Abstract
To address the rapid evolution of artificial intelligence in medical imaging, the authors present the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) 2024 Update.
Supplemental material is available for this article.
Summary
To address the rapid evolution of artificial intelligence in medical imaging, the authors present the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) 2024 Update.
Introduction
Since its introduction, the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) has sought to promote complete and consistent reporting of artificial intelligence (AI) science in medical imaging (1). CLAIM has been adopted widely in several medical specialties that involve imaging and AI; as of February 2024, PubMed identified 275 articles that cite the original guideline, and Google Scholar identified 608 citations. CLAIM is one of several reporting guidelines developed to address AI and medical imaging (2). Although not designed as a scoring system, some authors have applied it as such and have found variable adherence among published articles (3–6). Some authors have identified opportunities to improve the guideline, such as separating complex items in the original guideline and accommodating rapidly evolving techniques (7).
The CLAIM Steering Committee sought to revise, improve, and formalize the guideline (8). The authors renewed CLAIM’s registration with the Enhancing the Quality and Transparency of Health Research (EQUATOR) Network, an organization that promotes the use of reporting guidelines to improve health research (https://www.equator-network.org) (9,10). The CLAIM Steering Committee developed and conducted a formal Delphi consensus survey process to review the appropriateness and importance of existing checklist items and to identify new content to reflect current science in AI. The authors recruited 73 volunteers, including physicians from a variety of medical imaging–related specialties, AI scientists, journal editors, and statisticians to form the CLAIM 2024 Update Panel; 72 members completed the two survey rounds and are listed as contributors to this work.
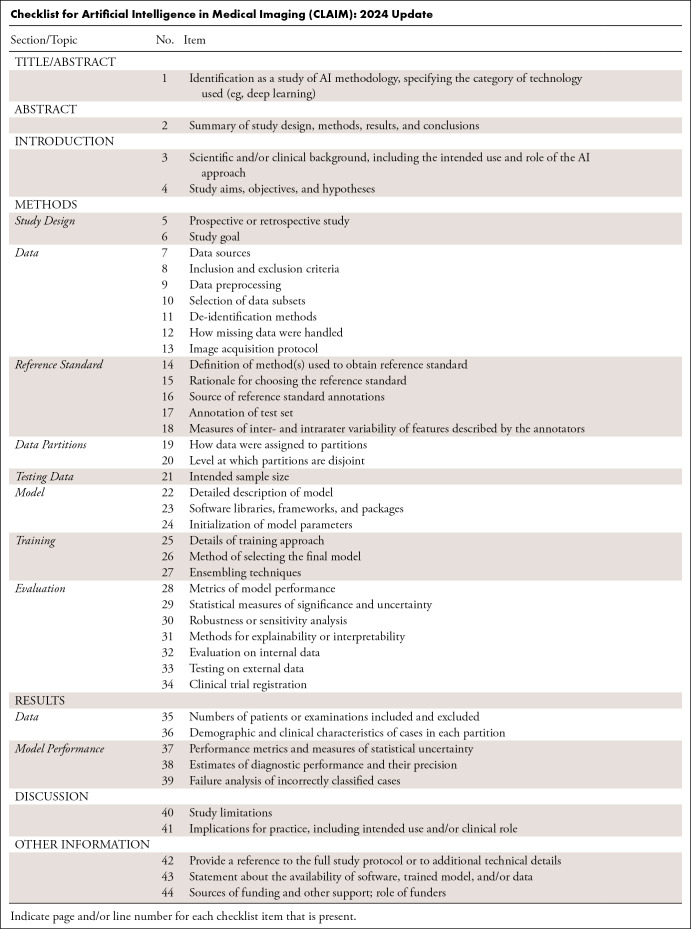
To address AI’s rapid scientific evolution, this article presents the CLAIM 2024 Update (see Table and see Appendix for downloadable Word document). Based on an expert-panel Delphi process, the guideline’s recommendations promote consistent reporting of scientific advances of AI in medical imaging to build trust in published results and enable clinical translation. This guideline serves as an educational tool for both authors and reviewers; it offers a best practice checklist to promote transparency and reproducibility of medical imaging AI research.
Checklist for Artificial Intelligence in Medical Imaging (CLAIM): 2024 Update
Key Features of the 2024 Update
Each CLAIM item now has three options: Yes, No, and Not Applicable (NA). For items marked Yes, authors are encouraged to indicate the page and line numbers in the manuscript. For items marked No or NA, authors are encouraged to explain why. The NA option was added at the suggestion of several panel members who indicated that some CLAIM items might not be applicable to all research studies.
The term “reference standard” has been adopted in lieu of “ground truth.” A detailed explanation appears below.
Because the term “validation” is ambiguous, the CLAIM team discourages its use. Authors are encouraged to use “internal testing” and “external testing” to describe the process of testing against a held-out subset of training data and a completely external dataset such as one from another institution, respectively.
Authors no longer need to define the data elements used as input or output of their model and need not link them to a reference data dictionary, such as common data elements. Item 11 from the initial version of the guideline has been deleted.
Item 13 has been added to ask authors to provide details about the image acquisition protocol.
Based on consensus of the Update Panel members, the checklist has not been extended to apply to research on imaging biomarkers (radiomics, pathomics, etc).
Reference Standard
The CLAIM Steering Committee and Update Panel strongly encourage the use of the term “reference standard” to indicate the benchmark against which an AI model’s performance is measured. The term is used widely in technology assessment and health services research when measuring the performance of a diagnostic test.
“Ground truth” is a term commonly used in statistics and machine learning that refers to the correct or “true” answer to a specific problem or question; typically, it is a label applied by expert human annotation. The term is borrowed from meteorology, where ground truth refers to information observed directly on the ground instead of by remote sensing. Similarly, the term “gold standard” has been used to describe a diagnostic test that is regarded as definitive for a particular disease, and thereby becomes the ultimate measure for comparison. That term is borrowed from economics, where it refers to the assignment of a currency’s fixed value to an amount of metallic gold.
The term “reference standard” has several advantages. It avoids the connotations and ambiguity of jargon-like terms such as “ground truth” and “gold standard,” and aligns with the language of other scientific reporting guidelines, such as the Standards for Reporting of Diagnostic Accuracy Studies (STARD) statement (11). Ground truth suggests that one knows the absolute truth, whereas in biomedicine, the label assigned as ground truth may be uncertain. Similarly, many economies no longer have a “gold standard”; rather than denoting a fixed standard, it can be variable. For these reasons, authors should use “reference standard” in lieu of “ground truth” or “gold standard.” Note that the corresponding Methods subsection of this guideline has been renamed accordingly.
Testing Dataset
The CLAIM Steering Committee encourages authors to use the term “internal testing dataset” to indicate the held-out subset of data from the training data. The machine learning term “validation” can cause confusion among medical professionals, who may interpret the term to mean the testing of the model against what is “valid” or true. Authors are urged to use the term “validation” with caution and to consider using “tuning” or “model optimization.” The term “external testing” is preferred over “external validation” to describe testing on external data, such as from another site.
Manuscript Title or Abstract
Item 1. Identification as a study of AI methodology, specifying the category of technology used (eg, deep learning). Specify the AI techniques used in the study—such as “vision transformers” or “deep learning”—in the article’s title and/or abstract; use judgment regarding the level of specificity.
The Abstract
Item 2. Summary of study design, methods, results, and conclusions. The abstract should present a succinct structured summary of the study’s design, methods, results, and conclusions. Include relevant detail about the study population, such as data source and use of publicly available datasets, number of patients or examinations, number of studies per data source, modalities and relevant series or sequences. Provide information about data partitions and level of data splitting (eg, patient- or image-level). Clearly state if the study is prospective or retrospective and summarize the statistical analysis that was performed. The reader should clearly understand the primary outcomes and implication of the study’s findings, including relevant clinical impact. Indicate whether the software, data, and/or resulting model are publicly available (including where to find more details, if applicable).
The Introduction
Item 3. Scientific and/or clinical background, including the intended use and role of the AI approach. The current practice should be explicitly mentioned. Describe the study’s rationale, goals, and anticipated impact. Present a focused summary of the pertinent literature to describe current practice and highlight how the investigation changes or builds on that work. Guide readers to understand the underlying science, assumptions underlying the methodology, and nuances of the study.
Item 4. Study aims, objectives, and hypotheses (if not a data-driven approach). Define clearly the clinical or scientific question to be answered; avoid vague statements or descriptions of a process. Limit the chance of post hoc data dredging by specifying the study’s hypothesis a priori. The study’s hypothesis and objectives should guide appropriate statistical analyses, sample size calculations, and whether the hypothesis will be supported or not.
The Methods Section
Describe the study’s methodology in a clear, concise, and complete manner that could allow a reader to reproduce the described study. If a thorough description exceeds the journal’s word limits, summarize pertinent content in the Methods section and provide additional details in a supplement.
Study Design
Item 5. Prospective or retrospective study. Indicate if the study is prospective or retrospective. Evaluate AI models in a prospective setting, if possible.
Item 6. Study goal. Describe the study’s design and goals, such as model creation, exploratory study, feasibility study, or noninferiority trial. For classification systems, state the intended use, such as risk assessment, triage, diagnosis, screening, staging, monitoring, surveillance, prediction, or prognosis. Describe the type of predictive modeling to be performed, the target of predictions, and how it will solve the clinical or scientific question.
Data
Item 7. Data sources. State the source(s) of data, including publicly available datasets and/or synthetic images; provide links to data sources and/or images, if available. Describe how well the data align with the intended use and target population of the model. Provide links to data sources and/or images, if available. Authors are strongly encouraged to deposit data and/or software used for modeling or data analysis in a publicly accessible repository.
Item 8. Inclusion and exclusion criteria. Specify inclusion and exclusion criteria, such as location, dates, patient-care setting, demographics (eg, age, sex, race), pertinent follow-up, and results from prior tests. Define how, where, and when potentially eligible participants or studies were identified. Indicate whether a consecutive, random, or convenience series was selected.
Item 9. Data preprocessing. Describe preprocessing steps to allow other investigators to reproduce them. Specify the use of normalization, resampling of image size, change in bit depth, and/or adjustment of window/level settings. If applicable, state whether the data have been rescaled, threshold-limited (“binarized”), and/or standardized. Specify processes used to address regional formatting, manual input, inconsistent data, missing data, incorrect data type, file manipulations, and missing anonymization. State any criteria used to remove outliers (11). When applicable, include description for libraries, software (including manufacturer name and location and version numbers), and all option and configurations settings.
Item 10. Selection of data subsets. State whether investigators selected subsets of raw extracted data during preprocessing. For example, describe whether investigators selected a subset of the images, cropped portions of images, or extracted segments of a report. If this process is automated, describe the tools and parameters used. If performed manually, describe the training of the personnel and criteria used in their instruction. Justify how this manual step would be accommodated in context of the clinical or scientific problem, describing methods of scaling processes, when applicable.
Item 11. De-identification. Describe the methods used to de-identify data and how protected health information has been removed to meet U.S. (HIPAA), EU (AI Act, EU Health Data Space, GDPR), or other relevant regulations (12,13).
Item 12. Missing data. Clearly describe how missing data were handled. For example, describe processes to replace them with approximate, predicted, or proxy values. Discuss biases that imputed data may introduce.
Item 13. Image acquisition protocol. Describe the image acquisition protocol, such as manufacturer, MRI sequence, ultrasound frequency, maximum CT energy, tube current, slice thickness, scan range, and scan resolution; include all relevant parameters to enable reproducibility of the stated methods.
Reference Standard
Item 14. Definition of method(s) used to obtain the reference standard. Include a clear, detailed description of methods used to obtain the reference standard; readers should be able to replicate the reference standard based on this description. Include specific, standard guidelines provided to all annotators. Avoid vague descriptions, such as “white matter lesion burden,” and use precise definitions, such as “lesion location (periventricular, juxtacortical, infratentorial), size measured in three dimensions, and number of lesions as measured on T2/FLAIR MR brain images.” Provide an atlas of examples to annotators to illustrate subjective grading schemes (eg, mild, moderate, severe) and make that information available for review.
Item 15. Rationale for choosing the reference standard. Describe the rationale for choice of the reference standard versus any alternatives. Include information on potential errors, biases, and limitations of that reference standard.
Item 16. Source of reference standard annotations. Specify the source of reference standard annotations, citing relevant literature if annotations from existing data resources are used. Specify the number of human annotators and their qualifications (eg, level of expertise, subspecialty training). Describe the instructions and/or training given to annotators; include training materials as a supplement.
Item 17. Annotation of test set. Detail the steps taken to annotate the test set with sufficient detail so that another investigator could replicate the annotation. Include any standard instructions provided to annotators for a given task. Specify software used for manual annotation, including the version number. Describe if and how imaging labels were extracted from imaging reports or electronic health records using natural language processing or recurrent neural networks. This information should be included for any step involving manual annotation, in addition to any semiautomated or automated annotation.
Item 18. Measures of inter- and intrarater variability of features described by the annotators. Describe the methods to measure inter- and intrarater variability, reduce or mitigate variability, and/or resolve discrepancies between annotators.
Data Partitions
Item 19. How data were assigned to partitions. Specify how data were partitioned for training, model optimization (often termed “tuning” or “validation”), and testing. Indicate the proportion of data in each partition (eg, 80/10/10) and justify that selection. Indicate if there are any systematic differences between the data in each partition, and if so, why and how potential class imbalance was addressed. If using openly available data, use established splits to improve comparison to the literature. If freely sharing data, provide data splits so that others can perform model training and testing comparably.
Item 20. Level at which partitions are disjoint. Describe the level at which the partitions are disjoint (eg, patient-, series-, image-level). Sets of medical images generally should be disjoint at the patient level or higher so that images of the same patient do not appear in more than one partition.
Testing Data
Item 21. Intended sample size. Describe the size of the testing set and how it was determined. Use traditional power calculation methods, if applicable, to estimate the required sample size. For classification problems, in cases where there is no algorithm-specific sample size estimation method available, sample size can be estimated for a given area under the curve and confidence interval width (14).
Model
Item 22. Detailed description of model. If novel model architecture is used, provide a complete and detailed structure of the model, including inputs, outputs, and all intermediate layers, in sufficient detail that another investigator could exactly reconstruct the network. For neural network models, include all details of pooling, normalization, regularization, and activation in the layer descriptions. Model inputs must match the form of the preprocessed data. Model outputs must correspond to the requirements of the stated clinical problem, and for supervised learning should match the form of the reference standard annotations. If a previously published model architecture is employed, cite a reference that meets the preceding standards and fully describe every modification made to the model. Cite a reference for any proprietary model described previously, as well. In some cases, it may be more convenient to provide the structure of the model in code as supplemental data.
Item 23. Software libraries, frameworks, and packages. Specify the names and version numbers of all software libraries, frameworks, and packages used for model training and inference. A detailed hardware description may be helpful, especially if computational performance benchmarking is a focus of the work.
Item 24. Initialization of model parameters. Indicate how the parameters of the model were initialized. Describe the distribution from which random values were drawn for randomly initialized parameters. Specify the source of the starting weights if transfer learning is employed to initialize parameters. When there is a combination of random initialization and transfer learning, make it clear which portions of the model were initialized with which strategies.
Training
Item 25. Details of training approach. Describe the training procedures and hyperparameters in sufficient detail to enable another investigator to replicate the experiment. To fully document training, a manuscript should: (a) describe how training data were augmented (eg, types and ranges of transformations for images), (b) state how convergence of training of each model was monitored and what the criteria for stopping training were, and (c) indicate the values that were used for every hyperparameter, including which of these were varied between models, over what range, and using what search strategy. For neural networks, descriptions of hyperparameters should include at least the learning rate schedule, optimization algorithm, minibatch size, dropout rates (if any), and regularization parameters (if any). Discuss what objective function was employed, why it was selected, and to what extent it matches the performance required for the clinical or scientific use case. Define criteria used to select the best-performing model. If some model parameters are frozen or restricted from modification, for example in transfer learning, clearly indicate which parameters are involved, the method by which they are restricted, and the portion of the training for which the restriction applies. It may be more concise to describe these details in code in the form of a succinct training script, particularly for neural network models when using a standard framework.
Item 26. Method of selecting the final model. Describe the method and metrics used to select the best-performing model among all the models trained for evaluation against the held-out test set. If more than one model was selected, justify why this was appropriate.
Item 27. Ensembling technique. If the final algorithm involves an ensemble of models, describe each model comprising the ensemble in complete detail in accordance with the preceding recommendations. Indicate how the outputs of the component models are weighted and/or combined.
Evaluation
Item 28. Metrics of model performance. Describe the metrics used to assess the model’s performance and indicate how they address the performance characteristics most important to the clinical or scientific problem (15,16). Compare the presented model to previously published models.
Item 29. Statistical measures of significance and uncertainty. Indicate the uncertainty of the performance metrics’ values, such as with standard deviation and/or confidence intervals. Compute appropriate tests of statistical significance to compare metrics. Specify the statistical software, including version.
Item 30. Robustness or sensitivity analysis. Analyze the robustness or sensitivity of the model to various assumptions or initial conditions.
Item 31. Methods for explainability or interpretability. If applied, describe the methods that allow one to explain or interpret the model’s results and provide the parameters used to generate them. Describe how these methods were validated in the current study.
Item 32. Evaluation on internal data. Document and describe evaluation performed on internal data. If there are systematic differences in the structure of annotations or data between the training set and the internal test set, explain the differences, and describe the approach taken to accommodate the differences. Document whether there is consistency in performance on the training and internal test sets.
Item 33. Testing on external data. Describe the external data used to evaluate the completed algorithm. If no external testing is performed, note and justify this limitation. If there are differences in structure of annotations or data between the training set and the external testing set, explain the differences, and describe the approach taken to accommodate the differences.
Item 34. Clinical trial registration. If applicable, comply with the clinical trial registration statement from the International Committee of Medical Journal Editors (ICMJE). ICMJE recommends that all medical journal editors require registration of clinical trials in a public trials registry at or before the time of first patient enrollment as a condition of consideration for publication (17). Registration of the study protocol in a clinical trial registry, such as ClinicalTrials.gov or WHO Primary Registries, helps avoid overlapping or redundant studies and allows interested parties to contact the study coordinators.
The Results Section
Present the outcomes of the experiment in sufficient detail. If the description of the results exceeds the word count or other journal requirements, the data can be offered in a supplement to the manuscript.
Data
Item 35. Numbers of patients or examinations included and excluded. Document the numbers of patients, examinations, or images included and excluded based on each of the study’s inclusion and exclusion criteria. Include a flowchart or alternative diagram to show selection of the initial patient population and those excluded for any reason.
Item 36. Demographic and clinical characteristics of cases in each partition and dataset. Specify the demographic and clinical characteristics of cases in each partition and dataset. Identify sources of potential bias that may originate from differences in demographic or clinical characteristics, such as sex distribution, underrepresented racial or ethnic groups, phenotypic variations, or differences in treatment.
Model Performance
Item 37. Performance metrics and measures of statistical uncertainty. Report the final model’s performance. Benchmark the performance of the AI model against the reference standard, such as histopathologic identification of disease or a panel of medical experts with an explicit method to resolve disagreements. State the performance metrics on all data partitions and datasets, including any demographic subgroups.
Item 38. Estimates of diagnostic performance and their precision. For classification tasks, include estimates of diagnostic accuracy and their measures of statistical uncertainty, such as 95% confidence intervals. Apply appropriate methodology such as receiver operating characteristic analysis and/or calibration curves. When the direct calculation of confidence intervals is not possible, report nonparametric estimates from bootstrap samples. State which variables were shown to be predictive of the response variable. Identify the subpopulation(s) for which the prediction model worked most and least effectively. If applicable, recognize the presence of class imbalance (uneven distribution across data classes within or between datasets) and provide appropriate metrics to reflect algorithm performance (18,19).
Item 39. Failure analysis of incorrect results. Provide information to help understand incorrect results. If the task entails classification into two or more categories, provide a confusion matrix that shows tallies for predicted versus actual categories. Consider presenting examples of incorrectly classified cases to help readers better understand the strengths and limitations of the algorithm. Provide sufficient detail to frame incorrect results in the appropriate medical context.
The Discussion Section
This section provides four pieces of information: summary, limitations, implications, and future directions.
Item 40. Study limitations. Identify the study’s limitations, including those involving the study’s methods, materials, biases, statistical uncertainty, unexpected results, and generalizability. This discussion should follow succinct summarization of the results with appropriate context and explanation of how the current work advances our knowledge and the state of the art.
Item 41. Implications for practice, including intended use and/or clinical role. Describe the implications for practice, including the intended use and possible clinical role of the AI model. Describe the key impact the work may have on the field, including changes from current practice. Envision the next steps that one might take to build upon the results. Discuss any issues that would impede successful translation of the model into practice.
Other Information
Item 42. Provide a reference to the full study protocol or to additional technical details. State where readers can access the full study protocol or additional technical details if this description exceeds the journal’s word limit. For clinical trials, include reference to the study protocol text referenced in item 34. For experimental or preclinical studies, include reference to details of the AI methodology, if not fully documented in the manuscript or supplemental material. This information can help readers evaluate the validity of the study and can help researchers who want to replicate the study.
Item 43. Statement about the availability of software, trained model, and/or data. State where the reader can access the software, model, and/or data associated with the study, including conditions under which these resources can be accessed. Describe the algorithms and software in sufficient detail to allow replication of the study. Authors should deposit all computer code used for modeling and/or data analysis into a publicly accessible repository.
Item 44. Sources of funding and other support; role of funders. Specify the sources of funding and other support and the exact role of the funders in performing the study. Indicate whether the authors had independence in each phase of the study.
Conclusion
The 2024 Update of the CLAIM guideline provides a best practice checklist to promote transparency and reproducibility of medical imaging AI research. Its recommendations, derived from an expert panel through a Delphi consensus process, promote consistent reporting of scientific advances of AI in medical imaging.
A.S.T., M.E.K., and A.A.G. contributed equally to this work.
Members of the CLAIM 2024 Update Panel are listed at the end of this article.
Authors declared no funding for this work.
Disclosures of conflicts of interest: A.S.T. Trainee editorial board member for Radiology: Artificial Intelligence. M.E.K. Meeting attendance support from Bayer; trainee editorial board member for Radiology: Artificial Intelligence. A.A.G. CIHR Postdoctoral Fellowship; systems and methods for segmenting an image patent number: 11080857; shareholder in NeuralSeg, GeminiOV, ACLIP, and NodeAI; trainee editorial board member for Radiology: Artificial Intelligence. J.T.M. Grants from Siemens and Bunker Hill Health to university; royalties from GE (payments paid to author through university); support for attending meetings from Gibson Dune and RSNA; serves on RSNA AI Committee; stock/stock options in Annexon Biosciences; spouse employment at Annexon Biosciences, Bristol Myers Squibb; associate editor of Radiology: Artificial Intelligence; unpaid consulting for Nuance/Microsoft. L.M. Salary and travel support from Radiological Society of North America (RSNA) paid to employer for service as Editor of Radiology; grants from Siemens, Gordon and Betty Moore Foundation, Mary Kay Foundation, and Google; consulting fees from Lunit Insight, ICAD, Guerbet, and Medscape; payment or honoraria from ICAD, Lunit, and Guerbet; support for travel from British Society of Breast Radiology, European Society of Breast Imaging, and Korean Society of Radiology; participation on board of ACR DSMB; Society of Breast Imaging board; stock/stock options in Lunit. S.H.P. Honoraria from Bayer and Korean Society of Radiology; support for travel from Korean Society of Radiology. C.E.K. Salary and travel support from Radiological Society of North America (RSNA) paid to employer for service as Editor of Radiology: Artificial Intelligence; travel expenses (Sectra USA Radiology Advisory Panel); US Patent Pending re. AI orchestrator platform.
CLAIM 2024 Update Panel: Sunhy Abbara, Saif Afat, Udunna C. Anazodo, Anna Andreychenko, Folkert W. Asselbergs, Aldo Badano, Bettina Baessler, Bayarbaatar Bold, Sotirios Bisdas, Torkel B. Brismar, Giovanni E. Cacciamani, John A. Carrino, Julius Chapiro, Michael F. Chiang, Tessa S. Cook, Renato Cuocolo, John Damilakis, Roxana Daneshjou, Carlo N. De Cecco, Hesham Elhalawani, Guillermo Elizondo-Riojas, Andrey Fedorov, Benjamin Fine, Adam E. Flanders, Judy Wawira Gichoya, Maryellen L. Giger, Safwan S. Halabi, Sven Haller, William Hsu, Krishna Juluru, Jayashree Kalpathy-Cramer, Apostolos H. Karantanas, Felipe C. Kitamura, Burak Kocak, Dow-Mu Koh, Elmar Kotter, Elizabeth A. Krupinski, Curtis P. Langlotz, Cecilia S. Lee, Mario Maas, Anant Madabhushi, Lena Maier-Hein, Kostas Marias, Luis Martí-Bonmatí, Jaishree Naidoo, Emanuele Neri, Robert Ochs, Nikolaos Papanikolaou, Thomas Papathomas, Katja Pinker-Domenig, Daniel Pinto dos Santos, Fred Prior, Alexandros Protonotarios, Mauricio Reyes, Veronica Rotemberg, Jeffrey D. Rudie, Emmanuel Salinas-Miranda, Francesco Sardanelli, Mark E. Schweitzer, Luca Maria Sconfienza, Ronnie Sebro, Prateek Sharma, An Tang, Antonios Tzortzakakis, Jeroen van der Laak, Peter M. A. van Ooijen, Vasantha K. Venugopal, Jacob J. Visser, Bradford J. Wood, Carol C. Wu, Greg Zaharchuk, Marc Zins
The opinions expressed in this article are those of the authors and not necessarily the view of the U.S. Food and Drug Administration (FDA), National Institutes of Health (NIH), Department of Health and Human Services (DHHS), or the United States government. This article is not endorsed or approved by the FDA, NIH, DHHS, or the U.S. government. The mention of commercial products, their source, or their use in connection with material reported herein is not to be construed as an actual or implied endorsement by the United States government.
Abbreviations:
- AI
- artificial intelligence
- CLAIM
- Checklist for Artificial Intelligence in Medical Imaging
Contributor Information
Charles E. Kahn, Jr, Email: ckahn@rsna.org.
Collaborators: Sunhy Abbara, Saif Afat, Udunna C. Anazodo, Anna Andreychenko, Folkert W. Asselbergs, Aldo Badano, Bettina Baessler, Bayarbaatar Bold, Sotirios Bisdas, Torkel B. Brismar, Giovanni E. Cacciamani, John A. Carrino, Julius Chapiro, Michael F. Chiang, Tessa S. Cook, Renato Cuocolo, John Damilakis, Roxana Daneshjou, Carlo N. De Cecco, Hesham Elhalawani, Guillermo Elizondo-Riojas, Andrey Fedorov, Benjamin Fine, Adam E. Flanders, Judy Wawira Gichoya, Maryellen L. Giger, Safwan S. Halabi, Sven Haller, William Hsu, Krishna Juluru, Jayashree Kalpathy-Cramer, Apostolos H. Karantanas, Felipe C. Kitamura, Burak Kocak, Dow-Mu Koh, Elmar Kotter, Elizabeth A. Krupinski, Curtis P. Langlotz, Cecilia S. Lee, Mario Maas, Anant Madabhushi, Lena Maier-Hein, Kostas Marias, Luis Martí-Bonmatí, Jaishree Naidoo, Emanuele Neri, Robert Ochs, Nikolaos Papanikolaou, Thomas Papathomas, Katja Pinker-Domenig, Daniel Pinto dos Santos, Fred Prior, Alexandros Protonotarios, Mauricio Reyes, Veronica Rotemberg, Jeffrey D. Rudie, Emmanuel Salinas-Miranda, Francesco Sardanelli, Mark E. Schweitzer, Luca Maria Sconfienza, Ronnie Sebro, Prateek Sharma, An Tang, Antonios Tzortzakakis, Jeroen van der Laak, Peter M. A. van Ooijen, Vasantha K. Venugopal, Jacob J. Visser, Bradford J. Wood, Carol C. Wu, Greg Zaharchuk, and Marc Zins
References
- 1. Mongan J , Moy L , Kahn CE Jr . Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers . Radiol Artif Intell 2020. ; 2 ( 2 ): e200029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klontzas ME , Gatti AA , Tejani AS , Kahn CE Jr . AI reporting guidelines: How to select the best one for your research . Radiol Artif Intell 2023. ; 5 ( 3 ): e230055 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belue MJ , Harmon SA , Lay NS , et al . The low rate of adherence to Checklist for Artificial Intelligence in Medical Imaging criteria among published prostate MRI artificial intelligence algorithms . J Am Coll Radiol 2023. ; 20 ( 2 ): 134 – 145 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhandari A , Scott L , Weilbach M , Marwah R , Lasocki A . Assessment of artificial intelligence (AI) reporting methodology in glioma MRI studies using the Checklist for AI in Medical Imaging (CLAIM) . Neuroradiology 2023. ; 65 ( 5 ): 907 – 913 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Si L , Zhong J , Huo J , et al . Deep learning in knee imaging: a systematic review utilizing a Checklist for Artificial Intelligence in Medical Imaging (CLAIM) . Eur Radiol 2022. ; 32 ( 2 ): 1353 – 1361 . [DOI] [PubMed] [Google Scholar]
- 6. Sivanesan U , Wu K , McInnes MDF , Dhindsa K , Salehi F , van der Pol CB . Checklist for Artificial Intelligence in Medical Imaging reporting adherence in peer-reviewed and preprint manuscripts with the highest Altmetric attention scores: A meta-research study . Can Assoc Radiol J 2023. ; 74 ( 2 ): 334 – 342 . [DOI] [PubMed] [Google Scholar]
- 7. Kocak B , Keles A , Akinci D’Antonoli T . Self-reporting with checklists in artificial intelligence research on medical imaging: a systematic review based on citations of CLAIM . Eur Radiol 2024. ; 34 ( 4 ): 2805 – 2815 . [DOI] [PubMed] [Google Scholar]
- 8. Tejani AS , Klontzas ME , Gatti AA , et al . Updating the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) for reporting AI research . Nat Mach Intell 2023. ; 5 ( 9 ): 950 – 951 . [Google Scholar]
- 9. Altman DG , Simera I , Hoey J , Moher D , Schulz K . EQUATOR: reporting guidelines for health research . Lancet 2008. ; 371 ( 9619 ): 1149 – 1150 . [DOI] [PubMed] [Google Scholar]
- 10. Simera I , Moher D , Hirst A , Hoey J , Schulz KF , Altman DG . Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR Network . BMC Med 2010. ; 8 ( 1 ): 24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen JF , Korevaar DA , Altman DG , et al . STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration . BMJ Open 2016. ; 6 ( 11 ): e012799 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gasser U . An EU landmark for AI governance . Science 2023. ; 380 ( 6651 ): 1203 . [DOI] [PubMed] [Google Scholar]
- 13. European Commission . European Health Data Space . https://health.ec.europa.eu/ehealth-digital-health-and-care/european-health-data-space_en. Accessed March 2023 .
- 14. Kohn MA , Senyak J . Sample size calculators . https://sample-size.net/. Accessed February 2024 . [Google Scholar]
- 15. Reinke A , Tizabi MD , Baumgartner M , et al . Understanding metric-related pitfalls in image analysis validation . Nat Methods 2024. ; 21 ( 2 ): 182 – 194 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maier-Hein L , Reinke A , Godau P , et al . Metrics reloaded: recommendations for image analysis validation . Nat Methods 2024. ; 21 ( 2 ): 195 – 212 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International Committee of Medical Journal Editors . Clinical Trials . http://www.icmje.org/recommendations/browse/publishing-and-editorial-issues/clinical-trial-registration.html. Accessed April 2024 .
- 18. Megahed FM , Chen YJ , Megahed A , Ong Y , Altman N , Krzywinski M . The class imbalance problem . Nat Methods 2021. ; 18 ( 11 ): 1270 – 1272 . [DOI] [PubMed] [Google Scholar]
- 19. Fu GH , Yi LZ , Pan J . Tuning model parameters in class-imbalanced learning with precision-recall curve . Biom J 2019. ; 61 ( 3 ): 652 – 664 . [DOI] [PubMed] [Google Scholar]