Abstract
The nonstructural protein NSP2 is a component of rotavirus replication intermediates and accumulates in cytoplasmic inclusions (viroplasms), sites of genome RNA replication and the assembly of subviral particles. To better understand the structure and function of the protein, C-terminally His-tagged NSP2 was expressed in bacteria and purified to homogeneity. In its purified form, the protein did not exist as a monomer but rather was present as an 8S-10S homomultimer consisting of 6 ± 2 subunits of recombinant NSP2 (rNSP2). As shown by gel mobility shift assays, the rNSP2 multimers bound to RNA in discrete cooperative steps to form higher-order RNA-protein complexes. The RNA-binding activity of the rNSP2 multimers was determined to be nonspecific and to have a strong preference for single-stranded RNA over double-stranded RNA, for which it displayed little affinity. Enzymatic analysis revealed that rNSP2 possessed an associated nucleoside triphosphatase (NTPase) activity in vitro, which in the presence of Mg2+ catalyzed the hydrolysis of each of the four NTPs to NDPs with equal efficiency. Evidence indicating that the hydrolysis of NTP resulted in the covalent linkage of the γ-phosphate to rNSP2 was obtained. Additional experiments showed that NSP2 expressed transiently in MA014 cells is phosphorylated. We propose that NSP2 functions as a molecular motor, catalyzing the packaging of viral mRNA into core-like replication intermediates through the energy derived from its NTPase activity.
Rotaviruses, members of the family Reoviridae, are an important cause of diarrheal disease in humans and many species of animals (7). The mature virion is an icosahedron that consists of three concentric layers of protein and contains a genome of 11 segments of double-stranded RNA (dsRNA). The major component of the innermost layer is VP2, a nonspecific RNA-binding protein that can self-assemble into T=1 structures (19, 35). VP2, the minor core proteins VP1 and VP3, and the viral genome form the core of the virion (3). VP1 is the viral RNA-dependent RNA polymerase (29, 42), and VP3 is responsible for capping of viral mRNAs (20, 26, 33). Cores which are purified from virions and disrupted by exposure to hypotonic buffer (open cores) have replicase activity that catalyzes the synthesis of dsRNA from viral mRNA in vitro (6). Cores surrounded by VP6, the only component of the intermediate layer of the virion, form double-layered particles (DLPs) (35). DLPs have transcriptase activity and direct the synthesis of viral mRNA in infected cells (8). The outer layer of the virion is made up of the glycoprotein VP7 and the spike protein VP4 (35).
Rotavirus dsRNA synthesis and the morphogenesis of cores and DLPs are believed to occur in cytoplasmic inclusions termed viroplasms (32). Free dsRNA has not been detected in infected cells, suggesting that dsRNA is synthesized following packaging of the mRNA template into subviral particles (SVPs) (27). Indeed, characterization of replication intermediates isolated from infected cells indicates that only after the association of mRNA with core-like structures does the synthesis of dsRNA occur (11, 27). Evidence that VP2 and the core play an essential role in dsRNA synthesis has been provided by analysis of a rotavirus mutant with a temperature-sensitive defect in VP2 assembly and by reconstitution of replicase activity in vitro with VP1 and VP2 (21, 25, 29). While cell-free systems containing only core proteins that support the synthesis of dsRNA have been developed (6), the dsRNA product of these systems is not packaged. So far, the only cell-free system which supports RNA replication and packaging that has been described is one that contains not only the core proteins but nonstructural proteins as well (24). Thus, although not required for dsRNA synthesis, the nonstructural proteins may be essential for RNA packaging. The nonstructural proteins most likely to be involved in the packaging process are NSP2 (NS35) and NSP5 (NS26), since both of these proteins are components of intracellular replication intermediates that can direct the synthesis of dsRNA in vitro through an associated replicase activity (11, 14).
NSP2 (35 kDa) is a conserved basic protein that is expressed to high levels in the infected cell. Immunofluorescence studies have shown that NSP2 accumulates in viroplasms (32), a feature that is consistent with its suspected role in genome packaging and replication. Additional insights into the function of NSP2 have come from studies on tsE, an SA11 mutant with a temperature-sensitive lesion in the NSP2 gene (gene 8) (36). Cells infected with tsE and maintained at nonpermissive temperatures contain few viroplasms, lack replication intermediates with replicase activity, and produce virus particles that are mostly empty, i.e., lack dsRNA (5, 37). Results from the tsE studies have led to the suggestion that NSP2 may play an essential role in RNA packaging and may coordinate packaging with virion assembly. Two other lines of evidence also suggest that NSP2 has a role in packaging and replication: (i) NSP2 has nonspecific affinity for single-stranded RNA (ssRNA) in vitro and is bound to ssRNA and partially replicated viral RNA in vivo (2, 17), and (ii) cross-linking experiments have established that NSP2 forms complexes with VP1 and possibly VP3 in vivo (2, 16).
To gain a better understanding of the function of NSP2 in rotavirus replication, we have analyzed the structural and enzymatic properties of purified recombinant NSP2 (rNSP2). Consistent with earlier reports, rNSP2 was found to have nonspecific affinity for ssRNA but little affinity for dsRNA. In its purified form the protein was found to exist not as a monomer but rather as a stable homomultimer consisting of four to eight subunits. Enzymatic analysis showed that rNSP2 has an associated nucleoside triphosphatase (NTPase) activity which, in the presence of Mg2+, catalyzes the hydrolysis of all four NTPs to NDPs. Evidence was obtained indicating that the hydrolysis of NTP results in the phosphorylation of NSP2 both in vitro and in vivo. The NTPase activity of NSP2 may provide the energy necessary for the protein to function as a molecular motor that directs the packaging of viral mRNA.
MATERIALS AND METHODS
Cell culture and viruses.
Fetal rhesus monkey kidney (MA104) cells were maintained in medium 199 (Gibco) supplemented with 5% fetal bovine serum (HyClone), penicillin (100 IU/ml), and streptomycin (100 μg/ml). Simian rotavirus SA11 and the reassortant virus DxRRV were propagated and titered in MA104 cells. With the exception of the segment encoding VP7, which originates from the human rotavirus D, the genome segments of DxRRV are from rhesus rotavirus RRV (22).
Construction of the NSP2 expression vector.
Gene 8 was amplified from pSP65g8 (17) by using the Ampli Taq system (Life Technologies) and the positive-sense primer CCGAAACCatggctgagctag (NcoI site is underlined) and the negative-sense primer CGGAGATCTacgccaacttgagaaac (BglII site is underlined). Virus-specific sequences in primers are in lowercase. The amplification conditions were as follows: 94°C for 5 min, 1 cycle; 94°C for 1 min, 37°C for 1 min, and 72°C for 2.5 min, 10 cycles; 94°C for 1 min, 45°C for 1 min, and 72°C for 2.5 min, 20 cycles. The 998-bp product was gel purified and ligated into the PCR cloning vector pT7Blue (Novagen). Following transformation of Escherichia coli DH5, bacteria containing the appropriate plasmid (pT7Bg8) were identified based on antibiotic resistance, plasmid size, and restriction enzyme digestion. The sequence accuracy of the gene 8 insert in pT7B-g8 was confirmed by automated sequencing with an ABI PRISM 310 genetic analyzer (PE Applied Biosystems). To express rNSP2, pT7Bg8 was digested with NcoI and BglII, and the gene 8 fragment was ligated into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pQE60 (Qiagen), similarly digested with NcoI and BglII. Following transformation into E. coli DH5α, bacteria with the appropriate plasmid (pQE60g8) were identified and the plasmid was purified. pQE60g8 was then electroporated into E. coli M15 carrying the pREP4 repressor plasmid. Appropriate transformants were identified based on antibiotic resistance, restriction enzyme digestion, and expression of rNSP2. In pQE60g8, the open reading frame (ORF) for NSP2 is situated immediately upstream from six in-frame codons for His. Thus, rNSP2 expressed from pQE60g8 is tagged at its C terminus with six His residues.
Expression and purification of rNSP2.
E. coli M15[pREP4] containing pQE60g8 were grown to an optical density at 600 nm of 0.5 in Terrific Broth (Quality Biologics), and the expression of NSP2 was induced by adding IPTG to a final concentration of 1 mM. After incubation for 4 to 5 h at 37°C, the bacteria were recovered by centrifugation at 4,000 × g for 10 min, and the His6-tagged rNSP2 was purified under native conditions on a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen) according to the manufacturer’s protocols. The final eluate was dialyzed against low-salt buffer (LSB; 2 mM Tris-HCl [pH 7.2], 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT]) for 48 h at 4°C. The concentration of the purified protein was determined by Bradford assay using bovine serum albumin as the protein standard and by comparison with known amounts of bovine serum albumin coelectrophoresed on sodium dodecyl sulfate (SDS)-polyacrylamide gels and Coomassie blue staining. Purified rNSP2 was adjusted to a concentration of 0.5 mg per ml and stored at 4°C. The same protocol was used to express and purify His-tagged recombinant dihydrofolate reductase (DHFR). The expression plasmid containing the DHFR gene was provided by Qiagen.
Purified 35S-labeled rNSP2 was produced as described above except that rNSP2 expression was induced in Cys- and Met-free Dulbecco’s modified Eagle’s medium (MEM) containing 20 μCi of 35S-amino acids (35S-Express; 1,175 Ci/mmol; NEN) per ml of medium.
In vitro synthesis of RNAs.
The DNA template for synthesis of the Luc72 RNA probe was generated by amplifying a portion of the luciferase gene in plasmid pGL2 (Promega) with Taq polymerase (Life Technologies) and the positive-sense primer TAATACGACTCACTATACCATGGAAGACGCCAAAAACATAAAGAAAGG (the T7 core promoter sequence is underlined), and negative-sense primer GTTGCTCTCCAGCGGTTC. 32P-labeled Luc72 probe was transcribed from the amplified DNA with an Ambion MEGAshortscript kit according to the protocol of the manufacturer except that the concentration of cold (unlabeled) UTP was reduced by one-fourth and 50 μCi of [α-32P]UTP was included per 20 μl of reaction mixture (26). The 32P-labeled RNA probes were purified by electrophoresis on and elution from 8% polyacrylamide gels containing 7 M urea (25).
The DNA template for synthesis of green fluorescent protein (GFP) plus-sense RNA was amplified from the plasmid pGreen Lantern-1 (Life Technologies) by using the positive-sense primer AGAATGTATGTTATTGAATAT and the negative-sense primer, ACATCATACAACTATAACTTC. To produce gene 8 plus- and minus-sense RNAs, plasmids pSP65g8(+) and pSP65g8(−), respectively, were linearized with SacII, blunt-ended by treatment with T4 DNA polymerase, and transcribed by using the Ambion MAXIscript system (31). After runoff transcription, the RNA products were purified by phenol-chloroform extraction and isopropanol precipitation. The quality of GFP and gene 8 plus- and minus-sense RNAs was assessed by electrophoresis on 5% polyacrylamide gels containing 7 M urea (25). RNA concentrations were calculated from optical densities at 260 nm.
To generate gene 8 dsRNA, equal amounts of gene 8 plus- and minus-sense RNAs were annealed for 18 h at 42°C in 50 mM Tris-HCl (pH 8.3)–6 mM MgCl2–55 mM KCl. The sample was electrophoresed on a 1.5% agarose gel, and the desired 1-kb gene 8 dsRNA was recovered with a QIAquick gel extraction kit (Qiagen).
Gel shift assays.
The procedure used for analysis of rNSP2-RNA interactions by gel shift assay was similar to that described earlier (25). Typically, 32P-labeled probe (1 to 10 pmol) was incubated with rNSP2 (1 to 25 pmol) in LSB in a final volume of 15 to 35 μl for 30 min at room temperature in the presence or absence of competitor RNA. The reaction mixtures were analyzed by electrophoresis for 3 to 4 h at 175 V on nondenaturing 6% polyacrylamide gels containing 50 mM Tris-HCl and 50 mM glycine (pH 8.8) (25). Protein-probe complexes were detected on the gel by autoradiography, and intensities of the radiolabeled bands were quantitated with a phosphorimager.
Rate zonal centrifugation.
35S-labeled rNSP2, 32P-labeled probe, 35S-labeled rNSP2-RNA probe complexes, and protein standards were layered onto 12-ml 5 to 20% (wt/vol) sucrose gradients in LSB and centrifuged at 200,000 × g for 16 h in a Beckman SW40Ti rotor at 4°C. The proteins used as size markers in the gradients were thyroglobulin (650 kDa, 19S), catalase (250 kDa, 11.3S), and γ-globulin (156 kDa, 7S). One-milliliter fractions were collected from the gradients and analyzed for protein content by electrophoresis on 12% polyacrylamide gels containing SDS (SDS-12% PAGE) (18) and for RNA content by electrophoresis on 8% polyacrylamide gels containing 7 M urea (25).
Preparation of subviral particles.
SVPs were purified from MA104 cells infected with 10 to 20 PFU of DXRRV per cell as previously described (24). The infected cells were harvested at 6 h postinfection (p.i.) by washing and resuspension in cold hypotonic buffer (dilute reticulocyte standard buffer; 3 mM Tris-HCl [pH 8.1], 0.5 mM MgCl2, 3 mM NaCl) and then lysed by Dounce homogenization. Nuclei and large cellular debris were removed by centrifugation for 10 min at 10,000 × g, and SVPs were recovered from the supernatant by centrifugation at 200,000 × g for 2 h through 3-ml 15 to 30% (wt/vol) sucrose gradients in dilute reticulocyte standard buffer. The pellets were resuspended in HGD buffer (10 mM HEPES-HCl [pH 7.6], 10% glycerol, 2 mM DTT) and stored on ice until used. Approximately 100 μl of SVP suspension was prepared from 5 × 107 infected cells.
NTPase assay.
Typically, reaction mixtures for the NTPase assay contained 1 to 2 μg of rNSP2, 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2 and 10 μCi of α-32P-labeled ATP, GTP, CTP, or UTP (3,000 Ci/mmol; Amersham) in a final volume of 20 μl. After incubation at 37°C for 1 h, 1 μl of 0.5 mM EDTA was added to the reaction mixtures, and the samples were then deproteinized by phenol-chloroform extraction. One-microliter aliquots of each were spotted onto polyethyleneimine-cellulose F sheets (EM Science), and NTP, NDP, and Pi were resolved by ascending thin-layer chromatography (TLC) in 1.2 M LiCl. Radiolabeled spots on the sheets were detected by autoradiography and quantitated with a phosphorimager. NDP and NMP markers were made by limited hydrolysis of the radiolabeled NTP with 5 U of tobacco acid pyrophosphatase (Epicentre, Madison, Wis.).
In vitro phosphorylation of rNSP2 and NSP2 in SVPs.
The standard reaction mixture for in vitro phosphorylation contained 2 μg of rNSP2 or 2 μl of SVPs, 20 μCi of [γ-32P]NTP (800 to 3,000 Ci/mmol), 50 mM Tris-HCl (pH 7.5), and 5 mM MgCl2 in a final volume of 20 μl and was incubated at 37°C for 1 h. One-half of each reaction mixture was then combined with 10 μl of a solution containing 10 U of calf intestinal phosphatase (CIP; New England Biolabs), 50 mM Tris-HCl (pH 7.9), 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT and incubated at 37°C for 1 h. The phosphorylation and dephosphorylation assays were terminated by adding SDS sample buffer and heating to 100°C for 2 min. The phosphorylation and dephosphorylation of proteins was evaluated by SDS–12% PAGE and autoradiography.
In vivo phosphorylation of NSP2.
To determine whether NSP2 made during infection was phosphorylated, MA104 cells were infected at a multiplicity of infection of 10 to 50 with SA11 rotavirus. At 2 h p.i., the inoculum was replaced with MEM containing 5 μg of actinomycin D per ml. At 5 h p.i., the medium was replaced with either Cys- and Met-free MEM or phosphate-free MEM (LifeTechnologies). At 6 h p.i., 25 μCi of 35S-amino acids per ml of Cys- and Met-free MEM and 25 μCi of [32P]orthophosphate (150 mCi/ml, NEN) per ml of phosphate-free MEM were added. After incubation for 2 h, the cells were washed with phosphate-buffered saline and lysed by resuspension in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.5] 0.5% SDS, 1% Triton X-100, 1% sodium deoxycholate, 20 mM EDTA, 2 μg of aprotinin and 0.5 μg of leupeptin per ml). The lysate was clarified by centrifugation at 10,000 × g for 2 min.
The recombinant vaccinia virus vTF7.3 (10) was used to transiently express NSP2 in vivo. MA104 cells were grown to near confluency and infected with vTF7.3 at a multiplicity of infection of 10. At 1 h p.i., the inoculum was removed and replaced with transfection mixture made up of 4% Lipofectamine and 4 μg of pSP65g8 (17) per ml in medium 199. Beginning at 18 h p.i., the cells were labeled with 35S-amino acids and [32P]orthophosphate and harvested as described above.
Immunoprecipitation of NSP2 and Western blot analysis.
Polyclonal anti-NSP2 and anti-NSP5 antisera, were produced in guinea pigs by immunizing initially with 1 mg of rNSP2 and rNSP5, respectively, in Freund’s complete adjuvant and then immunizing at weeks 2 and 4 with 1 mg of rNSP2 and rNSP5 in Freund’s incomplete adjuvant. For immunoprecipitation, the polyclonal NSP2 and NSP5 antisera were added to clarified cell lysates in RIPA buffer at a dilution of 1:250, and the samples were gently mixed for 18 h at 4°C. After addition of protein A-Sepharose, the samples were incubated for an additional hour under the same conditions. The beads were recovered by low-speed centrifugation and washed three times with RIPA buffer. The immunoprecipitated proteins were released from the beads by boiling in SDS sample buffer and were identified by SDS–12% PAGE and autoradiography.
For Western blot analysis, proteins were resolved by SDS–12% PAGE and then electroblotted onto nitrocellulose sheets (Millipore). The blots were blocked by soaking in phosphate-buffered saline containing 5% skim milk suspension. Subsequently, the blots were incubated with either guinea pig anti-NSP2 polyclonal antisera, mouse anti-NSP2 monoclonal antibody (kindly provided by H. B. Greenberg), or mouse anti-Penta-His monoclonal antibody (Qiagen) at a dilution of 1:500. Goat anti-guinea pig and goat anti-mouse horseradish peroxidase-conjugated antibodies were used as secondary antibodies at a dilution of 1:5,000. The blots were developed by the Sigma Fast system.
RESULTS
Expression of rNSP2.
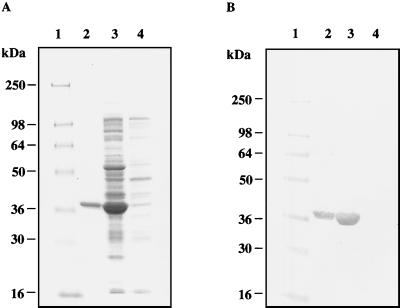
To study the structure and enzymatic properties of NSP2 in the absence of other viral proteins, a cDNA containing the NSP2 ORF of SA11 rotavirus was cloned into the IPTG-inducible bacterial expression vector pQE60. The NSP2 ORF was engineered into the vector such that the recombinant protein contained a C-terminal tag of six His residues. IPTG induction resulted in the expression of high levels of soluble rNSP2 in E. coli which could be purified by Ni-NTA affinity chromatography to near homogeneity as evaluated by SDS-PAGE and Coomassie blue staining (Fig. 1A). The identity of the rNSP2 was confirmed by Western blot analysis with an anti-NSP2 monoclonal antibody (Fig. 1B).
FIG. 1.
Expression and purification of rNSP2. Proteins were resolved by SDS-PAGE and stained with Coomassie blue (A) or blotted onto nitrocellulose and probed with anti-NSP2 antibody (B). Lane 1, protein standards; lane 2, His-tagged rNSP2 eluted from Ni2+-NTP agarose column; lane 3, bacterial lysate after induction with 1 mM IPTG; lane 4, bacterial lysate prior to induction.
Specificity of the RNA-binding activity of rNSP2.
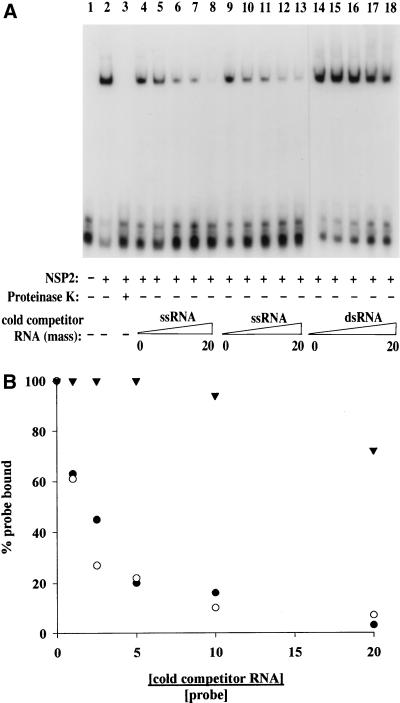
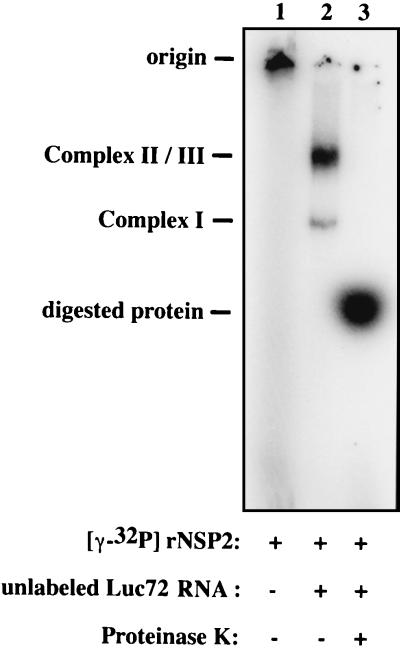
NSP2 synthesized in rotavirus-infected cells has been shown to have nonspecific RNA-binding activity (2, 17). To determine if rNSP2 had such an activity, the purified protein was incubated with 32P-labeled Luc72, an RNA probe of 72 nucleotides made from a gene encoding luciferase. rNSP2-probe complexes in the mixture were identified by electrophoresis on a nondenaturing 6% polyacrylamide gel in Tris-glycine buffer. The gel mobility shift assay indicated that rNSP2 bound the probe to produce a complex migrating in the upper one-third of the gel (Fig. 2A, lane 2). The suspected complex was not detected when the rNSP2-probe mixture was treated with proteinase K, confirming the presence of a protein component (Fig. 2A, lane 3). The presence of rNSP2 in complex eluted from the gel was verified by SDS-PAGE and Western blot assay (data not shown). These results demonstrated that rNSP2 possessed a sequence-independent RNA-binding activity.
FIG. 2.
Specificity of the RNA-binding activity of rNSP2. (A) rNSP2 (175 ng) and 32P-labeled Luc72 RNA (32 ng) were incubated alone or with a 1-, 2.5-, 5-, 10-, or 20-fold excess (in mass) of cold competitor RNA over probe RNA. The cold competitor RNAs were full-length rotavirus gene 8 mRNA (lanes 4 to 8), GFP mRNA (lanes 9 to 13), and rotavirus gene 8 dsRNA (14 to 18). In one case, after incubation of rNSP2 and probe, 40 μg of proteinase K was added and the sample was incubated for 15 min at room temperature (lane 3). Probe-protein complexes were detected by electrophoresis on a 6% nondenaturing polyacrylamide gel and autoradiography. The quantity of probe in the shifted band was determined with a phosphorimager. The amount of probe in the protein-RNA complexes of the competition reactions was normalized to the amount of probe in the protein-RNA complexes formed in the absence of cold competitor RNA (100%). (B) The extent of competition was assessed by plotting the ratio of cold competitor RNA over probe RNA (●, gene 8 ssRNA; ○, GFP ssRNA; ▾, gene 8 dsRNA) versus the percentage of probe bound to rNSP2.
The specificity of the RNA-binding activity of rNSP2 was addressed further by incubating rNSP2 and the 32P-labeled Luc72 probe with three cold competitor RNAs (gene 8 plus-sense RNA [mRNA], GFP RNA, and gene 8 dsRNA) and using the gel mobility shift assay to monitor the formation of rNSP2-probe complexes. In the presence of 1- to 2.5-fold (mass)-excess gene 8 mRNA and GFP RNA, the amounts of rNSP2-probe complex formed were reduced to 60% and 25 to 45%, respectively, of the amount of complex formed in the absence of competitor RNA (Fig. 2). With the addition of 10- to 20-fold-excess gene 8 mRNA and GFP RNA, the amounts of complex formed were reduced to 10 to 20% and <10%, respectively, of the amount formed by the control. In contrast, rNSP2 binding to the probe was not affected by the presence of up to 5-fold-excess cold gene 8 dsRNA and was only slightly affected (<10%) by the presence of 10-fold-excess dsRNA (Fig. 2). At 20-fold-excess dsRNA, the formation of the rNSP2-probe complex was reduced by only 25%. Overall, these results demonstrated that rNSP2 has sequence independent affinity for ssRNA and comparatively weak affinity for dsRNA.
Multiple subunits of rNSP2 bind to RNA.
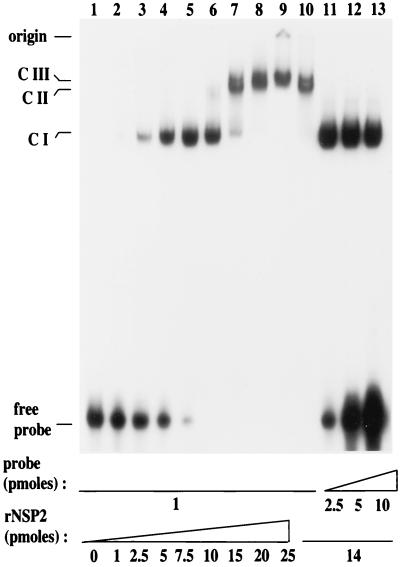
The gel mobility shift assays described above were performed under conditions of probe excess. To investigate whether higher-order rNSP2-RNA complexes could form under conditions where probe was limiting, increasing amounts of rNSP2 were titrated with fixed amounts of the 32P-labeled Luc72. The formation of rNSP2-probe complexes in the reaction mixtures was analyzed by gel mobility shift assay. The results showed that when 1 to 7.5 pmol of rNSP2 was added to 1 pmol of RNA probe (Fig. 3, lanes 2 to 5), probe was in excess and only a single rNSP2-probe complex (complex I) was detected. Complex I corresponds to the rNSP2-probe complex described above (Fig. 2). An increase of rNSP2 to 10 pmol resulted in the formation of low levels of another rNSP2-probe complex, designated complex II, which migrated in the gel more slowly than complex I. In the presence of 10 pmol of rNSP2, all of the probe was in the bound form (Fig. 3, lane 6). A further increase in rNSP2 from 10 to 15 pmol resulted in the loss of complex I and the concomitant formation of complexes II and III, the latter two migrating close to one another (Fig. 3, lane 7). In the presence of even greater amounts of rNSP2 (20 to 25 pmol), only complex III was detected (Fig. 3, lanes 8 and 9), indicating a complete conversion of complexes I and II to complex III. The addition of more than 25 pmol of rNSP2 to the reaction mixture resulted in the appearance of probe in the well and a partial or complete loss of complex III (data not shown). This finding suggests the formation of rNSP2-probe complexes even larger than complex III that cannot be resolved by these electrophoretic conditions. These data suggest that multiple copies of rNSP2 can bind to a single RNA probe, even one that is only 72 nucleotides in length.
FIG. 3.
Formation of higher-order complexes by rNSP2 and RNA. Complexes formed by incubating 1 pmol of 32P-labeled Luc72 RNA (probe) with 0 to 25 pmol of rNSP2 (lanes 1 to 9) were resolved by electrophoresis on a nondenaturing 6% polyacrylamide gel and detected by autoradiography. Reversibility of complex formation was evaluated by incubating 1 pmol of 32P-labeled Luc72 RNA with 14 pmol of rNSP2 for 30 min, then adding an additional 0, 1.5, 4, and 9 pmol of radiolabeled probe (final probe concentrations, 1 [lane 10], 2.5 [lane 11], 5 [lane 12], and 10 [lane 13] pmol), and incubating the mixture for another 30 min. The positions of unbound (free) probe, complex I (CI), complex II (CII), and complex III (CIII) are indicated.
The possibility that rNSP2-probe interactions were reversible was examined by adding 1 pmol of 32P-labeled Luc72 RNA to 14 pmol of rNSP2, incubating the mixture to allow probe-rNSP2 complexes to form, and then adding 1.5, 4, and 9 pmol of additional probe (final probe concentrations, 2.5, 5, and 10 pmol, respectively) to the reaction mixtures. As shown in Fig. 3 (lane 10), coincubation of 1 pmol of probe with 14 pmol of rNSP2 (14 pmol) resulted in the formation of complexes II and III but little or no complex I. In this reaction mixture, no free probe was present, indicating that probe was limiting (lane 10). The addition of greater amounts of probe led to the appearance of complex I and the loss of complexes II and III (Fig. 3, lanes 11 to 13), indicating that the interaction between protein and RNA in complexes II and III is reversible.
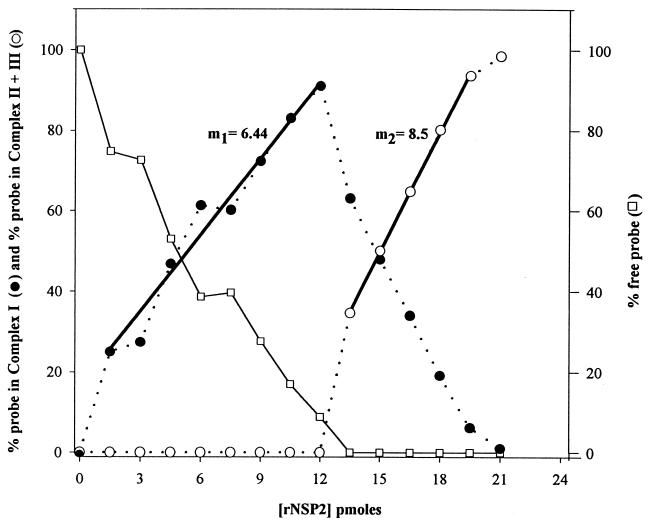
The transient nature of complex II (Fig. 3, lanes 7 and 10) indicated that cooperativity between free (unbound) rNSP2 and rNSP2 which was bound to RNA could be a factor in the formation of higher-order rNSP2-probe complexes, i.e., complexes II and III. Hence, as defined by the following binding reactions, the formation of complexes II and III would be more efficient than the formation of complex I: step I, rNSP2 + probe → complex I; step II, rNSP2 + complex I → complex II + complex III. To test this possibility, we performed an experiment whereby increasing amounts of rNSP2 (0 to 24 pmol), in steps of 1.5 pmol, were added to a constant amount (3 pmol) of the 32P-labeled Luc72 probe. The complexes were resolved by electrophoresis, and the percentage of probe in complex I and in complex II and complex III combined (complex II + III) in comparison to total probe in the reaction mixtures was determined. The results showed that when the rNSP2 concentration in the reaction mixtures was increased between 0 to 12 pmol, a corresponding and near linear increase was detected in the formation of complex I while the level of free probe decreased proportionally (Fig. 4). When the rNSP2 concentration was increased to 13.5 pmol, free probe was no longer detected, and of the bound probe, ∼65% was in complex I and ∼35% was in complex II + III. Further increase in rNSP2 resulted in a decrease in the level of complex I to the point that it was no longer detected (21 pmol) and resulted in a corresponding and near linear increase in the accumulation of complex II + III (Fig. 4). Comparison of the slopes calculated for the linear portion of the curves representing the formation of complex I and complex II + III showed that over a constant step increase of rNSP2 concentration, complex II + III was formed more efficiently than complex I. This finding suggests a positive cooperativity between rNSP2 subunits as they sequentially bind to RNA.
FIG. 4.
Cooperativity between rNSP2 multimers in forming higher-order RNA-probe complexes. 32P-labeled Luc72 RNA (3 pmol) was incubated with 0 to 21 pmol of rNSP2, added in steps of 1.5 pmol, and the complexes in the mixtures were resolved by electrophoresis on a 6% nondenaturing gel and quantitated with a phosphorimager. The formation of complex I (●) and complex II + III (○) in the reaction mixtures was compared by plotting the percent probe bound in the final product of each step [for step I, complex I; for step 2, complex II + III) as a function of the amount of rNSP2 in each reaction mixture. Slopes were determined for the linear portions of the curves representing the formation of complex I (m1) and of complex II + III (m2). The percent free probe (□) in each reaction mixture was also plotted as a function of rNSP2 concentration.
rNSP2 is a 10S homomultimer.
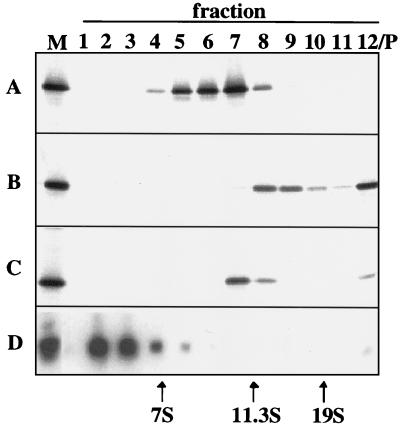
To determine the multimeric state of rNSP2 and rNSP2-RNA complexes, purified 35S-labeled rNSP2 in the absence and presence of Luc72 RNA and protein size markers was sedimented through linear 5 to 20% sucrose gradients. Electrophoretic analysis of the gradient fractions revealed that in the absence of RNA, nearly all of the rNSP2 migrated with a size of 8 to 10S and with a molecular mass of 140 to 250 kDa (Fig. 5A). From this information, we calculated that rNSP2 self-assembles into homomultimers, each made up of 6 ± 2 monomers. Notably, no rNSP2 was detected in fractions of the gradient expected to contain the protein monomer.
FIG. 5.
Sedimentation analysis of rNSP2 and rNSP2-RNA complexes. The following samples were centrifuged through 5 to 20% sucrose gradients: 35S-labeled rNSP2 (1.5 nmol) (A), 35S-labeled rNSP2 (1.5 nmol) incubated with cold Luc72 RNA (200 pmol) of which one-half was directly applied to the gradient (B) and the other half was treated with 5 μg of RNase A for 10 min at room temperature prior to being applied (C), and 32P-labeled Luc72 RNA (50 pmol) (D). Fractions (1 ml) from the gradients were analyzed by SDS–12% PAGE and autoradiography (A to C) and by electrophoresis on 8% polyacrylamide gels containing 7 M urea and autoradiography (D). 35S-labeled rNSP2 (A to C) and 32P-labeled Luc72 (D) were coelectrophoresed as markers (M). The pellet (P) is contained within fraction 12.
In the presence of unlabeled Luc72 RNA, one-half the rNSP2 migrated in the sucrose gradient with a size of 12 to 15S and with a molecular mass of 280 to 500 kDa. The other half of the rNSP2 was detected in the pellet fraction of the gradient, suggesting the formation of even larger protein-RNA complexes (Fig. 5B). Addition of RNase A to the rNSP2-RNA sample prior to sedimentation caused most of the 35S-labeled rNSP2 to shift back in size to 10S (Fig. 5C). By itself, Luc72 RNA barely sedimented into the gradient (Fig. 5D). Hence, the observed migration of the rNSP2-RNA to 12 to 15S and to the pellet fraction is probably a result of multiple copies of the 8S-10S rNSP2 homomultimer binding to the same RNA molecule and not due to the increase in mass contributed by the RNA. Aliquots of the fractions containing rNSP2-RNA complexes (Fig. 5B, fractions 8, 9, and pellet), when analyzed by gel mobility shift assay, revealed the presence of complexes I and III (data not shown). This finding is consistent with the idea that complex III and the 12 to 15S material both contain RNA molecules to which multiple copies of the rNSP2 are bound.
rNSP2 has an NTPase activity.
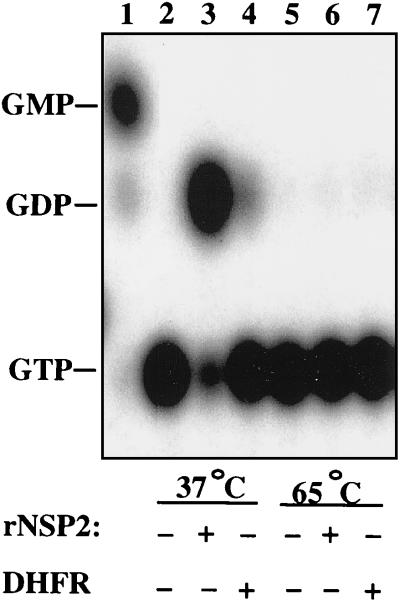
Earlier studies have suggested that NSP2 may be required for the assembly of replication intermediates with replicase activity, possibly functioning to package viral mRNA into viral cores where the RNA undergoes replication (2, 5, 27). As packaging of viral nucleic acids into capsids is thought to represent an energy-dependent process, we postulated that NSP2 might have an associated NTPase activity from which it could derive energy. To test rNSP2 for NTPase activity, we incubated the purified recombinant protein at 37°C with [α-32P]GTP and MgCl2 and resolved the products of the reaction by TLC. As shown in Fig. 6 (lane 3), in the presence of rNSP2, [α-32P]GTP was hydrolyzed to [α-32P]GDP, suggesting that rNSP2 possessed NTPase activity. Two control assays were performed to exclude the possibility that a copurifying bacterial protein, and not rNSP2, was responsible for the NTPase activity. First, a similar volume of His-tagged DHFR, expressed and purified in the same manner as rNSP2, was assayed for NTPase activity; second, purified rNSP2 and DHFR were each assayed for NTPase activity at 65°C, since characteristically bacterial NTPases are thermoresistant. The NTPase activity displayed by rNSP2 at 37°C was completely inhibited at 65°C, and no such activity was detected for DHFR at either temperature (Fig. 6). These results demonstrated that rNSP2 was responsible for the NTPase activity observed in the assay.
FIG. 6.
rNSP2 possesses an NTPase activity. Reaction mixtures containing no added protein or containing 1 μg of rNSP2 or 2 μg of DHFR and 10 μCi of [α-32P]GTP were incubated for 1 h at 37 or 65°C. The products of the reaction mixtures were resolved by TLC and detected by autoradiography. The positions of GDP and GMP were determined by cochromatography of markers prepared by partial digestion of [α-32P]GTP with tobacco acid pyrophosphatase (lane 1).
Properties of the NTPase activity of rNSP2.
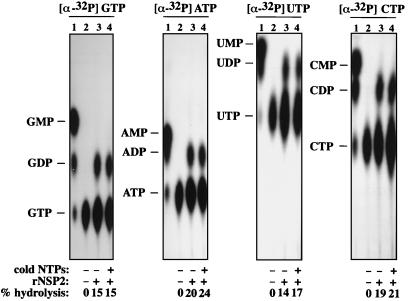
To determine if the NTPase activity of rNSP2 preferentially hydrolyzed one or more of the NTPs, we incubated rNSP2 with α-32P-labeled ATP, CTP, GTP, or UTP and analyzed the products by TLC. The specificity was further addressed by adding rNSP2 to reaction mixtures containing one of the radiolabeled NTPs and a 10-fold excess of each of the three other (cold) NTPs. The results showed that rNSP2 hydrolyzed all four NTPs to NDPs and did so to similar extents both in the assays containing only a single radiolabeled NTP and in the assays containing a single radiolabeled NTP along with cold competitor NTPs (Fig. 7). Thus, the NTPase activity of rNSP2 is nonspecific. Remarkably, the extent of hydrolysis in the assays ranged between 15 and 25%, regardless of whether 0.625 or 6.25 μM total NTPs were present in the reaction mixtures (Fig. 7). The most likely explanation for this phenomenon is that the products of NTP hydrolysis interacted with the NTPase to prevent an additional net increase in the accumulation of NDP and Pi when the ratio of substrate to product reached ∼4:1. Although the enzyme kinetics of the NTPase of rNSP2 have yet to be fully examined, these results suggest that the NTPase activity was being affected by the products of hydrolysis through uncompetitive or noncompetitive feedback inhibition.
FIG. 7.
Specificity of the NTPase activity of rNSP2. Reaction mixtures contained no protein or 1 μg of rNSP2 and 0.625 μM one of the four [α-32P]NTPs. In some cases, 6.25 μM each of the three (cold) NTPs not represented by the radiolabeled NTP were also added to the reaction mixture. After incubation for 1 h at 37°C, the products of the reactions were resolved by TLC, detected by autoradiography, and quantitated with a phosphorimager. Percent hydrolysis = (quantity of [32P]NDP)/(quantity of [32P]NDP and [32P]NTP) × 100. Markers were generated by partial digestion of [α-32P]NTPs with tobacco acid pyrophosphatase.
Analysis of the cofactor requirement for NTP hydrolysis showed that Mg2+ was essential and that NTPase activity was maximal at 1 to 5 mM Mg2+ (data not shown). Higher concentrations (20 mM) of Mg2+ inhibited the NTPase activity of rNSP2. Ca2+ did not serve as a cofactor for NTP hydrolysis (data not shown).
NTP hydrolysis results in rNSP2 phosphorylation.
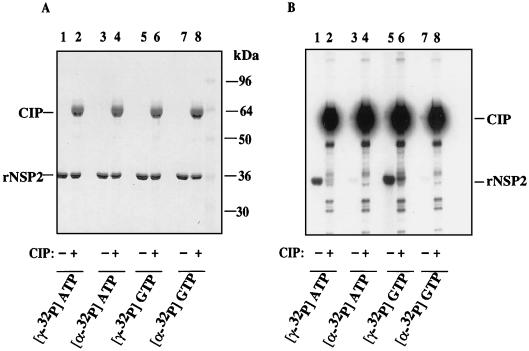
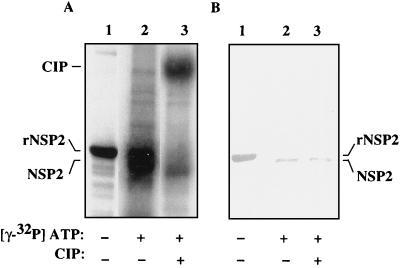
To examine the effect of NTP hydrolysis on the status of rNSP2, the protein was incubated with α-32P-labeled ATP or GTP or γ-32P-labeled ATP or GTP and then analyzed by SDS-PAGE and Coomassie blue staining. The results showed that the NTPase assay neither affected the migration nor caused the degradation of rNSP2 (Fig. 8A). However, autoradiography revealed that rNSP2 became radiolabeled in the NTPase assay when γ-32P-labeled ATP or GTP was present and not when α-32P-labeled ATP or GTP was present (Fig. 8B). This finding indicated that rNSP2 was phosphorylated as a result of NTP hydrolysis and that the cleaved γ-phosphate of NTP was transferred to the protein. The fact that the protein was not radiolabeled when incubated with α-32P-labeled ATP or GTP ruled out the possibility that uncleaved NTPs were being linked to the protein. The nature of the bond between the protein and the γ-phosphate was further examined by treatment of γ-32P-labeled rNSP2 with phosphatase. As shown in Fig. 8A and B (lanes 2 and 6), exposure to phosphatase did not affect the integrity of rNSP2 but did decrease the extent of phosphorylation. Thus, rNSP2 catalyzed the hydrolysis of NTPs to NDP, and in doing so became phosphorylated by the covalent linkage of the cleaved γ-phosphates.
FIG. 8.
Autophosphorylation of rNSP2. Purified rNSP2 was incubated with α-32P-labeled ATP or GTP or with γ-32P-labeled ATP or GTP for 1 h at 37°C. Afterwards, 10 U of CIP was added to some reaction mixtures, which were then incubated for 1 h at 37°C. Proteins in the samples were detected by SDS–12% PAGE and staining with Coomassie blue (A). 32P-labeled proteins in the gel shown in (A) were identified by autoradiography (B). CIP becomes radiolabeled due to its affinity for NTPs.
Phosphorylation of NSP2 expressed in vivo.
While there are no reports in the literature of the phosphorylation of NSP2 in infected cells, the results presented above led us to reassess this possibility. As a first approach, we examined whether the NSP2 that is associated with SVPs isolated from rotavirus-infected cells could be phosphorylated in vitro under the conditions used to assay rNSP2 for phosphorylation. The results showed that SVP NSP2 was radiolabeled when incubated with [γ-32P]ATP and that the 32P label was lost upon treatment with phosphatase (Fig. 9A, lanes 2 and 3). The identity of the protein in the 32P-labeled band was confirmed to be NSP2 by Western blot assay (Fig. 9B). NSP2 expressed in vivo had a slightly greater mobility upon electrophoresis than did rNSP2 (Fig. 9); this is because the His6 tag of rNSP2 increases its molecular mass by approximately 700 Da. Together, these data demonstrated that NSP2 made in infected cells, like rNSP2, can be a substrate for phosphorylation.
FIG. 9.
Phosphorylation of NSP2 from infected cells. Subviral particles were incubated with 10 μCi of [γ-32P]ATP for 1 h at 37°C. One-half of the reaction mixture was treated at room temperature with 10 U of CIP for 1 h at 37°C, and then the untreated (lane 2) and treated (lane 3) portions of the reaction mixture were resolved by SDS–12% PAGE. Radiolabeled proteins in the gel were detected by autoradiography (A). The position of NSP2 in the gel was confirmed by Western blot assay with polyclonal anti-NSP2 antibody (B). 35S-labeled rNSP2 was coelectrophoresed in lane 1.
As another approach for examining whether NSP2 expressed in vivo could be phosphorylated, MA104 cells were programmed to transiently express NSP2 with the vaccinia virus vTF7.3 expression system or were infected with SA11 rotavirus. The cells were labeled with either 35S-amino acids or [32P]orthophosphate; following harvesting and lysis in RIPA buffer, immunoprecipitates were prepared from the total cell lysates with NSP2-specific antisera. Transient expression of NSP2 in the presence of 35S-amino acids and [32P]orthophosphate showed that the protein was produced in the cells and that the protein, when expressed in the absence of other rotavirus proteins in vivo, was phosphorylated (Fig. 10A, lanes 3 and 5).
FIG. 10.
Phosphorylation of NSP2 synthesized in vivo. MA104 cells either were infected with vTF7.3 and programmed to express NSP2 by transfection with plasmid SP65g8 or were infected with SA11 rotavirus. After labeling with 35S-amino acids or [32P]orthophosphate, NSP2 was recovered from lysates of the cells by immunoprecipitation with anti-NSP2 (A) or anti-NSP5 (B) polyclonal antisera. The immunoprecipitates were analyzed by SDS–12% PAGE and autoradiography. (A) NSP2 immunoprecipitates from mock-infected cells (lane 2), transfected cells (lanes 3 and 5), and SA11-infected cells (lanes 4 and 6) that were maintained in 35S-amino acids (lanes 2 to 4) or [32P]orthophosphate (lanes 5 and 6). An immunoprecipitate of 35S-labeled rNSP2 was coelectrophoresed as a control (lane 1). (B) NSP5 immunoprecipitates from SA11-infected cells maintained in 35S-amino acids (lane 1) and [32P]orthophosphate (lane 2).
Likewise, labeling with 35S-amino acids showed that NSP2 was produced in rotavirus-infected cells (Fig. 10A, lane 4). However, labeling with [32P]orthophosphate did not reveal the presence of 32P-labeled NSP2 (Fig. 10A, lane 6), even though in the same cell lysate, 32P-labeled NSP5 was detected by immunoprecipitation with NSP5-specific antisera (Fig. 10B, lane 2). In summary, our results suggest that the NTPase activity of NSP2 leads to the phosphorylation of the protein, but that in the infected cell, either the level of NTPase activity is lower, and thus the extent of phosphorylation lower, or phosphorylation is more transient due to the interaction of NSP2 with other viral components.
Phosphorylated rNSP2 retains RNA-binding activity.
To examine the possibility that phosphorylation of rNSP2 prevented the protein from binding to RNA, 32P-labeled rNSP2 was prepared by incubating rNSP2 with [γ-32P]ATP. The 32P-labeled rNSP2 was then incubated with unlabeled Luc72 RNA probe, and the mixture was analyzed for rNSP2-RNA complexes by gel mobility shift assay. As shown in Fig. 11, 32P-labeled rNSP2 was able to bind to the RNA probe, resulting in the formation of radiolabeled complex I and complexes II and/or III (lane 2). Hence, phosphorylated rNSP2 retains RNA-binding activity. Interestingly, 32P-labeled rNSP2 in the absence of any added RNA failed to migrate into the gel (lane 1). This probably stems from the high pI of rNSP2 (9.02), which is greater than the pH 8.8 running buffer of the electrophoretic system. By interaction with an RNA probe, the protein gains a net negative charge sufficient to allow it to migrate into the gel.
FIG. 11.
Phosphorylated rNSP2 retains RNA-binding activity. 32P-labeled rNSP2 was prepared by incubating 360 pmol (∼13 μg) with 10 μCi of [γ-32P]ATP for 1 h at 37°C. Sixty picomoles of 32P-labeled rNSP2 was incubated for 30 min either alone (lane 1) or with 14 pmol of unlabeled Luc72 RNA (lane 2 and 3). Afterwards, 40 μg of proteinase K was added to one of the reaction mixtures containing 32P-labeled rNSP2 and unlabeled Luc72, and the mixture was incubated for 15 min at room temperature (lane 3). Probe-protein complexes were detected by electrophoresis on a 6% nondenaturing polyacrylamide gel and autoradiography.
DISCUSSION
The aim of this study was to define structural and enzymatic features of NSP2 that would provide a clearer understanding of the protein’s function in rotavirus replication. This was made possible by expressing NSP2 as a recombinant protein with a C-terminal His tag in bacteria and then successfully purifying large amounts of soluble rNSP2 by adsorption to Ni2+ columns. rNSP2, like its native counterpart, formed stable homomultimers consisting of an estimated four to eight subunits, suggesting that it retained the same fold and structural organization. These multimers were shown to bind ssRNA nonspecifically and to have an associated nonspecific NTPase activity. In addition, rNSP2 was found to be phosphorylated in vitro and in vivo. To our knowledge, this is the first report that demonstrates any enzymatic activity for NSP2 and that establishes that the protein undergoes phosphorylation. The only other rotavirus nonstructural protein known to be phosphorylated is NSP5, the product of gene 11 of rotavirus (4, 34, 40). The fact that rNSP2 has NTPase activity suggests that the protein has an energy-dependent function in the viral replication process.
NSP2 was shown previously to be a nonspecific RNA-binding protein (2, 17); however, these analyses were carried out under conditions where other cellular and viral proteins and RNAs may have been present and therefore may have influenced the activity of the protein. Consequently, we reanalyzed NSP2-RNA interactions with purified rNSP2 by two approaches: (i) sedimentation gradient analysis and (ii) gel mobility shift assay. Sedimentation analysis revealed that rNSP2 exists nearly exclusively as 8-to-10S homomultimers and that these multimers, or multiples of them, have affinity for ssRNA. The results were consistent with those of Kattoura et al. (16), who showed that NSP2 derived from infected cells when subjected to sedimentation gradient analysis formed a ∼10S species capable of binding RNA when immunoadsorbed onto protein A-Sepharose beads. As shown by the gel mobility shift assays, RNA binding to rNSP2 occurred in discrete cooperative steps to form higher-order RNA-protein complexes. The formation of these complexes was driven solely by increasing the concentration of rNSP2 relative to RNA. Based on the amount of rNSP2 and RNA probe needed to drive all the probe into complex I, and for the complete conversion of complex I to complex II and of complex II to complex III, RNA binding to rNSP2 was estimated to occur with a stoichiometry of 1:6 ± 2 (RNA:protein) for complex I, 1:12 ± 4 for complex II, and 1:18 ± 6 for complex III. Only three RNA-protein complexes were detected by the gel mobility shift assay using the Luc72 RNA probe. The fact that at even higher concentrations of rNSP2, complexes were found not to migrate out of the well of the nondenaturing gel used in the mobility shift assay suggests that RNA-rNSP2 complexes larger than complex III may exist (data not shown). Likewise, the detection of rNSP2-Luc72 complexes in the pellet of gradients used for sedimentation analysis suggests that such larger structures may exist. It remains unclear how multiple copies of the relatively large 8S-10S rNSP2 homomultimer can bind simultaneously to a short RNA of 72 nucleotides; however, the length of the RNA would be expected to limit the number of higher-order RNA-rNSP2 complexes that could be formed.
In the cell, NSP2 has been reported to exist in close association with ssRNA and with viral dsRNA (2, 17). Our efforts to quantitate the affinity of NSP2 for ssRNA and dsRNA by competition assay showed that rNSP2 has a significant preference for ssRNA over dsRNA. The affinity of rNSP2 for dsRNA was indeed so low as to bring into question whether in vivo there is any biological relevance to the interaction of NSP2 with dsRNA. Experiments showing that NSP2 can be cross-linked to dsRNA also indicated that the dsRNA was partially single stranded and therefore likely represented a partially replicated RNA (2). Since NSP2 is a component of replication intermediates with replicase activity, it is conceivable that NSP2 is bound to the mRNA template for minus-strand synthesis, and as RNA replication occurs, the protein falls off the template and dissociates from the intermediate as a result of its reduced affinity for the newly formed dsRNA or as a result of displacement by another protein with higher affinity for the template.
Several studies have shown that NSP2 is a component of intracellular replication intermediates that possess replicase activity and direct the synthesis of dsRNA (11, 14, 27). Analysis of core-like replication intermediates recovered from infected cells indicates that the mRNA templates for replication move from outside to inside the core-like structure as dsRNA synthesis occurs. The two findings most supportive of this model are (i) that the mRNA template associated with core-like replication intermediates is sensitive to RNase digestion whereas the dsRNA product is not (27) and (ii) that replication intermediates undergo a continuous decrease in overall size as dsRNA is synthesized (28). The reduction in size that occurs during dsRNA synthesis can be mimicked by treating the intermediates with RNase to remove their associated mRNA templates. Since packaging of viral mRNA into core-like structures would be an entropically unfavorable process, we postulate that NSP2 may function as a molecular motor, by binding viral mRNA and catalyzing its packaging through the energy generated by its NTPase activity.
We cannot rule out the possibility that NSP2 simply functions as an unwindase during packaging, with the NTPase of the protein being used to relax the secondary structure of the mRNA template for dsRNA synthesis. However, given that the viral RNA polymerase can both replicate and transcribe in vitro in the absence of NSP2, it seems less likely that the primary function of NSP2 is as an unwindase. In addition to its NTPase activity, we showed that NSP2 is autophosphorylated following cleavage of the γ-phosphate from NTP. Removal of the linked phosphate by CIP clearly demonstrated that the phosphate linkage was through a covalent bond. Phosphorylated rNSP2 retained the ability to bind to RNA hence it is unlikely that the phosphorylation of NSP2 serves as a molecular switch for RNA binding.
NSP2 expressed in vivo in the absence of other rotavirus proteins was phosphorylated. In contrast, we could not detect the presence of phosphorylated NSP2 in infected cells. This suggests that during viral replication, phosphorylation of NSP2 is extremely transient or that other viral proteins interact with NSP2 to alter its NTPase activity and thereby decrease its extent of phosphorylation. Several lines of evidence suggest that NSP2 and the viral protein kinase NSP5 have collaborative functions during viral replication and that NSP5 could affect the phosphorylation status of NSP2: (i) NSP2 and NSP5 colocalize to viroplasms in infected cells and only when transiently expressed together in vivo will coassemble to form viroplasm-like inclusions (9), (ii) NSP2 and NSP5 are both components of replication intermediates with replicase activity (11, 27), and (iii) transient expression of NSP2 in vivo induces the hyperphosphorylation of NSP5 (1). The molecular basis of NSP5 hyperphosphorylation has not been defined but potentially may involve a cascade of events initiated by the NTPase activity of NSP2 causing the autophosphorylation of the protein. In this scenario, the interaction of NSP5 with phosphorylated NSP2 would catalyze the transfer of the γ-phosphate from NSP2 to NSP5 and cause the hyperphosphorylation of NSP5.
NSP2 shares a number of interesting parallels with other proteins encoded by segmented dsRNA viruses whose functions have been implicated in packaging and assembly. These include the nonstructural proteins of two other members of the family Reoviridae, ςNS of reovirus and NS2 of bluetongue virus. Like NSP2, ςNS and NS2 form homomultimeric complexes and have nonspecific affinity for ssRNA (12, 15, 39), and like NSP2, NS2 accumulates in cytoplasmic inclusions (38). Although NS2 is phosphorylated in vivo, neither NS2 nor ςNS has been demonstrated to possess an NTPase activity. Except that it is a structural protein, the P4 protein of the dsRNA phage φ6 shares two important features with NSP2: it has nonspecific NTPase activity and forms homomultimers consisting of similar numbers of subunits (23).
ACKNOWLEDGMENTS
We appreciate the assistance of Melinda Jones and Vladimir Chizhikov on this project. We also thank Robert Chanock, Albert Kapikian, and Kim Green for critically reviewing the manuscript.
REFERENCES
- 1.Afrikanova I, Fabretti E, Miozzo M C, Burrone O R. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J Gen Virol. 1998;79:2679–2686. doi: 10.1099/0022-1317-79-11-2679. [DOI] [PubMed] [Google Scholar]
- 2.Aponte C, Poncet D, Cohen J. Recovery and characterization of a replicase complex in rotavirus-infected cells using a monoclonal antibody against NSP2. J Virol. 1996;70:985–991. doi: 10.1128/jvi.70.2.985-991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bican P, Cohen J, Charpilienne A, Scherrer R. Purification and characterization of bovine rotavirus cores. J Virol. 1982;43:1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackhall J, Fuentes A, Hansen K, Magnusson G. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J Virol. 1997;71:138–144. doi: 10.1128/jvi.71.1.138-144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D J L, Gombold, Ramig R F. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology. 1990;178:143–151. doi: 10.1016/0042-6822(90)90387-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen D Y, Zeng C Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen M S. Human viral gastroenteritis. Clin Microbiol Rev. 1989;2:51–89. doi: 10.1128/cmr.2.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J. Ribonucleic acid polymerase activity associated with purified calf rotavirus. J Gen Virol. 1977;36:395–402. doi: 10.1099/0022-1317-36-3-395. [DOI] [PubMed] [Google Scholar]
- 9.Fabbretti E, Afrikanova I, Vascotto F, Burrone O R. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol. 1999;80:333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst R T, Niles G E, Studier W F, Moss B. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesized bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallegos C O, Patton J T. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 12.Gillian A L, Nibert M L. Amino terminus of reovirus nonstructural protein ςNS is important for ssRNA binding and nucleoprotein complex formation. Virology. 1998;240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 13.Gonzales S A, Burrone O. Rotavirus NS26 is modified by addition of a single O-linked residue of N-acetylglucosamine. Virology. 1991;182:8–16. doi: 10.1016/0042-6822(91)90642-o. [DOI] [PubMed] [Google Scholar]
- 14.Helmberger-Jones M, Patton J T. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology. 1986;155:655–665. doi: 10.1016/0042-6822(86)90225-4. [DOI] [PubMed] [Google Scholar]
- 15.Huismans H, Joklik W K. Reovirus-coded polypeptides in infected cells: Isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology. 1972;70:411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- 16.Kattoura M D, Chen X, Patton J T. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology. 1994;202:803–813. doi: 10.1006/viro.1994.1402. [DOI] [PubMed] [Google Scholar]
- 17.Kattoura M D, Clapp L L, Patton J T. The rotavirus nonstructural protein, NS35, is a nonspecific RNA-binding protein. Virology. 1992;191:698–708. doi: 10.1016/0042-6822(92)90245-k. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of the structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lawton J A, Zeng C Q-Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lui M, Mattion N M, Estes M K. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- 21.Mansell E A, Patton J T. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J Virol. 1990;64:4988–4996. doi: 10.1128/jvi.64.10.4988-4996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Midthun K, Greenberg H B, Hoshino Y, Kapikian A Z, Wyatt R G, Chanock R M. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J Virol. 1985;53:949–954. doi: 10.1128/jvi.53.3.949-954.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paatero A O, Syvaoja J E, Bamford D H. Double-stranded RNA bacteriophage φ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J Virol. 1995;69:6729–6734. doi: 10.1128/jvi.69.11.6729-6734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton J T. Synthesis of simian rotavirus SA11 double stranded RNA in a cell-free system. Virus Res. 1986;6:217–223. doi: 10.1016/0168-1702(86)90071-7. [DOI] [PubMed] [Google Scholar]
- 25.Patton J T. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patton J T, Chen D. RNA-binding and capping activities of proteins in rotavirus open cores. J Virol. 1999;73:1382–1391. doi: 10.1128/jvi.73.2.1382-1391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton J T, Gallegos C O. Structure and protein composition of the rotavirus replicase particle. Virology. 1988;166:358–365. doi: 10.1016/0042-6822(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 28.Patton J T, Gallegos C O. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J Gen Virol. 1990;71:1087–1094. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- 29.Patton J T, Jones M T, Kalbach A N, He Y W, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton J T, Salter-Cid L, Kalbach A, Mansell A E, Kattoura M D. Nucleotide and amino acid sequence analysis of the rotavirus non-structural RNA binding protein NS35. Virology. 1993;192:438–446. doi: 10.1006/viro.1993.1059. [DOI] [PubMed] [Google Scholar]
- 31.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNAs. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrie B L, Greenberg H B, Graham D Y, Estes M K. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1984;1:133–152. doi: 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- 33.Pizarro J L, Sandino A M, Pizarro J M, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 34.Poncet D, Lindenbaum P, Haridon R L, Cohen J. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J Virol. 1997;71:34–41. doi: 10.1128/jvi.71.1.34-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad B V V, Wang G J, Clerx J P M, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 36.Ramig R F. Isolation and genetic characterization of temperature-sensitive mutants of simian rotavirus SA11. Virology. 1982;120:93–105. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 37.Ramig R F, Petrie B L. Characterization of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J Virol. 1984;49:665–673. doi: 10.1128/jvi.49.3.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas C P, Booth T F, Roy P. Synthesis of bluetongue virus encoded phosphoprotein and formation of inclusion bodies by recombinant baculovirus in insect cells: it binds the single stranded RNA species. J Gen Virol. 1990;71:2073–2083. doi: 10.1099/0022-1317-71-9-2073. [DOI] [PubMed] [Google Scholar]
- 39.Uitenweerde J M, Theron J, Stoltz M A, Huismans H. The multimeric nonstructural proteins of bluetongue virus, African horsesickness virus, and epizootic haemorrhagic disease virus differ in their single-stranded RNA binding ability. Virology. 1995;209:624–632. doi: 10.1006/viro.1995.1294. [DOI] [PubMed] [Google Scholar]
- 40.Welch S K, Crawford S E, Estes M K. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J Virol. 1989;63:3974–3982. doi: 10.1128/jvi.63.9.3974-3982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeager M, Dryden K A, Olsen N H, Greenberg H B, Baker T S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng C Q-Y, Wentz M J, Estes M K, Ramig R F. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]