Abstract
The nonstructural protein NS5A of hepatitis c virus (HCV) has been demonstrated to be a phosphoprotein with an apparent molecular mass of 56 kDa. In the presence of other viral proteins, p56 is converted into a slower-migrating form of NS5A (p58) by additional phosphorylation events. In this report, we show that the presence of NS3, NS4A, and NS4B together with NS5A is necessary and sufficient for the generation of the hyperphosphorylated form of NS5A (p58) and that all proteins must be encoded on the same polyprotein (in cis). Kinetic studies of NS5A synthesis and pulse-chase experiments demonstrate that fully processed NS5A is the substrate for the formation of p58 and that p56 is converted to p58. To investigate the role of NS3 in NS5A hyperphosphorylation, point and deletion mutations were introduced into NS3 in the context of a polyprotein containing the proteins from NS3 to NS5A. Mutation of the catalytic serine residue into alanine abolished protease activity of NS3 and resulted in total inhibition of NS5A hyperphosphorylation, even if polyprotein processing was allowed by addition of NS3 and NS4A in trans. The same result was obtained by deletion of the first 10 or 28 N-terminal amino acids of NS3, which are known to be important for the formation of a stable complex between NS3 and its cofactor NS4A. These data suggest that the formation of p58 is closely connected to HCV polyprotein processing events. Additional data obtained with NS3 containing the 34 C-terminal residues of NS2 provide evidence that in addition to NS3 protease activity the authentic N-terminal sequence is required for NS5A hyperphosphorylation.
Hepatitis C virus (HCV) is the major agent of non-A, non-B hepatitis, with more than 100 million infected individuals worldwide (58). It belongs to the Flaviviridae family and contains a single-stranded, positive-sense RNA genome that is translated into a long viral polyprotein (8, 30, 53). Proteolytic processing between C-E1, E1-E2, E2-p7 and p7-NS2 is performed by host cell proteases (23, 49, 52), whereas viral proteases cleave between the nonstructural proteins NS2-NS3, NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B. The NS2-NS3 junction is cleaved by a zinc-dependent autoproteinase associated with NS2 and the N-terminal region of the NS3 protein (21, 25, 41, 50, 57). All cleavage sites downstream of NS3 are processed by the NS3 protease either in cis between NS3 and NS4A or in trans at the cleavage sites NS4A/4B, NS4B/NS5A, and NS5A/5B (4, 11, 22, 24, 33, 56). NS4A has been demonstrated to function as a cofactor for the NS3 serine protease and is strictly required for the cleavage of the NS3/4A and NS4B/NS5A sites (5, 15, 36, 54). The interaction between NS3 and its cofactor NS4A is mediated by the extreme N-terminal amino acids of NS3 (16, 32, 51).
Transient expression assays of NS5A from genotypes 1a, 1b, and 2a in mammalian cells in the presence of [32P]orthophosphate and phosphoamino acid analysis indicate that NS5A is a phosphoprotein which is preferentially modified at serine residues (28, 45). Phosphorylation has been observed to occur in the absence of other viral proteins, suggesting either that NS5A acts as an autokinase or that it is phosphorylated by one or more cellular kinases. In support of the latter hypothesis, coimmunoprecipitation and affinity purification experiments have shown a tight association of cellular kinase(s) with NS5A (26, 45). The effect of different protein kinase inhibitors on NS5A phosphorylation classified the kinase as a member of the CMGC group of serine/threonine kinases (casein kinase II and proline-directed kinases such as the mitogen-activated protein kinase, cyclin-dependent kinases and glycogen synthase kinase 3). In addition, Escherichia coli-expressed glutathione S-transferase–NS5A could be phosphorylated in vitro by a purified preparation of cyclic AMP-dependent protein kinase A-α catalytic subunit (26). Phosphorylation of NS5A by stably associated kinases is not restricted to HCV but has been observed also in bovine viral diarrhea virus and yellow fever virus, members of the other two established genera in the family Flaviviridae (46).
Mutations within a discrete region of NS5A, termed the interferon sensitivity-determining region, have been associated with the resistance to interferon treatment (7, 12, 13, 35, 47). This phenotype might be due to the inhibition of PKR, a protein kinase involved in the cellular antiviral and antiproliferative response induced by interferon, by direct interaction with NS5A (18, 19). A possible correlation between NS5A phosphorylation and interferon resistance has not yet been established.
In addition to the phosphorylated form of NS5A (p56), a slower-migrating form of NS5A (p58) has been described (28). It is generally accepted that p58 is a hyperphosphorylated form of NS5A p56; however, the requirements necessary for its formation are controversial. Earlier results suggested that the formation of p58 was enhanced in the presence of NS4A, and mutagenesis experiments identified three serine residues (S-2197, S-2201, and S-2204) in the central region of NS5A as phosphorylation sites responsible for the formation of p58 (55). However, none of the sites has directly been identified by amino acid analysis. The NS5A region from amino acids 2135 to 2139 was found to be important for NS4A-dependent phosphorylation (2). A recent report indicated that NS5A, which was expressed in the context of a polyprotein expressing proteins from NS3 to NS5B, could be hyperphosphorylated when functional NS2 generated from a NS2-NS3 precursor protein was supplied in trans (43).
To get more insight into the mechanisms that lead to the formation of p58, we introduced several deletion and point mutations into the polyprotein of HCV and analyzed electrophoretic migration and phosphorylation patterns of NS5A expressed from these constructs. Our results indicate that the polyprotein expressing the proteins from NS3 to NS5A was necessary and sufficient for the formation of p58. Time course labelling and pulse-chase experiments demonstrated that NS5A released from the polyprotein migrated as p56, which then served as a substrate for the formation of the hyperphosphorylated form p58. NS4A and NS4B are required, but they cannot be supplied in trans with respect to NS5A. In addition, we observed that the formation of p58 strictly depends on the presence of an active NS3 serine protease with its authentic N terminus. This latter observation implies a correlation between NS3-dependent polyprotein processing and the formation of NS5A p58.
MATERIALS AND METHODS
Tissue culture.
Hep3B cells were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. Vaccinia virus vTF7-3 (17) was amplified in RK13 cells cultured in minimum essential medium containing 10% fetal calf serum.
Construction of recombinant plasmids.
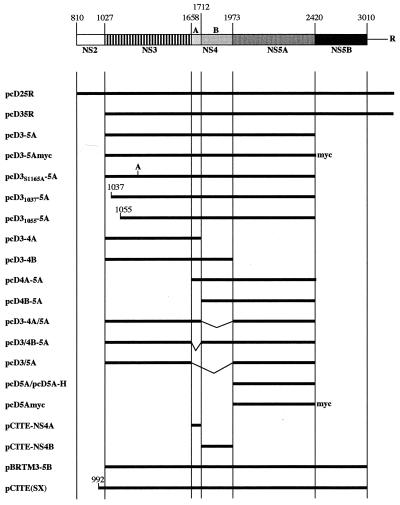
Unless otherwise indicated, all HCV-coding regions were derived from cDNA clones of the HCV BK strain and introduced into the polylinker of the eukaryotic expression vector pcDNA3 (Invitrogen). Appropriate synthetic oligonucleotides and PCR were used to introduce artificial initiation and termination codons. All DNA fragments obtained by PCR were verified by DNA sequence analysis. Standard recombinant DNA technology (48) was used for construction of the plasmids described below (numbers in parentheses indicate first and last amino acids of the expressed proteins). pcD25R (810-3010) and pcD35R (1027-3010) contain, in addition to the indicated coding region, the entire 3′ untranslated region (UTR) of HCV and the hepatitis δ antigenomic ribozyme (27) at its extreme 3′ end. pcD3-5A (1027-2419), pcD5A (1973-2419), pcD3-4A (1027-1711), pcD3-4B (1027-1972), pcD4A-5A (1658-2419), and pcD4B-5A (1712-2419) were derived from pcD25R but do not contain the HCV 3′ UTR, like all following constructs. pcD5Amyc and pcD3-5Amyc are identical to pcD5A and pcD3-5A, respectively, except that a Myc epitope (-FEEQKLISEEDL-stop [14]) was added in frame to the C terminus of NS5A. The N-terminal deletion mutants pcD31037-5A and pcD31055-5A as well as the point mutant pcD3S1165A-5A were obtained by PCR using appropriate synthetic oligonucleotides. pcD3/5A contains an internal deletion of NS4A-4B (1658-1972) and results in an in-frame fusion of NS3 with NS5A. The last amino acid of NS3 was mutated from threonine to cysteine (T1657C). Deletion mutants pcD3-4A/5A and pcD3/4B-5A contain internal deletions of NS4B (1712-1972) and NS4A (1658-1711), respectively. The construct pcD3/4B-5A contains the T1657C mutation in NS3. NS4A (1658-1711) and NS4B (1712-1972) were cloned into the expression vector pCITE-1 (Novagen). The construction of the plasmid pCITE(SX) has been described earlier (56). The expression vector pBRTM/HCVNS3-5B (1027-3010) contains the cDNA of the HCV-H strain and was a kind gift from C. M. Rice. pcD5A-H (1973-2419) contains the coding regions derived from pBRTM/HCVNS3-5B cloned into the pcDNA3 vector.
Transient expression of HCV proteins and preparation of labelled extracts.
DNA transfection and metabolic labelling of cells was performed as described previously (56). Briefly, Hep3B cells (5.5 × 105 in 60-mm-diameter dishes) were infected with vaccinia virus vTF7-3 for 1 h at 37°C and then transfected with 20 μg of recombinant plasmid by the calcium phosphate precipitation technique (20). In cotransfection experiments, 10 μg of each plasmid were used. After 4 h of transfection, cells were starved for 1 h in minimal essential medium without methionine (GibcoBRL) and labelled for 3 h with 100 μCi of 35S-labelled methionine (Promix; Amersham) per ml. For [32P]orthophosphate labelling, cells were washed once after transfection with Dulbecco’s modified Eagle’s medium without phosphate (ICN) and labelled for 4 h in the same medium containing 500 μCi of [32P]orthophosphate (285.5 Ci/mg; NEN) per ml. Cells were harvested, and cell extract was prepared in 150 μl of lysis buffer (25 mM sodium phosphate [pH 7.5], 20% glycerol, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol [DTT], 2 mM phenylmethylsulfonyl fluoride).
For pulse-chase experiments, Hep3B cells were starved for 1 h in minimal essential medium without methionine (GibcoBRL) and labelled for 15 min with 100 μCi of 35S-labelled methionine per ml. Cells were either collected (time zero) or washed twice with phosphate-buffered saline and further incubated with complete Dulbecco’s modified Eagle’s medium supplemented with 4.6 mM cold cysteine and 2.3 mM cold methionine. Cells were collected at different time points as indicated in the figure legends.
Immunoprecipitation.
For immunoprecipitation under denaturing conditions, 20 μl of extract was heated at 95°C for 4 min in the presence of 2% sodium dodecyl sulfate (SDS) and 10 mM DTT. Five microliters of HCV-specific antisera (for description, see references 42 and 56) or 10 μl of Myc-specific monoclonal antibodies (a kind gift from P. Delmastro) was incubated with 50 μl of protein A-Sepharose (PAS) for 1 h at 4°C in 300 μl of immunoprecipitation buffer (IPB150; 20 mM Tris-HCl [pH 8], 150 mM NaCl, 1% Triton X-100), washed once with IPB150, and incubated with the extract for 1 h at 4°C in a volume of 500 μl of IPB150. All subsequent procedures were as described previously (56). For immunoprecipitation experiments under native conditions, the immunoprecipitation buffer was changed to IPB-phosphate (20 mM sodium phosphate buffer [pH 7.5], 150 mM NaCl, 10% glycerol, 0.5% Triton X-100); 20 μl of extract was incubated with antisera bound to PAS in 300 μl of IPB-phosphate. After incubation for 1 h at 4°C, the PAS was layered on 0.5× NDETmod (0.4% sodium deoxycholate, 0.5% Triton X-100, 10 mM Tris-HCl [pH 7.5], 10 mM EDTA) containing 30% sucrose and pelleted by centrifugation for 5 min at 5,000 × g. The pellet was washed once with 500 μl of NDETmod and once with 500 μl of IPB-phosphate. Protein was detached from the PAS-resin by boiling in SDS sample dye.
In vitro dephosphorylation.
Cell extract was immunoprecipitated under native conditions as described above. Protein bound to PAS was washed once with 500 μl of Tris-buffered saline (10 mM Tris-HCl [pH 8], 150 mM NaCl), washed once with 100 μl of phosphatase buffer (50 mM Tris [pH 7.5], 0.1 mM EDTA, 5 mM DTT, 0.01% Brij 35, 2 mM MnCl2), and incubated for 1 h at 37°C with 200 U of λ-phospatase (Biolabs) in 100 μl of phosphatase buffer. After incubation, the resin was washed twice with 500 μl of Tris-buffered saline, and protein was detached from the resin by boiling in SDS loading dye.
RESULTS
The minimal HCV polyprotein sufficient for the formation of NS5A p58 contains NS3, NS4A/B, and NS5A.
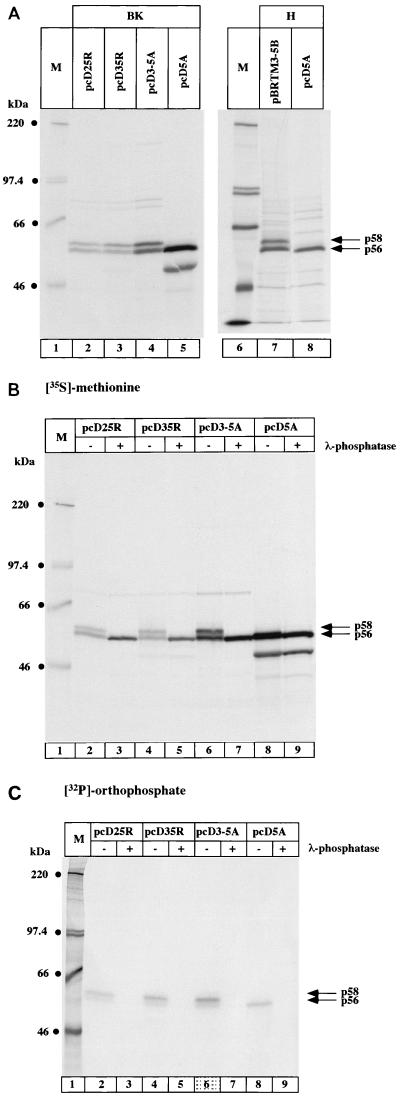
NS5A has been demonstrated to be a phosphoprotein migrating in SDS-polyacrylamide gel electrophoresis (PAGE) with an apparent molecular mass of 56 kDa. It was shown that in the presence of other viral proteins, p56 is converted into a slower-migrating form of NS5A (p58) by additional phosphorylation events. To understand which viral proteins are necessary for NS5A hyperphosphorylation, we expressed all HCV nonstructural proteins from the BK strain (genotype 1b) and different deletion mutants (schematized in Fig. 1) in Hep3B cells, using the recombinant vaccinia virus T7 infection/transfection system. Cells were metabolically labelled, and proteins of interest were immunoprecipitated and analyzed by SDS-PAGE. Expression of the construct pCD25R resulted in the formation of p56 and p58 (Fig. 2A, lane 2), whereas no p58 was visible when NS5A alone was expressed (lane 5). The faster-migrating band visible in lane 5 was presumably due to the usage of an internal ATG as start codon. Expression of polyproteins in which either NS2 (lane 3) or both NS2 and NS5B (lane 4) were deleted still resulted in the production of p58. It can thus be concluded that the polyprotein starting from NS3 and ending with NS5A is sufficient for the generation of NS5A p58. NS5A hyperphosphorylation was not restricted to genotype 1b, as p58 was also formed in the H strain (genotype 1a). The expression of H NS5A alone resulted in the formation of p56 (lane 8), whereas the expression of NS5A in the context of the polyprotein from NS3 to NS5B produced the hyperphosphorylated form p58 (lane 7). To verify that the slower-migrating form of NS5A was phosphorylated, NS5A labelled with either [35S]methionine (Fig. 2B) or [32P]orthophosphate (Fig. 2C) was immunoprecipitated and treated with λ-phosphatase prior to loading for SDS-PAGE. When NS5A was labelled with [35S]methionine, the slower-migrating form (p58) disappeared while the intensity of p56 increased (Fig. 2B, lanes 2 to 7). In contrast, no change of migration or intensity was detectable when [35S]methionine-labelled NS5A alone was expressed (lanes 8 and 9). In the case of [32P]orthophosphate-labelled NS5A, the bands corresponding to both forms of NS5A disappeared (Fig. 2C, lanes 2 to 9). These results prove that both isoforms of NS5A (p56 and p58) are phosphoproteins and that phosphorylation of p56 occurs in the absence of other viral proteins.
FIG. 1.
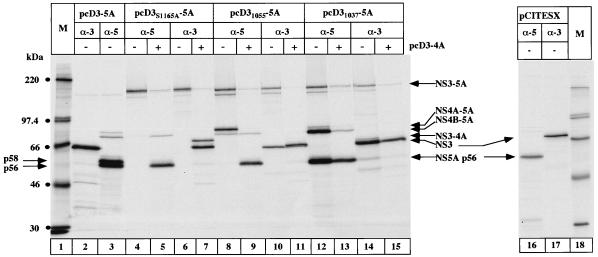
Schematic representation of the expression plasmids used in this study. The organization of the nonstructural region of the viral polyprotein is shown at the top. Each numbers above the bar indicates the position of the N-terminal amino acid of the following protein within the viral polyprotein. The horizontal line indicates the 3′ UTR. R, ribozyme sequence. The vertical bars indicate the boundaries between the different proteins. The HCV polyprotein portions expressed by the different constructs are shown below. myc, epitope with the amino acid sequence EQKLISEEDL; A, the mutation S1165A in NS3.
FIG. 2.
Characterization of minimal requirements for NS5A hyperphosphorylation. Hep3B cells were transfected with the indicated constructs and labelled with either [35S]methionine (A and B) or [32P]orthophosphate (C) for 3 h. NS5A was immunoprecipitated with NS5A antiserum, and protein was loaded onto an SDS–7.5% polyacrylamide gel. (B and C) Immunoprecipitated proteins were incubated with (+) or without (−) λ-phosphatase as described in Materials and Methods prior to loading for SDS-PAGE. NS5A p56 and p58 are indicated by arrows on the right; the sizes of molecular weight marker proteins (M) are indicated on the left. BK, HCV proteins derived from the BK strain; H, HCV proteins derived from the H strain.
Kinetics of NS5A hyperphosphorylation.
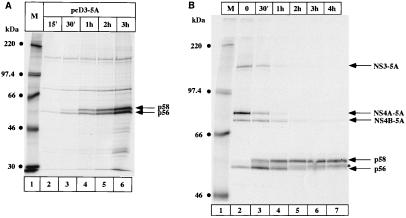
Next we investigated the biogenesis of p56 and p58 from the NS3-5A precursor protein by pulse or pulse-chase experiments. In Fig. 3A, the construct pcD3-5A was labelled with [35S]methionine for the indicated times and NS5A was immunoprecipitated. The p56 band could already be detected after 15 min of labelling, and its intensity increased with labelling time. p58 could be detected with a delay of 15 min (lane 3) and reached the same intensity as p56 after 2 h of labelling (lane 5). This result indicates that the first event to occur is the release of mature NS5A (p56) from the precursor protein, which then serves as a substrate for the hyperphosphorylation event(s) and the formation of p58.
FIG. 3.
Time course of NS5A synthesis and phosphorylation. (A) Pulse-labelling of NS5A expressed from the construct pcD3-5A. Hep3B cells were transfected with pcD3-5A and labelled with [35S]methionine for the indicated times. (B) Pulse-chase analysis of NS5A phosphorylation. Cells were transfected as described above, labelled for 15 min with [35S]methionine, and chased for the indicated times as described in Materials and Methods. NS5A was immunoprecipitated with NS5A antiserum, and proteins were loaded onto an SDS–7.5% polyacrylamide gel. NS5A p56 and p58 proteins as well as the precursor proteins NS3-5A, NS4A-5A, and NS4B-5A are indicated by arrows on the right. M, molecular weight marker proteins.
The time course experiment shown in Fig. 3A demonstrates that a constant ratio of p56 and p58 was reached after 2 h. The sustained presence of p56 could be explained by continued processing of newly synthesized polyproteins or by the fact that part of the produced NS5A molecules was in a conformation unfavorable for hyperphosphorylation events leading to p58. We therefore performed a pulse-chase experiment (Fig. 3B) to follow the fate of the p56 molecules. After 15 min of labelling, 35S-labelled methionine was chased with an excess of cold methionine, and cells were collected after the indicated times. At time zero, most of the protein was still contained in several precursor proteins, the most abundant of which was NS4A-NS5A. Only a fraction of the molecules migrated as p56 (Fig. 3B, lane 2). The hyperphosphorylated form of NS5A became evident after 30 min of chase and reached a plateau after 2 h of chase (lanes 5–7). Interestingly, an additional band migrating slightly slower than p56 (marked with an asterisk in Fig. 3B) became visible after 1 h of chase and increased in intensity with time. The same band was detected when pulse-chase experiments were performed using the construct pcD5A (data not shown). In both cases, this slower-migrating band was sensitive to phosphatase treatment (data not shown). We concluded that this band is the result of modification events of p56 independent of the presence of other viral proteins. These results demonstrate that p56 is a maturation intermediate of NS5A which is converted to p58 with a half-life of approximately 30 min to 1 h.
NS3 must be encoded on the same polyprotein as NS5A.
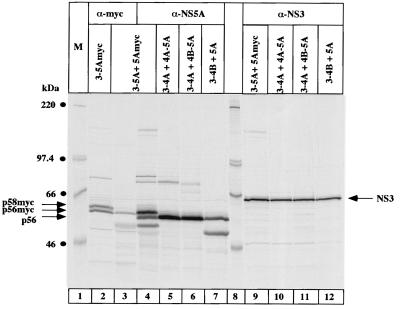
Having demonstrated that the polyprotein consisting of HCV proteins from NS3 to NS5A is sufficient for NS5A hyperphosphorylation, we wanted to analyze whether hyperphosphorylation also occurred when the individual proteins were translated from different RNA templates (Fig. 4). To this end, we constructed the series of expression plasmids described in Fig. 1. The first question addressed was whether the presence of an NS3-5A polyprotein enables hyperphosphorylation of NS5A added in trans. To discriminate between NS5A expressed by the polyprotein and NS5A added in trans, a Myc epitope was added to the C-terminal end of NS5A (pCD5Amyc). Immunoprecipitation was performed with a Myc-specific monoclonal antibody. As shown in Fig. 4, lane 3, no p58 could be detected. The Myc tag did not inhibit NS5A hyperphosphorylation because NS5Amyc expressed in the context of the polyprotein (pcD3-5Amyc) was clearly hyperphosphorylated (lane 2). The addition of the Myc epitope resulted in a slower migration of the NS5A band. Thus, it appeared that p56-Myc comigrates with p58 of the untagged NS5A molecule (compare lanes 3 and 4).
FIG. 4.
Analysis of NS5A hyperphosphorylation with NS5A and NS3 expressed in trans. Cells were cotransfected with the following plasmids: lane 2, pcD3-5Amyc alone; lanes 3, 4, and 9, pcD3-5A plus pcD5Amyc; lanes 5 and 10, pcD4A-5A plus pcD3-4A; lanes 6 and 11, pcD4B-5A plus pcD3-4A; lanes 7 and 12, pcD3-4B plus pcD5A. Proteins were immunoprecipitated with Myc-specific monoclonal antibodies (α-myc) NS5A antiserum (α-NS5A), or with NS3 antiserum (α-NS3) and loaded onto an SDS–7.5% polyacrylamide gel. NS5A p56, p58, and NS3 are indicated by arrows on the right; p56-Myc and p58-Myc are indicated by arrows on the left. M, molecular weight marker proteins.
We then performed a series of experiments where NS3, NS4A, NS4B, and NS5A were all present but not arising from the same polyprotein. In these experiments, NS3 and NS5A were always provided in trans, while NS4A and/or NS4B were generated in cis or in trans with respect to NS5A (lanes 5 to 7). In all cases, NS5A was correctly processed, but no p58 was visible. The same result was obtained in an additional experiment, in which the construct pcD4A-5A was cotransfected with a construct expressing only NS3. The only difference to the result shown in Fig. 4 was a reduced efficiency of processing (data not shown). The levels of expression of NS3 were comparable in all experiments (lanes 9 to 12). We conclude from these results that NS3 must be encoded on the same polyprotein as NS5A (in cis) to enable NS5A hyperphosphorylation.
NS4A and NS4B are required in cis for NS5A hyperphosphorylation.
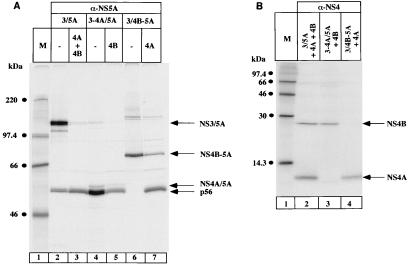
The results in Fig. 4 demonstrate that NS3 must be expressed in cis with NS5A. We next investigated whether the presence of NS3 in cis with NS5A is sufficient for the formation of p58. To this end, we fused NS3 directly to NS5A, at the same time changing the terminal amino acid of NS3 from threonine to cysteine (pcD3/5A). The cysteine in position P1 was presumed to facilitate the cis cleavage between NS3 and NS5A (34). This construct produces mature NS5A without coexpression of NS4A, indicating that the cis cleavage between NS3 (T1657C) and NS5A takes place even in the absence of NS4A. However, most of the protein was present as a nonprocessed fusion protein NS3/NS5A (Fig. 5A, lane 2). Addition of NS4A and NS4B in trans resulted in complete cleavage of the fusion protein into mature NS5A and NS3 (lane 3), but no formation of p58 was found. We then constructed internal deletion mutants retaining either NS4A (construct pCD3-4A/5A [lanes 4 and 5]) or NS4B (construct pcD3/4B-5A, [lanes 6 and 7]) in cis with NS3 and NS5A. The deleted proteins were then supplied in trans as indicated in Fig. 5A. The fusion protein NS3-NS4A/NS5A was completely processed, resulting in mature NS5A (lane 4), which was not hyperphosphorylated even when NS4B was added in trans (lane 5). The slower-migrating band visible in lanes 4 and 5 does not correspond to NS5A p58 but represents the precursor protein NS4A/NS5A, which could be immunoprecipitated with an anti-NS4 antiserum (data not shown). Using the construct pcD3/4B-5A, we detected an NS4A-independent cis cleavage between NS3 (T1657C) and NS4B. In this case, the anti-NS5A antiserum immunoprecipitated the NS4B-NS5A precursor protein (lane 6). Addition of NS4A in trans resulted in a partial cleavage between NS4B and NS5A, but no formation of p58 was observed (lane 7).
FIG. 5.
NS4A/B is required in cis for the formation of p58. Cells were cotransfected with the indicated constructs. (A) Proteins were immunoprecipitated with NS5A antiserum (α-NS5A) and loaded onto an SDS–7.5% polyacrylamide gel. (B) Proteins were immunoprecipitated with NS4 antiserum (α-NS4) and loaded onto an SDS–15% polyacrylamide gel. Arrows on the right indicate HCV-specific proteins. 3/5A, pcD3/5A, 3-4A/5A, pcD3-4A/5A; 3/4B-5A, pcD3/4B-5A; 4A, pCITE4A; 4B, pCITE4B; M, molecular weight marker proteins.
Expression of the NS4A and NS4B proteins supplied in trans was controlled in Fig. 5B. Constructs expressing NS4A or NS4B supplied in trans were transfected in molar excess with respect to the construct expressing the fusion protein and appear therefore as more intense protein bands after immunoprecipitation. The slightly faster migration of NS4A derived from processing of the fusion protein (Fig. 5B, lane 3) can be explained by the absence of the artificial methionine that had been added to the constructs used for expression in trans.
These data suggest that NS5A hyperphosphorylation occurs only when NS3, NS4A, and NS4B are present in cis with NS5A, even though one cannot rule out the possibility that trans complementation of NS4A and NS4B is prevented by the presence of an artificial N-terminal methionine residue.
The formation of p58 requires an active NS3 protease with its authentic N-terminal sequence.
Having obtained evidence that NS5A can be hyperphosphorylated only when expressed as a precursor comprising NS3, NS4A, and NS4B, we addressed the question whether NS3 must be active as a protease or whether the presence of the NS3 protein sequence as such is sufficient for the production of p58. We constructed an NS3 point mutant and different NS3 deletion mutants of known phenotype in the context of the polyprotein and tested their effects on NS5A hyperphosphorylation (Fig. 6). Proteolytic activity was abolished by changing the catalytic serine residue into alanine (pcD3S1165A-5A). As shown in Fig. 6, the construct pcD3S1165A-5A expressed a nonprocessed polyprotein containing proteins from NS3 to NS5A which could be immunoprecipitated with an NS5A-specific antiserum (lane 4) as well as with an NS3-specific antiserum (lane 6). Addition of NS3-4A in trans resulted in the formation of mature but not hyperphosphorylated NS5A (lane 5). It can therefore be concluded that NS5A hyperphosphorylation requires an active NS3 protease encoded on the same polyprotein molecule. Immunoprecipitation with anti-NS3 antibodies showed a double band (lane 7) corresponding to mature NS3 and the uncleaved NS3-NS4A precursor. It has been demonstrated that the cleavage between NS3 and NS4A occurs only in cis (56). For this reason, NS3 and NS4A encoded on the polyprotein NS3S1165A-5A migrated as a nonprocessed precursor protein, whereas the faster-migrating band corresponds to mature NS3 which had been added in trans. To exclude the possibility that the proteolytic release of NS3 encoded in cis is necessary for the formation of p58, we tested an N-terminal deletion mutant of NS3 (pcD31055-5A). This mutant is still capable of cis cleavage between NS3 and NS4A (16) (Fig. 6, lane 10) but cannot cleave efficiently the downstream NS4A/NS4B and NS4B/NS5A trans-cleavage sites (lane 8). Adding NS3-4A in trans produced mature NS5A (lane 9) but no p58.
FIG. 6.
NS5A hyperphosphorylation requires an active NS3 protease. Cells were transfected with the indicated plasmids in the presence (+) or absence (−) of plasmid pcD3-4A. Proteins were immunoprecipitated with NS5A (α-5) or NS3 (α-3) antiserum and loaded onto an SDS–7.5% polyacrylamide gel. HCV-specific proteins are indicated by arrows. The positions of NS5A p56 and p58 expressed from the control plasmid pcD3-5A are indicated on the left. M, molecular weight marker proteins.
The experiments described thus far suggest that the existence of mature but proteolytically defective NS3 in cis is not sufficient to promote the hyperphosphorylation of NS5A. However, it is worth pointing out that the NS31055 protein, which lacks the N-terminal 28 amino acids, is not able to cleave at the NS4B-NS5A site because it cannot interact with the serine protease NS4A cofactor (16). This finding raises the possibility that not only a proteolytically active NS3 proteinase must be in cis with NS5A in order to promote formation of p58 but also the formation of a stable NS3/NS4A complex is required. To test this hypothesis, we deleted the first 10 N-terminal amino acids from NS3 (pcD31037-5A). This NS3 mutant is expected to be activated by the NS4A cofactor but is no longer able to form an NS3/NS4A complex stable enough to be immunoprecipitated (16, 32). Our data obtained with the construct pcD31037-5A are in agreement with these previously published observations. NS3 expressed from this construct was capable of cleaving both cis- and trans-cleavage sites of the polyprotein, although with reduced efficiency (Fig. 6, lanes 12 and 14). The presence of significant amounts of precursor protein NS4B/NS5A (lane 12) is consistent with the finding that the interaction between NS3 and NS4A was not as stable as that with the wild-type protein (compare lanes 3 and 12). Interestingly, the fraction of NS5A which had been generated was not hyperphosphorylated.
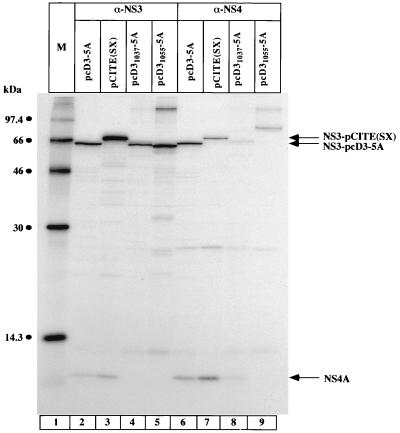
The latter results obtained with the N-terminal deletion mutants of NS3 are in line with the idea that formation of a stable complex between NS3 and NS4A is a prerequisite for the formation of p58. An alternative but not exclusive interpretation is that the correct N-terminal sequence of NS3 protein is required for the generation of p58 for reasons that are not related—or are additional—to the formation of an NS3-NS4A complex. Use of the construct pCITE(SX) (Fig. 1) helped us address the question of how crucial the authentic N terminus of NS3 is for NS5A hyperphosphorylation. NS3 expressed from this construct begins at amino acid 992 (NS3992) and thus contains the 34 C-terminal amino acids of NS2. Proteolytic activity of NS3993 was comparable with that of wild-type NS3 (Fig. 6; compare lanes 2 and 3 with lanes 16 and 17); interestingly, however, no hyperphosphorylation of NS5A was detected (lane 16). The stability of the complex between NS3 and NS4A expressed from the constructs pcD3-5A, pCITE(SX), and the N-terminal deletion mutants of NS3 was determined by native coimmunoprecipitation experiments (Fig. 7). Immunoprecipitation with anti-NS3 antiserum clearly demonstrated that NS4A coimmunoprecipitated with NS3 expressed from the constructs pcD3-5A and pCITE(SX) (lanes 2 and 3). No NS4A was present when the constructs pcD31037-5A and pcD31055-5A were used (lanes 4 and 5), as described previously (16). Lanes 6 to 9 show the results of the immunoprecipitation using anti-NS4 antiserum. The amount of coimmunoprecipitated NS3 was clearly lower in the case of pCITE(SX) than with pcD3-5A (compare lanes 6 and 7), even though the total amount of NS4A from pCITE(SX) was higher. These results suggest that the stability of the complex between NS3 and NS4A is lower when expressed from pCITE(SX) than from pcD3-5A; however, a significant amount of stable NS3/NS4A complex was detected.
FIG. 7.
Comparison of NS3/NS4A complex stability. Cells were transfected with the indicated plasmids, and proteins were immunoprecipitated under native conditions as described in Materials and Methods with either NS3 (α-NS3) or NS4 (α-NS4) antiserum. Immunoprecipitated proteins were then loaded onto an SDS–13.5% polyacrylamide gel. NS4, NS3 expressed from pCITE(SX), and NS3 expressed from pcD3-5A are indicated by arrows on the right. M, molecular weight marker proteins.
Together, these data demonstrate that the NS3 protein present in cis with NS5A must contain a protease domain with wild-type activity in order to facilitate the formation of p58. The activity of NS3, however, is dependent on the presence of a complex between NS3 and NS4A, which in turn requires a correct folding of the N-terminal sequence of NS3 (10). It is difficult to conclude whether the requirement of an authentic N-terminal sequence of NS3 influences NS5A hyperphosphorylation via its effect on the formation of a tight NS3/NS4A complex or through other, still unknown mechanisms.
DISCUSSION
Due to the lack of reliable tissue culture models for sustained HCV replication, it is not possible to identify the isoform(s) of NS5A important for the viral life cycle. However, transient transfection experiments (28), as well as human cell lines inducibly expressing all HCV structural and nonstructural proteins (38), showed two isoforms of NS5A with apparent molecular masses of 56 and 58 kDa. Here we demonstrate that transfection of NS5A alone resulted in the production of p56, whereas the formation of p58 required the presence of a polyprotein consisting of the nonstructural proteins from NS3 to NS5A (Fig. 2). Treatment of 32P-labelled NS5A with λ-phosphatase resulted in the loss of radioactive label of both isoforms, indicating that both proteins are phosphoproteins. The existence of phosphorylated NS5A/NS5 proteins is not unique to HCV but was also observed in bovine viral diarrhea virus, yellow fever virus, tick-borne encephalitis virus, and dengue virus type 2 (29, 39, 46), other members of the Flaviviridae family.
Characterization of the requirements necessary for the formation of p58 demonstrated that (i) NS5A has to be encoded on the same polyprotein with NS3 (Fig. 4) and (ii) the nonstructural proteins NS4A and NS4B have to be present in cis together with NS3 and NS5A (Fig. 5). These observations differ from what has been published previously by Shimotohno and colleagues (2, 55), who reported that the only viral protein necessary for NS5A hyperphosphorylation is NS4A which can be supplied in trans. Interestingly, this group used NS5A derived from the HCV J strain, while we and others (43) who found that NS4A is not sufficient for the production of p58 used the HCV BK strain.
Further characterization of the role of NS3 for NS5A hyperphosphorylation suggests that NS3 has to be active as a protease that cleaves all downstream cleavage sites present in the HCV polyprotein consisting of NS3 to NS5A. Mutations which abolish the activity of the serine-protease or which reduce the activity on cis- or trans-cleavage sites result in prevention of NS5A hyperphosphorylation (Fig. 6). It has been shown previously that the protease activity of NS3 depends on the interaction of NS3 with its cofactor NS4A. The results shown in Fig. 7 demonstrate that N-terminal deletion mutants of NS3 do not form an NS3/NS4A protein complex stable enough to be immunoprecipitated. This result is in agreement with the hypothesis that the formation of p58 requires a stable NS3/NS4A complex necessary for NS3 activity. The situation is different when the construct pCITE(SX) is used. The activity of NS3 seems to be comparable with that of wild-type NS3, and the NS3/NS4A complex in pCITE(SX) is present in reduced but still significant amounts. One can draw two conclusions from these observations. (i) Even though NS3 protease activity and NS3/NS4A complex stability seem to be comparable with that of wild-type NS3, at least in our experimental conditions, there might still be differences in the kinetic parameters of processing which could be significant for the production of p58. More detailed kinetic studies are needed for a better understanding of the correlation between NS3 processing and NS5A hyperphosphorylation. (ii) The correct N-terminal sequence of NS3 is important for NS5A hyperphosphorylation. Qingyan et al. (43) have reported that the HCV polyprotein containing the region from NS3 to NS5B is not sufficient for the formation of p58 but that also the presence of NS2 is necessary. In their case, however, NS3 did not initiate with the correct N-terminal amino acid sequence. NS2 may thus be required in their case to generate the authentic NS3 N terminus via the action of the NS2/3 protease.
The presence of the authentic N terminus of NS3 might be required for protein-protein interaction either directly between NS5A and NS3 or indirectly via additional viral or cellular proteins. Protein-protein interactions between HCV NS4A, NS4B, and NS5A have been demonstrated (37), and direct interactions between NS3 and NS5 have been shown for dengue virus type 2 (29). In rotavirus, protein-protein interaction between the phosphoprotein NSP5, the double-stranded RNA (dsRNA)- and single-stranded RNA (ssRNA)-binding protein NSP2, and the viral polymerase VP1 has been detected (1). In this regard, we note that the helicase domain of NS3 (31) contains ssRNA- as well as dsRNA-binding activity. Direct interaction between HCV proteins NS5A, NS3, and NS5B has not yet been demonstrated; however, it is possible that this protein complex is not stable enough to be coimmunoprecipitated, and further experiments are required to determine whether these interactions occur.
Kinetic studies of NS5A hyperphosphorylation demonstrated that there is a time lag between the release of NS5A from the precursor protein containing proteins from NS3 to NS5A and the formation of p58. This result suggests that p56 is the substrate for the formation of p58. The conversion of p56 to p58 could indicate that p58 functions after completion of polyprotein processing and is involved in later events of viral replication which involve the proteins NS3, NS4A, and NS4B and possibly other cellular proteins.
The role and mechanism of NS5A phosphorylation can only be speculated. It has been shown that NS5A is phosphorylated by a tightly associated cellular kinase (26, 45, 46). The consequence of NS5A phosphorylation could be the induction of conformational changes in NS5A that make it susceptible to additional phosphorylation either by the same, already associated kinase or by a second kinase. Conformational changes of viral proteins induced by phosphorylation have been demonstrated for other RNA viruses such as vesicular stomatitis virus (VSV) (9) and chandipura virus (44). In VSV, a viral protein present in different phosphoisoforms has been demonstrated to have different functions during viral replication (3, 40). The authors showed that sequential phosphorylation by a cellular kinase followed by a viral kinase produced a phosphoprotein required for RNA transcription, whereas the same protein in a nonphosphorylated state was necessary for viral genome replication. In the case of HCV, the proteins NS3, NS4A, and NS4B could induce or stabilize conformational changes of NS5A or promote the association of additional kinases and/or other cellular proteins necessary for the formation of p58. Another explanation would be that the viral proteins NS3, NS4A, and NS4B protect additional phosphorylation sites in p58 from fast dephosphorylation by cellular phosphatases. It was demonstrated for rotavirus (6) that hyperphosphorylated isoforms of the nonstructural protein NSP5, when transfected in the absence of other viral proteins, could be detected only in the presence of okadaic acid, a specific inhibitor of the major cytosolic phosphatases PP1 and PP2A.
From our data, we conclude that the events resulting in the formation of NS5A p58 are closely connected to HCV polyprotein processing and require at least the viral proteins NS3, NS4A, and NS4B and possibly additional cellular proteins. Sequential phosphorylation events are not unique to HCV but have been demonstrated also in negative-strand RNA viruses (VSV) and dsRNA viruses (rotavirus). These observations suggest that phosphorylation is important for successful virus propagation and/or pathogenesis. Further analysis is required to elucidate the role of NS5A and its different phosphoisoforms in the viral life cycle.
ACKNOWLEDGMENTS
We express our gratitude to L. Tomei, P. Gallinari, and C. Steinkühler for numerous helpful discussions, G. Migliaccio for critical reading of the manuscript, and M. Emili for photographic assistance.
REFERENCES
- 1.Afrikanova I, Fabbretti E, Miozzo M C, Burrone O R. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J Gen Virol. 1998;79:2679–2686. doi: 10.1099/0022-1317-79-11-2679. [DOI] [PubMed] [Google Scholar]
- 2.Asabe S-I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–796. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik S, Banerjee A. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J Virol. 1992;66:1109–1118. doi: 10.1128/jvi.66.2.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analysis of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackhall J, Mu∼noz M, Fuentes A, Magnusson G. Analysis of rotavirus nonstructural protein NSP5 phosphorylation. J Virol. 1998;72:6398–6405. doi: 10.1128/jvi.72.8.6398-6405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 8.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das T, Gupta A K, Sims P W, Gelfand C A, Jentoft J E, Banerjee A K. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA polymerase of vesicular stomatitis virus. J Biol Chem. 1995;270:24100–24107. doi: 10.1074/jbc.270.41.24100. [DOI] [PubMed] [Google Scholar]
- 10.De Francesco R, Steinkühler C. Structure and function of the hepatitis C virus NS3-NS4A serine protease. Curr Top Microbiol Immunol. 1999;242:149–170. doi: 10.1007/978-3-642-59605-6_8. [DOI] [PubMed] [Google Scholar]
- 11.Eckart M R, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo Q L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murankami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Maruno F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 14.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Failla C, Tomei L, De Francesco R. An amino-terminal domain of the hepatitis C virus NS3 proteinase is essential for interaction with NS4A. J Virol. 1995;69:1769–1777. doi: 10.1128/jvi.69.3.1769-1777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale M, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 19.Gale M, Jr, Blakely C M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hapatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-α catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 27.Jeng K-S, Daniel A, Lai M M C. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J Virol. 1996;70:2403–2410. doi: 10.1128/jvi.70.4.2403-2410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko T, Tanji Y, Satoh S, Hijikata M, Shinichi S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of human hepatitis C virus genome from Japanese patients with non-A non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 32.Koch J O, Lohmann V, Herian U, Bartenschlager R. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology. 1996;221:54–66. doi: 10.1006/viro.1996.0352. [DOI] [PubMed] [Google Scholar]
- 33.Komoda Y, Hijikata M, Tanji Y, Hirowatari Y, Mizushima H, Kimura K, Shimotohno K. Processing of hepatitis C viral polyprotein in Escherichia coli. Gene. 1994;145:221–226. doi: 10.1016/0378-1119(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 34.Komoda Y, Hijikata M, Sato S, Asabe S-I, Kimura K, Shimotohno K. Substrate requirements of hepatitis C virus serine proteinase for intermolecular polypeptide cleavage in Escherichia coli. J Virol. 1994;68:7351–7357. doi: 10.1128/jvi.68.11.7351-7357.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-β therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 36.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin C, Wu J-W, Hsiao K, Su M S-S. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J Virol. 1997;71:6465–6471. doi: 10.1128/jvi.71.9.6465-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moradpour D, Kary P, Rice C M, Blum H E. Continous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 39.Morozova O V, Tsekhanovskaya N A, Maksimova T G, Bachvalova V N, Matveeva V A, Kit Y Y. Phosphorylation of tick-borne encephalitis virus NS5 protein. Virus Res. 1997;49:9–15. doi: 10.1016/s0168-1702(96)01433-5. [DOI] [PubMed] [Google Scholar]
- 40.Pattnaik A K, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee A K. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J Virol. 1997;71:8167–8175. doi: 10.1128/jvi.71.11.8167-8175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pieroni L, Santolini E, Fipaldini C, Pacini L, Migliaccio G, La Monica N. In vitro study of the NS2-3 protease of hepatitis C virus. J Virol. 1997;71:6373–6380. doi: 10.1128/jvi.71.9.6373-6380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizzi E, Tramontano A, Tomei L, La Monica N, Failla C, Sardana M, Wood T, De Francesco R. Molecular model of the specificity of the hepatitis C virus protease: implications for substrate recognition. Proc Natl Acad Sci USA. 1994;91:888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qingyan L, Bhat R A, Prince A M, Zhang P. The hepatitis C virus NS2 protein generated by NS2-3 autocleavage is required for NS5A phosphorylation. Biochem Biophys Res Commun. 1999;254:572–577. doi: 10.1006/bbrc.1998.9986. [DOI] [PubMed] [Google Scholar]
- 44.Raha T, Chattopadhyay D, Chattopadhyay D, Roy S. A phosphorylation-induced major structural change in the N-terminal domain of the P protein of chandipura virus. Biochemistry. 1999;38:2119–2116. doi: 10.1021/bi980734c. [DOI] [PubMed] [Google Scholar]
- 45.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7181–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed K E, Gorbalenya A E, Rice C M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72:6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sáiz J-C, López-Labrador F-X, Ampurdanés S, Dopazo J, Forns X, Sánchez-Tapias J-M, Rodés J. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J Infect Dis. 1998;177:839–847. doi: 10.1086/515243. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santolini E, Pacini L, Fipaldini C, Migliaccio G, La Monica N. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J Virol. 1995;69:7461–7471. doi: 10.1128/jvi.69.12.7461-7471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satoh S, Tanji Y, Hijikata M, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus nonstructural protein 3 (NS3) is essential for stable complex formation with NS4A. J Virol. 1995;69:4255–4260. doi: 10.1128/jvi.69.7.4255-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimotohno K, Tanji Y, Hirowatari Y, Komoda Y, Kato N, Hijikata M. Processing of the hepatitis C virus precursor protein. J Hepatol. 1995;22:87–92. [PubMed] [Google Scholar]
- 53.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onoshi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urbani A, De Francesco R, Steinkühler C. Proteases of the hepatitis C virus. In: Dunn B, editor. Proteases of infectious agents. San Diego, Calif: Academic Press; 1999. pp. 61–91. [Google Scholar]
- 58.World Health Organization. Global surveillance and control of hepatitis C. J Viral Hepatitis. 1999;6:35–47. [PubMed] [Google Scholar]