Abstract
Atopic dermatitis (AD) is a common yet complex skin disease, posing a therapeutic challenge with increasingly recognized different phenotypes among variable patient populations. Because therapeutic response may vary on the basis of heterogeneous clinical and molecular phenotypes, a shift toward precision medicine approaches may improve AD management. Herein, we will consider biomarkers as potential instruments in the toolbox of precision medicine in AD and will review the process of biomarker development and validation, the opinion of AD experts on the use of biomarkers, types of biomarkers, encompassing biomarkers that may improve AD diagnosis, biomarkers reflecting disease severity, and those potentially predicting AD development, concomitant atopic diseases, or therapeutic response, and current practice of biomarkers in AD. We found that chemokine C-C motif ligand 17/thymus and activation-regulated chemokine, a chemoattractant of TH2 cells, has currently the greatest evidence for robust correlation with AD clinical severity, at both baseline and during therapy, by using the recommendations, assessment, development, and evaluation approach. Although the potential of biomarkers in AD is yet to be fully elucidated, due to the complexity of the disease, a comprehensive approach taking into account both clinical and reliable, AD-specific biomarker evaluations would further facilitate AD research and improve patient management.
Keywords: Atopic dermatitis, biomarker, International Eczema Council, CCL17/TARC, IgE, eosinophils, CCL22/MDC, CCL26/eotaxin-3, CCL27/CTACK, CCL18/pulmonary and activation-regulated chemokine, IL-13, IL-22
Atopic dermatitis (AD) is a complex disorder in which gene-gene and gene-environment interactions contribute to generate a highly heterogeneous clinical phenotype.1 This heterogeneity likely reflects yet-to-be-defined mechanisms, coupled with clinical relevance we are only beginning to grasp. Progress in our understanding of the role of microbiome, epidermal barrier function, and different cytokines and other immune mediators underlying the chronic AD inflammation has led to an unprecedented number of new compounds in clinical development, for both the topical and systemic therapy of AD.2 However, thus far none of the therapeutic approaches can be considered a magic bullet, or a “one-size-fits-all” agent. When using stringent end points such as percent of patients reaching investigator’s global assessment 0/ 1 with a 2-grade decrease or Eczema Area and Severity Index-90 in a monotherapy study design (ie, without adding topical/systemic anti-inflammatory medications), it appears that both biologics that specifically target cytokines or their receptors and broad-acting Janus kinase inhibitors fail to fully control AD in most patients.3–6 Hence, particularly considering the complexity of AD, there is a need to shift toward precision medicine approaches to improve AD management.
BIOMARKERS: DEFINITION, SUBTYPES, AND OTHER REGULATORY ASPECTS
Biomarkers have always existed for different purposes in medicine, principally as a diagnostic tool. However, AD diagnosis and treatment, as opposed to many other chronic diseases, relies completely on clinical scores rather than biochemical markers. Thus, a reliable biomarker will reduce observatory differences. Herein, we will consider biomarkers as potential instruments in the toolbox of precision medicine in AD. Biomarkers may have tremendous implications in prevention strategies and, most importantly, in strategies used for the development of upcoming new compounds on the background of stringent regulatory landscapes. In this regard, the definition of a biomarker given by regulatory organizations is particularly helpful but obviously not universal. The Food and Drug Administration (FDA) has adopted a rather broad definition: “A defined characteristic that is measured as an indicator of normal biologic processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions.” The FDA also adds the following comment: “Molecular, histologic, radiographic, or physiologic characteristics are types of biomarkers. A biomarker is not an assessment of how an individual feels, functions, or survives.” Interestingly, the European Medicines Agency has another, more restrictive definition: “A biological molecule found in blood, other body fluids, or tissues that can be used to follow body processes and diseases in humans and animals.”
In the process of biomarker discovery, one should distinguish between the kind of biologic material (or its origin) on one hand and the purpose/value of the biomarker on the other hand. For the first group, a wide range of biologic material can be used such as (1) genomic information (eg, specific gene sequences or epigenetic modification of genes), (2) transcriptomic profiles obtained by analysis of mRNA and miRNA, (3) proteins such as cytokines and other mediators from body fluids (whole blood, serum, plasma, tissue fluids) or tape stripping, and (4) morphological information (immunohistochemical staining and pictures thereof).
This is to be distinguished from the purpose/value of biomarkers with 7 different subtypes as defined by the FDA-NIH Biomarker Working Group (www.ncbi.nlm.nih.gov/books/NBK326791/): (1) susceptibility/risk, (2) diagnostic, (3) monitoring/severity, (4) prognostic, (5) predictive, (6) pharmacodynamic/response, and (7) safety. All these subtypes could potentially be of importance in the context of the management of AD.
Unfortunately, the literature and the classical understanding thereof in the scientific community has generated the idea that a biomarker can be easily described and used in the context of disease management. In reality, bringing a given biomarker from discovery to clinical practice and regulatory acceptance in clinical development and/or as a companion diagnostic is a rather complex procedure, widely underestimated by most scientists, which is often comparable to a drug development process. There are several crucial steps in the evolution of a biomarker before it reaches the status of qualification in clinical practice.
In a nutshell, the life of a biomarker starts with its discovery, which can be either by chance or the product of a hypothesis-driven biomarker discovery program based on a patient registry collecting high-quality phenotypical data linked to a biobank with several hundreds of specimens from these patients. The next step is a first (internal) analytical and clinical validation in a limited number of clinical cases. Thereafter, the biomarker must undergo another (external) validation step, ideally from independent institutions, using a large cohort of patients where the reproducibility is key. Once this goal of internal and external validation is reached, the biomarker is subjected to a complex process of regulatory qualification, which is supported by a number of guidance documents from the regulatory agencies (FDA, European Medicines Agency). Thus, developing a newly discovered biomarker to the stage of an accepted companion diagnostic for the management of a disease is a complex and demanding process.
THE GRADING OF RECOMMENDATIONS, ASSESSMENT, DEVELOPMENT, AND EVALUATION APPROACH TO ASSESS EVIDENCE STRENGTH
The Grading of Recommendations, Assessment, Development, and Evaluation (GRADE) approach offers a system for rating quality of evidence, with a structured process for developing and presenting evidence summary.7 Herein, we searched for AD-related publications that included correlation analysis and found a significant (P < .05) correlation coefficient of greater than or equal to 0.4 between AD clinical severity and blood/skin potential biomarkers, both at baseline and during various AD treatments, and across both pediatric and adult patients. Biomarkers that were found to robustly correlate with AD clinical severity in more than 3 publications were included in this review. Next, we summarized these findings using the GRADE approach, in which accumulated evidence per each potential biomarker (separated by pediatric and adults, and at baseline and during topical and systemic treatments) was graded on the basis of strength of the overall published data, given our inclusion threshold.
BIOMARKERS IN AD INTERNATIONAL ECZEMA COUNCIL SURVEY RESULTS
The International Eczema Council (IEC) consists of more than 100 councilors and associates (https://www.eczemacouncil.org/), all experts in AD. Before the IEC meeting at the Society of Investigative Dermatology meeting in 2019 in Chicago, an invitation to an internet-based survey on biomarkers for AD was sent by email to all IEC councilors and associates to examine their opinion regarding biomarkers in AD (Table I; for detailed questions, see this article’s Online Repository at www.jacionline.org). Monkey survey software was used for data collection. Overall, the experts believe AD is a heterogeneous disease with at least 3 different phenotypes, that biomarkers may help to stratify patients by phenotypes and improve patient management and treatment compliance, and that future developments should focus on their use as predictors of therapeutic response.
TABLE I.
Results of the biomarkers survey by IEC AD experts
| Question | Yes (N) | No (N) | Follow-up questions (N) |
|---|---|---|---|
| Do you think that AD is a heterogeneous disease? | 97.52% (41) | 2.38% (1) | How many different AD phenotypes are there? (38)
|
| Are you using blood tests/biomarkers for the diagnosis of AD? | 29.55% (13) | 70.45% (31) | Which are you using? (13)
|
| Do you think that blood tests/biomarkers are useful for assessing the severity of AD? | 59.09% (25) | 40.91% (18) | Why not?
|
| Could blood test/biomarkers be useful for assessing treatment compliance? | 76.74% (33) | 23.26% (10) | Which biomarkers would you suggest? (17)
|
| How would you prioritize the needs for blood test/biomarkers? | Top-rated development priorities: (40)
|
||
EASI, Eczema Area and Severity Index; LDH, lactate dehydrogenase; TEWL, transepidermal water loss.
TYPES OF BIOMARKERS IN AD
Potential biomarkers may be subdivided on the basis of their suggested use.
Biomarkers differentiating AD from psoriasis
Some biomarkers seem to reliably distinguish between AD and psoriasis (namely NOS2 and chemokine C-C motif ligand [CCL] 27/cutaneous T-cell–attracting chemokine [CTACK]),8–10 thus potentially improving the diagnosis and management of patients with psoriasiform dermatitis (Table II).
TABLE II.
Biomarkers as disease classifiers, potentially improving diagnosis by differentiating AD and psoriasis
| Biomarker | Full name | Functional effect | Abnormalities |
|---|---|---|---|
| NOS2 | Inducible nitric oxidase synthase | Catalyzing the production of nitric oxide, a toxic defense molecule against infections, and a regulator of functional activity, growth, and death of immune cells including T cells, antigen-presenting cells, mast cells, neutrophils, and natural killer cells | Upregulated in psoriasis, downregulated in AD |
| CCL27/CTACK | Chemokine (C-C motif) ligand 27/cutaneous T-cell–attracting chemokine | Expressed by keratinocytes. Mediates the migration of lymphocytes into the skin by binding to CCR10 | Upregulated in AD, downregulated in psoriasis |
Biomarkers correlating with clinical severity
These include markers related with general inflammation such as serum lactate dehydrogenase,11–14 C-reactive protein,11 along with markers related with allergy (eg, peripheral eosinophil counts).11,12,15–19 Because AD is TH2/TH22-centered, cytokines and chemokines related with these immune pathways and correlated with disease severity in untreated or posttreated tissues (either skin or serum) were also investigated as possible biomarkers (Tables III and IV and Fig 1). Such cytokines include the key TH2 marker IL-1341,58,59,66,67,95,100 and the key TH22-related cytokine IL-22.41,58,59,64,84 TH2-related chemokines correlated with AD severity include CCL17/thymus and activation-regulated chemokine (TARC),13,14,20,24,27–33,35–39,94,101–104 CCL26/eosinophil-attracting chemokine (eotaxin-3),43,58,68,85,91,95,105 CCL27/CTACK,29,30,33,74,75 CCL18/pulmonary and activation-regulated chemokine,65,68,83,84 and CCL22/macrophage-derived chemokine (MDC).26,28,40,44,45,90 Of note, circulating AD-related biomarkers are found in moderate to severe patients, whereas mild patients may not consistently display upregulation of AD-related biomarkers in their serum.25
TABLE III.
Potential biomarkers reported to strongly and significantly correlate with clinical severity indices of AD (correlation coefficient ≥0.4, P < .05)
| Biomarker (no. of publications) | Serum | Skin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author | Lab method | Corr method | Year | Cohort (n) | Author | Lab method | Corr method | Year | Cohort (n) | |
| CCL17/TARC (>20) | Kakinuma et al20 | E | S | 2001 | 40 | Morita et al21* (LS-TS) | IF | S | 2010 | 33 |
| Horikawa et al22 | E | P | 2002 | 52 | McAleer et al23† (NL-TS) | ECL | S | 2019 | 66 | |
| Fujisawa et al24 | E | S | 2002 | 29 | He et al25 (LS-B) | PCR | P | 2020 | 61 | |
| Leung et al26† | E | S | 2003 | 20 | ||||||
| Hijnen et al27‡ | E | S | 2004 | 177 | ||||||
| Jahnz-Rozyk et al28 | E | P | 2005 | 43 | ||||||
| Song et al29† | E | S | 2006 | 157 | ||||||
| Nakazato et al30† | E | S | 2008 | 34 | ||||||
| Fujisawa et al31† | E | S | 2009 | 27 | ||||||
| van Velsen et al32† | E | P/S | 2010 | 60 | ||||||
| Morita et al21 | E | S | 2010 | 33 | ||||||
| Kou et al13 | E | S | 2012 | 121 | ||||||
| Machura et al33† | E | S | 2012 | 26 | ||||||
| Furue et al34§ | E | S | 2012 | 61 | ||||||
| Mizawa et al14 | NA | S | 2013 | 30 | ||||||
| Kataoka35∥ | NA | NA | 2014 | 96 | ||||||
| Landheer et al36‡ | E | S | 2014 | 320 | ||||||
| Ahrens et al37† | E | S | 2015 | 128 | ||||||
| Gu et al38 | E | S | 2015 | 73 | ||||||
| Hulshof et al39† | L | S | 2018 | 41 | ||||||
| CCL22/MDC (>10) | Kakinuma et al40 | E | S | 2002 | 45 | Tintle et al41 | PCR | S | 2011 | 12 |
| Fujisawa et al24 | E | S | 2002 | 29 | Suarez-Farinas et al42 (LS/NL-B) | PCR | S | 2011 | 15 | |
| Leung et al26† | E | S | 2003 | 20 | Wen et al43 (NL-B) | PCR | P | 2018 | 12 | |
| Jahnz-Rozyk et al28 | E | P | 2005 | 43 | ||||||
| Gunther et al44 | E | P | 2005 | 36 | ||||||
| Angelova-Fischer et al45 | E | P | 2006 | 21 | ||||||
| Hashimoto et al46 | E¶ | NA | 2006 | 11 | ||||||
| Nakazato et al30† | E | S | 2008 | 34 | ||||||
| Wen et al43∥ | ECL | P | 2018 | 15 | ||||||
| Brunner et al47†‡ | O | S | 2019 | 30 | ||||||
| McAleer et al23† | ECL | S | 2019 | 47 | ||||||
| IgE (>10) | Tsuboi et al48# | IRMA | P | 1998 | 17 | NA | ||||
| Yoshizawa et al49 | NA | S | 2002 | 26 | ||||||
| Kaminishi et al50# | FEIA | P | 2002 | 20 | ||||||
| Jahnz-Rozyk et al28 | E | P | 2005 | 43 | ||||||
| Aral et al51† | N | S | 2006 | 20 | ||||||
| Salomon and Baran52 | E | P/S | 2008 | 49 | ||||||
| Wu et al53† | F | P | 2011 | 48 | ||||||
| Suarez-Farinas et al54 | E | P | 2013 | 42 | ||||||
| Zedan et al55† | E | NA | 2015 | 50 | ||||||
| Glatz et al56 | E | S | 2015 | 67 | ||||||
| Rosinska-Wieckowicz et al57 | F | S | 2016 | 102 | ||||||
| E | P | 2017 | 25 | |||||||
| Ungar et al58# | E | P | 2018 | 15 | ||||||
| Wen et al43 | E | S | 2019 | 15 | ||||||
| Sanyal et al59 | ||||||||||
| CCL22/MDC (>10) | Kakinuma et al40 | E | S | 2002 | 45 | Tintle et al41 | PCR | S | 2011 | 12 |
| Fujisawa et al24 | E | S | 2002 | 29 | Suarez-Farinas et al42 (LS/NL-B) | PCR | S | 2011 | 15 | |
| Leung et al26† | E | S | 2003 | 20 | Wen et al43 (NL-B) | PCR | P | 2018 | 12 | |
| Jahnz-Rozyk et al28 | E | P | 2005 | 43 | ||||||
| Gunther et al44 | E | P | 2005 | 36 | ||||||
| Angelova-Fischer et al45 | E | P | 2006 | 21 | ||||||
| Hashimoto et al46 | E# | NA | 2006 | 11 | ||||||
| Nakazato et al30† | E | S | 2008 | 34 | ||||||
| Wen et al43∥ | ECL | P | 2018 | 15 | ||||||
| Brunner et al47†‡ | O | S | 2019 | 30 | ||||||
| McAleer et al23† | ECL | S | 2019 | 47 | ||||||
| Eosinophils/ECP (>10) | Mukai et al60 | C | NA | 1990 | 30 | NA | ||||
| Czech et al19‡ | RIA | S | 1992 | 19 | ||||||
| Kagi et al16 | RIA | S | 1992 | 37 | ||||||
| Halmerbauer et al61† | RIA | K | 1997 | 20 | ||||||
| Tsuboi et al48 | C | P | 1998 | 17 | ||||||
| Yoshizawa et al49 | NA | S | 2002 | 26 | ||||||
| Raap et al62† | IC | S | 2012 | 60 | ||||||
| Kaminishi et al50 | C | P | 2002 | 20 | ||||||
| Angelova-Fischer et al45 | FEIA | P | 2006 | 21 | ||||||
| Morishima et al12 | C | S | 2010 | 58 | ||||||
| Wu et al53† | FEIA | P | 2011 | 48 | ||||||
| Ungar et al58 | C | P | 2017 | 25 | ||||||
| Chen et al63 | NA | S | 2019 | 12 | ||||||
| IL-22 (>5) | Nograles et al64 | F | LRA | 2009 | 12 | Tintle et al41 (LS-B)∥ | PCR | S | 2011 | 12 |
| Ungar et al58 | EMD | P | 2017 | 25 | Suarez-Farinas et al42 (LS/NL-B) | PCR | S | 2011 | 12 | |
| Esaki et al65†§ (LS-B) | PCR | P | 2016 | 19 | ||||||
| Ungar et al58 (LS/NL-B) | PCR | P | 2017 | 25 | ||||||
| Wen et al43 (NL-B)∥ | PCR | P | 2018 | 15 | ||||||
| Sanyal et al59 (LS-B) | PCR | S | 2019 | 15 | ||||||
| IL-13 (>5) | Koning et al66† | E | S | 1997 | 15 | Tintle et al41 (LS-B)∥ | PCR | S | 2011 | 12 |
| Ungar et al58 | E | P | 2017 | 25 | Suarez-Farinas et al42 (LS-B) | PCR | S | 2011 | 12 | |
| Szegedi et al67 (LS-ISF) | L | P/S | 2015 | 16 | ||||||
| Wen et al43 (NL-B) | PCR | P | 2018 | 15 | ||||||
| Guttman-Yassky et al68†** (LS-TS) | PCR | S | 2019 | 21 | ||||||
| Sanyal et al59 (LS/NL-B) | PCR | S | 2019 | 15 | ||||||
| He et al25 (LS-B) | PCR | P | 2020 | 61 | ||||||
| IL-18 (>5) | Hon et al69† | E | S | 2004 | 19 | Inoue et al70 (LS/NL-TS) | E | S | 2011 | 95 |
| Aral et al51† | E | S | 2006 | 20 | McAleer et al23*† (NL-TS) | ECL | S | 2019 | 66 | |
| Park and Youn71 | NA | NA | 2007 | 65 | Pavel et al72 (LS-B) | O | S | 2020 | 20 | |
| Kou et al13 | E | S | 2012 | 121 | ||||||
| Zedan et al55† | E | NA | 2015 | 50 | ||||||
| Suwarsa et al73 | E | P | 2017 | 20 | ||||||
| CCL27/CTACK (>5) | Kakinuma et al74 | E | S | 2003 | 50 | — | — | — | — | |
| Hon et al75*† | E | S | 2004 | 37 | ||||||
| Hijnen et al27‡ | E | S | 2004 | 76 | ||||||
| Song et al29† | E | S | 2006 | 157 | ||||||
| Nakazato et al30† | E | S | 2008 | 34 | ||||||
| Machura et al33† | E | S | 2012 | 26 | ||||||
| S100A7/12 (>5) | — | — | — | — | — | Suarez-Farinas et al42 (LS-B) | PCR | S | 2011 | 12 |
| Tintle et al41 (LS-B)∥ | PCR | S | 2011 | 12 | ||||||
| Suarez-Farinas et al54 (LS-B) | PCR | P | 2013 | 7 | ||||||
| Ungar et al58 (LS+/NL-B) | PCR | P | 2017 | 25 | ||||||
| Guttman-Yassky et al68† (LS-TS) | PCR | S | 2019 | 21 | ||||||
| Sanyal et al59 (LS-B) | PCR | S | 2019 | 15 | ||||||
| He et al25 (LS-B) | PCR | P | 2020 | 61 | ||||||
| E-selectin (>5) | Morita et al76 | E | NA | 1995 | 23 | — | — | — | — | — |
| Yamashita et al77 | E | K | 1997 | 53 | ||||||
| Wolkerstorfer et al†78 | E | S | 2003 | 15 | ||||||
| Angelova-Fischer et al45 | E | P | 2006 | 21 | ||||||
| Brunner et al79 | O | S | 2017 | 59 | ||||||
| Brunner et al47†‡ | O | S | 2019 | 30 | ||||||
| MMP12 (>5) | Brunner et al79 | O | S | 2017 | 59 | Suarez-Farinas et al42 (LS-B) | PCR | S | 2011 | 12 |
| Brunner et al47†‡ | O | S | 2019 | 30 | Suarez-Farinas et al54 (LS-B) | PCR | P | 2013 | 7 | |
| He et al80 | O | P | 2020 | 71 | Ungar et al58 (NL-B) | PCR | P | 2017 | 25 | |
| Pavel et al81†** (NL-TS) | R | S | 2020 | 19 | ||||||
| LDH (>5) | Mukai et al60 | NA | NA | 1990 | 80 | NA | ||||
| Tsuboi et al48 | P | 1998 | 17 | |||||||
| Morishima et al12 | S | 2010 | 58 | |||||||
| Kou et al13 | S | 2012 | 121 | |||||||
| Mizawa et al14 | S | 2013 | 30 | |||||||
| Kataoka35∥ | NA | 2014 | 96 | |||||||
| Olesen et al82 | S | 2019 | 43 | |||||||
| CCL18/PARC (≥5) | Hon et al83† | E | P | 2011 | 108 | Suarez-Farinas et al42 (NL-B) | PCR | S | 2011 | 12 |
| Gittler et al84 (NL-B) | PCR | S | 2012 | 10 | ||||||
| Esaki et al65† (LS-B) | PCR | P | 2016 | 19 | ||||||
| Guttman-Yassky et al68†** (LS-TS) | PCR | S | 2019 | 21 | ||||||
| Eotaxin-3/CCL26 (≥5) | Kagami et al85 | E | S | 2003 | 30 | Zhou et al86 (LS-B) | PCR | S | 2019 | 27 |
| Wen et al43 | ECL | P | 2018 | 15 | Guttman-Yassky et al68†§∥ (LS-TP) | PCR | S | 2019 | 21 | |
| IL-19 (≥5) | Oka et al87 | E | S | 2017 | 21 | Esaki et al65† (LS-B) | PCR | P | 2016 | 19 |
| Konrad et al88∥ | E | S | 2019 | 124 | Guttman-Yassky et al68† (LS-TS)∥ | PCR | S | 2019 | 21 | |
| Pavel et al81†‡ (LS-TS)∥ | R | S | 2020 | 19 | ||||||
B, Skin biopsy; C, cell count; Corr, correlation; ECP, eosinophil cationic protein; E, ELISA; ECL, electrochemiluminescence immunoassay; EMD, Erenna immunoassay; F, flow cytometry; FEIA, fluorescent enzyme immunoassays; IC, ImmunoCap system; IF, immunofluorescence; IRMA, immunoradiometric assay; ISF, interstitial fluid; K, Kendall rank correlation; L, Luminex; LDH, lactate dehydrogenase; LRA, linear regression analysis; LS, lesional; N, nephelometric method; NA, not applicable/available; NL, nonlesional; O, OLINK proteomics; PARC, pulmonary and activation-regulated chemokine; R, RNA-sequencing; RIA, ECP radioimmunoassay; SCORAD, SCORing of Atopic Dermatitis; TEWL, transepidermal water loss; TS, tape-strips.
Correlated with SCORAD components and not with the total SCORAD.
Pediatric cohort.
Correlated with Six Area, Six Sign AD/body surface area/Leicester severity score/scoring system as described by Costa et al.89
Correlated with TEWL.
P ≤.1.
Performed on monocyte-derived circulating dendritic cells.
# Log2(IgE) was correlated with SCORAD.
Correlated with pruritus.
TABLE IV.
Potential biomarkers reported to strongly and significantly correlate with clinical therapeutic response in AD (correlation coefficient ≥0.4, P < .05)
| Biomarker (no. of publications) | Serum | Skin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author | Lab method | Corr method | Year | Cohort (n) | Author | Lab method | Corr method | Year | Cohort (n) | |
| CCL17/TARC (>5) | Furukawa et al90 | E | NA | 2004 | 15 | Khattri et al91 (LS-B, P) | PCR | S | 2014 | 19 |
| Kwon et al92 | E | LRA | 2010 | 20 | Koppes et al93 (LS-TS) | E | S | 2016 | 21 | |
| Beck et al94* | E | NA | 2014 | 55 | Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | |
| Ungar et al58† | ECL | P | 2017 | 25 | ||||||
| MDC/CCL22 (>5) | Furukawa et al90 | E | NA | 2004 | 15 | Khattri et al91 (LS-B) | PCR | S | 2014 | 19 |
| Kwon et al92 | E | LRA | 2010 | 20 | Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | |
| Guttman-Yassky et al96 (LS-B)† | PCR | S | 2019 | 54 | ||||||
| IL-13 (≥5) | Ungar et al58 | E | P | 2017 | 25 | Khattri et al91 (LS-B) | PCR | S | 2014 | 19 |
| Ungar et al58 (LS-B) | PCR | P | 2017 | 25 | ||||||
| Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | ||||||
| Guttman-Yassky et al96 (LS-B)† | PCR | S | 2019 | 54 | ||||||
| S100A7/8/12 (≥5) | — | — | — | — | — | Tintle et al41 (LS-B)† | PCR | S | 2011 | 12 |
| Khattri et al91 (LS-B) | PCR | S | 2014 | 19 | ||||||
| Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | ||||||
| Bissonnette et al97‡ (LS-B) | PCR | S | 2019 | 40 | ||||||
| Guttman-Yassky et al96 (LS-B) | PCR | S | 2019 | 54 | ||||||
| IL-22 (>3) | — | — | — | — | — | Tintle et al41 (LS-B) | PCR | S | 2011 | 12 |
| Khattri et al91 (LS-B) | PCR | S | 2014 | 19 | ||||||
| Ungar et al58 (LS/NL-B) | PCR | P | 2017 | 25 | ||||||
| Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | ||||||
| CCL13/MCP-4 (>3) | Ungar et al58 | ECP | P | 2017 | 25 | Hamilton et al98 (LS-B)† | PCR | P | 2014 | 18 |
| Ungar et al58 (NL-B) | PCR | P | 2017 | 25 | ||||||
| Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | ||||||
| He et al99 (LS-TS) | O | S | 2020 | 26 | ||||||
| Eotaxin-3/CCL26 (≥3) | — | — | — | — | — | Hamilton et al98 (LS-B) | PCR | P | 2014 | 18 |
| Khattri et al91 (LS-B) | PCR | S | 2014 | 19 | ||||||
| Pavel et al95 (LS-B) | PCR | S | 2019 | 36 | ||||||
| CCL18/PARC (≥3) | Guttman-Yassky et al96 | E | S | 2019 | 54 | Khattri et al91 (LS-B) | PCR | S | 2014 | 19 |
| Ungar et al58 (LS/NL-B) | PCR | P | 2017 | 25 | ||||||
B, Skin biopsy; Corr, correlation; E, ELISA; ECL, electrochemiluminescence immunoassay; LRA, linear regression analysis; LS, lesional; N, nephelometric method; NL, nonlesional; PARC, pulmonary and activation-regulated chemokine; TS, tape-strips.
Correlated with pruritus.
P ≤.1.
Correlated with Leicester severity score/Investigator’s Static Global Assessment.
FIG 1.
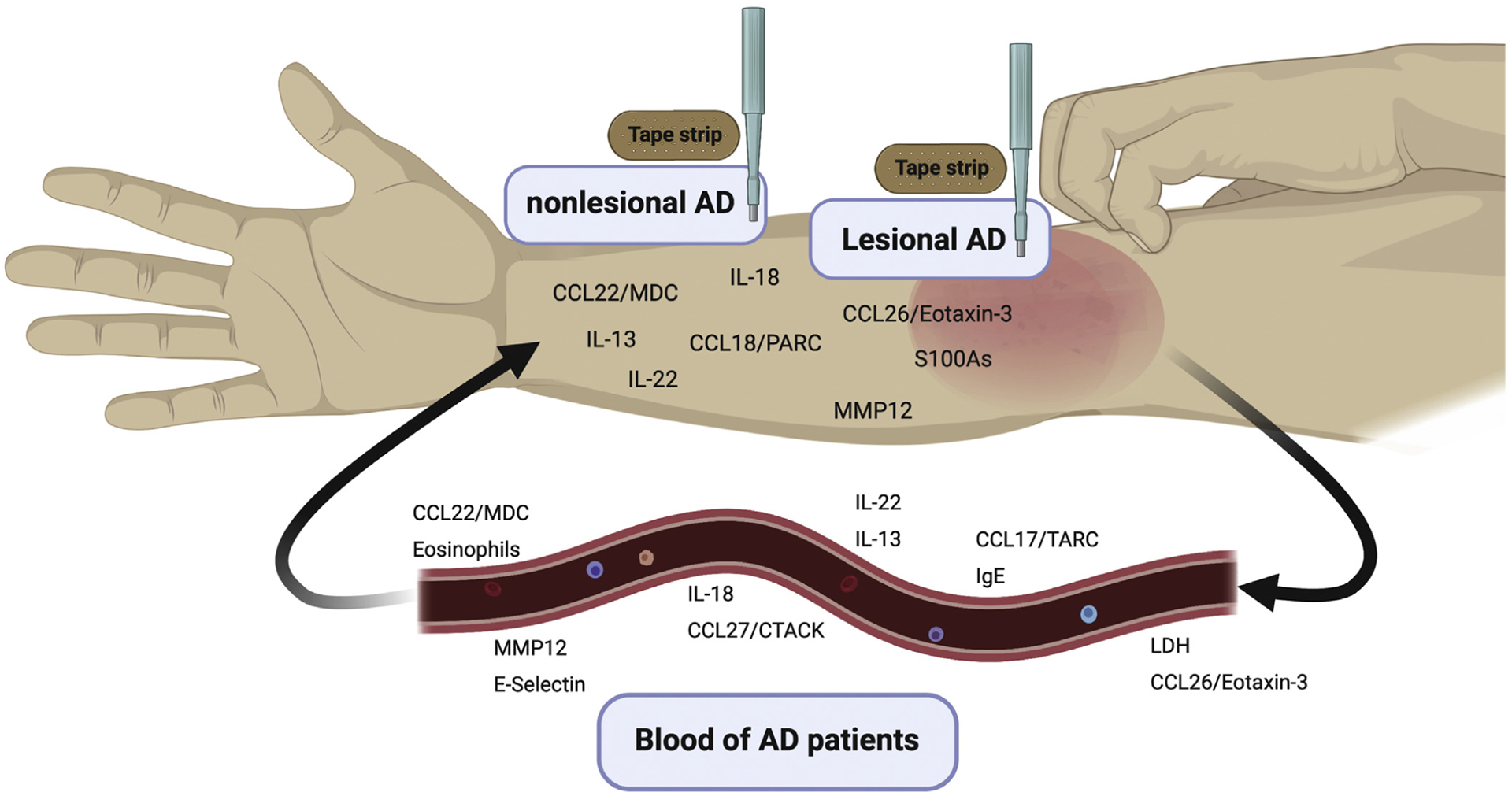
Potential biomarkers for AD in nonlesional and lesional AD skin (using both biopsies and tape-strips, top) as well as circulating potential biomarkers in blood of patients with AD (bottom). LDH, Lactate dehydrogenase.
Barrier-related potential biomarkers, including filaggrin (FLG), loricrin, and natural moisturizing factor, may inversely correlate with disease severity.106
Because of the complexity of AD pathogenesis, a few reports modeled a combination of biomarkers to better reflect molecular changes correlating with clinical severity.39,58,107 The current evidence from the literature, including only those reports in which a significant and robust correlation between AD clinical severity and a tissue biomarker was found (r ≥ 0.4; P < .05), is summarized using the GRADE approach in Table V.7
TABLE V.
GRADE evidence profile: Accumulated data on potential biomarkers correlating with disease severity in AD*
| Biomarker No. of studies; No. of subjects included | Weighted average of correlation strength (r)* | Limitation | Inconsistency | Indirectness/ imprecision/publication bias | Overall evidence for biomarker generalizability (highest achieved) |
|---|---|---|---|---|---|
| Biomarkers correlating with severity in nontreated adult AD | |||||
| CCL17/TARC 14; 1,136 | 0.58 | No serious limitations for blood; for skin—limited number of studies were found by our criteria | Not all studies correlated the biomarker with EASI or SCORAD as scores for AD clinical severity | No serious indirectness or imprecision; no publication bias detected | Very high (in blood) |
| IgE 11; 421 | 0.62 | No serious limitations | No serious inconsistency among these reports, but IgE levels are not consistently correlated with AD severity in multiple other reports | No serious indirectness or imprecision; no publication bias detected | High |
| CCL22/MDC 9; 227 | 0.62 | No serious limitations for blood; for skin—limited evidence exists by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | High (in blood) |
| LDH 7; 445 | 0.52 | No serious limitations | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | High |
| IL-18 5; 321 | 0.63 | No serious limitations | Variable laboratory methods in skin | No serious indirectness or imprecision; no publication bias detected | High |
| Eosinophils/ ECP 9; 253 | 0.6 | No serious limitations | Variable laboratory methods were reported. Different aspects of eosinophil upregulation/activation were analyzed | No serious indirectness or imprecision; no publication bias detected | Moderate-high |
| IL-22 7; 116 | 0.52 | Sparse reports in blood, with limited number of patients | Variable laboratory methods in blood | No serious indirectness or imprecision; no publication bias detected | Moderate (in skin) |
| IL-13 6; 144 | 0.54 | Sparse data in blood by our criteria. Limited number of patients in both skin and blood studies | Variable laboratory methods in blood | No serious indirectness or imprecision; no publication bias detected | Moderate (in skin) |
| E-selectin 4; 159 | 0.53 | No serious limitations | Laboratory and correlation methods and varied | No serious indirectness or imprecision; no publication bias detected | Moderate |
| MMP12 5; 174 | 0.46 | No serious limitations for skin. Only proteomic data were reported in blood by our criteria | Laboratory and correlation methods and varied | No serious indirectness or imprecision; no publication bias detected | Moderate |
| S100A7/12 6; 132 | 0.49 | No serious limitations | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate |
| CCL27/CTACK 2; 126 | 0.59 | Limited evidence in blood, no evidence in skin by our criteria | Different AD severity scores were used | No serious indirectness or imprecision; no publication bias detected | Moderate-low (in blood) |
| CCL26/eotaxin-3 3; 72 | 0.53 | Very limited data in skin | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-low (in blood) |
| CCL18/ PARC 2; 22 | 0.63 | Very limited data in adult skin; no data from adult blood by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Low |
| IL-19 2; 136 | 0.59 | No data were reported in adult skin by our criteria. The largest study in adult blood only achieved P < .1 | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Low (in blood) |
| Biomarkers correlating with severity in nontreated pediatric AD | |||||
| CCL17/TARC 9; 559 | 0.56 | No serious limitations in blood. No data were reported in pediatric skin by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | High (in blood) |
| CTACK/CCL27 4; 254 | 0.66 | No serious limitations in blood. No data were reported in pediatric skin by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | High (in blood) |
| IgE, 3; 118 IL-18, 4; 155, CCL22/MDC, 4; 131 | 0.76, 0.64, 0.46 (respectively) | Limited number of studies by our criteria. For CCL22/MDC, correlation was found only in blood | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-high (in blood) |
| E-selectin, 2; 45 | 0.45 | Limited evidence in pediatric blood; no data from pediatric skin by our criteria | Variable laboratory methods | No serious indirectness or imprecision; no publication bias detected | Moderate-low (in blood) |
| Eosinophils/ ECP 3; 128 | 0.71 | Limited number of studies by our criteria | Variable laboratory methods | No serious indirectness or imprecision; no publication bias detected | Moderate-low |
| CCL18/PARC 3; 148 | 0.47 | Limited number of studies by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-low |
| IL-22, 1; 19 IL-13, 1; 21 S100A7/12, 1; 21 | NA | Very limited number of studies and subjects in pediatric skin; no evidence in pediatric blood by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Low (in skin) |
| MMP12 2; 49 | 0.5 | Very limited number of studies and subjects in both skin and blood by our criteria | In blood, correlation was found with body surface area. In skin, correlation was found with pruritus | No serious indirectness or imprecision; no publication bias detected | Low |
| IL-19 3; 59 | 0.43 | Limited evidence in pediatric skin; no evidence in pediatric blood by our criteria. Tape-stripped pediatric skin only achieved P < .1 | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Low (in skin) |
| Biomarkers showing decreased levels in correlation with clinical improvement in longitudinal, topical treatment studies | |||||
| CCL17/TARC, CCL22/MDC, 1; 20 (for both) | NA | Limited data and only with an emollient. Correlation was found only in patients with moderate AD | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Low† |
| S100A7/8/12 1; 40 | NA | Limited data and only with crisaborole | Unlike other potential biomarkers, correlation was found with Investigator’s Static Global Assessment and not EASI/SCORAD | No serious indirectness or imprecision; no publication bias detected | Low |
| Biomarkers showing decreased levels in correlation with clinical improvement in longitudinal, systemic treatment studies | |||||
| CCL17/TARC 6; 170 | 0.55 | No serious limitations | During dupilumab treatment, biomarker reduction was correlated with pruritus | No serious indirectness or imprecision; no publication bias detected | Moderate-high (in blood) |
| CCL13/MCP-4, 5; 130, IL-13, 5; 159, CCL22/MDC, 4; 124 | 0.54, 0.56, 0.49 (respectively) | Sparse data in blood by our criteria | During dupilumab treatment, correlation with biomarker reduction only achieved P < .1. For CCL13/MCP-4, variable laboratory methods were reported | No serious indirectness or imprecision; no publication bias detected | Moderate-high (in skin)† |
| S100A7/8/12 4; 121 | 0.54 | No serious limitations | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate |
| IL-22 4; 92 | 0.56 | Limited evidence in skin; no evidence in blood | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-low |
| CCL18/PARC 3; 98 | 0.56 | Limited evidence in skin; sparse evidence in blood by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-low |
| CCL26/ eotaxin-3 3; 73 | 0.62 | No evidence in blood by our criteria | No serious inconsistency | No serious indirectness or imprecision; no publication bias detected | Moderate-low (in skin) |
EASI, Eczema Area and Severity Index; ECP, eosinophil cationic protein; LDH, lactate dehydrogenase; NA, not applicable because only 1 report was included by our criteria; PARC, pulmonary and activation-regulated chemokine; SCORAD, SCORing of Atopic Dermatitis.
Based on Tables III and IV, only studies with a significant positive correlation with a correlation coefficient of ≥0.4 were included.
Baseline CCL22/MDC expression in skin correlated with future clinical improvement in a report analyzing data across multiple studies (using both topical and systemic therapies) at various time points.108
Biomarkers that failed to show consistent correlation with severity
Although total serum IgE levels (particularly in extrinsic AD)11,13,28,49,52–54,56,59,109–111 are elevated in AD, these are not consistently correlated with disease severity or only weakly correlated,12,51,90,112 and in dupilumab studies, responses of patients with AD are regardless of their baseline IgE levels.113 Thus, it is likely that IgE is a bystander in AD pathogenesis, rather than a treatment target.114,115 Although periostin is implicated in the pathogenesis of AD and was suggested as a potential AD biomarker by some reports, evidence for a correlation with disease severity is weak.18,116,117 Curiously, despite some reports on the correlation of the “itch cytokine,” IL-31, with disease severity,62,118 more evidence is accumulating on the lack of such correlation.102,119–121
Predictive biomarkers
Tissue biomarkers predicting disease onset include TARC and IgE in the umbilical cords of newborns122,123 and natural moisturizing factor levels in neonates’ skin,124 which are known to strongly correlate with transepidermal water loss,23 another predictor of AD development in newborns.125,126 Moisturizers can prevent AD in high-risk infants,127 and were shown to alter skin microbiome and reduce skin pH in this population.128 Other biomarkers may predict AD persistence (eg, low serum vascular endothelial growth factor [VEGF]).129
Because of the heterogeneous nature of AD pathophysiology, AD therapies targeting individual pathways are unlikely to result in high levels of response by all patients with AD,130,131 as seen in psoriasis with IL-17/IL-23 targeting.132 Thus, using biomarkers that can identify patient subsets that are more likely to respond to individual drugs or pathway antagonism would be beneficial. AD clinical trials have increasingly incorporated mechanistic analyses to assess potential biomarkers in both skin and blood (Table IV).96,133,134 Also, as AD symptoms fluctuate over time, biomarkers have the potential to provide objective insights into patient response to treatment and elucidate the mechanism of action of a drug. Biomarkers can either be common to all treatments (“disease response biomarkers”) or can be specific to individual treatments (“treatment-specific biomarkers”). For example, data from the phase 2 tralokinumab (IL-13 blocker) trial in AD have identified dipeptidyl peptidase-4 as a potential treatment response biomarker for IL-13 inhibition,4 and in asthma, periostin was identified as a response biomarker to IL-13 inhibition.135,136 Another example is the mAb inhibiting IL-22, fezakinumab, to which patients with AD with high IL-22 levels in skin biopsies at baseline were significantly more likely to respond.133 Other treatment-specific predictive biomarkers include CXCL9 (TH1/interferon-related) for cyclosporine and CXCL2 (TH17-related) for dupilumab.137 Although broad immune-suppressors (eg, methotrexate or azathioprine) were also studied in AD, no decreases in individual cytokine levels significantly correlated with response across agents.138
Recently, CCL22/MDC was found as the best biomarker of disease response across studies using different therapeutics,137 as baseline CCL22/MDC expression correlated with future clinical improvement across multiple studies at various time points, including topical treatment (crisaborole), systemic immunosuppressant (cyclosporine), and targeted treatment (fezakinumab).137
CCL17/TARC AS A BIOMARKER OF AD
Since CCL17/TARC was introduced to the field, primarily in Japan, it was reported as the most reliable biomarker studied,15 sensitive to fluctuations in clinical findings.20,24,27,28,38,101–103 CCL17/TARC is a CC chemokine discovered in 1996 by Imai et al,139 constitutively expressed in the thymus and a member of the TH2 chemokine family that attracts CC chemokine receptor 4–positive cells. Of note, thymus size also correlates with AD activity, and thymectomy may reduce the risk of AD.140,141 TH2-type cells and related products are significantly upregulated across various AD populations.43,59,65,142–144 In AD lesional skin, CCL17/TARC is expressed on keratinocytes in the epidermis, vascular endothelial cells, T cells, and dendritic cells.20,142 In Japan, serum CCL17/TARC levels have been measured commercially under health insurance support since 2008. Currently, after more than a decade of experience in patients with AD, CCL17/TARC has become a useful clinical biomarker for monitoring the efficacy of treatment and for ensuring successful treatment outcomes in the Japanese population.101
The normal level of serum CCL17/TARC in healthy adults is less than 450 pg/mL; its level in healthy children differs depending on age.31 Several investigations have also confirmed a high correlation between the AD severity and serum CCL17/TARC levels in pediatric patients.27,29,31,33,37, Moreover, increased CCL17/TARC levels from umbilical cord blood may even predict AD in infancy.122 Reports on the correlations between CCL17/TARC levels in the skin and clinical severity are sparse.21,25,145
Monitoring of serum CCL17/TARC levels could also be harnessed as an educational tool, improving patients’ adherence to treatment regimens. Patients can view their own disease activity as an objective number, and a rapid fall in the initially high serum CCL17/TARC levels due to adequate treatment can surely enhance compliance, a known pitfall in AD management.146 Moreover, patients receiving proactive treatment showed decreased but still high serum CCL17/TARC levels and were thus motivated to receive continuous therapy. CCL17/TARC is also reliable for assessing nonvisible/subclinical yet active AD-related inflammation, and high levels of CCL17/TARC may suggest frequent AD relapses even after clinical resolution, where a relatively thorough proactive treatment may be recommended.35
Nevertheless, the limitations of CCL17/TARC as an AD biomarker should also be acknowledged. Elevated serum CCL17/TARC level is not specific to AD, and could also be found in bullous pemphigoid, scabies, polymorphic prurigo, cutaneous T-cell lymphoma, drug eruption, pustular dermatosis, and other skin diseases,147–149 as well as in hypereosinophilic syndrome, Hodgkin lymphoma, and other internal disorders.150,151 Also, some patients with severe AD and patients with nodular prurigo or longstanding severe chronic lichenified lesions occasionally show normal or even low serum CCL17/TARC levels. Such cases may be explained by the heterogeneric pathomechanisms of AD. In addition, the added benefit of CCL17/TARC beyond a surrogate of clinical severity, for example, as a predictor of therapeutic response or as a reliable biomarker for clinical trials, still needs to be validated in future studies, using repeated testing.
AD BIOMARKERS ACROSS DISEASE PHENOTYPES
Two T-cell subsets—TH2 and TH22—are commonly activated across AD subtypes, yet specific biomarkers vary among different populations. Some examples of AD subtypes where phenotypic features may be explained by biomarker-related findings follow.
Patients with AD in different ethnicities
The Asian AD phenotype is characterized by greater expression of TH17-related markers (IL-17A, IL-19, CCL20) along with upregulation of IL-22 and the IL-17/IL-22–induced S100A12 in comparison to European-American patients with AD, but not to the levels found in psoriasis.43,144,152 This is particularly significant given most Asian patients with AD have extrinsic AD (high IgE levels), which tends to be associated with lower TH17 expression than intrinsic AD.144 These data suggest that Asian patients with AD present an immune dysregulation that is between European-American AD and psoriasis, and correlates well with the clinical phenotype of Asian AD, characterized by relatively well demarcated, psoriasiform lesions.143 Black patients with AD largely lack FLG mutations, in parallel with TH2/TH22 predominance and TH1/TH17 attenuation.59 H These may contribute to the lower rate of transepidermal water loss in black AD and to the atypical lichenified phenotype commonly seen in black patients with AD, potentially resulting from TH22 overexpression.143,153
Age-related changes in AD
Elderly patients (≥61 years old) with AD present a relative decrease in TH2/TH22 biomarkers with parallel increase in TH1/TH17 biomarkers, and a less pronounced barrier defect.86 The latter finding may contribute to the clinical observation of allergic sensitization as part of the atopic march, following AD initiation only at young age, and supports the notion that impaired barrier likely plays a major role in this process.
In addition, early-onset AD in infants is molecularly similar to psoriasis with a relatively dominant TH17-related skewing,65 in line with the extensor distribution of lesions in this age group, resembling that of psoriasis.
Biomarkers in association with AD comorbidities
In pediatric patients with AD, KRT5, KRT14, KRT16, FLG breakdown products, and AD clinical severity were predictive of concomitant food allergy.154 FLG mutations with suppressed levels of FLG expression predispose to AD, but are also associated with other diseases including asthma, irritant and allergic contact dermatitis, and alopecia areata.155 High levels of IgE and dysfunctional/low levels of FLG may predispose patients with AD to food allergy as part of the atopic march.156
Presence of Staphylococcus aureus colonization in AD
Finally, patients with AD colonized with S aureus have higher levels of type 2 biomarkers (including eosinophils, IgE, CCL17/TARC, and CCL26/eotaxin-3) and lactate dehydrogenase, along with more severe clinical parameters, including all severity scores, barrier dysfunction (by transepidermal water loss), and greater allergen sensitization.157
MINIMALLY INVASIVE BIOMARKERS
Through studies of skin biopsies, biomarkers of the immune milieu and barrier alterations of AD have been defined and facilitated therapeutic development. The inflammatory profile of AD is characterized by TH2 and TH22 skewing, with variable TH1 and TH17 components, depending on the disease subset (as detailed above).43,59,143,144 The barrier defects of AD include abnormalities in epidermal differentiation (FLG, loricrin, etc), tight junction (claudins), and lipid products (elongation of very long chain fatty acids-like 3 [ELOVL3], fatty acid 2-hydroxylase [FA2H], etc). Skin biopsies were also instrumental and sensitive in providing useful information on early and late changes with various treatments. Treatment response biomarkers provide important information of how well a certain drug is able to inhibit its direct target as well as other immune axes, and what is the relationship between inhibition of certain immune pathways/products and restoration of the barrier abnormalities characterizing AD, as well as clinical measures of the disease. Blood represents an easier accessible source of biomarkers, due to the relatively easy collection by blood withdrawal, in contrast to the invasive skin biopsy necessary for the assessment of biomarkers in the skin. In addition, blood levels may more objectively represent overall skin involvement, whereas skin biopsy represents only the skin where the biopsy is performed. Unfortunately, although skin biopsies accurately reflect disease severity and robust changes can be found early in the skin of patients with AD with various treatments, changes in blood may be more subtle and/or may take longer to occur.20,158 In addition, some key AD biomarkers in skin (ie, CCL26/exotoxin-3)84 are not well detected in blood, limiting the use of blood as a surrogate to skin biopsies. Biopsies collected from skin could be further divided into lesional and nonlesional samples. Perhaps counterintuitively, mRNA expression levels of markers from nonlesional skin samples of untreated patients with AD show higher and more significant correlations with SCORing of Atopic Dermatitis, including general inflammatory (metalloproteinase 12 [MMP12])- and proliferation (keratin 16 [KRT16])-related markers, as well as markers related to TH22 (IL-22), TH17 (CXCL1), and TH17/TH22 (S100A9).58 Moreover, nonlesional untreated skin data better correlate with serum data as compared with lesional skin, whereas correlations between lesional skin and serum are sparse.43,58 A possible explanation is that lesional AD skin bears a highly inflamed background, making the AD-specific biomarkers harder to identify and dissect due to a dilution phenomenon of innate cytokines. Furthermore, because nonlesional AD skin is not normal and yet not as inflamed as lesional skin, it provides a unique window for assessing AD dissemination to apparently uninvolved skin, and interventions that normalize nonlesional skin have the hypothetic potential to also prevent AD development.
Nevertheless, although skin biopsies are feasible in proof-of-concept studies in which it is crucial to understand the mechanism of action, and are highly informative, biopsies may be associated with significant discomfort and complications, making it difficult to use them in the setting of large-scale clinical trials and longitudinal studies, as well as in pediatric studies. Furthermore, incorporating skin and blood biomarker testing into large clinical trials, longitudinal studies, and in the clinic may be challenging and, if it is to be adopted in the future, will require very simple testing methods.
Consequently, there is a large unmet need for development of minimally invasive cutaneous biomarkers that capture the AD profile of lesional and nonlesional skin. Recently, tape-strips studies, collecting stratum corneum proteins from both adults and children with AD, showed promise in defining key disease features.68,106,159–161 These include studies of predefined sets of proteins and genes, as well as a limited-scope RNAseq.68,106,159,160,162 Similar to mRNA data from skin biopsies, tape-strips from both lesional and nonlesional skin show significant correlations with disease severity.68,93,159,160 Comparisons of variable aspects of tape-strips and biopsies are presented in Table VI, including disadvantages that may limit the use of tape-strips in settings of clinical trials or longitudinal studies. Recently, transcriptomic studies by tape-strip collection in young children and adults with AD showed improved detection rates of close to 100% per sample and per marker, perhaps enabling this approach in larger-scale studies, without losing data.9,68 This may indicate that in the future it may be feasible to use tape-strips in larger clinical trials and longitudinal studies, and even in the clinic.
TABLE VI.
Comparison of biomarker assessment by tape-strips and full-thickness skin biopsies
| Parameter | Tape-strips | Skin biopsies |
|---|---|---|
| Detection rates | Limited sample detection rates of 50% or even less in some studies23,154,159,160 | Typically, very high |
| Depth of tissue sampled | Stratum corneum and some of the stratum granulosum | Entire epidermis and dermis (when punch biopsies are used) |
| Detection of key TH2/TH22, AD-related biomarkers | Limited in some studies (eg, IL-4/IL-13 and IL-5,23 IL-3123,68,93,154,159,160,163) | Usually captured well |
| Detection of epidermal barrier-related biomarkers | Captured well (eg, terminal differentiation markers such as FLG and LOR, lipid-related biomarkers such as ELOVL1–7).9,23,68,154,159,163 Expression of some biomarkers was correlated with biopsies in the same individuals.164 A recent report suggested tape-stripped skin may even capture barrier-related changes better than biopsied skin in early disease81 | Usually captured well. Barrier-related changes can be located at specific areas of the skin |
| Advantages | Minimally invasive, nonscarring, allows repeated testing even in pediatric patients | Provides enough tissue for various laboratory studies, including for full-thickness immunohistochemistry studies revealing structural changes |
| Disadvantages | Tissue processing is time-consuming and technically challenging; potential differences in depth of tape-stripped skin, location of biomarkers, and structural changes (eg, epidermal thickness) within the skin cannot be captured. Hyperlasia-related biomarkers (eg, K16 and Ki67) are not well captured | Painful, scarring (including hypertrophic/keloids), might be complicated by infections, poor healing |
ELOVL, Elongation of very long chain fatty acids protein; LOR, loricrin.
Conclusions
The accessibility of the skin makes it the perfect tissue for investigation of disease mechanisms, and bench-to-bedside translational approaches are rapidly facilitating the development of novel therapeutics for inflammatory skin diseases. Tissue-derived biomarkers may further accelerate clinical trials and allow better reproducibility and rigor.165 Nevertheless, the discovery of a novel, validated disease-related biomarker is demanding and requires multiple steps, from the first detection of the potential tissue-derived factor to the final confirmation and acceptance by regulatory organizations.
AD, a common yet complex skin disease, stemming from immune dysregulations as well as epidermal barrier abnormalities, still poses a therapeutic challenge. The “one-size-fits-all” approach does not always apply for AD, because diverse disease phenotypes have been recognized, and therapeutic response may vary on the basis of their clinical and molecular differences. Indeed, a survey of AD specialists (IEC councilors and associates) strongly supports the combination of clinical evaluation with biomarkers’ assessments for stratification of patients with AD due to the large heterogeneity of the disease. Despite the relatively easy inspection of the skin by physical examination, clinical observations may not fully appreciate skin abnormalities, and are not entirely objective. This is emphasized by the relatively normal clinical appearance of AD nonlesional skin, while tissue assessments unearth significant immune and barrier dysregulations, resembling lesional skin.42,166 As we move toward more targeted therapies, AD biomarkers are important to appreciate patient-specific molecular dysregulations that differ between various AD subtypes. Because biologics are expensive, characterization of biomarkers that predict which patients will likely benefit most from these targeted biologics is essential. Ideally, a validated set of reliable biomarkers using minimally invasive methods will allow the implementation of precision medicine in AD, improve patient management, and expedite the development of novel therapeutics.
Because a biomarker should be assessed repeatedly, especially in the context of treatment monitoring or longitudinal studies, the preference of less-invasive methods over skin biopsies is well understood. Furthermore, skin biopsies are even harder to obtain in the pediatric population, in which the burden of AD is most significant. It is thus not surprising that alternative methods for skin sampling are emerging, with tape-strips, a minimally invasive method sampling only superficial epidermal layers, showing promise in both adult and pediatric AD.9,68,81,154,159,160
A biomarker should be biologically relevant and linked to disease mechanism.165 AD is characterized by robust systemic and cutaneous immune activation,167 with a dominant TH2-skewing that is shared across AD subtypes.168 Currently, some clinicians are already assessing few potential AD-related blood biomarkers to complement the physical examination and assess severity more accurately. These include nonspecific markers of inflammation and atopy. Nevertheless, the chemokine with the greatest evidence-based support to become a potential AD biomarker, at both baseline and following therapy, is CCL17/ TARC, a chemoattractant of TH2 cells. Although CCL17/TARC is implicated in other atopic diseases as well, including asthma and allergic rhinitis,169,170 correlation with clinical severity was established only in patients with AD.27 Moreover, we were able to find more than 20 publications supporting the robust correlation of serum CCL17/TARC with AD clinical severity, mainly SCORing of Atopic Dermatitis, in both children and adults. Additional emerging potential biomarkers include other TH2-related chemokines, such as CCL18/pulmonary and activation-regulated chemokine, CCL22/MDC (recently reported to consistently predict therapeutic response across different treatments),137 CCL26/eotaxin-3, CCL27/CTACK, and the key TH2 and TH22 cytokines, that is, IL-13 and IL-22, respectively. In comparison to TH2-related chemokines, cytokines were less commonly reported as blood biomarkers for AD and were mostly found to correlate with severity when assessed in skin.
In conclusion, the potential of biomarkers in AD is yet to be fully elucidated. The significant burden of the disease, its heterogeneity with increasingly recognized various subtypes, and the challenges of developing a “magic bullet” that benefits all patients despite the progression of multiple novel therapies advocate for a precision medicine approach. This approach would benefit from a set of disease-specific biomarkers that will further facilitate AD research and improve patient management; however, as demonstrated by our GRADE-based evaluation, evidence on biomarkers is still lacking.
New studies using more minimal techniques such as tape-strips, including pretreatment and posttreatment assessments, in which biomarker dynamics are closely monitored in relation to therapeutic response, are needed to improve the validity and relevance of biomarkers in AD. Large-scale clinical trials with extensive biomarker evaluation, including patients with variable AD phenotypes (eg, variable races and ages), are critical to establish the potential role of biomarkers in AD management. These may lead in the future to a clinical approach using biomarkers as a practical clinical tool where AD treatment will be personally tailored.
Disclosure of potential conflict of interest:
E. Guttman-Yassky is an employee of Mount Sinai and has received research funds (grants paid to the institution) from AbbVie, Celgene, Eli Lilly, Janssen, Medimmune/Astra Zeneca, Novartis, Pfizer, Regeneron, Vitae, Glenmark, Galderma, Asana, Innovaderm, Dermira, and UCB and is also a consultant for Sanofi Aventis, Regeneron, Stiefel/GlaxoSmithKline, MedImmune, Celgene, Anacor, AnaptysBio, Dermira, Galderma, Glenmark, Novartis, Pfizer, Vitae, LEO Pharma, AbbVie, Eli Lilly, Kyowa, Mitsubishi Tanabe, Asana Biosciences, and Promius. J. P. Thyssen has been an investigator/speaker or advisor for Sanofi-Genzyme, Regeneron, Eli Lilly & Co, Pfizer, AbbVie, and LEO Pharma. The rest of the authors declare that they have no relevant conflicts of interest.
Y.R.-Y. was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health, through Rockefeller University (grant no. UL1TR001866).
Abbreviations used
- AD
Atopic dermatitis
- CCL
Chemokine C-C motif ligand
- CTACK
Cutaneous T-cell–attracting chemokine
- FDA
Food and Drug Administration
- FLG
Filaggrin
- GRADE
Grading of Recommendations, Assessment, Development, and Evaluation
- IEC
International Eczema Council
- MDC
Macrophage-derived chemokine
- TARC
Thymus and activation-regulated chemokine
Footnotes
The figure was created with BioRender.com.
REFERENCES
- 1.Bieber T, Traidl-Hoffmann C, Schappi G, Lauener R, Akdis C, Schmid-Grendlmeier P. Unraveling the complexity of atopic dermatitis: the CK-CARE approach towards precision medicine. Allergy 2020;75:2936–8. [DOI] [PubMed] [Google Scholar]
- 2.Renert-Yuval Y, Guttman-Yassky E. What’s new in atopic dermatitis. Dermatol Clin 2019;37:205–13. [DOI] [PubMed] [Google Scholar]
- 3.Han Y, Chen Y, Liu X, Zhang J, Su H, Wen H, et al. Efficacy and safety of dupilumab for the treatment of adult atopic dermatitis: a meta-analysis of randomized clinical trials. J Allergy Clin Immunol 2017;140:888–91.e6. [DOI] [PubMed] [Google Scholar]
- 4.Wollenberg A, Howell MD, Guttman-Yassky E, Silverberg JI, Kell C, Ranade K, et al. Treatment of atopic dermatitis with tralokinumab, an anti-IL-13 mAb. J Allergy Clin Immunol 2019;143:135–41. [DOI] [PubMed] [Google Scholar]
- 5.Simpson EL, Flohr C, Eichenfield LF, Bieber T, Sofen H, Taieb A, et al. Efficacy and safety of lebrikizumab (an anti-IL-13 monoclonal antibody) in adults with moderate-to-severe atopic dermatitis inadequately controlled by topical corticosteroids: a randomized, placebo-controlled phase II trial (TREBLE). J Am Acad Dermatol 2018;78:863–71.e11. [DOI] [PubMed] [Google Scholar]
- 6.Guttman-Yassky E, Silverberg JI, Nemoto O, Forman SB, Wilke A, Prescilla R, et al. Baricitinib in adult patients with moderate-to-severe atopic dermatitis: a phase 2 parallel, double-blinded, randomized placebo-controlled multiple-dose study. J Am Acad Dermatol 2019;80:913–21.e9. [DOI] [PubMed] [Google Scholar]
- 7.Guyatt G, Oxman AD, Akl EA, Kunz R, Vist G, Brozek J, et al. GRADE guidelines, 1: introduction—GRADE evidence profiles and summary of findings tables. J Clin Epidemiol 2011;64:383–94. [DOI] [PubMed] [Google Scholar]
- 8.Garzorz-Stark N, Krause L, Lauffer F, Atenhan A, Thomas J, Stark SP, et al. A novel molecular disease classifier for psoriasis and eczema. Exp Dermatol 2016;25:767–74. [DOI] [PubMed] [Google Scholar]
- 9.He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, et al. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol 2021;147:199–212. [DOI] [PubMed] [Google Scholar]
- 10.Quaranta M, Knapp B, Garzorz N, Mattii M, Pullabhatla V, Pennino D, et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med 2014;6:244ra90. [DOI] [PubMed] [Google Scholar]
- 11.Vekaria AS, Brunner PM, Aleisa AI, Bonomo L, Lebwohl MG, Israel A, et al. Moderate-to-severe atopic dermatitis patients show increases in serum C-reactive protein levels, correlating with skin disease activity. F1000Res 2017;6: 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morishima Y, Kawashima H, Takekuma K, Hoshika A. Changes in serum lactate dehydrogenase activity in children with atopic dermatitis. Pediatr Int 2010;52: 171–4. [DOI] [PubMed] [Google Scholar]
- 13.Kou K, Aihara M, Matsunaga T, Chen H, Taguri M, Morita S, et al. Association of serum interleukin-18 and other biomarkers with disease severity in adults with atopic dermatitis. Arch Dermatol Res 2012;304:305–12. [DOI] [PubMed] [Google Scholar]
- 14.Mizawa M, Yamaguchi M, Ueda C, Makino T, Shimizu T. Stress evaluation in adult patients with atopic dermatitis using salivary cortisol. Biomed Res Int 2013;2013:138027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thijs J, Krastev T, Weidinger S, Buckens CF, de Bruin-Weller M, Bruijnzeel-Koomen C, et al. Biomarkers for atopic dermatitis: a systematic review and meta-analysis. Curr Opin Allergy Clin Immunol 2015;15:453–60. [DOI] [PubMed] [Google Scholar]
- 16.Kagi MK, Joller-Jemelka H, Wuthrich B. Correlation of eosinophils, eosinophil cationic protein and soluble interleukin-2 receptor with the clinical activity of atopic dermatitis. Dermatology 1992;185:88–92. [DOI] [PubMed] [Google Scholar]
- 17.Ariens LFM, van der Schaft J, Bakker DS, Balak D, Romeijn MLE, Kouwenhoven T, et al. Dupilumab is very effective in a large cohort of difficult-to-treat adult atopic dermatitis patients: first clinical and biomarker results from the Bio-Day registry. Allergy 2020;75:116–26. [DOI] [PubMed] [Google Scholar]
- 18.Kou K, Okawa T, Yamaguchi Y, Ono J, Inoue Y, Kohno M, et al. Periostin levels correlate with disease severity and chronicity in patients with atopic dermatitis. Br J Dermatol 2014;171:283–91. [DOI] [PubMed] [Google Scholar]
- 19.Czech W, Krutmann J, Schopf E, Kapp A. Serum eosinophil cationic protein (ECP) is a sensitive measure for disease activity in atopic dermatitis. Br J Dermatol 1992;126:351–5. [DOI] [PubMed] [Google Scholar]
- 20.Kakinuma T, Nakamura K, Wakugawa M, Mitsui H, Tada Y, Saeki H, et al. Thymus and activation-regulated chemokine in atopic dermatitis: serum thymus and activation-regulated chemokine level is closely related with disease activity. J Allergy Clin Immunol 2001;107:535–41. [DOI] [PubMed] [Google Scholar]
- 21.Morita E, Takahashi H, Niihara H, Dekio I, Sumikawa Y, Murakami Y, et al. Stratum corneum TARC level is a new indicator of lesional skin inflammation in atopic dermatitis. Allergy 2010;65:1166–72. [DOI] [PubMed] [Google Scholar]
- 22.Horikawa T, Nakayama T, Hikita I, Yamada H, Fujisawa R, Bito T, et al. IFN-gamma-inducible expression of thymus and activation-regulated chemokine/ CCL17 and macrophage-derived chemokine/CCL22 in epidermal keratinocytes and their roles in atopic dermatitis. Int Immunol 2002;14:767–73. [DOI] [PubMed] [Google Scholar]
- 23.McAleer MA, Jakasa I, Hurault G, Sarvari P, McLean WHI, Tanaka RJ, et al. Systemic and stratum corneum biomarkers of severity in infant atopic dermatitis include markers of innate and T helper cell-related immunity and angiogenesis. Br J Dermatol 2019;180:586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujisawa T, Fujisawa R, Kato Y, Nakayama T, Morita A, Katsumata H, et al. Presence of high contents of thymus and activation-regulated chemokine in platelets and elevated plasma levels of thymus and activation-regulated chemokine and macrophage-derived chemokine in patients with atopic dermatitis. J Allergy Clin Immunol 2002;110:139–46. [DOI] [PubMed] [Google Scholar]
- 25.He H, Del Duca E, Diaz A, Kim HJ, Gay-Mimbrera J, Zhang N, et al. Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities [published online ahead of print October 1, 2020]. J Allergy Clin Immunol. 10.1016/j.jaci.2020.08.041. [DOI] [PubMed] [Google Scholar]
- 26.Leung TF, Ma KC, Hon KL, Lam CW, Wan H, Li CY, et al. Serum concentration of macrophage-derived chemokine may be a useful inflammatory marker for assessing severity of atopic dermatitis in infants and young children. Pediatr Allergy Immunol 2003;14:296–301. [DOI] [PubMed] [Google Scholar]
- 27.Hijnen D, De Bruin-Weller M, Oosting B, Lebre C, De Jong E, Bruijnzeel-Koomen C, et al. Serum thymus and activation-regulated chemokine (TARC) and cutaneous T cell-attracting chemokine (CTACK) levels in allergic diseases: TARC and CTACK are disease-specific markers for atopic dermatitis. J Allergy Clin Immunol 2004;113:334–40. [DOI] [PubMed] [Google Scholar]
- 28.Jahnz-Rozyk K, Targowski T, Paluchowska E, Owczarek W, Kucharczyk A. Serum thymus and activation-regulated chemokine, macrophage-derived chemokine and eotaxin as markers of severity of atopic dermatitis. Allergy 2005;60:685–8. [DOI] [PubMed] [Google Scholar]
- 29.Song TW, Sohn MH, Kim ES, Kim KW, Kim KE. Increased serum thymus and activation-regulated chemokine and cutaneous T cell-attracting chemokine levels in children with atopic dermatitis. Clin Exp Allergy 2006;36:346–51. [DOI] [PubMed] [Google Scholar]
- 30.Nakazato J, Kishida M, Kuroiwa R, Fujiwara J, Shimoda M, Shinomiya N. Serum levels of Th2 chemokines, CCL17, CCL22, and CCL27, were the important markers of severity in infantile atopic dermatitis. Pediatr Allergy Immunol 2008;19:605–13. [DOI] [PubMed] [Google Scholar]
- 31.Fujisawa T, Nagao M, Hiraguchi Y, Katsumata H, Nishimori H, Iguchi K, et al. Serum measurement of thymus and activation-regulated chemokine/CCL17 in children with atopic dermatitis: elevated normal levels in infancy and age-specific analysis in atopic dermatitis. Pediatr Allergy Immunol 2009;20:633–41. [DOI] [PubMed] [Google Scholar]
- 32.van Velsen SG, Knol MJ, Haeck IM, Bruijnzeel-Koomen CA, Pasmans SG. The Self-administered Eczema Area and Severity Index in children with moderate to severe atopic dermatitis: better estimation of AD body surface area than severity. Pediatr Dermatol 2010;27:470–5. [DOI] [PubMed] [Google Scholar]
- 33.Machura E, Rusek-Zychma M, Jachimowicz M, Wrzask M, Mazur B, Kasperska-Zajac A. Serum TARC and CTACK concentrations in children with atopic dermatitis, allergic asthma, and urticaria. Pediatr Allergy Immunol 2012;23:278–84. [DOI] [PubMed] [Google Scholar]
- 34.Furue M, Matsumoto T, Yamamoto T, Takeuchi S, Esaki H, Chiba T, et al. Correlation between serum thymus and activation-regulated chemokine levels and stratum corneum barrier function in healthy individuals and patients with mild atopic dermatitis. J Dermatol Sci 2012;66:60–3. [DOI] [PubMed] [Google Scholar]
- 35.Kataoka Y. Thymus and activation-regulated chemokine as a clinical biomarker in atopic dermatitis. J Dermatol 2014;41:221–9. [DOI] [PubMed] [Google Scholar]
- 36.Landheer J, de Bruin-Weller M, Boonacker C, Hijnen D, Bruijnzeel-Koomen C, Rockmann H. Utility of serum thymus and activation-regulated chemokine as a biomarker for monitoring of atopic dermatitis severity. J Am Acad Dermatol 2014;71:1160–6. [DOI] [PubMed] [Google Scholar]
- 37.Ahrens B, Schulz G, Bellach J, Niggemann B, Beyer K. Chemokine levels in serum of children with atopic dermatitis with regard to severity and sensitization status. Pediatr Allergy Immunol 2015;26:634–40. [DOI] [PubMed] [Google Scholar]
- 38.Gu CY, Gu L, Dou X. Serum levels of thymus and activation-regulated chemokine can be used in the clinical evaluation of atopic dermatitis. Int J Dermatol 2015;54:e261–5. [DOI] [PubMed] [Google Scholar]
- 39.Hulshof L, Overbeek SA, Wyllie AL, Chu M, Bogaert D, de Jager W, et al. Exploring immune development in infants with moderate to severe atopic dermatitis. Front Immunol 2018;9:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakinuma T, Nakamura K, Wakugawa M, Mitsui H, Tada Y, Saeki H, et al. Serum macrophage-derived chemokine (MDC) levels are closely related with the disease activity of atopic dermatitis. Clin Exp Immunol 2002;127:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tintle S, Shemer A, Suarez-Farinas M, Fujita H, Gilleaudeau P, Sullivan-Whalen M, et al. Reversal of atopic dermatitis with narrow-band UVB phototherapy and biomarkers for therapeutic response. J Allergy Clin Immunol 2011;128: 583–93.e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol 2011;127:954–64.e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen HC, Czarnowicki T, Noda S, Malik K, Pavel AB, Nakajima S, et al. Serum from Asian patients with atopic dermatitis is characterized by TH2/TH22 activation, which is highly correlated with nonlesional skin measures. J Allergy Clin Immunol 2018;142:324–8.e11. [DOI] [PubMed] [Google Scholar]
- 44.Gunther C, Bello-Fernandez C, Kopp T, Kund J, Carballido-Perrig N, Hinteregger S, et al. CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J Immunol 2005;174:1723–8. [DOI] [PubMed] [Google Scholar]
- 45.Angelova-Fischer I, Hipler UC, Bauer A, Fluhr JW, Tsankov N, Fischer TW, et al. Significance of interleukin-16, macrophage-derived chemokine, eosinophil cationic protein and soluble E-selectin in reflecting disease activity of atopic dermatitis–from laboratory parameters to clinical scores. Br J Dermatol 2006; 154:1112–7. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto S, Nakamura K, Oyama N, Kaneko F, Tsunemi Y, Saeki H, et al. Macrophage-derived chemokine (MDC)/CCL22 produced by monocyte derived dendritic cells reflects the disease activity in patients with atopic dermatitis. J Dermatol Sci 2006;44:93–9. [DOI] [PubMed] [Google Scholar]
- 47.Brunner PM, He H, Pavel AB, Czarnowicki T, Lefferdink R, Erickson T, et al. The blood proteomic signature of early-onset pediatric atopic dermatitis shows systemic inflammation and is distinct from adult long-standing disease. J Am Acad Dermatol 2019;81:510–9. [DOI] [PubMed] [Google Scholar]
- 48.Tsuboi H, Kouda K, Takeuchi H, Takigawa M, Masamoto Y, Takeuchi M, et al. 8-hydroxydeoxyguanosine in urine as an index of oxidative damage to DNA in the evaluation of atopic dermatitis. Br J Dermatol 1998;138:1033–5. [DOI] [PubMed] [Google Scholar]
- 49.Yoshizawa Y, Nomaguchi H, Izaki S, Kitamura K. Serum cytokine levels in atopic dermatitis. Clin Exp Dermatol 2002;27:225–9. [DOI] [PubMed] [Google Scholar]
- 50.Kaminishi K, Soma Y, Kawa Y, Mizoguchi M. Flow cytometric analysis of IL-4, IL-13 and IFN-gamma expression in peripheral blood mononuclear cells and detection of circulating IL-13 in patients with atopic dermatitis provide evidence for the involvement of type 2 cytokines in the disease. J Dermatol Sci 2002;29: 19–25. [DOI] [PubMed] [Google Scholar]
- 51.Aral M, Arican O, Gul M, Sasmaz S, Kocturk SA, Kastal U, et al. The relationship between serum levels of total IgE, IL-18, IL-12, IFN-gamma and disease severity in children with atopic dermatitis. Mediators Inflamm 2006;2006:73098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomon J, Baran E. The role of selected neuropeptides in pathogenesis of atopic dermatitis. J Eur Acad Dermatol Venereol 2008;22:223–8. [DOI] [PubMed] [Google Scholar]
- 53.Wu KG, Li TH, Chen CJ, Cheng HI, Wang TY. Correlations of serum interleukin-16, total IgE, eosinophil cationic protein and total eosinophil counts with disease activity in children with atopic dermatitis. Int J Immunopathol Pharmacol 2011; 24:15–23. [DOI] [PubMed] [Google Scholar]
- 54.Suarez-Farinas M, Dhingra N, Gittler J, Shemer A, Cardinale I, de Guzman Strong C, et al. Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol 2013;132:361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zedan K, Rasheed Z, Farouk Y, Alzolibani AA, Bin Saif G, Ismail HA, et al. Immunoglobulin E, interleukin-18 and interleukin-12 in patients with atopic dermatitis: correlation with disease activity. J Clin Diagn Res 2015;9:WC01–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glatz M, Buchner M, von Bartenwerffer W, Schmid-Grendelmeier P, Worm M, Hedderich J, et al. Malassezia spp.-specific immunoglobulin E level is a marker for severity of atopic dermatitis in adults. Acta Derm Venereol 2015;95:191–6. [DOI] [PubMed] [Google Scholar]
- 57.Rosinska-Wieckowicz A, Czarnecka-Operacz M, Adamski Z. Selected immunological parameters in clinical evaluation of patients with atopic dermatitis. Postepy Dermatol Alergol 2016;33:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ungar B, Garcet S, Gonzalez J, Dhingra N, Correa da Rosa J, Shemer A, et al. An integrated model of atopic dermatitis biomarkers highlights the systemic nature of the disease. J Invest Dermatol 2017;137:603–13. [DOI] [PubMed] [Google Scholar]
- 59.Sanyal RD, Pavel AB, Glickman J, Chan TC, Zheng X, Zhang N, et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/TH17 attenuation. Ann Allergy Asthma Immunol 2019;122:99–110.e6. [DOI] [PubMed] [Google Scholar]
- 60.Mukai H, Noguchi T, Kamimura K, Nishioka K, Nishiyama S. Significance of elevated serum LDH (lactate dehydrogenase) activity in atopic dermatitis. J Dermatol 1990;17:477–81. [DOI] [PubMed] [Google Scholar]
- 61.Halmerbauer G, Frischer T, Koller DY. Monitoring of disease activity by measurement of inflammatory markers in atopic dermatitis in childhood. Allergy 1997;52:765–9. [DOI] [PubMed] [Google Scholar]
- 62.Raap U, Weissmantel S, Gehring M, Eisenberg AM, Kapp A, Folster-Holst R. IL-31 significantly correlates with disease activity and Th2 cytokine levels in children with atopic dermatitis. Pediatr Allergy Immunol 2012;23:285–8. [DOI] [PubMed] [Google Scholar]
- 63.Chen YL, Gutowska-Owsiak D, Hardman CS, Westmoreland M, MacKenzie T, Cifuentes L, et al. Proof-of-concept clinical trial of etokimab shows a key role for IL-33 in atopic dermatitis pathogenesis. Sci Transl Med 2019;11:eaax2945. [DOI] [PubMed] [Google Scholar]
- 64.Nograles KE, Zaba LC, Shemer A, Fuentes-Duculan J, Cardinale I, Kikuchi T, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol 2009;123:1244–52.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esaki H, Brunner PM, Renert-Yuval Y, Czarnowicki T, Huynh T, Tran G, et al. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol 2016;138:1639–51. [DOI] [PubMed] [Google Scholar]
- 66.Koning H, Neijens HJ, Baert MR, Oranje AP, Savelkoul HF. T cell subsets and cytokines in allergic and non-allergic children, I: analysis of IL-4, IFN-gamma and IL-13 mRNA expression and protein production. Cytokine 1997;9:416–26. [DOI] [PubMed] [Google Scholar]
- 67.Szegedi K, Lutter R, Res PC, Bos JD, Luiten RM, Kezic S, et al. Cytokine profiles in interstitial fluid from chronic atopic dermatitis skin. J Eur Acad Dermatol Venereol 2015;29:2136–44. [DOI] [PubMed] [Google Scholar]
- 68.Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, et al. Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol 2019;155: 1358–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hon KL, Leung TF, Ma KC, Wong CK, Wan H, Lam CW. Serum concentration of IL-18 correlates with disease extent in young children with atopic dermatitis. Pediatr Dermatol 2004;21:619–22. [DOI] [PubMed] [Google Scholar]
- 70.Inoue Y, Aihara M, Kirino M, Harada I, Komori-Yamaguchi J, Yamaguchi Y, et al. Interleukin-18 is elevated in the horny layer in patients with atopic dermatitis and is associated with Staphylococcus aureus colonization. Br J Dermatol 2011;164:560–7. [DOI] [PubMed] [Google Scholar]
- 71.Park DS, Youn YH. Clinical significance of serum interleukin-18 concentration in the patients with atopic dermatitis [in Korean]. Korean J Lab Med 2007;27: 128–32. [DOI] [PubMed] [Google Scholar]
- 72.Pavel AB, Zhou L, Diaz A, Ungar B, Dan J, He H, et al. The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J Am Acad Dermatol 2020;82:690–9. [DOI] [PubMed] [Google Scholar]
- 73.Suwarsa O, Adi S, Idjradinata P, Sutedja E, Avriyanti E, Asfara A, et al. Inter-leukin-18 correlates with interleukin-4 but not interferon-gamma production in lymphocyte cultures from atopic dermatitis patients after staphylococcal enterotoxin B stimulation. Asian Pac J Allergy Immunol 2017;35:54–9. [DOI] [PubMed] [Google Scholar]
- 74.Kakinuma T, Saeki H, Tsunemi Y, Fujita H, Asano N, Mitsui H, et al. Increased serum cutaneous T cell-attracting chemokine (CCL27) levels in patients with atopic dermatitis and psoriasis vulgaris. J Allergy Clin Immunol 2003;111: 592–7. [DOI] [PubMed] [Google Scholar]
- 75.Hon KL, Leung TF, Ma KC, Li AM, Wong Y, Fok TF. Serum levels of cutaneous T-cell attracting chemokine (CTACK) as a laboratory marker of the severity of atopic dermatitis in children. Clin Exp Dermatol 2004;29:293–6. [DOI] [PubMed] [Google Scholar]
- 76.Morita H, Kitano Y, Kawasaki N. Elevation of serum-soluble E-selectin in atopic dermatitis. J Dermatol Sci 1995;10:145–50. [DOI] [PubMed] [Google Scholar]
- 77.Yamashita N, Kaneko S, Kouro O, Furue M, Yamamoto S, Sakane T. Soluble E-selectin as a marker of disease activity in atopic dermatitis. J Allergy Clin Immunol 1997;99:410–6. [DOI] [PubMed] [Google Scholar]
- 78.Wolkerstorfer A, Savelkoul HF, de Waard van der Spek FB, Neijens HJ, van Meurs T, Oranje AP. Soluble E-selectin and soluble ICAM-1 levels as markers of the activity of atopic dermatitis in children. Pediatr Allergy Immunol 2003; 14:302–6. [DOI] [PubMed] [Google Scholar]
- 79.Brunner PM, Suarez-Farinas M, He H, Malik K, Wen HC, Gonzalez J, et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci Rep 2017;7:8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He H, Li R, Choi S, Zhou L, Pavel A, Estrada YD, et al. Increased cardiovascular and atherosclerosis markers in blood of older patients with atopic dermatitis. Ann Allergy Asthma Immunol 2020;124:70–8. [DOI] [PubMed] [Google Scholar]
- 81.Pavel AB, Renert-Yuval Y, Wu J, Del Duca E, Diaz A, Lefferdink R, et al. Tape-strips from early-onset pediatric atopic dermatitis highlight disease abnormalities in non-lesional skin. Allergy 2021;76:314–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olesen CM, Holm JG, Norreslet LB, Serup JV, Thomsen SF, Agner T. Treatment of atopic dermatitis with dupilumab: experience from a tertiary referral centre. J Eur Acad Dermatol Venereol 2019;33:1562–8. [DOI] [PubMed] [Google Scholar]
- 83.Hon KL, Ching GK, Ng PC, Leung TF. Exploring CCL18, eczema severity and atopy. Pediatr Allergy Immunol 2011;22:704–7. [DOI] [PubMed] [Google Scholar]
- 84.Gittler JK, Shemer A, Suarez-Farinas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQ, et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol 2012;130:1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kagami S, Kakinuma T, Saeki H, Tsunemi Y, Fujita H, Nakamura K, et al. Significant elevation of serum levels of eotaxin-3/CCL26, but not of eotaxin-2/CCL24, in patients with atopic dermatitis: serum eotaxin-3/CCL26 levels reflect the disease activity of atopic dermatitis. Clin Exp Immunol 2003;134:309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou L, Leonard A, Pavel AB, Malik K, Raja A, Glickman J, et al. Age-specific changes in the molecular phenotype of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 2019;144:144–56. [DOI] [PubMed] [Google Scholar]
- 87.Oka T, Sugaya M, Takahashi N, Nakajima R, Otobe S, Kabasawa M, et al. Increased interleukin-19 expression in cutaneous T-cell lymphoma and atopic dermatitis. Acta Derm Venereol 2017;97:1172–7. [DOI] [PubMed] [Google Scholar]
- 88.Konrad RJ, Higgs RE, Rodgers GH, Ming W, Qian YW, Bivi N, et al. Assessment and clinical relevance of serum IL-19 levels in psoriasis and atopic dermatitis using a sensitive and specific novel immunoassay. Sci Rep 2019;9:5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costa C, Rilliet A, Nicolet M, Saurat JH. Scoring atopic dermatitis: the simpler the better? Acta Derm Venereol 1989;69:41–5. [PubMed] [Google Scholar]
- 90.Furukawa H, Takahashi M, Nakamura K, Kaneko F. Effect of an antiallergic drug (Olopatadine hydrochloride) on TARC/CCL17 and MDC/CCL22 production by PBMCs from patients with atopic dermatitis. J Dermatol Sci 2004;36: 165–72. [DOI] [PubMed] [Google Scholar]
- 91.Khattri S, Shemer A, Rozenblit M, Dhingra N, Czarnowicki T, Finney R, et al. Cyclosporine in patients with atopic dermatitis modulates activated inflammatory pathways and reverses epidermal pathology. J Allergy Clin Immunol 2014;133: 1626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwon YS, Oh SH, Wu WH, Bae BG, Lee HJ, Lee MG, et al. CC chemokines as potential immunologic markers correlated with clinical improvement of atopic dermatitis patients by immunotherapy. Exp Dermatol 2010;19:246–51. [DOI] [PubMed] [Google Scholar]
- 93.Koppes SA, Brans R, Ljubojevic Hadzavdic S, Frings-Dresen MH, Rustemeyer T, Kezic S. Stratum corneum tape stripping: monitoring of inflammatory mediators in atopic dermatitis patients using topical therapy. Int Arch Allergy Immunol 2016;170:187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 2014;371:130–9. [DOI] [PubMed] [Google Scholar]
- 95.Pavel AB, Song T, Kim HJ, Del Duca E, Krueger JG, Dubin C, et al. Oral Janus kinase/SYK inhibition (ASN002) suppresses inflammation and improves epidermal barrier markers in patients with atopic dermatitis. J Allergy Clin Immunol 2019;144:1011–24. [DOI] [PubMed] [Google Scholar]
- 96.Guttman-Yassky E, Bissonnette R, Ungar B, Suarez-Farinas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol 2019;143: 155–72. [DOI] [PubMed] [Google Scholar]
- 97.Bissonnette R, Pavel AB, Diaz A, Werth JL, Zang C, Vranic I, et al. Crisaborole and atopic dermatitis skin biomarkers: an intrapatient randomized trial. J Allergy Clin Immunol 2019;144:1274–89. [DOI] [PubMed] [Google Scholar]
- 98.Hamilton JD, Suarez-Farinas M, Dhingra N, Cardinale I, Li X, Kostic A, et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 2014;134:1293–300. [DOI] [PubMed] [Google Scholar]
- 99.He H, Olesen CM, Pavel AB, Clausen ML, Wu J, Estrada Y, et al. Tape-strip proteomic profiling of atopic dermatitis on dupilumab identifies minimally invasive biomarkers. Front Immunol 2020;11:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simon D, Vassina E, Yousefi S, Kozlowski E, Braathen LR, Simon HU. Reduced dermal infiltration of cytokine-expressing inflammatory cells in atopic dermatitis after short-term topical tacrolimus treatment. J Allergy Clin Immunol 2004;114: 887–95. [DOI] [PubMed] [Google Scholar]
- 101.Yasukochi Y, Nakahara T, Abe T, Kido-Nakahara M, Kohda F, Takeuchi S, et al. Reduction of serum TARC levels in atopic dermatitis by topical anti-inflammatory treatments. Asian Pac J Allergy Immunol 2014;32:240–5. [DOI] [PubMed] [Google Scholar]
- 102.Kyoya M, Kawakami T, Soma Y. Serum thymus and activation-regulated chemokine (TARC) and interleukin-31 levels as biomarkers for monitoring in adult atopic dermatitis. J Dermatol Sci 2014;75:204–7. [DOI] [PubMed] [Google Scholar]
- 103.Gohar MK, Atta AH, Nasr MM, Hussein DN. Serum thymus and activation regulated chemokine (TARC), IL-18 and IL-18 gene polymorphism as associative factors with atopic dermatitis. Egypt J Immunol 2017;24:9–22. [PubMed] [Google Scholar]
- 104.Bogaczewicz J, Malinowska K, Sysa-Jedrzejowska A, Wozniacka A. Medium-dose ultraviolet A1 phototherapy and mRNA expression of TSLP, TARC, IL-5, and IL-13 in acute skin lesions in atopic dermatitis. Int J Dermatol 2016;55:856–63. [DOI] [PubMed] [Google Scholar]
- 105.Kagami S, Saeki H, Komine M, Kakinuma T, Tsunemi Y, Nakamura K, et al. Interleukin-4 and interleukin-13 enhance CCL26 production in a human keratinocyte cell line, HaCaT cells. Clin Exp Immunol 2005;141:459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kezic S, O’Regan GM, Lutter R, Jakasa I, Koster ES, Saunders S, et al. Filaggrin loss-of-function mutations are associated with enhanced expression of IL-1 cytokines in the stratum corneum of patients with atopic dermatitis and in a murine model of filaggrin deficiency. J Allergy Clin Immunol 2012;129:1031–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thijs JL, Drylewicz J, Fiechter R, Strickland I, Sleeman MA, Herath A, et al. EASI p-EASI: utilizing a combination of serum biomarkers offers an objective measurement tool for disease severity in atopic dermatitis patients. J Allergy Clin Immunol 2017;140:1703–5. [DOI] [PubMed] [Google Scholar]
- 108.Glickman JW, Dubin C, Renert-Yuval Y, Dahabreh D, Kimmel GW, Auyeung K, et al. Cross-sectional study of blood biomarkers of patients with moderate to severe alopecia areata reveals systemic immune and cardiovascular biomarker dysregulation. J Am Acad Dermatol 2021;84:370–80. [DOI] [PubMed] [Google Scholar]
- 109.Laske N, Niggemann B. Does the severity of atopic dermatitis correlate with serum IgE levels? Pediatr Allergy Immunol 2004;15:86–8. [DOI] [PubMed] [Google Scholar]
- 110.Shaheen MA, Attia EA, Louka ML, Bareedy N. Study of the role of serum folic acid in atopic dermatitis: a correlation with serum IgE and disease severity. Indian J Dermatol 2011;56:673–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neuber K, Schwartz I, Itschert G, Dieck AT. Treatment of atopic eczema with oral mycophenolate mofetil. Br J Dermatol 2000;143:385–91. [DOI] [PubMed] [Google Scholar]
- 112.Thijs JL, Knipping K, Bruijnzeel-Koomen CA, Garssen J, de Bruin-Weller MS, Hijnen DJ. Immunoglobulin free light chains in adult atopic dermatitis patients do not correlate with disease severity. Clin Transl Allergy 2016;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hamilton JD, Ungar B, Guttman-Yassky E. Drug evaluation review: dupilumab in atopic dermatitis. Immunotherapy 2015;7:1043–58. [DOI] [PubMed] [Google Scholar]
- 114.Heil PM, Maurer D, Klein B, Hultsch T, Stingl G. Omalizumab therapy in atopic dermatitis: depletion of IgE does not improve the clinical course—a randomized, placebo-controlled and double blind pilot study. J Dtsch Dermatol Ges 2010;8: 990–8. [DOI] [PubMed] [Google Scholar]
- 115.Holm JG, Agner T, Sand C, Thomsen SF. Omalizumab for atopic dermatitis: case series and a systematic review of the literature. Int J Dermatol 2017;56:18–26. [DOI] [PubMed] [Google Scholar]
- 116.Uysal P, Birtekocak F, Karul AB. The relationship between serum TARC, TSLP and POSTN levels and childhood atopic dermatitis. Clin Lab 2017;63: 1071–7. [DOI] [PubMed] [Google Scholar]
- 117.Sung M, Lee KS, Ha EG, Lee SJ, Kim MA, Lee SW, et al. An association of periostin levels with the severity and chronicity of atopic dermatitis in children. Pediatr Allergy Immunol 2017;28:543–50. [DOI] [PubMed] [Google Scholar]
- 118.Raap U, Wichmann K, Bruder M, Stander S, Wedi B, Kapp A, et al. Correlation of IL-31 serum levels with severity of atopic dermatitis. J Allergy Clin Immunol 2008;122:421–3. [DOI] [PubMed] [Google Scholar]
- 119.Ozceker D, Bulut M, Ozbay AC, Dilek F, Koser M, Tamay Z, et al. Assessment of IL-31 levels and disease severity in children with atopic dermatitis. Allergol Immunopathol (Madr) 2018;46:322–5. [DOI] [PubMed] [Google Scholar]
- 120.Neis MM, Peters B, Dreuw A, Wenzel J, Bieber T, Mauch C, et al. Enhanced expression levels of IL-31 correlate with IL-4 and IL-13 in atopic and allergic contact dermatitis. J Allergy Clin Immunol 2006;118:930–7. [DOI] [PubMed] [Google Scholar]
- 121.Nygaard U, Hvid M, Johansen C, Buchner M, Folster-Holst R, Deleuran M, et al. TSLP, IL-31, IL-33 and sST2 are new biomarkers in endophenotypic profiling of adult and childhood atopic dermatitis. J Eur Acad Dermatol Venereol 2016;30: 1930–8. [DOI] [PubMed] [Google Scholar]
- 122.Miyahara H, Okazaki N, Nagakura T, Korematsu S, Izumi T. Elevated umbilical cord serum TARC/CCL17 levels predict the development of atopic dermatitis in infancy. Clin Exp Allergy 2011;41:186–91. [DOI] [PubMed] [Google Scholar]
- 123.Wen HJ, Wang YJ, Lin YC, Chang CC, Shieh CC, Lung FW, et al. Prediction of atopic dermatitis in 2-yr-old children by cord blood IgE, genetic polymorphisms in cytokine genes, and maternal mentality during pregnancy. Pediatr Allergy Immunol 2011;22:695–703. [DOI] [PubMed] [Google Scholar]
- 124.Ni Chaoimh C, Nico C, Puppels GJ, Caspers PJ, Wong X, Common JE, et al. In vivo Raman spectroscopy discriminates between FLG loss-of-function carriers vs wild-type in day 1–4 neonates. Ann Allergy Asthma Immunol 2020; 124:500–4. [DOI] [PubMed] [Google Scholar]
- 125.Kelleher M, Dunn-Galvin A, Hourihane JO, Murray D, Campbell LE, McLean WH, et al. Skin barrier dysfunction measured by transepidermal water loss at 2 days and 2 months predates and predicts atopic dermatitis at 1 year. J Allergy Clin Immunol 2015;135:930–5.e1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Horimukai K, Morita K, Narita M, Kondo M, Kabashima S, Inoue E, et al. Transepidermal water loss measurement during infancy can predict the subsequent development of atopic dermatitis regardless of filaggrin mutations. Allergol Int 2016;65:103–8. [DOI] [PubMed] [Google Scholar]
- 127.Dominguez-Huttinger E, Christodoulides P, Miyauchi K, Irvine AD, Okada-Hatakeyama M, Kubo M, et al. Mathematical modeling of atopic dermatitis reveals “double-switch” mechanisms underlying 4 common disease phenotypes. J Allergy Clin Immunol 2017;139:1861–72.e7. [DOI] [PubMed] [Google Scholar]
- 128.Glatz M, Jo JH, Kennedy EA, Polley EC, Segre JA, Simpson EL, et al. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS One 2018;13:e0192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lauffer F, Baghin V, Standl M, Stark SP, Jargosch M, Wehrle J, et al. Predicting persistence of atopic dermatitis in children using clinical attributes and serum proteins [published online ahead of print August 13, 2020]. Allergy. 10.1111/all.14557. [DOI] [PubMed] [Google Scholar]
- 130.Blauvelt A, de Bruin-Weller M, Gooderham M, Cather JC, Weisman J, Pariser D, et al. Long-term management of moderate-to-severe atopic dermatitis with dupilumab and concomitant topical corticosteroids (LIBERTY AD CHRONOS): a 1-year, randomised, double-blinded, placebo-controlled, phase 3 trial. Lancet 2017;389:2287–303. [DOI] [PubMed] [Google Scholar]
- 131.Simpson EL, Bieber T, Guttman-Yassky E, Beck LA, Blauvelt A, Cork MJ, et al. Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med 2016;375:2335–48. [DOI] [PubMed] [Google Scholar]
- 132.Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. N Engl J Med 2014; 371:326–38. [DOI] [PubMed] [Google Scholar]
- 133.Brunner PM, Pavel AB, Khattri S, Leonard A, Malik K, Rose S, et al. Baseline IL22 expression in atopic dermatitis patients stratifies tissue responses to fezakinumab. J Allergy Clin Immunol 2019;143:142–54. [DOI] [PubMed] [Google Scholar]
- 134.Khattri S, Brunner PM, Garcet S, Finney R, Cohen SR, Oliva M, et al. Efficacy and safety of ustekinumab treatment in adults with moderate-to-severe atopic dermatitis. Exp Dermatol 2017;26:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Piper E, Brightling C, Niven R, Oh C, Faggioni R, Poon K, et al. A phase II placebo-controlled study of tralokinumab in moderate-to-severe asthma. Eur Respir J 2013;41:330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Emson C, Pham TH, Manetz S, Newbold P. Periostin and dipeptidyl peptidase-4: potential biomarkers of interleukin 13 pathway activation in asthma and allergy. Immunol Allergy Clin North Am 2018;38:611–28. [DOI] [PubMed] [Google Scholar]
- 137.Glickman JW, Han J, Garcet S, Krueger JG, Pavel AB, Guttman-Yassky E. Improving evaluation of drugs in atopic dermatitis by combining clinical and molecular measures. J Allergy Clin Immunol Pract 2020;8:3622–5.e19. [DOI] [PubMed] [Google Scholar]
- 138.Roekevisch E, Szegedi K, Hack DP, Schram ME, Res P, Bos JD, et al. Effect of immunosuppressive treatment on biomarkers in adult atopic dermatitis patients. J Eur Acad Dermatol Venereol 2020;34:1545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Imai T, Yoshida T, Baba M, Nishimura M, Kakizaki M, Yoshie O. Molecular cloning of a novel T cell-directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J Biol Chem 1996;271:21514–21. [DOI] [PubMed] [Google Scholar]
- 140.Thyssen JP, Andersen YMF, Zhang H, Gislason G, Skov L, Egeberg A. Incidence of pediatric atopic dermatitis following thymectomy: a Danish register study. Allergy 2018;73:1741–3. [DOI] [PubMed] [Google Scholar]
- 141.Olesen AB, Andersen G, Jeppesen DL, Benn CS, Juul S, Thestrup-Pedersen K. Thymus is enlarged in children with current atopic dermatitis. A cross-sectional study. Acta Derm Venereol 2005;85:240–3. [DOI] [PubMed] [Google Scholar]
- 142.Vestergaard C, Bang K, Gesser B, Yoneyama H, Matsushima K, Larsen CG. A Th2 chemokine, TARC, produced by keratinocytes may recruit CLA1CCR41 lymphocytes into lesional atopic dermatitis skin. J Invest Dermatol 2000;115: 640–6. [DOI] [PubMed] [Google Scholar]
- 143.Kaufman BP, Guttman-Yassky E, Alexis AF. Atopic dermatitis in diverse racial and ethnic groups—variations in epidemiology, genetics, clinical presentation and treatment. Exp Dermatol 2018;27:340–57. [DOI] [PubMed] [Google Scholar]
- 144.Noda S, Suarez-Farinas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol 2015;136:1254–64. [DOI] [PubMed] [Google Scholar]
- 145.Zheng X, Nakamura K, Furukawa H, Nishibu A, Takahashi M, Tojo M, et al. Demonstration of TARC and CCR4 mRNA expression and distribution using in situ RT-PCR in the lesional skin of atopic dermatitis. J Dermatol 2003;30:26–32. [DOI] [PubMed] [Google Scholar]
- 146.Patel N, Feldman SR. Adherence in atopic dermatitis. Adv Exp Med Biol 2017; 1027:139–59. [DOI] [PubMed] [Google Scholar]
- 147.Saeki H, Tamaki K. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J Dermatol Sci 2006;43:75–84. [DOI] [PubMed] [Google Scholar]
- 148.Murayama T, Nakamura K, Tsuchida T. Eosinophilic pustular folliculitis with extensive distribution: correlation of serum TARC levels and peripheral blood eosinophil numbers. Int J Dermatol 2015;54:1071–4. [DOI] [PubMed] [Google Scholar]
- 149.Motegi S, Hattori M, Shimizu A, Abe M, Ishikawa O. Elevated serum levels of TARC/CCL17, eotaxin-3/CCL26 and VEGF in a patient with Kimura’s disease and prurigo-like eruption. Acta Derm Venereol 2014;94:112–3. [DOI] [PubMed] [Google Scholar]
- 150.Cuccaro A, Annunziata S, Cupelli E, Martini M, Calcagni ML, Rufini V, et al. CD681 cell count, early evaluation with PET and plasma TARC levels predict response in Hodgkin lymphoma. Cancer Med 2016;5:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dallos T, Heiland GR, Strehl J, Karonitsch T, Gross WL, Moosig F, et al. CCL17/thymus and activation-related chemokine in Churg-Strauss syndrome. Arthritis Rheum 2010;62:3496–503. [DOI] [PubMed] [Google Scholar]
- 152.Chan TC, Sanyal RD, Pavel AB, Glickman J, Zheng X, Xu H, et al. Atopic dermatitis in Chinese patients shows TH2/TH17 skewing with psoriasiform features. J Allergy Clin Immunol 2018;142:1013–7. [DOI] [PubMed] [Google Scholar]
- 153.Gupta J, Grube E, Ericksen MB, Stevenson MD, Lucky AW, Sheth AP, et al. Intrinsically defective skin barrier function in children with atopic dermatitis correlates with disease severity. J Allergy Clin Immunol 2008;121:725–30.e2. [DOI] [PubMed] [Google Scholar]
- 154.Leung DYM, Calatroni A, Zaramela LS, LeBeau PK, Dyjack N, Brar K, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med 2019;11:eaav2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med 2011;365:1315–27. [DOI] [PubMed] [Google Scholar]
- 156.Tham EH, Leung DY. Mechanisms by which atopic dermatitis predisposes to food allergy and the atopic march. Allergy Asthma Immunol Res 2019;11:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with atopic dermatitis colonized with Staphylococcus aureus have a distinct phenotype and endotype. J Invest Dermatol 2018;138:2224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bissonnette R, Maari C, Forman S, Bhatia N, Lee M, Fowler J, et al. The oral Janus kinase/spleen tyrosine kinase inhibitor ASN002 demonstrates efficacy and improves associated systemic inflammation in patients with moderate-to-severe atopic dermatitis: results from a randomized double-blind placebo-controlled study. Br J Dermatol 2019;181:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dyjack N, Goleva E, Rios C, Kim BE, Bin L, Taylor P, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol 2018;141: 1298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hulshof L, Hack DP, Hasnoe QCJ, Dontje B, Jakasa I, Riethmuller C, et al. A minimally invasive tool to study immune response and skin barrier in children with atopic dermatitis. Br J Dermatol 2019;180:621–30. [DOI] [PubMed] [Google Scholar]
- 161.Engebretsen KA, Bandier J, Kezic S, Riethmuller C, Heegaard NHH, Carlsen BC, et al. Concentration of filaggrin monomers, its metabolites and corneocyte surface texture in individuals with a history of atopic dermatitis and controls. J Eur Acad Dermatol Venereol 2018;32:796–804. [DOI] [PubMed] [Google Scholar]
- 162.Clausen ML, Slotved HC, Krogfelt KA, Agner T. Measurements of AMPs in stratum corneum of atopic dermatitis and healthy skin-tape stripping technique. Sci Rep 2018;8:1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Berdyshev E, Goleva E, Bronova I, Dyjack N, Rios C, Jung J, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 2018;3: e98006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim BE, Goleva E, Kim PS, Norquest K, Bronchick C, Taylor P, et al. Side-byside comparison of skin biopsies and skin tape stripping highlights abnormal stratum corneum in atopic dermatitis. J Invest Dermatol 2019;139:2387–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Aronson JK, Ferner RE. Biomarkers—a general review. Curr Protoc Pharmacol 2017;76:9.23.1–9.23.17. [DOI] [PubMed] [Google Scholar]
- 166.Brunner PM, Emerson RO, Tipton C, Garcet S, Khattri S, Coats I, et al. Nonlesional atopic dermatitis skin shares similar T-cell clones with lesional tissues. Allergy 2017;72:2017–25. [DOI] [PubMed] [Google Scholar]
- 167.Czarnowicki T, Malajian D, Shemer A, Fuentes-Duculan J, Gonzalez J, Suarez-Farinas M, et al. Skin-homing and systemic T-cell subsets show higher activation in atopic dermatitis versus psoriasis. J Allergy Clin Immunol 2015; 136:208–11. [DOI] [PubMed] [Google Scholar]
- 168.Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol 2019; 143:1–11. [DOI] [PubMed] [Google Scholar]
- 169.Berin MC. The role of TARC in the pathogenesis of allergic asthma. Drug News Perspect 2002;15:10–6. [DOI] [PubMed] [Google Scholar]
- 170.Takeuchi H, Yamamoto Y, Kitano H, Enomoto T. Changes in thymus- and activation-regulated chemokine (TARC) associated with allergen immunotherapy in patients with perennial allergic rhinitis. J Investig Allergol Clin Immunol 2005; 15:172–6. [PubMed] [Google Scholar]


