Highlights
-
•
The immunohistochemical expression of several potentially actionable proteins (MAGE-A4, NY-ESO-1, YAP1, TAZ, BRD9 and CXCR4) was explored in 91 synovial sarcoma samples using a tissue microarray.
-
•
All evaluated markers were expressed in a clinically relevant proportion of cases.
Keywords: Synovial sarcoma, Tissue microarray, Immunohistochemistry, Actionable targets
Abstract
Background
Synovial sarcoma (SynSa) is one of the most common translocation-related soft tissue sarcomas. Patients with metastatic SynSa have limited treatment options and a very poor prognosis. Several novel experimental therapies are currently being explored in clinical trials, including T cell-based therapies targeting cancer testis antigens such as New York esophageal squamous cell carcinoma 1 (NY-ESO-1) or melanoma-associated antigen A4 (MAGE-A4), and degraders targeting bromodomain-containing protein 9 (BRD9). Preclinical studies investigate inhibitors of Yes associated protein 1 (YAP1), transcriptional co-activator with PDZ-binding motif (TAZ) and inhibitors of chemokine receptor 4 (CXCR4).
Methods
We explored the immunohistochemical expression of these targets using a tissue microarray (TMA) constructed from 91 clinical SynSa samples and correlated these findings with corresponding clinicopathological data.
Results
Expression of MAGE-A4 and NY-ESO-1 was found in 69 % and 56 % of the samples, respectively. NY-ESO-1 was statistically higher expressed in samples from metastatic lesions as compared to samples from primary tumors. Nuclear expression of YAP1 and TAZ was observed in 92 % and 51 % of the samples, respectively. CXCR4 was expressed in the majority of the samples (82 %). BRD9 was highly expressed in all specimens. No prognostic role could be identified for any of the investigated proteins.
Conclusion
This study is a comprehensive study providing real-world data on the expression of several actionable proteins in a large proportion of SynSa samples. All evaluated markers were expressed in a clinically meaningful proportion of cases represented in our TMA, supporting the relevance of ongoing preclinical and clinical research with novel agents directed against these targets.
Introduction
Synovial sarcoma (SynSa) is one of the most common types of translocation-related soft tissue sarcoma (STS), but it is considered as a rare cancer with an incidence of 1,7 cases/106/year [1]. The pathognomonic chromosomal translocation that characterizes SynSa involves the SS18 gene on chromosome 18 and one of the SSX genes on chromosome X (most frequently SSX1 or SSX2, in rare cases SSX4). This results in the formation of SS18-SSX fusion oncogenes, which are the well-described drivers of this malignancy [2]. SynSa typically affects young adults with a median age of 34 years (range 2–94 years) [3]. For localized disease, SynSa treatment typically consists of surgery, in some cases combined with pre- and/or postoperative chemotherapy. Adjuvant radiotherapy is used in the case of large, deep-seated tumors [4]. Despite aggressive treatment of localized disease, about half of the patients with SynSa will develop metastases, most commonly localized in the lung. Patients with metastatic disease are usually treated with systemic chemotherapy, often with palliative intent [5]. SynSa is generally perceived as a more chemotherapy-sensitive sarcoma entity in comparison with other STS subtypes and agents such as doxorubicin, ifosfamide, trabectedin and pazopanib seem to have preferential activity in this disease [5]. Even though SynSa can respond quite impressively to systemic therapy, the general treatment outcome of patients with metastatic disease remains very poor with a 5-year overall survival (OS) rate of around only 10 % [6]. As such, there is a high unmet clinical need to develop more effective treatment options for this malignancy.
More insight on altered signaling pathways and expression profiles of potential targets in SynSa is needed to facilitate the development of novel treatment strategies. Analyses of archival tumor tissue with tissue microarrays (TMAs) can be used for tumor biology studies and parallel screening of novel targets in multiple cases of SynSa. TMAs typically contain multiple small tissue cores from different donors, arranged on a single paraffin block. These arrays can be used to study several biological markers and potential drug targets simultaneously under uniform conditions. This saves time and resources, while reducing overall costs and the amount of archival tissue required [7]. We created two TMAs consisting of 91 clinical SynSa samples from 78 individual donors and used them to analyze the epidemiology of selected novel, potentially actionable targets which are currently being explored in clinical and preclinical settings.
Firstly, the cancer testis antigens (CTAs) were investigated. These proteins are expressed in healthy tissue during embryonic development, but can only be detected in testis and placenta after birth [8]. Additionally, they are also highly expressed in some cancers, including SynSa. New York esophageal squamous cell carcinoma 1 (NY-ESO-1) and melanoma-associated antigen A4 (MAGE-A4) are the CTAs which are the best characterized in the context of SynSa [5]. Expression in this disease ranges from 53- 82 % for MAGE-A4 and 70–80 % for NY-ESO-1 [[9], [10], [11], [12], [13], [14]]. These proteins are very attractive targets as they are highly immunogenic and can elicit strong cellular and humoral antitumoral responses [15]. A number of vaccines and engineered T-cell receptor (TCR) therapies are currently under investigation in preclinical and clinical trials. Engineered TCR T-cells recognize processed peptides presented by human leukocyte antigen (HLA)-A*02, an allele subtype presented in 50 % of the Caucasian population, and this interaction led to further immune responses [16]. NY-ESO-1-specific TCR T-cells have shown antitumoral efficacy in clinical SynSa cases in phase I/II studies (www.clinicaltrials.gov NCT00670748 and NCT01343043) [17,18]. Overall response rates of 50 % in patients with unresectable or metastatic SynSa have been reported after treatment with T-cells expressing a modified TCR recognizing NY-ESO-1 [18]. Genetically engineered TCRs against MAGE-A4 showed comparable efficacy: a phase I study (NCT03132922) showed an overall response rate of 44 % in 16 heavily pre-treated metastatic SynSa patients. The median OS of 58.1 weeks is much better than the outcome reported with the current standard-of-care therapies [14]. A subsequent phase II study (Spearhead-1, NCT04044768) showed an overall response rate of 39 % in 44 heavily pretreated SynSa patients with a tolerable safety profile [13]. Previous clinical trials using immunotherapy, such as immune checkpoint inhibitors, have failed for SynSa and this highlights the significance of these clinical responses [19,20].
Another target of interest tested in clinical trials is bromodomain-containing protein 9 (BRD9). Recent research showed that BRD9 supports oncogenic mechanisms underlying the SS18-SSX fusion in SynSa. Degradation of BRD9 inhibits tumor progression in a murine SynSa model [21]. This work led to the development of two BRD9 inhibitors which are currently being tested in two phase I trials, which include patients with SynSa. CFT8634 is an oral degrader causing proteasomal degradation of BRD9 (NCT05355753) and FHD-609 is an intravenous BRD9 degrader (NCT04965753) [22,23]. The frequency of BRD9 expression using immunohistochemistry (IHC) has not been reported before in SynSa samples.
Next, we investigated some other, more novel and potentially actionable targets, which are currently being tested only in preclinical studies. The expression of Yes associated protein 1 (YAP1) and transcriptional co-activator with PDZ-binding motif (TAZ) are proteins involved in the Hippo pathway. This signaling pathway modulates proliferation, differentiation, and survival of cells and plays an important role in the development and homeostasis of organs. Dysregulation of the Hippo pathway can cause a variety of diseases, including cancer, and plays a role in tumor initiation, progression, metastasis and drug resistance. In the inactive state of the Hippo signaling cascade, non-phosphorylated YAP1 and TAZ translocate into the nucleus and can interact with certain transcription factors inducing the expression of target genes that play a role in e.g. cell proliferation. In the active state of the Hippo pathway, YAP1 and TAZ are phosphorylated and accumulate in the cytoplasm, which results in degradation of these proteins. As such, the nuclear presence of YAP1 and TAZ corresponds with the transcriptionally active pool of these proteins [24]. YAP1 and TAZ have gained interest as important oncoproteins in many epithelial cancers, including breast, colon, liver, lung and pancreas malignancies while the role of YAP1 and TAZ in mesenchymal tumors still needs to be investigated in more depth [25]. Isfort et al. demonstrated that activation of YAP/TAZ signals is a common pattern in SynSa and is functionally dependent on the chimeric SS18-SSX fusion protein [26]. They investigated a large cohort of STS and bone sarcoma samples by IHC and showed that YAP1 nuclear expression was the most prevalent in SynSa (78 %) and myxoid liposarcoma (91 %) [27]. Disruption of YAP1 and TAZ-mediated signal transduction might be an effective and novel therapeutic approach for SynSa [26].
A final target of interest is the chemokine receptor 4 (CXCR4), a transmembrane G protein-coupled receptor involved in cell migration, invasion, and angiogenesis [28]. In osteosarcoma, CXCR4 is higher expressed at the metastatic site as compared with the primary site, however this could not be confirmed by CXCR4 mRNA expression in STS [[29], [30], [31]]. Its nuclear expression in samples from patients with localized SynSa is associated with a worse 5-year OS, especially in patients treated with adjuvant chemotherapy. This might imply that CXCR4 plays a role in the development of chemoresistance [28]. The use of a CXCR4 antagonist, which is currently under preclinical investigation, could be a potential treatment option for patients with metastatic SynSa [32,33].
With protein expression data from our SynSa-specific TMA we provide comprehensive real-world data on the prevalence of all these targets. Previous studies looking into the expression of these proteins of interest had some weaknesses, as they explored multiple STS subtypes simultaneously, had a low sample size and did not correlate the expression pattern of these proteins with clinical data.
Material and methods
The SynSa TMAs were constructed from biological material collected from SynSa patients treated in the University Hospitals Leuven, Belgium between 01-12–1990 and 31-12-2020 as previously described [7]. The TMAs had one tothree assessable cores with a diameter of 1 mm per sample, for 98 % of the samples three cores were included. The diagnosis of SynSa in the samples of origin had been previously confirmed by a reference pathologist (RS) as part of the routine diagnostic process. The presence of the chromosomal translocation was proven by FISH (fluorescence in situ hybridization), RT-PCR (reverse transcription polymerase chain reaction), karyotyping or IHC for SSX and SS18-SSX. The creation of these TMAs from archival formalin-fixed paraffin-embedded (FFPE) samples was approved by the Ethics Committee of the University Hospitals Leuven (project S59181). Corresponding clinical data were collected in the LECTOR (Leuven Connective Tissue Oncology Repository) database (S51495) and included patient demography, tumor diagnosis and disease characteristics, prior therapies and follow-up data concerning local relapse, metastases, death and cause of death. The analysis of TMAs and subsequent clinical correlations with pseudonymized clinical data was approved by the Ethics Committee of University Hospitals Leuven (S68108).
Expression of the target proteins was investigated using IHC. TMA sections were pre-treated with Ultra Clear (VWR, Radnor, US) and 100 % ethanol (Thermo Scientific, Waltham, US) for deparaffinization and rehydration and were incubated in a solution of methanol (Thermo Scientific, Waltham, US) with hydrogen peroxidase (Merck, Darmstadt, Germany) for blocking endogenous peroxidase. Antigen retrieval was achieved by sections incubated in a citrate or tris-ethylenediamine tetra-acetic acid (EDTA) buffer in a preheated water bath at 95 °C for 30 min, followed by the incubation with serum-free protein block (DAKO A/S X0909, Glostrup, Denmark) [7]. Slides were stained with the respective primary antibodies mentioned in Supplementary Table 1 during an overnight incubation. After incubation with the secondary antibodies (Supplementary Table 1), visualization was done by 3,3′-Diaminobenzidine Substrate Chromogen System (DAKO A/S K34681, Glostrup, Denmark). After counterstaining with hematoxylin (VWR, Radnor, US), slides were dehydrated in a 100 % ethanol solution, cleared in Ultra Clear (VWR, Radnor, US) and mounted. As the CXCR4 antibody detects only unphosphorylated CXCR4, the slides were treated with lambda protein phosphatase (New England Biolabs, Ipswich, US). After antigen retrieval the sections were incubated with 800 units of lambda protein phosphatase in protein metallophosphatases buffer with MnCl2 provided by the manufacturer for 1 hour at 30 °C. After the phosphatase treatment, sections were rinsed in phosphate-buffered saline three times, followed by regular IHC processing.
IHC stainings were analyzed using a brightfield microscope (Olympus CH30, Tokyo, Japan). Each core was scored by two independent researchers (LDC and FP), blinded to the corresponding clinicopathological data at the moment of the evaluation. Expression was scored using an H-score, defined as: (% stained cells at 0 intensity) × 0 + (% stained cells at 1+ intensity) × 1 + (% stained cells at 2+ intensity) × 2 + (% stained cells at 3+ intensity) × 3. For YAP1 and TAZ scoring was limited to nuclear expression (equivalent to the transcriptionally active pool of YAP1 and TAZ) [27]. The average of the H-scores from multiple cores of the same donor sample was used for subsequent analyses. Furthermore, H-score per sample of the two readers was averaged. Based on previous published data we defined criteria for (high and low) positive expression for each of the proteins of interest (Table 1), with the exception of BRD9 as the cut-off to define positive expression in SynSa was not previously reported. Homogenous expression was defined as consistent positive or negative expression in at least two of the three cores corresponding to the same tumor sample.
Table 1.
Criteria to define positive expression of the proteins of interest based on previous reports.
For the cancer testis antigens: NY-ESO-1 and MAGE-A4 high and low positive expression was defined based on the current literature. In the Spearhead-I trial the cut-off for patient screening/enrollment was initially defined as a staining intensity of ≥1+ in ≥ 10 % of cells, but was later changed to ≥2+ or 3+ in ≥ 30 % of cells [34]. For BRD9 no cut-off was previously defined in literature.
| Target protein | (High) positive expression | (Low) positive expression |
|---|---|---|
| MAGE-A4 | ≥ 30 % of cells with ≥ 2+ staining intensity in at least 2 cores [13,14] | ≥ 10 % of cells with 1+ staining intensity, but < 2+ or 3+ in ≥ 30 % of cells [34] |
| NY-ESO-1 | ≥ 50 % of cells with ≥ 2+ staining intensity in at least 2 cores [18] | ≥ 1 % of cells with 1+ staining intensity, but < 2+ or 3+ in ≥ 50 % of cells [35] |
| YAP1 | Nuclear staining in ≥ 30 % of cells with a staining intensity of ≥ 2+ in at least 2 cores [27] | |
| TAZ | ||
| CXCR4 | ≥ 1+ staining intensity in ≥ 1 % of cells in at least 2 cores [28] | |
| BRD9 | No cut-off defined in literature | |
| SSX | > 5 % of cells with a staining intensity of ≥ 2+ in at least 2 cores [36] | |
| SS18-SSX | ||
Abbreviations: MAGE-A4: melanoma-associated antigen A4, NY-ESO-1: New York esophageal squamous cell carcinoma 1, YAP1: Yes associated protein 1, TAZ: transcriptional co-activator with PDZ-binding motif, CXCR4: chemokine receptor 4, BRD9: bromodomain-containing protein 9.
The expression of all evaluated proteins was correlated with clinical-pathological factors including the origin of the sample (primary tumor, metastasis, or local relapse), morphology on histopathological examination (biphasic versus monophasic SynSa), size of the primary tumor (≤5 cm versus >5 cm) and tumor grade according to the FNCLCC (Fédération Nationale des Centres de Lutte Contre le Cancer). Morphology, size and grade were only considered for primary tumor samples from treatment-naive patients. Results were analyzed using GraphPad Prism (GraphPad Software Inc., version 10.0.0 for Windows, US) and Statistical Package for the Social Sciences (SPSS; International Business Machines Corporation, version 29.0.2 for Windows, US). All tests of statistical significance were two-sided, a p-value < 0.05 was considered as statistically significant. Because we included partially paired samples (e.g. sample from primary tumor and metastasis from the same patient), results were analyzed using an appropriate linear mixed model (LMM) with random intercept for the continuous variable (H-score) and a logistic regression model based on generalized estimating equations (GEE) for the categorical variables (low/high positive versus negative expression). Only the results with a significant difference for both analyses are discussed further. An exception was made for BRD9, for which only continuous variables were available. Spearman's correlation was applied to analyze the relationship between biologically related proteins (YAP1 and TAZ, MAGE-A4 and NY-ESO-1).
We investigated the prognostic impact of targets in primary tumors only, since recurrent and metastatic tumors were more heterogeneous with respect to disease course, treatment and outcome. Survival data were analyzed with Cox regression and the Kaplan-Meier method, using the log-rank test for comparisons. OS was calculated as the interval between the date of sample resection and the date of death in years. For patients who were alive at the time of the analysis, the date of last follow-up was used to censor for OS. Patient lost to follow-up were censored at the moment of the last visit. Disease-free survival (DFS) was defined as the interval between the date of sample collection to the date of local and/or distant recurrence in years.
Results
Our SynSa TMA consisted of 270 cores from 91 clinical samples from archival, paraffin-embedded material originating from 78 individual donors with a male-to-female ratio of 1.17. Patients had a median age of 36 years at diagnosis. There were 12 patients from whom more than one sample was included in the TMA (15 %), we collected two samples from 11 patients and three samples from one patient from different stages of the disease. The majority of samples were collected from the primary tumor (64 %), followed by samples from a metastatic lesion (23 %) or a local recurrence (13 %). The diagnosis of SynSa was verified by a reference pathologist (RS) in the donor sample. The chromosomal translocation was proven in 43 % of the cases by FISH, by karyotyping (in 37 % of the samples) or RT-PCR (12 %). For some samples (8 %), FISH, karyotyping or RT-PCR could not be performed in the routine diagnostics. In these cases, IHC for SSX and SS18-SSX was used to confirm the diagnosis of SynSa (Table 2). Overall, all 91 samples of the TMA stained positive for SSX and 96 % of the samples stained positive for SS18-SSX. Samples that stained negative for SS18-SSX (4 %), had a proven SS18-SSX fusion by RT-PCR or by karyotyping in the past.
Table 2.
Clinicopathological information of samples included in synovial sarcoma tissue microarray and used for further analyses.
| Total | Percentage | |
|---|---|---|
| Number of patients | 78 | |
| Number of samples | 91 | |
| Number of samples/patient | ||
| 1 sample | 66/78 | 85 % |
| 2 samples | 11/78 | 14 % |
| 3 samples | 1/78 | 1 % |
| Clinical information of patients | ||
| Gender | ||
| Male | 42/78 | 54 % |
| Female | 36/78 | 46 % |
| Median age at diagnosis in years with range | 36 years (12–85 years) | |
| Development of disease recurrence (metastasis/local relapse) | 49/78 | 63 % |
| Outcome | ||
| Death of disease | 37/78 | 47 % |
| No evidence of disease | 34/78 | 44 % |
| Alive with disease | 3/78 | 4 % |
| Lost to follow-up | 4/78 | 5 % |
| Median overall survival in months with range | 75.5 months (7–452 months) | |
| Clinicopathological information of the samples | ||
| Presence of the translocation was confirmed with | ||
| FISH | 39/91 | 43 % |
| Karyotyping | 34/91 | 37 % |
| RT-PCR | 11/91 | 12 % |
| IHC, positivity for SS18-SSX staining | 7/91 | 8 % |
| Origin of sample | ||
| Primary tumor | 58/91 | 64 % |
| Metastasis | 21/91 | 23 % |
| Local recurrence | 12/91 | 13 % |
| Grading (FNCLCC) – only including primary tumors | ||
| Grade 3 | 21/58 | 36 % |
| Grade 2 | 15/58 | 26 % |
| Not specified | 15/58 | 26 % |
| Not applicable because of neoadjuvant therapy | 7/58 | 12 % |
| Morphology – only including primary tumors | ||
| Monophasic | 25/58 | 43 % |
| Biphasic | 16/58 | 28 % |
| Not specified | 17/58 | 29 % |
| Median tumor size– only including primary tumors (range) | 5.9 cm (0.5–28.5 cm) | |
| Localization of sample | ||
| Lower extremities | 42/91 | 46 % |
| Thoracic region | 29/91 | 32 % |
| Upper extremities | 9/91 | 10 % |
| Abdominal region | 6/91 | 7 % |
| Vertebrae | 4/91 | 4 % |
| Brain | 1/91 | 1 % |
| Pre-treatment of sample | ||
| Treatment-naive | 65/91 | 72 % |
| Systemic therapy alone | 21/91 | 23 % |
| Isolated limb perfusion | 2/91 | 2 % |
| Systemic therapy+ radiotherapy on site of sampling | 2/91 | 2 % |
| Radiotherapy on site of sampling | 1/91 | 1 % |
Abbreviations: FISH: fluorescence in situ hybridization RT-PCR: reverse transcription polymerase chain reaction, IHC: immunohistochemistry, FNCLCC: Fédération Nationale des Centres de Lutte Contre le Cancer.
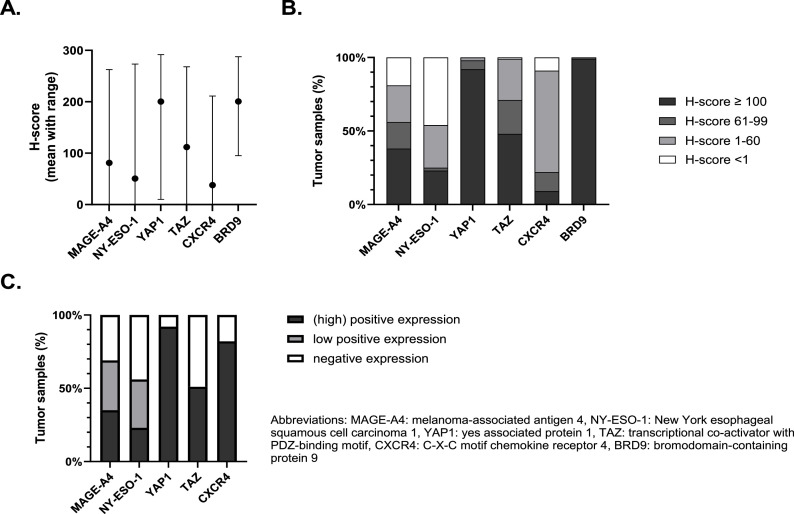
An example of a core stained for the different proteins of interest is shown in Supplementary Figure 1. MAGE-A4 and NY-ESO-1 were localized in nucleus and cytoplasm. YAP1 and TAZ were localized in nucleus and cytoplasm, however as previously mentioned only nuclear localization was counted. CXCR4 staining was predominantly membranous, only 4 samples showed nuclear localization of CXCR4. BRD9 staining was mainly nuclear, sometimes combined with a weaker cytoplasmatic signal. SSX and SS18-SSX staining was observed in the nucleus only. The expression of the proteins of interest in the cores was initially assessed using an H-score, resulting in a continuous variable ranging from 0 to 300. The average H-score of MAGE-A4, NY-ESO-1, TAZ and CXCR4 was 81, 51, 112 and 38, respectively. Our SynSa samples clearly expressed higher levels of YAP1 and BRD9 as compared to the other proteins with an average H-score of 200 and 201, respectively (Fig. 1A). For YAP1, BRD9 and TAZ the majority of the samples had an H-score ≥ 100. In contrast, for CXCR4 most samples had an H-score between 1 and 60. A relevant proportion of the samples did not express MAGE-A4 (19 %) and NY-ESO-1 (46 %), (Fig. 1B). Supplementary Figure 2A shows a scatterplot of the proteins of interest that are biologically related. There was a strong and significant correlation between the H-score of both CTAs: NY-ESO-1 and MAGE-A4 (Spearman correlation coefficient 0.56; p-value < 0.0001). Similarly, there was a strong and significant correlation between the H-score of YAP1 and TAZ, both proteins are important modulators of the Hippo pathway (Spearman correlation coefficient 0.49; p-value < 0.0001).
Fig. 1.
Overview of the H-scores distribution to assess the expression of proteins of interest in a synovial sarcoma tissue microarray (A and B). Expression profile of the target proteins using cut-offs previously defined in literature (C). A. The mean H-scores of the proteins of interest with ranges are summarized as bar charts. B. The distribution of the H-scores of proteins of interest divided in 4 categories (<1, 1–60, 61–99 and ≥100). C. Expression using categorical cut-offs previously defined in literature.
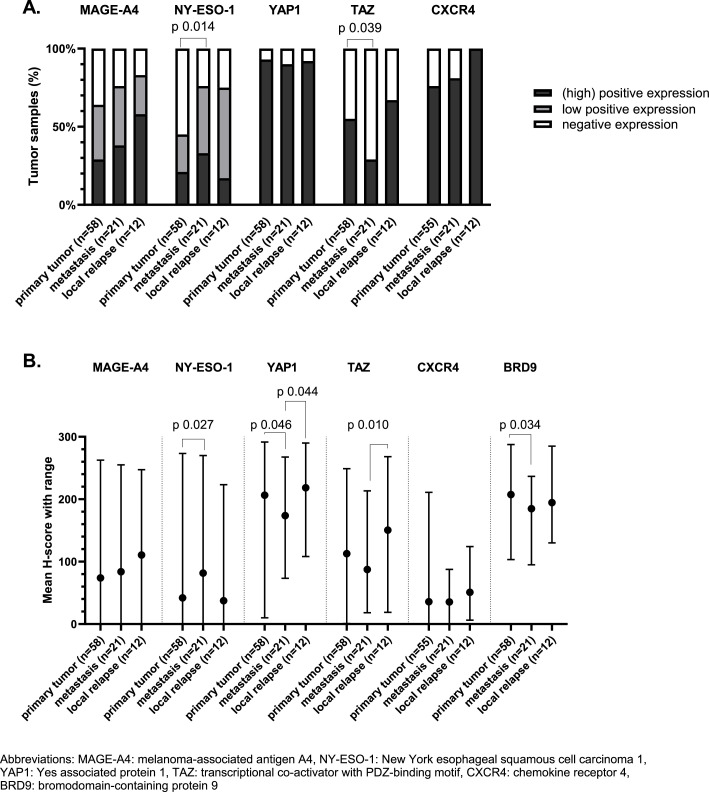
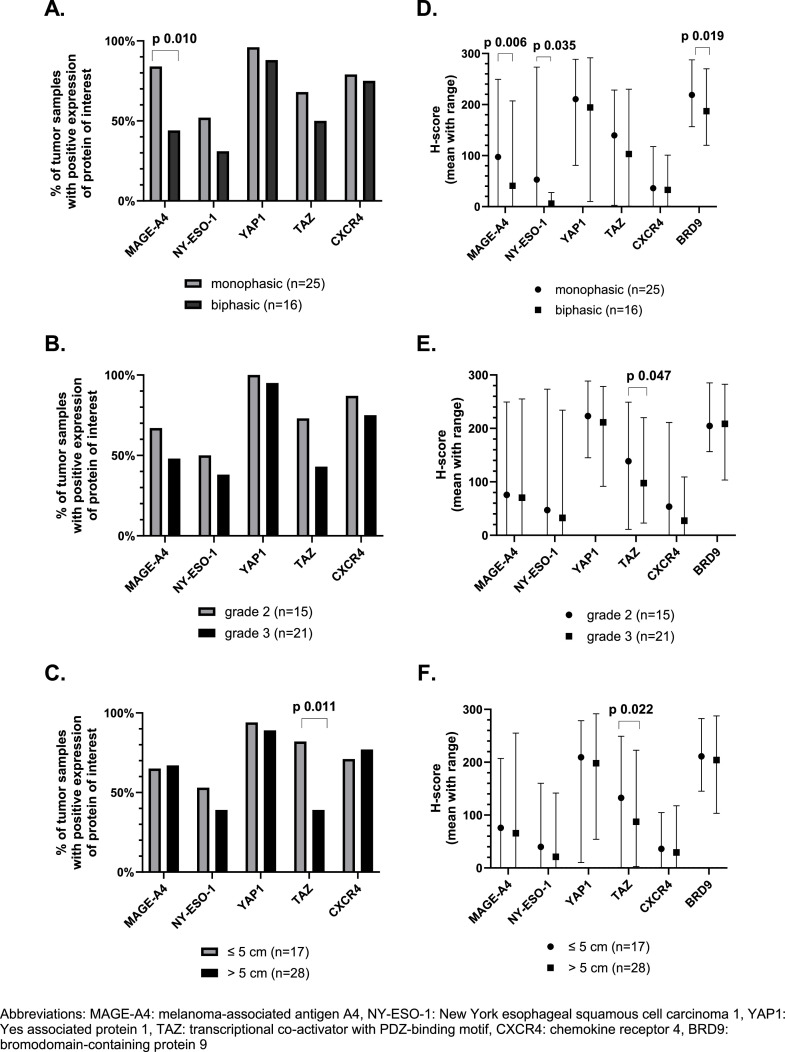
If we use the predefined cut-offs to assess expression, we noticed that positive (high and low) expression of MAGE-A4 and NY-ESO-1 was found in 69 % and 56 % of TMA samples. High expression was shown in 35 % and 23 % of all cases for MAGE-A4 and NY-ESO-1, respectively (Fig. 1c). In 77 % of the samples there was at least one of these CTAs expressed. Additionally, it is important to notice that NY-ESO-1 and MAGE-A4 were homogeneously expressed within the cores originating from the same donor sample (supplementary figure 2B). NY-ESO-1 was significantly higher expressed in samples collected from metastatic lesions as compared to samples from primary tumors (Fig. 2a and 2b). Spearman's correlation between the first and second sample from the same patient showed a positive correlation for NY-ESO-1 expression, however this was not statistically significant (Supplementary Figure 3). The expression of MAGE-A4 was slightly higher in samples originating from a metastasis or local relapse as compared to samples from a primary tumor, however this could not be statistically proven (Fig. 2a). There was a statistically significant and strong positive correlation between the expression of MAGE-A4 in the first and second sample (Spearman's correlation coefficient 0.65; (95 % confidence interval: 0.11; 0.90); p-value 0.025) (Supplementary Figure 3). The effect of size, grade and morphology on the expression of MAGE-A4 and NY-ESO-1 in primary tumors was evaluated and it should be noted that MAGE-A4 was significantly lower expressed in samples with a biphasic tumor morphology (Fig. 3a and 3d).
Fig. 2.
Protein expression correlated with origin of the sample (primary tumor, metastasis or local relapse), using cut-offs previously defined in literature (A) and H-score (B). A. Expression profile of the target proteins in our synovial sarcoma tissue microarray in correlation with origin of sample (primary tumor, metastasis or local relapse), using cut-offs previously defined in literature. Generalized estimating equations were used for statistical analyses. Only statistically significant differences are displayed (p-value < 0.05). B. Expression of proteins of interested assessed using H-score. The mean H-score with range of the proteins of interest is shown in correlation with origin of sample. Linear mixed model was used for the statistical analyses, only statistically significant differences are marked (p-value < 0.05).
Fig. 3.
Expression profile of the target proteins of interest in synovial sarcoma tissue microarrays using cut-offs previously described in literature (A-C) and mean H-score with range (D-F) and correlation with clinicopathological features of treatment-naive primary tumor samples (morphology, grade and size). A-C. Expression profile of the target proteins in our synovial sarcoma tissue microarrays using cut-offs described in literature in correlation with morphology, grade and size. Only treatment-naïve primary tumor samples are included. Generalized estimating equations were used for statistical analyses and only statistically significant differences are marked (p-value < 0.05). D-F. Expression of proteins of interest assessed using H-score. The mean H-score with range of the proteins of interest is displayed. Linear mixed model was used for the statistical analyses, only statistically significant differences are marked (p-value < 0.05).
Regarding the components of the Hippo pathway, YAP1 was predominantly expressed in SynSa. Positive nuclear expression of YAP1 was seen in 92 % of the samples. Positive nuclear expression of TAZ was seen in 51 % of the samples (Fig. 1c). YAP1 was more homogeneously expressed within the cores corresponding to the same sample as compared to TAZ (Supplementary Figure 2B). The average H-score of YAP1 and TAZ was lower in metastatic samples as compared to samples from a primary tumor or local relapses, however this was not statistically significant for both analyses (Fig. 2a and 2b). There was no statistically significant correlation between the expression of YAP1 and TAZ in the first and second sample (Supplementary Figure 3). Regarding the characteristics of the primary tumor, the expression of TAZ was significantly lower in samples originating from larger tumors (Fig. 3c and 3f).
BRD9 was highly expressed in all tumor samples (with a minimum H-score of 95). No categorical cut-off has been described in literature for this target, the H-score as a continuous variable was used to assess correlations with clinicopathological features. The average H-score of BRD9 in metastatic samples was significantly lower than the H-score in primary tumor samples (p-value 0.034). Additionally, the H-score of BRD9 in biphasic tumors was significantly lower as compared to tumors with a monophasic morphology (p-value 0.019). Positive expression of CXCR4 was seen in 82 % of the samples, but the average H-score of CXCR4 was low (38) (Fig. 1). No correlation with the investigated clinicopathological features was observed.
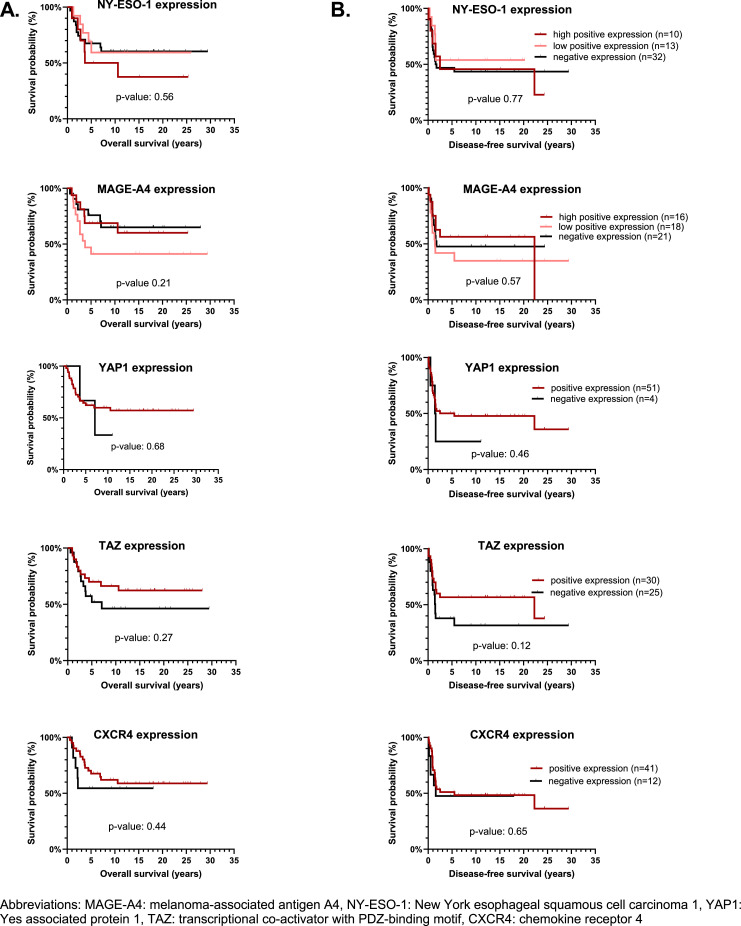
Expression of NY-ESO-1, MAGE-A4, YAP1, TAZ, BRD9 and CXCR4 was not significantly associated with survival (OS or DFS) and thus no prognostic role could be identified for any of the investigated proteins (Fig. 4 and Supplementary Table 2).
Fig. 4.
Overall survival (A) and disease-free survival curves (B) in regard to the expression of the proteins of interest. We investigated the prognostic impact of targets in primary tumors only, since recurrent and metastatic tumors were more heterogeneous with respect to disease course and treatment. Only patients treated with curative intent (n = 55) were included. Survival data were analyzed with the Kaplan-Meier method, using log-rank test to compare. For none of the proteins of interest a statistically significant difference between the 2 curves was observed. For CXCR4, samples from 2 patients were unevaluable (= no tissue was present in the cores on the tissue microarray slide) and as such not included.
Discussion
SynSa is one of the more common subtypes of STS and accounts for about 10 % of all STS. Although it is considered as more sensitive to chemotherapy as compared to other subtypes of STS, the outcome of patients with metastatic disease is poor [5,6]. We need more insight on altered signaling pathways and expression profiles of potential targets in SynSa to develop new therapies. Our SynSa TMA analyses provides real-world data on the expression of novel, potentially actionable targets which are currently being tested in clinical and preclinical settings.
We created a TMA consisting of 91 clinical samples from archival, paraffin-embedded material originating from 78 individual donors, with proven diagnosis of SynSa. The presence of the specific SS18-SSX fusion protein was also checked with IHC using two recently developed antibodies: the SS18-SSX fusion-specific and SSX C-terminus antibodies, which are both highly sensitive and specific for SynSa. Although they are not yet used in clinical practice, IHC using the SS18-SSX antibody could replace molecular genetic or cytogenetic testing in most cases. In our TMA, the SSX staining was positive in all samples and SS18-SSX staining was positive in 96 % of the samples. This is comparable with previously reported data [36].
The novel developed engineered TCR therapies targeting MAGE-A4 and NY-ESO-1 are promising. However, little is known about the variability in expression in SynSa tumors. Our observations in a large cohort of tumors confirmed that the CTAs MAGE-A4 and NY-ESO-1 are highly expressed in SynSa. In previous reports expression of MAGE-A4 ranged from 53 to 82 % [[9], [10], [11],14]. It is important to mention that the antibodies used to assess the expression of MAGE-A4 in previous reports varied. MAGE-A4 antibody (E701U) proved to only bind to MAGE-A4. On the other hand, the frequently used MAGE-A4 antibody (OTI1F9) cross-reacts with MAGE-A3, -A6, -A8, -A10, -A11 and -A12 [37]. We used the E701U clone, which can explain the relatively low positive expression in our cohort (69 %). For NY-ESO-1 previously reported expression rates ranged between 70 and 80 % [9,10,12,18]. Compared to previous reports the expression of NY-ESO-1 in our data set was relatively low (56 %). This can be explained by the fact that our cohort mainly consisted of primary tumors. As we described in our cohort, NY-ESO-1 was statistically higher expressed in metastatic samples as compared to samples of the primary tumor. In other tumor types such as melanoma or renal cell carcinoma, it was also shown that NY-ESO-1 is higher expressed in metastatic tumors in comparison to primary lesions [[38], [39], [40]]. The increasing expression of NY-ESO-1 in metastatic tumors is clinically relevant as these patients would benefit the most from these novel therapies. It is important to mention that in 77 % of the samples there was at least one CTA expressed, which implies many patients with SynSa might be potential candidates for one of the novel therapies targeting these CTAs. Apart from the expression of CTAs, patients should also express HLA-A*02. We could not study the HLA subtype, however Rosenbaum et al. screened patients with metastatic SynSa and 39 % of the patients had an HLA-A*02 subtype (29/75) [41].
The expression of MAGE-A4 and NY-ESO-A in SynSa is considered homogenous [42] and our data confirmed this observation. This offers the advantage that the expression of these proteins can be accurately assessed by a biopsy. Interestingly, in contradiction with the expression of NY-ESO-1 in SynSa, in the majority of epithelial cancer types this marker is expressed more heterogeneously, which can limit the treatment response to NY-ESO-1 targeted therapy in other tumor types [43]. Researchers of the NCT01343043 trial investigated the value of NY-ESO-1 as a predictive biomarker. Post-hoc analyses showed no significant impact of NY-ESO-1 expression on response, however patients in this cohort were only included if their tumor sample expressed NY-ESO-1 (defined as high or low positive expression), so no patients of which the sample did not express NY-ESO-1 were evaluated [35]. Contradictory results regarding the prognostic role of NY-ESO-1 and MAGE-A4 expression in different subtypes of STS have been described and these observations are limited by the small sample size [11,44]. No prognostic value could be identified for NY-ESO-1 and MAGE-A4 in our cohort, however this needs to be validated on a larger group of SynSa specimens. It will be important to clarify this question if a phase III study with these TCR therapies would start. If the novel therapy would show a survival benefit, the question might rise if this is due to the treatment effect or the protein expression.
The largest cohort of YAP1 and TAZ expression in STS was examined by Isfort et al. Nuclear YAP1 IHC-positivity was most prevalently detected in SynSa (78 %; 51/65) and myxoid liposarcoma (91 %; 77/85) samples. Nuclear TAZ immunoreactivity was detected in 34 % (22/65) of the SynSa tissue specimens. In our cohort YAP1 and TAZ were expressed in 92 % and 51 % of the samples, respectively. YAP1 was shown to be the predominantly activated Hippo signaling effector in SynSa [27]. The aberrant YAP1/TAZ activity in SynSa might suggest a therapeutic target in this sarcoma subtype. Several preclinical studies are currently investigating YAP1/TAZ inhibition in vitro and in vivo [26,27]. In our cohort, the expression of TAZ was significantly lower in samples originating from larger primary tumors. A correlation of YAP1/TAZ expression with clinical data has not been reported before in SynSa, so no conclusion can be drawn from this observation. The role of YAP1/TAZ in STS and more specifically SynSa needs to be further investigated.
BRD9 forms a complex with the SS18-SSX fusion protein in the nucleus and is highly expressed in all SynSa samples. There are no previous reports available describing the BRD9 expression assessed by IHC in SynSa samples. The average H-score of BRD9 in our TMAs is lower in metastatic tumors as compared to primary tumors and in tumor samples with a biphasic morphology as compared to monophasic SynSa samples. Also, these findings need to be validated on a larger sample cohort.
CXCR4 was expressed in 82 % of the SynSa samples, however the expression level was low with an average H-score of 38. We could not find a difference in the frequency of expression between primary and metastatic site. The expression on samples from a local relapse was higher than the expression on primary tumors or metastasis, however this could not be statically proven as the sample sizes were small (n = 12). Previous studies investigating the CXCR4 expression in osteosarcoma and STS used CXCR4 antibodies resulting in nuclear and cytoplasmatic staining. This expression pattern is contradictory with the current understanding of CXCR4 signaling and its role in migration of cancer cells [28,31]. We used the previously validated monoclonal anti-CXCR4 antibody UMB2, which led to highly effective plasma membrane staining of the tumor cells [45]. The role of CXCR4 in chemoresistance could not be studied as only a limited number of patients included in the TMA were treated with adjuvant chemotherapy (n = 3).
A key advantage of using TMAs to study the protein expression in SynSa is the possibility to investigate multiple SynSa samples at the same time under uniform experimental conditions. This allows us to rapidly evaluate proteins of interest in many cases and saving precious tissue samples of this rare disease [7]. To ensure the heterogeneity of the tumor was captured and avoid sampling bias, multiple cores were taken from different regions of the tumor sample. IHC is the easiest and the most commonly used approach to evaluate protein expressions. Additionally, IHC also provides information on the localization of the protein. However, several factors may affect the reproducibility and comparability of IHC including tissue fixation, age, storage of the FFPE blocks, the choice of the antibody, the subjectivity of scoring the staining and the threshold used for interpretation of positive immunostaining [[46], [47], [48]]. The defined threshold for positive immunostaining also varies within different publications, it would be beneficial if researchers use a semiquantitative score, such as the H-score, which might facilitate the comparison of the results. Additionally, it should be noted that our samples are heterogeneous. Some of them were pretreated, which might influence the biology and the expression of the investigated proteins. Ideally, all results must be confirmed on a larger cohort of samples. However, we need to consider that such study may not be feasible because SynSa is a rare disease and the availability of samples is limited.
In conclusion, this study is one of the first to provide real-world data on the expression of several targetable proteins in a wide range of SynSa samples. MAGE-A4, NY-ESO-1, YAP1, TAZ, CXCR4 and BRD9 were expressed in a clinically relevant proportion of SynSa cases represented in our TMA, which supports the utility of these antigens as targets for novel therapies in this disease. No prognostic role could be identified for any of the investigated proteins and these findings need to be validated on a larger sample size.
CRediT authorship contribution statement
Lore De Cock: Writing – review & editing, Writing – original draft, Visualization, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Flavia Paternostro: Writing – review & editing, Validation, Investigation, Formal analysis. Ulla Vanleeuw: Writing – review & editing, Methodology. Karo Wyns: Writing – review & editing, Methodology. Annouschka Laenen: Writing – review & editing, Validation, Formal analysis. Che-Jui Lee: Writing – review & editing, Methodology. Raf Sciot: Writing – review & editing, Validation, Resources, Methodology, Conceptualization. Agnieszka Wozniak: Writing – review & editing, Validation, Supervision, Resources, Methodology, Conceptualization. Patrick Schöffski: Writing – review & editing, Validation, Supervision, Resources, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to acknowledge the Translational Research Unit of the University of Bern, Switzerland for their help in the TMA construction. The authors are grateful for the donation of a generous gift from a private donor to support our research on the topic of SynSa. We want to thank the following surgeons involved in sample collection: I. De Wever, I. Samson†, D. Van Raemdonck, F. Sinnaeve, D. Hompes, M. Stas, P. De Leyn and H. Wafa. We are grateful for the help of the following geneticists involved in the diagnostic analyses of the samples: M. Debiec-Rychter, B. Peeters, and I. Vanden Bempt.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102057.
Appendix. Supplementary materials
References
- 1.de Pinieux G., Karanian M., Le Loarer F., Le Guellec S., Chabaud S., Terrier P., et al. Nationwide incidence of sarcomas and connective tissue tumors of intermediate malignancy over four years using an expert pathology review network. PLoS. One. 2021;16(2) doi: 10.1371/journal.pone.0246958. February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thway K., Fisher C. Synovial sarcoma: defining features and diagnostic evolution. Ann. Diagn. Pathol. 2014;18(6):369–380. doi: 10.1016/j.anndiagpath.2014.09.002. December. [DOI] [PubMed] [Google Scholar]
- 3.Sultan I., Rodriguez-Galindo C., Saab R., Yasir S., Casanova M., Ferrari A. Comparing children and adults with synovial sarcoma in the Surveillance, Epidemiology, and End Results program, 1983 to 2005: an analysis of 1268 patients. Cancer. 2009;115(15):3537–3547. doi: 10.1002/cncr.24424. Aug 1. [DOI] [PubMed] [Google Scholar]
- 4.Song S., Park J., Kim H.J., Kim I.H., Han I., Kim H.S., et al. Effects of adjuvant radiotherapy in patients with synovial sarcoma. Am. J. Clin. Oncol. 2017;40(3):306–311. doi: 10.1097/COC.0000000000000148. June. [DOI] [PubMed] [Google Scholar]
- 5.Blay J.Y., von Mehren M., Jones R.L., Martin-Broto J., Stacchiotti S., Bauer S., et al. Synovial sarcoma: characteristics, challenges, and evolving therapeutic strategies. ESMO Open. 2023;8(5) doi: 10.1016/j.esmoop.2023.101618. October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riedel R.F., Jones R.L., Italiano A., Bohac C., Thompson J.C., Mueller K., et al. Systemic anti-cancer therapy in synovial sarcoma: a systematic review. Cancers. (Basel) 2018;10(11) doi: 10.3390/cancers10110417. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C.J., Wozniak A., Van Cann T., Timmermans I., Wellens J., Vanleeuw U., et al. Establishment of an Academic Tissue Microarray Platform as a Tool for Soft Tissue Sarcoma Research. Sarcoma. 2021 doi: 10.1155/2021/6675260. August. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fratta E., Coral S., Covre A., Parisi G., Colizzi F., Danielli R., et al. The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol. Oncol. 2011;5(2):164–182. doi: 10.1016/j.molonc.2011.02.001. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iura K., Maekawa A., Kohashi K., Ishii T., Bekki H., Otsuka H., et al. Cancer-testis antigen expression in synovial sarcoma: NY-ESO-1, PRAME, MAGEA4, and MAGEA1. Hum. Pathol. 2017;61:130–139. doi: 10.1016/j.humpath.2016.12.006. March. [DOI] [PubMed] [Google Scholar]
- 10.Iura K., Kohashi K., Ishii T., Maekawa A., Bekki H., Otsuka H., et al. MAGEA4 expression in bone and soft tissue tumors: its utility as a target for immunotherapy and diagnostic marker combined with NY-ESO-1. Virchows Arch. 2017;471(3):383–392. doi: 10.1007/s00428-017-2206-z. September. [DOI] [PubMed] [Google Scholar]
- 11.Kakimoto T., Matsumine A., Kageyama S., Asanuma K., Matsubara T., Nakamura T., et al. Immunohistochemical expression and clinicopathological assessment of the cancer testis antigens NY-ESO-1 and MAGE-A4 in high-grade soft-tissue sarcoma. Oncol. Lett. 2019;17(4):3937–3943. doi: 10.3892/ol.2019.10044. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai J.P., Rosenberg A.Z., Miettinen M.M., Lee C.C. NY-ESO-1 expression in sarcomas: a diagnostic marker and immunotherapy target. Oncoimmunology. 2012;1(8):1409–1410. doi: 10.4161/onci.21059. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Angelo S.P., Araujo D.M., Abdul Razak A.R., Agulnik M., Attia S., Blay J.Y., et al. Afamitresgene autoleucel for advanced synovial sarcoma and myxoid round cell liposarcoma (SPEARHEAD-1): an international, open-label, phase 2 trial. Lancet. 2024;403(10435):1460–1471. doi: 10.1016/S0140-6736(24)00319-2. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong D.S., Van Tine B.A., Biswas S., McAlpine C., Johnson M.L., Olszanski A.J., et al. Autologous T cell therapy for MAGE-A4(+) solid cancers in HLA-A*02(+) patients: a phase 1 trial. Nat. Med. 2023;29(1):104–114. doi: 10.1038/s41591-022-02128-z. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Necochea-Campion R., Zuckerman L.M., Mirshahidi H.R., Khosrowpour S., Chen C.S., Mirshahidi S. Metastatic biomarkers in synovial sarcoma. Biomark. Res. 2017 doi: 10.1186/s40364-017-0083-x. February;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Galarza F.F., McCabe A., Santos E., Jones J., Takeshita L., Ortega-Rivera N.D., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic. Acids. Res. 2020;48(1):783–788. doi: 10.1093/nar/gkz1029. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins P.F., Kassim S.H., Tran T.L., Crystal J.S., Morgan R.A., Feldman S.A., et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 2015;21(5):1019–1027. doi: 10.1158/1078-0432.Ccr-14-2708. Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Angelo S.P., Melchiori L., Merchant M.S., Bernstein D., Glod J., Kaplan R., et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov. 2018;8(8):944–957. doi: 10.1158/2159-8290.CD-17-1417. August. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tawbi H.A., Burgess M., Bolejack V., Van Tine B.A., Schuetze S.M., Hu J., et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017;18(11):1493–1501. doi: 10.1016/S1470-2045(17)30624-1. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maki R.G., Jungbluth A.A., Gnjatic S., Schwartz G.K., D'Adamo D.R., Keohan M.L., et al. A pilot study of anti-CTLA4 antibody Ipilimumab in patients with synovial sarcoma. Sarcoma. 2013 doi: 10.1155/2013/168145. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brien G.L., Remillard D., Shi J., Hemming M.L., Chabon J., Wynne K., et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife. 2018 doi: 10.7554/eLife.41305. November;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins M., Wan J., Garbitt-Amaral V., et al. Preclinical validation of target engagement assay and investigation of mechanistic impacts of FHD-609, a clinical-stage BD9 degrader being developed for the treatment of synovial sarcoma. Connective Tissue Oncology Society Annual Meeting; Vancouver, Canada; 2022. [Google Scholar]
- 23.Jackson K.L., Agafonov R.V., Carlson M.W., Chaturvedi P., Cocozziello D., Cole K., et al. Abstract ND09: the discovery and characterization of CFT8634: a potent and selective degrader of BRD9 for the treatment of SMARCB1-perturbed cancers. Cancer Res. 2022;82(12_Supplement) doi: 10.1158/1538-7445.Am2022-nd09. [DOI] [Google Scholar]
- 24.Fu M., Hu Y., Lan T., Guan K.L., Luo T., Luo M. The Hippo signalling pathway and its implications in human health and diseases. Signal. Transduct. Target. Ther. 2022;7(1) doi: 10.1038/s41392-022-01191-9. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fullenkamp C.A., Hall S.L., Jaber O.I., Pakalniskis B.L., Savage E.C., Savage J.M., et al. TAZ and YAP are frequently activated oncoproteins in sarcomas. Oncotarget. 2016;7(21):30094–30108. doi: 10.18632/oncotarget.8979. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isfort I., Cyra M., Elges S., Kailayangiri S., Altvater B., Rossig C., et al. SS18-SSX-dependent YAP/TAZ signaling in synovial sarcoma. Clin. Cancer Res. 2019;25(12):3718–3731. doi: 10.1158/1078-0432.CCR-17-3553. June. [DOI] [PubMed] [Google Scholar]
- 27.Isfort I., Elges S., Cyra M., Berthold R., Renner M., Mechtersheimer G., et al. Prevalence of the Hippo effectors YAP1/TAZ in tumors of soft tissue and bone. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-56247-8. December. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmerini E., Benassi M.S., Quattrini I., Pazzaglia L., Donati D., Benini S., et al. Prognostic and predictive role of CXCR4, IGF-1R and Ezrin expression in localized synovial sarcoma: is chemotaxis important to tumor response? Orphanet. J. Rare Dis. 2015;10(1) doi: 10.1186/s13023-014-0222-5. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda Y., Tateishi N., Matono H., Matsuura S., Yamamaoto H., Tamiya S., et al. Chemokine receptor CXCR4 expression is correlated with VEGF expression and poor survival in soft-tissue sarcoma. Int. J. Cancer. 2009;124(8):1852–1859. doi: 10.1002/ijc.24128. April. [DOI] [PubMed] [Google Scholar]
- 30.Kim R.H., Li B.D., Chu Q.D. The role of chemokine receptor CXCR4 in the biologic behavior of human soft tissue sarcoma. Sarcoma. 2011 doi: 10.1155/2011/593708. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda Y., Yamamoto H., Tamiya S., Matsuda S., Tanaka K., Yokoyama R., et al. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod. Pathol. 2006;19(5):738–745. doi: 10.1038/modpathol.3800587. May. [DOI] [PubMed] [Google Scholar]
- 32.Debnath B., Xu S., Grande F., Garofalo A., Neamati N. Small molecule inhibitors of CXCR4. Theranostics. 2013;3(1):47–75. doi: 10.7150/thno.5376. February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S.Y., Lee C.H., Midura B.V., Yeung C., Mendoza A., Hong S.H., et al. Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the development of murine pulmonary metastases. Clin. Exp. Metastasis. 2008;25(3):201–211. doi: 10.1007/s10585-007-9133-3. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T., Navenot J.M., Stavros R., et al. Abstract LB001: identifying MAGE-A4-positive tumors for SPEAR T-cell therapies in HLA-A*02-eligible patients. Cancer Res. 2022;82 doi: 10.1158/1538-7445.Am2022-lb001. [DOI] [Google Scholar]
- 35.Gyurdieva A., Zajic S., Chang Y.F., Houseman E.A., Zhong S., Kim J., et al. Biomarker correlates with response to NY-ESO-1 TCR T cells in patients with synovial sarcoma. Nat. Commun. 2022;13(1) doi: 10.1038/s41467-022-32491-x. September. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranov E., McBride M.J., Bellizzi A.M., Ligon A.H., Fletcher C.D.M., Kadoch C., et al. A Novel SS18-SSX Fusion-specific Antibody for the Diagnosis of Synovial Sarcoma. Am. J. Surg. Pathol. 2020;44(7):922–933. doi: 10.1097/PAS.0000000000001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giacomazzi G., Liivrand M., Hieta R., Dupuis N., Rondas D., Swatkowski P., et al. 200P Precise tumor & patient selection for CDR404: a bispecific & bivalent MAGE-A4 T cell engager. Annal. Oncol. 2023;34:261–262. doi: 10.1016/j.annonc.2023.09.2923. October. [DOI] [Google Scholar]
- 38.Park T.S., Groh E.M., Patel K., Kerkar S.P., Lee C.C., Rosenberg S.A. Expression of MAGE-A and NY-ESO-1 in primary and metastatic Cancers. J. ImmunOther. 2016;39(1):1–7. doi: 10.1097/CJI.0000000000000101. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giesen E., Jilaveanu L.B., Parisi F., Kluger Y., Camp R.L., Kluger H.M. NY-ESO-1 as a potential immunotherapeutic target in renal cell carcinoma. Oncotarget. 2014;5(14):5209–5217. doi: 10.18632/oncotarget.2101. July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aung P.P., Liu Y.C., Ballester L.Y., Robbins P.F., Rosenberg S.A., Lee C.C. Expression of New York esophageal squamous cell carcinoma-1 in primary and metastatic melanoma. Hum. Pathol. 2014;45(2):259–267. doi: 10.1016/j.humpath.2013.05.029. February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenbaum E., Seier K., Bandlamudi C., Dickson M., Gounder M., Keohan M.L., et al. HLA genotyping in synovial sarcoma: identifying HLA-A*02 and its association with clinical outcome. Clin. Cancer Res. 2020;26(20):5448–5455. doi: 10.1158/1078-0432.CCR-20-0832. October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell G., Pollack S.M., Wagner M.J. Targeting cancer testis antigens in synovial sarcoma. J. ImmunOther Cancer. 2021;9(6) doi: 10.1136/jitc-2020-002072. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas R., Al-Khadairi G., Roelands J., Hendrickx W., Dermime S., Bedognetti D., et al. NY-ESO-1 based immunotherapy of cancer: current perspectives. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.00947. May;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto K., Nishimura S., Ito T., Akagi M. Clinicopathological assessment of cancer/testis antigens NY-ESO-1 and MAGE-A4 in highly aggressive soft tissue sarcomas. Diagnostics. (Basel) 2022;12(3) doi: 10.3390/diagnostics12030733. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer T., Nagel F., Jacobs S., Stumm R., Schulz S. Reassessment of CXCR4 chemokine receptor expression in human normal and neoplastic tissues using the novel rabbit monoclonal antibody UMB-2. PLoS. One. 2008;3(12) doi: 10.1371/journal.pone.0004069. December. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S.W., Roh J., Park C.S. Immunohistochemistry for pathologists: protocols, pitfalls, and tips. J. Pathol. Transl. Med. 2016;50(6):411–418. doi: 10.4132/jptm.2016.08.08. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tauber Z., Stetkova I., Cizkova K. Influence of fixation method and duration of archiving on immunohistochemical staining intensity in embryonic and fetal tissues. Ann. Anat. 2019;224:55–61. doi: 10.1016/j.aanat.2019.03.013. July. [DOI] [PubMed] [Google Scholar]
- 48.Grillo F., Bruzzone M., Pigozzi S., Prosapio S., Migliora P., Fiocca R., et al. Immunohistochemistry on old archival paraffin blocks: is there an expiry date? J. Clin. Pathol. 2017;70(11):988–993. doi: 10.1136/jclinpath-2017-204387. November. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.