Abstract
Herpes simplex virus (HSV) entry is dependent on the interaction of virion glycoprotein D (gD) with one of several cellular receptors. We previously showed that gD binds specifically to two structurally dissimilar receptors, HveA and HveC. We have continued our studies by using (i) a panel of baculovirus-produced gD molecules with various C-terminal truncations and (ii) a series of gD mutants with nonoverlapping 3-amino-acid deletions between residues 222 and 254. Binding of the potent neutralizing monoclonal antibody (MAb) DL11 (group Ib) was unaffected in forms of gD containing residues 1 to 250 but was greatly diminished in molecules truncated at residue 240 or 234. Both receptor binding and blocking of HSV infection were also affected by these C-terminal truncations. gD-1(234t) bound weakly to both HveA and HveC as determined by enzyme-linked immunosorbent assay (ELISA) and failed to block infection. Interestingly, gD-1(240t) bound well to both receptors but blocked infection poorly, indicating that receptor binding as measured by ELISA is not the only gD function required for blocking. Optical biosensor studies showed that while gD-1(240t) bound HveC with an affinity similar to that of gD-1(306t), the rates of complex formation and dissociation were significantly faster than for gD-1(306t). Complementation analysis showed that any 3-amino-acid deletion between residues 222 and 251 of gD resulted in a nonfunctional protein. Among this set of proteins, three had lost DL11 reactivity (those with deletions between residues 222 and 230). One of these proteins (deletion 222–224) was expressed as a soluble form in the baculovirus system. This protein did not react with DL11, bound to both HveA and HveC poorly as shown by ELISA, and failed to block HSV infection. Since this protein was bound by several other MAbs that recognize discontinuous epitopes, we conclude that residues 222 to 224 are critical for gD function. We propose that the potent virus-neutralizing activity of DL11 (and other group Ib MAbs) likely reflects an overlap between its epitope and a receptor-binding domain of gD.
The herpes simplex virus (HSV) genome codes for at least 11 glycoproteins, most of which are detectable in the virion envelope (50). Infection of susceptible cells is initiated by the attachment of virions, via glycoprotein C (gC) and/or gB, to cell surface heparan sulfate proteoglycans (21, 22, 59). This is followed by the interaction of gD with a cellular receptor. Then, pH independent fusion occurs between the virus envelope and the host cell plasma membrane (58); gB, gD, and the gH-gL complex have all been implicated in this step (50, 52).
Recently, expression cloning was used to identify several human genes whose products convert the normally nonpermissive Chinese hamster ovary cells into cells that are permissive for HSV type 1 (HSV-1) and HSV-2 entry (9, 19, 40, 53). These mediators of HSV entry are known as HveA, HveB, and HveC. HveA is a member of the tumor necrosis factor receptor superfamily of proteins (40) and interacts with both lymphotoxin α and LIGHT (38). HveB (also called PRR2) and HveC (also called PRR1) are closely related members of the immunoglobulin superfamily of proteins (36.1% amino acid sequence identity within the predicted extracellular domains) which share 53.2 and 33.9% amino acid sequence identities, respectively, with the poliovirus receptor extracellular domain (14, 19, 37, 53). The normal cellular functions of these proteins remain unknown, although recent data suggest that the murine homolog of HveB may be a cell-cell adhesion molecule (1). A splice variant of HveC, called HIgR, can also mediate HSV infection of nonpermissive cells (9). Soluble forms of gD have been shown to bind directly to soluble forms of HveA, HveC, and HIgR but not to HveB (8, 9, 31, 54, 55). In addition, antibodies to the receptors have been shown to block infection by HSV (9, 40, 53). Thus, it is clear that HSV can utilize several different and structurally unrelated cell surface proteins as receptors and that two of these receptors bind directly to HSV gD.
Two approaches were used in previous studies to try to define the relationship between gD structure and function: (i) examination of the properties of a panel of monoclonal antibodies (MAbs) to gD (11, 12, 23, 41, 43) and (ii) examination of the properties of a panel of gD mutants (7, 17, 42). First, the antigenic site I of gD was defined by seven MAbs, all of which possess potent virus-neutralizing activity in the absence of complement (41). Although all group I MAbs block the binding of other group I antibodies to gD, further subdivision of these MAbs into groups Ia and Ib was done on the basis of studies with truncated and other mutant forms of gD. Two group Ia MAbs, HD1 and LP2 (11), bind to gD truncated at amino acid residue 233, whereas DL11 and four other group Ib antibodies do not (11, 43). More recently, we showed that, whereas DL11 blocks the binding of soluble HveA or HveC to HSV virions, HD1 blocks the binding of HveC but not of HveA to HSV (31, 44). On the other hand, MAbs in group VII blocked the binding of HveA but not of HveC to HSV (31, 44). Taken together, these results suggest that the binding of gD to each of these receptors involves both a common region as well as unique portions of the gD molecule. Furthermore, information about the location of epitopes within antigenic sites I and VII has provided important clues as to the portions of gD involved in the binding of each receptor. For example, since group VII MAbs recognize a continuous epitope within amino acids 11 to 19 (10, 26), it is likely that residues near the amino terminus of gD are important for its interaction with HveA. In support of this hypothesis, a mutant form of gD with a change at amino acid 27 fails to bind to HveA but still interacts with HveC (31, 54). Not surprisingly, viruses with this change in gD are unable to utilize HveA as an entry receptor (40).
Using gD mutants and complementation analysis, we previously identified four distinct regions within the gD primary structure that are important for HSV infection which were designated functional regions I through IV (7). Several observations can be made regarding the relationship between antigenic site Ib (discussed above) and functional regions II and III. First, all of the linker insertions within functional region II abolished or greatly diminished binding by the group Ib MAb, DL11. Second, functional region II (residues 125 to 161) encompasses residues previously shown to affect the binding of certain group Ib antibodies (residues 132 and 140). Third, functional region III (residues 225 to 246) includes residues known to be required for group Ib antibody binding. These observations taken together suggest that functional regions II and III may be closely positioned within the folded (tertiary) structure of gD and may, together, form a functional domain.
Here we address the contribution of gD residues between 222 and 275 to the formation of both antigenic site Ib and a functional (receptor-binding) domain. To accomplish this, we constructed two sets of HSV-1 gD mutants. The first group is a nested set of C-terminal truncations consisting of molecules truncated at residues 234, 240, 250, 260, 285, and 306. The second set of constructs is a panel of 11 gD mutants containing adjacent, nonoverlapping, 3-amino-acid deletions within functional region III. Our results support our hypothesis that there is an overlap between antigenic site Ib and a domain involved in binding to the HSV receptors, HveA and HveC.
MATERIALS AND METHODS
Cells and virus.
HeLa and Vero cells were obtained from the ATCC and grown in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 5% fetal bovine serum (FBS). Sf9 (Spodoptera frugiperda) cells (GIBCO BRL) were grown in Sf900II medium (GIBCO BRL). The HSV1(KOS) recombinant, hrR3 (20), in which the lacZ gene, under the control of the ICP6 promoter, has been inserted into the ICP6 locus, was propagated in D14 cells as described by Goldstein and Weller (20) and titers were determined on Vero cells. COS-1 cells were grown in DMEM supplemented with 5% FBS. VD60 cells were grown in DMEM containing 5% FBS and 1 mM histidinol (35). The isolation and propagation of the gD-null virus, F-gDβ, has been described previously (35). The HSV-1 strain KOS was used where indicated.
Construction of baculovirus recombinants expressing truncated forms of gD.
The strategy employed in the construction of a baculovirus recombinant expressing gD-1(306t) has been described in detail elsewhere (49). The construction of bac-gD-1(285t) and bac-gD-1(234t) has also been described (47). Briefly, PCR primers were synthesized in order to amplify and modify the gD ectodomain coding region for cloning into the pVT-Bac transfer vector plasmid and expression in a recombinant baculovirus. The upstream primer, 5′-TTTTGGATCCCAAATATGCCTTGGCGGATG-3′, hybridized to the noncoding strand of the gD open reading frame (ORF) immediately beyond the predicted signal sequence coding region and incorporated a BamHI restriction enzyme cleavage site (underlined). Three different downstream primers were used separately with the upstream primer to generate ORFs coding for gD truncated after residue 260, 250, or 240. The downstream primer used to amplify the PCR fragment for gD-1(260t) cloning and expression was 5′-GGCGAATTCAGTGGTGGTGGTGGTGGTGGGTCTCGGACAGCTCCGGGGGCAG-3′ and incorporated an EcoRI restriction enzyme cleavage site (underlined). The downstream primer used to amplify the PCR fragment for gD-1(250t) cloning and expression was 5′-GGCGAATTCAGTGGTGGTGGTGGTGGTGGCTCGTGTATGGGGCCTT-3′ and incorporated an EcoRI restriction enzyme cleavage site (underlined). The downstream primer used to generate the PCR fragment for gD-1(240t) cloning and expression was 5′-GGCGAATTCAGTGGTGGTGGTGGTGGTGCCCGGCGATCTTCAAGCTGTATA-3′ and incorporated an EcoRI restriction enzyme cleavage site (underlined). The primer used to generate the PCR fragment for gD-1(Δ222–224, 306t) cloning and expression was 5′-TTTTCTGCAGTTAATGATGATGATGATGATGGTAAGGCGTCGCGG-3′ and incorporated a PstI restriction enzyme cleavage site (underlined). The PCR-amplified DNA fragments coded for gD lacking its natural signal sequence so that the melittin signal sequence, coded for by pVT-Bac, would replace the missing gD signal sequence. The downstream PCR primers were also designed to append six histidine codons prior to the termination codon to allow for purification of the recombinant proteins by nickel agarose chromatography. The PCR-amplified products were then digested with BamHI and either EcoRI or PstI and cloned into pVT-Bac which had been digested with the same enzymes. Once cloned into pVT-Bac, the resulting plasmid constructs were cotransfected with baculovirus DNA (Baculogold; Pharmingen) into Sf9 cells growing in monolayer culture. After 4 days, the culture supernatant (containing recombinant progeny virus) was replated onto Sf9 cell monolayers under Grace’s insect cell medium containing 1% agarose. Recombinant virus plaques were picked, amplified, and screened for the expression of secreted gD by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis of the culture medium from Sf9 cells infected with recombinant virus picks. Virus clones expressing gD were plaque purified two times, and protein expression from individual virus clones was verified at each stage by SDS-PAGE and immunoblot analysis. The plaque-purified baculovirus recombinant selected for routine use in production of gD-1 truncated at residue 260 was named bac-gD-1(260t). The soluble protein produced by bac-gD-1(260t) is referred to as gD-1(260t). The nomenclature for the 250 and 240 truncations followed the same pattern. These designations indicate that the secreted gD produced is truncated after the indicated amino acid residue of the predicted mature (signal sequence removed) protein (in this numbering system, the initiator methionine residue of gD occurs at position −25).
Production and purification of recombinant baculovirus-produced proteins.
Production and purification of gD-1(306t), gD-1(285t), gD-1(234t), HveA(200t), and HveC(346t) have been described (31, 47, 49, 54, 56). Production and purification of gD-1(260t), gD-1(250t), gD-1(240t), and gD-1(Δ222–224, 306t) were carried out as described previously for HveA(200t) [also called HVEM(200t) (54)].
ELISA.
Soluble receptor proteins HveA(200t) or HveC(346t) in phosphate-buffered saline (PBS) were bound to 96-well enzyme-linked immunosorbent assay (ELISA) plates for 3 h at room temperature. The plates were washed three times with PBS–0.2% Tween 20 and incubated in blocking solution (PBS, 5% nonfat milk, 0.2% Tween 20) for 30 min at 25°C. Plates were then washed three times with PBS–0.2% Tween 20 and incubated with truncated forms of gD at various concentrations in blocking solution for 16 h at 4°C. Plates were then washed three times with PBS–0.2% Tween 20 and incubated for 30 min with R7 (a rabbit polyclonal antiserum against gD) diluted 1 to 1,000 in blocking solution. After three washes with PBS–0.2% Tween 20, the plates were incubated in horseradish peroxidase-conjugated goat anti-rabbit antibody (Boehringer Mannheim) diluted 1/1,000 in blocking solution. Plates were washed three times with PBS–0.2% Tween 20 and then once with 20 mM sodium citrate (pH 4.5). After removal of the citrate buffer, ABTS substrate solution (Moss, Inc.) was added, and the absorbance at 405 nm in individual wells was read by using a Perkin-Elmer HTS 7000 Bio-Assay Reader. Finally, absorbance was plotted against the concentration of gD used.
Antibodies.
R7 is a rabbit polyclonal antiserum raised against native, full-length gD-2 isolated from virus-infected cells (26). R69 is a rabbit polyclonal antiserum raised against denatured, full-length gB-1 isolated from virus-infected cells (16). 1D3 is a group VII MAb recognizing gD residues 11 to 19 (13, 18). DL6 is a group II MAb recognizing residues 272 to 279 (15, 26). MAbs HD1 (group Ia), DL11 (group Ib), D2 (group Ib), DL2 (group VI), and ABD (group III) recognize discontinuous epitopes (11, 23, 41, 46, 48).
Blocking of HSV-1 entry into mammalian cells with soluble proteins.
The blocking of HSV entry into cells with soluble gD was carried out as described previously (27) and as modified by Nicola et al. (45).
SDS-PAGE.
Purified glycoproteins were separated by SDS-PAGE under “native” (0.1% SDS, no reducing agent, no boiling [11]) or denaturing (samples boiled 10 min in 2.5% SDS–350 mM β-mercaptoethanol) conditions in precast Tris-glycine gels (Novex). After SDS-PAGE, separated proteins were stained with silver nitrate (Pharmacia) or transferred to nitrocellulose, probed with antibodies, and visualized by enhanced chemiluminescence (Amersham).
Construction of gD-1 3-amino-acid deletion series.
Oligonucleotide-directed mutagenesis was carried out by using the method of Zoller and Smith (60), as modified by Kunkel et al. (33, 34), to generate the series of plasmid constructs expressing gD containing sequential, nonoverlapping 3-amino-acid deletions spanning residues 222 through 254. The template for mutagenesis was an M13mp18 construct containing the gD-1 (Patton) ORF (cloned into the unique HindIII site) which was excised from plasmid pRE4 (12) by HindIII digestion. The specific mutagenic primers used were as follows: Δ222–224, 5′-TGGTTCTCGGGGGGCAGCATC-3′; Δ225–227, 5′-ACGGTGCGCTGGATGAAGCGGGGC-3′; Δ228–230, 5′-GTATACGGCGACGTTCTCGGGGAT-3′; Δ231–233, 5′-CTTCAAGCTGTAGGTGCGCTGGTT-3′; Δ234–236, 5′-CCCGGCGATCTTTACGGCGACGGT-3′; Δ237–239, 5′-CCCGTGCCACCCCAAGCTGTATAC-3′; Δ240–242, 5′-GGCCTTGGGCCCGGCGATCTTCAAGC-3′; Δ243–245, 5′-CGTGTATGGGGCGTGCCACCCGGC-3′; Δ246–248, 5′-CAGGGTGCTCGTCTTGGGCCCGTGC-3′; Δ249–251, 5′-TCCGGGGGCAGCAGGTATGGGGCCTT-3′; Δ252–254, 5′-GGACAGCTCCGGGGTGCTCGTGTAT-3′.
After mutagenesis, the gD ORFs were excised (HindIII) from M13 replicative-form DNA and transferred into the mammalian expression plasmid, pRSVnt-EPA (5). Plasmids containing the gD ORF in the desired orientation were sequenced by using the method of Chen and Seeburg (6) to confirm the presence of the anticipated mutations (nine nucleotide deletions) within the gD ORF.
Antigenic analysis of mutants.
Transfection of COS cells and the subsequent preparation of cytoplasmic extracts were performed as previously described (12, 41).
Immunoperoxidase staining.
This procedure, which was performed as previously described (43), is a modification of that of Holland et al. (24) and Kousoulas et al. (30). Surface staining of transfected cells was studied with unfixed cells; for detection of intracellular antigens, the cells were fixed with 5% methanol in PBS before incubation with the MAbs.
Complementation assay.
The assay was performed essentially as previously described (43), except that COS cells were used instead of Vero cells. Briefly, cells were transfected with DNA-calcium phosphate precipitates for 16 h at 37°C and then washed and incubated in DMEM–5% FBS for 8 h at 37°C. Each dish of cells was subsequently infected at room temperature with 106 PFU of F-gDβ virus, followed by the addition of 5 ml of DMEM–5% FBS and incubation for 1 h at 37°C. The medium was then removed, and extracellular virus was inactivated by incubating the monolayer for 1 min in glycine-saline (pH 3.0) (4, 25). After 18 h in DMEM–5% FBS at 37°C, the medium was removed and stored at −70°C for subsequent determination of the virus titers. The cells were lysed by freeze-thawing and sonication with a Microson cell disruptor. Nuclei were then pelleted by low-speed centrifugation, and the supernatant was stored at −70°C for subsequent determination of the virus titers. Both intracellular and extracellular virus titers were determined on VD60 cells. Transfection with salmon sperm DNA was used as the negative control. One hundred percent complementation is defined as the titer obtained after transfection with plasmid pRE4, which expresses wild-type gD (12). Complementation with a mutant is then defined by the following formula: % complementation = 100 × (titer with mutant plasmid − titer with carrier DNA)/(titer with pRE4 − titer with carrier DNA).
Optical biosensor experiments.
Biosensor experiments were carried out on a Biacore X optical biosensor (Biacore AB) at 25°C as previously described (32, 47). Biosensor data were analyzed by using a global fitting routine with BIAevaluation software, version 3.0 (2). Model curve fitting was carried out by using a 1:1 Langmuir interaction with drifting baseline. This models the simple interaction between ligand (L) and receptor (R) as follows: L + R ⇌ LR. The rate of association (kon) was measured from the forward reaction, and koff was measured from the reverse reaction. For gD-1(234t), a maximum koff was estimated as previously described (47) by using the equation ln(R0/Rn) = koff(tn − t0), where R0 is the response at time zero (t0) of dissociation and Rn is the response at time n (tn) (29). Scatchard analyses of the gD-1(234t)–receptor complexes were performed as previously described (47).
RESULTS
C-terminal truncations.
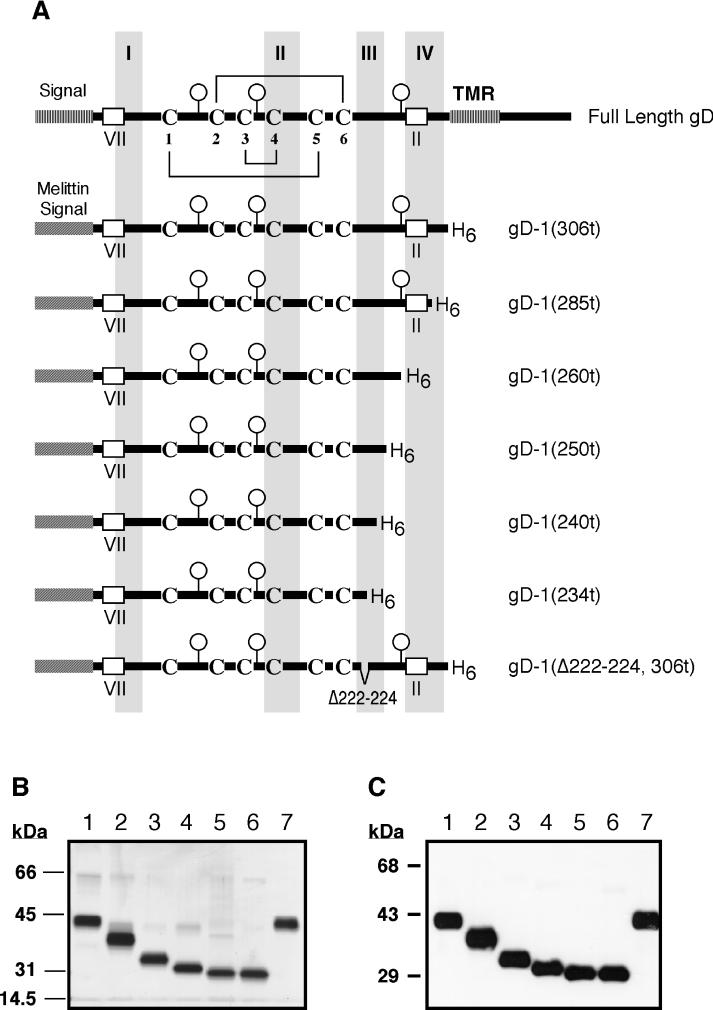
Krummenacher et al. (31) and Rux et al. (47) showed that, compared to gD truncated at residue 306 [gD-1(306t)], molecules truncated at residues 285 [gD-1(285t)] and 275 [gD-1(275t)] exhibited enhanced receptor binding. In contrast, a form consisting of residues 1 to 234 [gD-1(234t)] exhibited greatly diminished receptor binding. gD-1(234t) was shown to retain much of the native structure of the full-length molecule in that most MAbs recognizing discontinuous epitopes of gD reacted with gD-1(234t) (47). One exception was that the group Ib MAb, DL11, bound poorly to gD-1(234t). Since gD-1(234t) lacks a significant portion of functional region III (7) (residues 225 to 246), we reasoned that the diminished receptor binding of gD-1(234t) was consistent with the idea that functional region III is directly involved in receptor binding. To define more precisely the C-terminal gD residues required for receptor binding as well as for the binding of MAb DL11, we expressed three additional forms of gD in the baculovirus system. These gD molecules were truncated after residues 260 [gD-1(260t)], 250 [gD-1(250t)], and 240 [gD-1(240t)]. Stick diagrams of these, as well as other recombinant baculovirus products, are shown in Fig. 1A. Each truncated form of gD was constructed such that six histidine residues were present at the C terminus to allow for purification by nickel chromatography. The gD truncation mutants were purified by immunoaffinity chromatography [gD-1(306t) and gD-1(285t)] or by nickel chromatography (all other forms of gD). To assess the purity of the recombinant proteins, similar amounts were loaded onto an SDS–10% polyacrylamide gel, electrophoresed, and stained with silver nitrate. All of the proteins were purified to near homogeneity and were of the expected sizes (Fig. 1B). Western blot analysis with the group VII MAb 1D3 (Fig. 1C) confirmed that all of these purified proteins retained the correct N terminus of gD (Fig. 1A).
FIG. 1.
Recombinant baculovirus-produced proteins. (A) Stick diagrams of full-length HSV gD and the truncated forms used in this study (produced in recombinant baculovirus-infected cells). Functional regions I to IV as defined by Chiang et al. (7) are shown as shaded regions. The positions of linear epitopes for group II and group VII are indicated. The positions of consensus sites for N-glycosylation are marked by balloons. The disulfide bond pattern for the six cysteine residues located in the extracellular domain of gD (36) is shown on the full-length gD stick diagram. (B) Silver-stained SDS-polyacrylamide gel (10%) showing the purified recombinant baculovirus-produced proteins used in this study. Lane 1, gD-1(306t); lane 2, gD-1(285t); lane 3, gD-1(260t); lane 4, gD-1(250t); lane 5, gD-1(240t); lane 6, gD-1(234t); lane 7, gD-1(Δ222–224, 306t). (C) Western blot of the purified proteins shown in panel B probed with MAb 1D3 (group VII).
Antigenic analysis of C-terminal gD truncations.
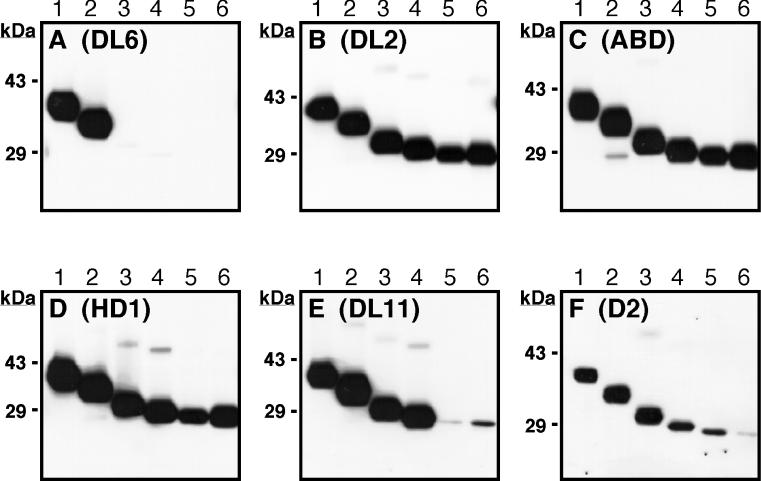
To assess the antigenic structure of the recombinant gD molecules, the proteins were separated on nondenaturing (“native”) SDS-polyacrylamide gels, and blots were probed with various MAbs. The blot shown in Fig. 2A was reacted with the group II MAb DL6, which recognizes a linear epitope (residues 272 to 279) (26) (Fig. 1A). The expected pattern of reactivity with DL6 was observed in that proteins smaller than gD-1(285t) were not reactive. The blots shown in Fig. 2B to D were reacted with MAbs DL2, ABD, and HD1, each of which recognizes a separate discontinuous epitope on gD. All of the truncated proteins reacted similarly with these MAbs, indicating that the native structure of gD was not grossly altered by the truncations. The blots shown in Fig. 2E and F were reacted with two group Ib MAbs, DL11 and D2. Although DL11 bound strongly to gD-1(306t), gD-1(285t), gD-1(260t), and gD-1(250t), it bound weakly to gD-1(240t) and gD-1(234t). In previous studies, we showed that DL11 competed with soluble HveA and HveC for binding to gD in HSV virions, suggesting that it binds within or near a region of gD involved in receptor interaction (31, 44). According to the data presented in Fig. 2E, gD residues immediately upstream of 250 contribute to the DL11 epitope. MAb D2, like DL11, bound weakly to gD-1(240t) and gD-1(234t) but also exhibited reduced reactivity with gD-1(250t) when compared with molecules truncated after residues 260, 285, and 306. These results confirm and extend previous work mapping residues critical to the formation of antigenic site Ib (11, 12, 41–43).
FIG. 2.
Antigenic analysis of baculovirus-produced gD truncation mutants. Purified proteins were separated by “native” SDS-PAGE, transferred to nitrocellulose, and probed with gD-specific MAbs. Lane 1, gD-1(306t); lane 2, gD-1(285t); lane 3, gD-1(260t); lane 4, gD-1(250t); lane 5, gD-1(240t); lane 6, gD-1(234t). (A) Blot probed with DL6 (group II MAb). (B) Blot probed with DL2 (group VI MAb). (C) Blot probed with ABD (group III MAb). (D) Blot probed with HD1 (group Ia MAb). (E) Blot probed with DL11 (group Ib MAb). (F) Blot probed with D2 (group Ib MAb).
ELISA.
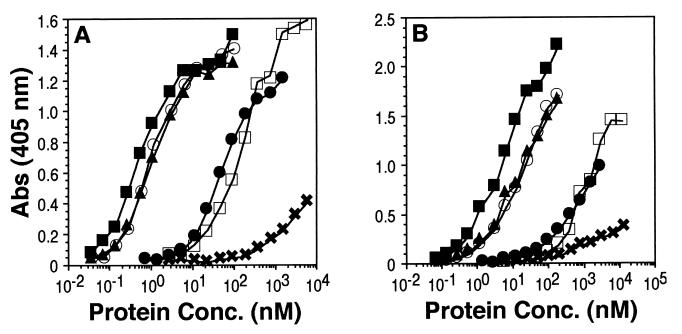
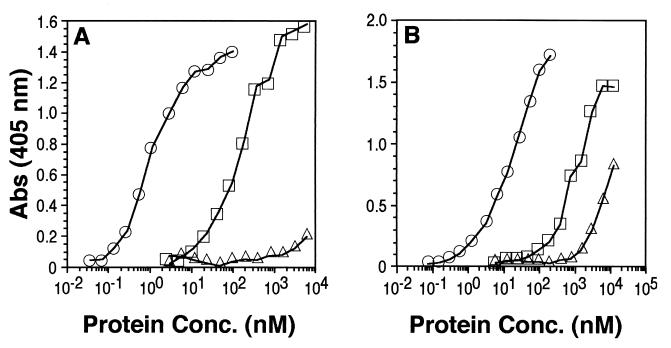
Previous studies showed that, compared to gD-1(306t), molecules truncated after residues 285 and 275 bound to HveA and HveC with increased affinity, while a molecule truncated after residue 234 bound with reduced affinity (31, 47). To assess the effect of the C-terminal truncations on receptor interaction, we analyzed their binding to truncated forms of HveA [HVEM(200t)] and [HveC (HveC(346t)] by ELISA (Fig. 3). Figure 3A shows the binding of truncated forms of gD to HveA, while Fig. 3B shows their binding to HveC. As previously reported (31, 47), gD-1(285t) bound to both HveA and HveC, as seen by ELISA, ca. 100-fold better than did gD-1(306t). gD-1(260t) and gD-1(250t) bound to both receptors as well as gD-1(285t). However, gD-1(240t) bound to both receptors about as well as gD-1(306t), whereas the binding of gD-1(234t) was nearly undetectable. We conclude from these observations that gD residues between positions 234 and 240 are critical for receptor binding and that residues between positions 240 and 250 may also be involved (since the receptor-binding activity of gD-1(240t), though not eliminated, is reduced relative to larger forms of gD). It is of interest to note that gD-1(240t), which showed diminished reactivity with the MAb DL11, still bound to both receptors.
FIG. 3.
Analysis of gD truncation mutants for receptor binding by ELISA. The wells of an ELISA plate were coated with an excess of HveA(200t) or HveC(346t) and incubated with increasing concentrations (shown on the x axis) of gD truncation mutants. Bound gD was detected by incubating sample wells with a rabbit antiserum raised against gD (R7), followed by peroxidase-conjugated goat anti-rabbit antibody and then peroxidase substrate. (A) Binding to HveA(200t). (B) Binding to HveC(346t). Symbols: □, gD-1(306t); ■, gD-1(285t); ▴, gD-1(260t); ○, gD-1(250t); ●, gD-1(240t); ✖, gD-1(234t).
Biosensor analysis of gDt binding to HveA and HveC.
Previously, we used optical biosensor technology to show that the increased affinity of gD-1(285t) for both HveA and HveC relative to gD-1(306t) resulted almost exclusively from a faster rate of complex formation (47). In contrast, gD-1(234t) exhibited a faster rate of complex dissociation (koff) with HveA compared to gD-1(306t), suggesting that some gD residues involved in HveA binding had been removed. Here we found that the binding kinetics and affinities of gD-1(285t), gD-1(260t), and gD-1(250t) for both receptors were quite similar (Table 1). In each case, the higher affinity was due primarily to a faster rate of complex formation (kon). gD-1(240t) exhibited binding kinetics and an affinity similar to gD-1(306t) in its interaction with HveA, consistent with its similar binding properties as determined by ELISA. Although gD-1(240t) exhibited an overall affinity for HveC similar to that of gD-1(306t), the kon was 10-fold faster and the koff was 4-fold faster than gD-1(306t). Thus, in contrast to the ELISA results, the optical biosensor enabled us to distinguish the binding of gD-1(306t) versus gD-1(240t) to HveC. As observed previously (47), gD-1(234t) bound to HveAt, but the data failed to fit a 1:1 Langmuir binding model. A similar result was obtained when gD-1(234t) binding to HveC was examined. Because of this, the binding kinetics for gD-1(234t) could not be analyzed by using the global fitting routine of the instrument software. Instead, maximum koff values were estimated by plotting ln(R0/Rn) versus time for the initial part of the dissociation phase. For both HveA and HveC, the maximum koff for gD-1(234t) was approximately 10-fold faster than for gD-1(306t). Finally, equilibrium dissociation constants (KD) for gD-1(234t) binding to HveA and HveC were calculated from data collected under conditions of binding equilibrium by using Scatchard analysis. For gD-1(234t) binding to HveA this calculation yielded a KD approximately sixfold lower than gD-1(306t), while for binding to HveC the KD was nearly identical to that of gD-1(306t).
TABLE 1.
Kinetic and affinity values for gD-receptor complex formation
| Immobilized receptor | gD | kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M) (koff/kone) |
|---|---|---|---|---|
| HveAt | 306ta | 6.1 ± 0.6b | 2.0 ± 0.2b | 3.2 ± 0.6 |
| 285ta | 300 | 1.1 | 0.037 | |
| 260t | 130 | 0.75 | 0.058 | |
| 250t | 520 | 0.94 | 0.018 | |
| 240t | 18 | 1.6 | 0.89 | |
| 234t | NDc | 20d | 20f | |
| HveCt | 306ta | 2.6 ± 0.7b | 0.71 ± 0.09b | 2.7 ± 0.2 |
| 285ta | 190 | 0.73 | 0.038 | |
| 260t | 280 | 0.71 | 0.025 | |
| 250t | 440 | 0.66 | 0.015 | |
| 240t | 25 | 2.9 | 1.2 | |
| 234t | NDc | 10d | 2f |
Values for gD-1(306t) (32, 57), gD-1(285t) (32, 47), and the HveA–gD-1(234t) complex (47) were reported previously and are shown here for comparison.
Average ± the standard deviation from at least three separate experiments.
ND, not determined.
Estimated maximum koff. As a control, this method was also used to calculate a koff for the dissociation of gD-1(306t) from HveA and HveC. The values obtained were 1.5 × 10−2 s−1 and 0.66 × 10−2 s−1, respectively.
χ2 values for the global fits ranged from 0.7 to 4.5.
From Scatchard analysis.
Blocking of virus entry by soluble forms of gD.
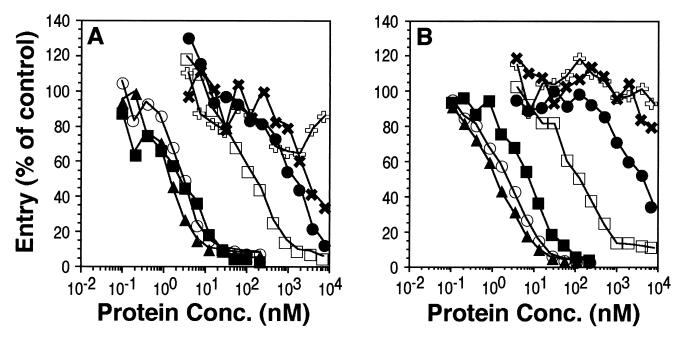
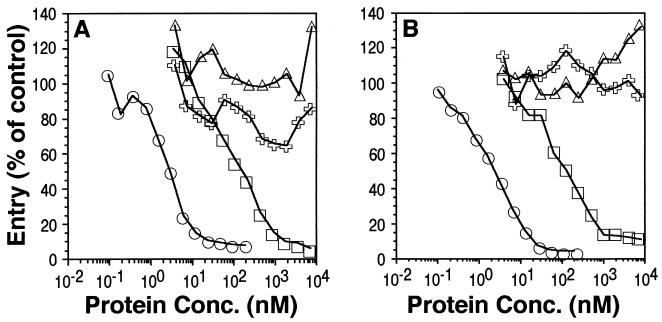
Soluble gD blocks HSV entry into susceptible cells, presumably by binding to and occupying cell surface receptors (28, 45). We tested the abilities of truncated gD to block infection of HeLa and Vero cells (Fig. 4A and B). gD-1(285t), gD-1(260t), and gD-1(250t) blocked HSV infection at similar concentrations (50% inhibition occurred between 1 and 10 nM) and were more potent than gD-1(306t) (50% inhibition occurred between 100 and 200 nM). These results were consistent with the ELISA and biosensor data showing that gD-1(285t), gD-1(260t), and gD-1(250t) bound to both HveA and HveC with greatly increased affinities relative to gD-1(306t). Similarly, the weak ability of gD-1(234t) to block HSV infection is consistent with its diminished capacity to bind HveA or HveC relative to gD-1(306t) (at least by ELISA). Surprisingly, gD-1(240t) blocked HSV infection much less effectively than gD-1(306t) (50% inhibition occurred at approximately 5 μM). This result was unexpected because both the ELISA and biosensor data indicated that this protein bound to both receptor molecules with an affinity similar to gD-1(306t). We have repeated these experiments several times with similar results. These data suggest that gD-1(240t) may lack a portion of a gD functional domain.
FIG. 4.
Analysis of gD truncation mutants for blocking of HSV entry. Cells in 96-well tissue culture plates were incubated with increasing concentrations (shown on the x axis) of various forms of gD prior to infection with HSV-1(KOS) carrying a β-galactosidase reporter gene (hrR3). At 5 h postinfection, cells were lysed and assayed for β-galactosidase activity. (A) HeLa cells. (B) Vero cells. Symbols: □, gD-1(306t); ■, gD-1(285t); ▴, gD-1(260t); ○, gD-1(250t); ●, gD-1(240t); ✖, gD-1(234t); , BSA.
Antigenic structure of 3-amino-acid deletion mutants.
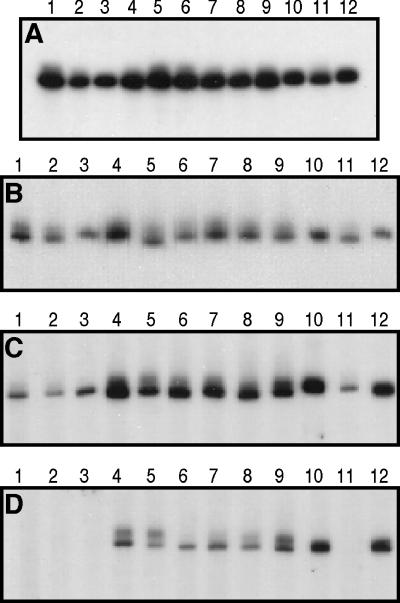
In order to characterize further the region of gD encompassing functional region III, as well as the residues involved in group Ib MAb binding, we used site-directed mutagenesis to generate a series of plasmids encoding full-length gD with nonoverlapping 3-amino-acid deletions spanning amino acid residues 222 through 254 (Fig. 5). COS cells were transfected separately with plasmids expressing each of the gD mutants. With the exception of the Δ240–242 deletion mutant, all of the mutated proteins were transported to the surface of transfected cells, as detected by immunoperoxidase staining (data not shown). Cytoplasmic extracts were then prepared from COS cells 40 h after transfection. As controls, plasmids pRE4, which expresses wild-type gD-1, and pWW17, which expresses the Δ234–244 deletion mutant (12), were included. To quantitate relative gD expression, equal volumes of each extract were electrophoresed on a denaturing polyacrylamide gel, followed by transfer to nitrocellulose and probing with MAb DL6 (26). Based on this result, the volumes of extract loaded on subsequent gels were normalized so as to give approximately equal signals with DL6 (Fig. 6A). No protein could be detected for mutant Δ240–242, even after repetition of the mutagenesis, so it was omitted from further analysis. Each extract was then electrophoresed on a native polyacrylamide gel with no comb, and strips of nitrocellulose were cut from the resulting Western blot and probed with various MAbs (Fig. 6B to D). MAbs HD1 (panel B) and ABD (panel C), which recognize discontinuous epitopes in antigenic sites Ia and III, respectively (41–43), bound to all of the mutant proteins. The binding of DL11 (panel D) was eliminated by deletion of residues 222 to 224, 225 to 227, and 228 to 230 (lanes 1 to 3, respectively), suggesting that residues 222 to 230 contribute to antigenic site Ib.
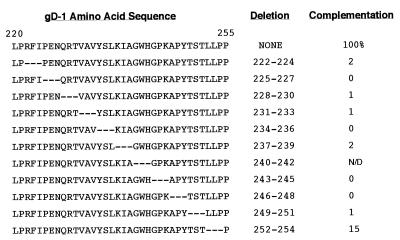
FIG. 5.
Complementation analysis of 3-amino-acid deletion mutants of HSV-1 gD. The predicted amino acid sequence for residues 220 to 255 of HSV-1(KOS) gD is shown at the top left (numbering is based on the assignment of residue number 1 to the lysine at the N terminus of the signal-peptidase-processed molecule, which is the 26th residue of the primary translation product). HSV-2(strain 333) has two amino acid changes relative to gD from the KOS strain in this region of the protein, V→L at residue 233 and A→P at residue 246. Below the wild-type sequence, the corresponding sequences of the 3-amino-acid deletion mutants are shown. The level of complementation of a gD-null virus is shown for each construct in the column at the right and is expressed as a percentage of that achieved with the wild-type construct (the mutant lacking residues 240 to 242 was never detected in transfected cells [see text]).
FIG. 6.
Antigenic analysis of HSV-1 gD 3-amino-acid deletion mutants. Replicate cultures of COS cells were transfected separately with plasmid constructs expressing wild-type gD, gD lacking residues 234 to 244, and each of the 3-amino-acid deletion mutants. Detergent extracts were prepared from transfected cells after 40 h, subjected to SDS-PAGE, and transferred to nitrocellulose (Western blot). Blots were then probed with MAbs against HSV gD. (A) Blot probed with DL6. (B) Blot probed with ABD. (C) Blot probed with HD1. (D) Blot probed with DL11. Lane 1, gD-1(Δ222–224); lane 2, gD-1(Δ225–227); lane 3, gD-1(Δ228–230); lane 4, gD-1(Δ231–233); lane 5, gD-1(Δ234–236); lane 6, gD-1(Δ237–239); lane 7, gD-1(Δ243–245); lane 8, gD-1(Δ246–248); lane 9, gD-1(Δ249–251); lane 10, gD-1(Δ252–254); lane 11, gD-1(Δ234–244); lane 12, wild-type gD-1.
Activity of deletion mutants in a complementation assay.
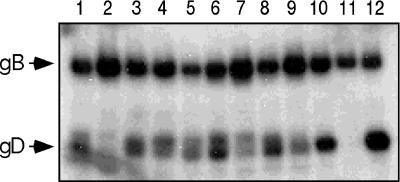
Each mutant was tested for its ability to complement the production of infectious F-gDβ virus in COS cells by using quantities of plasmid DNA that result in similar numbers of gD-expressing cells. F-gDβ lacks a gD gene and produces infectious virus only when functional gD protein is provided in trans (35). With the exception of Δ252–254, none of the mutants was able to complement F-gDβ, as found previously for the Δ234–244 mutant (42) (Fig. 5). To address the possibility that lack of complementation was due to failure of the mutated proteins to be incorporated into virions, extracellular complemented F-gDβ virus was centrifuged through a 10% sucrose cushion at 84,000 × g for 2 h at 4°C. The pellet was solubilized in denaturing SDS-PAGE sample buffer, electrophoresed on a 7.5% polyacrylamide gel, Western blotted, and probed with polyclonal antibodies against gD and gB (Fig. 7). Although each of the mutant proteins was detected in virions, the Δ225–227 and Δ243–245 proteins were incorporated inefficiently and may explain their complementation-negative phenotype. Taken together, these results suggest that a region encompassing at least residues 222 to 251 is required for gD function.
FIG. 7.
Detection of gD in F-gDβ virus complemented with the 3-amino-acid gD deletion mutants. Extracellular, F-gDβ virus which had been complemented separately with each of the 3-amino-acid deletion mutants was prepared as described in the text and analyzed by SDS-PAGE followed by Western blotting. The resulting blot was probed with a mixture of two polyclonal antibodies, R7 (raised against gD) and R69 (raised against gB). Lane 1, virus complemented with gD-1(Δ222–224); lane 2, virus complemented with gD-1(Δ225–227); lane 3, virus complemented with gD-1(Δ228–230); lane 4, virus complemented with gD-1(Δ231–233); lane 5, virus complemented with gD-1(Δ234–236); lane 6, virus complemented with gD-1(Δ237–239); lane 7, virus complemented with gD-1(Δ243–245); lane 8, virus complemented with gD-1(Δ246–248); lane 9, virus complemented with gD-1(Δ249–251); lane 10, virus complemented with gD-1(Δ252–254); lane 11, cells transfected with salmon sperm DNA; lane 12, virus complemented with wild-type gD-1.
Expression of gD-1(Δ222–224) and gD-1(Δ231–233) as soluble forms in the baculovirus system.
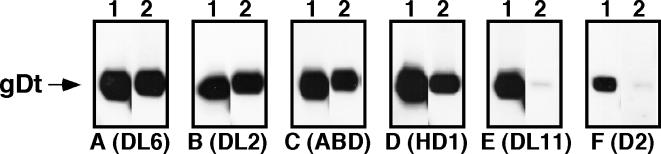
In order to examine the receptor-binding properties of a subset of the 3-amino-acid gD deletion mutants, we constructed two baculovirus recombinants expressing gD-1 truncated after residue 306 and lacking residues 222 to 224 (DL11 negative) or residues 231 to 233 (DL11 positive). While both proteins were detected in recombinant baculovirus-infected insect cells, only the Δ222–224 protein [gD-1(Δ222–224, 306t)] was secreted. The baculovirus-produced gD-1(Δ222–224, 306t) was purified by nickel-agarose chromatography (Fig. 1), and its reactivity with a panel of MAbs was analyzed by native Western blot (Fig. 8). As anticipated from the data shown in Fig. 5, this mutant form of gD reacted with MAbs ABD and HD1. The deletion mutant also reacted as well as gD-1(306t) with MAbs DL6 and DL2, further validating its structural integrity. In contrast, gD-1(Δ222–224, 306t) failed to react with either of two group Ib MAbs tested, DL11 and D2. These results were consistent with the antigenic properties of the full-length form of this protein expressed in mammalian cells (see Fig. 6).
FIG. 8.
Antigenic analysis of gD-1(Δ222–224, 306t) produced in recombinant-baculovirus-infected cells. Purified proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the gD-specific MAbs indicated below each panel. Lane 1, gD-1(306t); lane 2, gD-1(Δ222–224, 306t). The gD-1(306t) bands shown in each panel correspond to those shown in Fig. 2A to F.
gD-1(Δ222–224, 306t) binds poorly to HveA and HveC and fails to block HSV infection.
To assess the receptor-binding properties of gD-1(Δ222–224, 306t), we examined its ability to bind to HveA(200t) and to HveC(346t) by ELISA. As shown in Fig. 9A, the binding of gD-1(Δ222–224, 306t) to HveA(200t) was barely detectable, even at concentrations as high as 6 μM. The binding to HveC(346t) was diminished by ca. 10-fold relative to that of gD-1(306t). Consistent with its reduced binding to both HveA and HveC, gD-1(Δ222–224, 306t) failed to block virus infection of HeLa or Vero cells (Fig. 10). The failure of gD-1(Δ222–224, 306t) to bind well to either of two known HSV receptors most likely explains the inability of the full-length form of this protein to block virus infection or to complement the infectivity of a gD-null virus.
FIG. 9.
Analysis of gD-1(Δ222–224, 306t) for receptor binding by ELISA. The wells of an ELISA plate were coated with an excess of HveA(200t) or HveC(346t) and incubated with increasing concentrations (shown on the x axis) of gD-1(306t), gD-1(250t), or gD-1(Δ222–224, 306t). Bound gD was detected by incubating sample wells with a rabbit antiserum raised against gD (R7), followed by peroxidase-conjugated goat anti-rabbit antibody and then peroxidase substrate. (A) Binding to HveA(200t). (B) Binding to HveC(346t). Symbols: □, gD-1(306t); ○, gD-1(250t); ▵, gD-1(Δ222–224, 306t).
FIG. 10.
Analysis of gD-1(Δ222–224, 306t) for blocking of HSV entry. Cells in 96-well tissue culture plates were incubated with increasing concentrations (shown on the x axis) of gD-1(306t), gD-1(250t), or gD-1(Δ222–224, 306t) prior to infection with HSV-1(KOS) carrying a β-galactosidase reporter gene (hrR3). At 5 h postinfection, cells were lysed and assayed for β-galactosidase activity. (A) HeLa cells. (B) Vero cells. Symbols: □, gD-1(306t); ○, gD-1(250t); ▵, gD-1(Δ222–224, 306t); , bovine serum albumin.
DISCUSSION
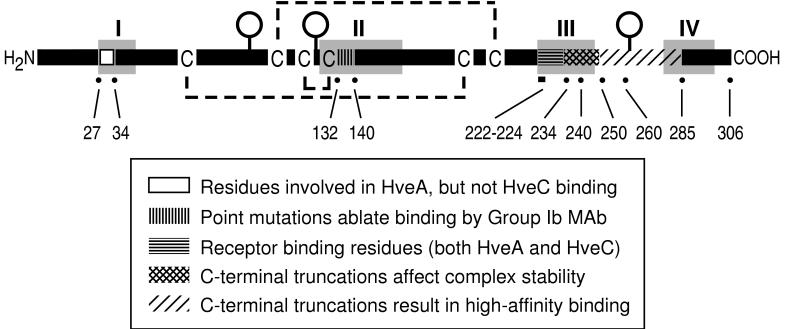
During the past decade, numerous studies have examined how the structure of HSV gD relates to its function. Some studies focused on a discontinuous antigenic site which was bound by several type-common, complement-independent neutralizing MAbs (antigenic site I). On the basis of additional characteristics, group I MAbs were subdivided into subgroups Ia and Ib (41). For example, the group Ia MAb, HD1, binds to gD truncated at amino acid residue 233, whereas the group Ib antibodies, such as DL11, do not. Separate studies demonstrated the involvement of residues upstream of 233 in antigenic site Ib as well (39, 43). Single-amino-acid changes were identified which allowed gD to function during virus replication while conferring resistance to neutralization by certain group I antibodies. Specifically, two group Ib antibodies failed to neutralize a virus expressing gD with a Ser-to-Asn change at residue 140, and a third group Ib antibody failed to neutralize a virus expressing gD with a Gln-to-Leu change at residue 132 (Fig. 11).
FIG. 11.
Stick diagram of HSV gD showing features relevant to this study. The amino- and carboxy-terminal ends of the gD ectodomain are indicated by H2N and COOH, respectively. Cysteine residues are denoted by C, and the disulfide bond pattern (36) is indicated by dashed lines. Functional regions I to IV as defined by Chiang et al. (7) are shaded in gray. The positions of individual residues relevant to the present study are marked by a dot and labeled with the residue number.
Evidence of an overlap between antigenic site Ib and a putative functional region of gD was provided by Muggeridge et al. (42), who examined seven gD mutants containing N-terminal, internal, or C-terminal amino acid deletions for their ability to complement a gD-null virus. gD lacking residues 234 to 244 was expressed on the surface of transfected cells and, although not globally altered in structure, failed to rescue the infectivity of a gD-null virus. Interestingly, this mutant protein failed to react with DL11, suggesting that antigenic site Ib, as well as a functional region of gD, was disrupted by this 11-amino-acid deletion. In a similar study, Feenstra et al. (17) found that deletion of residues 231 to 235 resulted in a protein which also failed to complement a gD-null virus but retained reactivity with DL11. More recently, Nicola et al. (44) showed that DL11 blocked the interaction of soluble HveA with gD or with HSV virions, and Krummenacher et al. (31) showed that DL11 blocked the interaction of soluble HveC with HSV virions. Finally, gD truncated at residue 234 was bound by DL11 weakly and bound HveA with a markedly lower affinity (KD) than molecules truncated at residue 275, 285, or 306 (47).
Linker-scanning mutational analysis of HSV gD (7) identified four distinct functional regions within the gD primary structure wherein linker insertions did not cause global structural alterations but greatly diminished or eliminated the protein’s ability to complement the infectivity of a gD-null virus (shaded areas designated I through IV in Fig. 11). This study also revealed a relationship between antigenic site Ib and regions II and III. First, linker insertions within region II abolished or greatly diminished binding by DL11. Second, region II (residues 125 to 161) encompasses residues previously shown to affect the binding of group Ib antibodies (residues 132 and 140; see Fig. 11). Third, region III (residues 225 to 246) includes residues required for group Ib antibody binding. These observations suggest that regions II and III may be closely positioned within the folded structure of gD and may, together, form a functional (receptor-binding) domain.
In the present study we addressed the contribution of gD residues between 222 and 275 to the formation of both antigenic site Ib as well as a receptor-binding domain. We constructed three baculovirus recombinants expressing gD truncated after residues 240, 250, and 260 and analyzed them along with previously described gD truncations (after residues 234, 285, and 306) for antigenic structure, receptor binding, and virus-blocking activity. All of the truncated proteins were bound by several MAbs recognizing discontinuous epitopes. In contrast, reactivity with DL11 was not exhibited by all of the truncation mutants. Although gD-1(250t) reacted strongly with DL11, gD-1(240t) and gD-1(234t) had significantly diminished reactivity. Thus, we conclude that the C-terminal limit for full DL11 binding occurs between residues 240 and 250.
Analysis of the C-terminal truncation mutants for receptor binding revealed a pattern somewhat similar to that seen for DL11 binding. Using the activity of gD-1(306t) as a reference point, gD-1(285t), gD-1(260t), and gD-1(250t) exhibited enhanced binding to both HveAt and HveCt (Fig. 11), a property previously demonstrated for gD-1(285t) and gD-1(275t) (31, 47). The higher affinity of gD-1(285t) and gD-1(275t) for HveA was shown by optical biosensor studies to result from a faster kon. Here we found that gD-1(260t) and gD-1(250t) bound to both HveA and HveC with kinetics and KD values very similar to those of gD-1(285t). The calculated KD values for the interactions of gD-1(240t) with HveA and HveC were quite similar to those for gD-1(306t). Interestingly, the kon and koff values obtained for the interaction of gD-1(240t) with HveC were significantly faster than those obtained for gD-1(306t). From these experiments it is clear that the loss of high-affinity receptor binding by gD is first evident in gD-1(240t), the same truncation point at which full DL11 reactivity is lost. The shorter truncation, gD-1(234t), exhibited an approximately 10-fold faster koff for both HveA and HveC relative to gD-1(306t), perhaps due to deletion of gD residues which stabilize the complexes (Fig. 11).
The ability of soluble gD to bind receptor molecules should theoretically correspond to its ability to block HSV infection of cells bearing those receptors. In the case of both HeLa and Vero cells, the blocking activities of all but one of the proteins closely matched their receptor-binding properties as seen by ELISA. Interestingly, gD-1(240t), which bound both HveA and HveC with a KD very similar to gD-1(306t), was much less effective in blocking HSV infection than gD-1(306t). Perhaps the membrane-bound forms of HveA and HveC recognize gD somewhat differently than the truncated forms used in the ELISA and biosensor studies. This result might also suggest that there is yet another receptor in HeLa and Vero cells to which gD-1(240t) binds with lower affinity than gD-1(306t). This explanation seems unlikely, at least for HeLa cells, since Cocchi et al. (8) showed that a MAb to HIgR (HveC) effectively blocks HSV infection of this cell line. Alternatively, the difference in blocking ability between gD-1(306t) and gD-1(240t) may reflect the different rates of gD-HveC complex formation and or dissociation observed in the optical biosensor studies discussed above.
The observation that gD-1(234t) bound HveA, albeit in an unstable manner, indicated that at least some receptor-binding residues were present upstream of 234. To extend our analysis of the group Ib epitope and functional region III, we generated a panel of plasmid constructs expressing full-length HSV-1 gD with sequential, nonoverlapping, 3-amino-acid deletions extending from residue 222 through residue 254. The altered gD molecules expressed in transiently transfected cells were first analyzed for function by using a complementation assay. Only one of the 3-amino-acid gD deletion mutants (Δ252–254) complemented the infectivity of a gD-null virus, confirming the conclusions of Chiang et al. (7). All forms of gD retained the folded structure necessary for reactivity with group III and group Ia MAbs, and each was detected on the surface of transfected cells. However, gD lacking residues 222 to 224, 225 to 227, or 228 to 230 failed to react with DL11, indicating that even small deletions in this region of gD disrupt antigenic site Ib. Earlier results, examined in connection with data presented here, suggest that gD residues 222 through 230 are critical for proper formation of the DL11 epitope, whereas residues between positions 231 and 250 may be important for proper presentation of the DL11 epitope but are not directly involved in antibody binding.
To examine the receptor-binding properties of some of the 3-amino-acid deletion mutants, we cloned and expressed two of these proteins (Δ222–224 and Δ231–233) as truncated forms in the baculovirus system. Although both of these molecules were detected in recombinant-baculovirus-infected cells, only gD-1(Δ222–224, 306t) could be purified from the culture medium. gD-1(Δ222–224, 306t) reacted with several MAbs (but not DL11, as expected), bound weakly to HveA and HveC, and failed to block HSV infection of mammalian cells. This result showed that gD-1(Δ222–224) is nonfunctional due, at least in part, to its greatly diminished binding to cellular receptors. Once again, these data support the concept of an overlap between a receptor-binding domain of gD and the DL11 epitope.
Antibodies directed to viral proteins can neutralize virus infectivity by binding to and occupying the site on a virion protein which interacts with a cellular receptor during virus attachment and entry. This mechanism of neutralization by certain MAbs has been demonstrated for several different viruses, including influenza virus (3, 51). Whether DL11 (and other group Ib MAbs) neutralizes HSV infectivity by occupying part of its receptor binding domain has yet to be conclusively demonstrated. The data presented here and in previous publications are clearly consistent with this interpretation, although it is formally possible that the receptor-binding site on gD is spatially distinct from the group Ib antigenic site. To address this and other questions, we are currently attempting to determine the crystal structure of gD alone, as well as gD complexed with each of its two known receptors or complexed with the DL11 MAb.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants AI-18289 to G.H.C. and R.J.E. from the National Institute of Allergy and Infectious Diseases and grants NS-30606 and NS-36731 to R.J.E. and G.H.C. from the National Institute of Neurologic Diseases and Stroke. C.K. was supported by a fellowship (823A-053464) from the Swiss National Science Foundation.
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Biacore, Inc. BIAevaluation software handbook, version 3.0. Uppsala, Sweden: Biacore, Inc.; 1997. [Google Scholar]
- 3.Bizebard T, Gigant B, Rigolet P, Rasmussen B, Diat O, Bosecke P, Wharton S, Skehel J, Knossow M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 4.Cai W, Person S, DebRoy C, Gu B. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 5.Carswell S, Resnick J, Alwine J C. Construction and characterization of CV-1P cell lines which constitutively express the simian virus 40 agnoprotein: alteration of plaquing phenotype of viral agnogene mutants. J Virol. 1986;60:415–422. doi: 10.1128/jvi.60.2.415-422.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen E U, Seeburg P H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 7.Chiang H-Y, Cohen G H, Eisenberg R J. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attribute of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen G H, Dietzschold B, Ponce de Leon M, Long D, Golub E, Varrichio A, Pereira L, Eisenberg R J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates production of neutralizing antibody. J Virol. 1984;49:102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen G H, Wilcox W C, Sodora D L, Long D, Levin J Z, Eisenberg R J. Expression of herpes simplex virus type 1 glycoprotein D deletion mutants in mammalian cells. J Virol. 1988;62:1932–1940. doi: 10.1128/jvi.62.6.1932-1940.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietzschold B, Eisenberg R J, Ponce de Leon M, Golub E, Hudecz F, Varrichio A, Cohen G H. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol. 1984;52:431–435. doi: 10.1128/jvi.52.2.431-435.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberlé F, Dubreuil P, Mattei M-G, Devilard E, Lopez M. The human PRR2 gene, related to the poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg R J, Long D, Ponce de Leon M, Matthews J T, Spear P G, Gibson M G, Lasky L A, Berman P, Golub E, Cohen G H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985;53:634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg R J, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Hastings J C, Cohen G H. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog. 1987;3:423–435. doi: 10.1016/0882-4010(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 17.Feenstra V, Hodaie M, Johnson D C. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J Virol. 1990;64:2096–2102. doi: 10.1128/jvi.64.5.2096-2102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman H M, Cohen G H, Eisenberg R J, Seidel C A, Cines D B. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature (London) 1984;309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 19.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZinsertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 22.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Highlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland T C, Marlin S D, Levine M, Glorioso J. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J Virol. 1983;45:672–682. doi: 10.1128/jvi.45.2.672-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang A, Wagner R. Penetration of herpes simplex virus into human epidermoid cells. Proc Soc Exp Biol Med. 1964;116:863–869. doi: 10.3181/00379727-116-29392. [DOI] [PubMed] [Google Scholar]
- 26.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D C, Gregory T, Burke R L. Inhibition of HSV entry into cells by purified HSV glycoprotein D and characterization of cell surface receptors for HSV. Nyborg Strand, Denmark: 14th International Herpesvirus Workshop; 1989. p. 237. [Google Scholar]
- 29.Karlsson R, Michaelsson A, Mattson A. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 30.Kousoulas K G, Pellett P E, Pereira L, Roizman B. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and tsdomains of HSV-1(F) gB gene. Virology. 1984;135:379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- 31.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 35.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long D, Wilcox W C, Abrams W R, Cohen G H, Eisenberg R J. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J Virol. 1992;66:6668–6685. doi: 10.1128/jvi.66.11.6668-6685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez M, Eberlé F, Mattei M-G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 38.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 39.Minson A C, Hodgman T C, Digard P, Hancock D C, Bell S E, Buckmaster E A. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 41.Muggeridge M I, Isola V J, Byrn R A, Tucker T J, Minson A C, Glorioso J C, Cohen G H, Eisenberg R J. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, using deletion mutants and monoclonal antibody-resistant mutants. J Virol. 1988;62:3274–3280. doi: 10.1128/jvi.62.9.3274-3280.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muggeridge M I, Wilcox W C, Cohen G H, Eisenberg R J. Identification of a site on herpes simplex virus type 1 gD that is essential for infectivity. J Virol. 1990;64:3617–3626. doi: 10.1128/jvi.64.8.3617-3626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muggeridge M I, Wu T-T, Johnson D C, Glorioso J C, Eisenberg R J, Cohen G H. Antigenic and functional analysis of a neutralization site of HSV-1 glycoprotein D. Virology. 1990;174:375–387. doi: 10.1016/0042-6822(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 44.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira L, Klassen T, Baringer J R. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect Immun. 1980;29:724–732. doi: 10.1128/iai.29.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rux A H, Willis S H, Nicola A V, Hou W, Peng C, Lou H, Cohen G H, Eisenberg R J. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J Virol. 1998;72:7091–7098. doi: 10.1128/jvi.72.9.7091-7098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seigneurin J M, Desgranges C, Seigneurin D, Paire J, Renversez J C, Jacquemont B, Micouin C. Herpes simplex virus glycoprotein D: human monoclonal antibody produced by bone marrow cell line. Science. 1983;221:173–175. doi: 10.1126/science.6304881. [DOI] [PubMed] [Google Scholar]
- 49.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 51.Stewart P, Nemerow G. Recent structural solutions for antibody neutralization of viruses. Trends Microbiol. 1997;5:229–233. doi: 10.1016/S0966-842X(97)01049-4. [DOI] [PubMed] [Google Scholar]
- 52.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warner M S, Martinez W, Geraghty R J, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 54.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the TNFR superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitbeck, J. C., C. Peng, G. H. Cohen, and R. J. Eisenberg. Unpublished data.
- 56.Willis S H, Peng C, Ponce de Leon M, Nicola A V, Rux A H, Cohen G H, Eisenberg R J. Expression and purification of secreted forms of herpes simplex virus glycoproteins from baculovirus-infected insect cells. In: Brown M S, MacLean A R, editors. Methods in molecular medicine: herpes simplex virus protocols. Vol. 10. Clifton, N.J: Humana Press; 1997. pp. 131–156. [DOI] [PubMed] [Google Scholar]
- 57.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Cohen G H, Eisenberg R J. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittels M, Spear P G. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 1990;18:271–290. doi: 10.1016/0168-1702(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 59.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982;10:6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]