Abstract
We showed previously that a human rhinovirus 14 (HRV14) 3′ untranslated region (3′ UTR) on a poliovirus genome was able to replicate with nearly wild-type kinetics (J. B. Rohll, D. H. Moon, D. J. Evans, and J. W. Almond, J. Virol 69:7835–7844, 1995). This enabled the HRV14 single 3′ UTR stem-loop structure to be studied in combination with a sensitive reporter system, poliovirus FLC/REP, in which the capsid coding region is replaced by an in-frame chloramphemicol acetyltransferase (CAT) gene. Using such a construct, we identified a mutant (designated mut4), in which the structure and stability of the stem were predicted to be maintained, that replicated very poorly as determined by its level of CAT activity. The effect of this mutant 3′ UTR on replication has been further investigated by transferring it onto the full-length cDNAs of both poliovirus type 3 (PV3) and HRV14. Virus was recovered with a parental plaque phenotype at a low frequency, indicating the acquisition of compensating changes, which sequence analysis revealed were, in both poliovirus- and rhinovirus-derived viruses, located in the active-site cleft of 3D polymerase and involved the substitution of Asn18 for Tyr. These results provide further evidence of a specific interaction between the 3′ UTR of picornaviruses and the viral polymerase and also indicate similar interactions of the 3′ UTR of rhinovirus with both poliovirus and rhinovirus polymerases.
The picornaviruses are small, nonenveloped RNA viruses, classified into six genera, of which the enteroviruses (including poliovirus and coxsackievirus) and the human rhinoviruses are members. These two genera, the most closely related of the Picornaviridae, are similar with respect to genomic organization, polyprotein structure, and processing pattern, and they produce viral proteins that function in essentially the same manner (24, 27). The single-stranded positive-sense RNA genome contains all of the signals required for translation of the viral polyprotein and replication of the genome within the cell cytoplasm. The polyprotein is cleaved posttranslationally into three regions; P1 produces the capsid proteins (VP1 to VP4), while P2 (2A to 2C) and P3 (3A to 3D) are the precursors of proteins involved in polyprotein maturation and RNA replication. The poliovirus RNA-dependent RNA polymerase, the 3D gene product, is responsible for the synthesis of negative strands from the input RNA, which are then copied to produce new positive-sense RNA. However, all of the P2 and P3 proteins, including several cleavage intermediates, have been implicated in some aspect of replication (for a review see reference 33).
The most 5′ proximal of the RNA structures, the poliovirus cloverleaf (CL; nucleotides [nt] 1 to 88), is the site of ribonucleoprotein complex formation required in the positive sense for replication (2). Complex formation involves the viral protease 3CD binding to the CL, either in conjunction with 3AB or following the binding of a cellular protein of 36 kDa which has been identified as the poly(rC) binding protein PCBP2 (1, 7, 17). The remainder of the unusually long 5′ untranslated region (UTR) forms an internal ribosome entry site involved in translation (28).
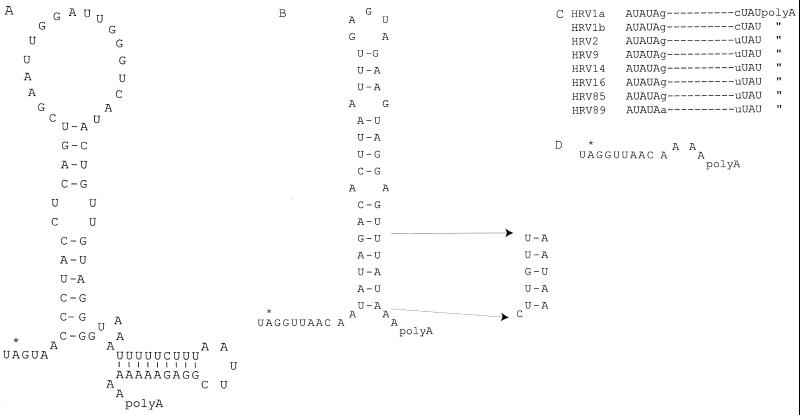
Whereas similar 5′ CLs are present in all entero- and rhinoviruses, the structures of the 3′ UTRs are diverse with regard to length and structure (20). Despite this variation, the 3′ structures formed fall into three main groups, the primary and secondary structures within each group being very highly conserved (23). Experimental evidence suggests that the 3′ UTR plays a key role in replication, as demonstrated by the inability to replace it with nonviral structures (26) or by the recovery of mutants that prevent tertiary interactions within this region and require reversions or compensating mutations to restore viability (13, 15, 21). However, this role in replication is difficult to correlate with the observed ability to interchange picornavirus 3′ UTRs with grossly different structures; e.g., the 3′ UTRs of coxackievirus B4 and human rhinovirus 14 (HRV14), with three and one stem-loops, respectively, can functionally replace the 3′ UTR of poliovirus type 3 (PV3), which has two stem-loops (26).
Evidence from cross-linking and electrophoretic mobility shift assays have shown that, in common with CL, the 3′ UTRs may be involved in ribonucleoprotein complex formation. The poliovirus proteins 3AB and 3CD have been shown to bind to this region, as have cellular proteins which are distinct from those which bind CL (7, 14, 29, 34). More recently, both polio- and rhinoviruses with partial or complete deletions of their 3′ UTRs have been recovered, indicating that these structures may not be absolute requirements for virus replication (31).
In a previous study, we showed that a chimeric poliovirus genome bearing an HRV14 3′ UTR replicated with nearly wild-type (wt) kinetics (26). This enabled the single stem-loop structure of HRV14, the most basic 3′ UTR, to be studied in the context of a subgenomic replicon (designated FLC/REP 19) that expresses a chloramphenicol acetyltransferase (CAT) reporter gene. A highly conserved base-paired motif at the base of the rhinovirus 3′ stem (23) was mutagenized, and one mutant (mut4), predicted to have a stem structure with a stability similar to that of the unmodified structure, showed levels of CAT expression only marginally above that of a replicon bearing a deletion within 3D polymerase, reflecting a significant defect in replication (26).
In this work, we have studied the effects of this replication-defective mutant on virus viability and replication efficiency. The HRV14 mut4 3′ UTR was transferred to full-length PV3 and HRV14 cDNAs, and virus was recovered from both constructs. We have investigated the effect that this mutation has on the infectivity of in vitro-transcribed RNA, the requirement for compensating mutations elsewhere in the genome, and the phenotype of recovered viruses. We suggest that although not absolutely required for replication, the function of the 3′ UTR involves an interaction with the active-site cleft of the viral polymerase.
MATERIALS AND METHODS
Construction of replicons and full-length cDNA clones.
pT7/FLC, an infectious clone of PV3 (19), has already been reported, as have derivatives in which the 3′ UTR was replaced by HRV14 sequences (26). The HRV14 mut4 3′ UTR was transferred from the corresponding poliovirus replicon (26) to pT7/FLC on a 1.2-kb XbaI-SalI fragment [nt 6265 to 3′ of the poly(A) tract] to generate pT7/FLC.mut4.
A T7 promoter was engineered preceding an infectious cDNA clone of HRV14 (27) in the vector pJM1, a pAT153 derivative in which the tetracycline gene was replaced with a kanamycin resistance cassette (14a), to generate pT7/HRV14. pT7/HRV14.mut4 was constructed in several stages due to the presence of duplicated restriction sites. The HRV14 3′ UTR was amplified from pT7/FLC.mut4 (primers 01-0280 and 06-0051; all primers are listed in Table 1), and the unique PstI-SalI fragment was cloned into pSL301 (Invitrogen). The adjacent HRV14-derived XbaI-HpaI fragment (nt 6525 to 7167) was introduced into the latter plasmid digested with SpeI and HpaI (pSL301 to nt 7167), and the mut4 3′ UTR was excised and rebuilt into pT7/HRV14, using a unique SphI site (nt 6632) and an AatII site present in both vectors after the poly(A) tail.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| 01-0280 | TACCATCTGCAGTTAATACGACTCACTATAGGGTAGGTTAACACTTGTTACACTTAAT | 3′ HRV14 mut4 plus-sense primer |
| 01-0285 | AACAAAAAAAAAAAAAAAAAAAAAAAAAGTACTTT | HRV14 3′ with deleted stem-loop |
| 01-0289 | AGGGGTTGGAATGGGTGTCC | PV3 plus-sense sequencing primer, binds 4180 |
| 01-0290 | GCCACTGCATAGTCAAACCC | PV3 minus-sense sequencing primer, binds 5435 |
| 01-0291 | TCATTCTTTGTGAGGACTGCTG | PV3 minus-sense sequencing primer, binds 6096 |
| 01-0292 | TTGGCACAACGGGGAAGAAG | PV3 plus-sense 3′ UTR sequencing primer |
| 01-0293 | GATCACGTCCGATCATTATGC | HRV14 plus-sense 3′ UTR sequencing primer |
| 01-0294 | CCAGACAAATCTGAAACTTTTAC | HRV14 plus-sense primer to amplify 3′ UTR |
| 01-0295 | AGGTTAACGTCGACTTTTTTTTTTTTTT | HRV14 minus-sense primer to amplify 3′ UTR |
| 01-0299 | CATTCCCCTTGTACTTAG | HRV14 minus-sense primer to sequence 3D/3C, binds 5960 |
| 01-0300 | TGATCTGAACCAAAATCC | HRV14 plus-sense primer to amplify EcoRV fragment |
| 01-0301 | TATTTTCAGTGGGTTCCG | HRV14 minus-sense primer to amplify EcoRV fragment |
| 01-0302 | TTCTGTGTCCTGGGTCTC | HRV14 minus-sense primer, EcoRV fragment from 6181 |
| 06-0051 | TTCGCGAGGTTAACGTCGACTTTTTTTTTTTTTT | 3′ UTR minus-sense primer, includes SalI site |
| 34-0042 | ATGAGACCATCAAAGGAGGC | PV3 plus-sense primer to amplify 3D region |
| 34-0086 | TGAGATCGGTCGACGGAGTGTGCCAATCGG | PV3 plus-sense primer to amplify 3′ UTR |
| 43-0006 | TCCAAATGCCATTCTCATGGCC | PV3 minus-sense primer to amplify HindIII fragment |
| 50-0005 | ATATATGCTAGCCATATGGGTGATAGTTGGTTGAAAAAATTTAC | PV3 plus-sense primer to amplify HindIII fragment |
| 3Dseq | TACTTCACTCAGAGCCAA | PV3 plus-sense sequencing primer, binds 5960–5977 |
The construction of a 3′ UTR deletion of HRV14 was made in an AseI-ClaI (nt 6568 to vector) subclone in a similarly cut preparation of pAT153. Oligonucleotide 285 (Table 1) was self-annealed, and the ends were filled in with Klenow prior to ScaI digestion. The resulting product was built into HpaI-ScaI (nt 7167 to vector)-digested and phosphatase-treated pAT153-derived subclone, and the modified 3′ region was returned to pT7/HRV14 on unique SphI-ClaI (nt 6632 to vector) sites to generate pT7/HRV14.Δ3′.
Reverse transcription-PCR-amplified fragments from recovered viruses were rebuilt into cDNA (pT7/FLC.mut4 or pT7/HRV14.mut4) to map the locations of compensating mutations. Viral RNA (vRNA) was extracted from plaque-purified viruses as described previously (25) and reverse transcribed by using primer 06-0051. Primer pairs 34-0042–06-0051 and 50-0005–43-0006 were used to amplify poliovirus nt 5993 to 3′ of the poly(A) tract and nt 4115 to 6553, respectively, which were rebuilt into pT7/FLC.mut4 on XbaI/SalI [nt 6265 to 3′ of the poly(A) tract] or HindIII [nt 4241 to 6507] fragments, to create pT7/FLC.X/S and pT7/FLC.mut4.H. The HindIII fragment was further dissected by using the NarI site at nt 5815, enabling the NarI-XhoI (nt 5815 to 6050) and XmaI-NarI (nt 2766 to 5815) fragments to be cloned independently and reciprocally from the parental and FLC.mut4-derived clones into pSL310. The resulting individual mutations were rebuilt into pT7/FLC.mut4 on the unique XmaI-XhoI fragment (nt 2766 to 6050) to yield plasmids pT7/FLC.mut4.3C and pT7/FLC.mut4.3D, respectively. The 3D mutation was subsequently transferred into pT7/FLC.3′HRV14 to yield plasmid pT7/FLC.3′HRV14.3D.
The 3CD region of HRV14 was amplified by using primers 01-300 and 01-0301 and cloned on an EcoRV fragment (nt 5490 to 6048) into the pT7/HRV14.mut4 cDNA, to generate pT7HRV14.mut4.E.
In vitro transcriptions and transfections.
Poliovirus- and rhinovirus-derived cDNAs were linearized with SalI and ClaI, respectively, prior to T7-mediated in vitro transcription as described previously (32). Samples of the transcription reactions were fractionated by agarose gel electrophoresis to assess yields, and similar quantities were serially diluted 10-fold prior to transfection (5), with the modification that the transfection buffer was Hanks balanced salt solution (10× stock contains 50 g of HEPES, 80 g of NaCl, 3.7 g of KCl, and 1.25 g of Na2HPO4 · 2H2O/liter, pH 7.05), plus final concentrations of 1 g of glucose and 0.5 g of dextran/liter.
Sequencing of recovered virus.
All DNA sequencing was performed on a Pharmacia ALF Express, using suitable Cy5-labelled primers. Direct RNA sequencing was carried out as described previously (5). Poliovirus- or rhinovirus-derived RNA was reverse transcribed with primer 06-0051 prior to PCR amplification of the required region. Primers 06-0051 and 34-0086 were used to amplify the 3′ UTR of FLC.mut4, which was sequenced with primer 01-0292. Primers 01-0289, 01-0290, 01-0291, and 3Dseq were used to sequence the amplified and cloned HindIII fragment (nt 4241 to 6507), and the identified mutations were confirmed by RNA sequencing with primer 01-0291.
Primer pairs 01-0294 and 01-0295 were used to amplify the 3′ UTR of HRV14.mut4, and the sequence determined with primer 01-0293. The RNA encoding the amino terminus of HRV14 3D was sequenced by using primer 01-0299, and the HRV14-derived EcoRV fragment containing the mutation (nt 5495 to 6052) was amplified with primers 01-0300 and 01-0301, sequenced with 01-0302, and rebuilt by using standard protocols.
Growth characteristics.
The growth of plaque-purified, recovered viruses was assessed in a one-step growth curve as described previously (26). Virus yields were quantified by plaque assay for the poliovirus- and HRV14-based mutant viruses and by 50% tissue culture infective dose for the HRV14 3′-deleted viruses (5).
RESULTS
Recovery of poliovirus with an HRV14 mut4 3′ UTR.
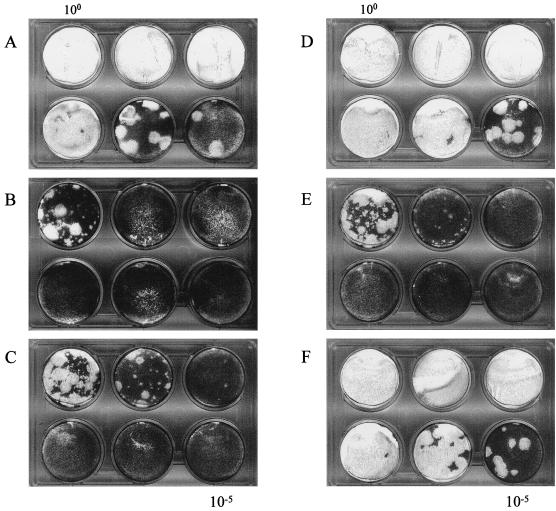
We have used a poliovirus subgenomic replicon (designated FLC/REP), in which a CAT reporter gene is fused in frame with VP2 of PV3 (19), to determine the effect of 3′ UTR substitutions and mutations on genome replication (26). Replacement of the poliovirus 3′ UTR of FLC/REP with the HRV14 3′ UTR (to generate FLC/REP.3′HRV14) produced a chimeric replicon that exhibited almost wt levels of replication (26), despite the complete lack of homology between these 3′ UTRs (Fig. 1A and B). A derivative of FLC/REP.3′HRV14, designated FLC/REP.mut4, bearing a modification of the conserved base of the stem (Fig. 1B) exhibited only background levels of CAT activity, despite having a structure and predicted stability close to that of the native HRV14 3′ UTR. To assess its effect on virus viability, the mut4 3′ UTR was transferred from the replicon to a full-length PV3 cDNA (pT7FLC) to generate pT7FLC.mut4. Subconfluent Ohio HeLa cells were transfected with RNA transcribed in vitro, and plaques were observed at a frequency ∼3 log10 below that for the control pT7FLC.3′HRV14 RNA (compare Fig. 2A and B). This reduction in specific infectivity (routinely measured in parallel experiments at ∼106 PFU/μg for pT7FLC-derived viruses) suggests that viability requires the acquisition of a mutation by reversion either at a key sequence within the mut4 3′ UTR or elsewhere in the genome. Although the majority of plaques obtained were very small, some exhibited a phenotype similar to that of FLC.3′HRV14 (visible in the 100 dilution in Fig. 2B).
FIG. 1.
Structures and sequences of 3′ UTRs. (A) Secondary structure of the PV3 3′ UTR. (B) Structure of the HRV14 3′ UTR showing the sequence change at the base of the stem to generate mut4. (C) Conserved sequence at the base of the 3′ stem of human rhinoviruses. (D) The 3′ UTR sequence remaining after removal of the stem-loop to generate HRV14.Δ3′.
FIG. 2.
In vitro-transcribed RNA from poliovirus-based cDNAs serially diluted (100 to 10−5), transfected into Ohio HeLa cells, and overlaid with semisolid agar. pT7FLC.3′HRV14 (A) has a wt HRV14 3′UTR, and pT7FLC.mut4 (B) has a stem mutant 3′ UTR. Other constructs, based on pT7FLC.mut4, include reverse-transcribed and PCR-amplified fragments from recovered virus. pT7FLC.mut4.X/S (C) and pFLC.mut4.H (D) contain the XbaI/SalI and HindIII fragments, respectively. pT7FLC.mut4.3C (E) and pT7FLC.mut4.3D (F) contain the identified 3C and 3D mutations respectively.
Identification of compensating mutations within the poliovirus genome.
To determine whether the large-plaque variant of FLC.mut4 retained the original mut4 3′ UTR mutation, two independent plaques were picked and subjected to two rounds of plaque purification, and a 180-nt fragment spanning the entire 3′ UTR was PCR amplified and sequenced. The sequences obtained from both plaques were identical to that of the pT7FLC.mut4 cDNA, indicating that the compensating mutation(s) must be located elsewhere in the genome.
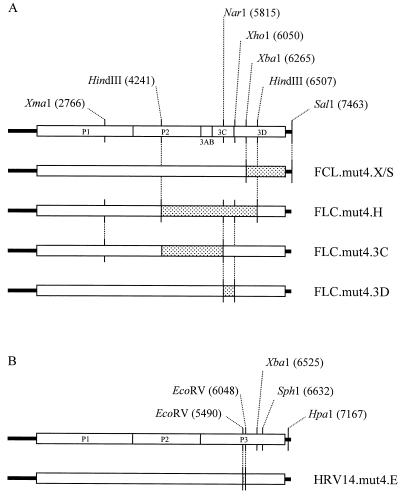
The P3 region of the genome was thought to be the most likely location for compensating mutations. It has been reported that the 3′ UTR of poliovirus binds P3-encoded proteins in cross-linking assays (7), and there are results to support the formation of a pseudoknot between the 3′ UTR of enteroviruses and the coding region (9), although no evidence for a similar tertiary structure was found for HRV14 (30). vRNA from one of the purified viruses was isolated, and two overlapping fragments covering the P3 region were amplified by PCR. Both the XbaI-SalI [nt 6265 to 3′ of the poly(A) tract] and HindIII (nt 4241 to 6507) fragments were independently rebuilt into pFLC.mut4 to generate pT7FLC.mut4.X/S and pT7FLC.mut4.H, respectively (Fig. 3A). Whereas the infectivity of the RNA transcribed in vitro from pT7FLC.mut4.X/S was identical to the original pT7FLC.mut4, that bearing the HindIII fragment, pT7FLC.mut4.H, was indistinguishable by infectivity and plaque morphology from pT7FLC.3′HRV14 (Fig. 2A, C, and D). Any compensating mutations required to restore wt levels of replication must therefore be located within this 2.2-kb HindIII fragment. Direct sequencing of the entire 2.2-kb HindIII fragment in pT7FLC.mut4H resulted in the identification of only two nucleotide substitutions: a noncoding change of thymidine 5779 to cytidine (T5779C) within the region encoding 3C, and A6029T, which introduces a coding change of asparagine to tyrosine at amino acid 18 of 3D (Fig. 4A). Although 3D, as part of 3CD, binds to the poliovirus 3′ UTR, the reported pseudoknot between the 3′ UTR and coding region (9) and recent reports of cis-acting replication signals within picornavirus genomes (5a, 11, 12) meant that the T5779C noncoding change in 3C could not be discounted from contributing to the phenotype of the revertant virus. The two changes were introduced independently into pT7FLC.mut4 to generate plasmids pT7FLC.mut4.3C and pT7FLC.mut4.3D, respectively. In vitro-transcribed RNA bearing the 3C noncoding change alone recovered as the original pT7FLC.mut4, whereas pT7FLC.mut4.3D was indistinguishable from FLC.mut4H and FLC.3′HRV14 (Fig. 2E and F). Direct RNA sequencing confirmed that both original purified plaques harbored the 3D mutation of A6029T but that only one had the 3C mutation. Therefore, the single coding change in poliovirus 3D polymerase is sufficient to compensate for the HRV14 3′ UTR mut4 stem mutation present in FLC.mut4.
FIG. 3.
Construction of PV3- and HRV14-based clones. Shaded areas are reverse-transcribed and PCR-amplified fragments from recovered mut4 virus. (A) All constructs are based on PV3 (Leon) with an HRV14 mut4 3′ UTR (FLC.mut4); the restriction endonuclease sites used are indicated, and the constructs generated are shown. (B) HRV14 cDNAs were constructed by using the restriction sites shown.
FIG. 4.
(A) Alignment of amino acids 8 to 36 of the PV3 and HRV14 polymerases. The observed nucleotide and amino acid changes are shown for both. (B to D) Tenfold serial dilutions (100 to 10−5) of RNA generated in vitro from pT7HRV14 (B), pT7HRV14.mut4 (C), and pT7HRV14.mut4.E (D) with substitution of the EcoRV fragment from recovered virus, transfected into Ohio HeLa cells, and overlaid directly with agar.
Recovery of the mut4 3′ UTR on the homologous human rhinovirus.
Alignment of PV3 and HRV14 3D sequences indicates that residue 18, which is conserved as an asparagine in the enteroviruses, hepatoviruses, and HRV14, marks the amino terminus of a region of significant homology between the two proteins (Fig. 4A) and so may represent a functionally conserved domain. We were interested to determine the phenotype of rhinovirus bearing the mut4 mutation in the 3′ UTR and, if it was less fit than the parental virus, to identify any compensating mutations that are required to restore wt growth characteristics. The mut4 3′ UTR was engineered in place of the native sequence in a full-length HRV14 cDNA to generate pT7HRV14.mut4.
Transfection of in vitro-produced pT7HRV14.mut4 RNA generated small, indistinct plaques after 4 days (Fig. 4C), and infectivity was reduced ∼10-fold compared to RNA generated from pT7HRV14 (compare Fig. 4B and C). Therefore, although pT7HRV14.mut4 RNA was infectious, virus replication was significantly impaired compared to that of wt HRV14. Our pT7HRV14 cDNA is routinely used to generate RNA with a specific infectivity of ∼104 PFU/μg, but we were unable to identify revertants with a large-plaque phenotype by direct transfection. Therefore, recovery of the in vitro-transcribed RNA on cells with a liquid overlay was attempted. Virus in the supernatant, transferred to fresh cells with an agar overlay, generated a mixture of large and small plaques (data not shown), suggesting that one or more compensating mutations are required for efficient replication of HRV14 with the mut4 3′ UTR.
Identification of a compensating mutation within the HRV14 genome.
A recovered virus with a large-plaque phenotype was subjected to two rounds of plaque purification, the vRNA was isolated, and a 380-nt fragment spanning the 3′ UTR was amplified by PCR, sequenced directly, and confirmed to be identical to the cDNA from which the virus was recovered. A further fragment of 1.5 kb spanning the amino terminus of 3D was amplified and sequenced; it contained a single substitution of A to T at nucleotide 5833, within the codon for residue 18 of HRV14 3D. To confirm that this mutation confers the wt phenotype and to exclude the possibility that the recovered virus contained additional compensating mutations, a fragment was amplified from the reverse-transcribed material to include the EcoRV sites at nt 5490 and 6048. This 558-nt fragment was reintroduced into the original pT7HRV14.mut4 cDNA (Fig. 3). In vitro-transcribed RNA was transfected into Ohio HeLa cells and shown to be as infectious as T7-generated HRV14 RNA and to produce plaques of comparable phenotype (Fig. 4D). The DNA sequence of the 558-bp EcoRV fragment in pHRV14.mut4.E was determined and found to contain only the single substitution of A5833T.
The effect of 3D Tyr18 on FLC.3′HRV14 replication.
3D polymerase N18Y has been identified as the only mutation required to compensate for the mut4 mutation on either a poliovirus or HRV14 genome. We were interested to investigate the effect of 3D tyrosine 18 on the phenotype of poliovirus in the absence of the modified mut4 3′ UTR. Recovery of virus from this construct (pT7FLC.3′HRV14.3D) showed that the specific infectivity of in vitro-transcribed RNA was similar to that of pT7FLC.3′HRV14 but fractionally larger plaques were produced (data not shown). To characterize this virus further, we compared it to FLC.3′HRV14 and FLC.mut4.3D in a single-step growth curve. It grew to a titer about 0.5 log10 higher than titers of the other two viruses (Fig. 5). Therefore, the 3D polymerase mutation that is absolutely required to compensate for the defective HRV14 mut4 3′ UTR on a chimeric poliovirus is capable of replicating the FLC.3′HRV14.3D genome at or above the rate of FLC.3′HRV14.
FIG. 5.
Plaque-purified PV3-HRV14 chimeric viruses were used to infect Ohio HeLa cells at a multiplicity of infection of 10 for a one-step growth curve to compare N18Y viruses. Mut4 has the 3′ stem mutant, and the 3D N18Y mutation is present where indicated. ■, FLC.3′HRV14; ▴, FLC.3′HRV14.3D; ⧫, FLC.mut4.3D.
If N18Y acted by nonspecifically increasing the activity of the polymerase, we would expect it to be equally effective in compensating for other debilitating mutations of the 3′ UTR. We have generated a 3′ UTR deletion of HRV14 essentially similar to that recently published by Todd et al. (31) which retains only six nucleotides between the polyprotein termination codon and the poly(A) tract (Fig. 1D). Transfection of in vitro-transcribed RNA from pT7/HRV14.Δ3′ generated a viable virus only following passage of the transfection supernatant onto a fresh cell monolayer. Further serial passage to optimize the replication potential was undertaken, and the growth kinetics at passage 5 and 13 were analyzed by single-step growth curve. In agreement with the results of Todd et al., viruses purified by limit dilution grew to a final titer ∼1 log10 less than that of the parental HRV14 (Fig. 6). The extended period required to observe replicating virus following transfection is indicative of the accumulation of several compensating mutations. Full analysis of these viruses will be published elsewhere; however, sequencing of the 3′ UTR and the region encoding the amino terminus of 3D confirmed that there were no modifications to the UTR and that residue 18 of 3D remained an asparagine.
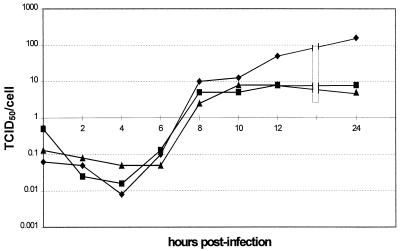
FIG. 6.
One-step growth curve to compare wt HRV14 with HRV14.Δ3′ recovered virus following 5 or 13 passages in tissue culture. ■, HRV14.Δ3′, passage 13; ▴, HRV14.Δ3′, passage 5; ⧫, HRV14. TCID50, 50% tissue culture infective dose.
DISCUSSION
The poliovirus, rhinovirus, and coxsackievirus 3′ UTRs are distinctly different in sequence and structure but remain functionally interchangeable, in both chimeric poliovirus subgenomic replicons and infectious cDNAs (26). The high level of 3′ UTR sequence conservation within each of these three groups and variation between them suggests a conserved and specific role for this region of the genome. However, although it is clear that nonviral synthetic stem-loop structures cannot replace the 3′ UTR without destroying viability (26), the recent demonstration that the 3′ UTRs of poliovirus and rhinovirus can be removed entirely (this study and reference 31) complicates the understanding of the function of this region.
We have extended our analysis of the 3′ UTR by studying a chimeric poliovirus bearing a mutagenized rhinovirus 3′ UTR. The mutation, described in an earlier report (26), was designed to disrupt a highly conserved base paired motif at the base of the single stem-loop within the HRV14 3′ UTR (Fig. 1B and c). As a subgenomic replicon, this resulted in background levels of replication. However, when the mutation was engineered onto a full-length poliovirus cDNA and transfected, plaques with wild-type phenotype arose at a frequency consistent with the acquisition of a single compensating mutation (Fig. 2). Characterization of two independent virus isolates demonstrated the presence of an A-to-U substitution at nt 6029. The importance of the A6029U substitution in compensating for the mut4 modification of the HRV14 3′ UTR was confirmed by rebuilding this mutation alone into the parental pT7/FLC.mut4 and demonstrating the acquisition of a plaque phenotype indistinguishable from that of pT7/FLC.3′HRV14 (Fig. 2). The A6031U substitution introduces a tyrosine in place of asparagine at position 18 of the 3D polymerase. It is interesting to note that 3D polymerase residue 18 is an asparagine in the enteroviruses, hepatitis A virus, and HRV14, which include all of the 3′ UTRs that we have previously demonstrated confer viability to a poliovirus replicon, but is a histidine in all other picornaviruses (including other rhinoviruses). Whether this residue plays a key role in the replication of these chimeric replicons or simply reflects the evolutionary relationships between the picornaviruses remains to be determined. Although not characterized further, we would speculate that the relatively abundant small plaque phenotype viruses also observed during the recovery of the poliovirus N18Y mutants represent the acquisition of partially compensating transitional mutations; these are known to occur far more frequently than transversions in poliovirus (3, 10) but presumably could not fully restore efficient replication to the virus.
To confirm that the N18Y substitution in poliovirus 3D polymerase is not selected as a consequence of the chimeric nature of the FLC.mut4 genome, we investigated the phenotype of HRV14 bearing an identical mut4 3′ UTR. The recovery of an identical N18Y substitution within the rhinovirus 3D polymerase, arising from a single A5833U mutation, emphasizes both the similarity of the two viruses and the importance of the amino acid substitution in 3D polymerase in restoring virus replication. This finding also strongly suggests that it is the amino acid substitution, rather than the underlying alteration in nucleotide sequence, that is responsible for the change in phenotype. Alignment of the nucleotide sequences encoding the first 25 amino acids of the poliovirus and rhinovirus 3D polymerase shows only 36% identity, with no evidence for any conserved higher-order structures. This conclusion is in agreement with the structural analysis of the 3′ UTR of rhinovirus by Todd and Semler (30), who could find no evidence for interactions of the coding and noncoding regions.
Assuming that it is the N18Y substitution that is critical for replication competence, the mechanism by which this is achieved remains to be determined. One possibility is that substitution directly effects the interaction of polymerase and substrate and that this specific mutation is required to accommodate the 3′ UTR bearing the mut4 modification. The location of residue 18 in the poliovirus 3D polymerase structure certainly does not preclude this possibility. Although the N terminus of 3D polymerase was largely disordered in the crystal structure, the location of residues 12 to 37 can be visualized as forming the N-terminal strand of the thumb subdomain (6) occupying one side of the active-site cleft. Although the R factor is insufficient to be confident about side chain orientation, Asn18 is located toward the base of the cleft, exposed to the palm domain, which contains the four consensus motifs conserved in all classes of polymerase (16). However, although Asn18 is clearly located within the active-site cleft, it is not clear how it might contribute to polymerization activity.
The limited number of mutations generated in the N terminus of poliovirus 3D polymerase (for a review, see reference 16) throw little light on the function of this region of the protein. Deletion of residues 1 to 6, or of Trp5 alone (6, 22), inactivates the enzyme, and charged-to-alanine substitutions at Lys36 and Glu29, which are located at the top of the thumb domain in a region of ordered structure, prevent the recovery of viable virus (4). One intriguing possibility, suggested by the crystal structure, is that the N terminus of 3D polymerase is involved in oligomerization. Residues 12 to 37 are situated on the opposite side of the polymerase from the next ordered segment, residues 67 to 97. It is unlikely that the intervening disordered residues (38 to 66) span almost 45 Å directly across the active site of the polymerase, and it is therefore probable that residues 12 to 37 visible in a polymerase monomer are actually derived from an adjacent molecule (6). Polymerase-polymerase interactions have been demonstrated by independent techniques (8, 18), and in vitro analysis suggests cooperativity in RNA binding and polymerization activity (18). This experimental evidence is supported by analysis of the packing of the polymerase molecules in crystals, which suggests that two significant interfaces are present (6). One of these (interface II, using the nomenclature of Hansen et al. [6]) involves a trans interaction of two polymerase proteins, in which residues 12 to 37 and 67 to 97 make contact. This suggests that the N18Y substitution may mediate its effect, either directly by a contribution to RNA interactions within the active site cleft or indirectly by affecting the interaction and oligomerization of polymerase monomers into higher-order structures.
One possibility, addressed in this study, is that the N18Y mutation acts by increasing polymerase activity and so compensates for a rate-limiting step introduced by modification of the 3′ UTR. This interpretation is supported by the growth characteristics of FLC/3′HRV14.3D (Fig. 5). In our previous study, we demonstrated that FLC/3′HRV14 accumulates to a final titer approximately 0.5 log10 lower than that of unmodified poliovirus FLC (26). The introduction of the mut4 mutation and acquisition of the N18Y substitution (to yield FLC.mut4.3D) restores replication to levels comparable to those for FLC.3′HRV14 (Fig. 5). In contrast, introduction of the N18Y substitution to FLC.3′HRV14 (generating FLC/3′HRV14.3D) produces a virus that replicates slightly better than either FLC.3′HRV14 or FLC.mut4.3D, at levels comparable to those for FLC (Fig. 5). However, if this were the case, one would expect there to be a selective pressure to acquire an N18Y substitution in other instances of debilitating modifications to the 3′ UTR. A severe test of this hypothesis would be the recovery of a virus in which the entire 3′ UTR was deleted. The recovery and preliminary analysis of HRV14.Δ3′ suggest that the N18Y mutation is not required to restore viability to this virus. This, in turn, may suggest that the N18Y mutation is specifically required to compensate for a defective 3′ UTR, rather than the total absence of such a sequence, possibly strengthening the case that this region of the polymerase is involved in direct RNA interactions.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 3.Delatorre J C, Giachetti C, Semler B L, Holland J J. High-frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc Natl Acad Sci USA. 1992;89:2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond S E, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans D J, Almond J W. Design, construction and characterization of poliovirus antigen chimeras. Methods Enzymol. 1991;203:387–400. doi: 10.1016/0076-6879(91)03022-9. [DOI] [PubMed] [Google Scholar]
- 5a.Goodfellow, I. G. Y. Chaudhry, A. Richardson, J. M. Meredith, J. W. Almond, W. S. Barclay, and D. J. Evans. Identification of a cis-acting replication element (CRE) within the poliovirus coding region. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 6.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 7.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 8.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuge S, Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J Virol. 1987;61:1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melchers W J G, Hoenderop J G J, Slot H J B, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellits K H, Meredith J M, Rohll J B, Evans D J, Almond J W. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J Gen Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 14a.Meredith, J. M., and D. J. Evans. Unpublished data.
- 15.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 17.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 18.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 19.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilipenko E V, Maslova S V, Sinyakov A N, Agol V I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs—a proposal for the existence of tert-RNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilipenko E V, Poperechny K V, Maslova S V, Melchers W J G, Bruins Slot H J, Agol V I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 22.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poyry T, Kinnunen L, Hyypia T, Brown B, Horsnell C, Hovi T, Stanway G. Genetic and phylogenetic clustering of enteroviruses. J Gen Virol. 1996;77:1699–1717. doi: 10.1099/0022-1317-77-8-1699. [DOI] [PubMed] [Google Scholar]
- 24.Racaniello V R, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rico Hesse R, Pallansch M A, Nottay B K, Kew O M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987;160:311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- 26.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanway G, Hughes P J, Mountford R C, Minor P D, Almond J W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart S R, Semler B L. RNA determinants of picornavirus cap-independent translation initiation. Semin Virol. 1997;8:242–255. [Google Scholar]
- 29.Todd S, Nguyen J H C, Semler B L. RNA-protein interactions directed by the 3′ end of human rhinovirus genomic RNA. J Virol. 1995;69:3605–3614. doi: 10.1128/jvi.69.6.3605-3614.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd S, Semler B L. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′-noncoding region. Nucleic Acids Res. 1996;24:2133–2142. doi: 10.1093/nar/24.11.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimmer E, Hellen C U T, Cao X M. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 34.Xiang W, Paul A V, Wimmer E. RNA signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]