Abstract
Castration-resistant prostate cancer (CRPC) is a frequently occurring disease with adverse clinical outcomes and limited therapeutic options. Here, we identify methionine adenosyltransferase 2a (MAT2A) as a critical driver of the androgen-indifferent state in ERG fusion-positive CRPC. MAT2A is upregulated in CRPC and cooperates with ERG in promoting cell plasticity, stemness and tumorigenesis. RNA, ATAC and ChIP-sequencing coupled with histone post-translational modification analysis by mass spectrometry show that MAT2A broadly impacts the transcriptional and epigenetic landscape. MAT2A enhances H3K4me2 at multiple genomic sites, promoting the expression of pro-tumorigenic non-canonical AR target genes. Genetic and pharmacological inhibition of MAT2A reverses the transcriptional and epigenetic remodeling in CRPC models and improves the response to AR and EZH2 inhibitors. These data reveal a role of MAT2A in epigenetic reprogramming and provide a proof of concept for testing MAT2A inhibitors in CRPC patients to improve clinical responses and prevent treatment resistance.
Subject terms: Cancer epigenetics, Cancer, Prostate cancer
The therapeutic options for castration-resistant prostate cancer (CRPC) remain limited. Here, the authors highlight the role of methionine adenosyltransferase 2a (MAT2A) in epigenetic reprogramming and suggest its therapeutic targeting in CRPC.
Introduction
Prostate cancer (PC) is a leading cause of cancer-related mortality1,2. Although localized PC is curable, many patients progress to metastatic and castration-resistant prostate cancer (CRPC), which can be rapidly lethal2. Androgen deprivation therapy (ADT) remains the first-line therapy for patients with metastatic PC2,3. However, resistance to ADT invariably emerges and represents the main barrier to curing metastatic PC patients2,3. Increased cell plasticity with the acquisition of stem-like properties, AR indifference, and neuroendocrine (NE) differentiation characterizes the most aggressive and highly metastatic variants of CRPC. Understanding the underlying mechanism sustaining the androgen indifferent state is fundamental to designing alternative therapeutic strategies to reverse this deadly disease.
The TMPRSS2:ERG gene rearrangement occurs in over half of PC4. ERG requires interaction with multiple additional cofactors to promote a fully oncogenic transcriptional program. We recently reported that ERG coopts EZH2 in a feedforward loop that supports the evolution of ERG fusion-positive PC towards a more aggressive and metastatic state5. EZH2 is the catalytic component of the PRC2 complex inducing histone H3 lysine 27 trimethylation (H3K27me3) and has a prominent role in PC progression6–8. EZH2 also has PRC2-independent non-canonical functions, interacting directly with transcription factors and acting as a transcriptional co-activator9. We recently found that EZH2 activated ERG via a post-translational modification at lysine 362 methylation (K362) and the formation of ERG/EZH2 chromatin complexes5. Notably, the cooperation between ERG and EZH2 led to the overexpression of a distinct set of ERG/EZH2 co-regulated genes associated with the transition to a stem-like and AR-indifferent state5.
Methionine adenosyltransferase 2a (MAT2A), the catalytic subunit of the S-adenosylmethionine synthetase complex, was one of the top ERG/EZH2 co-activated genes in ERG fusion-positive prostate tumor models10. MAT2A catalyzes the formation of SAM from methionine and ATP, providing the primary methyl donor for the methylation of DNA, RNA, histone, and non-histone proteins11.
Here, we report the function of MAT2A in sustaining cancer cell stemness and plasticity and promoting an androgen-indifferent state in CRPC. RNA, ATAC and ChIP sequencing coupled with histone post-translation modification analysis revealed a broad impact of MAT2A on the transcriptional and epigenetic landscape in ERG fusion-positive CRPC models. Mechanistically, MAT2A enhances H3K4me2, a histone mark associated with activating pro-tumorigenic non-canonical AR target genes. Furthermore, MAT2A promotes the ERG and EZH2-induced transcriptome re-programming by modulating the ERG level and activity. Genetic and pharmacological inhibition of MAT2A in multiple preclinical models promotes luminal epithelial differentiation and reduces stemness and tumorigenesis in vivo. Consistently, MAT2A inhibition improves the response to AR antagonists and EZH2 inhibitors. This study demonstrates the role of MAT2A and an enhanced vulnerability of ERG-positive CRPCs to MAT2A inhibitors. These data provide a mechanistic rationale for implementing MAT2A inhibitors for treating androgen-indifferent prostate tumors.
Results
MAT2A is overexpressed in CRPC and promotes stem-like and tumorigenic properties
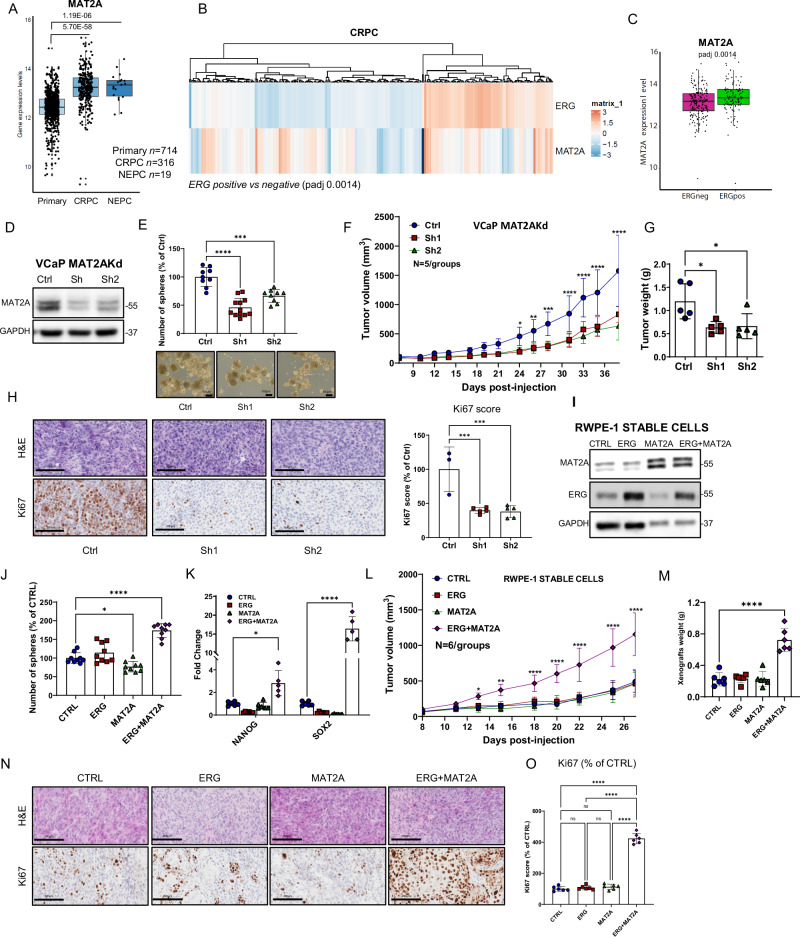
To understand the clinical relevance of MAT2A in prostate cancer, we evaluated a database including primary prostate tumors (n = 714), CRPC (n = 316) and NEPC (n = 19). MAT2A expression was significantly higher in CRPC and NEPC compared to primary tumors (Fig. 1A). MAT2A-expressing CRPC were prevalently ERG positive (Padj value: 0.0014) (Fig. 1B, C), in line with a link between ERG activation and MAT2A expression. However, MAT2A level was independent of the androgen receptor (AR) and was equally elevated in high (AR-high) and low (AR-low) AR expressing CRPC (Supplementary Fig. 1A, B). Consistent with a prevalent transcriptional upregulation of the gene, the frequency of MAT2A gene amplification in PC is less than 4% (Supplementary Fig. 1C), unlike other tumor types (Supplementary Fig. 1D).
Fig. 1. MAT2A is elevated in aggressive prostate tumors and its ablation reverts transformation in vitro and in vivo.
A Gene expression level of MAT2A in prostate cancer patients indicated subgroups (Primary, n = 714; CRPC, n = 316; NEPC, n = 19). p-values are indicated. B Heatmap of ERG and MAT2A expression values in the cohort of CRPC patients described in A. (Padj value 0.0014). C Gene expression level of MAT2A in CRPC described in A and divided in ERG positive (n = 113) and negative (n = 203). Adjusted p-value 0.0014. D Immunoblot of VCaP control (Ctrl) and MAT2Akd VCaP cells (Sh1, Sh2) with indicated Ab (n = 3 independent experiments). E Sphere formation assay (SFA) in indicated cell lines (n = 3 independent experiments with n = 4 (for sh1) and n = 3 (for ctrl and sh2) biological replicates. Bottom, representative images of tumor spheres. Scale bars, 100 µm. F Growth curve of VCaP xenograft from VCaP control (Ctrl) and VCaP with stable MAT2A knockdown (Sh1 and Sh2) were engrafted and growth was monitored by caliper every 2 days (n = 5 mice /group). G Tumor weight of indicated tumor xenografts (n = 5 mice/group). H Representative sections from the indicated tumor xenografts. Scale bars, 200 µm. Right, Immunoscore of Ki67 by Aperio tool (n = 3 mice Ctrl, n = 5 mice Sh1, n = 5 mice Sh2). I Immunoblot of RWPE-1 cells with stable expression of ERG, MAT2A and ERG + MAT2A with indicated Ab (n = 3 independent experiments). J SFA in indicated cell lines (n = 3 independent experiments with 3 biological replicates). K Fold change of mRNA levels of NANOG and SOX2 in RWPE-1 cells with stable expression of ERG, MAT2A and ERG + MAT2A. (n = 5-6 biological replicates). L Growth curve of RWPE-1 xenografts. Indicated cell lines were engrafted and growth was monitored by caliper every 2 days (n = 6 mice/group). M Tumor weight of indicated tumor xenografts (n = 6). N Representative sections from the indicated tumor xenografts. Scale bars are 200 µm. O Immunoscore of Ki67 using the Aperio tool (n = 6 mice/group). Molecular weights are indicated in kilodaltons (kDa). All error bars, mean ± SD. For box-and-whisker plots in A and C, the line inside the box shows the median value. The bounds of the box represent the 25th–75th percentiles, with whiskers at minimum and maximum values. One-way-ANOVA was used to test significant differences between groups in all panels, except for (K) where 2-way-ANOVA was used. Data presented in F and L are independent replicates derived from individual mice. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001.
We hypothesized that, as a potential cofactor of multiple epigenetic effectors, MAT2A could cooperate with ERG and EZH2 and support the growth of ERG fusion-positive PC and the evolution of primary tumors in CRPC and NE-CRPC. Hence, we knocked down MAT2A in ERG fusion-positive VCaP cells using short-hairpin RNA (shRNAs) (Fig. 1D). MAT2A-depleted (MAT2Akd) cells exhibited a reduced ability to form tumor-spheres in stem cell-selective conditions (Fig. 1E), a phenotype associated with cell plasticity and the induction of cell stemness5. Compared to control cells, MAT2Akd cells also had a reduced ability to grow as tumor xenografts in mice (Fig. 1F–G) and exhibited reduced proliferation as indicated by Ki67 immunostaining (Fig. 1H). These data supported the fact that MAT2A sustains stemness and tumor-initiating capability in VCaP cells and that ERG and MAT2A cooperate in ERG fusion-positive tumors. To test this, we generated normal prostate epithelial RWPE-1 cells with stable overexpression of MAT2A, ERG, or both (Fig. 1I). These epithelial cells are non-transformed, non-tumorigenic, and non-invasive and, therefore, provide a relevant model to study the oncogenic effect of ERG alone and in combination with MAT2A12–14. Expression of ERG or MAT2A alone did not affect the formation of tumor spheres (Fig. 1J). However, the co-expression of ERG and MAT2A significantly increased tumor-sphere growth compared to parental RWPE-1 cells, supporting the relevance of the cooperation between ERG and MAT2A in the induction of stemness (Fig. 1J). Consistently, we found enhanced expression of the stem cell markers NANOG and SOX2 in ERG and MAT2A co-expressing cells (Fig. 1K). Next, we tested the ability of ERG and MAT2A, alone or in combination, to induce tumor initiation in vivo by implanting RWPE1 cells in NRG mice (Fig. 1L, M). Parental RWPE1 cells are non-tumorigenic and, when injected subcutaneously in mice with Matrigel and in the presence of urogenital sinus mesenchymal cells (UGSM), form normal-like acinar structures without signs of transformation and invasion14–16. Consistently, control RWPE1, RWPE1-ERG, and RWPE1-MAT2A had moderate growth ability and did not form tumors (Fig. 1L, M). In line with our hypothesis, RWPE-1 cells co-expressing ERG and MAT2A (RWPE1-ERG + MAT2A) formed tumors (Fig. 1L, M). Histopathological examination revealed disrupted acinar structures and multiple malignant foci in ERG + MAT2A xenografts and only normal-like acinar structures in the other groups (Fig. 1N). Acquisition of tumorigenic properties was supported by a robust increase in Ki67 staining in ERG/MAT2A xenografts compared to the other groups (Fig. 1N, O). Collectively, these data demonstrated that ERG and MAT2A can cooperate to promote prostate tumorigenesis.
MAT2A promotes cancer-associated transcriptional features in ERG fusion-positive tumors
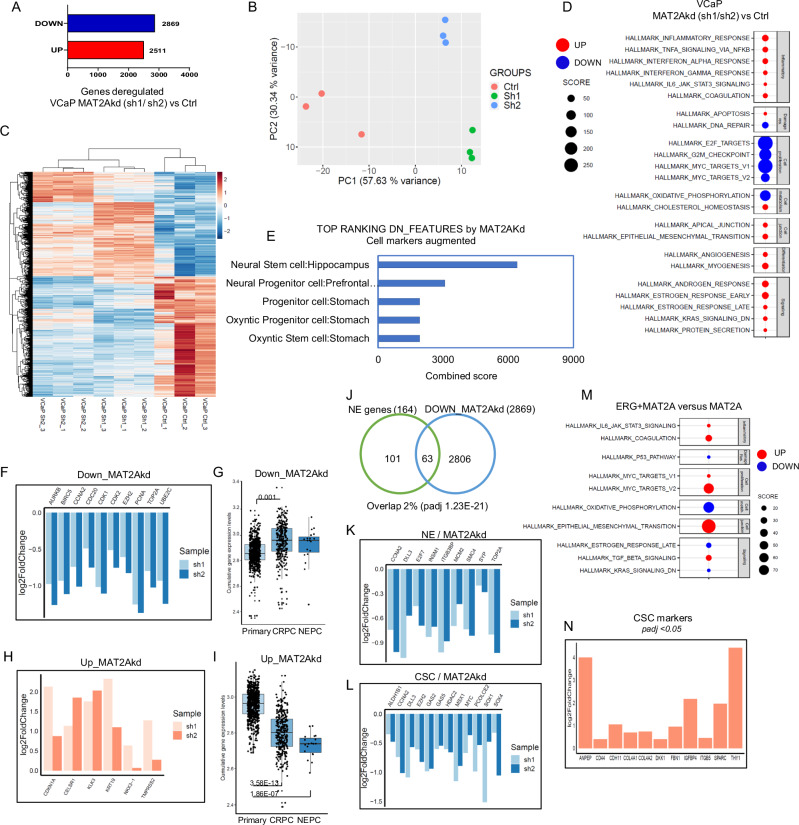
MAT2A could act as a central node for transcriptional and epigenetic reprogramming during malignant transformation and evolution of ERG fusion-positive PC. Therefore, we investigated the transcriptional changes induced by MAT2A depletion in VCaP cells by RNA-seq. Knockdown of MAT2A led to robust changes in gene expression, with similar numbers of activated and repressed genes (Fig. 2A). Principal component analysis (PCA) and unsupervised clustering supported the substantial divergence between control and MAT2A-depleted cells (Fig. 2B, C). Functional annotation analysis showed that MAT2A knockdown led to the preferential attenuation of genes controlled by c-Myc and E2F and genes associated with mitosis, cell cycle, and oxidative phosphorylation (Fig. 2D). Notably, the features repressed by MAT2A depletion also included genes related to stem and progenitor cell properties (e.g., neural stem and progenitor cells) (Fig. 2E). The stem/progenitor-related genes affected by MAT2A depletion comprised key oncogenic drivers like EZH2, TOP2A, CCNA2, BIRC5, AURKB, and PCNA (Fig. 2F). These data supported the hypothesis that MAT2A contributes to dedifferentiation, cell plasticity, and stem-like properties and promotes progression to the castration-resistant phenotype. Assessment of RNA-seq data of a large cohort of prostate cancer patients (n = 1041) revealed that CRPC and NE-CRPC had higher expression of the genes attenuated upon MAT2A knockdown (i.e., genes activated by MAT2A) compared to primary prostate tumors (Fig. 2G), in line with a role of these MAT2A-modulated genes in tumor progression.
Fig. 2. MAT2A promotes cancer-associated transcriptional features in ERG fusion-positive cells.
A Number of genes affected by knockdown of MAT2A (Sh1 and Sh2) in VCaP cells (p-value ≤ 0.05). B Principal component analysis (PCA) plot of VCaP cell lines, colored according to the condition (Ctrl, Sh1, Sh2). C Heatmap of differentially expressed genes in MAT2Akd (VCaP Sh1 and VCaP Sh2) versus control VCaP cells (VCaP Ctrl). Replicates (n = 3 biological replicates) are indicated. D Hallmark enrichment analysis of MAT2Akd VCaP cells (Sh1 and Sh2) compared to control cells, performed with cameraPR function. Down-regulated pathways are colored in blue, up-regulated pathways are colored in red. E Functional annotation of top-ranking features of MAT2Akd VCaP cells (Sh1 and Sh2) performed with enrichR (cell markers augmented). F Fold changes of selected oncogenic drivers in MAT2Akd cells (Sh1 and Sh2) versus VCaP control, evaluated by RNA-seq. G Cumulative gene expression level of down-regulated genes in MAT2Akd VCaP cells (Sh1 and Sh2) in the cohort of primary (n = 714), CRPC (n = 316) and NEPC (n = 19) patients. H Fold changes of selected canonical androgen genes in MAT2Akd (Sh1 and Sh2) versus VCaP control cells evaluated by RNA-seq. I Cumulative gene expression level of up-regulated genes in MAT2Akd VCaP cells (Sh1 and Sh2) in the cohort of primary (n = 714), CRPC (n = 316) and NEPC (n = 19) patients. J Venn diagram showing the overlap between down-regulated genes in VCaP MAT2Akd cells (Sh1 and Sh2) and gene set associated with NEPC. Test: Fisher exact test. Fold changes of selected NEPC (K) and cancer stem cell genes (L), repressed by MAT2Akd in VCaP cells (Sh1 and Sh2). M Hallmark enrichment analysis of ERG + MAT2A RWPE-1 cells compared to RWPE-1 MAT2A performed with cameraPR function. Down-regulated pathways are colored in blue, up-regulated pathways are colored in red. N Fold change of selected CSC- and EMT-related genes up-regulated in RWPE-1 ERG + MAT2A versus RWPE-1 MAT2A. For box-and-whisker plots in G and I, the line inside the box shows the median value. The bounds of the box represent the 25th–75th percentiles, with whiskers at minimum and maximum values. One-way-ANOVA was used to test significant differences between groups.
MAT2A knockdown also activated several genes that MAT2A likely repressed in VCaP cells (Fig. 2A). Functional annotation revealed the enrichment of androgen- and estrogen-response genes among the MAT2Akd-activated genes (Fig. 2D). Relevantly, MAT2A knockdown reactivated a set of canonical AR target genes, like NKX3.1, KLK3, and TMPRSS2, known to be repressed by ERG and EZH2 in ERG fusion-positive tumor cells (Fig. 2H)5,17–19. Intriguingly, CRPC and NE-CRPC had an attenuated expression of the genes enhanced by MAT2A knockdown (i.e., genes repressed by MAT2A) compared to primary PC (Fig. 2I). Thus, MAT2A might influence the expression of distinct sets of genes in both CRPC and NE-CRPC. Focusing on the NE-CRPC subtype, we found a significant convergence between the genes attenuated in MAT2A-kd cells and a previously defined NE gene signature20 (Fig. 2J, K). Moreover, we identify a set of cancer stem cell genes21 that were significantly downregulated by MAT2A ablation (Fig. 2L) further supporting the role of MAT2A in promoting stemness. Interestingly, we observed a decrease in selected cancer stem cell and NE markers also at the protein level (Supplementary Fig. 2A). The transcriptomic profiling of MAT2A-depleted VCaP cells indicated that MAT2A promoted transcription of pro-tumorigenic and stemness-related genes and repressed canonical androgen-responsive and pro-differentiation genes. These data identified MAT2A as a critical cofactor in the oncogenic reprogramming of the transcriptome of ERG fusion-positive PC. To evaluate the relevance of the synergistic impact between ERG and MAT2A on the transcriptome, we performed RNA-seq analysis of RWPE-1 cells with stable expression of ERG, MAT2A, and ERG/MAT2A. We observed a robust impact of MAT2A and ERG/MAT2A on the overall transcriptome with significant divergence in PCA and unsupervised clustering analysis (Supplementary Fig. 2B, C). In contrast, ERG had a minor effect, consistent with previous reports showing a weak impact of ERG in normal prostate epithelial cells5,18. We examined the distinctive features deregulated by the combined expression of ERG/MAT2A and MAT2A alone. Functional annotation revealed that the most enriched hallmarks in combined ERG/MAT2A cells were epithelial to mesenchymal transition (EMT) and Myc targets (Fig. 2M), two important hallmarks of aggressive prostate tumors and highly related to induction of cancer stem cell properties. In support of the induction of stemness, several CSC markers were upregulated by ERG + MAT2A in RWPE-1 cells compared to MAT2A alone (Fig. 2N). These data support the enhanced tumor sphere formation and induction of tumor-initiating properties observed in cells with combined ERG and MAT2A compared to MAT2A alone (Fig. 1J). Next, we performed a convergence analysis between the genes modulated by the overexpression of ERG and MAT2A in RWPE-1 cells and MAT2A knockdown in VCaP cells. Several genes were significantly convergent, including upregulated and downregulated features (Supplementary Fig. 2E, F). Hallmark pathway analysis revealed an enrichment of pro-proliferative genes, mitotic checkpoints, E2F and Myc targets among the convergent genes induced by ERG and MAT2A (Supplementary Fig. 2G). Hypoxia, apoptosis, and heme metabolism emerged among the convergent genes repressed by ERG and MAT2A (Supplementary Fig. 2H). Furthermore, compared to primary tumors, CRPC and NE-CRPC had increased and decreased expression of the genes either induced or repressed by the combined expression of ERG and MAT2A, respectively (Supplementary Fig. 2I, J). Thus, the analysis of transcriptomic data in both VCAP and RWPE1 stably expressing ERG and MAT2A, reinforced the notion of oncogenic cooperation between ERG and MAT2A and their contribution to the transcriptional landscape associated with the advanced stages of PC.
MAT2A controls chromatin accessibility and promotes an androgen-independent transcriptional program
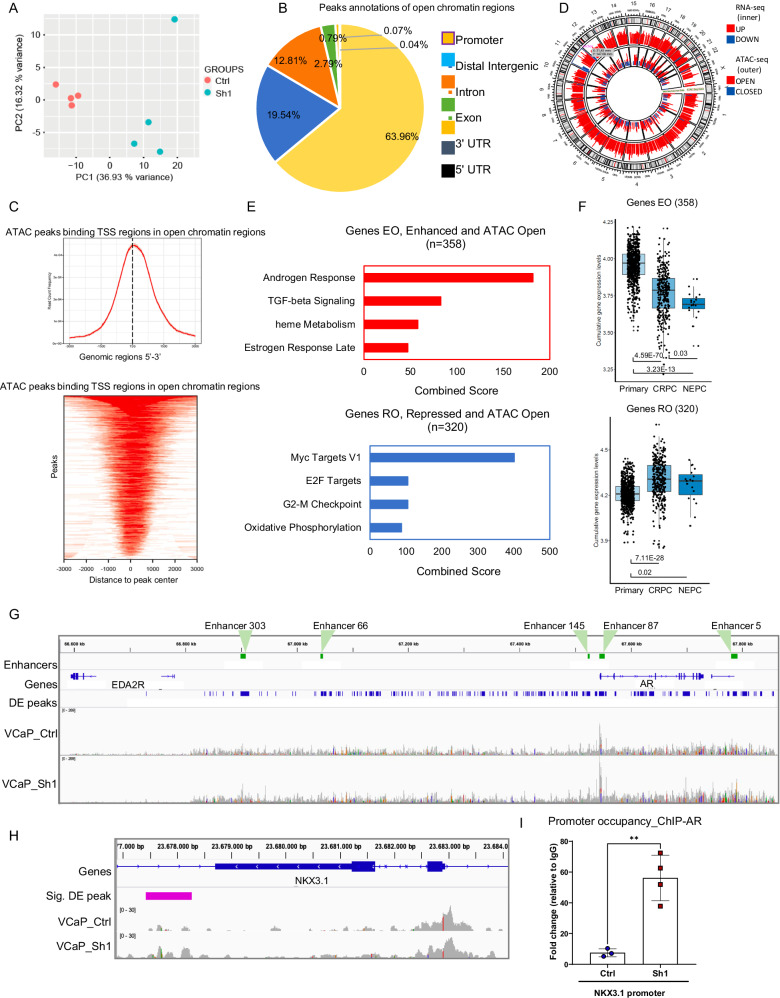
We investigated the consequence of MAT2A depletion on chromatin organization by probing chromatin-accessible sites in VCaP cells using the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq). We observed a substantial effect on chromatin upon MAT2A ablation, with a significant divergence in the PCA among the groups (Fig. 3A). There was increased chromatin accessibility in MAT2A-depleted cells with multiple open sites (n = 2795, p-value ≤ 0.05). The accessible sites were in promoters (63%), along with distal intergenic (20%) and intronic sites (12%) (Fig. 3B). Consistently, opened chromatin peaks were within a relatively short distance (0.5–1 kb) from the transcriptional start sites (TSS) of genes (Fig. 3C). Very few regions (n = 81, p-value ≤ 0.05) were closed with reduced chromatin accessibility in MAT2A-depleted cells and were distributed almost equally between promoters and distal intergenic regions (26 and 19%, respectively) (Supplementary Fig. 3A). Peaks of closed chromatin also had a distinctive distribution compared to open sites, being mainly distal and downstream to the genes TSS, consistent with the higher percentage of peaks (~50%) in exon, intron and 3’UTR sequences (Supplementary Fig. 3B).
Fig. 3. MAT2A controls chromatin accessibility at the AR enhancer.
A PCA plot showing VCaP samples examined through ATAC-seq (VCaP Ctrl and MAT2Akd Sh1). B Diagram showing the distribution of the annotated peaks in the open regions. C Top, ATAC-seq peaks binding TSS regions in open chromatin regions. Bottom peak location in TSS regions. D Circular plot of the integrative analysis ATAC-seq/RNA-seq showing the distribution of the differentially expressed peaks (outer layer) and the significantly expressed genes (inner layer). E Functional annotation of the genes located in the regions affected by chromatin changes upon MAT2Akd. Top, pathways activated by MAT2Akd. Bottom, pathways repressed by MAT2Akd. F Box plots of cumulative gene expression of the genes emerged from the integrative RNA-seq/ATAC-seq analyses. Top, genes with enhanced expression after opening of the chromatin. Bottom, genes with reduced expression after chromatin opening. Primary, n = 714; CRPC, n = 316; NEPC, n = 19. For box-and-whisker plots, the line inside the box shows the median value. The bounds of the box represent the 25th–75th percentiles, with whiskers at minimum and maximum values. G ATAC-seq peaks at AR gene and enhancer. Peak profiles of VCaP MAT2Akd and VCaP control samples. Plot was generated through IGV software. Significantly differentially expressed peaks, with a Log2FC ≥ 1.5, between VCaP MAT2Akd and control are represented in blue. Enhancer regions of AR are highlighted in green. H ATAC-seq peaks at the NKX3.1 gene. Peak profiles from ATAC-seq of VCaP control (Ctrl) and VCaP MAT2Akd (Sh1) cells. Plot was generated as described above. I AR occupancy at the NKX3.1 promoter in VCaP control (n = 3 for Input and IgG and 4 for AR biological replicates) and MAT2Akd cells(Sh1) (n = 4 for Input and AR and n = 3 for IgG biological replicates) All error bars, mean ± s.d. **p < 0.01, two-sided t-test was used to test significant differences between groups.
Integration of ATAC-seq and RNA-seq data identified genes that were transcriptionally modulated and associated with opened chromatin sites upon MAT2A depletion (Fig. 3D). Specifically, 371 genes were activated and associated with opened chromatin sites (genes enhanced and opened, EO), and 329 genes were near opened areas and repressed (genes repressed and opened, RO) by MAT2A knockdown. Notably, pathway enrichment analysis revealed that the genes with opened chromatin and enhanced expression were prevalently androgen-responsive genes (Fig. 3E, top). Thus, MAT2A in VCaP cells contributes to the silencing of genes like KLK3, NKX3.1, and TMPRSS2, which are functionally related to the canonical androgenic response. Intriguingly, MAT2A upregulation also repressed genes involved in heme metabolism, a set of genes also affected by the combined expression of ERG and MAT2A in RWPE-1 cells. Consistently, analysis of patient data revealed that the MAT2A-repressed genes were silenced in CRPC and NE-CRPC and expressed in primary tumors (Fig. 3F, top). In contrast, the genes in open chromatin regions but repressed by MAT2A knockdown (i.e., RO genes) were related to cell growth and pro-tumorigenic pathways (Fig. 3E, bottom) and they were over-expressed in both CRPC and NE-CRPC compared to primary PC (Fig. 3F, bottom).
A critical event in the PC progression is the conversion from a canonical AR-regulated gene network to a non-canonical AR-driven transcriptional program, concomitantly with the loss of epithelial cell identity and the induction of stemness and cell plasticity19,22. MAT2A may represent a critical factor in this conversion by repressing canonical AR target genes and promoting cancer cell stemness. Intriguingly, ATAC-seq revealed broad areas of opened chromatin and clusters of multiple open peaks surrounding many genes (Supplementary Fig. 3C). Strikingly, the AR locus was one of the most affected areas, with several opened chromatin sites (Supplementary Fig. 3C). The most affected sites in the AR locus overlapped the AR gene and the enhancer elements, both proximal and distal to the gene (Fig. 3G and Supplementary Fig. 3D, E). The AR gene and the enhancer region are amplified frequently in mCRPC23. This region is also amplified in VCaP cells, leading to aberrant AR expression23,24. VCaP cells display an increment in the total number of chromatin interaction modules comprising the AR gene23. AR, ERG, and other transcription factors bind the AR enhancer and promote AR transcription23. Intriguingly, we found that the increased chromatin accessibility at the AR enhancer in MAT2A-depleted VCaP cells was associated with reduced AR transcript levels, as indicated by RNA-Seq and by qRT-PCR (Supplementary Fig. 3F). The co-occurrence of opened chromatin and gene silencing could appear contradictory. However, the increased chromatin opening might reflect the recruitment of transcriptional repressors, disruption of activating long-range enhancer-promoter interactions, or altered histone modification patterns with consequent inhibition of transcription24–26. Overall, MAT2A depletion may favor a more physiologic, normal-like regulation of the AR gene transcription. Interfering with MAT2A might impact the downstream AR signaling and redirect AR to canonical target genes, reversing the aberrant activation of non-canonical AR targets in CRPC cells. Consistently, we found increased chromatin opening in the body and enhancer region of the canonical AR target gene NKX3.1 upon MAT2A knockdown (Fig. 3H). ChIP-PCR demonstrated increased AR occupancy on the promoter of the NKX3.1 gene in MAT2A-depleted cells compared to control VCaP cells (Fig. 3I), indicating a more permissive local environment for the activation of canonical AR targets. Thus, MAT2A upregulation results in the repression of canonical androgenic signaling genes and promotes an aberrant AR-dependent transcriptional program with the acquisition of features of castration-resistant tumors. Interfering with MAT2A reverses this pattern and restores a canonical AR program.
MAT2A alters H3K4me2 distribution and activates an oncogenic transcriptional program
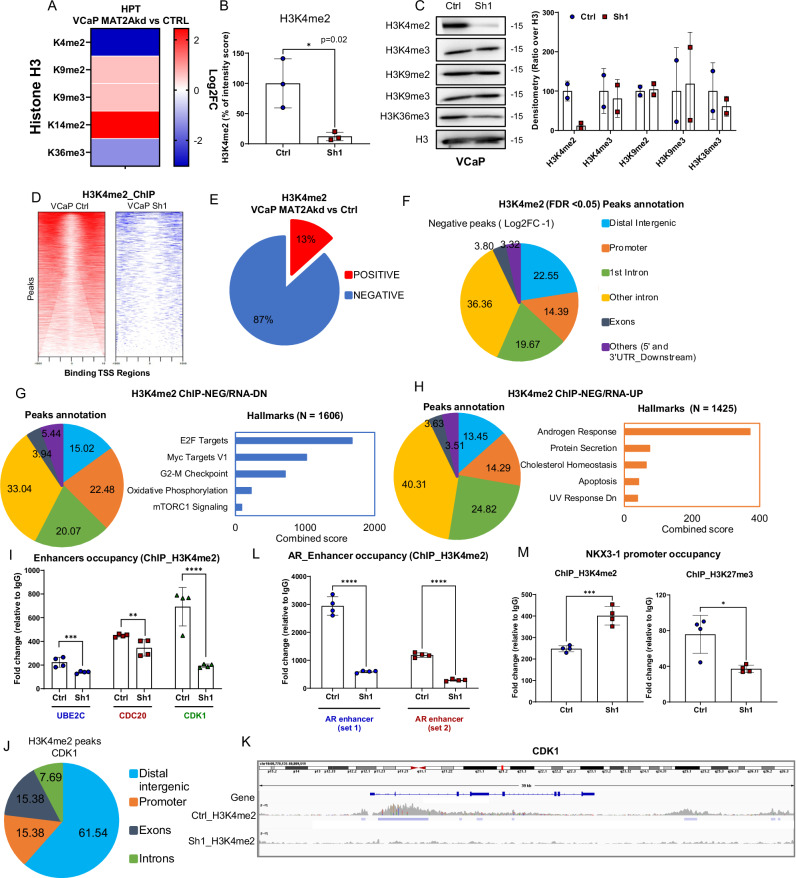
MAT2A can affect the activity of SAM-dependent chromatin-modifying enzymes and, consequently, multiple histone post-translational modifications (PTMs), ultimately impacting chromatin and transcriptional regulation27. To gain further insights into the epigenetic impact of MAT2A activity, we used high-resolution mass spectrometry (MS) to interrogate a broad spectrum of histone PTMs. Combining specific chemical histone derivatization procedures and high-resolution MS, we focused on changes in histone 3 (H3) lysine methylation in response to MAT2A silencing. This approach enabled direct and quantitative comparison of histone marks between paired samples (Fig. 4A and Supplementary Fig. 4A). Strikingly, histone H3 lysine 4 di-methylation (H3K4me2) was the most affected histone mark, decreasing significantly after MAT2A depletion (Fig. 4A, B and Supplementary Fig. 4A, B). The decrease in H3K4me2 was confirmed by immunoblotting (Fig. 4C). Of note, significant changes also were observed for H3K9me2, H3K9me3, H3K14me2, and H3K36me3 in response to MAT2A silencing (Fig. 4A and Supplementary Fig. 4C). The changes in H3K4me3, H3K27me3 and other histone marks were below the threshold of statistical significance (adjusted p-value ≤ 0.05) and were not confirmed by immunoblotting.
Fig. 4. MAT2A alters H3K4me2 distribution and activates an oncogenic transcriptional program.
A Heatmap generated by global profiling of histone H3 lysine methylation and including only the markers significantly changed in MAT2Akd (Sh1) versus VCaP control (Ctrl). Color coding in the heatmap represents the Log2FC of the intensity score for each modification in Sh1 versus Ctrl. P-values are also indicated. B Percentage of intensity score of H3K4me2 in indicated samples (n = 3 biological replicates); p-value = 0.02. C Representatives immunoblot of VCaP control (Ctrl) and MAT2Akd (Sh1) with indicated Ab. (n = 2 independent experiments). Right, densitometries of two independent experiments of indicated histone marks are shown. D Density plot of H3K4me2 peaks around the transcription start site of VCaP Ctrl (left) and VCaP Sh1 (right). The heatmaps represents all the peaks that were called for each condition. Only the peaks around the TSS (−1500, +1500) were considered. VCaP Ctrl samples (15018 peaks); VCaP Sh1(2203 peaks). E Percentage of peak changes in VCaP MAT2Akd cells compared to VCaP Ctrl. F Peak distribution of negative H3K4me2 peaks in VCaP MAT2Akd cells. G Right, peak annotation of the genes with negative ChIP-seq peak and enhanced expression in RNA-seq. Left, hallmarks of genes with negative ChIP-seq peak and enhanced expression in RNA-seq. H Right, peak annotation of the genes with negative ChIP-seq peak and reduced expression in RNA-seq. Left, hallmarks of genes with negative ChIP-seq peak and reduced expression in RNA-seq. I Occupancy of H3K4me2 on the enhancer of UBE2C, CDK1 and CDC20 in VCaP Ctrl and VCaP Sh1 (n = 4 biological replicates). J Peak distribution of H3K4me2 on CDK1 in VCaP Sh1 compared to VCaP Ctrl (FDR < 0.05; Log2FC ± 1). K ChIP-seq peaks profile at the CDK1 gene of VCaP Ctrl and VCaP Sh1 cells. Plot was generated as described in Fig. 3G. L Occupancy of H3K4me2 on two enhancer regions of AR in VCaP Ctrl and VCaP Sh1 cells (n = 4 biological replicates). M Occupancy of H3K4me2 (left) and of H3K27me3 (right) on the promoter of NKX3.1 in VCaP Ctrl and VCaP Sh1 cells (n = 4 biological replicates). Molecular weights are indicated in kilodaltons (kDa). All error bars, mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001. two-sided t-test was used to test significant differences between the two groups.
Notably, H3K4me2 is enriched at the enhancers of growth-promoting and pro-tumorigenic genes activated by AR in androgen-independent CRPC cells28. Indeed, the H3K4me2 accumulation promotes the binding of FoxA1 and AR to the enhancers and activates the transcription of the non-canonical AR targets in CRPC models28. Hence, we performed ChIP-seq to examine the genome-wide distribution of H3K4me2 in MAT2A-depleted and control VCaP cells (Fig. 4D). We observed a drastic reduction of H3K4me2 peaks (87%; negative peaks) with only a minority of sites with enhanced methylation (13%; positive peaks) (Fig. 4E). The sites with negative H3K4me2 peaks were prevalently located at intronic, distal intergenic and promoter sites (Fig. 4F), while the few positive peaks were prevalent in the promoters (Supplementary Fig. 4D).
Integrating H3K4me2 occupancy and RNA-seq data (Supplementary Fig. 4E), we observed that repressed and activated genes in MAT2A-depleted cells had a similar distribution of negative H3K4me2 peaks with a prevalence of distal intergenic and intronic regions (Fig. 4G, H) consistent with a major role of H3K4me2 at the level of enhancers. The positive peaks were prevalent in the promoter regions for both repressed and activated genes (Supplementary Fig. 4F, G).
Functional annotation of the genes associated with loss of H3K4me2 marks confirmed the overrepresentation of genes with a pro-tumorigenic and proliferative function among the repressed genes and of androgen-responsive genes among the upregulated ones (Fig. 4G, H). This trend was also observed for the few positive peaks, supporting that context-dependent selective loss of this histone mark might impact oncogenic activation and repression of pro-differentiation genes (Supplementary Fig. 4F, G). Furthermore, integrating ATAC-seq with H3K4me2 occupancy (Supplementary Fig. 4H) showed a robust overlap between reduced H3K4me2 and open chromatin sites extended to both activated and repressed genes (Supplementary Fig. 4I, J) and a prevalence of intronic and distal intergenic regions. Consistently, functional annotation of reduced H3K4me2 and open chromatin marked a similar set of pro-tumorigenic genes with reduced expression after MAT2A knockdown concomitantly with a derepressed set of androgen-related targets (Supplementary Fig. 4I). The overlap between gained H3K4me2 and open chromatin, albeit weaker, showed convergence with pro-differentiation, pro-apoptotic and canonical androgen-related targets prevalently activated upon MAT2A ablation (Supplementary Fig. 4J). Intriguingly, many genes with oncogenic functions (e.g., EZH2, AR and CDK1) and related to castration-resistance had multiple distal intergenic and intronic H3K4me2 peaks that were reduced in response to MAT2A knockdown and associated with transcriptional repression. Among these genes, CDK1, which is known to be associated with androgen independence and castration resistance, had reduced transcript levels (Fig. 2F) and multiple H3K4me2 peaks prevalently located in enhancer regions in MAT2A-depleted cells (Fig. 4I–K). Also, the AR gene showed significantly reduced H3K4me2 at the two enhancer regions tested by ChIP-PCR (Fig. 4L). In contrast, on the promoter of NKX3.1, ChIP-PCR revealed that the histone mark H3K4me2 increased, while H3K27me3 decreased in MAT2Akd VCaP cells (Fig. 4M). Thus, in the regulatory region of NKX3.1, we found the opening of chromatin, enhanced AR binding, and reduced repressive marks consistent with the gene activation upon MAT2A inhibition.
We further investigated how the opening of the chromatin and the loss of H3K4me2 deposition on the genome affected gene expression in MAT2Akd VCaP cells compared to controls. To this end, we integrated the data from ATAC-seq, ChIP-seq and RNA-seq (Supplementary Fig. 4K, L). We show examples of genes with known pro-tumorigenic properties acquiring open chromatin, loss of H3K4me2 peaks, and reduced expression (Supplementary Fig. 4M). Conversely, we found up-regulations of genes associated with a normal-like and pro-differentiation tumor suppressive role (Supplementary Fig. 4N). These data support a scenario in which MAT2A promotes H3K4me2 at enhancer sites and transcriptional activation of androgen-independent AR-regulated genes associated with tumor progression and castration resistance while favoring repression of canonical androgen-regulated genes. MAT2A inactivation drastically changes this scenario, reversing H3K4me2 accumulation and aberrant transcriptional activation of genes.
MAT2A interacts with ERG altering ERG/EZH2/AR crosstalk
Integrating RNA-seq data from MAT2A-depleted VCaP cells (this study) with publicly available ERG ChIP-seq data29, we found that the genes modulated upon MAT2A knockdown overlapped significantly with genes with ERG-bound at promoter sites (Supplementary Fig. 5A). Hence, MAT2A could directly influence ERG transcriptional activity. In keeping with this hypothesis, MAT2A-depleted cells had reduced activity of the ETS-responsive luciferase reporter compared to control VCaP cells (Supplementary Fig. 5B, left). Conversely, ETS reporter activity increased significantly in RWPE-1 cells expressing ERG and MAT2A (Supplementary Fig. 5B, right), in line with the ERG and MAT2A functional cooperation. Relevantly, immunoprecipitation experiments showed a direct interaction between ERG and MAT2A in VCaP cells (Supplementary Fig. 5C) and RWPE-1 cells co-expressing the two proteins (Supplementary Fig. 5D). Moreover, we confirmed the direct binding of MAT2A and ERG by microscale thermophoresis (MST) in a cell-free condition (Supplementary Fig. 5E). This evidence suggested that MAT2A could serve as a scaffold and transcriptional co-factor promoting ERG interactions with chromatin-bound protein partners and providing substrate to SAM-dependent enzymes, like EZH2. Intriguingly, we also observed reduced ERG protein in MAT2A-depleted cells (Supplementary Fig. 5F). Thus, the interaction with MAT2A could also control ERG protein stability. To understand the basis of this phenomenon, we treated control and MAT2Akd cells with the proteasome inhibitor PS341 (Supplementary Fig. 5G). Proteasome inhibition increased the ERG protein level, indicating MAT2A knockdown enhanced ERG degradation (Supplementary Fig. 5G). To further understand the molecular mechanisms of MAT2A-induced ERG stability, we transfected MAT2A along with a plasmid coding for full-length ERG or ERG mutated at the methylation site (pERG-K362A)5. We have shown that the mutation at lysine 362 (K362) prevents ERG methylation5. Following co-transfection and puromycin blockade of protein translation, we found that the co-transfection of MAT2A and ERG enhanced the stability of ERG. Conversely, ERG-K362A was not protected from degradation (Supplementary Fig. 5H). Thus, MAT2A, in addition to promoting histone PTMs, interacts and regulates ERG stability by enhancing methylation at K362. All these events foster functional crosstalk between ERG, EZH2, and AR, leading to transcriptional and chromatin reprogramming17–19.
To support this latter hypothesis, we performed a convergence analysis using the list of AR-bound genes at the promoter sites based on published AR ChIP-seq data29 and the ERG/EZH2 co-bound genes we had previously defined in VCaP cells5. Crossing the AR-bound genes with the MAT2A-modulated genes (n = 5379) revealed a significant convergence (p-value: 5.32E−103) (Supplementary Fig. 5I), confirming the relevance of MAT2A for transcriptional deregulation of AR target genes in prostate tumors.
In support of the disruption of the ERG/EZH2 crosstalk by MAT2A ablation, we also found reduced methylated ERG (active form) and EZH2 protein levels in MAT2A-depleted VCaP cells (Supplementary Fig. 5J). Furthermore, using a list of the ERG/EZH2 co-bound genes, we also found a high degree of convergence with the MAT2A-modulated genes (Supplementary Fig. 5K), supporting the impact of MAT2A on the ERG/EZH2 gene network. Strikingly, the ERG/EZH2/MAT2A convergent genes were also putatively AR-bound targets (Supplementary Fig. 5L). Specifically, among the shared targets, the MAT2A putatively repressed genes (UP_MAT2Akd) were enriched with androgen-responsive genes (Supplementary Fig. 5M, N). Overall, these data sustain the notion that MAT2A contributes to the ERG/EZH2-induced transcriptional reprogramming and deregulation of the AR-regulated gene network.
Antitumor activity and phenotypic reversion by MAT2A inhibitors in ERG fusion-positive preclinical models
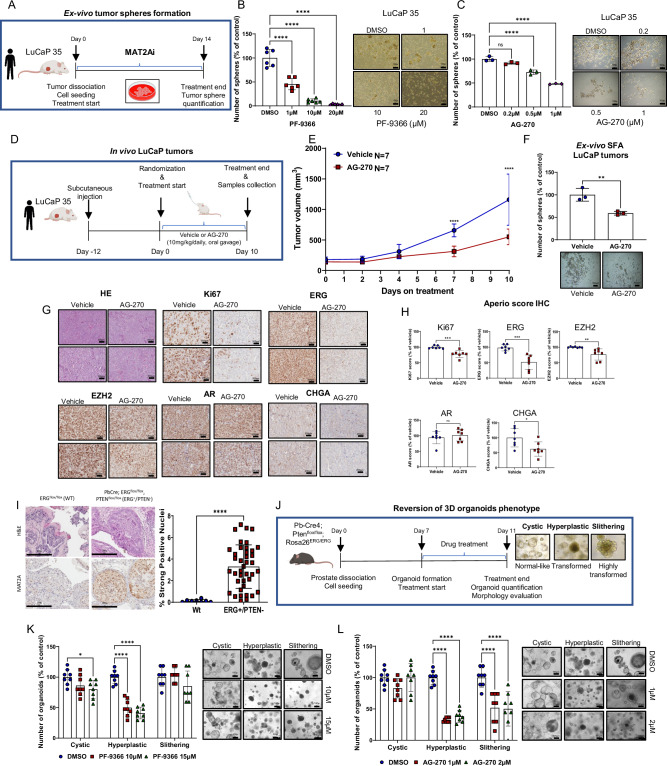
To date, MAT2A inhibitors are in clinical trials for treating tumors with genetic deletion of the methylthioadenosine phosphorylase (MTAP) gene, exploiting their increased vulnerability to SAM depletion due to inefficient methionine salvage pathways10,30,31. MTAP gene deletion occurs in approximately 15% of all cancers but is rare (≤0.01%) in prostate cancer (Supplementary Fig. 6A, B) and prostate cancer cell lines (Supplementary Fig. 6C). Our data, however, sustain that MAT2A could be an actionable target for CRPCs, particularly in those with ERG/EZH2 activation. To test this hypothesis, we used two chemical inhibitors of MAT2A, PF-9366 and AG-27032,33. Consistent with the genetic knockdown, both drugs strongly suppressed tumor sphere formation in VCaP cells, indicative of a substantial impact on stem-like tumor-initiating cells (Supplementary Fig. 6D, E). Consistent with the hypothesis of a direct link between MAT2A and ERG/EZH2 axis activation, MAT2A inhibition by both compounds, decreased ERG and EZH2 protein levels, and significantly reduced methylated ERG (mERG), the active form of ERG dependent on EZH2-induced lysine methylation (Supplementary Fig. 6F, left and right).
To evaluate the efficacy of this approach in a clinically relevant PC model, we used ex-vivo tumor spheroids generated from LuCaP 35, a patient-derived xenograft (PDX) established from a metastatic PC with the TMPRSS2:ERG gene fusion and PTEN deletion34 (Fig. 5A). Immunoblots and IHC showed high expression of both ERG and MAT2A in LuCaP 35 tumor xenografts (Supplementary Fig. 6G, H). Tumor spheroids generated from LuCaP 35 dissociated tumors are enriched of tumor-initiating stem-like cancer cells and represent excellent models to assess the efficacy of MAT2A inhibitors on prostate CSCs. Notably, PF-9366 and AG-270 inhibited the growth of LuCaP 35 tumor spheroids (Fig. 5B, C). Next, we assessed the impact of pharmacological inhibition of MAT2A in vivo in LuCaP 35 subcutaneously implanted in immunodeficient mice. Mice received either vehicle or AG-270 (10 mg/kg/daily) by oral gavage (Fig. 5D). Treatment with a daily low dose of AG-270 resulted in a significant tumor growth delay compared to the control group (Fig. 5E). Oral administration of AG-270 did not induce any toxic effect and did not affect mouse body weight (Supplementary Fig. 6I). Importantly, we found a significant reduction of the ex-vivo tumor-sphere forming ability of tumor cells dissociated from AG-270 treated xenografts compared to the control group (Fig. 5F). LuCaP 35 tumors in the AG-270 group showed a reduction of Ki-67 compared to the control group, which is compatible with the antitumor and anti-proliferative effect of the treatment (Fig. 5G). Relevantly, MAT2A inhibition by AG-270 significantly decreased ERG protein levels, supporting the impact of MAT2A on ERG levels and stability (Fig. 5G). We also found a significant decrease in EZH2 while the level of AR was unchanged. Notably, the expression of Chromogranin A, an NE marker, also decreased upon AG-270 treatment (Fig. 5G). These data supported a significant impact of the MAT2A inhibitor in vivo, particularly on the oncogenic ERG/EZH2 crosstalk.
Fig. 5. Pharmacologic inhibition of MAT2A reverses stemness in vitro and ex-vivo and significantly reduces growth of human PDX in vivo.
A Schematic representation of ex-vivo SFA assay, starting from LuCaP 35. B Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of PF-9366 Right, images of spheres at the indicated concentration (n = 6 biological replicates). Scale bars are 100 µm. C Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of AG-270. Scale bars are 100 µm. Right, images of spheres at the indicated concentration (n = 3 biological replicates). D Schematic representation of systemic in vivo treatment of LuCaP 35 tumors with vehicle or AG-270. E Growth curve of LuCaP 35 xenografts. Mice (n = 7/group) were treated by oral gavage with either vehicle or AG-270. Tumor growth was monitored every 2 days with caliper. F Ex-vivo SFA assay from explanted LuCaP 35 tumors (n = 3 tumors/group) after treatment with either vehicle or AG-270. Right, images of spheres of indicated treatment groups. Scale bars are 100 µm. G Representative sections from the indicated LuCaP tumors. Scale bars are 200 µm. H Immunoscore of Ki67, ERG, EZH2, AR and CHGA of tumor xenografts shown in (E) (n = 7 mice/group/;n = 7 biological replicates) using the Aperio tool. I Representative immunohistochemical evaluation of MAT2A (left) and quantification (right) in prostate tissue from WT (Pb-Cre;ERGflox/flox; n = 4 mice, n = 8 technical replicates) and ERG + /PTEN-(Pb-Cre4;Ptenflox/flox;Rosa26ERG/ERG; n = 6 mice, n = 40 technical replicates). Scale bars, 200 µm. J Schematic representation of 3D organoids assay derived from ERG/PTEN prostates to evaluate the reversion of the organoid phenotype. K Organoids treated with the indicated doses of PF-9366 (n = 8 biological replicates). Right, representative images are shown. Scale bars, 100 µm. L Organoids treated with the indicated doses of AG-270 (n = 8 biological replicates). Right, representative images are shown. Scale bars are 100 µm. All error bars, mean ± s.d. Ordinary one-way-ANOVA was used to test significant differences between groups in all panels, except for (K, L) where was used two-way-ANOVA. P-value was determined using unpaired t-test in F–H and I. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001. Panels A, B and J Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
We tested the efficacy of MAT2A inhibition in an additional preclinical model, tumor organoids derived from the Pb-Cre4;Ptenflox/flox;Rosa26ERG/ERG (ERG/PTEN) mice. These mice combine ERG overexpression and PTEN deletion and have a high degree of ERG/EZH2 activation5. Immunohistochemistry revealed that MAT2A was highly expressed in ERG/PTEN tumors and was enriched in the cell nuclei (Fig. 5I, left and right). Tumor organoids from ERG/PTEN mice reflect the malignant phenotype of their prostate tumors, generating cystic (normal-like) and more transformed full and hyperplastic structures (Fig. 5J). Following treatment with MAT2A inhibitors, we observed a significant reduction of dysmorphic and hyperplastic organoids (Fig. 5K, L), suggesting a reversion of the malignant cell phenotype. Relevantly, AG-270 significantly affected the formation of organoids with a slithering appearance, a phenotype that is linked to the ability to migrate and invade (Fig. 5L). Together, these data demonstrate the efficacy of MAT2A inhibitors in relevant in vitro and in vivo preclinical models of ERG fusion-positive prostate cancer.
MAT2A inhibitors reverse the transcriptional program of ERG fusion-positive CRPC cells
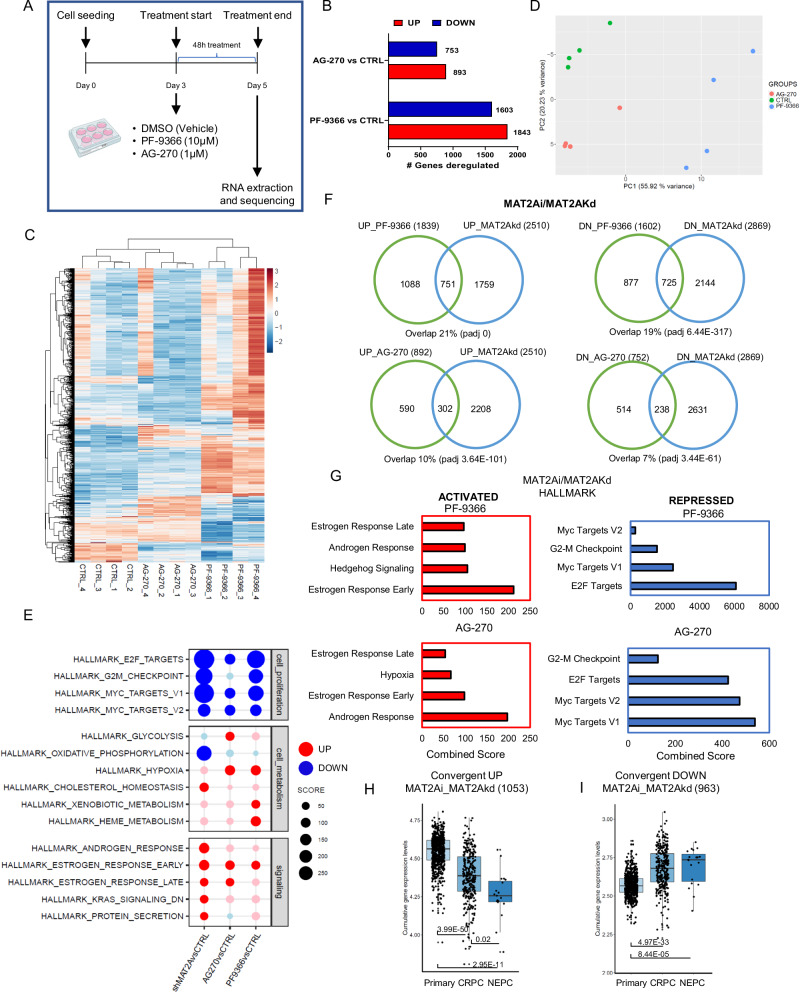
We assessed the transcriptional impact of MAT2A inhibition by performing RNA-seq in VCaP cells after the treatment with PF-9366 and AG-270 (Fig. 6A). Both compounds induced changes in the transcript levels of several genes (Fig. 6B). PCA and unsupervised clustering showed a substantial divergence between treated and control cells (Fig. 6C, D). Conversely, there was a significant overlap between the genes modulated by the two drugs, confirming the shared mechanism of action (Supplementary Fig. 7A). Functional annotation analysis showed the repression of pro-tumorigenic and proliferative pathways, like E2F and Myc targets, and the activation of steroid-responsive pathways with both compounds (Fig. 6E). We found a substantial similarity of the affected pathways between the chemical inhibitors and genetic depletion of MAT2A, supporting the compounds’ specificity and on-target activity. There was a significant sharing of the genes modulated by PF-9366 and AG-270 and MAT2A depletion (Fig. 6F). The common upregulated genes were functionally associated with steroid and androgenic responses (Fig. 6G). The convergent downregulated genes were related to proliferative and pro-tumorigenic pathways (Fig. 6G). Interestingly, the upregulated genes shared by drugs and genetic MAT2A knockdown were more expressed in primary tumors and repressed in both CRPCs and NEPCs (Fig. 6H, I).
Fig. 6. Pharmacological inhibition of MAT2A by PF-9366 and AG-270 reverses the malignant transcriptome.
A Schematic representation of the experimental procedure for RNA sequencing following drug treatment. B Number of genes deregulated in VCaP cells treated with PF-9366 or AG-270, compared to VCaP control cells. C Heatmap of differentially expressed genes in VCaP cells treated with either PF-9366 or AG-270, versus control VCaP cells. D PCA plot of VCaP cells, colored according to the conditions (CTRL, PF-9366 and AG-270). E Hallmark enrichment analysis of VCaP MAT2Akd (Sh1 and Sh2), VCaP treated with either PF-9366 or AG-270 versus respective control samples. Down-regulated pathways are colored in blue, up-regulated pathways are colored in red. F Convergence between MAT2Ai and MAT2Akd deregulated targets with threshold on Log2FC ± 0.1 (up-regulated and down-regulated genes as indicated). Top, overlap with genes deregulated by PF-9366. Bottom, overlap with genes deregulated by AG-270. G Functional annotation of convergent genes deregulated by AG-270, PF-9366 and MAT2Akd. Pathways upregulated and downregulated are shown in the left and right panel, respectively. Adjusted p-value ≤ 0.01. H Cumulative gene expression level of convergent up-regulated genes (1053) by PF-9366 and AG-270 and MAT2Akd in VCaP cells, in the cohort of primary, CRPC and NEPC patients. I Cumulative gene expression level of convergent down-regulated genes (963) by PF-9366 and AG-270, and MAT2Akd in VCaP cells, in the cohort of primary, CRPC and NEPC patients. Primary, n = 714; CRPC, n = 316; NEPC, n = 19. For box-and-whisker plots in H and I, the line inside the box shows the median value. The bounds of the box represent the 25th–75th percentiles, with whiskers at minimum and maximum values. One-way-ANOVA was used to test significant differences between groups. Panel A Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
These results were in line with the notion that MAT2A inhibition reverses the malignant phenotype by reprogramming the AR transcriptome from an androgen-independent to an androgen-dependent gene network. This is achieved by activating pro-differentiation canonical androgenic responsive genes and suppressing pro-tumorigenic androgen-independent genes modulating H3K4me2 genomic levels and distribution. In support of this hypothesis, treatment with MAT2A inhibitors significantly reduced the H3K4me2 level in VCaP (Supplementary Fig. 7B). We also found a substantial overlap between the genes modulated by the MAT2A inhibitors and the ERG/EZH2 co-bound and co-regulated targets (Supplementary Fig. 7C). Thus, consistent with the changes induced by the genetic MAT2A knockdown, MAT2A inhibitors affect the ERG/EZH2 oncogenic axis and the associated pathways. Overall, these data supported the efficacy and specificity of the two MAT2A inhibitors in targeting MAT2A-induced transcriptional alterations in ERG-positive CRPCs.
MAT2A inhibitors synergize with AR and EZH2 antagonists in CRPC preclinical models
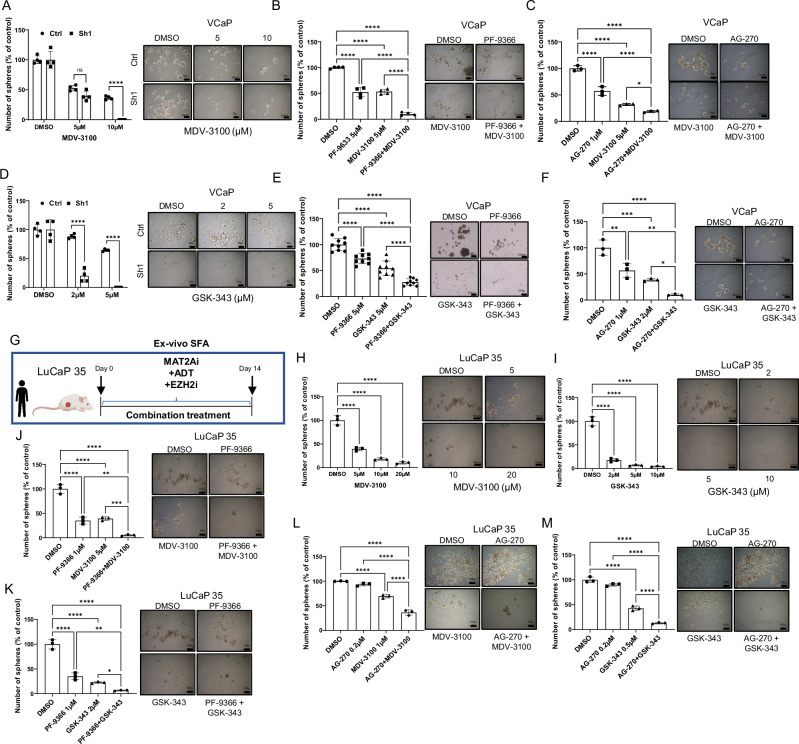
Based on the impact of MAT2A genetic and pharmacological inhibition on the AR and ERG/EZH2 gene network, we reasoned that MAT2A inhibitors could be highly effective in co-targeting strategies to prevent or reverse the progression to CRPC. Specifically, the central role of MAT2A in the crosstalk between ERG, EZH2, and AR, unveiled in this study, suggested that MAT2A inhibitors could disrupt these interactions and synergize with inhibitors of AR and EZH2. MAT2A inhibitors could reactivate canonical AR signaling and increase the sensitivity of CRPC cells to AR antagonists, like enzalutamide. Furthermore, inhibition of MAT2A could synergize with EZH2 inhibitors and suppress the activation of the ERG/EZH2 complexes in ERG fusion-positive cancer cells. Consistently, MAT2A genetic depletion (Fig. 7A) and inhibition by PF-9366 and AG-270 (Fig. 7B, C) increased the response of VCaP cells to the AR antagonist enzalutamide with a significant reduction of tumor sphere growth. In line with our hypothesis, MAT2A genetic depletion (Fig. 7D) and inhibition by PF-9366 and AG-270 also substantially enhanced the response of VCaP cells to the EZH2 inhibitor GSK-343 (Fig. 7E, F) in tumor sphere assays, supporting the relevance of the ERG/EZH2/MAT2A interaction in promoting the growth of stem-like cancer cells. We performed proliferation assays in normal prostate epithelial RWPE-1 cells. Increasing doses of PF-9366 and AG-270 did not have any effect on normal cell proliferation (Supplementary Fig. 7D), supporting the selectivity of MAT2A inhibitors toward cancer cells with high expression of MAT2A.
Fig. 7. Pharmacologic inhibition of MAT2A in ERG positive models reverses stemness and restores the sensitivity to enzalutamide and EZH2 inhibition.
A SFA in VCaP MAT2Akd cells with the indicated concentration of enzalutamide (MDV-3100) (n = 4 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 50 µm. B SFA in VCaP cells under treatment with the indicated concentration of PF-9366 and MDV-3100, alone or in combination (n = 4 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 50 µm. C SFA in VCaP cells under treatment with the indicated concentration of AG-270 and MDV-3100, alone or in combination (n = 3 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 50 µm. D SFA in VCaP MAT2Akd cells with indicated concentration of GSK-343 (n = 4 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 50 µm. E SFA in VCaP cells under treatment with the indicated concentration of PF-9366 and GSK-343, alone or in combination (n = 3 independent experiments, with 3 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 200 µm. F SFA in VCaP cells under treatment with the indicated concentration of AG-270 and GSK-343, alone or in combination (n = 3 biological replicates for each drug concentration). Right, representative images of spheres. Scale bars are 50 µm. G Schematic representation of ex-vivo SFA assay, starting from LuCaP 35. H Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of MDV-3100 (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars are 100 µm. I Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of GSK-343 (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars, 100 µm. J Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of PF-9366 and MDV-3100, alone and in combination (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars, 100 µm. K Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of PF-9366 and GSK-343, alone and in combination (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars are 100 µm. L Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of AG-270 and MDV-3100, alone and in combination (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars, 100 µm. M Ex-vivo SFA from dissociated LuCaP 35 under treatment with the indicated concentration of AG-270 and GSK-343, alone and in combination (n = 3 biological replicates). Right, images of spheres at the indicated concentration. Scale bars are 100 µm. All error bars, mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001. ns = no significant. One-way-ANOVA was used to test significant differences between groups in all panels, except for A and D where two-sided t-test was used. Panel G Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
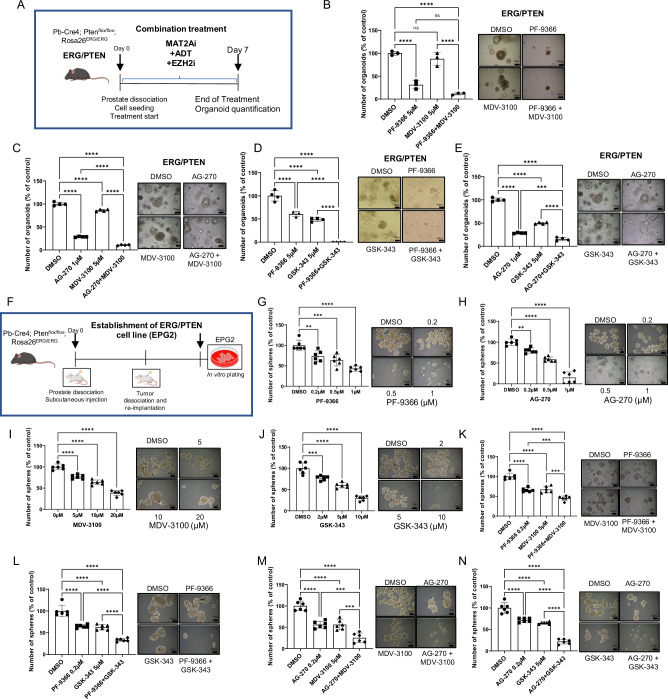
Next, we tested the MAT2A, AR and EZH2 inhibitors in ex-vivo generated tumor spheres of the LuCaP 35 PDX (Fig. 7G). Enzalutamide and GSK-343 had a relevant effect on LuCaP 35 tumor spheroids (Fig. 7H, I). Nevertheless, the MAT2A inhibitor PF-9366 (Fig. 7J, K) and AG-270 (Fig. 7L, M) combined with enzalutamide or GSK-343 had a synergistic effect. Primary organoid cultures established from ERG/PTEN mouse prostates were also incubated with PF-9366 and AG-270 alone or in combination with enzalutamide and GSK-343 (Fig. 8A). We observed a significant reduction of ERG/PTEN organoids in the presence of PF-9366 and AG-270 (Fig. 8B–E). Moreover, there was a substantially enhanced response by combining PF-9366 and AG-270 with the AR antagonist enzalutamide (Fig. 8B, C) or the EZH2 inhibitor GSK-343 (Fig. 8D, E).
Fig. 8. Pharmacological inhibition of MAT2A alone and in combination significantly reverse malignant phenotype and restore drug sensitivity in ERG/PTEN preclinical models.
A Schematic representation of 3D organoid formation assay, starting from ERG/PTEN mouse prostates. B Organoids formation from dissociated prostates of ERG/PTEN mice under treatment with the indicated concentration of PF-9366 and MDV-3100, alone and in combination (n = 3 biological replicates). Right, images of organoids at the indicated concentrations. C Organoids formation from dissociated prostates of ERG/PTEN mice under treatment with the indicated concentration of AG-270 and MDV-3100, alone and in combination (n = 4 biological replicates). Right, images of organoids at the indicated concentrations. D Organoids formation from dissociated prostates of ERG/PTEN mice under treatment with the indicated concentrations of PF-9366 and GSK-343, alone and in combination (n = 4 biological replicates). Right, images of organoids at the indicated concentrations. E Organoids formation from dissociated prostates of ERG/PTEN mice under treatment with the indicated concentrations of AG-270 and GSK-343, alone and in combination (n = 4 biological replicates). Right, images of organoids at the indicated concentrations. F Schematic representation of the generation of ERG/PTEN prostate-derived cell line (EPG2). G SFA assay in EPG2 cells treated with indicated concentration of PF-9366 (n = 6 biological replicates). Right, representative images are shown. H SFA assay in EPG2 cells treated with indicated concentration of AG-270. Right, representative images are shown (n = 6 biological replicates). I SFA assay in EPG2 cells treated with indicated concentration of MDV-3100 (n = 6 biological replicates). Right, representative images are shown. J SFA assay in EPG2 cells treated with indicated concentration of GSK-343 (n = 6 biological replicates). Right, representative images are shown. K SFA assay in EPG2 cells treated with indicated concentration of PF-9366 and MDV-3100, alone and in combination (n = 6 biological replicates). Right, representative images are shown. L SFA assay in EPG2 cells treated with indicated concentration of PF-9366 and GSK-343, alone and in combination (n = 6 biological replicates). Right, representative images are shown. M SFA assay in EPG2 cells treated with indicated concentration of AG-270 and MDV-3100, alone and in combination (n = 6 biological replicates). Right, representative images are shown. N SFA assay in EPG2 cells treated with indicated concentration of AG-270 and GSK-343, alone and in combination (n = 6 biological replicates). Right, representative images are shown. Scale bars indicated in all images are 50 µm. All error bars, mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001, two-sided t test. ns = no significant. One-way-ANOVA was used to test significant differences between groups in all panels. Panels A and F Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
To further explore the efficacy of these MAT2A-based combinations, we took advantage of a murine cell line (EPG2) established in our laboratory from the prostatic tumors generated in ERG/PTEN mice and stabilized by in vivo passaging and then subculturing in vitro (Fig. 8F). The murine EPG2 cells are highly tumorigenic, are enriched tumor-initiating stem-like cells, and grow in vitro as tumor spheroids. Furthermore, like the original ERG/PTEN tumors, EPG2 cells are ERG positive and PTEN negative and express high levels of MAT2A (Supplementary Fig. 8A). MAT2A inhibitors counteracted ERG and EZH2 activation in EPG2 cells, drastically reducing ERG, mERG, and EZH2 levels (Supplementary Fig. 8B). Importantly, we demonstrated the MAT2A and ERG interaction in EPG2 cells by co-immunoprecipitation (Supplementary Fig. 8C). Additionally, MAT2A inhibition with AG-270 strongly reduced the H3K4me2 level (Supplementary Fig. 8D). These data further confirmed the importance of MAT2A in promoting ERG-induced transcriptional reprogramming. PF-9366 and AG-270 affected tumor sphere growth (Fig. 8G, H) and colony formation (Supplementary Fig. 8E, F), consistent with high vulnerability of these cells to MAT2A inhibition. Notably, EPG2 cells displayed an AR-independent phenotype and were only moderately sensitive to enzalutamide and GSK-343 in tumor sphere (Fig. 8I, J) and colony-forming (Supplementary Fig. 8G, H) assays. Combined therapy with MAT2A inhibitors with either enzalutamide (Fig. 8K, L and Supplementary Fig. 8I, J) or GSK-343 (Fig. 8M, N and Supplementary Fig. 8K, L) led to a substantially greater response than each treatment alone. Thus, the activation of ERG and the consequent MAT2A-assisted reprogramming of the EZH2 and AR gene networks render these cells particularly sensitive to MAT2A inhibition.
Collectively, these data support the efficacy of targeting MAT2A to revert tumorigenic and stem-like properties in different human and mouse models of ERG fusion-positive CRPC. Moreover, the combination of MAT2A inhibitors with AR antagonists and EZH2 inhibitors may represent an effective strategy for treating advanced CRPC. Blocking MAT2A reverts the transcriptional and epigenetic reprogramming linked to cancer cell stemness and plasticity that underlies the evolution of PC to CRPC.
High MAT2A expression confers sensitivity to MAT2A inhibitors in ERG-negative CRPC
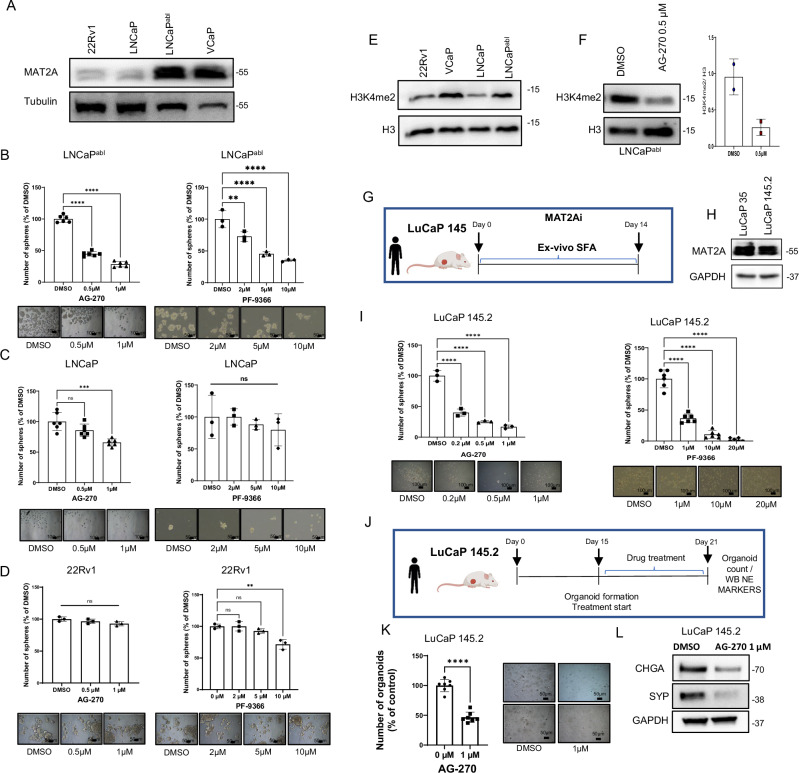
Our data support the efficacy of targeting MAT2A in human and mouse CRPC models. All the models tested so far had the TMPRSS2:ERG gene fusion and expressed high levels of both ERG and MAT2A. Next, we aimed to understand the efficacy and specificity of this approach independently of ERG gene fusion. Indeed, a significant number of advanced PCs had high levels of MAT2A in the absence of ERG overexpression. To this end, we examined the level of MAT2A protein expression in a panel of ERG fusion-negative cell lines (Fig. 9A). Interestingly, the castration-resistant LNCaPabl cells had significantly higher expression of MAT2A compared to the castration-sensitive LNCaP cells (Fig. 9A), supporting the notion of MAT2A upregulation also in ERG-negative CRPC. Next, we tested the effects of MAT2A inhibitors in LNCaP and LNCaPabl cells as representative models of ERG-negative castration-sensitive (MAT2Alow) and castration-resistant counterpart (MAT2Ahigh) prostate cancer, respectively. MAT2A inhibitors significantly reduced the growth of tumor spheres of LNCaPabl cells (Fig. 9B), whereas LNCaP cells were minimally affected (Fig. 9C). We also tested the response of 22RV1 cells. These CRPC cells are ERG-negative and have a low level of MAT2A (Fig. 9A). 22RV1 cells were not affected by the MAT2A inhibitors (Fig. 9D). Together, these data support the association of MAT2A with the CRPC phenotype and the vulnerability of MAT2Ahigh-expressing CRPC cells to its inhibition, even in the absence of ERG gene fusion. Relevantly, the level of H3K4me2 histone mark correlated with MAT2A expression in these cell lines. Specifically, H3K4me2 was higher in the castration-resistant LNCaPabl cells compared to LNCaP and 22RV1 cells (Fig. 9E). Furthermore, inhibiting MAT2A in LNCaPabl cells reduced the H3K4me2 level, which is consistent with the specific role of MAT2A in controlling this histone modification in CRPC cells (Fig. 9F).
Fig. 9. High MAT2A expression confers sensitivity to MAT2A inhibitors in ERG-negative CRPC.
A Immunoblot of MAT2A in ERG positive and negative cell lines (n = 2 independent experiments). B SFA from LNCaPabl cells under treatment with the indicated concentration of AG-270 (Left) (n = 6 biological replicates for each drug concentration) or PF-9366 (Right) (n = 3 biological replicates for each drug concentration). Bottom, images of spheres at the indicated concentration. C SFA from LNCaP cells under treatment with the indicated concentration of AG-270 (Left) (n = 6 biological replicates for each drug concentration) or PF-9366 (Right) (n = 3 biological replicates for each drug concentration). Bottom, images of spheres at the indicated concentration. D SFA from 22Rv1 cells under treatment with the indicated concentration of AG-270 (Left) (n = 3 biological replicates for each drug concentration) or PF-9366 (Right) (n = 3 biological replicates for each drug concentration). Bottom, images of spheres at the indicated concentration. E Immunoblot of H3K4me2 in ERG positive and negative cell lines (n = 2 independent experiments). F Immunoblot with indicated antibodies in LNCaPabl cells after treatment with the indicated dose of AG-270. Right, densitometry analysis of the indicated markers (n = 2 independent experiments). G Schematic representation of ex-vivo SFA from dissociated patient-derived xenograft LuCaP 145.2. H Immunoblot of MAT2A in patient-derived xenograft LuCaP 145.2 (n = 2 independent experiments). I Ex-vivo SFA from dissociated LuCaP 145.2 under treatment with the indicated concentration of AG-270 (Left) (n = 3 biological replicates for each drug concentration) or PF-9366 (Right) (n = 6 biological replicates for each drug concentration). Bottom, images of spheres at the indicated concentration. J Schematic representation of organoid establishment from LuCaP 145.2 treatment with AG-270, and evaluation of NE markers. K Number and images of organoids at the indicated concentrations (n = 7 biological replicates). L Immunoblot of NE markers in LuCaP 145.2 organoids treated with AG-270. Molecular weights are expressed in kDa (n = 2 biological experiments). Scale bars indicated in all images are 50 µm except for B and I which are 100 µm. All error bars, mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001. ns = no significant. One-way-ANOVA was used to test significant differences between groups in all panels, except for K where two-sided t-test was used. Panels G and J Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Therefore, high MAT2A expression confers increased vulnerability to MAT2A inhibitors in various CRPC models. Next, we also investigated the impact of MAT2A inhibitors in an NE-CRPC model using tumor-spheroids derived ex vivo from LuCaP 145.2 patient-derived xenografts (Fig. 9G). Interestingly, LuCaP 145.2 xenografts had high expression of MAT2A (Fig. 9H). Treatment of LuCaP 145.2-derived tumor spheroids with MAT2A inhibitors reduced their growth (Fig. 9I). AG-270 reduced the number of tumor organoids generated by LuCaP 145.2 cells and, concomitantly, the expression of the NE markers CHGA and SYN (Fig. 9J–L). We also evaluated MAT2A expression and the impact of MAT2A inhibition in the NCI-H660 cells. These cells are ERG gene fusion-positive but have NE features and express low levels of ERG and AR. Notably, NCI-H660 expressed high levels of MAT2A (Supplementary Fig. 9A), and inhibiting MAT2A reduced tumor sphere growth (Supplementary Fig. 9B). Concomitantly, the expression of canonical NE markers was reduced by MAT2A inhibitors (Supplementary Fig. 9C). Intriguingly, AG270 also decreased the expression of these NE markers in VCAP cells (Supplementary Fig. 9D), in line with the data in MAT2Akd cells and suggesting a link between MAT2A, stem cell plasticity and phenotypic transitions in advanced PC. These data confirm the relevance of MAT2A in promoting cancer cell stemness and the evolution to castration resistance in the context of both AR-dependent and independent phenotypes and the vulnerability of CRPCs with high MAT2A expression to MAT2A inhibitors.
Discussion
Understanding the events promoting and sustaining the androgen-indifferent status is highly relevant due to the increasing frequency of therapy-resistant prostate tumor subtypes emerging under prolonged ADT and ARPI treatment2. Research in this area can create new opportunities for discovering effective therapeutic strategies to reverse or prevent lethal prostate cancers. Profound changes in the epigenetic and transcriptional landscape are associated with the acquisition of aggressive, metastatic, and treatment-resistant traits25,35–40. Epigenetic and transcriptional regulators are potentially excellent targets for developing new therapeutic strategies, alternative or complementary to AR-targeted drugs25,35–40.
An essential aspect of the rewiring of the epigenetic landscape in cancer cells is the need for a constant supply of the metabolites acting as substrates and cofactors, like acetyl-CoA or S-adenosylmethionine (SAM), to sustain the activity of the chromatin-modifying enzymes41,42. SAM is the universal methyl donor for the methylation of histone and non-histone proteins, DNA and RNA11,43,44. The intracellular level of SAM affects many cellular functions and the cell epigenome27,45,46. Notably, SAM biosynthesis is essential to maintain the self-renewal capability of normal stem cells and tumor-initiating stem-like cancer cells43,44,46,47. Accordingly, inhibition of SAM biosynthesis and methionine dietary restriction elicit antitumor effects in preclinical tumor models and may be an effective treatment strategy32,47–53.
MAT2A, the catalytic subunit of the S-adenosylmethionine synthetase 2, catalyzes the formation of SAM from methionine and ATP11. Increasing the SAM availability, MAT2A contributes actively to the epigenetic and transcriptional reprogramming in cancer cells10. Furthermore, MAT2A, present in the cell nucleus, can physically interact with epigenetic effector complexes and directly respond to the demands of DNA and chromatin-modifying enzymes54. This study identifies the role of MAT2A as a critical factor sustaining the epigenetic reprogramming associated with the androgen-indifferent state in prostate tumors and an actionable target to reverse CRPC.
Our data indicate that a) MAT2A is upregulated and associated with castration resistance in experimental models and clinical samples of ERG fusion-positive CRPCs; b) genetic ablation of MAT2A impairs stem-like capability of CRPC cells in vitro and in vivo; c) MAT2A affect the transcriptional and epigenetic landscape in ERG fusion-positive CRPC models; d) MAT2A enhances selectively H3K4me2, a histone modification favoring the activation of non-canonical AR target genes in androgen-independent CRPC; e) MAT2A interacts with ERG and reprograms the ERG, EZH2 and AR regulated gene networks; f) MAT2A inhibitors recapitulate the phenotypic and transcriptional effects of genetic ablation of MAT2A in CRPC cells; g) MAT2A inhibitors reverse tumor growth and malignant phenotype in relevant preclinical models of CRPCs; h) MAT2A inhibitors synergize with AR and EZH2 inhibitors in CRPC models.
We investigated the consequence of MAT2A depletion on the transcriptional and chromatin state in VCaP cells. Transcriptomic profiling revealed that MAT2A promotes the expression of pro-tumorigenic and stem cell-related genes while repressing canonical androgen-responsive and pro-differentiation genes. We found a striking association between the MAT2A-modulated genes and the clinical stage and subtype in PC patients, in line with a role in tumor progression to a castration-resistant state. These data were consistent with the hypothesis that MAT2A contributes to enhanced stemness and plasticity, driving prostate cancer cells toward de- and trans-differentiation and phenotypic transitions.
MAT2A depletion altered the epigenetic landscape and chromatin organization in VCaP cells, likely due to its influence on epigenetic processes27. Notably, the methylation of histones has a critical role in transcriptional regulation and chromatin organization55. We found many sites with increased chromatin accessibility in MAT2A-depleted cells. The enhanced accessible sites were prevalent (63%) in gene promoters, with fewer sites exhibiting reduced accessibility mainly (50%) in the gene body and distal to the TSS. Interestingly, integrating ATAC and RNA-seq data, we found that regions with increased accessibility were associated with both upregulated and downregulated genes. Whereas the increased chromatin accessibility would favor the binding of positive transcriptional regulators at activated genes, different mechanisms might be in play to explain the transcriptional repression. Intriguingly, the AR locus was one of the most affected genomic regions with multiple open chromatin sites. The increased chromatin accessibility in the AR locus was associated with reduced AR transcript levels in MAT2A-depleted cells. The CDK1 gene locus also had multiple open chromatin sites and was transcriptionally repressed in MAT2A-depleted cells. The co-occurrence of opened chromatin and gene silencing seemed contradictory. However, MAT2A upregulation might favor an aberrant deposition of histone marks and enhance AR gene transcription. Conversely, MAT2A depletion might restore a more normal-like pattern of histone modifications, favoring the recruitment of transcriptional repressors or the eviction of transcriptional activators, with consequent inhibition of the gene transcription24–26.
To address these questions, we performed an MS-based analysis of histone PTMs in control and MAT2A-depleted cells, which revealed a striking decrease of H3K4me2 upon MAT2A silencing. This finding is particularly intriguing. H3K4me2, along with H3K4me1, is a marker of active enhancers as opposed to H3K4me3, which marks active promoters56. H3K4me2 labels FoxA1 binding sites at cell type-specific enhancers57. Moreover, H3K4me2 was selectively enriched at enhancers controlling a set of pro-tumorigenic genes (e.g., CDK1) in androgen-independent CRPC cells but not in androgen-dependent PC models28. Intriguingly, H3K4me2 accumulation promoted Foxa1 binding and AR recruitment at these enhancers, leading to AR-dependent and androgen-independent expression of the pro-tumorigenic genes in CRPC cells28. Thus, the increased H3K4me2 distribution to non-canonical AR enhancers underlies the activation of the aberrant AR-driven transcriptional program in CRPCs28.
Accordingly, our histone PMT analysis and ChIP-seq data indicate that MAT2A promotes H3K4me2 accumulation and the acquisition of H3K4me2 marks at specific genomic locations in ERG fusion-positive CRPC tumors. The sites of H3K4me2 deposition could be defined by the functional interaction of MAT2A with ERG and EZH2, which repurpose the canonical pro-differentiation AR program towards the activation of pro-tumorigenic non-canonical AR-target genes. One of the relevant findings of the study is that MAT2A helps ERG to reprogram both the EZH2 function and AR gene network in ERG fusion-positive tumor cells. ERG overexpression alters the AR-regulated cistrome, driving a shift toward pro-proliferative and pro-tumorigenic target gene activation17,18. Here, we show that MAT2A cooperates with ERG to orchestrate the repression of genes, like KLK3, NKX3.1 and TMPRSS2, functionally related to the canonical androgen response pathway. MAT2A depletion reversed this pattern, modifying the transcriptional and chromatin profile and leading to the preferential reactivation of canonical androgen-responsive genes and repression of AR-regulated androgen-independent genes. Thus, MAT2A depletion favors a more physiologic, normal-like regulation of the AR signaling, modulating AR gene expression and function and promoting a rearrangement of chromatin and histone modifications. Moreover, inhibiting MAT2A affects AR downstream signaling and redirects AR to canonical target genes, reversing the aberrant transcriptional program activated in ERG fusion-positive tumors.
Collectively, our data point to MAT2A as a driver of castration resistance and an actionable therapeutic target. The availability of MAT2A inhibitors with a good profile and in an advanced stage of development renders these findings highly attractive for clinical translation of this concept. Various MAT2A inhibitors are currently available, with at least two compounds (AG-270 and IDE-397) in clinical trials for patients with advanced cancers but not with prostate cancers10,30,32. The tolerability of these drugs has been excellent, with minimal toxicity and a good safety profile. We show here that chemical inhibitors of MAT2A, like PF-9366 and AG-270, recapitulate the phenotypic effects of genetic MAT2A ablation in ERG fusion-positive PC cells, reducing CSC proliferation and tumor growth. Significantly, MAT2A inhibitors reduced the H3K4me2 mark, recapitulating the effect of the genetic knockdown. At the transcriptional level, there was a consistent overlap of the genes modulated by chemical inhibitors and MAT2A depletion, demonstrating their specificity and efficacy. The upregulated genes were androgen-responsive and functionally related to cell differentiation pathways, consistent with the reactivation of canonical AR signaling. In contrast, the downregulated genes belonged preferentially to growth-promoting and pro-tumorigenic pathways. We also found a substantial overlap between the genes modulated by the MAT2A inhibitors and the ERG/EZH2 co-regulated genes, confirming that MAT2A also influences the ERG/EZH2 oncogenic axis. Inhibiting MAT2A also resulted in reduced levels of ERG and K362 methylated ERG (mERG), the oncogenic active form of ERG in ERG fusion-positive PC5. These results were in line with the notion that MAT2A inhibition reprograms the ERG-orchestrated transcriptional program globally and is a promising approach for inhibiting ERG oncogenic activity.
Currently, the clinical development of MAT2A inhibitors has focused on tumors with MTAP gene deletion. Genetic deletion of MTAP is frequent in some tumor types and occurs in approximately 15% of all cancers10,30,31. The synthetic lethality effect caused by MAT2A inhibition in tumor cells is due to an inefficient methionine salvage pathway in MTAP-deleted cells. MTAP deletion is infrequent in prostate tumors, and prostate cancer patients were not included in past clinical trials. A relevant finding of this study is the demonstration of the high efficacy of MAT2A inhibitors in various PC models independent of the MTAP deletion. We show that CRPC models with high MAT2A expression are highly susceptible to MAT2A inhibitors, confirming the relevance of MAT2A in the evolution of PCs toward castration resistance in the context of both AR-dependent and independent phenotypes.
ERG fusion-positive CRPCs with enhanced ERG/EZH2 activation require MAT2A to sustain the ERG oncogenic program and are particularly vulnerable to MAT2A inhibition. ERG promotes the expression of MAT2A and physically and functionally interacts with MAT2A. Inhibiting MAT2A blocks the consequences of ERG overactivity in ERG fusion-positive CRPC cells. MAT2A inhibition counteracts the ERG/EZH2 functional cooperation, methylation-dependent ERG activation, histone H3K4me2 mark deposition, enhancers reprogramming, deregulation of the AR gene network, and the expression of genes associated with stemness, cell plasticity, and AR-independence. These findings suggest that clinical testing of the currently available MAT2A inhibitors should include patients with prostate cancer based on their tumor phenotypic characteristics. ERG fusion, along with MAT2A and EZH2 expression, could potentially be used as predictive biomarkers to select CRPC patients more likely to respond to MAT2A inhibitors in clinical studies. We also show that, in ERG-positive models, the inhibition of MAT2A has an immediate impact on ERG methylation and protein level, which could serve as effective biomarkers for pharmacodynamic studies assessing treatment response. In this context, we also explored combinatorial strategies with the MAT2A-targeted compounds and showed that MAT2A inhibition enhances the response to AR antagonists and EZH2 inhibitors in ERG fusion-positive CRPC models. In both human and murine models, we found that the combination of MAT2A inhibitors with either AR antagonists or EZH2 inhibitors had a greater effect in restraining tumor sphere formation and clonogenic capability than the single treatments.
In conclusion, our study supports the role of MAT2A and the feasibility and efficacy of targeting MAT2A alone or in combination with AR and EZH2 inhibitors for treating CRPCs. We have discovered an oncogenic axis connecting ERG and MAT2A, demonstrated its impact on prostate tumorigenesis, and showed how to counteract its consequences by disrupting MAT2A-ERG interaction with inhibitors of MAT2A in ERG-positive tumors. These results represent an important step toward understanding and managing the progression of PC to lethal castration-resistant tumors.
Methods
Our research complies with all relevant ethical regulations. Animal handling was carried out according to the protocols approved by the Swiss Cantonal Veterinary Authority (TI29/19, TI44/23, and TI04/20).
Statistic and reproducibility
All assays were performed in at least three independent experiments with sufficient biological and technical replicates to demonstrate statistical significance. Specifically, for in vitro and in vivo studies, no statistical method was used to predetermine sample size, and no data were excluded from the analyses. Sample sizes were determined based on pilot studies and previous similar studies with similar biologic conditions. For in vivo studies: animal experiments were conducted with 4–7 mice per groups. These sample sizes in previous and similar studies have given statistically significant results based on the variance of xenograft growth in control mice. For ethical reasons, the minimum number of animals necessary to achieve the scientific objectives was used. Statistical analyses were performed using GraphPad Prism (version 7.00) software. Unpaired two-tailed Student’s t-tests were used to analyze two unpaired samples. One-way ANOVA was used to analyze multiple unpaired samples. Paired two-tailed Student’s t-test was performed to analyze the statistical significance of matched tissue samples. Error bars represent mean ± SD. To evaluate the significance of overlap among gene lists we applied the hypergeometric test, considering the total number of genes that were tested for differential expression as background.
Cell culture
RWPE-1, VCaP, LNCaP and 22Rv1 cells were purchased from American Type Culture Collection and maintained in their respective media. LNCaPabl were purchased from Cellosaurus (cat. CVCL_4793). RWPE1 cells were cultured in keratinocyte serum-free growth medium (Gibco, cat. 131500 A) supplemented with human recombinant Epidermal Growth Factor (rEGF) (Gibco, cat. 10450-013) and Bovine Pituitary Extract (BPE) (Gibco, cat.13028-014). VCaP were cultured in DMEM (ATCC, cat. 30-2002) supplemented with 10% fetal bovine serum (FBS, Biowest, cat. S181B) and 1% Pen/Strep (Thermo Fisher, cat. 15070063). LNCaP cells were cultured in RPMI-1640 medium (Gibco™ 21875034), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Pen/Strep). LNCaPabl58, were maintained in RPMI-1640 no-phenol red, supplemented with 10% charcoal-stripped serum (CSS) and 1% Pen/Strep. Cells were regularly checked for mycoplasm contamination using MycoAlert Mycoplasma detection kit [Lonza]. UGSM-2 cells were purchased from Cellosaurus (cat. CVCL_LF83). UGSM-2 cells were maintained in DMEM supplemented with 5% FBS, 5% NuSerum IV, 1x glutamine, 1x penicillin/streptomycin solution, 0.01 μM (final concentration) of Dihydrotestosterone, 25ug/ml insulin. NCI-H660 were purchased from ATCC (CRL-5813). NCI-H660 cells were maintained in RPMl-1640 medium (cat. n. 30-2001), 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nM sodium selenite, 10 nM beta-estradiol, 10 nM hydrocortisone, extra 10 mM HEPES (final conc. of 20 mM), extra 2 mM L-glutamine (for final concentration. of 4 mM), 5% FBS. For cell line authentication, ATCC uses PCR based approaches, karyotyping, and morphology to confirm the identity of human cell lines. For LNCAPabl and UGSM-2 (Cellosaurus), authentication was performed by STR.
Establishment of stable cell lines
VCaP with stable knockdown of MAT2A were generated by viral infection of control (Ctrl) and MAT2A shRNAs (Sh1 and Sh2) (Sigma-Aldrich; Supplementary Table 1), followed by subsequent selection with Puromycin (Sigma-Aldrich). RWPE-1 cells were stably transfected with expression plasmids for ERG (kindly provided by S. Izraeli)5 and MAT2A (100521, Addgene) (Supplementary Table 1) using JetPRIME (Polyplus), according to manufacturer instructions.
Generation of ERG/PTEN prostate-derived cell line
To generate ERG/PTEN prostate-derived cell line (EPG2), we dissociated the prostates of three Pb-Cre4;PTENflox/flox;Rosa26ERG/ERG (ERG/PTEN) mice of 38 weeks of age. Prostates were dissected and resuspended in RPMI 16040 medium (Gibco™ 21875034), supplemented with Collagenase D (cat. 11088866001, Roche) for 45 minutes. Cell suspension was passed through a 40μm cell strainer to collect single cells. 2 × 106 of cells were then subcutaneously injected in NRG (NOD-Rag2-IL2rgTm1/Rj) mice with Matrigel (1:1). Tumor growth was monitored every 2 days with caliper. When tumors reached the maximum volume of 1000-1500mm3, mice were euthanised. Tumor xenografts were dissociated as described and 2 × 106 of cells were again subcutaneously injected in NRG mice with Matrigel (1:1). Tumor growth was monitored every 2 days with caliper. When tumors reached the maximum volume of 1000–1500mm3, mice were euthanised. Tumor xenografts were dissociated and cells were plated in 2D culture conditions in DMEM medium, supplemented with 10% FBS and 1% Pen/Strep. EPG2 cells were maintained in DMEM (ATCC, cat. 30-2002) supplemented with 10% fetal bovine serum (FBS, Biowest, cat. S181B) and 1% Pen/Strep (Thermo Fisher, cat. 15070063). Cells were regularly checked for mycoplasm contamination using MycoAlert Mycoplasma detection kit [Lonza].
RNA Extraction, Realtime PCR and expression analysis
Total RNA was extracted using Trizol (Invitrogen) and Direct-zol RNA-MiniPrep kit (Zymo Research), according to manufacturer’s instructions. qRT-PCR was performed using 20 ng of RNA as template for SYBR Green Fast One-step kit (Qiagen). RNA concentration was obtained by spectrophotometric analysis with Nanodrop 2000 [Thermo Fisher Scientific, cat. #ND-2000]. The level of each gene was calculated by normalizing the Ct value in the samples to the amount of β-actin (Supplementary Table 1). StepOneplus software was used to analyze the data.
RNA sequencing
For VCaP MAT2Akd cells and RWPE-1 cells over-expressing ERG and MAT2A, isolated RNA was marked with Illumina total prep 96 RNA amplification kit (Ambion). RNA sequencing was performed using Next Ultra II Directional RNA Library Prep Kit for Illumina starting from 800 ng of total RNA from each sample and sequenced on the Illumina NextSeq500 with single-end, 75 base pair long reads. Sequencing reads were aligned to the GRch38.p12 release of the human genome and quantified using STAR aligner59 and the quality was accessed through FastQC tool60. RNA-Seq analysis was carried out using the DESeq2 pipeline61 in R environment62. The Benjamini-Hochberg multiple test correction method was adopted.
For VCaP cells treated with MAT2A inhibitors (PF-9366 and AG-270), NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs Inc.) was employed with the NEBNext Multiplex Oligos for Illumina (New England BioLabs Inc.) and NEBNext® Poly(A) mRNA Magnetic Isolation Module for cDNA synthesis with the addition of barcode sequences. The sequencing of the pre-pools was performed using the NextSeq2000 (Illumina, San Diego, CA, USA) with the P2 reagents kit V3 (100cycles; Illumina). Samples were processed starting from stranded, single-ended 120bp-long sequencing reads.
In vitro treatment of VCaP cells with MAT2A inhibitors for RNA sequencing
For RNA-sequencing, 2 × 106 VCaP cells were plated in each well of a 6-well plate. After 72 h, cells were treated with either 10 µM PF-9366 (cat. HY-107778, MedChemExpress) or 1 µM AG-270 (cat. HY-138630, MedChemExpress). DMSO was used as control. Cell were collected after 48 h of treatment and RNA extraction was performed as described in the previous paragraph.
Downstream analysis of transcriptomic data
PCA and heatmap images were made with pcaplot and pheatmap R functions. Enrichment analysis was performed with the cameraPR function from limma package63: the accounted genes and proteins were weighted (statistic parameter) according to the Wald statistic (from “results” function by DESeq2 package). To functionally annotate lists of genes or proteins enrichR package was adopted64. In case of multiple comparisons, the Benjamini-Hochberg correction method was used.
ATAC-sequencing
The experimental design includes 4 replicates per condition (Control and shMAT2A n.1). Sample preparation involves the collection of 50,000 cells per replicate and resuspension of the pellet in TRIzol buffer. The library was prepared according to guidelines reported in Buenrostro et al.65. ATAC-sequencing was performed on the Illumina NextSeq500 with single-end, 75 base pair long reads. The overall quality of sequencing reads was determined using FastQC tool60.
Downstream analysis of ATAC-seq data
The analysis of data was carried out in 3 steps: Pre-processing, Core-Analysis, Downstream analysis. The pre-processing phase involves several steps including: Trimming, using TrimGalore66 to remove low-quality bases from sequence reads; alignment, using Bowtie267 for aligning sequencing reads to the reference human genome (GRCh38); removal of ChrM and blacklist regions (refer to encodeproject), using SAMtools68 and BEDtools69, respectively. The Core-Analysis was conducted in a bash environment removing PCR duplicates with SAMtools68. Furthermore, peak calling and peak count were performed using Genrich70 and featureCounts71, respectively. The downstream analysis was conducted in R environment62 using DESeq61, Homer72 and enrichR64 packages in order to perform the differential expression analysis, quantify and annotate peaks, and to perform the Gene Set Enrichment Analysis, respectively. For the visualization of the results, the following packages were exploit: ChIPseeker73, circlize74.
Mapping of the AR locus
To understand where the most affected regions in the AR locus mapped, we took advantage of the Integrative Genomics Viewer (IGV) tool75. The enhancer regions were identified employing the Human Super-Enhancer Database (http://www.licpathway.net/sedb/index.php) and are located both in proximity and at distal sites from the AR gene76,77.
Histone post-translational modification (PTM) analysis
For histone lysine-methylation analysis, 1 × 106 cells for each replicate were collected and processed with EpiQuikTM total histone extraction kit (Epigentek,cat. OP-0006), according to manufacturer instructions78. Briefly, cell pellets were resuspended and protein quantified. Then, acetone was added to allow histone precipitation, at −20 °C over-night. Chemical derivitization of histones was performed using propionic anhydride. Afterwards, trypsin was used for proteolytic digestion of the samples, at 37 °C over-night. The following day, a second step of propionylation of histone peptides at N-termini was performed. Dried peptides were resuspended in 2% acetonitrile (ACN) and 0.3% trifluoroacetic acid (TFA) and purified on C18 StageTips, from which peptides were eluted with 80% ACN and 0.5% acetic acid. Finally, the elution buffer was eliminated by vacuum centrifugation and purified peptides were resolved in 2% ACN with 0.5% acetic acid and 0.1% TFA. For LC-MS/MS analysis, 1 µg of purified peptides from each sample were injected as single-shot measurements.
LC-MS/MS analysis
Peptides were separated on an EASY-nLC 1200 HPLC system (Thermo Fisher Scientific) coupled online to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray source (Thermo Fisher Scientific). Peptides were loaded in buffer A (0.1% formic acid) into an in-house packed (ReproSil-Pur C18-AQ 1.9 µm resin, Dr. Maisch HPLC GmbH) 75 µm inner diameter, 50 cm length column, and eluted with a 150-min linear gradient of 5-30% buffer B (80% ACN, 0.1% formic acid) at a 250 nl/min flow rate. The Q Exactive HF operated in a data-dependent mode with a survey scan range of 300–1650 m/z, resolution of 60,000 at 200 m/z, maximum injection time of 20 ms and AGC target of 3e6. The ten most abundant ions with charge 2–5 were isolated with a 1.8 m/z isolation window and fragmented with higher-energy collisional dissociation (HCD) with a normalized collision energy of 27. MS/MS spectra were acquired with a resolution of 15,000 at 200 m/z, maximum injection time of 55 ms and AGC target of 1e5. Dynamic exclusion was set to 30 s to avoid repeated sequencing. Data were acquired with the Xcalibur software (Thermo Scientific).
LC-MS/MS data analysis
Xcalibur raw files were processed using the MaxQuant software v.1.6.7.079 as previously described80. Searches were performed against the UniProt human histone sequences (57 entries, including H1, H2A, H2B, H3, and H4) by the integrated Andromeda search engine81 to identify peptides and proteins with a false discovery rate of <1%. Enzyme specificity was set as “Trypsin_R”, which allows cleavages only after arginine, with a maximum of 2 missed cleavages and 6 as the minimum length required for peptides. Lysine propionylation, methyl_propionylation, di-methylation, tri-methylation and acetylation were set as variable modifications, while N-terminal peptide propionylation was set as a fixed modification. Match between runs was enabled to transfer identifications across samples, based on mass and normalized retention times, with a matching time window of 0.7 min and an alignment time window of 20 min. The minimum score for modified peptides was lowered to 0. The MaxQuant output table msms.txt was filtered for identifications with Delta Score ≥ 15 and localization probabilities of identified modifications ≥0.75. The resulting list was then imported into the Skyline environment, version 21.2.0.568 (https://skyline.ms/project/home/software/Skyline/begin.view)82 for peaks inspection and quantification. Peptides with ambiguous identifications were discarded, and only peaks supported by a MS/MS identification in all replicates of at least one experimental group, or 2/3 of both groups, were considered for manual inspection. The resulting peaks for each modified peptide across samples were quantified by TIC Normalized Peak Area. Finally, for each replicate all area values (intensity scores) for peptides including a specific modification in a given position were summed and the result used to compare the modification level between the experimental groups. Intensity scores were compared using Student’s t test for statistical significance. To determine the entity of the changes between experimental groups, we calculated the logarithm of the ratio between the averages (Log2FC). For this study only H3 identifications were analyzed.
Analysis in patient datasets
To investigate the expression level of sets of genes (such as MAT2A signatures) in prostate cancer patients, we took advantage from an integrate database of prostate cancer patients (n = 1049) including 714 primary prostate tumors, 316 CRPC, and 19 CRPC with neuroendocrine features (NE)83. Vst-normalized expression data along with its annotations were downloaded from Zenodo repository (10.5281/zenodo.5546618). A cumulative expression value was computed through Single Sample Gene Set Enrichment Analysis method84. For cumulative gene expression levels, we utilized the same cutoffs as those we used in RNA-seq analysis (p-value adjusted ≤0.05 and Log2FC ± 1). The cumulative gene expression levels were computed with gsva function with “ssgsea” mode from GSVA package84. Briefly, GSVA uses a non-parametric technique to measure the relative enrichment of gene sets across samples. GSVA converts a p-gene by n-sample gene expression matrix into a g-geneset by n-sample pathway enrichment matrix. Patients were divided into two categories: AR-high and AR-low. Observing the probability density distribution of the AR gene, a bimodal distribution can be identified, characterized by two peaks. AR-high patients are identified as the ones belonging to the first peak, AR-low to the second one. For the rigorous assignation of a patient to the corresponding category, a threshold is considered as the first local minimum of the distribution after the first peak. Patients were also classified in ERG-low and ERG-high following the same procedures adopted for AR.
Chromatin immunoprecipitation (ChIP)
Cell line samples (3 × 106 cells per IP) were exposed to formaldehyde (37%) to cross-link protein-DNA complexes and processed as previously described21,85. Chromatin extracts were immunoprecipitated with antibodies against AR (cat. 06-680, Millipore), H3K4me2 (cat. 9725 S, Cell Signaling), H3K27me3 (cat. 9733 S, Cell Signaling) and IgG (Millipore) as control. Quantitative real-time PCR was performed using KAPA SYBR Fast qPCR kit (KAPA Biosystems) and primers spanning the regions of interest (Supplementary Table 1). The amount of immunoprecipitated DNA was calculated in reference to a standard curve and normalized to input DNA or subsequently to IgG85. For comparison of test and control samples, the amount of DNA was then normalized to the control samples.
Chromatin immunoprecipitation with sequencing (ChIP-seq)
Library preparation for next-generation sequencing was performed using NebNext Ultra II DNA Library Prep with samples purification beads (Cat. no. #E7103S, NewEngland Biolabs) and Multiplex Oligos for Illumina (Dual Index Primers) (Cat. no. #E7600S, NewEngland Biolabs) according to manufacturer’s instructions. Briefly, libraries were prepared for 4 samples in biological triplicates (VCaP Ctrl input, VCaP Ctrl anti-H3K4me2, VCaP Sh1 input, VCaP Sh1 anti-H3K4me2). A total of 5 ng of purified chromatin was used for each library preparation, followed by adaptor ligation. Samples were then purified from unbound adaptors using magnetic beads, eluted, and amplified with a unique combination of dual-index primers. Amplified chromatin fragments were additionally purified using magnetic beads. Quality control was performed on Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and Qubit V4 (Invitrogen Thermo Fisher Scientific, Waltham, MA, USA). Next-generation sequencing was performed on NextSeq2000 (Illumina, San Diego, CA, USA) with the P2 reagents kit V3 (100 cycles; Illumina). Samples were processed starting from single-ended 120bp-long sequencing reads.
Downstream analysis of ChIP-seq data
The computational analysis of the ChIP-Seq data was carried out using a methodical pipeline and approach that involve quality control, alignment, processing, peak calling, differential expression analysis, peak annotation and visualization. Initial quality control and trimming of the raw reads were executed with Trim Galore (Martin, 2011), a tool that excises low-quality bases from the ends of the reads and removes adapter sequences, in order to reduce the potential for errors in the subsequent alignment phase. Post-trimming, the cleaned reads were aligned to the reference genome using Bowtie2 (https://www.nature.com/articles/nmeth.1923), which demonstrates excellent performance in terms of efficiency and speed when aligning sequencing reads to long reference sequences. Following the alignment, the GATK’s MarkDuplicates (McKenna et al., 2010) function was used to identify and mark potential PCR duplicates. The marked duplicates were then removed, and the resultant BAM files were indexed with SAMtools (Li et al., 2009), to minimize redundancy and prevent the overrepresentation of certain genomic regions. Once the BAM files were finalized, peak calling was performed using MACS3 (Zhang et al., 2008), in order to identify transcription factor binding sites and histone modification-enriched regions from ChIP-Seq data. The differential binding analysis was conducted using the DiffBind (Stark, R. & Brown, G.) package in R, a software that not only establishes a consensus of multiple peak callers, but also utilizes count data to effectively compute variations in binding affinity across diverse experimental conditions. The statistical significance of the differential binding was calculated using the DESeq2-based test, employing a model based on the negative binomial distribution. Lastly, the annotation of the identified peaks was carried out using the ChIPseeker73 R package designed specifically for the annotation of ChIP-Seq data.
Proteasome inhibition assay
For proteasome inhibition, cells were prepared as follows. VCaP Ctrl and MAT2Akd were plated in a 6-well plate at a concentration of 2 × 106 cells/well. After 72 h cells were treated with 10 µM PS341 for 0 h, 4 h and 6 h. After treatment, cells lysates were prepared for immunoblotting.
Co-transfection and puromycin treatment
For co-transfection experiments and subsequent treatment with puromycin, we plated 500’000 RWPE-1 cells per well in a 6-well plate. After 24 h, co-transfection experiments were performed using JetPRIME reagent (Polyplus), according to manufacturer’s instructions. Transfection was performed using 3 µg the following plasmids: MAT2A (Addgene), ERG5, ERG-K362A5. After 8 h from the transfection, treatment with 1 µg of puromycin (cat. P7255, Sygma-Aldrich) was performed at different time points (0 h, 10 h, 12 h, 14 h). After treatment, cell lysates were prepared for immunoblotting.
Immunoblotting
Cell lysate was prepared by resuspending cell pellets in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% Na-deoxycholate), 4 mM Na3VO4, 1 mM PMSF along with protease inhibitors (Complete mini, Roche) and phosphatase inhibitors (PhosphoStop, Roche). Proteins were quantified using the Pierce BCA analysis kit (Thermo Fisher Scientific, cat. 23228). The absorbance was measured at 562 nm with the Cytation 3 Cell Imaging Multi-Mode Reader. 40 µg of protein was resolved on a 10% polyacrylamide gel. After electrophoresis, proteins were transferred to a PVDF membrane. Immunoblotting was performed using primary antibody anti-MAT2A (1:500, cat. NB110-94158, Novus Biological), anti-ERG (1:3000, cat. ab-92513, Abcam), anti-AR (1:1000, cat. 06-680, Merk), anti-EZH2 (1:1000, cat. 612667, BD Biosciences), anti-mERG (1:1000, custom-made)5;, anti-chromogranin A (1: 1000, cat. ab-15160, Abcam), anti-synaptophysin (1: 1000, MA5-14532, ThermoFisher Scientific), anti-H3K4me2 (1:1000, cat. 9725 S, Cell Signaling), anti-H3K4me3 (1:1000, cat. 9751 S, Cell Signaling), anti-H3K9me2 (1:1000, cat. 4658 S, Cell Signaling), anti-H3K9me3 (1:1000, cat. 13969 S, Cell Signaling), anti-H3K36me3 (1:1000, cat. 4909 S, Cell Signaling), anti-H3 (1:1000, cat. 4499 S, Cell Signaling), anti-GAPDH (1:10,000, cat. sc-47724, Santa Cruz Biotechnology), anti-HSP90 (1:10,000, cat. 13171-1-AP, ProteinTech) and anti-Tubulin (1:10,000, cat. CP06, Calbiochem) in overnight incubation at 4 °C. Western Bright or Quantum ECL (Advasta-Witec cat. K-12045-D50; cat. K-12042-D10) was used for chemoluminescent detection. Membranes were analyzed and bands were quantified with FUSION SOLO S system (Vilber Smart Imaging).
MAT2A inhibition and immunoblots
For MAT2A inhibition, with subsequent immunoblotting, cells were prepared as follows. 2 × 106 VCaP cells were plated in each well of a 6-well plate. After 48 h or 72 h, cells were treated with 10 µM PF-9366 or 1 µM AG-270. DMSO was used as control. Cells were collected treatment and protein extraction was performed as described in the following paragraph. 1 × 106 EPG2 cells were plated in each well of a 6-well plate. After 48 h or 72 h, cells were treated with either 0.5 µM PF-9366 or 0.2 µM AG-270. DMSO was used as control. Cells were collected after treatment and protein extraction was performed as described above.
Immunoprecipitation
Immunoprecipitation experiments were performed using Protein G PLUS/Protein A-Agarose mixture (Calbiochem-Millipore), which was incubated with 100–300 µg of cell lysate and 1–2 μg of anti-MAT2A and anti-ERG antibodies, overnight at 4 °C. Proteins were resolved on 10% SDS PAGE and analyzed by western blot.
SRB proliferation assay
To assess cell proliferation, RWPE-1 cells were plated in 96-well tissue culture plates at a density of 2500 cells/well. The following day, RWPE-1 were treated with the two MAT2A inhibitors (PF-9366 and AG-270) with increasing concentration up to 62.5 µM. Plates were incubated at 37 °C, 5% CO2. After 72 h, cells were fixed with cold trichloroacetic acid 10% (TCA) (wt/vol) incubated at 4 °C for 1 hour, washed with slow-running tap water and stained with sulforhodamine B solution (SRB, 0.057%) (wt/vol, in acid acetic 1%) for 30 min. SRB was then solubilized with 10 mM Tris base solution (pH 10.5) for optical density (OD) measurement at 560 nm with Cytation 3 Cell Imaging Multi-Mode Reader.
Sphere forming assay (SFA)
For sphere-forming assay (SFA), cells were plated in Poly-HEMA (Poly-2-hydroxyethyl methacrylate, 1X) (Sigma-Aldrich) coated 6-well plates in serum-free Mammary Epithelial Basal Medium (MEBM, Cambrex) supplemented with B27 Factor (1X), Follicular Growth Factor (20 ng/ml), Epidermal Growth Factor (10 ng/ml), Insulin (0.4 mg/ml) and 1% Pen/Strep (Thermo Fisher, cat. 15070063). For SFA, we seeded 1,000 cells/mL of VCaP and EPG2 and 2000 cells/mL for RWPE-1 and NCI-H660 cells. VCaP and RWPE1 spheres were counted after 14 days and NCI-H660 spheres were counted after 10 days. EPG2 spheres were counted after 7 days. Representative pictures were taken using Zeiss Microscope with Canon EOS 450D. When indicated, spheres were treated with PF-9366, AG-270, MDV-3100 (cat. HY-70002, MedChemExpress) and GSK-343 (cat. HY-13500, MedChemExpress) at the indicated doses, using DMSO as control.
Colony formation assay
For colony formation assay, cells were plated at low-density in 12-well plates. A total of 250 EPG2 cells per well were plated per condition. Cells were treated with increasing concentration of PF-9366, AG-270, GSK-343 or MDV-3100. After 7 days, colonies were fixed with 10% (wt/vol) TCA and then colored with 0.057% (wt/vol) sulforhodamine B (SRB) solution. Colonies were then photographed using FUSION SOLO S equipment and counted with an automated colony counter (ImageJ).
3D organoids
For in vitro 3D organoid assay, single cell suspensions from ERG/PTEN mouse prostates was obtained as described in the previous paragraph. Cells were plated at a concentration of 5 × 103 cells/well, in non-tissue treated 96-well plate, in a Matrigel drop (80% Matrigel, 20% cells). Cells were cultured in adDMEM/F12, supplemented as previously described86. Organoids with a diameter greater than 50 µm were counted and organoid formation efficiency was evaluated. Organoids derived from ERG/PTEN prostates were evaluated after 7 days. Pharmacological treatments were performed by adding the indicated drug to the medium at the beginning of the assay. Cells were treated with different concentrations of PF-9366, AG-270, GSK-343, MDV-3100 or DMSO (as control). Treatments were performed by adding the indicated drugs to the medium at plating time. Each experiment was carried out in triplicate.
For reversion of 3D organoids phenotype, cells derived from ERG/PTEN mouse prostates were obtained and plated as previously described86. Cells were cultured for 7 days in adDMEM/F1286 to allow organoids formation. For reversion of the phenotype, organoids were treated with different concentrations of PF-9366, AG-270 or DMSO (as control) in regular growth medium depleted of EGF, as previously described87. Organoids with a diameter greater than 50 µm were counted, organoid formation efficiency and morphological changes was evaluated. Organoids were evaluated after 4 days post-treatment.
For in vitro 3D organoid assay, LuCaP 145.2 xenografts were dissected and resuspended in RPMI 16040 medium (Gibco™ 21875034), supplemented with Collagenase D (cat. 11088866001, Roche) for 45 minutes. Cell suspension was passed through a 40μm cell strainer to collect single cells and washed twice with PBS. Then, cells were resuspended with Red Blood Lysis buffer (cat. 11814389001, Roche) for 5 minutes. Cells were centrifuged and plated in non-tissue treated 96-well plate, in a Matrigel drop (80% Matrigel, 20% cells) at a concentration of 10,000 cells/well. Cells were cultured in adDMEM/F12, supplemented as previously described in organoid condition86. Medium was replenished every 5 days. After 15 days, organoids were treated with 1 µM of AG-270 or DMSO (as control) in regular growth medium depleted of EGF. After 6 days post-treatment, organoids with a diameter greater than 40 µm were counted and WB assay was performed.
Animals and tumor xenografts
Immuno-compromised (NOD.Cg-PrkdcSCID Il2rgtm1Wjl/SzJ) NSG and (NOD-Rag2-IL2rgTm1/Rj) NRG mice (8–10 weeks old) were purchased from Charles River Laboratories. Mice were maintained under pathogen-free conditions with food and water provided ad libitum and their general health status was monitored daily. The Pb-Cre4;PTENflox/flox;Rosa26ERG/ERG (ERG/PTEN) (24–26 weeks old) mouse was provided by Dr. Charles L. Sawyers88. Animals handling was carried out according to the protocol approved by the Swiss Cantonal Veterinary Authority (TI29/19). LuCaP patient derived xenograft 35 and 145.2 were established from specimens acquired at either radical prostatectomy or at autopsy, implanted, and maintained by serial passage in NSG mice34. Animals handling was carried out according to the protocol approved by the Swiss Cantonal Veterinary Authority (TI 04/2020). Tumor xenografts and animals handling were conducted in conformity with the institutional guidelines for animal experimentation and in compliance with national and international policies. The relative humidity was kept at 45 to 65%. Mouse rooms and cages were kept at the temperature range of 20–24 °C. The maximal tumour size/burden permitted by our license approved by ethics committee or institutional review board was 1500mm3 and maximal tumour size/burden was not exceeded. For VCAP subcutaneous tumor xenografts, 3 × 106 cells/mouse were inoculated with Matrigel (1:1) in the flank of NSG mice (n = 5/group). For RWPE1 subcutaneous xenografts, RWPE1 control and stably expressing ERG, MAT2A, ERG + MAT2A (3 × 106 cells/mouse) were inoculated in the flank of NSG mice (n = 6/group)with Matrigel (1:1) and UGSM cells (3 × 105 cells/mouse). Tumor growth was monitored every 2 days with a caliper. Animals were euthanised by inhalation of CO2 and by checking for cessation of respiration for two minutes and lack of reflexes. All animal experiments were carried out according to the protocol approved by the Swiss Cantonal Veterinary Authority (TI 04/2020). Animals were well-ventilated, softly lit, and subject to a light/dark cycle.
In vivo treatment with AG-270
For in vivo use, AG-270 was solubilized in 10% DMSO and 90% (20% SBE-β-CD in saline). Control mice received only the vehicle as 10% DMSO and 90% (20% SBE-β-CD in saline). For in vivo treatment with AG-270, 2 × 106 LuCaP 35 cells were subcutaneously injected in NRG mice with Matrigel (1:1). Tumor growth was monitored every 2 days with caliper. When tumor volume reached 100–200 mm3, mice were randomized into two groups: vehicle (n = 7) and AG-270 (n = 7). Vehicle or AG-270 were administered once a day by oral gavage, at a dosage of 10 mg/kg for 10 days. When control groups reached the maximum volume of 1000–1500 mm3, mice were euthanised, and tumor xenografts were collected for subsequent experiments.
Ex-vivo sphere-forming assay
For ex-vivo SFA, ERG/PTEN mouse prostates from 33-weeek-old mice were dissected and minced into small pieces in Hanks Balanced Salt solution (cat. H9394, Sigma Aldrich) supplemented with 1 mg/ml of collagenase (cat. C0130, Sigma Aldrich) for 1 h. Cell suspension was passed through a 40 μm cell strainer (cat. 352340, Falcon) to collect single cells. Then, cells were centrifuged (400× g for 5 minutes), washed (PBS, 2 times), and plated in Poly-HEMA coated 6-well plates in serum-free MEBM at a concentration of 5000 cells/well. Medium was replenished every 3 days. After 10 days, spheres with a diameter greater than 50μm were counted. For ex-vivo SFA, LuCaP 35 xenografts were dissected and resuspended in RPMI 16040 medium (Gibco™ 21875034), supplemented with Collagenase D (cat. 11088866001, Roche) for 45 minutes. Cell suspension was passed through a 40μm cell strainer to collect single cells and washed twice with PBS. Then, cells were resuspended with Red Blood Lysis buffer (cat. 11814389001, Roche) for 5 minutes. Cells were centrifuged and plated in Poly-HEMA coated 12-well plates in serum-free MEBM at a concentration of 5000 cells/well. Medium was replenished every 3 days. After 14 days, spheres with a diameter greater than 50 µm were counted. Treatments were performed by adding the indicated drugs to medium at plating time. Each condition was carried out in triplicate.
MicroScale Thermophoresis (MST)
For MST experiments, Histidine-tagged MAT2A protein (BPS Bioscience, cat 71401) was labelled with fluorescent dye using Monolith His-Tag Labeling Kit (cat MO-L008) (NanoTemper Technologies GmbH). Human recombinant protein ERG (OriGene, cat. TP308093) or BSA was incubated with labelled MAT2A (5 nM) at room temperature for 30 minutes. Microscale thermophoresis (MST) measurements were performed utilizing a Monolith NT.115 instrument (NanoTemper Technologies GmbH, Munchen, Germany). Binding affinity was measured by titrating different concentrations of ERG (200 nM to 0.0061 nM) into a fixed concentration of labeled MAT2A (5 nM). All measurements were performed using Premium capillaries at 12% LED excitation and high MST power.
Immunohistochemistry
Tissue sections from formalin-fixed paraffin-embedded samples were de-paraffinized and rehydrated with alcohol-based Ottix solutions: Ottix Plus for 5 minutes; Ottix Shaper for 3 minutes; dH2O2 for 5 minutes. Subsequently, antigen retrieval was achieved by heating the slides in water bath (98 °C) for 20 minutes with pH 6.0 Citrate buffer (DIAPATH T0050) or pH 8.0 EDTA buffer (DIAPATH T0100) or pH 9.0 EDTA buffer (DAKO K8024). After antigen retrieval, the sections were incubated with an anti-MAT2A (Novus Biological, cat. NBP1-92100), anti-ERG and anti-Ki67 (SP6, RTU Eupredia, cat. RM-9106-R7) antibodies at room temperature for 1 h. Next, sections were incubated with a biotinylated secondary antibody (LSAB2-DAKO) and then with streptavidin-HRP (Horseradish peroxidase) conjugate or by Vectastain ABC Kit (Vectorlabs, vector PK-6100) (A 1:150 B 1:150 in PBS) for 30 minutes at room temperature, followed by 3-3’Diaminobenzidine (DAB) chromogenic substrate for detection. Finally, sections were dehydrated with alcohol-based Ottix solutions: Ottix Shaper for 3 minutes, Ottix Plus for 5 minutes, then covered and stabilized with mounting medium.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We acknowledge the support from the Swiss Cancer League (grant KLS-4569-08-2018 and KLS-4899-08-2019), Swiss National Science Foundation (grant SNSF-310030_189081, SNSF-IZLSZ3_170898 and SNSF-310030L_170182), Fondation Nelia and Amadeo Barletta (FNAB), Fondazione Ticinese Ricerca sul Cancro, and Fondazione San Salvatore.
Author contributions
A.C. performed experiments, analyzed the data, and wrote the manuscript draft; D.S. performed experiments, analyzed the data, and wrote the manuscript draft. C.M. performed experiments, and analyzed the data; G.S. performed bioinformatics and statistical analysis; L.G. performed bioinformatics and statistical analysis D.A. performed experiments and analyzed data; G.C. performed experiments and established a cell line from GEMM; E.S. performed in vivo experiments; S.M. assisted in histological sample preparation and analysis and performed IHC; E.F. assisted with patient derived xenografts (PDX) establishment and performed experiments; A.F. performed experiments in vitro; M.I. assisted with the HPTM; A.R. assisted in RNA samples preparation and performed RNA-Seq and ATAC seq; M.P. performed and analyzed the HPTM experiments; R.G. supervised the HPTM experiments; M.B. supervised the bioinformatic analyses; C.V.C. supervised the experimental plans, interpreted results, provided insights, resources and funding for the project, wrote and reviewed the manuscript; G.M.C. conceived the project, supervised the experimental plans, mentored participants, analyzed and interpreted the results, provided resources and funding, and wrote and reviewed the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw RNA-seq data from the experiment of RWPE-1 and VCaP cell lines generated in this study are deposited in the ArrayExpress database with the following accession numbers: E-MTAB-12029 and E-MTAB-12031 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12029?query=E-MTAB-12029%20; https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12031?query=E-MTAB-12031%20). The raw ATAC-seq data generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12025. The raw RNA-seq data from the experiment of VCaP cells treated with MAT2A inhibitors generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12536 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12036?query=E-MTAB-12029%20. The Raw ChIP-seq data generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12990. The raw mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium89 via the PRIDE90,91 partner repository with the dataset identifier PXD042895. The publicly available RNA-seq data used in this study are available in GEO (Gene Expression Omnibus), SRA (Short Read Archive), and EMBL-EBI databases under accession codes and respective hyperlinks: GSE12079592 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120795), GSE12074193(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120741), GSE11843594 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118435),GSE12607895 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126078),PRJNA47744996 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA477449,PRJEB2109297 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB21092, and E-MTAB-965698 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-9656). The publicly available ERG ChIP-seq data used in this study are available in NCBI GEO repository under accession number GSE28951. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2895129. Source data are provided with this paper (Source data file). The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alessia Cacciatore, Dheeraj Shinde.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50908-7.
References
- 1.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. CA Cancer J Clin71, 7–33 (2021). 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Rebello, R. J. et al. Prostate cancer. Nat Rev Dis Primers7, 9 (2021). 10.1038/s41572-020-00243-0 [DOI] [PubMed] [Google Scholar]
- 3.Attard, G. et al. Prostate cancer. Lancet387, 70–82 (2016). 10.1016/S0140-6736(14)61947-4 [DOI] [PubMed] [Google Scholar]
- 4.Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science310, 644–648 (2005). 10.1126/science.1117679 [DOI] [PubMed] [Google Scholar]
- 5.Zoma, M. et al. EZH2-induced lysine K362 methylation enhances TMPRSS2-ERG oncogenic activity in prostate cancer. Nat Commun12, 4147 (2021). 10.1038/s41467-021-24380-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, K. H. & Roberts, C. W. Targeting EZH2 in cancer. Nat Med22, 128–134 (2016). 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge, R. et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol. 31, 470–479 (2020). 10.1016/j.annonc.2020.02.002 [DOI] [PubMed] [Google Scholar]
- 8.Kim, J. et al. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep25, 2808–2820 e2804 (2018). 10.1016/j.celrep.2018.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu, K. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science338, 1465–1469 (2012). 10.1126/science.1227604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, C. et al. Overview of methionine adenosyltransferase 2A (MAT2A) as an anticancer target: structure, function, and inhibitors. J. Med. Chem.65, 9531–9547 (2022). [DOI] [PubMed]
- 11.Maldonado, L. Y., Arsene, D., Mato, J. M. & Lu, S. C. Methionine adenosyltransferases in cancers: Mechanisms of dysregulation and implications for therapy. Exp. Biol. Med.243, 107–117 (2018). 10.1177/1535370217740860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Moreno, L., Carmena, M. J., Prieto, J. C., Schally, A. V. & Bajo, A. M. Tumorigenic transformation of human prostatic epithelial cell line RWPE-1 by growth hormone-releasing hormone (GHRH). Prostate82, 933–941 (2022). 10.1002/pros.24339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni, P. et al. Role of the PI3K/Akt pathway in cadmium induced malignant transformation of normal prostate epithelial cells. Toxicol. Appl. Pharmacol.409, 115308 (2020). 10.1016/j.taap.2020.115308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bello, D., Webber, M. M., Kleinman, H. K., Wartinger, D. D. & Rhim, J. S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis18, 1215–1223 (1997). 10.1093/carcin/18.6.1215 [DOI] [PubMed] [Google Scholar]
- 15.Webber, M. M., Bello, D., Kleinman, H. K. & Hoffman, M. P. Acinar differentiation by non-malignant immortalized human prostatic epithelial cells and its loss by malignant cells. Carcinogenesis18, 1225–1231 (1997). 10.1093/carcin/18.6.1225 [DOI] [PubMed] [Google Scholar]
- 16.Albino, D. et al. Circulating extracellular vesicles release oncogenic miR-424 in experimental models and patients with aggressive prostate cancer. Commun. Biol.4, 119 (2021). 10.1038/s42003-020-01642-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunderfranco, P. et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One5, e10547 (2010). 10.1371/journal.pone.0010547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu, J. et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell17, 443–454 (2010). 10.1016/j.ccr.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan, X. et al. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene33, 2815–2825 (2014). 10.1038/onc.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med.22, 298–305 (2016). 10.1038/nm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albino, D. et al. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res.72, 2889–2900 (2012). 10.1158/0008-5472.CAN-12-0212 [DOI] [PubMed] [Google Scholar]
- 22.Davies, A. et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol.23, 1023–1034 (2021). 10.1038/s41556-021-00743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramanand, S. G. et al. The landscape of RNA polymerase II-associated chromatin interactions in prostate cancer. J. Clin. Investig.130, 3987–4005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda, D. Y. et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell174, 422–432 e413 (2018). 10.1016/j.cell.2018.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, F. et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science376, eabe1505 (2022). 10.1126/science.abe1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt, F. et al. Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res.45, 54–66 (2017). 10.1093/nar/gkw1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mentch, S. J. et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab.22, 861–873 (2015). 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Q. et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell138, 245–256 (2009). 10.1016/j.cell.2009.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chng, K. R. et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J.31, 2810–2823 (2012). 10.1038/emboj.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalev, P. et al. MAT2A Inhibition Blocks the Growth of MTAP-Deleted Cancer Cells by Reducing PRMT5-Dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell39, 209–224.e211 (2021). 10.1016/j.ccell.2020.12.010 [DOI] [PubMed] [Google Scholar]
- 31.Marjon, K. et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep.15, 574–587 (2016). 10.1016/j.celrep.2016.03.043 [DOI] [PubMed] [Google Scholar]
- 32.Quinlan, C. L. et al. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat. Chem. Biol.13, 785–792 (2017). 10.1038/nchembio.2384 [DOI] [PubMed] [Google Scholar]
- 33.Konteatis, Z. et al. Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion. J. Med. Chem.64, 4430–4449 (2021). 10.1021/acs.jmedchem.0c01895 [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, H. M. et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease and Serve as Models for Evaluating Cancer Therapeutics. Prostate77, 654–671 (2017). 10.1002/pros.23313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cejas, P. et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat. Commun.12, 5775 (2021). 10.1038/s41467-021-26042-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baca, S. C. et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun.12, 1979 (2021). 10.1038/s41467-021-22139-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeronimo, C. et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur. Urol.60, 753–766 (2011). 10.1016/j.eururo.2011.06.035 [DOI] [PubMed] [Google Scholar]
- 38.Perry, A. S., Watson, R. W., Lawler, M. & Hollywood, D. The epigenome as a therapeutic target in prostate cancer. Nat. Rev. Urol.7, 668–680 (2010). 10.1038/nrurol.2010.185 [DOI] [PubMed] [Google Scholar]
- 39.Davies, A. H., Beltran, H. & Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol.15, 271–286 (2018). 10.1038/nrurol.2018.22 [DOI] [PubMed] [Google Scholar]
- 40.Rubin, M. A., Bristow, R. G., Thienger, P. D., Dive, C. & Imielinski, M. Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers. Mol. Cell80, 562–577 (2020). 10.1016/j.molcel.2020.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etchegaray, J. P. & Mostoslavsky, R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol. Cell62, 695–711 (2016). 10.1016/j.molcel.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boon, R., Silveira, G. G. & Mostoslavsky, R. Nuclear metabolism and the regulation of the epigenome. Nat. Metab.2, 1190–1203 (2020). 10.1038/s42255-020-00285-4 [DOI] [PubMed] [Google Scholar]
- 43.Yang, M. & Vousden, K. H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer16, 650–662 (2016). 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- 44.Sanderson, S. M., Gao, X., Dai, Z. & Locasale, J. W. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer19, 625–637 (2019). 10.1038/s41568-019-0187-8 [DOI] [PubMed] [Google Scholar]
- 45.Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science339, 222–226 (2013). 10.1126/science.1226603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiraki, N. et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab.19, 780–794 (2014). 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 47.Wang, Z. et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med.25, 825–837 (2019). 10.1038/s41591-019-0423-5 [DOI] [PubMed] [Google Scholar]
- 48.Kalev, P. et al. MAT2A inhibition blocks the growth of MTAP-deleted cancer cells by reducing PRMT5-dependent mRNA splicing and inducing DNA damage. Cancer Cell39, 209–224.e11 (2021). 10.1016/j.ccell.2020.12.010 [DOI] [PubMed] [Google Scholar]
- 49.Villa, E. et al. mTORC1 stimulates cell growth through SAM synthesis and m(6)A mRNA-dependent control of protein synthesis. Mol. Cell81, 2076–2093.e2079 (2021). 10.1016/j.molcel.2021.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho, S. et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Mol. Cell81, 2064–2075 (2021). 10.1016/j.molcel.2021.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Affronti, H. C. et al. Pharmacological polyamine catabolism upregulation with methionine salvage pathway inhibition as an effective prostate cancer therapy. Nat. Commun11, 52 (2020). 10.1038/s41467-019-13950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reina-Campos, M. et al. Increased Serine and One-Carbon Pathway Metabolism by PKClambda/iota Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell35, 385–400 e389 (2019). 10.1016/j.ccell.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao, X. et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature572, 397–401 (2019). 10.1038/s41586-019-1437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, X., Egervari, G., Wang, Y., Berger, S. L. & Lu, Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol.19, 563–578 (2018). 10.1038/s41580-018-0029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet.13, 343–357 (2012). 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet.39, 311–318 (2007). 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 57.Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell132, 958–970 (2008). 10.1016/j.cell.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Culig, Z. et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br. J. Cancer81, 242–251 (1999). 10.1038/sj.bjc.6690684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander, D. STAR: ultrafast universal RNA-seq aligner. in Bioinformatics (Oxford, England), Vol. 29 15-21 (2013). [DOI] [PMC free article] [PubMed]
- 60.Andrews, S. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 61.Anders, S. & Heidelberg, E. DESeq: differential gene expression analysis based on the negative binomial distribution (2019).
- 62.Team, R.C. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2020).
- 63.Smyth, G. et al. limma: Linear Models for Microarray Data (2020).
- 64.Jawaid, W. enrichR: Provides an R Interface to Enrichr (2021).
- 65.Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol109, 21 29 21-21 29 29 (2015). [DOI] [PMC free article] [PubMed]
- 66.Krueger, F. TrimGalore. Vol. 2021 TrimGalore https://zenodo.org/badge/DOI/10.5281/zenodo.5127899.svg (2021).
- 67.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience10, giab008 (2021). [DOI] [PMC free article] [PubMed]
- 69.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaspar, J. M. Genrich. https://github.com/jsh58/Genrich.
- 71.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 72.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell38, 576–589 (2010). 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics31, 2382–2383 (2015). 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- 74.Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics30, 2811–2812 (2014). 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- 75.Robinson, J. T., Thorvaldsdottir, H., Wenger, A. M., Zehir, A. & Mesirov, J. P. Variant review with the integrative genomics viewer. Cancer Res.77, e31–e34 (2017). 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang, Y. et al. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res.47, D235–D243 (2019). 10.1093/nar/gky1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Y. et al. SEdb 2.0: a comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 51, D280–D290 (2023). [DOI] [PMC free article] [PubMed]
- 78.Sidoli, S., Bhanu, N. V., Karch, K. R., Wang, X. & Garcia, B. A. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp. 54112 10.3791/54112 (2016). [DOI] [PMC free article] [PubMed]
- 79.Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol.26, 1367–1372 (2008). 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 80.Yuan, Z. F., Lin, S., Molden, R. C. & Garcia, B. A. Evaluation of proteomic search engines for the analysis of histone modifications. J. Proteome Res.13, 4470–4478 (2014). 10.1021/pr5008015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res.10, 1794–1805 (2011). 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 82.Marsh, A. N. et al. Skyline batch: an intuitive user interface for batch processing with skyline. J. Proteome Res.21, 289–294 (2022). 10.1021/acs.jproteome.1c00749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolis, M. et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat. Commun.12, 7033 (2021). 10.1038/s41467-021-26840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics14, 7 (2013). 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longoni, N. et al. ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-kappaB and drives prostate cancer progression. Cancer Res.73, 4533–4547 (2013). 10.1158/0008-5472.CAN-12-4537 [DOI] [PubMed] [Google Scholar]
- 86.Drost, J. et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc.11, 347–358 (2016). 10.1038/nprot.2016.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan, J. M. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science377, 1180–1191 (2022). 10.1126/science.abn0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen, Y. et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat. Med.19, 1023–1029 (2013). 10.1038/nm.3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deutsch, E. W. et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res.51, D1539–D1548 (2023). 10.1093/nar/gkac1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perez-Riverol, Y. et al. PRIDE inspector toolsuite: moving toward a universal visualization tool for proteomics data standard formats and quality assessment of ProteomeXchange datasets. Mol. Cell Proteomics15, 305–317 (2016). 10.1074/mcp.O115.050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suntsova, M. et al. Atlas of RNA sequencing profiles for normal human tissues. Sci. Data6, 36 (2019). 10.1038/s41597-019-0043-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stelloo, S. et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat. Commun.9, 4900 (2018). 10.1038/s41467-018-07270-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharp, A. et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Investig.129, 192–208 (2019). 10.1172/JCI122819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Labrecque, M. P. et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Investig.129, 4492–4505 (2019). 10.1172/JCI128212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oberhuber, M. et al. STAT3-dependent analysis reveals PDK4 as independent predictor of recurrence in prostate cancer. Mol. Syst. Biol.16, e9247 (2020). 10.15252/msb.20199247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramnarine, V. R. et al. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience7, giy050 (2018). [DOI] [PMC free article] [PubMed]
- 98.Karkampouna, S. et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat Commun12, 1117 (2021). 10.1038/s41467-021-21300-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data from the experiment of RWPE-1 and VCaP cell lines generated in this study are deposited in the ArrayExpress database with the following accession numbers: E-MTAB-12029 and E-MTAB-12031 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12029?query=E-MTAB-12029%20; https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12031?query=E-MTAB-12031%20). The raw ATAC-seq data generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12025. The raw RNA-seq data from the experiment of VCaP cells treated with MAT2A inhibitors generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12536 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12036?query=E-MTAB-12029%20. The Raw ChIP-seq data generated in this study are deposited in the ArrayExpress database with the accession number E-MTAB-12990. The raw mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium89 via the PRIDE90,91 partner repository with the dataset identifier PXD042895. The publicly available RNA-seq data used in this study are available in GEO (Gene Expression Omnibus), SRA (Short Read Archive), and EMBL-EBI databases under accession codes and respective hyperlinks: GSE12079592 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120795), GSE12074193(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120741), GSE11843594 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118435),GSE12607895 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126078),PRJNA47744996 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA477449,PRJEB2109297 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB21092, and E-MTAB-965698 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-9656). The publicly available ERG ChIP-seq data used in this study are available in NCBI GEO repository under accession number GSE28951. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2895129. Source data are provided with this paper (Source data file). The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.