Abstract
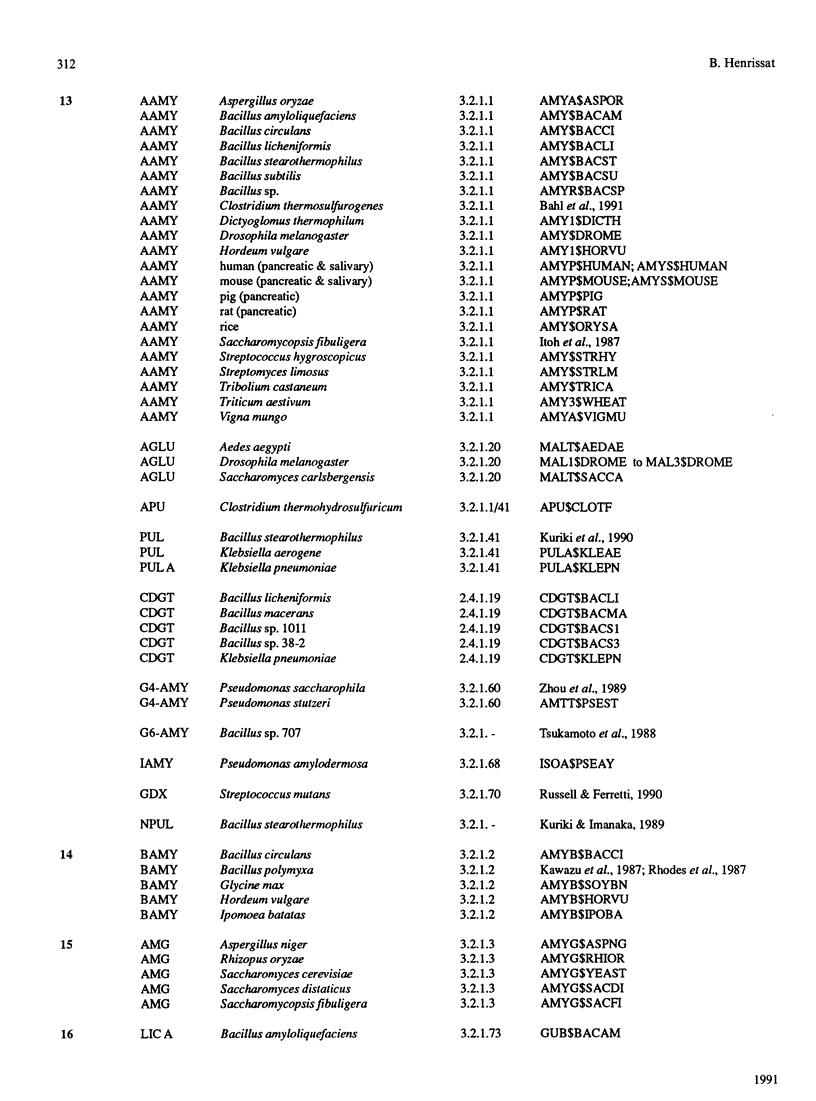
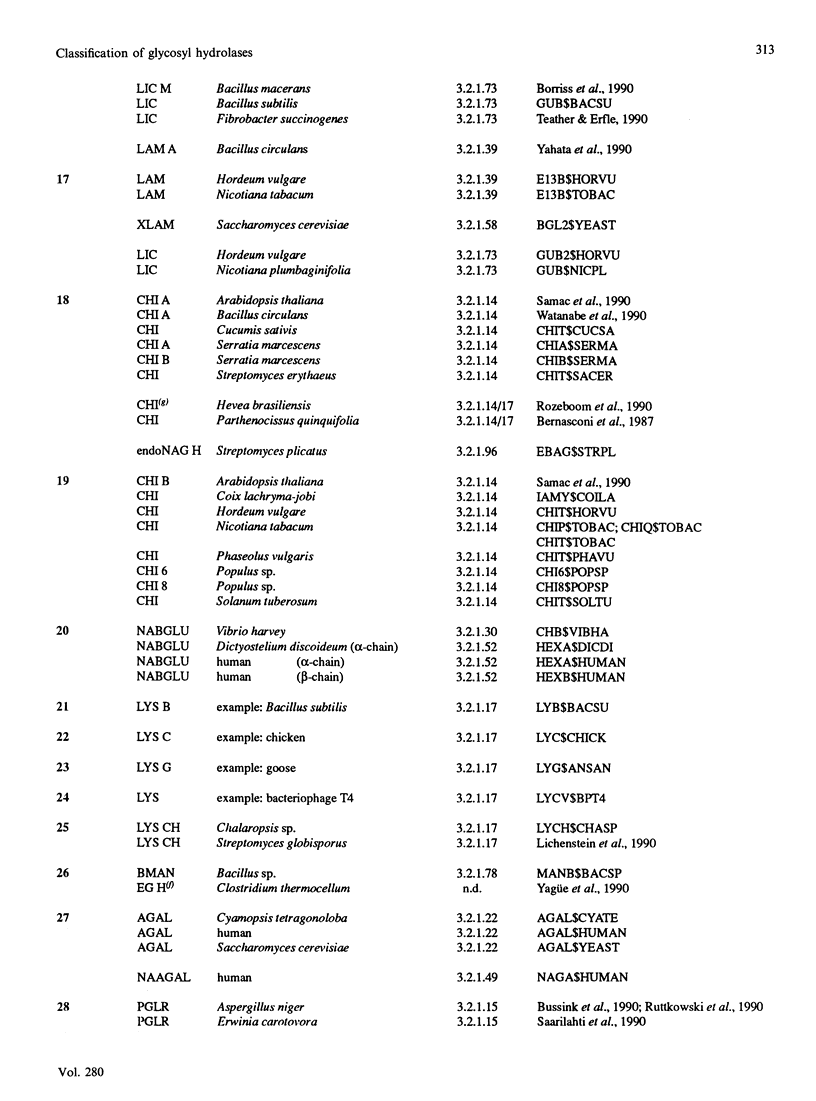
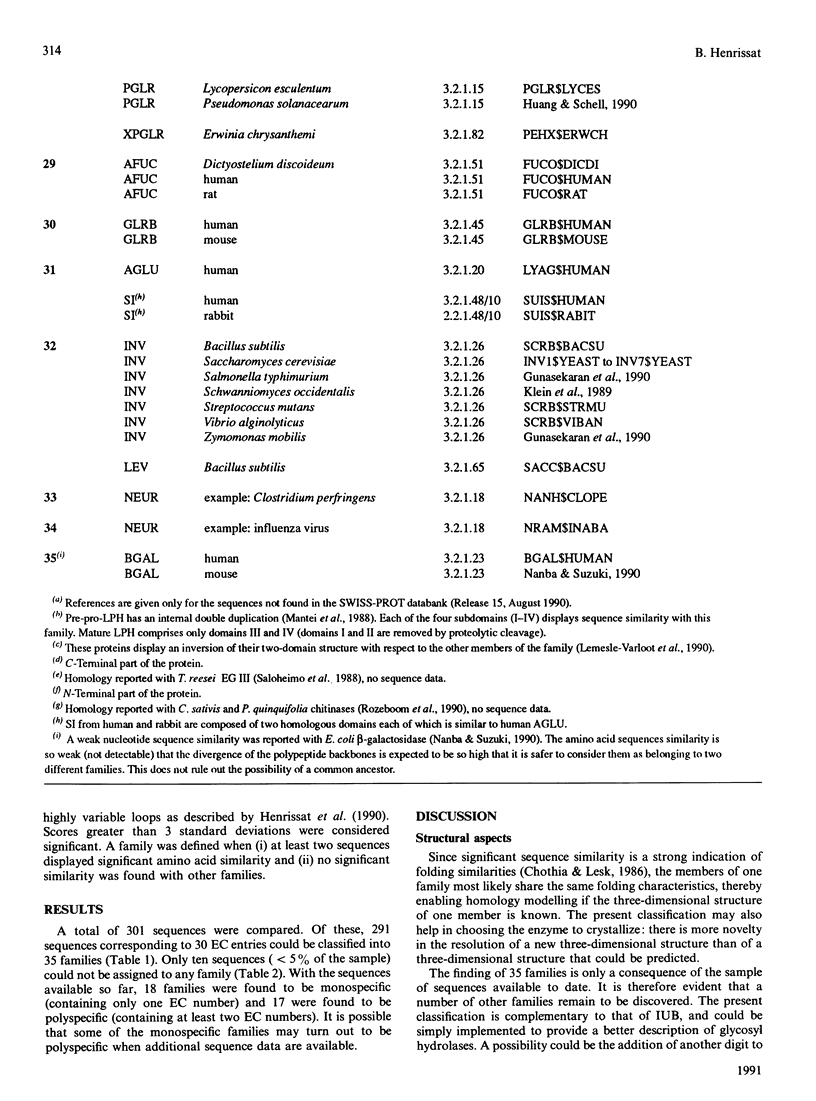
The amino acid sequences of 301 glycosyl hydrolases and related enzymes have been compared. A total of 291 sequences corresponding to 39 EC entries could be classified into 35 families. Only ten sequences (less than 5% of the sample) could not be assigned to any family. With the sequences available for this analysis, 18 families were found to be monospecific (containing only one EC number) and 17 were found to be polyspecific (containing at least two EC numbers). Implications on the folding characteristics and mechanism of action of these enzymes and on the evolution of carbohydrate metabolism are discussed. With the steady increase in sequence and structural data, it is suggested that the enzyme classification system should perhaps be revised.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AURICCHIO S., RUBINO A., LANDOLT M., SEMENZA G., PRADER A. ISOLATED INTESTINAL LACTASE DEFICIENCY IN THE ADULT. Lancet. 1963 Aug 17;2(7303):324–326. doi: 10.1016/s0140-6736(63)92991-x. [DOI] [PubMed] [Google Scholar]
- Aduse-Opoku J., Tao L., Ferretti J. J., Russell R. R. Biochemical and genetic analysis of Streptococcus mutans alpha-galactosidase. J Gen Microbiol. 1991 Apr;137(4):757–764. doi: 10.1099/00221287-137-4-757. [DOI] [PubMed] [Google Scholar]
- Bahl H., Burchhardt G., Spreinat A., Haeckel K., Wienecke A., Schmidt B., Antranikian G. alpha-Amylase of Clostridium thermosulfurogenes EM1: nucleotide sequence of the gene, processing of the enzyme, and comparison of other alpha-amylases. Appl Environ Microbiol. 1991 May;57(5):1554–1559. doi: 10.1128/aem.57.5.1554-1559.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-beta-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Baird S. D., Johnson D. A., Seligy V. L. Molecular cloning, expression, and characterization of endo-beta-1,4-glucanase genes from Bacillus polymyxa and Bacillus circulans. J Bacteriol. 1990 Mar;172(3):1576–1586. doi: 10.1128/jb.172.3.1576-1586.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E., Jones W. A., Jones D. T., Woods D. R. Cloning and sequencing of an endoglucanase (end1) gene from Butyrivibrio fibrisolvens H17c. Mol Gen Genet. 1989 Oct;219(1-2):193–198. doi: 10.1007/BF00261176. [DOI] [PubMed] [Google Scholar]
- Berger E., Jones W. A., Jones D. T., Woods D. R. Sequencing and expression of a cellodextrinase (ced1) gene from Butyrivibrio fibrisolvens H17c cloned in Escherichia coli. Mol Gen Genet. 1990 Sep;223(2):310–318. doi: 10.1007/BF00265068. [DOI] [PubMed] [Google Scholar]
- Borriss R., Buettner K., Maentsaelae P. Structure of the beta-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other beta-glucanases. Mol Gen Genet. 1990 Jul;222(2-3):278–283. doi: 10.1007/BF00633829. [DOI] [PubMed] [Google Scholar]
- Bueno A., Vazquez de Aldana C. R., Correa J., del Rey F. Nucleotide sequence of a 1,3-1,4-beta-glucanase-encoding gene in Bacillus circulans WL-12. Nucleic Acids Res. 1990 Jul 25;18(14):4248–4248. doi: 10.1093/nar/18.14.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink H. J., Kester H. C., Visser J. Molecular cloning, nucleotide sequence and expression of the gene encoding prepro-polygalacturonaseII of Aspergillus niger. FEBS Lett. 1990 Oct 29;273(1-2):127–130. doi: 10.1016/0014-5793(90)81066-w. [DOI] [PubMed] [Google Scholar]
- Cass L. G., Kirven K. A., Christoffersen R. E. Isolation and characterization of a cellulase gene family member expressed during avocado fruit ripening. Mol Gen Genet. 1990 Aug;223(1):76–86. doi: 10.1007/BF00315799. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R., East P. D., Watson K. endAFS, a novel family E endoglucanase gene from Fibrobacter succinogenes AR1. J Bacteriol. 1991 May;173(10):3265–3268. doi: 10.1128/jb.173.10.3265-3268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Tsukagoshi N., Udaka S. Nucleotide sequence of the cellobiohydrolase gene from Trichoderma viride. Nucleic Acids Res. 1990 Sep 25;18(18):5559–5559. doi: 10.1093/nar/18.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986 Apr;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubellis M. V., Rozzo C., Montecucchi P., Rossi M. Isolation and sequencing of a new beta-galactosidase-encoding archaebacterial gene. Gene. 1990 Sep 28;94(1):89–94. doi: 10.1016/0378-1119(90)90472-4. [DOI] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Faure E., Belaich A., Bagnara C., Gaudin C., Belaich J. P. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene. 1989 Dec 7;84(1):39–46. doi: 10.1016/0378-1119(89)90137-6. [DOI] [PubMed] [Google Scholar]
- Fukumori F., Kudo T., Sashihara N., Nagata Y., Ito K., Horikoshi K. The third cellulase of alkalophilic Bacillus sp. strain N-4: evolutionary relationships within the cel gene family. Gene. 1989;76(2):289–298. doi: 10.1016/0378-1119(89)90169-8. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Hall J., Hazlewood G. P., Ferreira L. M. The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol Microbiol. 1990 May;4(5):759–767. doi: 10.1111/j.1365-2958.1990.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Giorda R., Ohmachi T., Shaw D. R., Ennis H. L. A shared internal threonine-glutamic acid-threonine-proline repeat defines a family of Dictyostelium discoideum spore germination specific proteins. Biochemistry. 1990 Aug 7;29(31):7264–7269. doi: 10.1021/bi00483a015. [DOI] [PubMed] [Google Scholar]
- González-Candelas L., Ramón D., Polaina J. Sequences and homology analysis of two genes encoding beta-glucosidases from Bacillus polymyxa. Gene. 1990 Oct 30;95(1):31–38. doi: 10.1016/0378-1119(90)90410-s. [DOI] [PubMed] [Google Scholar]
- Gough C. L., Dow J. M., Keen J., Henrissat B., Daniels M. J. Nucleotide sequence of the engXCA gene encoding the major endoglucanase of Xanthomonas campestris pv. campestris. Gene. 1990 Apr 30;89(1):53–59. doi: 10.1016/0378-1119(90)90205-6. [DOI] [PubMed] [Google Scholar]
- Gunasekaran P., Karunakaran T., Cami B., Mukundan A. G., Preziosi L., Baratti J. Cloning and sequencing of the sacA gene: characterization of a sucrase from Zymomonas mobilis. J Bacteriol. 1990 Dec;172(12):6727–6735. doi: 10.1128/jb.172.12.6727-6735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K. R., Rockman E., Young C. A., Pearce L., Maddox I. S., Scott D. B. Expression and nucleotide sequence of the Clostridium acetobutylicum beta-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1991 May;173(10):3084–3095. doi: 10.1128/jb.173.10.3084-3095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Saloheimo M., Lavaitte S., Knowles J. K. Structural homology among the peroxidase enzyme family revealed by hydrophobic cluster analysis. Proteins. 1990;8(3):251–257. doi: 10.1002/prot.340080307. [DOI] [PubMed] [Google Scholar]
- Henrissat B. Sequence homology between a beta-galactosidase and some beta-glucosidases. Protein Seq Data Anal. 1991 Jul;4(1):61–62. [PubMed] [Google Scholar]
- Henrissat B. Weak sequence homologies among chitinases detected by clustering analysis. Protein Seq Data Anal. 1990 Dec;3(6):523–526. [PubMed] [Google Scholar]
- Hofmann B. E., Bender H., Schulz G. E. Three-dimensional structure of cyclodextrin glycosyltransferase from Bacillus circulans at 3.4 A resolution. J Mol Biol. 1989 Oct 20;209(4):793–800. doi: 10.1016/0022-2836(89)90607-4. [DOI] [PubMed] [Google Scholar]
- Huang J. H., Schell M. A. DNA sequence analysis of pglA and mechanism of export of its polygalacturonase product from Pseudomonas solanacearum. J Bacteriol. 1990 Jul;172(7):3879–3887. doi: 10.1128/jb.172.7.3879-3887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Yamashita I., Fukui S. Nucleotide sequence of the alpha-amylase gene (ALP1) in the yeast Saccharomycopsis fibuligera. FEBS Lett. 1987 Jul 27;219(2):339–342. doi: 10.1016/0014-5793(87)80248-x. [DOI] [PubMed] [Google Scholar]
- Jauris S., Rücknagel K. P., Schwarz W. H., Kratzsch P., Bronnenmeier K., Staudenbauer W. L. Sequence analysis of the Clostridium stercorarium celZ gene encoding a thermoactive cellulase (Avicelase I): identification of catalytic and cellulose-binding domains. Mol Gen Genet. 1990 Sep;223(2):258–267. doi: 10.1007/BF00265062. [DOI] [PubMed] [Google Scholar]
- Jørgensen P. L., Hansen C. K. Multiple endo-beta-1,4-glucanase-encoding genes from Bacillus lautus PL236 and characterization of the celB gene. Gene. 1990 Sep 1;93(1):55–60. doi: 10.1016/0378-1119(90)90135-e. [DOI] [PubMed] [Google Scholar]
- Kawazu T., Nakanishi Y., Uozumi N., Sasaki T., Yamagata H., Tsukagoshi N., Udaka S. Cloning and nucleotide sequence of the gene coding for enzymatically active fragments of the Bacillus polymyxa beta-amylase. J Bacteriol. 1987 Apr;169(4):1564–1570. doi: 10.1128/jb.169.4.1564-1570.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Poorman R. A., Favreau M. A., Shea M. H., Hatzenbuhler N. T., Nulf S. C. Cloning and sequence analysis of the gene encoding invertase from the yeast Schwanniomyces occidentalis. Curr Genet. 1989 Sep;16(3):145–152. doi: 10.1007/BF00391470. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989 Jun;135(6):1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- Lemesle-Varloot L., Henrissat B., Gaboriaud C., Bissery V., Morgat A., Mornon J. P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990 Aug;72(8):555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- Lichenstein H. S., Hastings A. E., Langley K. E., Mendiaz E. A., Rohde M. F., Elmore R., Zukowski M. M. Cloning and nucleotide sequence of the N-acetylmuramidase M1-encoding gene from Streptomyces globisporus. Gene. 1990 Mar 30;88(1):81–86. doi: 10.1016/0378-1119(90)90062-v. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Altschul S. F., Kececioglu J. D. A tool for multiple sequence alignment. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4412–4415. doi: 10.1073/pnas.86.12.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Jasmat N. B., Grayling R. A., Love D. R., Bergquist P. L. Cloning, sequence analysis, and expression in Escherichia coli of a gene coding for a beta-mannanase from the extremely thermophilic bacterium "Caldocellum saccharolyticum". Appl Environ Microbiol. 1991 Mar;57(3):694–700. doi: 10.1128/aem.57.3.694-700.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Love D. R., McAnulty J., Wallace C., Caughey P. A., Saul D., Bergquist P. L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Apr;56(4):1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor E. A., Svensson B. A super-secondary structure predicted to be common to several alpha-1,4-D-glucan-cleaving enzymes. Biochem J. 1989 Apr 1;259(1):145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M., Ohtsuki I., Fukui S., Yamashita I. Nucleotide sequences of Saccharomycopsis fibuligera genes for extracellular beta-glucosidases as expressed in Saccharomyces cerevisiae. Appl Environ Microbiol. 1988 Dec;54(12):3147–3155. doi: 10.1128/aem.54.12.3147-3155.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannarelli B. M., Evans S., Lee D. Cloning, sequencing, and expression of a xylanase gene from the anaerobic ruminal bacterium Butyrivibrio fibrisolvens. J Bacteriol. 1990 Aug;172(8):4247–4254. doi: 10.1128/jb.172.8.4247-4254.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantei N., Villa M., Enzler T., Wacker H., Boll W., James P., Hunziker W., Semenza G. Complete primary structure of human and rabbit lactase-phlorizin hydrolase: implications for biosynthesis, membrane anchoring and evolution of the enzyme. EMBO J. 1988 Sep;7(9):2705–2713. doi: 10.1002/j.1460-2075.1988.tb03124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita O., Russell J. B., Wilson D. B. Cloning and sequencing of a Bacteroides ruminicola B(1)4 endoglucanase gene. J Bacteriol. 1990 Jul;172(7):3620–3630. doi: 10.1128/jb.172.7.3620-3630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke A., Braun C., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Unusual sequence organization in CenB, an inverting endoglucanase from Cellulomonas fimi. J Bacteriol. 1991 Jan;173(1):308–314. doi: 10.1128/jb.173.1.308-314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moranelli F., Barbier J. R., Dove M. J., MacKay R. M., Seligy V. L., Yaguchi M., Willick G. E. A clone coding for Schizophyllum commune beta-glucosidase: homology with a yeast beta-glucosidase. Biochem Int. 1986 Jun;12(6):905–912. [PubMed] [Google Scholar]
- Nanba E., Suzuki K. Molecular cloning of mouse acid beta-galactosidase cDNA: sequence, expression of catalytic activity and comparison with the human enzyme. Biochem Biophys Res Commun. 1990 Nov 30;173(1):141–148. doi: 10.1016/s0006-291x(05)81033-2. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neufeld E. F., Lim T. W., Shapiro L. J. Inherited disorders of lysosomal metabolism. Annu Rev Biochem. 1975;44:357–376. doi: 10.1146/annurev.bi.44.070175.002041. [DOI] [PubMed] [Google Scholar]
- Ooi T., Shinmyo A., Okada H., Murao S., Kawaguchi T., Arai M. Complete nucleotide sequence of a gene coding for Aspergillus aculeatus cellulase (FI-CMCase). Nucleic Acids Res. 1990 Oct 11;18(19):5884–5884. doi: 10.1093/nar/18.19.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K., Shikata S., Kawai S., Ito S., Okamoto K. Molecular cloning and nucleotide sequence of a gene for alkaline cellulase from Bacillus sp. KSM-635. J Gen Microbiol. 1990 Jul;136(7):1327–1334. doi: 10.1099/00221287-136-7-1327. [DOI] [PubMed] [Google Scholar]
- Park S. H., Kim H. K., Pack M. Y. Characterization and structure of the cellulase gene of Bacillus subtilis BSE616. Agric Biol Chem. 1991 Feb;55(2):441–448. [PubMed] [Google Scholar]
- Poole D. M., Hazlewood G. P., Laurie J. I., Barker P. J., Gilbert H. J. Nucleotide sequence of the Ruminococcus albus SY3 endoglucanase genes celA and celB. Mol Gen Genet. 1990 Sep;223(2):217–223. doi: 10.1007/BF00265057. [DOI] [PubMed] [Google Scholar]
- Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991 Feb;4(3):325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- Raimbaud E., Buleon A., Perez S., Henrissat B. Hydrophobic cluster analysis of the primary sequences of alpha-amylases. Int J Biol Macromol. 1989 Aug;11(4):217–225. doi: 10.1016/0141-8130(89)90072-x. [DOI] [PubMed] [Google Scholar]
- Rhodes C., Strasser J., Friedberg F. Sequence of an active fragment of B. polymyxa beta amylase. Nucleic Acids Res. 1987 May 11;15(9):3934–3934. doi: 10.1093/nar/15.9.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozeboom H. J., Budiani A., Beintema J. J., Dijkstra B. W. Crystallization of hevamine, an enzyme with lysozyme/chitinase activity from Hevea brasiliensis latex. J Mol Biol. 1990 Apr 5;212(3):441–443. doi: 10.1016/0022-2836(90)90321-C. [DOI] [PubMed] [Google Scholar]
- Russell R. R., Ferretti J. J. Nucleotide sequence of the dextran glucosidase (dexB) gene of Streptococcus mutans. J Gen Microbiol. 1990 May;136(5):803–810. doi: 10.1099/00221287-136-5-803. [DOI] [PubMed] [Google Scholar]
- Ruttkowski E., Labitzke R., Khanh N. Q., Löffler F., Gottschalk M., Jany K. D. Cloning and DNA sequence analysis of a polygalacturonase cDNA from Aspergillus niger RH5344. Biochim Biophys Acta. 1990 Sep 10;1087(1):104–106. doi: 10.1016/0167-4781(90)90130-t. [DOI] [PubMed] [Google Scholar]
- Saarilahti H. T., Heino P., Pakkanen R., Kalkkinen N., Palva I., Palva E. T. Structural analysis of the pehA gene and characterization of its protein product, endopolygalacturonase, of Erwinia carotovora subspecies carotovora. Mol Microbiol. 1990 Jun;4(6):1037–1044. doi: 10.1111/j.1365-2958.1990.tb00676.x. [DOI] [PubMed] [Google Scholar]
- Sakka K., Shimanuki T., Shimada K. Nucleotide sequence of celC307 encoding endoglucanase C307 of Clostridium sp. strain F1. Agric Biol Chem. 1991 Feb;55(2):347–350. doi: 10.1271/bbb1961.55.347. [DOI] [PubMed] [Google Scholar]
- Saloheimo M., Lehtovaara P., Penttilä M., Teeri T. T., Ståhlberg J., Johansson G., Pettersson G., Claeyssens M., Tomme P., Knowles J. K. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63(1):11–22. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- Samac D. A., Hironaka C. M., Yallaly P. E., Shah D. M. Isolation and Characterization of the Genes Encoding Basic and Acidic Chitinase in Arabidopsis thaliana. Plant Physiol. 1990 Jul;93(3):907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson B. Regional distant sequence homology between amylases, alpha-glucosidases and transglucanosylases. FEBS Lett. 1988 Mar 28;230(1-2):72–76. doi: 10.1016/0014-5793(88)80644-6. [DOI] [PubMed] [Google Scholar]
- Teather R. M., Erfle J. D. DNA sequence of a Fibrobacter succinogenes mixed-linkage beta-glucanase (1,3-1,4-beta-D-glucan 4-glucanohydrolase) gene. J Bacteriol. 1990 Jul;172(7):3837–3841. doi: 10.1128/jb.172.7.3837-3841.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto A., Kimura K., Ishii Y., Takano T., Yamane K. Nucleotide sequence of the maltohexaose-producing amylase gene from an alkalophilic Bacillus sp. #707 and structural similarity to liquefying type alpha-amylases. Biochem Biophys Res Commun. 1988 Feb 29;151(1):25–31. doi: 10.1016/0006-291x(88)90554-2. [DOI] [PubMed] [Google Scholar]
- Tucker M. L., Milligan S. B. Sequence analysis and comparison of avocado fruit and bean abscission cellulases. Plant Physiol. 1991 Mar;95(3):928–933. doi: 10.1104/pp.95.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Suzuki K., Oyanagi W., Ohnishi K., Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990 Sep 15;265(26):15659–15665. [PubMed] [Google Scholar]
- Yagüe E., Béguin P., Aubert J. P. Nucleotide sequence and deletion analysis of the cellulase-encoding gene celH of Clostridium thermocellum. Gene. 1990 Apr 30;89(1):61–67. doi: 10.1016/0378-1119(90)90206-7. [DOI] [PubMed] [Google Scholar]
- Yahata N., Watanabe T., Nakamura Y., Yamamoto Y., Kamimiya S., Tanaka H. Structure of the gene encoding beta-1,3-glucanase A1 of Bacillus circulans WL-12. Gene. 1990 Jan 31;86(1):113–117. doi: 10.1016/0378-1119(90)90122-8. [DOI] [PubMed] [Google Scholar]
- Yoshigi N., Taniguchi H., Sasaki T. Cloning and sequencing of the endo-cellulase cDNA from Robillarda sp. Y-20. J Biochem. 1990 Sep;108(3):388–392. doi: 10.1093/oxfordjournals.jbchem.a123211. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T., Anraku Y. Nucleotide sequence of AMS1, the structure gene of vacuolar alpha-mannosidase of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1989 Sep 15;163(2):908–915. doi: 10.1016/0006-291x(89)92308-5. [DOI] [PubMed] [Google Scholar]
- Zhou J. H., Baba T., Takano T., Kobayashi S., Arai Y. Nucleotide sequence of the maltotetraohydrolase gene from Pseudomonas saccharophila. FEBS Lett. 1989 Sep 11;255(1):37–41. doi: 10.1016/0014-5793(89)81056-7. [DOI] [PubMed] [Google Scholar]
- Zvelebil M. J., Sternberg M. J. Analysis and prediction of the location of catalytic residues in enzymes. Protein Eng. 1988 Jul;2(2):127–138. doi: 10.1093/protein/2.2.127. [DOI] [PubMed] [Google Scholar]


