Abstract
Human cytomegalovirus (HCMV) infects a number of organs and cell types in vivo, leading to the hypothesis that HCMV disease and tissue tropism may be related to specific sequence variants. A potential component of HCMV variant strains is the UL144 open reading frame (ORF), which encodes a homologue of the herpesvirus entry mediator, HveA, a member of the tumor necrosis factor receptor superfamily. Sequence analysis of the UL144 ORF in 45 low-passage clinical isolates demonstrated significant strain-specific variability. In individual isolates, nucleotide substitutions occur at up to 21% of the 531 positions, resulting in approximately the same percentage of substitutions in the predicted 176-amino-acid sequence. Phylogenetic analysis indicated that the nucleotide and amino acid sequences diverge into three major groups. For genotypic comparison, the known hypervariable region encompassing the proteolytic cleavage site of the glycoprotein B (gB) gene was also sequenced. All of the isolates could be typed according to the four known gB groups; however, the gB and UL144 sequence groups appeared to be phylogenetically unlinked. The predicted UL144 product homology with tumor necrosis factor receptor family members, along with the unexpectedly high level of sequence variability of the UL144 ORF, suggests that the predicted product may play a role in HCMV infectivity and subsequent host disease.
Human cytomegalovirus (HCMV) infection leads to serious disease under the immunosuppressive conditions of AIDS and organ transplantation; however, the mechanisms of viral pathogenesis are not well understood. The diversity of organs and cell types infected by HCMV in vivo has led to the hypothesis that HCMV disease and tissue tropism may be related to sequence variation among strains (12, 14, 29, 30, 50). The most widely characterized variant gene is glycoprotein B (gB), for which there are four major sequence groups determined by the region surrounding the proteolytic cleavage site (12). A number of studies have attempted to correlate gB types with specific disease manifestations or sites of infectivity (6, 10, 14, 29, 30, 44, 50, 52, 56). The results have not established definitive associations between gB types and HCMV disease, leaving open the possibility that other variant HCMV-encoded products, possibly in combination with gB, may play a role in viral pathogenesis.
A new region of DNA containing at least 19 open reading frames (ORFs) was recently found (8) in the low-passage HCMV clinical strain Toledo and several other low-passage clinical isolates. This region, however, is not present in the HCMV laboratory strain AD169 (EMBL accession no. X17403). Thus, the products encoded by these ORFs can be deleted with no apparent effect on the viability of HCMV in cell culture. However, the fact that the clinical isolates retain the ORFs suggests that the predicted gene products may be essential for viral infection in vivo.
The possibility that these new ORFs may provide genetic markers for HCMV pathogenesis prompted us to investigate these genes in HCMV clinical isolates. The UL144 ORF is a prime candidate for a pathogenesis marker because it encodes a homologue of the herpes simplex virus entry mediator (HveA or HVEM) (4), a member of the tumor necrosis factor receptor (TNFR) superfamily (3, 15, 47, 55). The TNFR superfamily plays several roles in innate defenses and adaptive immune responses that are critical for defense against viral pathogens such as HCMV (7, 38). During our investigation of this ORF in low-passage clinical isolates, we discovered significant strain-specific sequence variability. In this report, we have determined the extent and nature of this variability for a large group of these isolates.
MATERIALS AND METHODS
Cells and virus.
All virus strains were propagated in human foreskin fibroblasts (HFF) (Viromed, Minneapolis, Minn.) in Eagle’s minimum essential medium (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum, 20 mM HEPES buffer, 2.5 μg of amphotericin B per ml, and 50 μg of gentamicin per ml. The laboratory strain AD169 was obtained from Karen Biron at Glaxo-Wellcome, Research Triangle Park, N.C. HCMV clinical strains were obtained through the Rush Medical Laboratories, Rush-Presbyterian-St. Luke’s Medical Center, Chicago, Ill., and the Clinical Microbiology Laboratory, Loyola University Medical Center, Maywood, Ill. Clinical strains were passaged as cell-associated virus five to seven times in HFF. All strains retained the cell-associated phenotype when the infected cells were harvested for virus stocks.
Southern hybridization.
Restriction enzyme digestion fragments of HCMV genomic DNA from both laboratory and clinical strains were separated by electrophoresis on 0.7% agarose gels and transferred to a Magna Charge membrane (Micron Separations Inc., Westborough, Mass.). A PCR-amplified product obtained with a DNA template from clinical isolate PT30 (Table 1) was radiolabeled with 32P-dCTP (Amersham, Arlington Heights, Ill.) by random priming (Gibco BRL) for use as a probe. Hybridization was carried out in 50% formamide at 42°C overnight as previously described (25).
TABLE 1.
HCMV clinical strains
| Strain | Clinical history | Sourcea | Target organb | Group
|
|
|---|---|---|---|---|---|
| gB | UL144 | ||||
| PT26 | Liver transplant | BAL | Lung | 2 | 1A |
| PT27 | Liver transplant | Blood | NA | 1 | 1A |
| NW17 | Lung transplant | BAL | Lung | 3 | 1A |
| NW24 | Lung transplant | BAL | Lung | 3 | 1A |
| PT20 | Kidney transplant | Urine | NA | 4 | 1A |
| PT21 | AIDS | Blood | NA | 3 | 1A |
| PT22 | Bone marrow transplant | BAL | Lung | 3 | 1A |
| PT23 | Lung transplant | BAL | Lung | 2 | 1A |
| PT24 | AIDS | Blood | None | 4 | 1A |
| PT25 | AIDS | Urine | None | 4 | 1A |
| NW4 | Heart transplant | Blood | NA | 3 | 1B |
| PT28 | Liver transplant | BAL | Lung | 1 | 1B |
| PT29 | Congenital HCMV | Urine | NA | 3 | 1B |
| PT30 | Heart transplant | Blood | NA | 1 | 1B |
| NW14 | Lung transplant | BAL | Lung | 1 | 1C |
| PT19 | AIDS | Oral lesion | None | 2 | 1C |
| NW13 | Lung transplant | BAL | Lung | 2 | 1C |
| NW23 | Lung transplant | BAL | Lung | 1 | 1C |
| PT16 | Liver transplant | BAL | Lung | 1 | 2 |
| PT17 | Lung cancer | Lung tissue | Lung | 4 | 2 |
| PT18 | AIDS | BAL | Lung | 1 | 2 |
| NW9 | Heart transplant | Blood | NA | 2 | 2 |
| PT1 | Kidney transplant | Urine | NA | 3 | 3 |
| PT2 | Lung cancer | BAL | Lung | 2 | 3 |
| PT3 | Heart transplant | Urine | NA | 2 | 3 |
| PT4 | Lung transplant | BAL | Lung | 1 | 3 |
| PT5 | Lung cancer | Lung tissue | Lung | 3 | 3 |
| PT6 | AIDS | BAL | Lung | 4 | 3 |
| PT7 | Lung transplant | GI tract lesion | GI tract | 1 | 3 |
| PT8 | Lung transplant | BAL | Lung | 1 | 3 |
| PT9 | Heart transplant | Blood | None | 1 | 3 |
| PT10 | Heart transplant | Blood | None | 1 | 3 |
| PT11 | AIDS | BAL | Lung/retina | 2 | 3 |
| PT12 | Bone marrow transplant | BAL | Lung | 1 | 3 |
| PT13 | Lung transplant | BAL | Lung | 2 | 3 |
| PT14 | Heart transplant | BAL | Lung | 2 | 3 |
| PT15 | AIDS | Blood | Retina | 3 | 3 |
| NW1 | Lung transplant | BAL | Lung | 1 | 3 |
| NW7 | Lung transplant | BAL | Lung | 1 | 3 |
| NW8 | Lung transplant | BAL | Lung | 1 | 3 |
| NW12 | Lung transplant | BAL | Lung | 3 | 3 |
| NW16 | Bone marrow transplant | Blood | NA | 1 | 3 |
| NW20 | Lung transplant | BAL | Lung | 1 | 3 |
| NW22 | Lung transplant | BAL | Lung | 2 | 3 |
| NW25 | Kidney transplant | BAL | Lung | 1 | 3 |
BAL, bronchial alveolar lavage; GI, gastrointestinal.
NA, not available.
PCR amplification.
Genomic viral DNA was extracted from cells infected with laboratory or clinical HCMV strains as previously described (25). The extracted DNA was diluted in water to optimal concentrations for use as templates for PCR amplification of specific regions of the HCMV genome.
In our initial study of the region containing the 19 ORFs homologous to the Toledo strain sequence (8), a 399-bp portion (nucleotides 6 to 405) of the UL144 coding sequence was amplified with the primer pair designated UL144-A: forward, 5′-GCCTCTGATAATGCTCATCTGC-3′; and reverse, 5′-GGCTAGAGTATGACGACCGCTT-3′. For subsequent studies of the complete UL144 coding sequence, a 737-bp product was amplified with the primer pair designated UL144-B: forward, 5′-TCGTATTACAAACCGCGGAGAGGAT-3′; and reverse, 5′-ACTCAGACACGGTTCCGTAA-3′. A 410-bp fragment encompassing the proteolytic cleavage site (nucleotides 1072 to 1482) of the HCMV gB gene (EMBL accession no. X04606) was amplified with the following primers: forward, 5′-TCCGAAGCCGAAGACTCGTA-3′; and reverse, 5′-GATGTAACCGCGCAACGTGT-3′. Amplification was carried out with a Perkin-Elmer Cetus DNA Thermal Cycler (Perkin-Elmer Instruments, Norwalk, Conn.). Ampliwax (Roche Molecular Systems, Branchburg, N.J.) was used to achieve a hot start. The conditions for amplification with all primer sets were 94°C for 5 min followed by 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The 30 cycles were followed by a single extension cycle at 72°C for 5 min. PCR products were purified and concentrated with Microcon 100 microconcentrators (Amicon, Beverly, Mass.) for use as sequencing templates.
DNA sequencing.
The concentrated products were sequenced directly with the BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, Calif.). Sequencing reactions were performed with a PE Applied Biosystems Geneamp PCR System 2400 or 9700 at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min for a total of 30 cycles. The sequencing products were analyzed on an ABI 377 automated sequencer (PE Applied Biosystems). Sequences were aligned with Align Plus version 3.0 (Scientific and Educational Software, Durham, N.C.).
RT-PCR.
For reverse transcription (RT)-PCR, total RNA was isolated from approximately 4 × 106 HCMV-infected HFF at 96 h postinfection. The cells were trypsinized, pelleted, and resuspended in 1 ml of Trizol (Gibco BRL). The Trizol suspension was extracted with chloroform, and the aqueous phase was precipitated with isopropanol. The pellet was washed in 75% ethanol, air dried, and resuspended in diethyl pyrocarbonate-treated water. The extracted RNA was treated twice with RNase-free DNase (Promega, Madison, Wis.), ethanol precipitated, and resuspended in diethyl pyrocarbonate-treated water at a concentration of 3 to 4 μg/μl.
The RNA was reverse transcribed with oligo(dT) or gene-specific reverse primers and 1 U of Superscript II reverse transcriptase (Gibco BRL). The reaction mixtures were incubated at 42°C for 60 min. The templates from the RT reactions were amplified with the UL144-B or gB primer pairs described above for PCR amplification. In addition, the following primers were used to amplify a 230-bp product from the β-actin gene for positive RNA and loading controls: forward, 5′-GGGTCAGAAGGATTCCTATG-3′; and reverse, 5′-TCTCAAACATGATCTGGGTC-3′. PCR was performed at 94°C for 2 min for 1 cycle followed by 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s for 30 cycles with a PE Applied Biosystems 9700 thermal cycler. Control reactions for residual DNA contamination were performed as described above but without the reverse transcriptase step.
Phylogenetic analysis.
Phylogenetic analysis was performed with program modules from the Phylip program package (version 3.57c; obtained from J. Felsenstein, University of Washington, Seattle). The program module SEQBOOT was used to generate 100 bootstrap data sets. Genetic distances were calculated from the bootstrap data with DNADIST, and the resulting values were used to generate phylogenetic trees with NEIGHBOR. A consensus tree was computed with CONSENSE. The final tree was rendered with TreeView (34). Amino acid sequences were aligned with Align Plus version 3.0. Protein homologues of the predicted UL144 product were identified by a BLAST (1) search of protein databases. Functional motifs were identified from the PROSITE database (2).
Nucleotide sequence accession numbers.
The UL144 sequences of the clinical strains have been assigned GenBank accession no. AF084976 to AF085005 and AF179196 to AF179210.
RESULTS
Profiles of clinical strains.
The original report of Cha et al. (8), which suggested that the newly described 19 ORFs could be responsible for HCMV virulence and tissue tropism, was based on the Toledo strain and only five additional clinical isolates. To demonstrate that these ORFs are consistently present in a large number of HCMV clinical strains, a total of 45 clinical strains were selected from strains collected for our repository over the past 5 years (Table 1). Strains were selected for diversity of underlying patient disease; however, the relatively large number of strains from lung transplant recipients reflects the frequency of HCMV infection in that patient population. The numbers of patients and various types of diseases or associated clinical conditions are as follows: 8 AIDS, 3 lung cancer, 7 heart transplant, 16 lung transplant, 3 bone marrow transplant, 3 kidney transplant, 4 liver transplant, and 1 congenital HCMV. The sample source of each strain is also listed in Table 1, along with the target organ, if known. Several patients had HCMV infections without apparent end organ disease.
Included in this study are strains PT7 and PT8, from two different lung transplant patients who received lungs from the same donor. These strains were previously shown to be identical by comparison of restriction digests and DNA sequence analysis (25) and were included for comparison of the effects of long-term infection of epidemiologically related strains in different hosts. The laboratory strain AD169 was assayed in parallel with the clinical strains.
Presence of a DNA region homologous to the Toledo sequence in clinical strains.
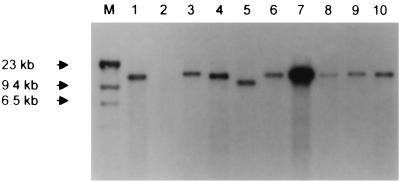
The initial approach in characterizing the strains was to determine whether sequences homologous to ORFs from the Toledo sequence (GenBank accession no. U33331) were present in each of the 45 clinical strains. DNA extracted from cells infected with each strain or AD169 was digested with EcoRI. Southern blots of these digests were probed with a radiolabeled PCR product derived from the amplification of strain PT30 with primer pairs targeting specific ORFs across the region. The UL144-A primer pair was used for the UL144 ORF. As predicted from the Toledo restriction map (8) (Fig. 1), EcoRI restrictions of all clinical strains showed a single large restriction fragment hybridizing with the UL144-A probe. A representative blot is shown in Fig. 2. The EcoRI digest of AD169 DNA (Fig. 2, lane 2), as predicted, showed no fragment hybridizing with the probe. Also shown in Fig. 2 are EcoRI digests of DNA from different clinical strains. All have a detectable large band, indicating that at least a portion of the UL144 ORF is present in all of the strains. There are differences in the sizes of the hybridizing fragments among the strains, a result which may be a reflection of sequence variability leading to gain or loss of EcoRI restriction sites. Alternatively, in some of the strains there may be short deletions and/or insertions within the region represented by the EcoRI fragment.
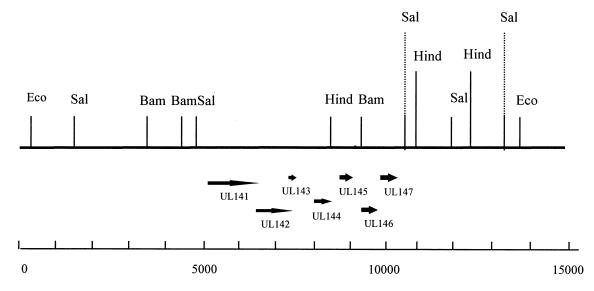
FIG. 1.
Restriction map of the region of HCMV strain Toledo not present in laboratory strains (8). The relative sizes and direction of the UL144 and adjacent ORFs are marked. The scale at the bottom shows numbers of base pairs, based on the Toledo sequence. Restriction enzymes: Eco, EcoRI; Sal, SalI; Bam, BamHI; Hind, HindIII.
FIG. 2.
Southern blot of EcoRI restriction digests of DNA from AD169 and clinical strains probed with a 32P-labeled PT30 UL144 PCR product (UL144-B primer pair). Lane M, lambda HindIII markers; lane 2, AD169; lanes 1 and 3 through 10, clinical strains.
Presence of the complete UL144 ORF in clinical strains.
To address the possibility that some ORFs could be deleted in some of the strains, the amplification of products from specific ORFs within the region of the EcoRI fragment was attempted. During the preliminary comparative PCR analysis, the UL144-A primer pair was used to detect the presence of the UL144 ORF. Less than half of the clinical strains tested with this primer pair yielded the predicted 400-bp product. To determine whether the lack of a product resulted from deletion or mutation of a primer binding site, the UL144-B primer pair was designed for subsequent PCR analysis. With this primer pair, which flanks the UL144 ORF, a product approximating the predicted size of 737 bp was obtained from all 45 clinical strains listed in Table 1. These results suggested that the failure to amplify a product with the UL144-A primer pair resulted from sequence variability rather than deletion. As expected, no product could be amplified from AD169 DNA with either primer set (24).
DNA sequence and phylogenetic analysis of UL144.
Primer binding can be disrupted by only a few base changes; therefore, lack of amplification gives no indication of the number of nucleotide changes between primer sites. To determine the extent of sequence variability for the entire UL144 ORF, the PCR product amplified with the UL144-B primer pair from each of the 45 clinical strains was sequenced directly. A comparison of the sequencing results showed the ORF to be hypervariable. Ten strains have sequences either identical or very similar to that of Toledo. However, over half of the strains have more than 70 nucleotide differences relative to Toledo. These changes are concentrated in the 5′ half of the gene, although some changes occur throughout the remainder of the coding sequence. In these strains, the binding site for the forward primer of the UL144-A primer pair has up to six base substitutions, and the binding site for the reverse primer has up to three base substitutions. This variability likely explains the inability to consistently obtain PCR products with the UL144-A primer pair.
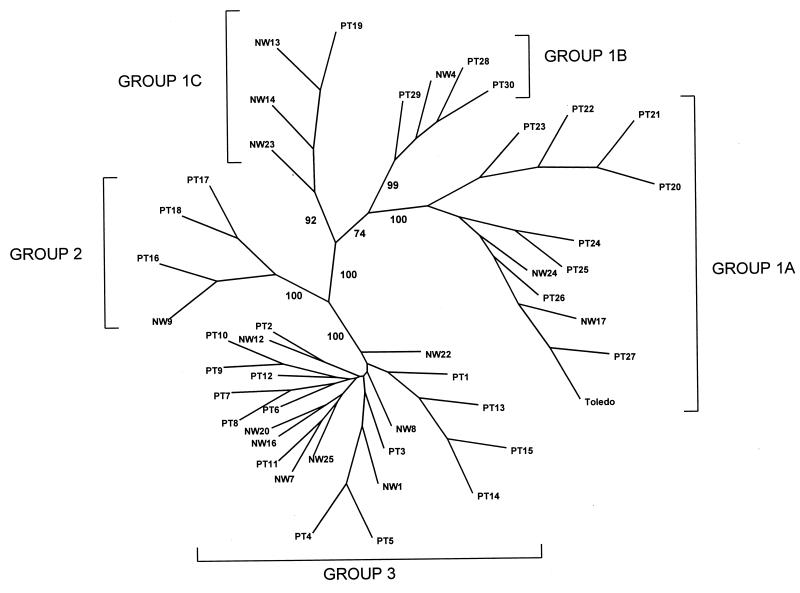
Visual inspection of the DNA sequences from all 45 strains indicated that there are groups of relatively conserved sequence variants. To confirm this observation, the sequences were subjected to phylogenetic analysis to generate an unrooted tree (Fig. 3). Definitive bootstrap values of 70% or higher for the major branch points (16) are shown. Based on this analysis, 11 strains, including Toledo (Table 1), form a major sequence cluster, which we have designated subgroup 1A. The DNA sequences of strains within this group differ from that of Toledo by no more than 7 nucleotides out of the 531 nucleotides in the complete ORF. Strains PT28, PT29, PT30, and NW4 form a second subgroup, 1B, which differs from Toledo by 15 to 20 nucleotides. Strains PT19, NW13, NW14, and NW23 form a third subgroup, 1C, which differs from Toledo by 16 to 31 nucleotides. Strains PT16, PT17, and PT18 form a second major sequence cluster, group 2, with 74 to 77 nucleotide substitutions relative to Toledo. The remaining 24 strains form group 3. The UL144 sequences of the strains in this group differ from that of Toledo by over 100 nucleotides. A comparison of all three sequence groups shows that with Toledo as the reference, at least 50% of the nucleotide substitutions are nonsynonymous. The exceptions are subgroup 1A strains PT26, PT27, and NW17, which are phylogenetically closest to Toledo and have either no substitutions or synonymous substitutions. In addition, subgroup 1B strains NW4 and PT28 have 20 and 15 nucleotide substitutions, respectively; however, 35 and 47%, respectively, from each strain are nonsynonymous. In contrast, averages of 57 and 65% of the nucleotide substitutions in group 2 and group 3 strains, respectively, are translated into amino acid changes.
FIG. 3.
Results of phylogenetic analysis of the 45 UL144 DNA sequences. An unrooted tree was generated from the Phylip program CONSENSE data with TreeView (34). Bootstrap values for major branches are shown.
Amino acid sequence analysis of the predicted UL144 protein.
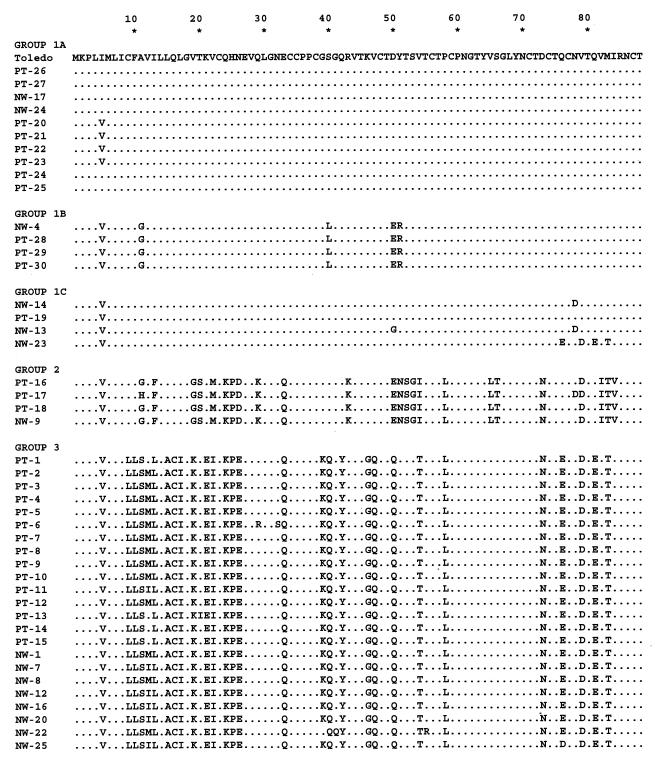
The large number of nonsynonymous nucleotide substitutions found in the sequences of strains of the different UL144 DNA sequence groups by definition leads to the large number of amino acid substitutions. This fact is illustrated by the amino acid sequence alignment of the 45 strains, which were aligned with Toledo as the arbitrary reference (Fig. 4). Visual inspection of the sequences supports the DNA phylogenetic analysis. Subgroup 1A strains differ from Toledo by a maximum of 3 amino acid substitutions, while subgroup 1B strains (PT28, PT29, PT30, and NW4) differ by 6 to 8 substitutions. Subgroup 1C strains have up to 14 substitutions relative to Toledo. The amino acid sequences of group 2 and group 3 strains reflect a significant DNA sequence divergence from Toledo. There are 29 to 32 amino acid substitutions in group 2 and 35 to 37 substitutions and one deletion in group 3, out of the total predicted 176 residues for the Toledo UL144 product. These values represent a range of 16 to 21% amino acid variability relative to Toledo, and the majority of the amino acid substitutions for both groups are nonconservative. Note that the amino acid sequences of the subgroup 1C strains from residues 97 through 161 resemble those of group 3 strains, while the N-terminal sequences are very similar to those of subgroup 1A and Toledo strains.
FIG. 4.
Alignment of amino acid sequences of the 45 clinical strains with the Toledo sequence as a reference. Numbers above the sequences represent amino acid residues. Dots indicate identity. Strains are listed according to sequence groups determined by phylogenetic analysis of the DNA sequences.
The amino acid sequence alignment in Fig. 4 also shows that strains from both group 2 and group 3 have highly conserved group-specific substitutions relative to Toledo and different from each other. These group-specific changes are concentrated in the N-terminal half of the gene, although a few occur in the C-terminal portion. Amino acid residues 137 to 176 are invariant among all strains, except for the conservative substitution of lysine for arginine at residue 161 in four strains. The alignment shows a high level of identity of the predicted amino acid sequences among strains within each group, with occasional substitutions also being maintained across sequence groups. For example, there is a deletion of glutamine 115 in all strains in group 3 as well as in the majority of strains in subgroup 1C. In addition, between residues 97 and 106, the subgroup 1C strains have substitutions in common with the group 3 strains.
The distribution of UL144 genotypes among the 45 strains, based on the major sequence groups and the three subgroups, is as follows: subgroup 1A, 22%; subgroup 1B, 9%; subgroup 1C, 9%; group 2, 9%; and group 3, 51%. It appears from the data summarized in Table 1 that no group is clearly associated with a single type of underlying condition or target organ. Of the 35 patients for whom clinical data were available, 28 had HCMV pneumonitis, with the following distribution among UL144 sequence groups: subgroup 1A, 5 (18%); subgroup 1B, 1 (3.6%); subgroup 1C, 3 (10.7%); group 2, 3 (10.7%); and group 3, 16 (57%). Two of the patients with AIDS had retinitis; however, strains PT11 and PT15 were obtained from blood and bronchial alveolar lavage cultures, respectively. Several strains were isolated from blood or urine samples, although the patients had no HCMV end organ disease when the virus was obtained. Included among this group of patients were two with AIDS (strains PT24 and PT25). The small number of strains from patients with known end organ disease other than the lung does not provide enough data to determine a possible association with specific UL144 sequence groups. In addition, the long-term clinical outcome of HCMV disease in each patient is not available; thus, no conclusions can be drawn concerning potential pathogenicity associated with particular UL144 sequence groups.
As noted above, the 45 strains are unrelated, except for PT7 and PT8. These strains were obtained from two lung transplant patients with the same donor and were previously shown to be identical, based on sequence comparisons of the DNA polymerase and UL97 phosphotransferase genes (25). The UL144 DNA sequences of PT7 and PT8 are also identical and fall into group 3. Therefore, the UL144 sequence was conserved during passage of the original donor strain in two different hosts. We also have observed conservation of the UL144 sequence in three other isolates collected over a period of 5 years from the patient from whom PT7 was isolated (24). In addition, we have comparative sequence data from serial isolates from two patients not included in this study (24) showing that the UL144 sequence is maintained in all isolates from the same patient. These data indicate that, despite the hypervariability among unrelated strains, the UL144 sequence does not change during in vivo passage in the same host and that sequence adaptation during passage in cell culture is unlikely.
Structural analysis of the predicted UL144 protein.
From the original Toledo sequence (8), the UL144 ORF is predicted to encode a type 1 transmembrane glycoprotein with features common to the TNFR superfamily; therefore, the effect of the strain-specific sequence variability on this homology was analyzed. Despite the observed hypervariability, the basic structural features of the TNFR superfamily are shared by all strains from all groups. The consensus sequences for the major UL144 groups are aligned in the top three lines of Fig. 5. All contain an N-terminal signal peptide (53), seven or eight N-linked glycosylation sites, depending on the sequence group (2), two cysteine-rich domains (CRDs) of 6 cysteine residues each, and a hydrophobic transmembrane domain followed by a short cytoplasmic domain at the C terminus.
FIG. 5.
(A) Amino acid sequences encoded by the UL144 ORF and representing each of the three major groups of strains. CRDs from TNFR superfamily members HveA and variola G2R (3, 25, 44) are aligned to show homology (from a BLAST search [1]). Numbers correspond to amino acid residues of either the predicted UL144 product or the TNFR superfamily member identified by use of BLAST. Potential functional features associated with growth factor receptors are designated as follows: single line, signal peptide; double line, CRDs; shading, conserved cysteines; broken line, transmembrane domain; arrows, N glycosylation sites of UL144 sequences identified by use of PROSITE (2). (B) Identity of the UL144 group 3 sequence with various members of the TNFR superfamily (4). The percent identities in the ectodomains (without leader sequences) and CRD2 were calculated from the fraction of identical residues following pairwise alignment.
The results of a BLAST search (1) of protein databases show homology between the amino acid sequences of all UL144 groups and several receptors of the TNFR superfamily. The homology is limited to the CRDs that are the defining feature of this family, although the predicted UL144 product contains only two of the three or more such domains usually present in other family members (3, 11, 15, 48).
The UL144 amino acid sequence has the highest homology with the cellular herpes simplex virus entry mediator, HveA (formerly HVEM) (31, 49), a recently identified member of the TNFR superfamily (GenBank accession no. U70321). A comparison of UL144 amino acid sequences representing each of the three groups with that of HveA (1) demonstrates 41% identity in the regions of the two conserved CRDs (Fig. 5A). Thus, the amino acid homology of UL144 groups 1, 2, and 3 with HveA is comparable in spite of the extensive sequence diversity. The two CRDs of the UL144 product are homologous to CRD1 and CRD2 of HveA, and the UL144 product is the first identified member of the TNFR superfamily to contain only these two domains. UL144 CRD2, which has the highest homology with HveA (46% identity), still displays significant homology to the ligand binding domain of TRAIL-R2 (43%) (Fig. 5B). The poxvirus variola TNFR homologue, G2R (GenBank accession no. U18339) (28), has a slightly lower CRD2 sequence identity (40%). Essentially all ligand-receptor contacts in the lymphotoxin α/TNFR (55 kDa) complex occur in CRD2 and CRD3. The lack of a CRD3-homologous region in UL144 suggests that the predicted product does not possess a complete ligand binding domain (4).
Expression of UL144.
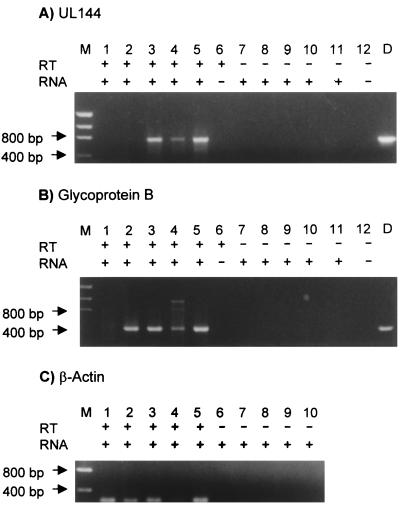
From a comparison of nucleotide and amino acid sequences of the HCMV strains, it is evident that the sequence variability does not affect conserved structural features of the predicted UL144 product. To examine the effect of the variability on UL144 transcription, RT-PCR analysis was used to detect UL144-specific transcripts produced during infection by isolates representing each major UL144 sequence group. Total RNA was extracted at 96 h postinfection from cells infected with one of three different clinical strains, PT26 (subgroup 1A), PT17 (group 2), and PT11 (group 3), or the laboratory strain AD169. Templates generated by the RT reaction were amplified with the UL144-B, gB, or β-actin primer pairs (Fig. 6). No product was amplified from uninfected-cell RNA (Fig. 6, lanes 1) or in the absence of RT (Fig. 6A and B, lanes 7 to 12, and Fig. 6C, lanes 6 to 10) with either the UL144-B or the gB primers. The UL144-B primers produced amplified products from RNA extracted from cells infected with each of the three clinical strains (Fig. 6A, lanes 3 to 5) but not AD169-infected cells (lane 2). The gB primers amplified products from RNA extracted from cells infected with AD169 as well as cells infected with each of the three clinical strains (Fig. 6B, lanes 2 to 5). These results indicate that UL144-specific transcripts are present in strains representing all three sequence groups. As predicted, there is no UL144-specific transcript in AD169-infected cells, and gB-specific transcripts can be detected in all strains, including AD169.
FIG. 6.
RT-PCR analysis of total RNA isolated from mock-infected HFF, AD169, or three clinical strains (PT11, PT17, and PT26). (A) UL144 ORF reactions with RT (+) (lanes 1 to 6) and without RT (−) (lanes 7 to 12). Lane M, size markers; lanes 1 and 7, mock-infected HFF RNA; lanes 2 and 8, AD169 RNA; lanes 3 and 9, PT11 RNA; lanes 4 and 10, PT17 RNA; lanes 5 and 11, PT26 RNA; lanes 6 and 12, no RNA; lane D, UL144 product amplified from a DNA template. (B) gB reactions from the region surrounding the cleavage site. Reagents and RNA templates are the same as in panel A. (C) β-Actin RT-PCR reactions. Lanes 1 to 5, with RT (+); lanes 6 to 10, without RT (−), lane M, size markers; lanes 1 and 6, mock-infected HFF; lanes 2 and 7, AD169; lanes 3 and 8, PT11; lanes 4 and 9, PT17; lanes 5 and 10, PT26.
Comparative sequence analysis of the hypervariability of gB in clinical strains.
The hypervariability of the gB gene among HCMV strains is well established (12, 29, 39). To determine whether UL144 groups correlate with gB groups, a 410-bp region surrounding the proteolytic cleavage site of gB in each of the 45 strains was sequenced. The results of the gB genotyping of these strains (Table 1) showed the following distribution: gB1, 19 strains (42%); gB2, 11 strains (24%); gB3, 10 strains (22%); and gB4, 5 strains (11%). The frequency of each group is similar to that reported by others (12, 42), and no gB group appears to be associated with any specific underlying disease or target organ. For example, among patients with lung disease, the distribution of gB groups is as follows: gB1, 46%; gB2, 29%; gB3, 18%; and gB4, 7%. This distribution essentially reflects the relative frequencies of the gB groups among these strains. As mentioned above for the UL144 sequence analysis, the clinical outcome in each patient is not available; therefore, potential pathogenicity associated with gB groups cannot be determined. Typing of the 45 strains based on gB did not correlate with that based on UL144 (Table 1). The lack of a phylogenetic linkage is most likely explained by the process of recombination, because the gB and UL144 ORFs are located more than 90 kb apart on the HCMV genome.
DISCUSSION
The large coding capacity and slow cell-associated replication cycle of HCMV suggest that pathogenesis involves a complex interaction of viral proteins with multiple host cell targets. Although the functions of many of the proteins in the viral replication cycle have yet to be determined, several products of HCMV ORFs, including US2, UL18, and US28, have recently been shown to have immunomodulatory functions, which are likely to play a role in HCMV pathogenesis (19, 36, 41). The TNFR homologue encoded by the UL144 ORF is another potential immunomodulatory product to be added to this list.
The significance of this TNFR homology for HCMV pathogenesis is supported by the fact that a number of viruses have been found to encode products that modulate apoptotic and proinflammatory signaling pathways induced by members of this family of receptors (5, 17, 18, 23, 28, 32, 40, 43, 46, 48). In addition, herpes simplex virus uses HveA, a TNFR superfamily member (31, 49), as an entry pathway to infect activated T cells.
What effect the UL144 product as a TNFR homologue would have on the HCMV replication cycle in vivo, however, is still in question. One possibility suggested by the predicted homology with both cellular and viral TNFR homologues is that the UL144 product could serve as either a soluble (3, 9, 17, 23, 28, 37, 47, 48, 55) or a membrane-bound (35, 45) decoy receptor. However, recent evidence argues against a decoy role for the UL144 product. Benedict et al. (4) reported that the UL144 protein is retained intracellularly during infection of fibroblasts and that when UL144 is expressed as an Fc fusion protein, it does not bind any of the known tumor necrosis factor-related ligands. The highly conserved cytoplasmic domain of the UL144 product in all the clinical strains suggests a critical role for this region in its mechanism of action. The C terminus of the UL144 product contains a conserved YXXZ motif that has been shown to function as a recognition sequence for interaction with adaptor complexes associated with clathrin-mediated receptor internalization at the plasma membrane and in the trans-Golgi network (27). These observations, along with the finding that the UL144 gene is expressed early after infection, raise the likely possibility that the action of the UL144 product is to modify expression and/or signaling pathways for members of the TNFR family that may allow escape from immune defenses (4).
In contrast to the conserved TNFR structural motifs found in all of the UL144 sequences from our clinical strains is the unexpected observation of high-level UL144 sequence variability among these same strains. This finding is strikingly different from the low degree of variation at the nucleotide level and the even lower degree of amino acid substitution observed for functionally important viral enzymes, such as the DNA polymerase and UL97 phosphotransferase (13, 25). The only product previously demonstrated to vary to a similar degree among unrelated strains is gB (12, 29, 39) which, aside from its role as an essential protein in virus penetration and cell fusion (33), has been hypothesized to function as a determinant of differential tissue tropism and disease (6, 14, 30, 33, 42, 44, 50). The data generated in support of this hypothesis, however, are inconclusive and, in some instances, contradictory. Other studies have found no relationship between gB groups and specific diseases (10, 52). The analysis of the strains from our cohort of patients similarly shows no obvious correlation of gB and UL144 sequence groups with underlying disease. The lack of available patient data related to clinical outcome, however, makes it impossible to conclude from our studies that there is no correlation between UL144 or gB sequence variants and HCMV pathogenesis.
Despite similar levels of sequence hypervariability of gB and UL144, we did not observe a phylogenetic linkage between the defined gB and UL144 sequence groups. However, the fact that gB and UL144 strain-specific sequences diverge as sequence groups and not random mutations throughout the coding region of each gene strongly suggests that there are selective conditions for each type of variant.
Recent reports of sequence hypervariability of ORFs in other herpesviruses indicate that this phenomenon is more widespread than previously thought (22, 51). Relevant to our findings for the predicted UL144 protein are parallel features described for the human herpesvirus 8 hypervariable gene K1 (21, 57). Both UL144 and K1 encode predicted transmembrane glycoproteins. The K1 sequences can be separated into four major groups, A to D, which differ from each other at the amino acid level by up to 30%. UL144 has three major sequence groups with amino acid substitutions of up to 21%. There is evidence for recombination between groups for both K1 and UL144. Notably, UL144 subgroup 1C may represent recombinants of group 1 and group 3 strains. Alternatively, putative recombinants of either K1 or UL144 may represent evolutionary intermediates between groups. Within the groups of both the K1 and the UL144 proteins, there are less distinct variant subgroups. However, a significant observation for both genes is that no change in DNA sequence occurred in serial samples from the same patient (24, 26, 57).
An important finding for K1 is the correlation of the clade pattern of the variant strains with patient geographic and ethnic backgrounds. There is some evidence suggesting that HCMV gB groups may be associated with geographic and demographic differences among patients (54, 56). Future prospective studies of clinical HCMV strains may show that a relationship also exists between UL144 sequence groups and patient demographics.
Thus, the question of the role of variant viral products in the course of HCMV infection and disease remains unanswered. From extensive work on gB, it is evident that sequence hypervariability can be superimposed on an otherwise structurally conserved protein without an apparent effect on the essential functions. It is possible, however, that the high level of amino acid variability affects the host antibody response; indeed, it has been reported (4, 20) that HCMV antibody-positive human sera vary in neutralizing activity against virus isolates from different hosts. Although gB is a major target of the antibody response to HCMV, there is preliminary evidence (20) that differences in neutralization titer against HCMV strains do not correlate with gB groups. Benedict et al. (4) have shown that HCMV antibody-positive sera have differential reactivity against cells expressing UL144, although there is no evidence yet that correlates the reactivity with UL144 sequence groups. The complex nature of the interaction of HCMV with the host immune response suggests that more than one variant gene may be involved in the observed differential seroreactivity. In support of this hypothesis, we have obtained preliminary results (26) that the products of the UL142, UL146, and UL147 ORFs adjacent to UL144 have amino acid sequence variability similar to that which we have observed for the UL144 product. Therefore, the infectivity and pathogenicity of an individual HCMV strain may be defined by the combination of multiple variant genes that it encodes. Our investigation of UL144 represents a first step in defining potential markers of HCMV pathogenesis. A larger prospective study of HCMV strains obtained from patients with different underlying diseases could determine the correlation of multiple strain-specific markers with demographic background, tissue tropism, and clinical outcome related to HCMV disease.
ACKNOWLEDGMENTS
We thank Joan Siegel and Kenneth Thompson for critical review of the manuscript; Harold Kessler, Staci Fischer, and Vijay Yeldandi for identifying patients; and Mary Hayden and Ronald Lollar for providing access to the clinical isolates.
This work was supported in part by grants from the American Lung Association of Metropolitan Chicago and the National Institutes of Health (grant AI38858 to N.S.L.), National Institutes of Health grants AI03368 and PO1CA69381 (to C.F.W.) and training grant AG00252 (to C.A.B.), and National Institutes of Health contract N01-AI35172 (to J.W.B.).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banchereau J, Bazan F, Blanchard D, Brière F, Galizzi J P, van Kooten C, Liu Y J, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 4.Benedict C A, Butrovich K D, Lurain N S, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware C F. A novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol. 1999;162:6967–6970. [PubMed] [Google Scholar]
- 5.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G-H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bongarts A, von Laer D, Vogelberg C, Ebert K, van Lunzen J, Garweg J, Vaith P, Hufert F T, Haller O, Meyer-König U. Glycoprotein B genotype of human cytomegalovirus: distribution in HIV-infected patients. Scand J Infect Dis. 1996;28:447–449. doi: 10.3109/00365549609037937. [DOI] [PubMed] [Google Scholar]
- 7.Brukiewicz R R, Welsh R M. Major histocompatibility complex class I antigens and the control of viral infections by natural killer cells. J Virol. 1995;69:3967–3971. doi: 10.1128/jvi.69.7.3967-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamuleau M E D, ten Berge I J M, Schellekens P T A, Wilmink J M, Hintzen R Q, van Lier R A W. Serum levels of soluble CD27 in renal transplant recipients. Transplantation. 1992;54:932–936. [PubMed] [Google Scholar]
- 10.Chern K C, Chandler D B, Martin D F, Kuppermann B D, Wolitz R A, Margolis T P. Glycoprotein B subtyping of cytomegalovirus (CMV) in the vitreous of patients with AIDS and CMV retinitis. J Infect Dis. 1998;178:1149–1153. doi: 10.1086/515672. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan A M, O’Rourke K, Yu G-L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Signal transduction by DR3, a death domain-containing receptor related to TNRF-1 and CD95. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 12.Chou S, Dennison K M. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229–1234. doi: 10.1093/infdis/163.6.1229. [DOI] [PubMed] [Google Scholar]
- 13.Chou S, Lurain N S, Weinberg A, Crumpacker C S. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob Agents Chemother. 1999;43:1500–1502. doi: 10.1128/aac.43.6.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fries B C, Chou S, Boeckh M, Torok-Storb B. Frequency distribution of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J Infect Dis. 1994;169:769–774. doi: 10.1093/infdis/169.4.769. [DOI] [PubMed] [Google Scholar]
- 15.Gruss H-J, Duyster J, Herrmann F. Structural and biological features of the TNF receptor and TNF ligand superfamilies: interactive signals in the pathobiology of Hodgkin’s disease. Ann Oncol. 1996;7(Suppl. 4):S19–S26. doi: 10.1093/annonc/7.suppl_4.s19. [DOI] [PubMed] [Google Scholar]
- 16.Hillis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic trees. Syst Biol. 1993;42:182–192. [Google Scholar]
- 17.Hu F-Q, Smith C A, Pickup D J. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Vincenz C, Buller M, Dixit V M. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 19.Jones T, Sun L. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J Virol. 1997;71:2970–2979. doi: 10.1128/jvi.71.4.2970-2979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein M, Schoppel K, Amvrossiadis N, Mach M. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J Virol. 1999;73:878–886. doi: 10.1128/jvi.73.2.878-886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Guo J, Li M, Choi J-K, DeMaria M, Rosenzweig M, Jung J U. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi’s sarcoma-associated herpesvirus. Mol Cell Biol. 1998;18:5219–5228. doi: 10.1128/mcb.18.9.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H, Trimble J J, Yoon D-W, Regier D, Desrosiers R C, Jung J U. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J Virol. 1997;71:3817–3825. doi: 10.1128/jvi.71.5.3817-3825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loparev V N, Parsons J M, Knight J C, Panus J F, Ray C A, Buller R M L, Pickup D J, Esposito J J. A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc Natl Acad Sci USA. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lurain, N. S. Unpublished data.
- 25.Lurain N S, Ammons H C, Kapell K S, Yeldandi V V, Garrity E R, O’Keefe J P. Molecular analysis of human cytomegalovirus strains from two lung transplant recipients with the same donor. Transplantation. 1996;62:497–502. doi: 10.1097/00007890-199608270-00012. [DOI] [PubMed] [Google Scholar]
- 26.Lurain N S, Kapell K S, Winkfield E, Paintsil J, Bremer J W. Abstracts of the 24th International Herpesvirus Workshop. 1999. Strain-specific sequence variability of human cytomegalovirus clinical strains, abstr. 11.027. [Google Scholar]
- 27.Marks M, Ohno H, Kirchhausen T, Bonifacino J. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 28.Massung R F, Esposito J J, Liu L I, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, Selivanov N A, Cavallaro K F, Kerlavage A R, Mahy B W J, Venter J C. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 29.Meyer-König U, Haberland M, von Laer D, Haller O, Hufert F T. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J Infect Dis. 1998;177:1162–1169. doi: 10.1086/515262. [DOI] [PubMed] [Google Scholar]
- 30.Meyer-König U, Vogelberg C, Bongarts A, Kampa D, Delbrück R, Wolff-Vorbeck G, Kirste G, Gaberland M, Hufert F T, von Laer D. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J Med Virol. 1998;55:75–81. [PubMed] [Google Scholar]
- 31.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 32.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 33.Navarro D, Pedro P, Tugizov S, Topp K, La Vail J, Pereira L. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology. 1993;197:143–158. doi: 10.1006/viro.1993.1575. [DOI] [PubMed] [Google Scholar]
- 34.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 35.Pan G, Ni J, Wei Y-F, Yu G-L, Gentz R, Dixit V M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 36.Pleskoff O, Tréboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 37.Prasad K V S, Ao Z, Yoon Y, Wu M X, Rizk M, Jacquot S, Schlossman S R. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci USA. 1997;94:6346–6351. doi: 10.1073/pnas.94.12.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinnan G V, Burns W H, Kirmani N, Rook A H, Manischewitz J, Jackson L, Santos G W, Saral R. HLA-restricted cytotoxic T lymphocytes are an early immune response and important defense mechanism in cytomegalovirus infections. Rev Infect Dis. 1984;6:156–163. doi: 10.1093/clinids/6.2.156. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen L, Hong C, Zipeto D, Morris S, Sherman D, Chou S, Miner R, Drew W L, Wolitz R, Dowling A, Warford A, Merigan T C. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J Infect Dis. 1997;175:179–184. doi: 10.1093/infdis/175.1.179. [DOI] [PubMed] [Google Scholar]
- 40.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1β converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 41.Reyburn H T, Mandelboim O, Valés-Gómez M, Davis D M, Pazmany L, Strominger J L. The class I MHC homologue of human cytomegalovirus inhibits attack by natural killer cells. Nature. 1997;386:514–517. doi: 10.1038/386514a0. [DOI] [PubMed] [Google Scholar]
- 42.Rosen H R, Corless C L, Rabkin J, Chou S. Association of cytomegalovirus genotype with graft rejection after liver transplantation. Transplantation. 1998;66:1627–1631. doi: 10.1097/00007890-199812270-00010. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber M, Sedger L, McFadden G. Distinct domains of M-T2, the myxoma virus tumor necrosis factor (TNF) receptor homolog, mediate extracellular TNF binding and intracellular apoptosis inhibition. J Virol. 1997;71:2171–2181. doi: 10.1128/jvi.71.3.2171-2181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shepp D H, Match M E, Ashraf A B, Lipson S M, Millan C, Pergolizzi R. Cytomegalovirus glycoprotein B groups associated with retinitis in AIDS. J Infect Dis. 1996;174:184–187. doi: 10.1093/infdis/174.1.184. [DOI] [PubMed] [Google Scholar]
- 45.Sheridan J P, Marsters S A, Pitti R M, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, Goddard A D, Godowski P, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 46.Shisler J, Yang C, Walter B, Ware C F, Gooding L. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 48.Smith C A, Hu F-Q, Smith T D, Richards C L, Smolak P, Goodwin R G, Pickup D J. Cowpox virus genome encodes a second soluble homologue of cellular TNF receptors, distinct from CrmB, that binds TNF but not LTα. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 49.Terry-Allison R R, Montgomery R I, Whitbeck J D, Xu R, Cohen G H, Eisenberg R J, Spear P. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torok-Storb B, Boeckh M, Hoy C, Leisenring W, Myerson D, Gooley T. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood. 1997;90:2097–2102. [PubMed] [Google Scholar]
- 51.Triantos E, Boulter A W, Leao J C, Di Albert L, Porter S R, Scully C M, Birnbaum W, Johnson N W, Teo C G. Diversity of naturally occurring Epstein-Barr virus revealed by nucleotide sequence polymorphism in hypervariable domains in the BamHI K and N subgenomic regions. J Gen Virol. 1998;79:2809–2817. doi: 10.1099/0022-1317-79-11-2809. [DOI] [PubMed] [Google Scholar]
- 52.Vogelberg C, Meyer-König U, Hufert F T, Kirste G, von Laer D. Human cytomegalovirus glycoprotein B genotypes in renal transplant recipients. J Med Virol. 1996;50:31–34. doi: 10.1002/(SICI)1096-9071(199609)50:1<31::AID-JMV7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 53.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 54.Wada K, Mizuno S, Kato K, Kamiya T, Ozawa K. Cytomegalovirus glycoprotein B sequence variation among Japanese bone marrow transplant recipients. Intervirology. 1997;40:215–219. doi: 10.1159/000150549. [DOI] [PubMed] [Google Scholar]
- 55.Ware C F, VanArsdale T L, Crowe P D, Browning J. The ligands and receptors of the lymphotoxin system. Curr Top Microbiol Immunol. 1995;198:175–218. doi: 10.1007/978-3-642-79414-8_11. [DOI] [PubMed] [Google Scholar]
- 56.Zipeto D, Hong C, Gerna G, Zavattoni M, Katzenstein D, Merigan T C, Rasmussen L. Geographic and demographic differences in the frequency of human cytomegalovirus gB genotypes 1-4 in immunocompromised patients. AIDS Res Hum Retroviruses. 1998;14:533–536. doi: 10.1089/aid.1998.14.533. [DOI] [PubMed] [Google Scholar]
- 57.Zong J-C, Ciufo D M, Alcendor D J, Wan X, Nicholas J, Browning P J, Rady P L, Tyring S K, Orenstein J M, Rabkin C S, Su I-J, Powell K F, Croxson M, Foreman K E, Nickoloff B J, Alkan S, Hayward G S. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]