Abstract
Semliki Forest virus (SFV) is an enveloped alphavirus that infects cells via a membrane fusion reaction triggered by acidic pH in the endocytic pathway. Fusion is mediated by the spike protein E1 subunit, an integral membrane protein that contains the viral fusion peptide and forms a stable homotrimer during fusion. We have characterized four monoclonal antibodies (MAbs) specific for the acid conformation of E1. These MAbs did not inhibit fusion, suggesting that they bind to an E1 region different from the fusion peptide. Competition analyses demonstrated that all four MAbs bound to spatially related sites on acid-treated virions or isolated spike proteins. To map the binding site, we selected for virus mutants resistant to one of the MAbs, E1a-1. One virus isolate, SFV 4-2, showed reduced binding of three acid-specific MAbs including E1a-1, while its binding of one acid-specific MAb as well as non-acid-specific MAbs to E1 and E2 was unchanged. The SFV 4-2 mutant was fully infectious, formed the E1 homotrimer, and had the wild-type pH dependence of infection. Sequence analysis demonstrated that the relevant mutation in SFV 4-2 was a change of E1 glycine 157 to arginine (G157R). Decreased binding of MAb E1a-1 was observed under a wide range of assay conditions, strongly suggesting that the E1 G157R mutation directly affects the MAb binding site. These data thus localize an E1 region that is normally hidden in the neutral pH structure and becomes exposed as part of the reorganization of the spike protein to its fusion-active conformation.
All enveloped animal viruses use membrane fusion to cross the barrier of the host cell membrane and deliver the virus genome into the cytoplasm. This critical membrane fusion reaction is mediated by the virus spike protein, which undergoes structural rearrangements that convert the protein into a fusion-active form. The general scheme of the structural rearrangements, although differing mechanistically for different groups of viruses, appears to involve the release of a hydrophobic fusion peptide from a previously hidden or inactive position within the spike protein and its insertion into the target membrane to trigger fusion. A key question is the mechanism of protein refolding from a fusion-inactive form to the fusion-active form that carries out fusion peptide insertion. Molecular understanding of this refolding reaction may lead to the development of novel strategies to block virus fusion and infection. For a group of diverse viruses exemplified by influenza virus, the fusogenic spike protein conformational change involves the formation of an extended α-helical coiled-coil domain that appears to be a key feature of the fusion mechanism (17, 36).
The alphavirus Semliki Forest virus (SFV) is a small, highly organized enveloped RNA virus whose fusion activity has been extensively studied (20, 21, 40). The SFV fusion reaction is triggered by low pH (<pH 6.2) during the endocytic uptake of the virus by cells. Fusion and infection are blocked by weak bases such as NH4Cl or specific inhibitors such as bafilomycin, which act to raise the pH within endocytic vesicles (14, 20). The SFV spike promoter is composed of the E1 and E2 transmembrane subunits, each ∼50 kDa and associated as a noncovalent heterodimer, and the E3 subunit, a peripheral polypeptide of ∼10 kDa. Each virus particle contains 240 copies of this spike promoter organized as 80 trimeric spikes, [E1-E2-E3]3. Fusion is mediated by the spike E1 subunit, which binds to target membranes and contains a highly conserved hydrophobic domain from amino acids 79 to 97 that is believed to be the fusion peptide (12, 20, 24, 26).
Studies of the SFV spike protein during fusion indicate that upon exposure to mildly acidic pH, the E1-E2 dimer dissociates. E1 then undergoes conformational changes that result in the exposure of previously masked epitopes for monoclonal antibody (MAb) binding and the formation of a highly stable, trypsin-resistant E1 homotrimer (20, 24). These E1 conformational changes occur with kinetics slightly faster than those of fusion (3, 19) and are enhanced by the presence of target membranes containing cholesterol and sphingolipid, two lipid components that are specifically required for SFV fusion (20, 21, 26, 49). E1 then associates with the target membrane by insertion of the fusion peptide, and membrane fusion is triggered.
Central questions in understanding SFV fusion include the mechanism of formation of the critical E1 homotrimer and the identities of the regions of the E1 protein that are involved in its fusogenic refolding. Structural predictions suggest that, unlike spike proteins of the influenza virus class, SFV E1 does not refold into an extended α-helical coiled coil during fusion (26). Thus, the formation of the fusion-active E1 trimer may represent a novel refolding mechanism. One tool in identifying regions of viral spike proteins that become exposed during fusion has been to localize the binding sites for MAbs that are specific for the fusion-active conformation of the spike (48). The fusion-active, low-pH-treated form of SFV E1 is specifically recognized by four MAbs that inefficiently recognize the pH 7 form of E1 (23, 45). Three of these MAbs were isolated and characterized by our laboratory (23) and are termed E1a-1, E1a-2, and E1a-3. The fourth MAb, anti-E1", was originally isolated by Boere et al. (1) and further characterized by Wahlberg and coworkers (3, 44, 45). Extensive evidence indicates that exposure of the epitopes recognized by these acid-specific MAbs is relevant to the fusion activity of SFV. First, the epitopes are exposed with the same pH and temperature dependence as are required for virus fusion (3, 19, 23). Detailed kinetic studies demonstrate that epitope exposure occurs slightly faster than fusion (3, 19). This is in contrast to other SFV MAbs that detect spike epitopes exposed after the fusion reaction is completed (19). SFV fusion is remarkably dependent on cholesterol and sphingolipid as well as low pH, and the conformational changes detected by the antibodies are strikingly dependent on both lipids when either the virus (4, 6) or the spike ectodomain (26) is assayed. The epitopes become exposed during the endocytic entry of the virus into the cell and, like fusion, epitope exposure is blocked by agents such as NH4Cl or monensin that interfere with endosome acidification (23, 28, 32, 38, 45). Several virus E2 mutants that shift the pH dependence of virus fusion by shifting the pH dependence of E1-E2 dimer dissociation also shift the pH dependence of epitope exposure (13, 33). A number of biochemical experiments demonstrate that the epitopes are not simply exposed as part of a general “loosening” of the tightly packed virus structure at low pH, since detergent solubilization, for example, did not make the epitopes accessible (23). Lastly, the same pool of E1 protein reacts with the antibodies, binds specifically to cholesterol- and sphingolipid-containing target liposomes, and forms the homotrimeric structure believed to be critical for the fusion reaction (26). Thus, the kinetics and properties of epitope exposure argue for the relevance of these conformational changes to fusion.
We set out to determine the functional effects of E1 binding by these four MAbs, to characterize the similarities and differences in their binding site(s), and in particular to localize the epitope(s) with which they interact. Our studies showed that the presence of the MAbs did not affect fusion and that all four MAbs bound to a spatially related site(s) on E1. The region of the spike protein recognized by three of the MAbs appears distinct from the fusion peptide and from the epitope recognized by anti-E1". Our results thus indicate a region of E1 other than the virus fusion peptide that becomes exposed during the SFV fusion reaction at acidic pH.
MATERIALS AND METHODS
Viruses and antibodies.
The wild-type (wt) virus used in these studies was a well-characterized plaque-purified stock virus (13, 42), and the SFV fus-1 mutant was previously isolated and characterized as a pH shift fusion mutant (13, 25). All virus stocks were prepared by propagation in BHK cells at low multiplicity in minimal essential medium plus 0.2% bovine serum albumin (BSA) and 10 mM HEPES (pH 7.4) and stored at −70°C (25). [35S]methionine-[35S]cysteine-labeled virus was prepared in BHK cells, purified by banding on a discontinuous sucrose gradient (25), and stored at −70°C in ∼45% (wt/wt) sucrose in TN buffer (50 mM Tris [pH 7.4], 100 mM NaCl). Unlabeled virus was prepared in BHK cells, purified on tartrate gradients, and stored at −70°C in TN buffer (25).
The E1 acid conformation-specific MAbs E1a-1, E1a-2, and E1a-3 and the non-acid-specific E1 and E2 MAbs E1-1 and E2-1 were previously isolated and characterized in our laboratory (23), and were prepared in large quantities for biotinylation and other experiments by growth in a CELLMAX (Gibco BRL, Gaithersburg, Md.) in the Einstein Hybridoma facility. MAb anti-E1" was obtained as a concentrated ascites preparation from Harm Snippe and has been previously characterized for its reactivity to the low-pH conformation of E1 (26, 45). Biotin conjugation was performed with sulfo-NHS-biotin (Pierce, Rockford, Ill.).
Functional effects of antibodies on virus fusion.
Liposome binding studies measured the cofloatation of radiolabeled SFV with liposomes on sucrose gradients as previously described (24), with pH treatment performed in the presence of the indicated concentrations of antibodies. Virus-liposome fusion was assayed by monitoring the decrease in the eximer peak of pyrene-labeled SFV upon fusion with unlabeled liposomes prepared by extrusion (7, 24). Pyrene-labeled wt SFV was prepared by propagation in one roller bottle of BHK cells in which the phospholipids were prelabeled by growth in the presence of 10 μg of pyrene (Molecular Probes, Eugene, Oreg.) per ml (3, 44). Labeled virus was purified by banding on a discontinuous sucrose gradient (25). Fusion assays were performed with pyrene-labeled virus at concentrations of 0.6 μM virus lipid, liposomes at concentrations of 200 μM lipid, and the indicated concentrations of antibodies; HNE buffer (5 mM HEPES [pH 7.0], 150 mM NaCl, 0.1 mM EDTA) was used at 37°C in a stirred cuvette with a Perkin-Elmer LS5 fluorometer (3, 44). Fusion was triggered by the addition of a pretitrated volume of 0.3 M morpholinoethanesulfonic acid (MES) (pH 4.8), to give a final pH of 5.5.
Fusion of virus with the plasma membrane of BHK cells was assayed essentially as previously described (42, 45). In brief, virus was bound on ice to BHK cells grown on 12-mm coverslips, and the coverslips were then inverted over a 75-μl drop of medium (RPMI without bicarbonate plus 0.2% BSA and 30 mM sodium succinate [pH 5.5]) containing the indicated antibody on a square of Parafilm and incubated at the indicated temperature for the indicated time (1 to 5 min). The coverslips were flooded with medium to neutralize and reinverted, and the cells were incubated overnight at 28°C in minimal essential medium containing 0.2% BSA and 10 mM HEPES (pH 7.4), plus 20 mM NH4Cl to prevent secondary infection. Infected cells were quantitated by immunofluorescence with a polyclonal antibody to the SFV spike protein (42).
Selection of antibody binding mutants.
The overall strategy was to pH treat the virus to expose the binding site for MAb E1a-1, remove MAb-reactive virus by immunoprecipitation, and repeat for multiple selections. This strategy is similar to that successfully used to isolate a Sindbis virus MAb-resistant mutant (30). A 150-μl volume of wt virus stock containing ∼109 PFU was adjusted to a final pH of ∼5.5 by dilution with an equal volume of buffer containing 50 mM succinate (pH 5.0) and 100 mM NaCl, incubated for 5 min on ice, and neutralized by the addition of a precalibrated volume of 0.5 N NaOH. This mild pH treatment did not reduce virus infectivity (3) and induced full reactivity with MAb E1a-1 as tested by enzyme-linked immunosorbent assay (ELISA) of whole virus (data not shown). The virus was then incubated overnight on ice with 3 μg of biotin-conjugated MAb E1a-1. Antibody-virus complexes were removed by gentle rotation for 1 h at 4°C with 100 μl of streptavidin-conjugated magnetic particles (PerSeptive Diagnostics, Inc., Cambridge, Mass.), and bound virus and particles were precipitated with a magnet-containing test tube rack (Dynal, Inc., Lake Success, N.Y.). The specific selection gave a ∼10-fold reduction in viral titer, and no reduction resulted when an unrelated biotin-conjugated MAb was substituted. Virus remaining in the supernatant was expanded by growth on four independent plates of BHK cells at a multiplicity of ∼1 PFU/cell for 6 to 7 h. The selection of these individual stocks was repeated for a total of nine cycles each, a point at which further selection gave little reduction in the titer of the virus stocks. The selected stocks were plaqued under agarose, and isolated plaques were picked and expanded by growth on BHK cells in 24-well trays until the cells displayed cytopathic effects. Virus isolates were treated at low pH with succinate buffer as above, and screened by the ELISA described below for reactivity with MAbs E2-1 and E1a-1. Isolates showing strong reactivity with the E2 MAb, indicating a high titer of virus, and reduced activity with the selecting MAb were expanded and further tested as described below. Other selections based on neutralization with acid conformation-specific MAb, second antibody, and complement or based on epitope exposure by heat treatment did not produce sufficient specific selection pressure and were not further pursued (data not shown).
ELISA analysis.
Antibody competition experiments with intact virus were performed by coating ELISA wells with 1 μg of MAb E2-1 per ml; this MAb was previously shown to bind to intact virus after either acidic or neutral pH treatment (23). The plates were blocked with 1% BSA, purified acidic- or neutral-pH-treated virus was added at a protein concentration of 3 μg/ml, and the mixture incubated overnight at 4°C. The tethered virus was then reacted for 1 to 2 h at 37°C with 60 to 100 ng of biotin-conjugated MAb E1a-1 or anti-E1" per ml, concentrations that were shown to be in the linear range of the assay, in the presence of the indicated concentrations of unlabeled competing MAb. The plate was developed by incubation with saturating concentrations of streptavidin-alkaline phosphatase (Fisher Scientific, Pittsburgh, Pa.) for 1.5 h at 37°C followed by substrate incubation. Antibody competition experiments with purified detergent-solubilized spike proteins were performed by directly coating ELISA wells with 0.5 μg of spike proteins per ml for 1 to 2 h at 37°C, blocking with BSA, and then incubating with biotin-MAb and competitors as above. Spike proteins were purified by Triton X-114 phase separation (2) of gradient-banded wt and mutant viruses.
To screen for mutants with reduced binding of MAb E1a-1, virus stocks were treated at pH 5.5 for 3 min at 37°C with succinate buffer as above, neutralized, and adjusted to 0.5% Triton X-100 to disrupt the virus. Samples were incubated overnight at 4°C in ELISA wells that had been precoated with 1 μg of either MAb E2-1 or MAb E1a-1 per ml. Bound spike proteins were detected with a rabbit antibody to the spike protein followed by goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma, St. Louis, Mo.). This assay could reliably detect pH-dependent epitope exposure in virus stocks with titers as low as 2 × 107 PFU/ml.
Virus sequence analysis.
RNA was prepared from virus grown at low multiplicity in one 75-cm2 flask of BHK cells (13). The sequence of the RNA encoding the structural proteins was determined by reverse-transcription (RT)-PCR amplification with Pfu polymerase (Stratagene, La Jolla, Calif.) and automated sequencing in the Einstein DNA sequencing facility, all as previously described (13, 42). Both strands of the cDNA were sequenced, and sequence changes were confirmed by analysis of an independent RT-PCR product. The sequence of our laboratory wt virus, the parent virus used for mutant selection, was found to contain a lysine (AAA) at position 85 of the capsid protein. This capsid residue is asparagine (AAC) in the wt infectious clone and published wt virus sequence (11).
Assays of the mutant phenotype.
The NH4Cl sensitivity of wt and mutant virus infection was assayed as previously described by infecting BHK cells in 24-well trays for 90 min with virus at a multiplicity of ∼1 PFU/cell in the presence of the indicated concentrations of NH4Cl and quantitating virus-specific RNA synthesis by labeling with [3H]uridine for 3.5 h (13). Low-pH-dependent conformational changes in the virus spike proteins were assayed by treating [35S]methionine-[35S]cysteine-labeled virus at the indicated pH for 10 min at 37°C in the presence of 1 mM liposomes prepared by extrusion as indicated. Formation of the E1 homotrimer in these samples was investigated by incubating samples in sodium dodecyl sulfate (SDS) gel sample buffer for 3 min at 30°C and analyzing them by SDS-PAGE (24, 45). E1 conversion to trypsin resistance was assayed by digestion of samples for 10 min at 37°C with 200 μg of tolysulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin per ml in 1% Triton X-100 in phosphate-buffered saline (PBS) containing calcium and magnesium (24, 35), and the presence of acid conformation-specific epitopes was determined by immunoprecipitation with the indicated MAbs (23) followed by SDS-PAGE analysis. The pH dependence of E1-E2 dimer dissociation was assayed by coimmunoprecipitation (8, 43). Fluorograms of SDS-gels were quantitated by PhosphorImager analysis (ImageQuant version 1.2; Molecular Dynamics, Inc., Sunnyvale, Calif.).
RESULTS
Functional effects of acid-specific MAbs on virus fusion.
We wished to compare the functional properties of our three acid conformation-specific MAbs with those of MAb anti-E1". Previous studies suggested that the presence of anti-E1" inhibited low-pH-induced fusion of SFV with cell plasma membranes (45) or liposomes (44) and low-pH-induced virus-liposome attachment (44). If the acid conformation-specific MAbs inhibit virus-cell fusion, this could be the basis of a selection for MAb-resistant virus mutants. To test for inhibition of SFV-plasma membrane fusion, virus was bound to BHK cells on ice and treated for 1 min at pH 5.5 to induce fusion, and the infected cells resulting from virus fusion were quantitated by immunofluorescence with a polyclonal antibody to the virus spike protein. Under these conditions, no virus infected the cells during a 1-min treatment at pH 7 and between 209 and 256 cells were infected by virus at pH 5.5 (Table 1). When the pH treatment was performed in the presence of either MAb anti-E1" or E2-1, a nonneutralizing MAb to the E2 subunit, little inhibition resulted from the presence of either MAb at ∼300 μg/ml (Table 1, experiment I), from MAb anti-E1" at higher concentrations (500 or 900 μg/ml) (experiment II), or from MAb anti-E1" when the conformational changes and fusion were slowed by incubation at either 20 or 4°C (data not shown). The previous results reported inhibition of virus-cell membrane fusion by anti-E1" at concentrations ranging from a 1:10 dilution of ascities fluid to 350 μg of MAb/ml (45). Our acid-specific MAbs E1a-1, E1a-2, and E1a-3 also had no effect on virus-cell fusion (experiment III). In contrast, a polyclonal rabbit antibody to the SFV spike specifically inhibited virus-cell fusion when used under conditions previously shown to neutralize virus infectivity (23) (experiment IV). Thus, the lack of inhibition by the acid conformation-specific MAbs is not due to an inherent inability to detect inhibition in this assay. Control ELISA experiments also demonstrated that all four acid-specific MAbs were fully active in spike protein binding even when maintained at pH 5.5 (data not shown) and thus were capable of binding the virus under low-pH conditions.
TABLE 1.
Effect of acid conformation-specific MAbs on SFV-cell fusion
| Expt | No. (%) of virus-infected cellsa after treatment with:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH7 | No Ab | E1a-1 | E1a-2 | E1a-3 | E1" | E1" (500 μg/ml) | E1" (900 μg/ml) | E2-1 | Unb | Prec | Polyd | |
| I | 0 | 229 (100) | 183 (80) | 180 (79) | 196 (86) | |||||||
| II | 0 | 213 (100) | 175 (82) | 224 (105) | 202 (95) | |||||||
| III | 230 (100) | 256 (111) | 253 (110) | 229 (100) | ||||||||
| IVe | 0 | 209 (100) | 208 (100) | 1 (0.5) | ||||||||
SFV was bound to BHK cells on ice, and fusion triggered by warming the cells to pH 5.5 for 1 min at 37°C. The pH treatment was performed in the presence of the indicated MAbs at 300 μg/ml unless otherwise noted. Infected cells resulting from virus fusion were quantitated by immunofluorescence. Values represent the average number of infected cells on duplicate coverslips, and the numbers in parentheses represent the percentage of control fusion in the absence of antibody.
MAb to an unrelated antigen.
Preimmune rabbit serum.
Polyclonal rabbit spike antibody.
Virus was preincubated for 2 h at room temperature and overnight at 4°C in the presence of 50 μg of antibody per ml as indicated. Fusion was then assayed in the continued presence of antibody.
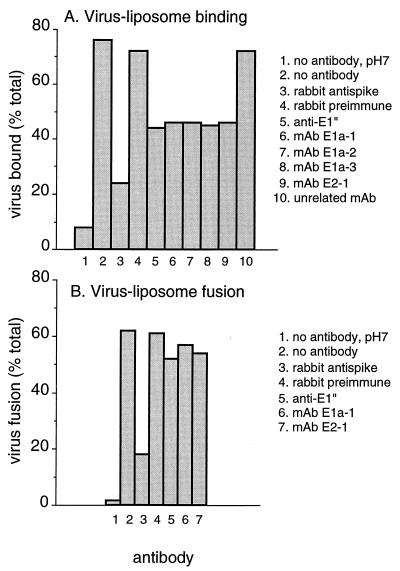
We examined the ability of the antibodies to inhibit low-pH-induced virus attachment to liposomes, a step that precedes fusion (Fig. 1A). After treatment at pH 5.5, about 75% of the added virus radioactivity cofloated with liposomes, compared with ∼8% after treatment at pH 7.0 (lanes 1 and 2). Inclusion of any of the acid conformation-specific E1 MAbs reduced the level of binding to about 45% (lanes 5 to 8). However, similar inhibition resulted with a MAb to the E2 subunit, E2-1 (lane 9). The rabbit polyclonal anti-spike antibody reduced binding to ∼24% of the added virus radioactivity (lane 3). Little inhibition resulted from inclusion of an unrelated MAb or rabbit antibody (lanes 4 and 10). Thus, moderate inhibition of radiolabeled virus binding to liposomes occurred with either the acid conformation-specific E1 MAbs or a MAb to E2, while these MAbs did not appreciably affect productive virus-cell fusion. We therefore tested the effects of these MAbs by using a sensitive assay for SFV fusion with liposomes.
FIG. 1.
Functional effects of MAbs on SFV membrane attachment and fusion. (A) Virus-liposome attachment. [35S]methionine-[35S]cysteine-labeled wt SFV was mixed with liposomes and the indicated antibodies at final concentrations of 1.2 mM lipid and 50 μg of antibody per ml. The samples were preequilibrated for 3 to 5 min at 37°C, adjusted to pH 5.5 for 10 min at 37°C unless otherwise indicated, and then returned to neutral pH. Virus association with liposomes was quantitated by flotation on sucrose gradients (see Materials and Methods). Data shown are the average of two experiments. (B) Virus-membrane fusion. Pyrene-labeled wt SFV (0.6 μM) was mixed with unlabeled liposomes (200 μM) in the presence of 50 μg of the indicated antibodies per ml, preequilibrated for 3 to 5 min at 37°C, and adjusted to pH 5.5, and the final extent of fusion was measured after ∼30 s at 37°C by quantitating the decrease in the pyrene eximer peak by spectrofluorometry (see Materials and Methods). Treatment with the rabbit preimmune and anti-spike antibodies was performed by overnight incubation at 4°C followed by incubation as above. Data shown are the average of two or three experiments.
We used a recently described fluorescence method based on biosynthetic labeling of virus with pyrene fatty acid (3, 44). Fusion of pyrene-labeled virus with unlabeled liposomes results in a reduction in the pyrene eximer peak, which can be monitored in a spectrofluorometer in real time (Fig. 1B). In the absence of MAbs, approximately 62% of the input virus fused at pH 5.5 (lane 2) and ∼2% fused at pH 7.0 (lane 1). Previous studies reported an SFV liposome fusion level of ∼50% at pH 5.5, which was reduced to ∼20% when 7 to 15 μg of anti-E1" per ml was included in the reaction mixture (44). In contrast, we found no significant inhibition of virus-liposome fusion by MAb anti-E1", E1a-1, or E2-1 at 50 μg/ml (lanes 5 to 7). Treatment with the neutralizing rabbit anti-spike antibody at this concentration reduced fusion to 18% (lane 3), while preimmune rabbit serum was without effect (lane 4).
Taken together, the data indicate that none of the acid conformation-specific MAbs to E1, including anti-E1", caused significant inhibition of virus fusion with cells or liposomes. Some inhibition of radiolabeled virus-liposome attachment resulted, but it occurred with a MAb to the E2 subunit as well as with the acid-specific E1 MAbs. It is not clear why our results differ from those of previous studies that reported inhibition, but it is possible that addition of solutions of MAbs caused changes in the buffering capacity of the fusion reactions, resulting in decreased fusion, a phenomenon that we observed with pH treatments close to the fusion threshold (data not shown). We adjusted all antibody stock solutions to the same protein and buffer concentrations to control for these effects. As reviewed in the discussion below, data from a variety of virus systems has shown that MAbs can detect relevant conformational changes during virus membrane fusion without directly inhibiting fusion.
Competition analysis of MAb binding to virus and spike proteins.
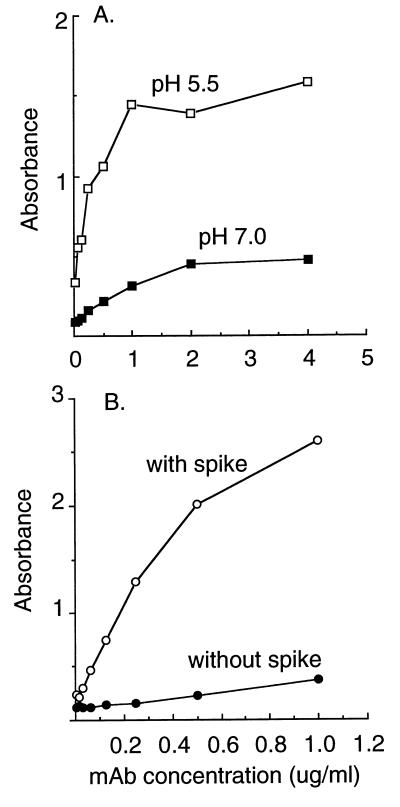
To use the acid conformation-specific MAbs as a tool to localize the conformational changes in E1, it is important to know if the MAbs recognize the same site on the SFV E1 polypeptide or identify several independent E1 sites that become accessible as part of overall refolding during fusion. All four MAbs are clearly independent (23), even in one case having been isolated by a different laboratory (1). However, the properties of the epitope(s) recognized by these four MAbs are indistinguishable in all of the assays that have been performed to date. Exposure of the epitope(s) has similar pH dependence (23), comparable kinetics in in vitro assays (3, 19) and in vivo during endocytic uptake of virus by cells (23, 32, 45), and a comparable cholesterol requirement (23, 26). All four MAbs are also nonneutralizing and do not recognize SDS-denatured E1 (1, 23). We therefore used competition analysis to characterize the locations of the MAb binding sites on E1. We first established two types of assay systems. One assay tested the recognition of whole virus particles by the MAbs. Purified virus was pretreated at acidic or neutral pH, returned to neutral pH, and tethered to ELISA plates with E2-1, a MAb to the E2 spike subunit. Tethered virus was reacted with biotin-conjugated MAb, and MAb binding was detected with streptavidin-alkaline phosphatase. Results for MAb E1a-1 are shown in Fig. 2A. The whole virus ELISA faithfully reproduced the increased MAb binding observed in previous assays of the acid-treated virus (23), with binding at nonsaturating MAb concentrations being five- to sixfold higher in the acid-treated samples. A second assay tested the reactivity of purified, detergent-solubilized spike proteins that were adsorbed directly to the ELISA plate. Similar to results from other low pH-dependent virus systems (47), direct adsorption of the spike protein caused sufficient protein unfolding to expose the E1a-1 epitope (Fig. 2B), and no further increase in MAb binding was observed when the spikes were pretreated at acidic pH (data not shown). Binding to isolated spike proteins was somewhat more efficient than binding to intact virus particles, since a comparable MAb concentration gave similar binding levels with about a sixfold-lower level of spike proteins in the isolated preparations. This probably represents steric hindrance in the intact virus, in keeping with the tight packing of the spike proteins. Similar results for both types of assays were obtained with MAb anti-E1" (data not shown).
FIG. 2.
MAb E1a-1 binding to whole virus and isolated spike proteins. (A) Binding to whole virus. Gradient-purified wt SFV was pretreated at pH 5.5 or 7.0 for 5 min at 37°C as indicated, returned to neutral pH, and adjusted to 3 μg/ml. Virus was tethered to ELISA plates that were precoated with a MAb to the E2 spike subunit (E2-1). Biotin-conjugated MAb E1a-1 was added at the indicated concentrations and allowed to react with the virus for 1 h at 37°C. Biotinylated MAb binding was detected by incubation with streptavidin conjugated to alkaline phosphatase followed by incubation with substrate, each for 1 h at 37°C. Data shown are a representative example of four experiments. (B) Binding to adsorbed spike proteins. Using gradient-purified wt SFV, spike proteins were prepared by Triton X-114 phase separation and adsorbed directly to ELISA plates at 0.5 μg/ml. Biotin-conjugated MAb E1a-1 was added at the indicated concentrations and allowed to react with immobilized spike proteins for 1 h at 37°C. MAb binding was detected by incubation with streptavidan conjugated to alkaline phosphatase followed by incubation with substrate, each for 1 h at 37°C. The control consists of biotin-conjugated MAb E1a-1 binding in the absence of adsorbed spike proteins. Similar low absorbance was observed for binding of an unrelated MAb to wells containing spike protein. Data shown are a representative example of two experiments.
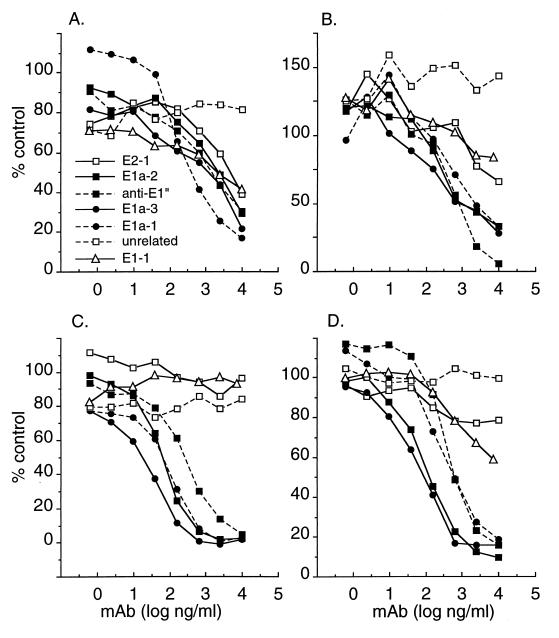
Having established assays for the binding of MAbs to acid-treated virus and isolated spike proteins, we used these assays to determine if various unlabeled MAbs could compete for the binding of biotin-labeled E1a-1 or anti-E1" (Fig. 3). The spike protein assay showed that all four acid conformation-specific MAbs competed for the binding of either MAb E1a-1 (Fig. 3C) or anti-E1" (Fig. 3D). In both cases, MAb E1a-3 showed the strongest competition. Negligible competition was observed with non-acid-specific MAbs against E1 or E2 (E1-1 and E2-1) or with an unrelated MAb. Competition analysis with acid-treated whole virus as the antigen likewise showed that all four acid-specific MAbs competed for the binding of either MAb E1a-1 (Fig. 3A) or MAb anti-E1" (Fig. 3B). When higher MAb concentrations were assayed in the whole-virus system, competition was also observed when non-acid-specific MAbs against E1 or E2 were used (Fig. 3A and B). This is in agreement with results from others indicating that assembly of the virus particle brings epitopes on E1 and E2 into close proximity (39). Taken together, results with the spike protein and whole-virus ELISAs demonstrate that all four acid conformation-specific MAbs bind to a spatially related site(s) on the SFV E1 subunit.
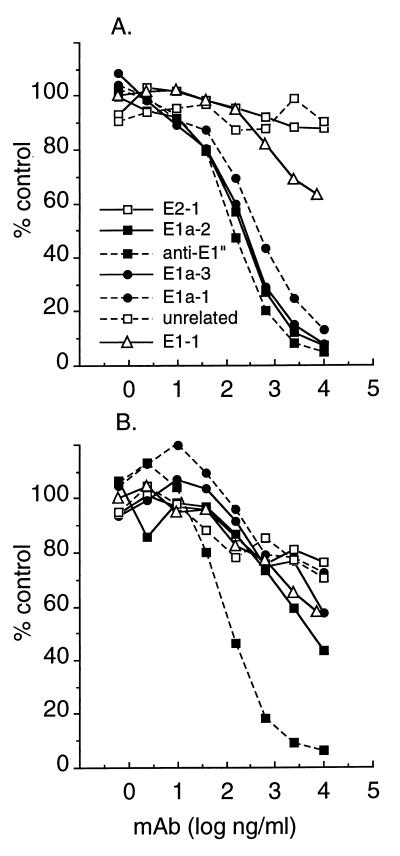
FIG. 3.
Antibody competition analysis. The ELISAs in Fig. 2 were used to analyze MAb binding to pH 5.5-treated intact wt virus (A and B) or purified wt spike proteins adsorbed to ELISA plates (C and D). The binding of biotin-conjugated MAb E1a-1 (A and C) or biotin-conjugated MAb anti-E1" (B and D) was quantitated at a constant biotin-MAb concentration of 100 ng/ml, in the presence of the indicated concentrations of competing, nonconjugated MAbs. Final binding results (in duplicate) were expressed as a percentage of the reactivity obtained with biotin-MAb in the absence of any competing MAb.
Selection and screening for MAb-resistant SFV mutants.
To specifically localize the binding site(s) for the acid conformation-specific MAbs, we wished to select for MAb-resistant virus mutants. The four acid conformation-specific MAbs are nonneutralizing (1, 23), do not inhibit virus-cell fusion (Table 1), and do not recognize linear sequences, precluding the use of synthetic peptides for mapping studies (1, 23). We therefore used an immunodepletion method to enrich for nonreactive viruses. A nonmutagenized virus stock was treated briefly at pH 5.5 on ice. The virus was then incubated with excess biotin-conjugated MAb E1a-1, antibody-reactive viruses were removed with streptavidin-conjugated magnetic particles, and the remaining virus was propagated on independent plates of BHK cells. The conditions of pH treatment were selected to obtain binding to the MAb (which according to all the published data represents an irreversible conformational change) without inactivating infectivity. Under these conditions, only a 10-fold decrease in virus titer resulted from the MAb treatment. The selection was therefore repeated for a total of nine cycles to increase the chances of significantly enriching for MAb-resistant mutants. Isolated plaques were then picked from the selected virus stocks, and the virus was expanded, treated at low pH, and screened by an ELISA for reactivity with both an E2 MAb and the selecting MAb. Two independent isolates, SFV 1-8 and SFV 4-2, were chosen for further study on the basis of their strong reactivity with MAb E2-1 and reduced (SFV 1-8) or negligible (SFV 4-2) reactivity with MAb E1a-1 after acid treatment.
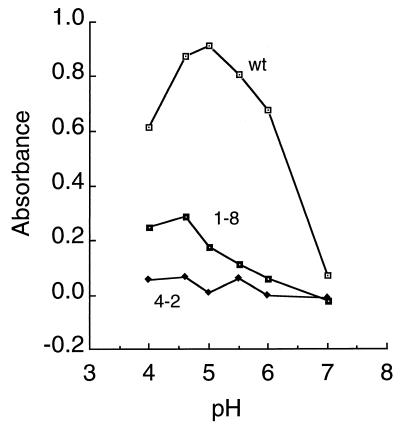
The SFV 1-8 and SFV 4-2 isolates were propagated on BHK cells at low multiplicity and yielded titers comparable to those of wt virus. These stocks were then diluted, treated for 5 min at 37°C at pH values ranging from 4.0 to 7.0, and tested in a sensitive sandwich ELISA for binding to MAb E1a-1 (Fig. 4). wt SFV showed strong binding following treatment at pH 6.0 to 4.0, with an increase of ∼13-fold in binding of pH 5.0-treated virus compared to pH 7.0-treated virus. The reduced binding observed in the pH 4.0-treated sample is probably due to acid inactivation (3). The SFV 1-8 isolate showed decreased but significant binding following treatment at pH 5.5 to 4.0, while binding by the SFV 4-2 isolate was not above background even after treatment at very low pH. Similar decreases in SFV 1-8 and SFV 4-2 binding to MAb E1a-1 were observed following pH treatment at 20°C (data not shown), conditions that favor efficient epitope exposure and decrease acid inactivation (3). Irrespective of pH treatment, both SFV 1-8 and SFV 4-2 showed high levels of binding to the anti-E2 MAb E2-1, comparable to those of wt virus (data not shown).
FIG. 4.
pH dependence of MAb E1a-1 binding to wt and mutant SFV. wt SFV and the 1-8 and 4-2 mutant virus isolates were diluted to a final titer of 108 PFU/ml in MES-saline-BSA buffer, and adjusted to the indicated pH for 5 min at 37°C. The samples were adjusted to neutral pH and 0.5% Triton X-100 and incubated in ELISA plates that were precoated with saturating concentrations of MAb E1a-1 (1 μg/ml). Bound spike protein was detected by the addition of a rabbit polyclonal antibody to the virus spike and an alkaline phosphatase-conjugated second antibody. Data shown are a representative example of four experiments.
The decreased MAb E1a-1 binding observed with SFV 1-8 and SFV 4-2 could be a direct consequence of amino acid changes in the MAb binding site or an indirect effect of a lack of response of the spike protein to low pH or of interfering mutations in E1, E2, or capsid protein. Both isolates grew efficiently to high titers, arguing that they must have functional spike proteins capable of carrying out virus-membrane fusion and responding to acidic pH. The spike protein ELISA described above (Fig. 2B) was used to test for steric interference by non-epitope-related sites. Epitope exposure in this assay is due to direct protein adsorption to the ELISA plate. Unlike the whole-virus ELISA, no interference with MAb E1a-1 binding was observed by either E2 or non-acid-specific E1 MAbs (Fig. 3), suggesting that sites on E1 and E2 are spatially separated in the adsorbed spike proteins. Purified spike proteins were prepared from wt virus, SFV 1-8, and SFV 4-2; bound to ELISA plates at 0.5 μg/ml; and reacted with a series of biotin-conjugated MAbs against E1 and E2. As shown in Fig. 5, the SFV 1-8 spike protein was similar to wt in binding MAbs to E2 and E1 and in binding the four acid conformation-specific MAbs to E1, including E1a-1. Thus, the decreased binding of MAb E1a-1 to SFV 1-8 observed in Fig. 4 is most probably due to steric interference by amino acid alterations not in the MAb binding site. Binding to the SFV 4-2 spike protein was unaltered for the non-acid-conformation-specific MAbs E1-1 and E2-1 and for the single-acid-conformation-specific MAb anti-E1" (Fig. 5). Strikingly, however, binding of SFV 4-2 by the three acid conformation-specific MAbs E1a-1, E1a-2, and E1a-3 was significantly impaired, ranging from 16% of the wt level with E1a-1 to 35% with E1a-3. Competition analysis was then performed for anti-E1" binding to purified spike proteins from SFV 1-8 (Fig. 6A) and SFV 4-2 (Fig. 6B). Anti-E1" binding to SFV 1-8 spike proteins was efficiently and comparably competed by all four acid conformation-specific MAbs, similar to the wt results previously shown in Fig. 3D. In contrast, while anti-E1" bound efficiently to spike proteins from SFV 4-2, this binding was competed only by anti-E1", not by any of the other acid-specific E1 MAbs, even at concentrations of 10 μg/ml. Taken together, these data suggest that the SFV 1-8 E1 protein has an unaltered MAb E1a-1 binding site while SFV 4-2 E1 carries an alteration that affects the binding of a subset of the E1 acid conformation-specific MAbs including E1a-1.
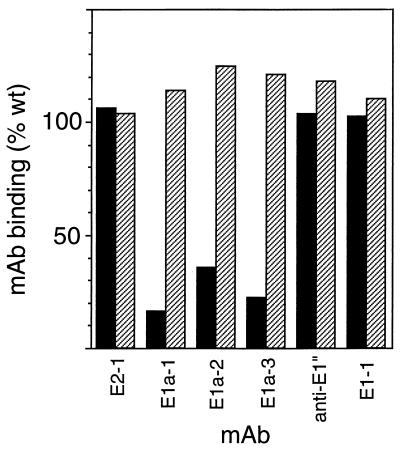
FIG. 5.
MAb binding to isolated mutant spike proteins. wt virus, SFV 1-8, and SFV 4-2 were purified by gradient sedimentation, and spike proteins were isolated by Triton X-114 phase separation. Spike proteins were adsorbed directly to ELISA plates at 0.5 μg/ml and reacted with 100 ng of the indicated biotin-conjugated MAbs per ml. This concentration was within the linear range of MAb binding to wt spike proteins (Fig. 2B). MAb binding was detected by reacting with streptavidin conjugated to alkaline phosphatase, and mutant spike protein reactivity was expressed as percentage of wt binding. SFV 4-2 binding is shown as solid bars, and SFV 1-8 binding is shown as hatched bars. Data are a representative example of three experiments.
FIG. 6.
Antibody competition analysis of isolated mutant spike proteins. Purified spike proteins were prepared from SFV 1-8 (A) and SFV 4-2 (B) and adsorbed to ELISA plates, and the binding of biotin-conjugated MAb anti-E1" was quantitated in the presence of the indicated concentrations of competing, nonconjugated MAbs, all as in Fig. 3D. Final binding results (in duplicate) were expressed as a percentage of the reactivity obtained with biotin-MAb in the absence of any competing MAb.
Biochemical and sequence analysis of SFV mutants.
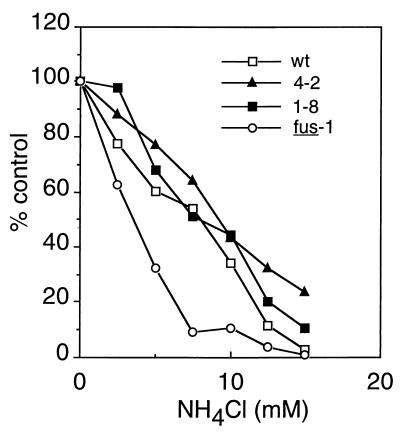
Given that our selection involved low-pH treatment of the virus, albeit under noninactivating conditions, we were concerned that our mutants might be low-pH resistant. To determine if the spike proteins of both mutants still mediated entry with a similar pH dependence to that of wt virus, we tested the sensitivity of virus infection to inhibition by the weak base NH4Cl (Fig. 7). Cells were infected with wt, SFV 1-8, SFV 4-2, or the pH shift SFV mutant fus-1 (13) in the presence of various concentrations of NH4Cl. Virus infection was quantitated by monitoring the incorporation of [3H]uridine into viral RNA (13). As previously observed (13, 25), both wt and fus-1 infections were inhibited by NH4Cl and fus-1 was sensitive to lower NH4Cl concentrations than was wt, in keeping with the relative acid resistance of the fus-1 fusion (pH threshold of ∼pH 5.5 for fus-1 compared to ∼pH 6.2 for wt). Both SFV 1-8 and SFV 4-2 showed NH4Cl sensitivities similar to that of wt SFV, indicating that both viruses had pH-dependent entry mechanisms with comparable pH requirements as wt SFV.
FIG. 7.
Sensitivity of wt and mutant virus infection to inhibition by NH4Cl. BHK cells in 24-well trays were infected with 1 PFU of wt or mutant SFV per cell in the presence of the indicated concentration of NH4Cl for 90 min. Infection was then quantitated by determining the incorporation of [3H]uridine into viral RNA, in the presence of 20 mM NH4Cl to prevent secondary infection. Data shown are a representative example of two experiments.
We examined several of the pH-dependent conformational changes in the mutant spike proteins. Upon exposure to low pH, the SFV spike dimer becomes more labile. Unlike wt SFV, both SFV 1-8 and SFV 4-2 mutants dissociated in nonionic detergent at pH 8.0 (data not shown), precluding the usual assays for pH-dependent dissociation of the dimer. Similar dimer dissociation in nonionic detergent was previously observed for two viruses with mutations in the E1 fusion peptide (8). The formation of the E1 homotrimer in SFV 1-8 and SFV 4-2 was assayed by using migration on SDS-gels (44) and trypsin resistance (24). The pH threshold for formation of the homotrimer appeared similar in the wt and the two mutants, although the efficiency of homotrimerization between pH 6.0 and 5.0 was somewhat lower in both mutants than in wt virus (data not shown; see Fig. 8 below). Overall, these data indicate that both mutants were able to infect cells and form the E1 homotrimer in a low-pH-dependent manner. Thus, the alterations in the antigenicity of SFV 1-8 and SFV 4-2 do not appear to cause functional effects in spike protein fusion activity.
FIG. 8.
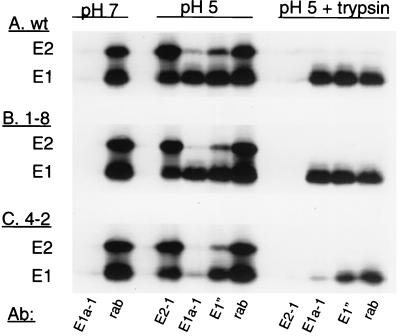
MAb reactivity of isolated E1 homotrimers from wt and mutant SFV. [35S]methionine-[35S]cysteine-labeled wt SFV (A), SFV 1-8 (B), or SFV 4-2 (C) was mixed with liposomes (final concentration, 0.8 mM), adjusted to the indicated pH for 10 min at 37°C, and returned to neutral pH. One set of samples (pH 5 plus trypsin) was then treated with 200 μg of trypsin per ml in 1% Triton X-100 in PBS for 10 min at 37°C to digest E2, capsid, and any nontrimerized E1, and the digestion was terminated by the addition of trypsin inhibitor. All other samples were treated with premixed trypsin and trypsin inhibitor in 1% Triton X-100 in PBS. Antibody reactivity was analyzed by immunoprecipitation with the indicated antibodies followed by SDS-PAGE. rab indicates a rabbit polyclonal antibody against the SFV E1 and E2 subunits.
To establish the amino acid alteration(s) responsible for the antigenic changes in SFV 4-2 and SFV 1-8, virus RNA was isolated and amplified by RT-PCR, and the sequence encoding the structural proteins was determined for each virus. The sequence of the SFV 1-8 mutant showed one amino acid change from that of the wt SFV parent, histidine to arginine at position 232 of E2 (H232R, CAT→CGT). This region from the wt virus RNA was sequenced and confirmed the difference between the wt and SFV 1-8 sequences. No amino acid changes occurred in the SFV 1-8 E1 protein sequence, in keeping with the unaltered antigenicity of the isolated spike protein (Fig. 5 and 6). The sequence of the SFV 4-2 mutant, surprisingly, contained the E2 H232R change. In addition, SFV 4-2 contained a single amino acid change on the E1 subunit, glycine 157 to arginine (G157R, GGG→AGG). Thus, the E1 G157R mutation appeared to be the mutation responsible for the loss of MAb E1a-1 binding in the SFV 4-2 mutant.
Our model for the effect of these sequence changes on the antigenicity of E1 was that the E2 H232R mutation in SFV 1-8 affected the MAb E1a-1 binding site through steric interference (Fig. 4), which was lost when the spike protein was isolated and partially denatured by direct binding to the ELISA plate (Fig. 5 and 6). In contrast, although SFV 4-2 contained the same E2 mutation, the G157R mutation on SFV 4-2 E1 directly prevented MAb E1a-1 binding due to its location in the MAb binding site. We tested the role of the E1 G157R mutation by assaying the immunoreactivity of isolated homotrimeric E1. Radiolabeled wt, SFV 1-8, and SFV 4-2 virus preparations were mixed with liposomes and treated at pH 5.0 to induce the formation of the E1 homotrimer. The virus mixtures were adjusted to neutral pH, and aliquots were incubated with trypsin in 1% Triton X-100 at 37°C, conditions that completely digest capsid, E2, and monomeric E1 but preserve the trypsin-resistant E1 homotrimer (22, 24). The samples were then analyzed by immunoprecipitation with a rabbit polyclonal anti-spike antibody which quantitates the total E1 and E2 (rab), a MAb against E2 (E2-1), and the acid conformation-specific MAbs E1a-1 and anti-E1". When treated at pH 5.0, the wt E1 subunit (Fig. 8A) became immunoreactive with MAbs E1a-1 and anti-E1" (54 and 69% of the total E1, respectively). When the low-pH-treated virus was digested with trypsin, the capsid and E2 subunits were degraded, as confirmed by the absence of precipitation with MAb E2-1 and loss of capsid protein (not shown on this gel), while the E1 subunit was resistant to trypsin (44% of the total E1). Immunoprecipitation analysis showed that 88 to 100% of the trypsin-resistant wt E1 was recognized by MAbs E1a-1 and anti-E1", respectively. Thus, trypsin digestion removed monomeric, non-acid-reactive E1, leaving almost exclusively the homotrimeric, acid-reacted conformation of E1, which was efficiently precipitated with both acid conformation-specific MAbs. Similar results were observed with low-pH-treated E1 from SFV 1-8 (Fig. 8B), which was recognized by MAbs E1a-1 and anti-E1" at 31 and 46% of the total E1, respectively, and became trypsin resistant (29% of the total E1). Similar to wt E1, the trypsin-resistant E1 from SFV 1-8 was completely precipitated by either MAb E1a-1 or anti-E1" (116 and 102%, respectively). In contrast, low-pH-treated E1 from the SFV 4-2 mutant (Fig. 8C) became trypsin resistant (31% of the total E1) and was recognized by anti-E1" (29% of the total E1) but was not efficiently precipitated by MAb E1a-1 (1% of the total E1). Even when E2, capsid, and monomeric E1 were removed by trypsin digestion, the remaining homotrimeric E1, although recognized by anti-E1" (59% of the trypsin-resistant E1), was not recognized by MAb E1a-1 (9% of the trypsin-resistant E1). Thus, in the absence of the E2 protein, the G157R mutation on SFV 4-2 E1 strongly interfered with the binding of MAb E1a-1 and to a small extent with that of anti-E1".
Comparison of alphavirus sequences in the E1 G157 region showed that this is a quite unconserved area of E1 that contains numerous charged amino acids. Position 157 is a glycine residue in SFV, in the closely related Ross River virus, and in eastern equine encephalitis virus. In many other alphaviruses, the E1 157 position contains a charged amino acid, and for western equine encephalitis virus, like SFV 4-2, this residue is arginine. Thus, it is reasonable that the introduction into SFV of the large, basic arginine side chain in place of the glycine hydrogen would act to disrupt the binding site for three acid conformation-specific MAbs but not have deleterious effects on the overall structure and function of E1.
DISCUSSION
Functional effects of the acid-specific MAbs.
Our studies showed that the four acid conformation-specific E1 MAbs do not have strong inhibitory effects on SFV-membrane fusion. SFV fusion is extremely rapid and is completed within seconds at 37°C (3). Cryoelectron microscopy studies have shown that spike protein conformational changes can be detected within the first 25 ms of low-pH treatment (10). The lack of effect of the MAbs on fusion thus may be due either to the rapid completion of fusion prior to recognition by MAb or to noninhibitory MAb binding prior to fusion. Our data do not differentiate between these two possibilities, but decreasing the rate of virus fusion with the cell plasma membrane by low pH incubation at either 20 or 4°C did not increase inhibition by added MAbs (see results above).
Results from several virus systems indicate that MAbs to spike protein epitopes involved in fusion exhibit a wide variety of properties in virus infection and fusion assays. For example, studies with the flavivirus West Nile virus have characterized a polyclonal antibody that inhibits virus-liposome fusion, although it is not clear if this antibody acts by steric interference, masking of the hydrophobic virus fusion peptide, or stabilizing the neutral-pH form of the spike protein (16). Acid conformation-specific MAbs to the influenza virus hemagglutinin (HA) have been informative and extensively used reagents to map regions of HA that become exposed during its fusogenic conformational changes (46–48). Some MAbs to HA block virus fusion within endosomes (18). An antibody to the membrane-proximal region of HA is nonneutralizing and does not recognize the low-pH conformation of HA. However, this MAb appears to inhibit both the low-pH-dependent conformational changes in HA and virus-induced cell-cell fusion (41). In contrast, and similar to our results with the SFV MAbs, several widely used acid conformation-specific MAbs to HA are nonneutralizing at either acidic or neutral pH and do not inhibit virus fusion (46).
It is clear that MAbs that recognize low pH-treated SFV spike proteins also can have a variety of properties (1, 23). The kinetics, pH dependence, cholesterol and sphingolipid dependence, and extensively characterized in vitro and in vivo properties of the acid conformation-specific epitope studied here argue strongly for its relevance to the infectious fusion reaction of SFV. Thus, the mapping of this epitope provides information on a biologically important spike protein conformational change.
Evidence for mapping of the E1a-1 epitope.
Competition studies on isolated wt spike proteins demonstrated that all four independent acid conformation-specific MAbs bound to sites on E1 that were either identical or sufficiently adjacent to produce binding interference. The SFV 4-2 mutation, E1 G157R, decreased the binding of the acid conformation-specific MAbs E1a-1, E1a-2, and E1a-3, while the binding of anti-E1" was essentially unaffected. Thus, while the competition data suggested close proximity of the anti-E1" binding site to those of the other acid conformation-specific MAbs, the SFV 4-2 mutation indicated that these epitopes were nevertheless distinct. It was possible that the SFV 4-2 mutation was acting indirectly to inhibit MAb binding by inducing a conformational change in a distant E1 epitope (31). We have tested the reactivity of E1 after triggering epitope exposure by acid treatment of virus particles and assay of the spike protein dimer (Fig. 4), by direct adsorption and denaturation of purified spike proteins on ELISA plates (Fig. 5 and 6), and by acid treatment of virus followed by isolation of homotrimeric E1 (Fig. 8). The decreased binding to SFV 4-2 E1 under all assay conditions strongly suggests that the 4-2 mutation acts by directly affecting the MAb binding site and therefore that the G157 region of E1 is masked in the neutral-pH structure and becomes accessible during fusion-permissive conditions.
In addition to the unique E1 G157D mutation, SFV 4-2 shares an additional mutation, E2 H232R, with the SFV 1-8 isolate. Since SFV 4-2 and SFV 1-8 were independently derived, the presence of this mutation in both isolates may suggest that a virus containing the shared mutation was present at nondetectable levels in the original plaque-purified parental virus stock. Alternatively, the mutation may have arisen spontaneously at a high frequency during selection and been rescued by its conferred effects on MAb recognition. The E2 H232R mutation does not have strong effects on the activity or pH dependence of the mutant fusion proteins (Fig. 7) but does cause the 1-8 mutant spike protein to be less well recognized by MAb E1a-1 when it is assayed under conditions that preserve the spike protein dimer (Fig. 4). In contrast, when assayed by direct binding to ELISA plates or as an isolated E1 homotrimer, the 1-8 mutant showed comparable MAb binding to that of wt SFV (Fig. 5, 6, and 8). The finding that MAb reactivity is decreased only when the intact SFV 1-8 spike dimer is assayed suggests that the E2 H232R mutation directly interferes with the accessibility of the MAb E1a-1 epitope. Alternatively, the H232R mutation could be acting indirectly to cause another site on E2 to mask the E1 epitope.
Comparisons with other alphavirus epitopes.
Anti-E1" was originally designated UM8.64 and characterized as nonneutralizing and nonprotective in mice but active in hemagglutination inhibition (1). This MAb defined an antigenic determinant termed E1d in competition analysis. Our competition analysis also places MAbs E1a-1, E1a-2, and E1a-3 in the E1d group with anti-E1". Thus, to date, all the acid conformation-specific MAbs against SFV E1 fall into this group, and it is not known if additional, noncompeting acid-specific antigenic determinants exist. However, although the competition analysis shows that all four acid conformation-specific MAbs have spatially related binding sites, the SFV 4-2 mutant data indicate that three of the current members of the E1d group bind an epitope different from that of anti-E1". The exact recognition site of anti-E1" has not been defined, except for its spatial relationship to G157 in competition analysis.
It is of interest to compare the G157 epitope to epitopes that were previously defined in the Sindbis virus E1 protein. Sindbis virus is another well-characterized member of the alphaviruses (40) that has been shown to enter cells by a membrane fusion reaction that is dependent on low pH (14) and cholesterol (27). Sindbis virus was selected for resistance to a neutralizing MAb that recognizes an E1 site accessible on the surface of native virus (34, 37). Interestingly, virus mutants resistant to this MAb had mutations of E1 glycine 132 to either arginine or glutamic acid (39). Thus, although this epitope is unrelated to the acid conformation-specific epitope described here, it provides another example of an E1 G→R mutation that blocks MAb binding, similar to the G157R mutation of the SFV 4-2 mutant.
Sindbis virus also has interesting E1 and E2 sites termed transitional epitopes, which are unmasked during virus penetration into the cell (9). Similar to the epitopes we have characterized here in SFV, the Sindbis E1 transitional epitopes are shielded and nonneutralizing in native Sindbis virus and can be exposed by treatment of virus particles with low pH, heat, or reducing agents or by direct adsorption to ELISA plates (29). However, unlike the acid conformation-specific SFV epitopes described here (23, 32), the in vivo exposure of the Sindbis virus transitional epitopes occurs following virus-receptor binding at the cell surface and is not blocked by agents that block endosomal acidification (9). Sindbis virus mutants were isolated based on their resistance to two E1 MAbs recognizing independent transitional epitopes. The mutations that confer resistance map to E1 residue 300 for one epitope and to E1 residue 361 or 381 for a second epitope (30). Both the locations and properties of these transitional epitopes thus appear distinct from the acid conformation-specific epitope mapped here. It is not known if the Sindbis E1 transitional epitopes become accessible following detergent solubilization of virus, which would suggest a possible analogy to several SFV E1 MAbs that recognize sites which are shielded in the intact virus particle (23).
Conclusions.
The data presented here suggest that a region of E1 defined by the G157R mutation becomes exposed or more accessible during the low-pH-induced fusion of SFV and can then be recognized by the acid conformation-specific MAbs E1a-1, E1a-2, and E1a-3. The acid-dependent refolding involving the E1 157 region is part of the overall series of conformational changes in the SFV spike protein during fusion. Kinetic studies of fusion (3) and results with SFV mutants with altered E1-E2 dimer dissociation (13, 33) indicate that acid conformation-specific epitope exposure is blocked until the E1-E2 dimer dissociates. Results with an SFV fusion peptide mutant indicate that the acid conformation-specific MAbs recognize a conformational change that can occur in the absence of E1 trimerization (24). Taken together, the results to date suggest a fusion model in which G157 epitope exposure occurs after E1-E2 dimer dissociation but prior to formation of the E1 homotrimer and insertion of the SFV fusion peptide into the target membrane. The kinetics of exposure of the G157 epitope are strongly enhanced by the presence of cholesterol and sphingolipid-containing liposomes (4, 6, 26), suggesting that E1 interacts in some way with the target membrane already during this relatively early stage of the fusion reaction. Future studies will address the role of lipid in the conformational changes that lead to epitope exposure. In addition, advances in cryoelectron microscopy (5, 10) may make it possible to localize the G157 region in the virus particle during exposure to low pH. As our knowledge of the SFV spike protein structure becomes more detailed, it may be possible to design virus mutations that could inhibit the rearrangement of the G157 region (15), thus potentially trapping an intermediate in virus-membrane fusion and permitting detailed characterization of its phenotype.
ACKNOWLEDGMENTS
We thank Matthew Scharff, Philippe Valadon, Susan Buhl, and other members of the Scharff laboratory and the Einstein hybridoma facility for their very generous help and insightful advice during this work. We thank Jan Wilschut and Yolande Smit for very helpful advice and protocols for the preparation and use of pyrene-labeled SFV, and we thank Harm Snippe for MAb anti-E1". We also thank the members of our laboratory for helpful discussions and suggestions and Duncan Wilson and the members of our laboratory for critical reading of the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (GM52929), by the Jack K. and Helen B. Lazar Fellowship in Cell Biology, and by Cancer Center Core Support Grant NIH/NCI P30-CA13330.
REFERENCES
- 1.Boere W A M, Harmsen T, Vinje J, Benaissa-Trouw B J, Kraaijeveld C A, Snippe H. Identification of distinct antigenic determinants on Semliki Forest virus by using monoclonal antibodies with different antiviral activities. J Virol. 1984;52:575–582. doi: 10.1128/jvi.52.2.575-582.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 3.Bron R, Wahlberg J M, Garoff H, Wilschut J. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, P. K., M. Vashishtha, and M. Kielian. Submitted for publication.
- 5.Cheng R H, Kuhn R J, Olson N H, Rossman M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corver J. Membrane fusion activity of Semliki Forest virus. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1998. [Google Scholar]
- 7.Corver J, Bron R, Snippe H, Kraaijeveld C, Wilschut J. Membrane fusion activity of Semliki forest virus in a liposomal model system: specific inhibition by Zn2+ ions. Virology. 1997;238:14–21. doi: 10.1006/viro.1997.8799. [DOI] [PubMed] [Google Scholar]
- 8.Duffus W A, Levy-Mintz P, Klimjack M R, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn D C, Meyer W J, Mackenzie J M, Johnston R E. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasma membrane as a consequence of early virus-cell interaction. J Virol. 1990;64:3643–3653. doi: 10.1128/jvi.64.8.3643-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 11.Garoff H, Frischauf A-M, Simons K, Lehrach H, Delius H. The capsid protein of Semliki Forest virus has clusters of basic amino acids and prolines in its amino-terminal region. Proc Natl Acad Sci USA. 1980;77:6376–6380. doi: 10.1073/pnas.77.11.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garoff H, Frischauf A-M, Simons K, Lehrach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature. 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 13.Glomb-Reinmund S, Kielian M. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J Virol. 1998;72:4281–4287. doi: 10.1128/jvi.72.5.4281-4287.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glomb-Reinmund S, Kielian M. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology. 1998;248:372–381. doi: 10.1006/viro.1998.9275. [DOI] [PubMed] [Google Scholar]
- 15.Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel J J, Wiley D C, Wharton S. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 16.Gollins S W, Porterfield J S. A new mechanism for the neutralization of enveloped viruses by antiviral antibody. Nature. 1986;321:244–246. doi: 10.1038/321244a0. [DOI] [PubMed] [Google Scholar]
- 17.Hughson F M. Structural characterization of viral fusion proteins. Curr Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- 18.Imai M, Sugimoto K, Okazaki K, Kida H. Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Res. 1998;53:129–139. doi: 10.1016/s0168-1702(97)00143-3. [DOI] [PubMed] [Google Scholar]
- 19.Justman J, Klimjack M R, Kielian M. Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol. 1993;67:7597–7607. doi: 10.1128/jvi.67.12.7597-7607.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 21.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. Specific role for lipids in virus fusion and exit: Examples from the alphaviruses. Subcell. Biochem., in press. [DOI] [PubMed]
- 22.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian M, Jungerwirth S, Sayad K U, DeCandido S. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian M C, Keränen S, Kääriäinen L, Helenius A. Membrane fusion mutants of Semliki Forest virus. J Cell Biol. 1984;98:139–145. doi: 10.1083/jcb.98.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimjack M R, Jeffrey S, Kielian M. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J Virol. 1994;68:6940–6946. doi: 10.1128/jvi.68.11.6940-6946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y E, Cassese T, Kielian M. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J Virol. 1999;73:4272–4278. doi: 10.1128/jvi.73.5.4272-4278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martys J L, Wjasow C, Gangi D M, Kielian M C, McGraw T E, Backer J M. Wortmannin-sensitive trafficking pathways in Chinese hamster ovary cells—differential effects on endocytosis and lysosomal sorting. J Biol Chem. 1996;271:10953–10962. doi: 10.1074/jbc.271.18.10953. [DOI] [PubMed] [Google Scholar]
- 29.Meyer W J, Gidwitz S, Ayers V K, Schoepp R J, Johnston R E. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J Virol. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer W J, Johnston R E. Structural rearrangement of infecting Sindbis virions at the cell surface: mapping of newly accessible epitopes. J Virol. 1993;67:5117–5125. doi: 10.1128/jvi.67.9.5117-5125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry N, Fox G, Rowlands D, Brown F, Fry E, Acharya R, Logan D, Stuart D. Structural and serological evidence for a novel mechanism of antigenic variation in foot-and-mouth disease virus. Nature. 1990;347:569–572. doi: 10.1038/347569a0. [DOI] [PubMed] [Google Scholar]
- 32.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmaljohn A L, Kokubun K M, Cole G A. Protective monoclonal antibodies define maturational and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology. 1983;130:144–154. doi: 10.1016/0042-6822(83)90124-1. [DOI] [PubMed] [Google Scholar]
- 35.Schmid S L, Fuchs R, Kielian M, Helenius A, Mellman I. Acidification of endosome subpopulations in wild-type Chinese hamster ovary cells and temperature-sensitive acidification-defective mutants. J Cell Biol. 1989;108:1291–1300. doi: 10.1083/jcb.108.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 37.Stec D S, Waddell A, Schmaljohn C S, Cole B A, Schmaljohn A L. Antibody-selected variation and reversion in Sindbis virus neutralization epitopes. J Virol. 1986;57:715–720. doi: 10.1128/jvi.57.3.715-720.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockert R J, Potvin B, Tao L, Stanley P, Wolkoff A W. Human hepatoma cell mutant defective in cell surface protein trafficking. J Biol Chem. 1995;270:16107–16113. doi: 10.1074/jbc.270.27.16107. [DOI] [PubMed] [Google Scholar]
- 39.Strauss E G, Stec D S, Schmaljohn A L, Strauss J H. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J Virol. 1991;65:4654–4664. doi: 10.1128/jvi.65.9.4654-4664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanlandschoot P, Beirnaert E, Barrere B, Millar B, Wharton S, Jou W M, Fiers W. An antibody which binds to the membrane-proximal end of influenza virus haemagglutinin (H3 subtype) inhibits the low-pH-induced conformational changes and cell-cell fusion but does not neutralize virus. J Gen Virol. 1998;79:1781–1791. doi: 10.1099/0022-1317-79-7-1781. [DOI] [PubMed] [Google Scholar]
- 42.Vashishtha M, Phalen T, Marquardt M T, Ryu J S, Ng A C, Kielian M. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J Cell Biol. 1998;140:91–99. doi: 10.1083/jcb.140.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahlberg J M, Boere W A M, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlberg J M, Garoff H. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster R G, Brown L E, Jackson D C. Changes in the antigenicity of the hemagglutinin molecule of H3 influenza virus at acidic pH. Virology. 1983;126:587–599. doi: 10.1016/s0042-6822(83)80015-4. [DOI] [PubMed] [Google Scholar]
- 47.White J M, Wilson I A. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza hemagglutinin. J Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 49.Wilschut J, Corver J, Nieva J L, Bron R, Moesby L, Reddy K C, Bittman R. Fusion of Semliki Forest virus with cholesterol-containing liposomes at low pH: a specific requirement for sphingolipids. Mol Membr Biol. 1995;12:143–149. doi: 10.3109/09687689509038510. [DOI] [PubMed] [Google Scholar]