I. & Thon, V. Neonatal diet and gut microbiome development after C-section during the first three months after birth: A systematic review. Front. Nutr.9, 941549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nardi, G. M. et al. Maternal and neonatal oral microbiome developmental patterns and correlated factors: A systematic review—does the apple fall close to the tree?. Int. J. Environ. Res. Public Health18, 5569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandall, J. et al. Short-term and long-term effects of caesarean section on the health of women and children. Lancet (London, England)392, 1349–57 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A.107, 11971–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med.10.1126/scitranslmed.aad7121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogacka, A. et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome5, 93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Money, D., Allen, V. M., Infectious Diseases Committee. The prevention of early-onset neonatal group B streptococcal disease. J. Obstet. Gynaecol. Can. JOGC35, 939–948 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Betran, A. P., Ye, J., Moller, A.-B., Souza, J. P. & Zhang, J. Trends and projections of caesarean section rates: Global and regional estimates. BMJ Glob. Health6, e005671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seale, A. C. et al. Estimates of the burden of group b streptococcal disease worldwide for pregnant women, stillbirths, and children. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am.65(Suppl 2), S200-19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trijbels-Smeulders, M. et al. Epidemiology of neonatal group B streptococcal disease in the Netherlands before and after introduction of guidelines for prevention. Arch. Dis. Child. Fetal Neonatal Ed.92, F271-276 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrag, S. J. & Verani, J. R. Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: Experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine31(Suppl 4), D20-26 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Zhu, Y. & Lin, X.-Z. Updates in prevention policies of early-onset group B streptococcal infection in newborns. Pediatr. Neonatol.62, 465–75 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Hasperhoven, G., Al-Nasiry, S., Bekker, V., Villamor, E. & Kramer, B. Universal screening versus risk-based protocols for antibiotic prophylaxis during childbirth to prevent early-onset group B streptococcal disease: A systematic review and meta-analysis. BJOG Int. J. Obstet. Gynaecol.127, 680–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dyke, M. K. et al. Evaluation of universal antenatal screening for group B streptococcus. N. Engl. J. Med.360, 2626–36 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Stearns, J. C. et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep.7, 16527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann, P. & Curtis, N. Effect of intrapartum antibiotics on the intestinal microbiota of infants: A systematic review. Arch. Dis. Child. Fetal Neonatal Ed.105, 201–8 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Hirsch, A. G. et al. Early life antibiotic use and subsequent diagnosis of food allergy and allergic diseases. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol.47, 236–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kummeling, I. et al. Early life exposure to antibiotics and the subsequent development of eczema, wheeze, and allergic sensitization in the first 2 years of life: The KOALA birth cohort study. Pediatrics119, e225-31 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Azad, M. B., Bridgman, S. L., Becker, A. B. & Kozyrskyj, A. L. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int. J. Obes.38, 1290–8 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Gomez Arango, L. et al. Antibiotic treatment at delivery shapes the initial oral microbiome in neonates. Sci. Rep.7, 43481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapiainen, T. et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep.9, 10635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, Y. Y. et al. Impact of maternal intrapartum antibiotics, and caesarean section with and without labour on bifidobacterium and other infant gut microbiota. Microorganisms9, 1847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azad, M. et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG Int. J. Obstet. Gynaecol.123, 983–93 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Aloisio, I. et al. Influence of intrapartum antibiotic prophylaxis against group B Streptococcus on the early newborn gut composition and evaluation of the anti-Streptococcus activity of Bifidobacterium strains. Appl. Microbiol. Biotechnol.98, 6051–60 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Zhou, P. et al. Perinatal antibiotic exposure affects the transmission between maternal and neonatal microbiota and is associated with early-onset sepsis. mSphere5, e00984-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavlova, T. et al. Urinary intermediates of tryptophan as indicators of the gut microbial metabolism. Anal. Chim. Acta987, 72–80 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Videnska, P. et al. Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform. Sci. Rep.9, 13837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas, C. A. et al. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci. Rep.10, 123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claassen, S. et al. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J. Microbiol. Methods94, 103–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotásková, I. et al. Actinotignum schaalii: Relation to concomitants and connection to patients’ conditions in polymicrobial biofilms of urinary tract catheters and urines. Microorganisms9, 669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tourlousse, D. M. et al. Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing. Nucleic Acids Res.45, e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci.108, 4516–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J.7, 1–8 (2013). [Google Scholar]
- 41.Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods7, 335–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res.41, D590-6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baldassarre, M. E. et al. Duration of meconium passage in preterm and term infants. Arch. Dis. Child. Fetal Neonatal Ed.95, F74-75 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome6, 226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pincus, R. & Aitchison, J. The Statistical Analysis of Compositional Data. Biom J.30, 794–794 (Chapman and Hall, London, New York, 1988).
- 46.R: The R Project for Statistical Computing. https://www.r-project.org/ (2023).
- 47.Van den Boogaart, K. & Tolosana-Delgado, R. Compositional data analysis with `R’ and the package ‘compositions’. Geol. Soc.264(1), 119–127 (2006). [Google Scholar]
- 48.Oksanen, J, et al. Vegan: Community Ecology Package. http://cran.r-project.org (2022).
- 49.Kassambara, A. & Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. https://CRAN.R-project.org/package=factoextra (2020).
- 50.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
- 51.Murray, M. & Blume, J. FDRestimation: Estimate, Plot, and Summarize False Discovery Rates. 10.12688/f1000research.52999.2 (2022).
- 52.Conway, J. & Gehlenborg N. UpSetR: A More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. 10.1093/bioinformatics/btx364 (2019).
- 53.Guo, H. et al. The dynamic communities of oral microbiome in neonates. Front. Microbiol.13, 1052525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe24, 133-145.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dzidic, M. et al. Oral microbiome development during childhood: An ecological succession influenced by postnatal factors and associated with tooth decay. ISME J.12, 2292–306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belizário, J. E. & Napolitano, M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol.6, 1050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, J., Fiscella, K. A. & Gill, S. R. Oral microbiome: Possible harbinger for children’s health. Int. J. Oral Sci.12, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampaio-Maia, B. & Monteiro-Silva, F. Acquisition and maturation of oral microbiome throughout childhood: An update. Dent. Res. J.11, 291–301 (2014). [PMC free article] [PubMed] [Google Scholar]
- 59.Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med.23, 314–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verma, D., Garg, P. K. & Dubey, A. K. Insights into the human oral microbiome. Arch. Microbiol.200, 525–40 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Lif Holgerson, P., Harnevik, L., Hernell, O., Tanner, A. C. R. & Johansson, I. Mode of birth delivery affects oral microbiota in infants. J. Dent. Res.90, 1183–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dzidic, M. et al. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy73, 2000–11 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Boix-Amorós, A., Collado, M. C. & Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol.7, 492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyoshi, T. et al. Gemella haemolysans inhibits the growth of the periodontal pathogen Porphyromonas gingivalis. Sci. Rep.11, 11742 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng, X. et al. Oral microbiota in human systematic diseases. Int. J. Oral Sci.14, 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li, H. et al. The impacts of delivery mode on infant’s oral microflora. Sci. Rep.8, 11938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurley, E. et al. The microbiota of the mother at birth and its influence on the emerging infant oral microbiota from birth to 1 year of age: A cohort study. J. Oral Microbiol.11, 1599652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kennedy, B. et al. Oral microbiota development in early childhood. Sci. Rep.9, 19025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dierikx, T. H. et al. The influence of prenatal and intrapartum antibiotics on intestinal microbiota colonisation in infants: A systematic review. J. Infect.81, 190–204 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Jauréguy, F. et al. Effects of intrapartum penicillin prophylaxis on intestinal bacterial colonization in infants. J. Clin. Microbiol.42, 5184–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arboleya, S., Saturio, S. & Gueimonde, M. Impact of intrapartum antibiotics on the developing microbiota: A review. Microbiome Res. Rep.1, 22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imoto, N. et al. Administration of β-lactam antibiotics and delivery method correlate with intestinal abundances of Bifidobacteria and Bacteroides in early infancy, in Japan. Sci. Rep.11, 6231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saturio, S. et al. Effect of intrapartum antibiotics prophylaxis on the bifidobacterial establishment within the neonatal gut. Microorganisms9, 1867 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corvaglia, L. et al. Influence of intrapartum antibiotic prophylaxis for group B streptococcus on gut microbiota in the first month of life. J. Pediatr. Gastroenterol. Nutr.62, 304 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Dierikx, T. et al. Influence of timing of maternal antibiotic administration during caesarean section on infant microbial colonisation: A randomised controlled trial. Gut71, 1803–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazzola, G. et al. Early gut microbiota perturbations following intrapartum antibiotic prophylaxis to prevent group B streptococcal disease. PLoS ONE11, e0157527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data set from 16S rRNA amplicon sequencing can be accessed at the National Center for Biotechnology Information (NCBI) under the accession number PRJNA1036118 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1036118). Other data are available upon request from the corresponding author.