Abstract
By using replication-defective vectors derived from human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency virus (SIVmac), and murine leukemia virus (MuLV), all of which were pseudotyped with the vesicular stomatitis virus (VSV) G glycoprotein, the efficiency of postentry, early infection events was examined in target cells of several mammalian species. Titers of HIV-1 vectors were significantly lower than those of SIVmac and MuLV vectors in most cell lines and primary cells from Old World monkeys. By contrast, most New World monkey cells exhibited much lower titers for the SIVmac vector compared with those of the HIV-1 vector. Prosimian cells were resistant to both HIV-1 and SIVmac vectors, although the MuLV vector was able to infect these cells. Cells from other mammalian species were roughly equivalent in susceptibility to the three vectors, with the exception of rabbit cells, which were specifically resistant to the HIV-1 vector. The level of HIV-1 vector expression was very low in transduced cells of rodent, rabbit, cow, and pig origin. Early postentry restriction of primate immunodeficiency virus infection exhibits patterns largely coincident with species borders and applies to diverse cell types within an individual host, suggesting the involvement of species-specific, widely expressed cellular factors.
The primate lentiviruses include the human immunodeficiency viruses types 1 and 2 (HIV-1 and HIV-2) and simian immunodeficiency viruses (SIV). HIV-1 and HIV-2 infect humans, HIV-1-like viruses infect chimpanzees, and SIV variants infect African macaques (4, 10, 12, 25, 31). Humans infected by HIV-1 and HIV-2 and Asian macaques infected by certain SIV strains often develop life-threatening immunodeficiency due to depletion of CD4-positive T lymphocytes (17).
The tropism of HIV and SIV isolates is determined by cell-type-specific and species-specific host factors. The entry of these viruses into host cells is dependent upon the surface expression of viral receptors, CD4, and members of the chemokine receptor family (1, 9, 11, 13–16, 18, 34, 43). These receptors bind the gp120 envelope glycoprotein of the virus, promoting virus attachment and fusion of the viral and cellular membranes (59). All primate immunodeficiency viruses can use the CCR5 chemokine receptor (1, 8, 9, 13, 15, 16, 33, 44), as well as other seven-transmembrane-spanning proteins specific to each virus.
The activation state of the host cell represents a second determinant that can influence the efficiency of infection by HIV-1 (54, 62). Although virus entry can occur in resting T lymphocytes expressing the receptors for the virus, subsequent events in the viral life cycle, reverse transcription, or migration of the preintegration complex to the nucleus have been reported to be inefficient in these cells. These steps in the replication of an oncoretrovirus, murine leukemia virus (MuLV), can be dominantly blocked by a host cell factor, the product of the Fv-1 allele (48, 50). In this case, a Gag-like product of the Fv-1 allele, which is apparently derived from an endogenous retroviral sequence, interferes with early, postentry events in MuLV infection (5).
Late events in HIV-1 and SIV replication have also been shown to be dependent upon cellular factors. Transcription of the provirus and nuclear-cytoplasmic transport of viral messages are modulated by cellular proteins that interact with the viral Tat and Rev proteins, respectively (6, 20, 28, 55, 58). Host proteins have also been implicated in the assembly of fully infectious virions. HIV-1 but not SIVmac, for example, selectively incorporates cyclophilin A into viral particles (19, 39, 56). Cyclosporins, functional antagonists of cyclophilin binding to the viral Gag proteins, block the ability of HIV-1, but not SIVmac, to complete an early step in the infection of new host cells (49, 56). The interaction of the viral Vif protein with a species-specific inhibitory cellular factor represents a second example of host involvement in the attainment of virion infectivity (23, 42, 52, 53, 57). Vif-deficient HIV-1 and SIVmac produced in cell types expressing the unidentified inhibitory factor do not efficiently complete reverse transcription in newly infected target cells (29, 52).
HIV-1 replication is restricted to human and ape cells in culture, and although chimpanzees and gibbon apes can be infected by HIV-1, only in rare instances does immunodeficiency result (2, 22, 24, 41). Although the CD4 glycoprotein and chemokine receptors of several monkey species support HIV-1 entry (8, 44), the products of HIV-1 reverse transcription are significantly reduced in rhesus macaque peripheral blood mononuclear cells (PBMC) compared with those in human PBMC (30). Consistent with this observation are studies of chimeric simian-human immunodeficiency viruses (SHIVs) indicating that HIV-1 regions in the 5′ half of the genome are responsible for restricted replication in macaque cells (38, 51). SHIVs containing the HIV-1 reverse transcriptase replicate efficiently in monkeys, indicating that restricted HIV-1 replication in monkeys is not determined by this viral protein (3). The available data do not support a role for Vif or cyclophilin A in this restriction (61).
MATERIALS AND METHODS
Cell lines and primary cell cultures.
In general, adherent cells were grown in Dulbecco modified Eagle medium (DMEM)–10% fetal calf serum (FCS) with antibiotics; suspension cells were grown in RPMI–10% FCS with antibiotics. The rhesus macaque T-lymphocyte cell line 221 (a kind gift from R. Desrosiers) was cultured in RPMI–10% FCS with antibiotics in the presence of 10 U of recombinant human interleukin-2 per ml. Cell lines purchased from the American Type Culture Collection (ATCC) were cultured in the recommended media.
Cell lines used in this study are described in Table 1.
TABLE 1.
Cell lines used in this study
| Group and cell line | Species | Tissue | Source |
|---|---|---|---|
| Humans | |||
| HEK293 | Human | Kidney | ATCC |
| Hut 78 | Human | T lymphocyte | ATCC |
| CEM | Human | T lymphocyte | ATCC |
| CEMx174 | Human | T/B-lymphocyte hybrid | NIH AIDS repository |
| Jurkat | Human | T lymphocyte | NIH AIDS repository |
| U937 | Human | Monocyte | ATCC |
| Raji | Human | B lymphocyte | ATCC |
| SupT1 | Human | T lymphocyte | ATCC |
| MT4 | Human | T lymphocyte | NIH AIDS repository |
| A201 | Human | T lymphocyte | NIH AIDS repository |
| C8166 | Human | T lymphocyte | NIH AIDS repository |
| Molt 4 Clone 8 | Human | T lymphocyte | NIH AIDS repository |
| Apes, CARL | Chimpanzee | B lymphocyte | ATCC |
| Old World monkeys | |||
| Cv-1 | African green monkey | Kidney | ATCC |
| Cos-7 | African green monkey | Kidney | ATCC |
| Vero | African green monkey | Kidney | ATCC |
| Bsc-1 | African green monkey | Kidney | ATCC |
| DBS-FCL1 | African green monkey | Lung | ATCC |
| DBS-FCL2 | African green monkey | Lung | ATCC |
| LCL8664 | Rhesus macaque | B lymphocyte | ATCC |
| 221-89 | Rhesus macaque | T lymphocyte | Ron Desrosiers |
| RF/6A | Rhesus macaque | Eye | ATCC |
| LLC-MK2 | Rhesus macaque | Kidney | ATCC |
| Rh Spleen | Rhesus macaque | Spleen | This study |
| Rh Lung | Rhesus macaque | Lung | This study |
| Rh Skin | Rhesus macaque | Skin | This study |
| Rh Thymus | Rhesus macaque | Thymus | This study |
| New World monkeys | |||
| OMK | Owl monkey | Kidney | ATCC |
| OML | Owl monkey | B lymphocyte | J. Scammell |
| Pindak | Squirrel monkey | Kidney | J. Scammell |
| DPSO | Squirrel monkey | Lung | ATCC |
| Sq Lung | Squirrel monkey | Lung | This study |
| Sq Intestine | Squirrel monkey | Intestine | This study |
| Sq Muscle | Squirrel monkey | Muscle | This study |
| Sq Liver | Squirrel monkey | Liver | This study |
| Sq Kidney | Squirrel monkey | Kidney | This study |
| NZP15 | Golden-headed lion tamarin | Kidney | This study |
| NZP52 | Black-tailed marmoset | Kidney | This study |
| NZP55 | Black-tailed marmoset | Lung | This study |
| NZP60 | Black-tailed marmoset | Kidney | L. Munson, ATCC |
| NZP59 | Goeldi’s marmoset | Lung | This study |
| NZP66 | Goeldi’s marmoset | Kidney | This study |
| Prosimians | |||
| KZ11 | Ring-tailed lemur | Lung | This study |
| KZ12 | Ring-tailed lemur | Kidney | This study |
| Scandents | |||
| NZP89 | Long-nosed tree shrew | Lung | This study |
| TR Brain | Tree shrew | Brain | This study |
| TR Heart | Tree shrew | Heart | This study |
| TR Kidney | Tree shrew | Kidney | This study |
| TR Lung | Tree shrew | Lung | This study |
| TR Spleen | Tree shrew | Spleen | This study |
| Others | |||
| MDCK | Dog | Kidney | ATCC |
| DH82 | Dog | Monocyte | ATCC |
| AK-D | Cat | Lung | ATCC |
| CRFK | Cat | Kidney | ATCC |
| LLCRK1 | Rabbit | Kidney | ATCC |
| AG 04676 | Rabbit | Epithelial | Coriell |
| AG 04677A | Rabbit | Epithelial | Coriell |
| AG 10187 | Rabbit | Epithelial | Coriell |
| ESK4 | Pig | Kidney | ATCC |
| AG 05056 | Pig | Epithelial | Coriell |
| AG 08113 | Pig | Fibroblast, skin | Coriell |
| AG 08114 | Pig | Fibroblast, lung | Coriell |
| AG 12119 | Pig | Fibroblast, skin | Coriell |
| BW5147 | Mouse | Lymphocyte | ATCC |
| BHK21 | Hamster | Kidney | ATCC |
| MBDK | Cow | Kidney | ATCC |
Primary tissue cultures were established by mincing dissected organs, followed by trypsinization for 15 to 30 min at 37°C. Cells were washed twice, filtered through a 40-μm-pore-size tissue culture filter mesh (Falcon) and plated in DMEM–10% FCS, 10 ng of human epidermal growth factor (Gibco) per ml, antibiotics, and antimycotics.
Viral vectors.
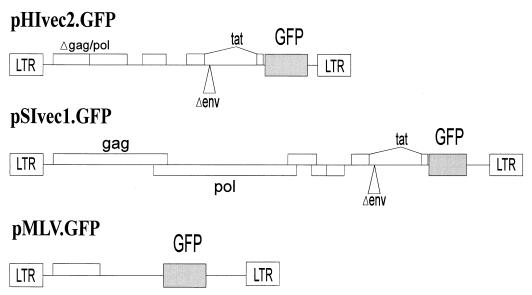
Defective retroviral vectors based on HIV-1, SIVmac, and MuLV, which were capable of expressing the green fluorescent protein (GFP), were constructed. The HIV-1 vector, pHIvec2.GFP, was derived from v653 rtatpC (47) by deleting env and vpu sequences but leaving the Rev-responsive element (HXBc2 nucleotide positions 6094 to 7655) intact and by introduction of a BamHI-XbaI fragment containing the EGFP gene (Clontech) into HXBc2 positions 8474 to 8624. Recombinant viruses were produced by cotransfection of 293T cells with pHIvec2.GFP, pCMVΔP1ΔenvpA (47), pHCMV-G (60), and a Rev-expressing plasmid in a 10:10:2:1 ratio. At 12 h after transfection, the medium was changed. Conditioned medium containing recombinant viruses was harvested and filtered (0.45-μm pore size) 24 h later.
The SIVmac vector, pSIvec1.GFP, was constructed by converting the unique SphI site in a full-length SIVmac239 proviral clone to a SalI site, by introducing a NotI site 3′ to the env stop codon, and then by inserting the SalI-NotI fragment of pHIvec2.GFP. Recombinant viruses were produced by transfection of 293T cells with a 20:2:1 ratio of pSIvec1.GFP, pHCMV-G, and a Rev-expressing plasmid.
The MuLV vector, pMLV.GFP, was made by introduction of the EGFP coding region (Clontech) into pMX (a kind gift from T. Kitamura, DNAX Corporation). Viral particles were produced by cotransfection of 293T cells with pMLV.GFP, pMDgag/pol (an MuLV packaging plasmid kindly supplied by Richard Mulligan), and pHCMV-G in a 10:10:2 ratio.
Infection of cells and detection of GFP expression.
Cells were plated at a density of 2 × 104 cells/well (adherent cells) or 105 cells/well (suspension cells) in 24-well plates. Medium containing recombinant HIV-1 or SIVmac vectors was normalized according to reverse transcriptase and added in serial dilutions to the cells, which were incubated for 5 days. Cells were then trypsinized if necessary, fixed in 3.7% formaldehyde, and analyzed by fluorescence-activated cell sorting (FACS; Becton Dickinson FACscan). The average HIV-1/SIVmac infectivity ratio (IRH/S) ratio, defined in the Results section, and the standard deviation were derived from at least two independent experiments.
For fluorescence microscopy, cells were infected with replication-defective HIV-1, SIVmac, and MuLV vectors expressing GFP. Five days later, fluorescence microscopy was performed by using a fluorescein isothiocyanate (FITC) filter set on a Nikon TE300 inverted microscope.
RESULTS
Infectibility of cells from various primate species by HIV-1, SIV, and MuLV vectors.
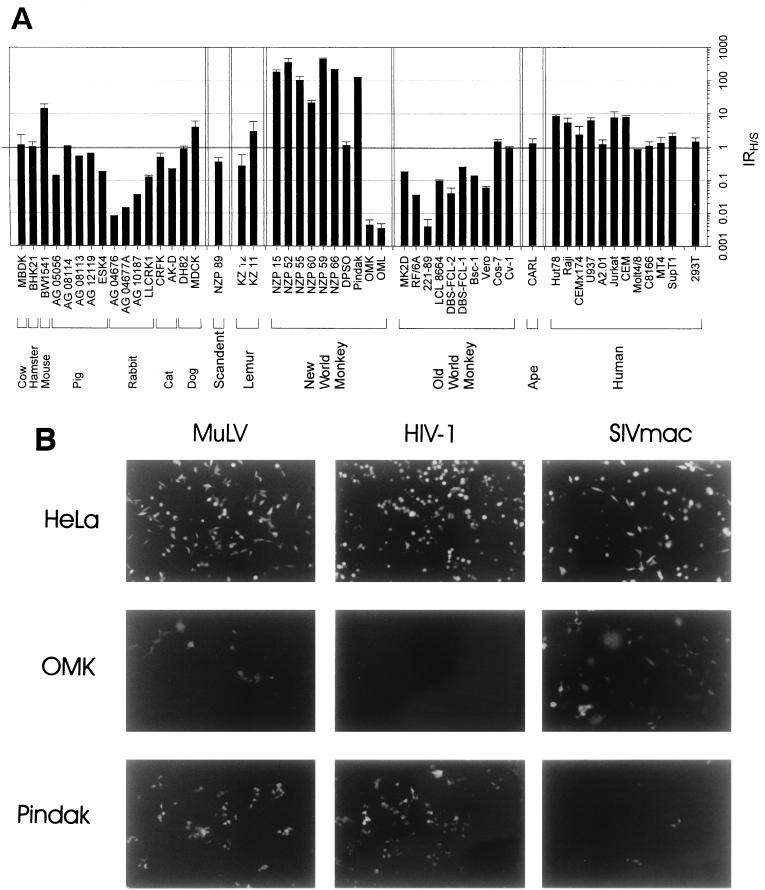
Here we analyze the ability of HIV-1 and SIV to mediate postentry, early steps in virus infection of cells from several mammalian species. Because many factors could potentially modulate the infectibility of diverse cells, we examined in parallel the ability of a MuLV vector to infect these cells, thus providing a standard of reference. We constructed replication-defective vectors containing core elements from HIV-1, SIVmac239, and MuLV strains that were capable of expressing GFP and that were pseudotyped with the G glycoprotein from the vesicular stomatitis virus (VSV) (Fig. 1). The use of GFP as a reporter gene in conjunction with FACS analysis allows the distinction between viral titer (GFP-positive cells/volume of virus-containing supernatant) and viral gene expression (intensity of fluorescence in a GFP-positive cell). The use of the G glycoprotein from the pantropic VSV presumably bypasses any potential restriction on vector entry, allowing postentry events to be studied. Because the overall efficiency of VSV G glycoprotein-mediated entry may differ among cell lines due to differences in the abundance of receptor molecules, the ratios of infectivities of the three viral vectors were examined, as well as the absolute infectivity titers. Serial experiments demonstrated that, even when the latter titers varied among experiments, the infectivity ratios remained relatively constant for a particular target cell. The infectivity ratio is designated IRH/S, which represents the percent GFP-positive cells for the HIV-1 vector divided by that for the SIVmac vector, when equal numbers of cells were exposed to equivalent concentrations of virus (in reverse transcriptase units/milliliter).
FIG. 1.
Vectors used in the study. The HIV-1, SIVmac, and MuLV vectors used in the study are shown. The construction of the vectors is described in Materials and Methods.
The IRH/S values for a number of mammalian cell lines, which are grouped according to species of origin, are shown in Fig. 2. In human cell lines, the IRH/S varied between 1 and 10. Although the A2.01 line is a CEM clone, the IRH/S values for these two lines differed by a factor of 10, indicating that some variation in the relative infectibility of cells by these viruses can occur during tissue culture passage. Nonetheless, the results generally agree with previous observations that human cells support both HIV-1 and SIVmac infection (45). The MuLV vector also efficiently infected the human cell lines on which it was tested (Fig. 3 and data not shown). A chimpanzee lymphocyte line, CARL, also exhibited roughly equivalent infectibility by HIV-1 and SIV vectors (Fig. 2).
FIG. 2.
Infection of cells from different mammals with HIV-1 and SIVmac vectors. (A) Medium containing recombinant HIV-1 or SIVmac vectors was normalized according to reverse transcriptase and added in serial dilutions to the indicated cells, which were incubated for 5 days. Cells were then trypsinized if necessary, fixed in 3.7% formaldehyde, and analyzed by FACS (Becton Dickinson FACscan). The average IRH/S ratio and standard deviation derived from at least two independent experiments are shown. (B) An example of the data used to generate Fig. 2A is shown. Human (HEK293), owl monkey (OMK), and squirrel monkey (Pindak) cells were infected with replication-defective HIV-1, SIVmac, and MuLV vectors expressing GFP. Five days later, fluorescence microscopy was performed.
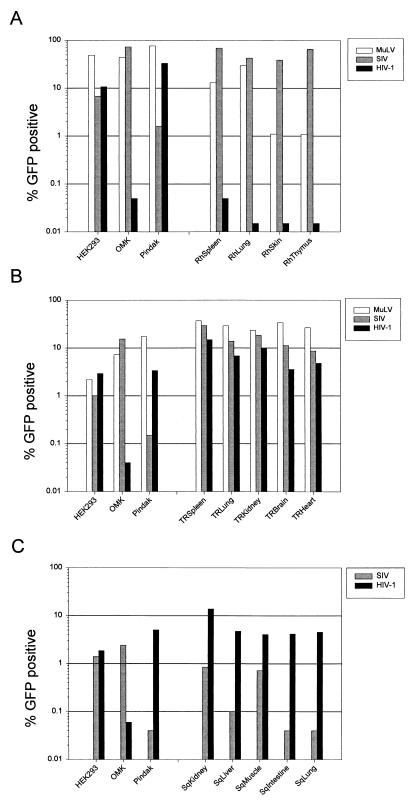
FIG. 3.
Infectibility of primary cells by HIV-1, SIVmac, and MuLV vectors. Primary cells from rhesus monkeys (A), tree shrews (B), and squirrel monkeys (C) were infected with HIV-1, SIVmac, and MuLV vectors as described in Materials and Methods, alongside control cell lines (HEK293, OMK, and Pindak). The infectious titer of each vector in a typical experiment is shown. Although the absolute values of infectivity varied between experiments, the relative infectivity of the vectors was comparable in different experiments.
In contrast to the results in human and chimpanzee cells, low IRH/S values (0.02 to 0.2) were observed in cell lines derived from rhesus macaques, which are Old World monkeys (Fig. 2). Both SIVmac and MuLV vectors infected primary cultures derived from the spleen, lung, skin, and thymus tissues of a rhesus macaque, whereas the infectivity of HIV-1 vectors in these cells was 100- to 8,000-fold lower (Fig. 3). Fluorescence microscopy and FACS analysis revealed that the cells in these early-passage primary cultures exhibited substantial heterogeneity in size, shape, and granularity and that successful infection with the SIVmac and MuLV vectors was not limited to specific subsets thereof (data not shown). These results support the concept that HIV-1 infection is specifically blocked in rhesus macaque cells and argue that any host cell factors governing this susceptibility and/or restriction are widely expressed in various cell types within the species. This block was observed when the HIV-1 vectors were produced in either human 293T cells, which were used for most of the experiments reported herein, or African green monkey COS-1 cells (data not shown).
Low IRH/S values (0.04 to 0.2) were seen in four cell lines derived from African green monkeys, another Old World monkey species (Fig. 2). These cell lines were derived from lung and kidney tissue of the monkeys. The only Old World monkey lines that did not demonstrate low IRH/S values were the African green monkey kidney line, CV-1, and its derivative, COS-1. Even though the CV-1 and COS-1 lines exhibited mitochondrial DNA sequences closely related to those of the other African green monkey lines (data not shown), the IRH/S values were close to 1 in the former cells. It is possible that certain African green monkey species or cell types exhibit variable expression of the factors modulating HIV-1 infectivity or that the expression or functional integrity of these factors was modified during culture of the cells.
All of the currently identified primate immunodeficiency viruses are believed to be of Old World, probably African, origin. To examine the infectibility of cells derived from monkeys not known to harbor lentiviruses, we determined the IRH/S values for several lines derived from New World (South American) monkeys. The IRH/S values were high (20 to 500) in most of the cell lines from squirrel monkeys, black-tailed marmosets, Goeldi’s marmosets, and golden-headed lion tamarins (Fig. 2). Primary cells derived from the kidney, liver, lung, small intestine, and skeletal muscle of a squirrel monkey embryo also exhibited high IRH/S values (Fig. 3). The extent of permissiveness for SIVmac and HIV-1 varied to some extent among the cells derived from different tissues but, in all cases, infection of these Old World monkey cells by SIVmac vectors was inefficient. MuLV infection was efficient in all squirrel monkey cell types that were tested, a finding consistent with the interpretation that a specific block to SIVmac infection exists in these New World monkey cells.
A kidney cell line, OMK, derived from one New World monkey species, the owl monkey, exhibited a very low IRH/S value of 0.006. Both MuLV and SIVmac vectors infected this cell line efficiently, indicating that OMK cells exhibit a restriction specific for HIV-1 infection. The owl monkey origin of the OMK line used in these studies was confirmed by PCR amplification and sequencing of a major histocompatibility complex class I gene fragment (data not shown). A second owl monkey cell line, OML, which was of B-lymphocyte origin, also exhibited a low IRH/S value (Fig. 2). Thus, cells derived from owl monkeys exhibit a different pattern of primate lentivirus restriction than that seen in cells from other New World monkey species.
Primary kidney and lung cells from ring-tailed lemurs, a prosimian species, were equally infectible by HIV-1 and SIVmac vectors, with IRH/S values near 1 (Fig. 2). However, unlike any of the aforementioned cell lines, both HIV-1 and SIVmac infection was very inefficient in these cells. By contrast, some lemur cells were infected efficiently by the MuLV vector. Thus, these prosimian cells exhibit specific restrictions against both HIV-1 and SIVmac infection.
Infectibility of cells from various nonprimate species by HIV-1, SIV, and MuLV vectors.
Because it has been shown that cells from some nonprimate species can be infected by HIV-1 vectors pseudotyped with heterologous envelope glycoproteins (32, 46), restriction against HIV-1 infection must not exist at some evolutionary distance from primates. Thus, we examined cells from tree shrews, which represent the mammalian family of scandents. Tree shrews have several primate characteristics, although most recent analyses recommend their separate classification (21, 40). A cell population derived from the lung of long-nosed tree shrews was found to be efficiently and equally infectible by both HIV-1 and SIVmac vectors (Fig. 2). Primary cell cultures derived from the kidney, spleen, lung, heart, and brain of a newborn tree shrew exhibited similar susceptibility to both HIV-1 and SIVmac vectors, albeit with slight variation in IRH/S values among cells of different tissues (Fig. 3C). As was observed in other primary cultures, a variety of morphologic cell types were capable of being infected by either vector (data not shown). Apparently, tree shrew cells efficiently support infection by primate immunodeficiency virus vectors.
Cell lines from other mammalian species were examined for infectibility by HIV-1 and SIVmac vectors. Several rabbit cell lines did not support high levels of HIV-1 infection, even though infection by the SIVmac and MuLV vectors was efficient. Most of the cells from other mammalian species supported both HIV-1 and SIVmac infection, although some variation in IRH/S values was apparent (Fig. 2).
Level of vector gene expression in target cells from different species.
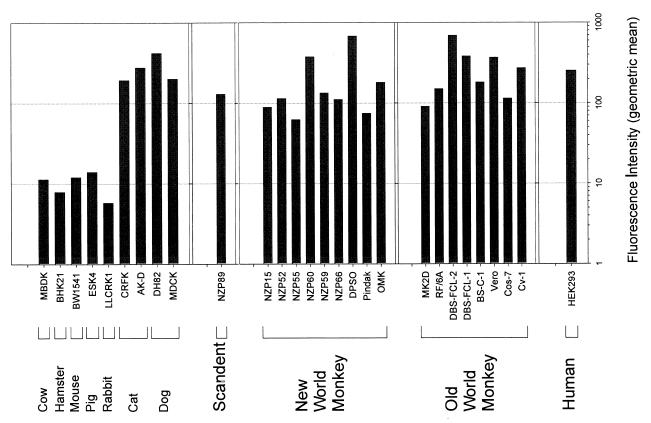
Although nonprimate mammalian cells infected by the HIV-1 vector often exhibited a high percentage of infected (GFP-positive) cells, the level of GFP expression within the infected cells was noted to be low in some cell types. Figure 4 shows the mean fluorescence intensity observed in populations of transduced cells derived from different species. The fluorescence intensity values were comparably high in cells derived from humans, primates, scandents, dogs, and cats (Fig. 4 and data not shown). Fluorescence intensity was more than 10-fold lower in cells from mice, hamsters, rabbits, pigs, and cows. Thus, the cells of certain mammalian species less efficiently support vector gene expression, a result consistent with previous studies of host cell factors (e.g., cyclin T) required for Tat-mediated activation of the viral long terminal repeat (58). Other previously described species-specific factors required for Rev-mediated enhancement of nuclear-cytoplasmic viral RNA transport would probably not apply to the multiply spliced GFP message produced by our vectors (6, 20, 55).
FIG. 4.
Fluorescence intensity of GFP in HIV-1 vector-infected cells. The indicated cells were infected with the HIV-1 vector as described in Materials and Methods. GFP-positive and -negative cells were gated separately, and positive values were normalized for background fluorescence. The values shown represent the geometric mean (fold above background).
Effect of multiplicity of infection on infectibility of restricted cells.
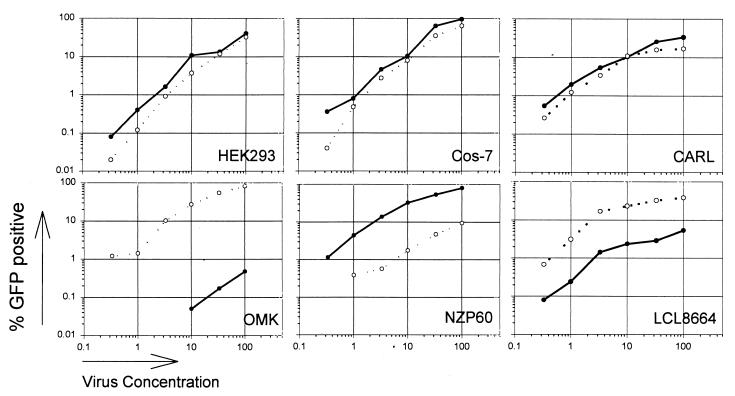
MuLV infection of Fv-1-restricted cells exhibits multiple-hit kinetics, whereas infection of permissive cells exhibits single-hit kinetics (48). Apparently, higher multiplicities of infection are required to overcome the restriction imposed by the Fv-1 product. To examine whether multiplicity of infection influenced the degree of restriction to HIV-1 and SIVmac vectors observed in this study, recombinant viruses were diluted over 3 orders of magnitude and used to infect a constant number of target cells (Fig. 5). For both restricted and permissive cells, a direct relationship between vector multiplicity of infection and number of GFP-positive cells was observed.
FIG. 5.
Effect of multiplicity of infection on the infectibility of cells. Cells were infected, as described in Materials and Methods, with various dilutions of the HIV-1 and SIVmac vectors. The infectious titer of each vector is indicated (HIV-1, solid lines and datum points; SIVmac, broken lines and open datum points).
DISCUSSION
Two types of postentry restrictions of HIV-1 and SIVmac infection were observed in this study. The first is specific for either HIV-1 or SIVmac and limits the number of virus-infected cells (i.e., the viral titer). The block to HIV-1 infection of rhesus macaque cells has been shown to limit successful reverse transcription (30). Our results indicate that cells from other Old World primates (e.g., African green monkeys) also exhibit blocks to HIV-1 infection. Although the inefficiency of HIV-1 replication in rhesus monkey cells was reported to be dependent upon the chemokine receptor used for virus entry (7), our results demonstrate that some restriction applies even when HIV-1 entry is mediated by heterologous envelope glycoproteins and receptors. Patterns of restriction were discernible, with most Old World monkey cells being resistant to HIV-1 and most New World monkey cells being resistant to SIVmac. Some exceptions to these generalizations were seen; for example, cells of the owl monkey, a New World species, were efficiently infected by SIVmac but not HIV-1 vectors. Within a species, comparable levels of restriction were observed in diverse cell types, suggesting the involvement of widely expressed, genetically determined cellular factors.
Postentry restrictions of primate immunodeficiency virus infection were also evident in other mammalian species. Prosimian cells did not support infection by either HIV-1 or SIVmac vectors. Rabbit cells exhibited specific restrictions against infection by HIV-1 but not SIVmac vectors. As was observed in monkey cells, several different cell lines derived from a single species exhibited similar patterns of restriction. In contrast to the results with primate immunodeficiency virus vectors, no species-specific blocks to MuLV infection were observed.
Cellular factors mediating resistance to primate immunodeficiency virus infection could be evolutionarily advantageous. Infection of feral monkeys and chimpanzees with these viruses is typically without pathogenic consequences (27, 35, 36). However, the introduction of these viruses into a new host species, including humans, often results in life-threatening immunodeficiency (12, 26, 27, 31, 37). Restricting cellular factors may have limited the range of species susceptible to the lentiviruses.
The species-specific pattern of restriction is complex and does not allow a simple model consistent with current concepts of mammalian evolution to be proposed. Indeed, the mechanisms and cellular factors responsible for the observed restrictions may differ in the different species studied herein. The available data do not allow us to distinguish whether the restrictions result from the absence of positive cellular factors, from the presence of inhibitory cellular factors, or from both. Unlike the situation seen with Fv-1 in mice, multiplicity of viral infection did not alter the relative degree of the early phase blocks observed for HIV-1 and SIV. The cellular factors responsible for the replication restrictions appear to be difficult to saturate. Future work will be needed to identify the host cell factors associated with the early, postentry blocks to HIV-1 and SIVmac infection.
The second restriction to viral replication observed in our study applied to both HIV-1 and SIVmac vectors and was associated with a decrease in the level of reporter gene expression in successfully infected cells. Because both vectors rely on Tat-activated long-terminal-repeat promoters, one likely basis for this restriction is the requirement for cellular cofactors for Tat (28, 58).
Our results have relevance for attempts to establish animal models for immunodeficiency virus infection. Dogs, cats, and tree shrews apparently support early, postentry events in HIV-1 infection efficiently. Furthermore, HIV-1 provirus-directed gene expression in cells of these species is robust, in contrast to several other mammalian species. At a minimum, dogs, cats, and tree shrews should be suitable hosts for the study of lentivirus vectors in vivo. In addition, if the assembly of infectious virions can occur in the cells of these species, completion of the entire HIV-1 life cycle except for virus entry might be achievable. Future studies might examine the potential of these mammalian hosts to support infection of HIV-1 variants.
ACKNOWLEDGMENTS
We thank Maris Handley, Justine Milligan, and Joseph O’Brien at the Dana-Farber Cancer Institute flow cytometry core for excellent technical support; Ted Friedmann, Richard Mulligan, and Ronald Desrosiers for reagents; Michael Farzan, Markus Koch, and F. C. Jensen for helpful discussions; and Yvette McLaughlin and Sheri Farnum for manuscript preparation.
This work was supported by National Institutes of Health grant HL 54785. Dana-Farber Cancer Institute is the recipient of Cancer Center and Center for AIDS Research awards from the National Institutes of Health. This work was also supported by the Mathers Charitable Foundation, the Friends 10, Douglas and Judy Krupp, and the late William F. McCarty-Cooper. W. Hofmann was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-α", MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;28:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J, Eichberg J W, Masur H, Saxinger W C, Gallo R, Macher A M, Lane H C, Fauci A S. Transmission of HTLV-III infection from human plasma to chimpanzees: man animal model for AIDS. Science. 1984;226:549–552. doi: 10.1126/science.6093251. [DOI] [PubMed] [Google Scholar]
- 3.Balzarini J, De Clercq E, Uberla K. SIV/HIV-1 hybrid virus expressing the reverse transcriptase gene of HIV-1 remains sensitive to HIV-1-specific reverse transcriptase inhibitors after passage in rhesus macaques. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:1–4. doi: 10.1097/00042560-199705010-00001. [DOI] [PubMed] [Google Scholar]
- 4.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 5.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Clavel F, Guyader M, Guetard D, Salle M, Montagnier L, Alizon M. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature. 1986;324:691–695. doi: 10.1038/324691a0. [DOI] [PubMed] [Google Scholar]
- 11.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 12.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 13.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 14.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 15.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 16.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 17.Fauci A S. The human immunodeficiency virus: infectivity and mechanisms of pathogenesis. Science. 1988;239:617–622. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 19.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 20.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs E. Tree shrews. In: Poole T, editor. UFAW handbook on the care and management of laboratory animals. 7th ed. Oxford, United Kingdom: Blackwell; 1999. pp. 235–245. [Google Scholar]
- 22.Fultz P N, McClure H M, Daugharty H, Brodie A, McGrath C R, Swenson B, Francis D P. Vaginal transmission of human immunodeficiency virus (HIV) to a chimpanzee. J Infect Dis. 1986;154:896–900. doi: 10.1093/infdis/154.5.896. [DOI] [PubMed] [Google Scholar]
- 23.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajdusek D C, Amyx H L, Gibbs C J, Jr, Asher D M, Rodgers-Johnson P, Epstein L G, Sarin P S, Gallo R C, Maluish A, Arthur L O, et al. Infection of chimpanzees by human T-lymphotropic retroviruses in brain and other tissues from AIDS patients. Lancet. 1985;i:55–56. doi: 10.1016/s0140-6736(85)91011-6. [DOI] [PubMed] [Google Scholar]
- 25.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, Palker T J, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature. 1992;358:459–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 27.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 28.Garber M E, Wei P, KewalRamani V N, Mayall T P, Hermann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Himathongkham S, Luciw P A. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 33.Kirchhoff F, Pohlmann S, Hamacher M, Means R E, Kraus T, Uberla K, Di Marzio P. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 35.Kurth R, Norley S. Simian immunodeficiency viruses of African green monkeys. In: Letvin N L, Desrosiers R C, editors. Simian immunodeficiency viruses. New York, N.Y: Springer-Verlag; 1994. pp. 21–33. [DOI] [PubMed] [Google Scholar]
- 36.Lackner A A. Simian immunodeficiency viruses of African green monkeys. In: Letvin N L, Desrosiers R C, editors. Simian immunodeficiency viruses. New York, N.Y: Springer-Verlag; 1994. pp. 35–64. [Google Scholar]
- 37.Letvin N L, Eaton K A, Aldrich W R, Sehgal P K, Blake B J, Schlossman S F, King N W, Hunt R D. Acquired immunodeficiency syndrome in a colony of macaque monkeys. Proc Natl Acad Sci USA. 1983;80:2718–2722. doi: 10.1073/pnas.80.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquir Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 39.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 40.Luckett P. Comparative biology and evolutionary relationships of tree shrews. New York, N.Y: Plenum Press; 1980. [Google Scholar]
- 41.Lusso P, Markham P D, Ranki A, Earl P, Moss B, Dorner F, Gallo R C, Krohn K J. Cell-mediated immune response toward viral envelope and core antigens in gibbon apes (Hylobates lar) chronically infected with human immunodeficiency virus-1. J Immunol. 1988;141:2467–2473. [PubMed] [Google Scholar]
- 42.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 44.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, et al. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 47.Parolin C, Taddeo B, Palu G, Sodroski J. Use of cis- and trans-acting viral regulatory sequences to improve expression of human immunodeficiency virus vectors in human lymphocytes. Virology. 1996;222:415–422. doi: 10.1006/viro.1996.0438. [DOI] [PubMed] [Google Scholar]
- 48.Pincus T, Hartley J W, Rowe W P. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J Exp Med. 1971;133:1219–1233. doi: 10.1084/jem.133.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenwirth B, Billich A, Datema R, Donatsch P, Hammerschmid F, Harrison R, Hiestand P, Jaksche H, Mayer P, Peichl P, et al. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob Agents Chemother. 1994;38:1763–1772. doi: 10.1128/aac.38.8.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe W P, Sato H. Genetic mapping of the Fv-1 locus of the mouse. Science. 1973;180:640–641. doi: 10.1126/science.180.4086.640. [DOI] [PubMed] [Google Scholar]
- 51.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J Virol. 1991;65:3514–3520. doi: 10.1128/jvi.65.7.3514-3520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon J H, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 54.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 56.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 57.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 59.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 60.Yee J K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin L, Boussard S, Allan J, Luban J. The HIV type 1 replication block in nonhuman primates is not explained by differences in cyclophilin A primary structure. AIDS Res Hum Retroviruses. 1998;14:95–97. doi: 10.1089/aid.1998.14.95. [DOI] [PubMed] [Google Scholar]
- 62.Zack J A, Cann A J, Lugo J P, Chen I S. HIV-1 production from infected peripheral blood T cells after HTLV-1 induced mitogenic stimulation. Science. 1988;240:1026–1029. doi: 10.1126/science.2835813. [DOI] [PubMed] [Google Scholar]