Abstract
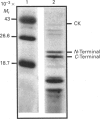
We have identified by protein microsequencing a glutamic acid residue (Glu-166) in a folding intermediate of chick muscle creatine kinase that is very sensitive to cleavage by staphylococcal proteinase V8. Most other glutamic acid residues, including Glu-168, are already partly protected from proteolytic attack at this stage. After the final stages of protein refolding, when enzyme activity is recovered, Glu-166 is also resistant to proteolysis.
Full text
PDF
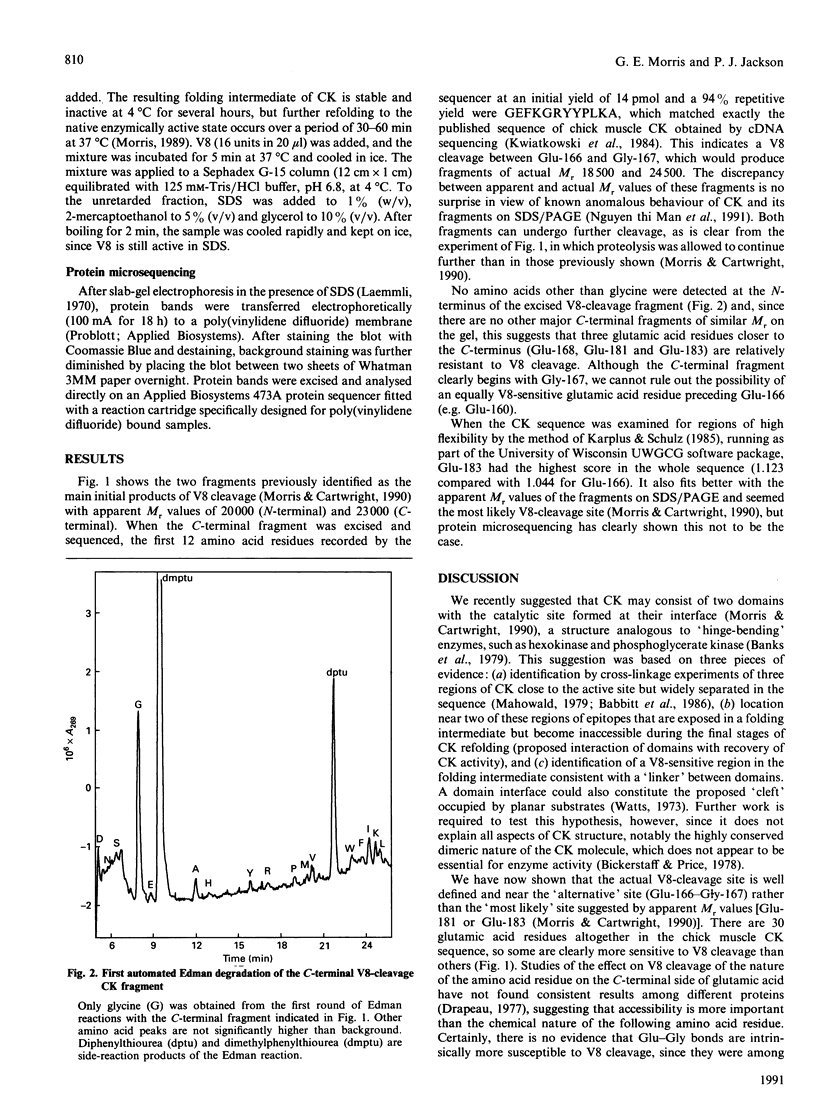

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banks R. D., Blake C. C., Evans P. R., Haser R., Rice D. W., Hardy G. W., Merrett M., Phillips A. W. Sequence, structure and activity of phosphoglycerate kinase: a possible hinge-bending enzyme. Nature. 1979 Jun 28;279(5716):773–777. doi: 10.1038/279773a0. [DOI] [PubMed] [Google Scholar]
- Betton J. M., Desmadril M., Yon J. M. Detection of intermediates in the unfolding transition of phosphoglycerate kinase using limited proteolysis. Biochemistry. 1989 Jun 27;28(13):5421–5428. doi: 10.1021/bi00439a016. [DOI] [PubMed] [Google Scholar]
- Bickerstaff G. F., Price N. C. Creatine kinase: a review of some recent work on the mechanism and subunit behaviour of the enzyme. Int J Biochem. 1978;9(1):1–8. doi: 10.1016/0020-711x(78)90128-3. [DOI] [PubMed] [Google Scholar]
- Drapeau G. R. Cleavage at glutamic acid with staphylococcal protease. Methods Enzymol. 1977;47:189–191. doi: 10.1016/0076-6879(77)47023-x. [DOI] [PubMed] [Google Scholar]
- Eppenberger H. M., Dawson D. M., Kaplan N. O. The comparative enzymology of creatine kinases. I. Isolation and characterization from chicken and rabbit tissues. J Biol Chem. 1967 Jan 25;242(2):204–209. [PubMed] [Google Scholar]
- Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49(2-3):117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski R. W., Schweinfest C. W., Dottin R. P. Molecular cloning and the complete nucleotide sequence of the creatine kinase-M cDNA from chicken. Nucleic Acids Res. 1984 Sep 25;12(18):6925–6934. doi: 10.1093/nar/12.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morris G. E., Cartwright A. J. Monoclonal antibody studies suggest a catalytic site at the interface between domains in creatine kinase. Biochim Biophys Acta. 1990 Jul 6;1039(3):318–322. doi: 10.1016/0167-4838(90)90265-h. [DOI] [PubMed] [Google Scholar]
- Morris G. E. Monoclonal antibody studies of creatine kinase. The ART epitope: evidence for an intermediate in protein folding. Biochem J. 1989 Jan 15;257(2):461–469. doi: 10.1042/bj2570461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen thi Man, Cartwright A. J., Osborne M., Morris G. E. Structural changes in the C-terminal region of human brain creatine kinase studied with monoclonal antibodies. Biochim Biophys Acta. 1991 Jan 29;1076(2):245–251. doi: 10.1016/0167-4838(91)90274-4. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Folding of protein fragments. Adv Protein Chem. 1981;34:61–92. doi: 10.1016/s0065-3233(08)60518-5. [DOI] [PubMed] [Google Scholar]
- Williamson J., Greene J., Chérif S., Milner-White E. J. Heterogeneity of rabbit muscle creatine kinase and limited proteolysis by proteinase K. Biochem J. 1977 Dec 1;167(3):731–737. doi: 10.1042/bj1670731. [DOI] [PMC free article] [PubMed] [Google Scholar]